Abstract
Glutathione S-transferases (GSTs) are one of the major families of detoxifying enzymes that detoxifies different chemical compounds including insecticides in different insect species. Among the GST subclasses, sigma GSTs are found to be the most abundant and conserved among different insect orders. These GSTs are found to play an important role in lipid peroxidation as well as detoxification. Cotton aphid, Aphis gossypii is the most damaging sucking pest with a wide range of hosts and vector of more than 50 plant viruses. Resistance to insecticides in A. gossypii is reported in India and in other countries. Glutathione S transferases (GSTs), an oxidative enzyme is understood to have a role in insecticide resistance and plant resistance breakdown. In relation to this, we have focused on the sigma 1 (GenBank Accession No: JN989964.1) and sigma 2 (GenBank Accession No: JN989965.1) GSTs of A. gossypii and their interaction with plant natural compounds and insecticides. Molecular screening of different insecticides (Chlorphinamidine, Mevinphos, Nitenpyrum, Piperonyl butoxide, Tetrachlorovinphos, Pyrethrins, Resmetrin, Pirimicarb and Dinotefuran) and known plant derived natural compounds (Catechin, Gossypol, Myrcene, Kaempferol, P-coumaric acid, Quercetin, Tannins, α-mangostin, Capsaicin, Cinnamic acid, Citronellal, Curcumin, Dicumarol, Ellagic acid, Eugenol, Geriniol, Isoeugenol, Juglone, Menadione, Methyl jasmonate, Morin, Myricetin, Myristicin, Piperine, Plumbagin, Tangitinin C, Thymol, Vanillin, Alpha pipene, α-terpineol Apigenin and β-Caryophyllene) with sigma 1 and sigma 2 GST protein models was completed using Maestro 9.3 (Schrodinger, USA). This exercise showed the binding of piperonyl butoxide with sigma 1 GST and tannin with sigma 2 GST for further consideration.
Background
In the plant–insect ecosystem, there is an arms race between the plant and insect to encounter each other through array of chemically mediated interactions. Plants produce different kinds of secondary metabolites that may negatively influence the behaviour and biology of those insects that interacts with the plants either for feeding, shelter or oviposition. Similarly, insects employ behavioural and physiological adaptations to encounter the chemical armory of the plants. Insect detoxification enzymes (esterase and glutathione S-transferase) and oxidoreductases (polyphenol oxidase, peroxidase, and catalase) are one of the important agents used by insects that involved in the resistance against the secondary plant metabolites found in host plants. Glutathione S-transferases (GSTs) belong to a protein family (EC No. 2.5.1.18) and are the important detoxifying enzymes found in bacteria, plants, vertebrates, insects and mammals [1]. These enzymes catalyze the conjugation of electrophilic molecules with reduced glutathione (GSH) and the convert the products to more water soluble and excretable form. In insects, the GST activity is found to be present in the midgut [2, 3], fat body, haemolymph and other tissues [4]. Among the GSTs, sigma class GSTs is one of the largest GST subfamilies which exhibit multiple functions and are found to be present in vertebrate as well as invertebrate animals [5]. Sigma class GST genes can be said as the most common or second most common genes among different insect order like hemiptera, orthoptera, coleoptera and hymenoptera [6]. The previous studies on Drosophila sigma GST had revealed that sigma GSTs were associated with an indirect flight muscle and only possess oxidant role [7]. However, it is observed that these GSTs also play a role in xenobiotic detoxification and insecticide resistance [8]. The cotton aphid, Aphis gossypii Glover (Hemiptera: Aphididae) is a cosmopolitan species colonizing more than 600 host plants. It is the vector of more than 50 plant viruses and is a major pest of many crops, including melon and other cultivated members of the Cucurbitaceae family [9]. A. gossypii has developed resistance against insecticides like monocrotophos, acephate, dimethoate, phosphamidon and triazophos in Guntur district of Andhra Pradesh in India [10]. It has been found that virus transmission can reduce the cotton yield up to 30-40%. To manage insecticide resistance, application of piperonyl butoxide (PBO) along with the insecticides is used [11]. The PBO is found to be a potential inhibitor of GST in combination with other insecticides [12]. Likewise, plant natural compounds have been identified to inhibit insect GSTs that includes plant phenols (quercetin, ellagic acid, juglone, menadione and plumbagin), ethacrynic acid, organotin compounds and hydroxyamic acids [13].
In context with the issue mentioned above, we have focused on the two GSTs viz., sigma 1 (GenBank Accession No: JN989964.1) and sigma 2 (GenBank Accession No: JN989965.1). We have carried out the molecular docking of sigma 1 and sigma 2 GST protein models of cotton aphid A. gossypii with different insecticides and previously studied plant natural compounds that act as GST inhibitors. As the structures were not deposited in the PDB, we have manually prepared protein structure models for these GSTs by using different bioinformatics tools and software which were then validated with standard range of parameters. The molecular docking was performed using Maestro 9.3 (Schrodinger, USA). Based on the glide score the potent GST inhibitors for both the GSTs were identified.
Methodology
Generation of 3D model of sigma 1 and sigma 2 GSTs of A. gossypii and their validation:
PDB structure of sigma 1(accession No: JN989964.1) and sigma 2 GST (accession No: JN989965.1) in A. gossypii were generated using Swiss model workspace [14] in automated mode by submission of protein FASTA sequences collected from GenBank database of National Center for Biotechnology Information (NCBI) [15]. The models were checked for quality by using Structural Analysis and Verification Server NIH MBI Laboratory for Structural Genomics and Proteomics [16] and resulting values were compared with the standard values of parameters like sequence identity, E-value, QMEANscore4, QMEAN Z score, ERRAT, Verify 3D and Ramachandran plot.
Active site residue prediction and ligand selection for docking:
Active site residues used for grid generation in docking of sigma 1 and sigma 2 GSTs were predicted using Q-Site Finder tool [17]. Ligands were selected based on the previously studied different GST activity inhibitors that include different insecticides (9), natural compounds and secondary metabolites in cotton as well as other plant sources (32). The chemical structures for these compounds were downloaded from PubChem database [18] in SDF format.
Sigma 1 and sigma 2 GST model structure preparation:
Protein model structure for sigma 1 and sigma 2 GSTs was prepared using protein preparation wizard in Schrodinger Maestro 9.3 software that corrects and optimize the protein structures. Energy minimization for models (sigma 1 and sigma 2 GST) was done using OPLS_2005 force field. The active sites of the proteins were defined for generating the grid.
Ligand structures preparation and grid generation:
The ligand structures obtained were prepared using Ligprep wizard in Schrodinger Maestro 9.3 by submission of ligands in SDF format as an input. Ligprep optimizes the ligand structures. The force field OPLS_2005 was used for minimization and the ionization states were generated at the default pH of 7.0 ± 2.5 using Epik. In Ligprep, desalt option was used to exclude other molecules from the ligand structure. Tautomers were generated using Generate Tautomer option. The option Retain specified chiralities were used to order to fix the irregularities of ligand. Active site residues obtained for both the GSTs were added in grid generation wizard of Maestro 9.3 and gird generation was performed.
Molecular docking of sigma 1 and sigma 2 GSTs:
In order to know the binding mode of inhibitors of sigma 1 and sigma 2 GSTs, molecular docking of sigma 1 and sigma 2 GSTs model was carried out using XP (extra precision) module in which model was docked in different orientations with the ligands. Performance of GST inhibitors was analyzed based on the parameters like Glide score, H-bonding score, H-bond interaction and bond distance (Å). For ligands with multiple Glide score in different orientations, the best scoring pose and XP Glide score were taken into consideration.
Result & Discussion
Generation of 3D PDB model of sigma 1 and sigma 2 GSTs of A. gossypii and quality check:
The 3D structure model for sigma 1 and sigma 2 GSTs of A. gossypii were downloaded from Swiss model work space in PDB format (Figure 1 a & b), which was further used for identification of active site residues and protein preparation in docking. When the sequence similarity between the target and the template sequence drops below 40%, the quality of model obtained is reduced [19]. The sigma 1 and sigma 2 GSTs had shared homology of 48% and 43% with Chain A crystal structure of the Drosophila GST-2 in complex with glutathione (PDB ID : 1M0U). Hence, our models were quite good in terms of similarity between target and template. The raw Q mean score values should be in the range from 0 to 1 for good models [20]. QMEANscore4 obtained from swiss model workspace had shown values of 0.722 and 0.714 for sigma 1 and sigma 2 GSTs respectively, which has satisfied the range of standard parameters. The standard value of QMEAN Z score should not be too much negative [21] is satisfied by QMEAN Z score of sigma 1 and sigma 2 GSTS which are -0.962 and -1.076 respectively. Generally structures with high resolution produce ERRAT values around 95% or higher, whereas for lower resolutions (2.5 to 3A) the average value is 91%. The values for sigma 1 and sigma 2 GSTs were 84.375 and 92.746 respectively which were not showing much difference with the standard parameters. Verify 3D output had shown that at least 80% of the amino acids have scored >= 0.2 in the 3D/1D profile which has satisfied the verified 3D test [22]. A good quality protein model should have above 90% of the residues in the “core” regions Ranchandran plot (Morris et al., 1992) as it is a measure of stereochemical quality of proteins. In Ramachandran plot analysis, sigma 1 and sigma 2 GSTs had shown 91.6% and 89.6% residues in most favored regions showing the good quality of our models Table 1 (see supplementary material). Active site residues (LEU, ILE, ASN, PHE, TYR, LYS, VAL and GLY) predicted for model sigma 1 and sigma 2 GSTs Table 2 (see supplementary material). The minimized energy for the optimized model structures of sigma 1 and sigma 2 GSTs using OPLS_2005 force field were found to be -389.522 kcal/mol and - 280.469 kcal/mol respectively. In molecular docking, the lower glide score with no hydrogen bonding were not taken into consideration. In Glide, the better binders possess more negative scores than loose or non binders. Among the insecticide class, Piperonyl butoxide was found to show the best glide score for the model sigma 1 GST (-6.8) with good H bond score (-4.6), whereas it was less in the case of sigma 2 GST (-5.5) for the same. Among the compounds in cotton plants, tannin was found to have the highest score in all the compound classes for both the sigma 1 (-8.9) and sigma 2 GST models (-8.4) with strong H-bonding score of -4.6 and -5.2 respectively (Figure 1 c, d, e & f). Likewise, among other plant natural compounds ellagic acid was found to show a good glide score for both the sigma 1 (-6.2) as well as sigma 2 (-6.1) GSTs Table 3, 4 (see supplementary material).
Figure 1.
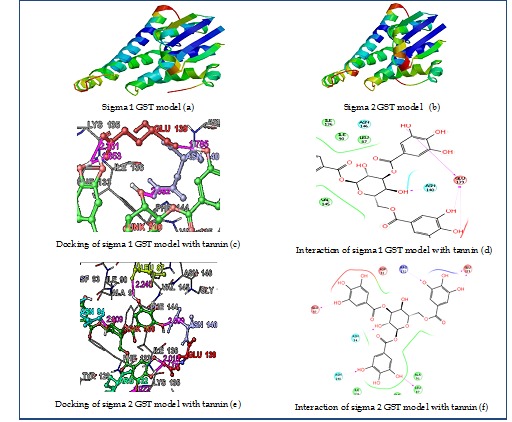
Structure of 3D models generated for sigma1 and sigma 2 GSTs using swiss model workspace
Conclusion
From the molecular docking results, based on the glide score obtained for sigma 1 GST, it was found that the compounds were in the order of Tannins> PBO> α-mangostin> Capsaicin> Ellagic acid> Myricetin> Catechin as GST inhibitors, whereas the order was Tannins> Gossypol> Ellagic acid> Quercetin> α- mangostin for sigma 2 GST.
Supplementary material
Footnotes
Citation:Gawande et al, Bioinformation 10(11): 679-683 (2014)
References
- 1.Singh M, et al. Gene. 2000;247:167. doi: 10.1016/s0378-1119(00)00102-5. [DOI] [PubMed] [Google Scholar]
- 2.Tate LG, et al. Comp Biochem Physio. 1982;72:75. doi: 10.1016/0306-4492(82)90207-6. [DOI] [PubMed] [Google Scholar]
- 3.Snyder MJ, et al. Insect Biochem Mol Biol. 1995;25:455. doi: 10.1016/0965-1748(94)00083-b. [DOI] [PubMed] [Google Scholar]
- 4.Franciosa H, Berge JB. Insect Biochem Mol Biol. 1995;25:311. doi: 10.1016/0965-1748(94)00053-k. [DOI] [PubMed] [Google Scholar]
- 5.Qin G, et al. J Insect Physiol. 2012;58:220. doi: 10.1016/j.jinsphys.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Zhou WW, et al. PLoS ONE. 2013;8:e56604. doi: 10.1371/journal.pone.0056604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agianian B, et al. J Mol Biol. 2003;326:151. doi: 10.1016/s0022-2836(02)01327-x. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto K, et al. Biosci Biotechnol Biochem. 2007;71:553. doi: 10.1271/bbb.60592. [DOI] [PubMed] [Google Scholar]
- 9.Ebert TA, Cartwright B. Southwest Entomol. 1997;22:116. [Google Scholar]
- 10.Kumar PSS, et al. Indian J Plant Protec. 2008;36:224. [Google Scholar]
- 11.Khan HA, et al. PLoS ONE. 2013;8:e60929. doi: 10.1371/journal.pone.0060929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young SJ, et al. Pest Manag Sci. 2005;61:397. doi: 10.1002/ps.996. [DOI] [PubMed] [Google Scholar]
- 13.Yu SJ, Abo-Elghar GE. Pestic Biochem Physiol. 2000;68:173. [Google Scholar]
- 14. http://swissmodel.expasy.org/workspace.
- 15. http://www.ncbi.nlm.nih.gov.
- 16. http://nihserver.mbi.ucla.edu.
- 17. http://www.modelling.leeds.ac.uk/qsitefinder.
- 18. http://www.ncbi.nlm.nih.gov/pccompound.
- 19.Schwede T, et al. Nucleic Acids Res. 2003;31:3381. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sohpal VK, et al. Int J Adv Res Sci Eng. 2013;2:56. [Google Scholar]
- 21.Demchuk OP, et al. Acta Biol Sinica. 2011;54:15. [Google Scholar]
- 22.Luthy RJ, et al. Nature. 1992;356:83. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


