Summary
The organization of individual respiratory chain complexes into supercomplexes or respirasomes has attracted great interest because of the implications for cellular energy conversion. Recently, it was reported that commonly used mouse strains harbor a short COX7a2l (SCAFI) gene isoform that supposedly precludes the formation of complex IV-containing supercomplexes. This claim potentially has serious implications for numerous mouse studies addressing important topics in metabolism, including adaptation to space flights. Using several complementary experimental approaches, we show that mice with the short COX7a2l isoform have normal biogenesis and steady-state levels of complex IV-containing supercomplexes and consequently have normal respiratory chain function. Furthermore, we use a mouse knockout of Lrpprc and show that loss of complex IV compromises respirasome formation. We conclude that the presence of the short COX7a2l isoform in the commonly used C57BL/6 mouse strains does not prevent their use in metabolism research.
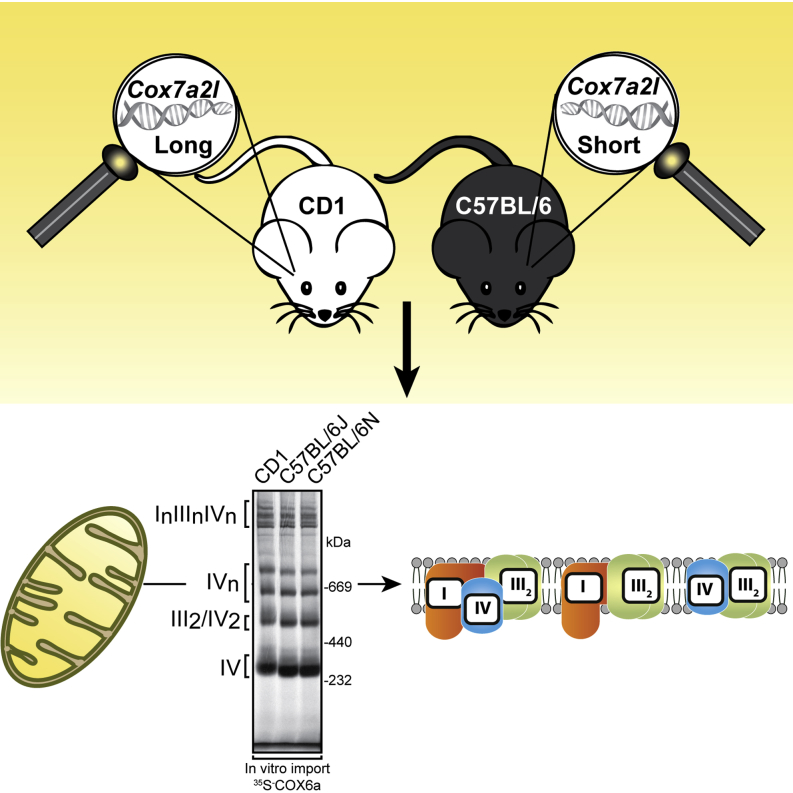
Graphical Abstract
Highlights
-
•
C57BL/6J and C57BL/6N mouse strains contain a short COX7a2l gene isoform
-
•
The short COX7a2l isoform does not impair respiratory chain function or oxidative capacity
-
•
Complex IV-containing supercomplexes exist in mice with the short COX7a2l isoform
-
•
The biogenesis of respirasomes is normal in mice with the short COX7a2l isoform
In contrast to a recent report, Mourier et al. show that the presence of the short COX7a2l isoform in the commonly used C57BL/6 mouse strains does not affect the supramolecular organization and function of the mitochondrial respiratory chain.
Introduction
Mitochondria are the cellular power plants that produce the bulk part of the energy currency ATP through energy conversion by the oxidative phosphorylation (OXPHOS) system. The OXPHOS system, which is located in the inner mitochondrial membrane, is composed of two functional entities, i.e., the respiratory chain and the phosphorylation system, which includes the ATP synthase and carriers, such as the ATP/ADP carrier and the phosphate carrier. One important feature of mitochondria is that they harbor their own genome, mtDNA, which encodes 13 of the subunits of the enzyme complexes constituting the OXPHOS system, whereas the remaining ∼90 subunits are encoded by nuclear genes and imported into mitochondria (Hällberg and Larsson, 2014). Historically, the mitochondrial respiratory chain has been defined as an ensemble of complexes I, II, III, and IV, some of which, i.e., complexes I, III, and IV, couple redox reactions to proton extrusion. The respiratory chain complexes are enriched in the tubular inner mitochondrial membrane invaginations called cristae (Busch et al., 2013, Gilkerson et al., 2003, Vogel et al., 2006) and were initially thought to be randomly distributed as independent entities (Hackenbrock et al., 1986). However, this view has been challenged based on a wealth of structural and functional analyses that support a model that the individual respiratory chain complexes interact to form stable supercomplexes (Genova and Lenaz, 2014, Schägger and Pfeiffer, 2000). One of the most important arguments for a higher-order organization of the respiratory chain was provided by the use of blue native polyacrylamide gel electrophoresis (BN-PAGE), which showed that respiratory chain complexes from a wide range of organisms can be extracted in supramolecular assemblies when mitochondria are solubilized using mild detergents (Acín-Pérez et al., 2008, Dudkina et al., 2010, Schägger, 2001). In mammalian mitochondria, supercomplexes consisting of complexes I, III, and IV provide the bulk part of the proton motive force and are termed respirasomes (Schägger and Pfeiffer, 2000). Although the supramolecular organization of the respiratory chain complexes has been extensively documented in different model organisms, the existence of factors mediating respirasome assembly or their stabilization have remained elusive (Schägger and Pfeiffer, 2000, Schägger and Pfeiffer, 2001). Interactions between respiratory chain complexes within supercomplexes were originally described to be dependent on cardiolipin-protein interactions, in contrast to the ATP synthase dimer complexes, which depend on specific protein-protein interactions (Genova and Lenaz, 2014, Pfeiffer et al., 2003). Two proteins, Rcf1/HIG2A and Rcf2, were recently reported to mediate supercomplex assembly in yeast and in mammals (Chen et al., 2012, Strogolova et al., 2012, Vukotic et al., 2012). In addition, the COX7a2-like protein (COX7a2l, also called supercomplex assembly factor I, SCAFI), originally identified as an estrogen-responsive element (Watanabe et al., 1998), was reported to be essential for formation of complex IV-containing supercomplexes (Lapuente-Brun et al., 2013) and stabilization of the respirasome (Ikeda et al., 2013).
It has been suggested that supercomplex formation is of key importance for the stability of the individual respiratory chain complexes and that it may be initiated already at the assembly stage of complex I (Moreno-Lastres et al., 2012, Ugalde et al., 2004). Furthermore, supercomplex formation has been proposed to reduce the diffusion distance of ubiquinone and cytochrome c, the two mobile electron carriers of the respiratory chain, thereby increasing the electron transport efficiency and reducing reactive oxygen species production (Genova and Lenaz, 2014, Maranzana et al., 2013). Despite these hypotheses, the functional roles of the supercomplex assemblies remain unclear. Contrary to what has been reported in yeast (Rigoulet et al., 2010), a recent report from the Enriquez laboratory (Lapuente-Brun et al., 2013) proposed that altered supercomplex assembly provides a mechanism for physiological regulation of energy metabolism in mammals by providing alternate paths for electrons derived from metabolism of specific substrates. When performing a single-nucleotide polymorphism (SNP) analysis, the authors discovered that some commonly used mouse strains, such as C57BL/6J and BALB/c, are homozygous for a six-base-pair deletion of the COX7a2l gene, which leads to the production of an unstable, short COX7a2l isoform that cannot support supercomplex formation (Lapuente-Brun et al., 2013). Consequently, mice with the short COX7a2l isoform were shown to lack complex IV-containing supercomplexes and to have aberrant respiratory chain function.
The report that COX7a2l is a novel supercomplex factor necessary for respirasome formation has attracted a lot of attention and was recently discussed in a Preview in Cell Metabolism (Barrientos and Ugalde, 2013). The results from the Enriquez laboratory (Lapuente-Brun et al., 2013) showing that C57BL/6 mice cannot form respirasomes have alarming consequences, as they challenge the interpretation of experimental results from a large number of mouse models generated in the C57BL/6 background. This mouse strain is widely used in metabolism research (Agostino et al., 2003, Chen et al., 2003, Diaz et al., 2005, Li et al., 1995), in studies of basic mechanisms regulating mitochondrial function (Cámara et al., 2011, Metodiev et al., 2009, Metodiev et al., 2014) and in aging research (Ross et al., 2013, Trifunovic et al., 2004). In fact, the C57BL/6 is one of the most widely used mouse strains, and it was the first one to have its genome sequenced (Waterston et al., 2002). Furthermore, the importance of the C57BL/6 mouse as a standard mammalian model organism was demonstrated by the fact that this mouse strain was recently sent to space to study the physiological and metabolic impact of long-term space flights (Sychev et al., 2014). In contrast to the report from the Enriquez laboratory (Lapuente-Brun et al., 2013), we have previously observed respirasomes when performing knockout studies in C57BL/6N mice (Milenkovic et al., 2013, Sterky et al., 2012). However, in these previous studies, we had not performed detailed analyses of supercomplex composition or assessed the COX7a2l genotype. To gain further insights into the role of COX7a2l isoforms in control of respiratory chain function and supercomplex formation, we extensively characterized the commonly used C57BL/6J and C57BL/6N mouse strains. We report here that both of these mouse strains indeed are homozygous for the short isoform of COX7a2l, but this circumstance does not affect their respiratory chain function or the maximal oxidative capacity. Furthermore, the steady-state levels of supercomplexes and respirasome formation are not affected in these mice. Therefore, we conclude that mice harboring the short COX7a2l isoform have no gross bioenergetic aberrations that prevent their use in metabolism research.
Results and Discussion
CD1 and C57BL/6 Mice Contain Different COX7a2l Alleles but Exhibit Very Similar Mitochondrial Bioenergetic Properties
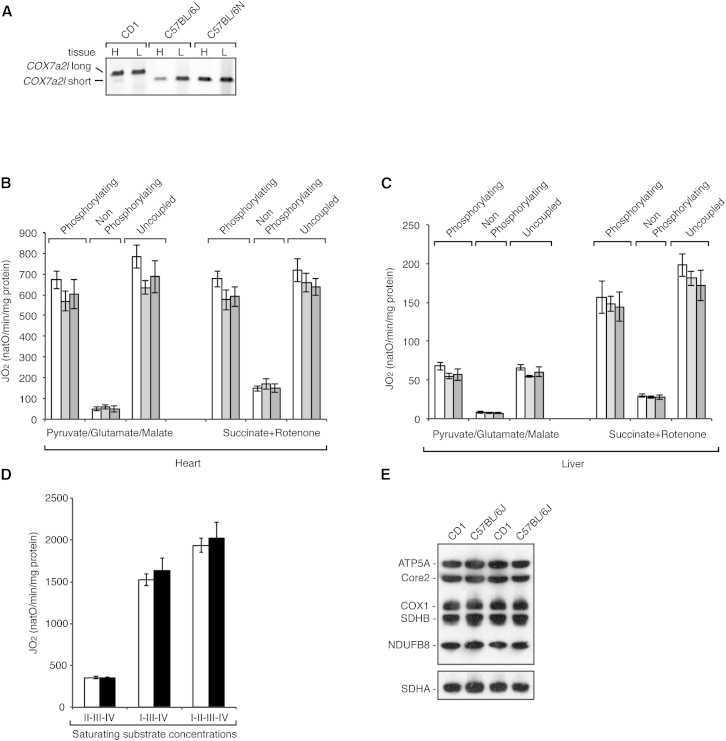
We analyzed the allelic variation of COX7a2l in genomic DNA extracted from CD1 and C57BL/6J mice and, in agreement with the previous report from the Enriquez laboratory (Lapuente-Brun et al., 2013), we found that C57BL/6J mice indeed are homozygous for a short isoform of the COX7a2l allele (Figure 1A). In addition, we analyzed the C57BL/6N mouse strain and found that it also contains the short COX7a2l allele in a homozygous state (Figure 1A). As the different COX7a2l alleles have been previously reported to modulate respiration driven by complexes I and II, we proceeded to investigate the mitochondrial bioenergetic properties in these different mouse strains. To this end, we assessed the oxygen consumption rate in isolated heart and liver mitochondria (Figures 1B and 1C). Freshly isolated heart and liver mitochondria were incubated with respiratory substrates whose metabolism results in delivery of electrons at the level of complexes I or II. In contrast to the previous report (Lapuente-Brun et al., 2013), the analyzed mouse strains exhibited similar oxygen consumption rates with complex I or complex II substrates under phosphorylating, nonphosphorylating, and uncoupled conditions (Figures 1B and 1C). Furthermore, as the respiration in intact mitochondria is known to depend on substrate transport and matrix dehydrogenase activities, we also measured respiration in permeabilized mitochondria incubated with saturating concentrations of substrates and cytochrome c. When succinate was added at saturating amounts, the respiration capacity (dependent on complexes II, III, and IV) was similar to the capacity found in intact mitochondria incubated with succinate (Figure 1D). However, the respiration stimulated by adding NADH (dependent on complexes I, III, and IV) or NADH and succinate (dependent on complexes I, II, III, and IV) was clearly increased, but there was no difference between mitochondria from CD1 and C57BL/6J mice (Figure 1D). Furthermore, western blot analyses of mitochondria isolated from the different strains showed indistinguishable steady-state levels of OXPHOS proteins (Figure 1E), which is in good agreement with the very similar respiration patterns (Figures 1B–1D).
Figure 1.
Different Cox7a2l Isoforms Have No Differential Effects on Respiratory Chain Activity
(A) PCR analysis of Cox7a2l alleles in heart (H) and liver (L) tissue from CD1, C57BL/6J, and C57BL/6N mouse strains.
(B) Oxygen consumption of heart mitochondria from CD1 (white bars), C57BL/6J (light gray bars), and C57BL/6N (dark gray bars) mouse strains at 10 weeks of age. n = 4; error bars indicate mean ± SEM.
(C) Oxygen consumption of liver mitochondria from CD1 (white bars), C57BL/6J (light gray bars), and C57BL/6N (dark gray bars) mouse strains at 10 weeks of age. n = 4; error bars indicate mean ± SEM.
(D) The maximal oxygen consumption rate assessed in permeabilized heart mitochondria from CD1 (white bars) and C57BL/6J (black bars) mice in the presence of saturating concentrations of cytochrome c, NADH (I-III-IV), succinate (II-III-IV), or both NADH and succinate (I-II-III-IV). n = 7; error bars indicate mean ± SEM.
(E) Steady-state levels of OXPHOS subunits in CD1 and C57BL/6J isolated heart mitochondria as determined by western blot analyses.
Respirasomes Are Present in Mice Containing the Short COX7a2l Isoform
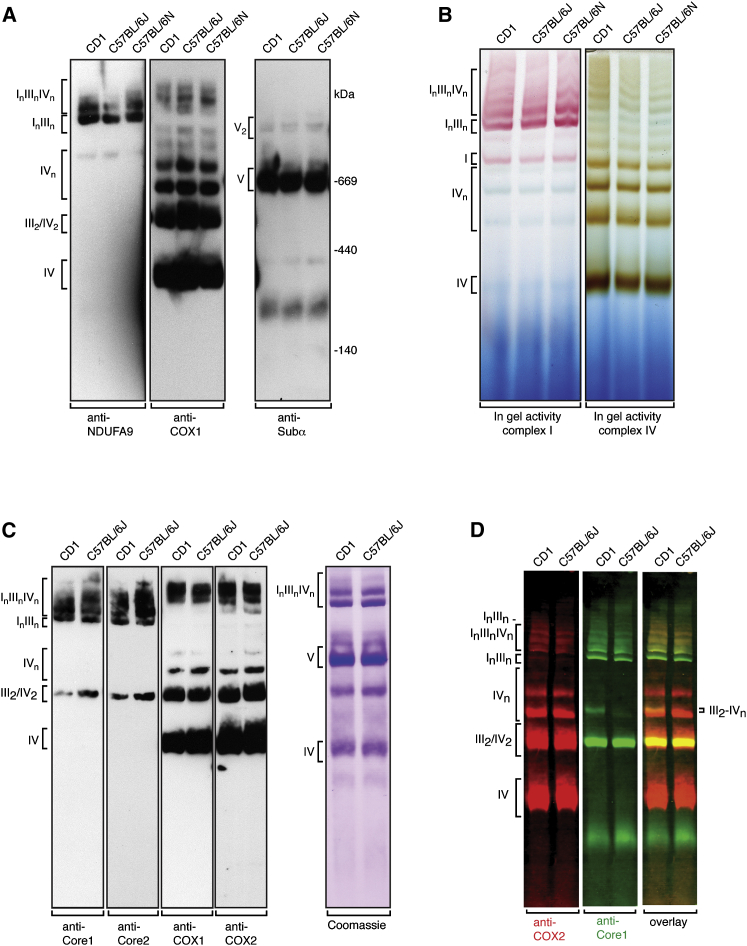
The finding that the respiratory chain capacity assessed in the presence of different substrate combinations was very similar in mouse strains harboring the short or long COX7a2l alleles (Figures 1B–1D) was in stark contrast to the results from the Enriquez laboratory (Lapuente-Brun et al., 2013). This surprising discrepancy prompted us to proceed to investigate the supramolecular organization of the respiratory chain by using standard BN-PAGE techniques. To this end, we solubilized mitochondria in mild detergent conditions (a ratio of digitonin to mitochondrial protein of ∼6 g/g), as digitonin/protein ratios between 4 and 8 g/g are the most commonly used (Dudek et al., 2013, Maranzana et al., 2013, Moreno-Lastres et al., 2012, Schägger and Pfeiffer, 2000, Sterky et al., 2012, Wittig et al., 2006). After extraction with detergent, the respiratory chain supercomplexes were separated by BN-PAGE and analyzed by western blotting (Figure 2A) or in-gel enzyme activity assays (Figure 2B). As expected under these mild solubilization conditions, we observed ATP synthase dimers (Figure 2A, right panel), thus showing that our results are in agreement with previous reports (Habersetzer et al., 2013). Remarkably, we found no alteration in the supramolecular organization of complexes I, III, and IV in CD1, C57BL/6J, and C57BL/6N mice when using western blot analyses (Figure 2A) or in-gel enzyme activity assays (Figure 2B), showing that the short and long COX7a2l isoforms do not influence supercomplex formation. To further validate our conclusions, we performed a set of western blots with additional antibodies directed against subunits of complex III and complex IV (Figures 2C and 2D). Consistent with many previous reports (Moreno-Lastres et al., 2012, Schägger, 2001, Schägger and Pfeiffer, 2000), we mainly found complex III and complex IV as free complexes or incorporated into respirasomes (Figures 2A–2C). To assess comigration of complexes more stringently and to exclude misalignment of gels, we used a two-color fluorescent labeling system allowing double immunodetection on the same western blot membrane (Figure 2D). When the migration of the labeled complex IV (Figure 2D, red) and the labeled complex III (Figure 2D, green) was compared, we found incorporation of both complexes into supercomplexes in CD1 as well as in C57BL/6J mice. The group of Ugalde (Moreno-Lastres et al., 2012) has quantitatively assessed that a minor amount of complex III (∼10%) and trace amounts of complex IV (<3%) are found in a III2-IVn complex. Consistent with their results, we observed such a III2-IVn complex in CD1 and C57BL/6J mice (Figure 2D). The proportion of complex III2 in the III2-IVn complex was reduced in the C57BL/6J strain in comparison with the CD1 strain (Figure 2D). This minor difference in complex III2-IVn abundance is unlikely to have any physiological impact, because CD1 and C57BL/6J mice show no difference in respiratory chain capacity or in levels of supercomplexes. Our results thus suggest that complex IV and complex III are only partly comigrating and, interestingly, that the amount of complex IV in this complex seems to be independent of complex III (Figure 2D). Furthermore, identical respirasome organization was observed using the BALB/c mouse strain also reported to be homozygous for the Cox7a2l short allele (Figures S1A and S1B). We thus conclude that the respirasome organization, as judged by the migration of complexes I, III, and IV, is indistinguishable in CD1, C57BL/6J, C57BL/6N, and BALB/c mouse strains when analyzed with BN-PAGE (Figures 2A–2D, S1A, and S1B).
Figure 2.
The Short Cox7a2l Isoform Does Not Affect the Supramolecular Organization of the Respiratory Chain
(A) Supramolecular organization of the OXPHOS system in different wild-type mouse strains. Heart mitochondria were extracted with a ratio of 6 g/g of digitonin to mitochondrial protein and analyzed by BN-PAGE. Immunodetection of subunits of complex I (NDUFA9), complex IV (COX1), and ATP synthase (subunit α) was performed after transfer of proteins from the BN-PAGE gel to a PVDF membrane. The positions of respiratory chain complexes and supercomplexes are indicated to the left. A representative image from five independent experiments is shown.
(B) In-gel enzyme activities of complexes I and IV in different wild-type mouse strains. The BN-PAGE conditions are as in (A). The data are representative of three independent experiments.
(C) Supramolecular organization of the respiratory chain in different wild-type mouse strains as determined by immune staining with antibodies against complex III (anti-Core1 and anti-Core2) and complex IV (anti-COX1 and anti-COX2). Coomassie Brilliant Blue staining of the gel was performed as a loading control. The BN-PAGE conditions are identical to the ones used in (A). The data are representative of three independent experiments.
(D) Supramolecular organization of the respiratory chain in different wild-type mouse strains. Western blot analyses were performed with double fluorescent detection of complex IV (anti-COX2, red color) and complex III (anti-Core2, green color). The BN-PAGE conditions are identical to the ones used in (A). The data are representative of four independent experiments.
Complex IV Is Essential for Respirasome Formation
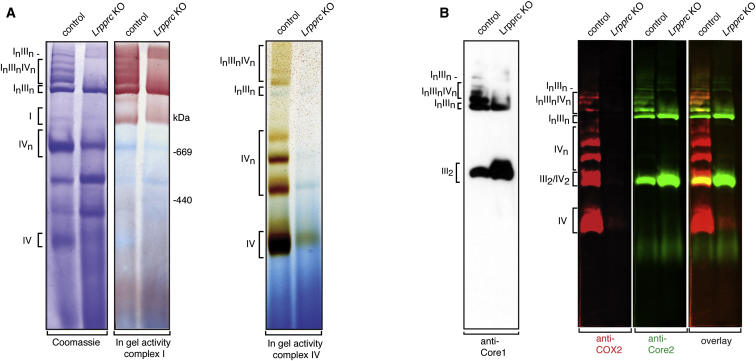
To rule out the possibility of a fortuitous comigration between putative complex IV multimers and other types of supercomplexes and to prove that complex IV physically interacts with complexes I–III, we utilized the conditional Lrpprc mouse knockout (Ruzzenente et al., 2012). These knockout mice have a severe complex IV deficiency but near-normal steady-state levels of the other respiratory chain complexes (Mourier et al., 2014, Ruzzenente et al., 2012). Heart mitochondria isolated from control and conditional Lrpprc knockout C57BL/6N mice were analyzed by BN-PAGE, and a drastic reduction of complex IV levels was found (Figures 3A and 3B). The low residual levels of complex IV were present in a free form (IV) in the knockout mice. As a consequence of the complex IV deficiency, we observed that most of the high-molecular-weight supercomplexes (In-IIIn-IVn) disappeared, and only supercomplexes containing complexes I and III (InIIIn) remained (Figures 3A and 3B). Moreover, we found accumulation of a complex III dimer (III2) in conditional Lrpprc knockout mice (Figure 3B), consistent with the observation that loss of complex IV prevents incorporation of complex III into supercomplexes (Figures 3A and 3B). In addition, it should be mentioned that the analysis of mitochondria from the conditional Lrpprc knockout mouse confirmed the specificity of the COX1 and COX2 antibodies (Mourier et al., 2014, Ruzzenente et al., 2012) and clearly showed that the observed complex IV in-gel enzyme activity assay indeed was dependent on the presence of complex IV (Figures 3A and 3B).
Figure 3.
Complex IV-Containing Respirasomes Are Present in Mouse Strains Harboring the Short Cox7a2l Isoform
(A) Supramolecular organization of complexes I, III, and IV in control and Lrpprc heart knockout mitochondria from C57BL/6N mice. Mitochondria were solubilized in digitonin (6 g/g mitochondrial protein) followed by Coomassie Brilliant Blue staining and in-gel enzyme activity staining for complexes I and IV. The data are representative of three independent experiments.
(B) BN-PAGE analysis of heart mitochondria isolated from control and Lrpprc heart knockout C57BL/6N mice. The BN-PAGE conditions are identical to the ones used in (A). The left panel shows western blot analysis of complex III (anti-Core1). The right panels show western blot analyses with double fluorescent detection of complex IV (anti-COX2, red color) and complex III (anti-Core2, green color). The data are representative of three independent experiments.
Complex IV Assembly Is Not Affected in Mice Containing the Short COX7a2l Allele
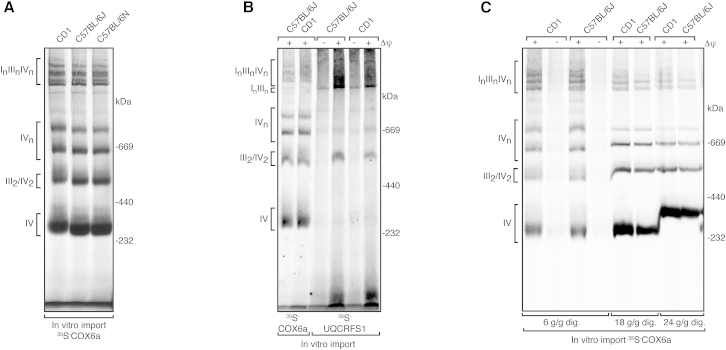
Next, we assessed complex IV assembly in CD1, C57BL/6J, and C57BL/6N mice by using an in vitro import assay on isolated mitochondria. For this purpose, we radiolabeled the COX6a subunit of complex IV and the UQCRFS1 subunit of complex III and performed import experiments in freshly isolated heart (Figures 4A–4C) and liver (Figure S2A) mitochondria. Import into the mitochondrial matrix is typically dependent on a potential across the inner mitochondrial membrane, and we therefore performed experiments in the presence and absence of the mitochondrial membrane potential in both heart (Figures 4B and 4C) and liver (Figure S2A) mitochondria. We followed the assembly pathway of imported UQCRFS1 and COX6a by performing BN-PAGE (Figures 4A–4C and S2A). There was no difference in the pattern of assembled complex III, complex IV, and related supercomplexes in CD1, C57BL/6J, and C57BL/6N mice (Figures 4A–4C). It has been argued that the detergent concentration is an important parameter when observing lack of supercomplexes in mice expressing the short COX7a2l isoform. To address this point, we investigated whether the supramolecular organization of complex IV is altered if mitochondria are solubilized at a wide range of digitonin concentrations prior to BN-PAGE analysis. In line with our previous results, we observed identical patterns of complex IV-containing supercomplexes in CD1 and C57BL/6J mice at various detergent concentrations (Figures 4C and S2B).
Figure 4.
The Assembly of Complex IV Containing Respirasomes Is Unaffected in Mouse Strains Harboring the Short Cox7a2l Isoform
(A) Import of the radiolabeled Cox6a precursor and subsequent incorporation into complex IV and respirasomes in intact heart mitochondria isolated from different wild-type strains. After 60 min incubation, the mitochondria were solubilized in digitonin (6 g/g mitochondrial protein) and analyzed by BN-PAGE. A representative image from eight independent experiments is shown.
(B) Import of the radiolabeled UQCRFS1 precursor and subsequent incorporation into complex III and supercomplexes in intact heart mitochondria isolated from different wild-type strains. After 60 min incubation, the mitochondria were solubilized in digitonin (6 g/g mitochondrial protein) and analyzed by BN-PAGE. A representative image from three independent experiments is shown.
(C) Import of the radiolabeled Cox6a precursor and subsequent incorporation into complex IV and respirasomes in intact heart mitochondria isolated from different wild-type strains. After 60 min incubation, the mitochondria were solubilized in buffers containing different digitonin concentrations (6–24 g/g mitochondrial protein). Import experiments were performed in the presence or absence of the mitochondrial membrane potential (Δψ). The data are representative of three independent experiments.
Conclusions
The use of inbred mouse strains allows reproducible experiments to be performed in independent laboratories and is an important tool to investigate regulation of metabolism in normal physiology, disease, and aging. Recently, it was claimed that mice harboring a short isoform of COX7a2l cannot form complex IV-containing supercomplexes and that this aberration leads to absence of respirasomes and clear bioenergetic aberrations (Lapuente-Brun et al., 2013). This finding would have profound implications for interpretation of results with mice on the C57BL/6J and C57BL/6N background, because both strains harbor the short COX7a2l isoform and are widely used in experimental mouse genetics. Here we demonstrate, using a variety of independent approaches, that the short COX7a2l gene isoform does not affect respiratory chain activity or respiratory chain supercomplex formation. Therefore, we conclude that mice harboring the short COX7a2l isoform unequivocally remain a suitable tool for metabolic and mitochondrial research. Furthermore, we provide evidence, using the Lrpprc knockout mouse, that complex IV is essential for respirasome formation in vivo.
Experimental Procedures
BN-PAGE
For BN-PAGE, 75 μg isolated mitochondria were lysed in 50 μl solubilization buffer (20 mM Tris [pH 7.4], 0.1 mM EDTA, 50 mM NaCl, 10% [v/v] glycerol) containing 1%–10% (w/v) digitonin (Calbiochem) and mixed with loading dye (5% [w/v] Coomassie Brilliant Blue G-250, 150 mM BIS-TRIS, and 500 mM ε-amino-n-caproic acid [pH 7.0]). BN-PAGE samples were resolved on 4%–10% gels and further subjected to Coomassie staining, western blot analysis, or in-gel activity staining for complexes I and IV. In-gel activity assays were performed as previously described (Wittig et al., 2007).
In Vitro Import
Radiolabeled human Cox6a and UQCRFS1 proteins were obtained by coupled transcription and translation in the presence of 35S-methionine (PerkinElmer) using TNT SP6 Quick Coupled System (Promega). For import experiments, freshly isolated mitochondria from heart and liver tissue were incubated with the radiolabeled proteins at 37°C in potassium-acetate import buffer (250 mM sucrose, 5 mM Mg-acetate, 80 mM K-acetate, 20 mM HEPES [pH 7.4], 10 mM Na-succinate, 1 mM ATP, and 1 mM DTT). To dissipate the membrane potential before import, 1 μM valinomycin was added. Samples were lysed in solubilization buffer and further analyzed on 4%–10% gradient gels. Signals were visualized by digital autoradiography.
Ethics Statement
This study was performed in accordance with the guidelines of the Federation of European Laboratory Animal Science Associations. The protocol was approved by the Landesamt für Natur, Umwelt und Verbraucherschutz in Nordrhein-Westfalen in Germany.
Acknowledgments
This study was supported by a European Research Council advanced investigator grant (268897) and by grants from the Deutsche Forschungsgemeinschaft (SFB829) and the Swedish Research Council (2013-2859) to N.-G.L. We thank Lysann Schmitz for technical assistance and Vera Kozjak-Pavlovic for providing Cox6a plasmid.
Published: December 2, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes Supplemental Experimental Procedures and two figures and can be found with this article online at http://dx.doi.org/10.1016/j.cmet.2014.11.005.
Contributor Information
Nils-Göran Larsson, Email: larsson@age.mpg.de.
Dusanka Milenkovic, Email: dmilenkovic@age.mpg.de.
Supplemental Information
References
- Acín-Pérez R., Fernández-Silva P., Peleato M.L., Pérez-Martos A., Enríquez J.A. Respiratory active mitochondrial supercomplexes. Mol. Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Agostino A., Invernizzi F., Tiveron C., Fagiolari G., Prelle A., Lamantea E., Giavazzi A., Battaglia G., Tatangelo L., Tiranti V., Zeviani M. Constitutive knockout of Surf1 is associated with high embryonic lethality, mitochondrial disease and cytochrome c oxidase deficiency in mice. Hum. Mol. Genet. 2003;12:399–413. doi: 10.1093/hmg/ddg038. [DOI] [PubMed] [Google Scholar]
- Barrientos A., Ugalde C. I function, therefore I am: overcoming skepticism about mitochondrial supercomplexes. Cell Metab. 2013;18:147–149. doi: 10.1016/j.cmet.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K.B., Deckers-Hebestreit G., Hanke G.T., Mulkidjanian A.Y. Dynamics of bioenergetic microcompartments. Biol. Chem. 2013;394:163–188. doi: 10.1515/hsz-2012-0254. [DOI] [PubMed] [Google Scholar]
- Cámara Y., Asin-Cayuela J., Park C.B., Metodiev M.D., Shi Y., Ruzzenente B., Kukat C., Habermann B., Wibom R., Hultenby K. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-C., Taylor E.B., Dephoure N., Heo J.-M., Tonhato A., Papandreou I., Nath N., Denko N.C., Gygi S.P., Rutter J. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F., Thomas C.K., Garcia S., Hernandez D., Moraes C.T. Mice lacking COX10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum. Mol. Genet. 2005;14:2737–2748. doi: 10.1093/hmg/ddi307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek J., Cheng I.-F., Balleininger M., Vaz F.M., Streckfuss-Bömeke K., Hübscher D., Vukotic M., Wanders R.J.A., Rehling P., Guan K. Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of Barth syndrome. Stem Cell Res. (Amst.) 2013;11:806–819. doi: 10.1016/j.scr.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Dudkina N.V., Kouril R., Peters K., Braun H.-P., Boekema E.J. Structure and function of mitochondrial supercomplexes. Biochim. Biophys. Acta. 2010;1797:664–670. doi: 10.1016/j.bbabio.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Genova M.L., Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochim. Biophys. Acta. 2014;1837:427–443. doi: 10.1016/j.bbabio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Gilkerson R.W., Selker J.M.L., Capaldi R.A. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 2003;546:355–358. doi: 10.1016/s0014-5793(03)00633-1. [DOI] [PubMed] [Google Scholar]
- Habersetzer J., Larrieu I., Priault M., Salin B., Rossignol R., Brèthes D., Paumard P. Human F1F0 ATP synthase, mitochondrial ultrastructure and OXPHOS impairment: a (super-)complex matter? PLoS ONE. 2013;8:e75429. doi: 10.1371/journal.pone.0075429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenbrock C.R., Chazotte B., Gupte S.S. The random collision model and a critical assessment of diffusion and collision in mitochondrial electron transport. J. Bioenerg. Biomembr. 1986;18:331–368. doi: 10.1007/BF00743010. [DOI] [PubMed] [Google Scholar]
- Hällberg B.M., Larsson N.-G. Making proteins in the powerhouse. Cell Metab. 2014;20:226–240. doi: 10.1016/j.cmet.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Shiba S., Horie-Inoue K., Shimokata K., Inoue S. A stabilizing factor for mitochondrial respiratory supercomplex assembly regulates energy metabolism in muscle. Nat Commun. 2013;4:2147. doi: 10.1038/ncomms3147. [DOI] [PubMed] [Google Scholar]
- Lapuente-Brun E., Moreno-Loshuertos R., Acín-Pérez R., Latorre-Pellicer A., Colás C., Balsa E., Perales-Clemente E., Quirós P.M., Calvo E., Rodríguez-Hernández M.A. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- Li Y., Huang T.T., Carlson E.J., Melov S., Ursell P.C., Olson J.L., Noble L.J., Yoshimura M.P., Berger C., Chan P.H. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat. Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- Maranzana E., Barbero G., Falasca A.I., Lenaz G., Genova M.L. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal. 2013;19:1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev M.D., Lesko N., Park C.B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C.M., Larsson N.-G. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Metodiev M.D., Spåhr H., Loguercio Polosa P., Meharg C., Becker C., Altmueller J., Habermann B., Larsson N.-G., Ruzzenente B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014;10:e1004110. doi: 10.1371/journal.pgen.1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D., Matic S., Kühl I., Ruzzenente B., Freyer C., Jemt E., Park C.B., Falkenberg M., Larsson N.-G. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum. Mol. Genet. 2013;22:1983–1993. doi: 10.1093/hmg/ddt051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Lastres D., Fontanesi F., García-Consuegra I., Martín M.A., Arenas J., Barrientos A., Ugalde C. Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 2012;15:324–335. doi: 10.1016/j.cmet.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourier A., Ruzzenente B., Brandt T., Kühlbrandt W., Larsson N.-G. Loss of LRPPRC causes ATP synthase deficiency. Hum. Mol. Genet. 2014;23:2580–2592. doi: 10.1093/hmg/ddt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer K., Gohil V., Stuart R.A., Hunte C., Brandt U., Greenberg M.L., Schägger H. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- Rigoulet M., Mourier A., Galinier A., Casteilla L., Devin A. Electron competition process in respiratory chain: regulatory mechanisms and physiological functions. Biochim. Biophys. Acta. 2010;1797:671–677. doi: 10.1016/j.bbabio.2010.01.030. [DOI] [PubMed] [Google Scholar]
- Ross J.M., Stewart J.B., Hagström E., Brené S., Mourier A., Coppotelli G., Freyer C., Lagouge M., Hoffer B.J., Olson L., Larsson N.G. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013;501:412–415. doi: 10.1038/nature12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzenente B., Metodiev M.D., Wredenberg A., Bratic A., Park C.B., Cámara Y., Milenkovic D., Zickermann V., Wibom R., Hultenby K. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H. Respiratory chain supercomplexes. IUBMB Life. 2001;52:119–128. doi: 10.1080/15216540152845911. [DOI] [PubMed] [Google Scholar]
- Schägger H., Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., Pfeiffer K. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J. Biol. Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- Sterky F.H., Hoffman A.F., Milenkovic D., Bao B., Paganelli A., Edgar D., Wibom R., Lupica C.R., Olson L., Larsson N.-G. Altered dopamine metabolism and increased vulnerability to MPTP in mice with partial deficiency of mitochondrial complex I in dopamine neurons. Hum. Mol. Genet. 2012;21:1078–1089. doi: 10.1093/hmg/ddr537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogolova V., Furness A., Robb-McGrath M., Garlich J., Stuart R.A. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol. Cell. Biol. 2012;32:1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sychev V.N., Ilyin E.A., Yarmanova E.N., Rakov D.V., Ushakov I.B., Kirilin A.N., Orlov O.I., Grigoriev A.I. [The BION-M1 project: overview and first results] Aviakosm. Ekolog. Med. 2014;48:7–14. [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Ugalde C., Janssen R.J.R.J., van den Heuvel L.P., Smeitink J.A.M., Nijtmans L.G.J. Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum. Mol. Genet. 2004;13:659–667. doi: 10.1093/hmg/ddh071. [DOI] [PubMed] [Google Scholar]
- Vogel F., Bornhövd C., Neupert W., Reichert A.S. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 2006;175:237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukotic M., Oeljeklaus S., Wiese S., Vögtle F.N., Meisinger C., Meyer H.E., Zieseniss A., Katschinski D.M., Jans D.C., Jakobs S. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15:336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Inoue S., Hiroi H., Orimo A., Kawashima H., Muramatsu M. Isolation of estrogen-responsive genes with a CpG island library. Mol. Cell. Biol. 1998;18:442–449. doi: 10.1128/mcb.18.1.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wittig I., Braun H.-P., Schägger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Wittig I., Carrozzo R., Santorelli F.M., Schägger H. Functional assays in high-resolution clear native gels to quantify mitochondrial complexes in human biopsies and cell lines. Electrophoresis. 2007;28:3811–3820. doi: 10.1002/elps.200700367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.