Abstract
Disassembly of focal adhesions (FAs) allows cell retraction and integrin detachment from the ECM, processes critical for cell movement. Growth of MT (microtubule) can promote FA turnover by serving as tracks to deliver proteins essential for FA disassembly. The molecular nature of this FA “disassembly factor”, however, remains elusive. By quantitative proteomics, we identified MAP4K4 (mitogen-activated protein kinase kinase kinase kinase 4) as a FA regulator that associates with MTs. Conditional knockout (cKO) of MAP4K4 in skin stabilizes FAs and impairs epidermal migration. By exploring underlying mechanisms, we further show that MAP4K4 associates with EB2, a MT binding protein, and IQSEC1, a guanine nucleotide exchange factor (GEF) specific for Arf6, whose activation promotes integrin internalization. Together, our findings provide critical insights into FA disassembly, suggesting that MTs can deliver MAP4K4 toward FAs through EB2, where MAP4K4 can in turn activate Arf6 via IQSEC1 and enhance FA dissolution.
Keywords: cytoskeleton, cell migration, microtubule, Focal adhesion, MAP4K4
INTRODUCTION
Cell migration is an essential process for developmental morphogenesis, wound healing and tumor metastasis. The intricate, multi-step process of directional cell movement requires integrated activities of the cytoskeleton, membrane, and cell/ECM adhesions (Lauffenburger and Horwitz, 1996; Rodriguez et al., 2003). Disassembly of FAs, organelles that connect the cytoskeletal network and ECM, is a critical part of this process. Interestingly, MTs have been observed to target peripheral FAs (Kaverina et al., 1999), a process mediated by mammalian spectraplakin protein, ACF7 (Wu et al., 2008; Wu et al., 2011). MT targeting promotes FA turnover, likely through MT-mediated delivery of key disassembly factors (Krylyshkina et al., 2002). Additional explorations have also posited roles for FAK (focal adhesion kinase) and dynamin in MT-mediated FA dynamics, suggesting involvement of endocytosis (Ezratty et al., 2005), but the molecular nature of the mysterious “disassembly factor” remains elusive.
Recent development of SILAC (stable isotope labeling by amino acids in cell culture) technology offers us an effective approach to quantitatively compare the FA proteome in normal vs. MT-depleted cells (Kuo et al., 2011; Ong et al., 2003). Our analysis revealed an intriguing candidate for the FA “disassembly factor”, MAP4K4, whose presence in FAs is dependent upon an intact MT network. MAP4K4 is a serine/threonine protein kinase that belongs to the germinal-center kinase (GCK)-IV group of yeast sterile 20 protein (Ste20) kinase family (Dan et al., 2001). It has been shown that MAP4K4 may regulate JNK signaling pathway in mammalian cells and mediate various cellular processes, including cell motility (Xue et al., 2001). The MAP4K4 homologs in Drosophila (Misshapen) and C. elegans (MIG-15) have also been implicated in cell movement (Chapman et al., 2008; Su et al., 1998). Recently, it has been shown that mutations of Msn (Misshapen) block migration of Drosophila ovary border cells (Lewellyn et al., 2013). Interestingly, Msn can decrease cell surface integrin level in Drosophila epithelial cells and facilitate detachment of cells’ trailing edges during cell migration (Lewellyn et al., 2013), implying a potential role of MAP4K4 in FA dynamics.
Mammalian skin provides an excellent platform to investigate cytoskeletal dynamics and cell migration in vivo (Blanpain and Fuchs, 2006; Wu et al., 2008; Wu et al., 2011). Here, we employ conditional gene targeting to ablate MAP4K4 expression in skin epidermis. Our results have uncovered essential roles for MAP4K4 in skin wound healing and epidermal migration, which we trace to its function in controlling dynamics of FAs. To probe deeper into the role of MAP4K4 in FA dynamics and decipher its connection with MT networks, we further identified two key binding partners of MAP4K4, EB2 (end binding 2) and IQSEC1 (IQ motif and SEC7 domain-containing protein 1).
The three MT end-binding proteins (EB1, EB2 and EB3) in mammalian cells can track the plus ends of growing MTs (Akhmanova and Steinmetz, 2008). It has been established that EB1 and EB3 together can regulate MT dynamics by promoting MT growth and suppressing catastrophe (Komarova et al., 2009; Komarova et al., 2005). In contrast, EB2 does not play a direct role in MT dynamic instability (Komarova et al., 2009). Little is known about EB2’s cellular function (Goldspink et al., 2013). Our results raise the intriguing possibility that EB2 can act as an adaptor protein to recruit MAP4K4 to MTs, thus promoting FA turnover and cell motility.
IQSEC1 is a guanine nucleotide exchange factor with reported specificity toward Arf6 (Someya et al., 2001), which is a small GTPase critically involved in endocytosis and vesicle recycling (D'Souza-Schorey and Chavrier, 2006). Arf6-mediated trafficking controls multiple steps that impinge upon cell migration. It has been demonstrated that Arf6 can regulate the internalization and trafficking of various membrane adhesion proteins, including integrin at FAs (Schweitzer et al., 2011). Depletion of IQSEC1 in mammalian cells leads to an accumulation of integrin receptors on cell surface and stabilization of cell adhesion to ECM (Dunphy et al., 2006; Hiroi et al., 2006). IQSEC1 has also been shown to participate in cancer cell invasion and phagocytosis of monocytic phagocytes via its Arf6 GEF activity (Morishige et al., 2008; Someya et al., 2010). Its interaction with MAP4K4 suggests that MT-delivered MAP4K4 can enhance FA turnover by subsequent activation of Arf6.
Taken together, our results both identify the potential FA “disassembly factor” delivered by MTs and unravel a signaling cascade centering on MAP4K4. Our results provide important mechanistic insights into how MT networks regulate FA turnover and directional cell movement.
RESULTS
Identification of MAP4K4 as a potential MT-dependent FA “Disassembly Factor”
Disruption of MT network with nocodazole leads to FA stabilization in various cell lines (Bershadsky et al., 1996; Ezratty et al., 2005; Wu et al., 2008). Like mouse primary keratinocytes, HaCaT cells (human keratinocyte cell line) adhere to fibronectin matrix and develop robust FAs at the cell periphery (Fig. 1A). Treatment with nocodazole dramatically reduced the rate of FA turnover in HaCaT cells (Supplementary Fig. 1A).
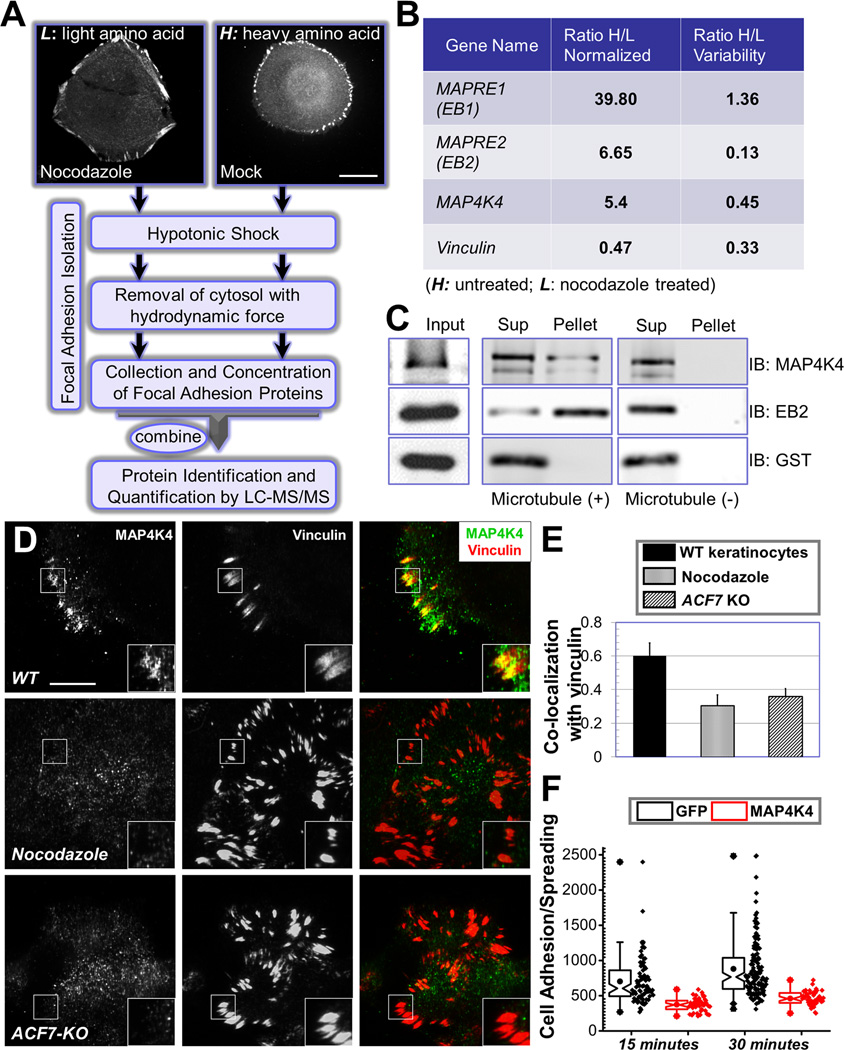
Figure 1. Identification of MAP4K4 as a MT-dependent FA protein.
(A) Flow diagram of the protocol used to isolate FA proteins for SILAC analysis. HaCaT cells were stained with α-vinculin antibody to visualize FAs (top two panels). Scale bar = 20 µm. (B) Candidates that exhibit most significant changes in protein levels upon nocodazole treatment. Ratios of each candidate protein retrieved from heavy or light cells (H/L) were shown together with the ratio variability. MAPRE1 or 2: microtubule associated protein, RP/EB family member 1 or 2. The level change of vinculin is included as a control. (C) Interaction with MTs was examined by co-sedimentation assay. Presence of protein in the original lysate (input), pellet of centrifugation as well as supernatant was determined by immunoblot with different antibodies as indicated. (D) WT mouse primary keratinocytes treated with or without nocodazole and ACF7 KO keratinocytes were fixed and subjected to immunofluorescence staining with different antibodies as indicated. Stained cells were examined by TIRF microscopy. Scale bar = 20 µm. (E) Co-localization between MAP4K4 and FAs was determined by Pearson correlation coefficient. N=10, and P<0.01. (F) GFP or GFP-MAP4K4 cells were harvested and replated onto fibronectin-coated surface for 15 or 30 minutes. Cells were then fixed and stained. Adhesion and spreading of cells (cell area) were quantified with Image J and shown as Box and Whisker plots. Box and whisker plots indicates the mean (empty square within the box), 25th percentile (bottom line of the box), median (middle line of the box), 75th percentile (top line of the box), 5th and 95th percentile (whiskers), 1st and 99th percentile (solid diamonds) and minimum and maximum measurements (solid squares), with actual data points shown at right. N>50 for each group, and P<0.01 between GFP and MAP4K4 for each time point.
In order to quantitatively assess the proteomic changes in FAs upon loss of MTs, we applied SILAC technology by metabolically labelling two populations of HaCaT cells with either regular lysine (light) or deuterium-substituted lysine (heavy). Cells were maintained in the labelling medium for more than 10 doublings, leading to a labelling efficiency greater than 99% (data not shown). FA proteins were then fractionated from these two populations by an established protocol (Kuo et al., 2011) with or without prior treatment of nocodazole (Fig. 1A and supplementary Fig. 1B). Isolated proteins were then combined and subjected to analysis by tandem mass spectrometry (LC-MS/MS). Interestingly, the two proteins that displayed most significant decrease upon nocodazole treatment were identified as EB1 and EB2 (Fig. 1B, and supplementary table 1). This finding is consistent with previous reports that MT plus ends can be targeted to FAs (Ezratty et al., 2005; Kaverina et al., 1999; Wu et al., 2008). Thus, plus end tracking proteins, such as EB1, are present in the FAs in untreated cells, but lost upon nocodazole treatment. Our analysis revealed another intriguing candidate, MAP4K4, whose abundance in FAs is consistently reduced (~5 fold) upon dissolution of MT networks (Fig. 1B).
Immunoblot analyses verified our proteomic results, showing diminished level of EB1, EB2, and MAP4K4 in FAs derived from nocodazole treated cells (Supplementary Fig. 1C). To examine the potential connection between MAP4K4 and MTs, we carried out a MT pull down assay. Like EB2, significant amount of MAP4K4 was co-precipitated with MTs (Fig. 1C). To determine whether MT can deliver MAP4K4 toward FAs, we examined localization of endogenous MAP4K4 by TIRF (total internal refection fluorescence) microscopy. Our results reveal significant enrichment of endogenous MAP4K4 at the tips of MT bundles that associate with FAs in cultured keratinocytes (Supplementary Fig. 1D). Lack of detectible MAP4K4 signal in cultured MAP4K4 KO cells confirmed specificity of our staining. Interestingly, co-localization between endogenous MAP4K4 and FA is markedly reduced in nocodazole treated cells or ACF7 null cells (Fig. 1D–E). ACF7 has been demonstrated to play an essential role to mediate targeting of MTs toward FAs (Wu et al., 2008). The reduction of MAP4K4 localization is specific to FA, as epi-fluorescence imaging of the same cells (Fig. 1D) show comparable level of MAP4K4 (Supplementary Fig. 1E), and immunoblots confirmed that nocodazole treatment or loss of ACF7 does not significantly change the level of endogenous MAP4K4 (Supplementary Fig. 1F). Together, these results suggest that MAP4K4 could serve as the potential candidate that mediates crosstalk between MTs and FAs.
In support of this hypothesis, accumulating evidence suggests that MAP4K4 and its homologues in other model organisms act as pro-migratory kinases (Chapman et al., 2008; Su et al., 1998; Xue et al., 2001). Mutation of Msn, the Drosophila homolog of MAP4K4, increases surface integrin level and impairs cell detachment (Lewellyn et al., 2013), strongly suggesting a role of MAP4K4 in FA turnover. In line with previous findings, we observed that exogenous expression of GFP-MAP4K4 but not GFP alone in mouse primary keratinocytes caused cell to round up and detach from the matrix (data not shown). When exposed to fibronectin, cells expressing GFP-MAP4K4 exhibited significantly delayed adhesion and spreading comparing with GFP-expressing cells (Fig. 1F). Taken together, our results identify MAP4K4 as a potential regulator of FA dynamics that associates with MTs.
Conditional Knockout of MAP4K4 Leads to Aberrant Wound Repair and Defects in Epidermal Cell Migration
While adult mouse skin serves as an ideal system to study cell movement in vivo (Wu et al., 2008), deletion of MAP4K4 in mice leads to early embryonic lethality (Xue et al., 2001). To circumvent this issue, we developed skin cKO of MAP4K4. By homologous recombination, two loxP sites were inserted in the 5’ UTR and the 2nd intron of MAP4K4 respectively. To conditionally target MAP4K4 in skin, we bred MAP4K4fl/fl mice with K14-Cre recombinase transgenic mice, which efficiently excised floxed exons by embryonic day E15.5 (Wu et al., 2008) (Fig. 2A). Neonatal mice genotypic for K14-Cre and MAP4K4fl/fl alleles (cKO) were born in the expected Mendelian numbers and grew to adulthood. Western blot confirmed the loss of MAP4K4 protein in cKO skin epidermis (Fig. 2A).
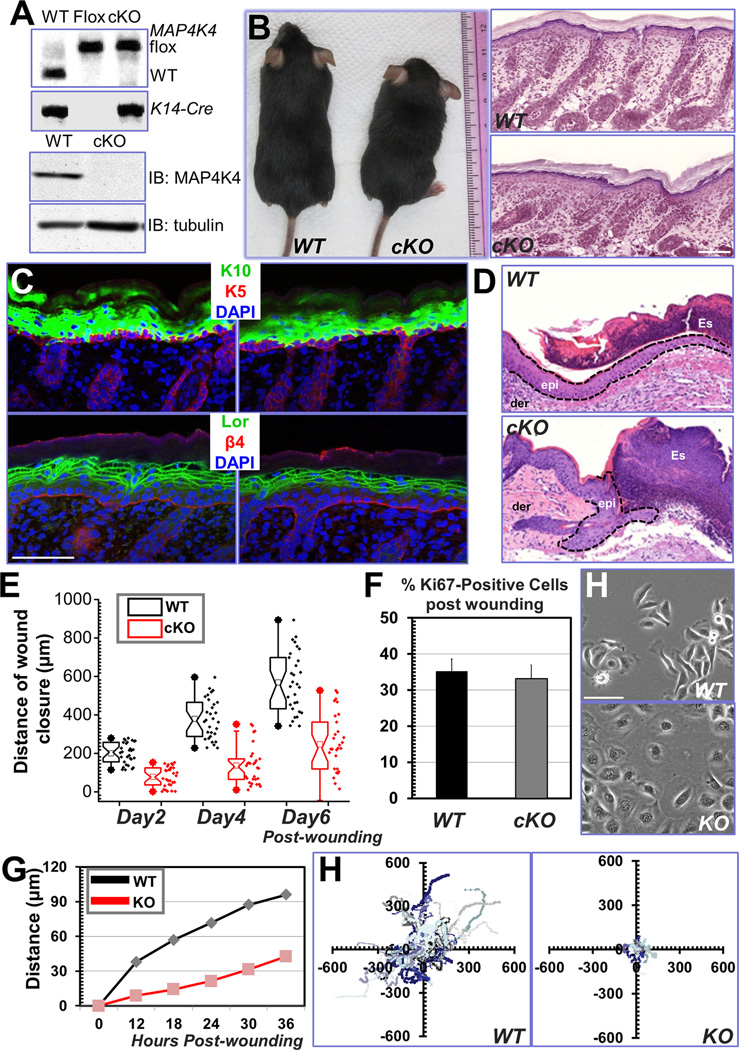
Figure 2. Cell migration defects in MAP4K4-deficient keratinocytes.
(A) PCR genotyping of MAP4K4 cKO mouse DNAs (top panel; WT: wildtype, Flox: MAP4K4fl/fl; cKO: MAP4K4fl/fl: K14-Cre). Immunoblot analysis of P0 epidermal extracts (20 µg total protein) probed with different antibodies as indicated. (B) MAP4K4 cKO mice can grow to adulthood (left panel). Histological analysis (hematoxylin/eosin staining) confirms rather normal skin and hair follicles in cKO skin (right panel). Scale bar = 50 µm. (C) Dorsal skins of neonatal mice were immunostained with different antibodies as indicated (K5: keratin 5, K10: keratin 10, Lor: Loricrin, β4: β4-integrin, CD104). Scale bar=50 µm. (D) Wound healing as monitored by histological staining of skin sections at the wound edges 4 days after injury. Halves of wound sections are shown. Epi: epidermis; der: dermis; Es: eschar. Dotted lines denote dermal–epidermal boundaries. Scale bar = 50 µm. (E) Quantification of the length of hyperproliferative epidermis generated at times indicated after wounding. Number of biological replicates (N) =30 for each group. P<0.01 for each time point. (F) Quantification of Ki67-positive cells present in wound HE. Error bars represent standard deviations (SD). N=10, P=0.22. (H) Morphology of primary keratinocytes isolated from WT or MAP4K4 cKO skin. Scale bar = 50 µm. (G) Migration of confluent monolayers of mouse keratinocytes cultured from MAP4K4 cKO and WT littermates was assessed by in vitro scratch-wound assays. The kinetics of in vitro wound healing was quantified. N=3, P<0.01. (H) Movements of individual keratinocytes were traced by videomicroscopy. Migration tracks of multiple cells for each group (WT or KO) are shown here as scatter plots. N=30.
Although MAP4K4 skin cKO animals usually appeared smaller comparing with their littermates, they manifested no gross morphological changes in skin or hair coat (Fig. 2B). Histologically, epidermal homeostasis appeared normal and immunofluorescence staining with antibodies against β4 integrin (basal surface of the epidermis), keratin 5 (basal layer), keratin10 (spinous layer), and loricrin (granular layer) all displayed localization patterns similar to WT (Wild Type) littermate’s skin (Fig. 2C).
Directional cell movement makes critical contribution to skin wound repair. When challenged to respond to injury, MAP4K4 cKO skin exhibited a significant delay in repairing full-thickness wounds as compared to WT skin. Histological analysis and quantification revealed that the area of hyperproliferative epithelium (HE) that typically proliferates and migrates into the wound site was diminished by more than 50% over 2–6 days following injury (Fig. 2D and E). Interestingly, despite the delay in wound-closure, no significant alterations were found in cell proliferation upon loss of MAP4K4, as judged by labeling for Ki67 (Fig. 2F). Thus, the delayed wound healing response in MAP4K4-deficient skin seemed more likely to be rooted in potential defects in cell migration.
To examine this hypothesis, we isolated and analyzed primary mouse keratinocytes from WT and cKO skin (Fig. 2H). Cells deficient for MAP4K4 exhibited a flatter and round morphology, resembling ACF7 null cells that develop abnormally stable FA and migrate poorly in vitro (Wu et al., 2008). We then monitored cell migration with isolated keratinocytes, during recovery of scratch wounds introduced into cell monolayers. Whereas WT keratinocytes usually closed the gap usually within 36 hours, KO keratinocytes moved only ~30% of this distance into the wound (Fig. 2G and supplementary Fig. 2A). Videomicroscopy permitted image and monitoring the velocities and directed migration of individual keratinocytes. Our results demonstrated that MAP4K4 KO cells moved significantly more slowly compared to their WT counterparts. Quantification of cell motility revealed a dramatic decrease in the average speed of KO keratinocytes (Figure 2H and supplementary Fig. 2B).
MAP4K4 Deficiency Leads to Aberrant Regulation of FA Dynamics
To determine the role of MAP4K4 in FA dynamics, we examined cell/ECM adhesions after MAP4K4 deletion. Immunofluorescence microscopy showed significantly enhanced labeling of FAs in MAP4K4 null keratinocytes relative to WT controls (Fig. 3A). Quantification of the presence of Vinculin showed a significant increase in the size of FAs in MAP4K4 null cells (Fig. 3B).
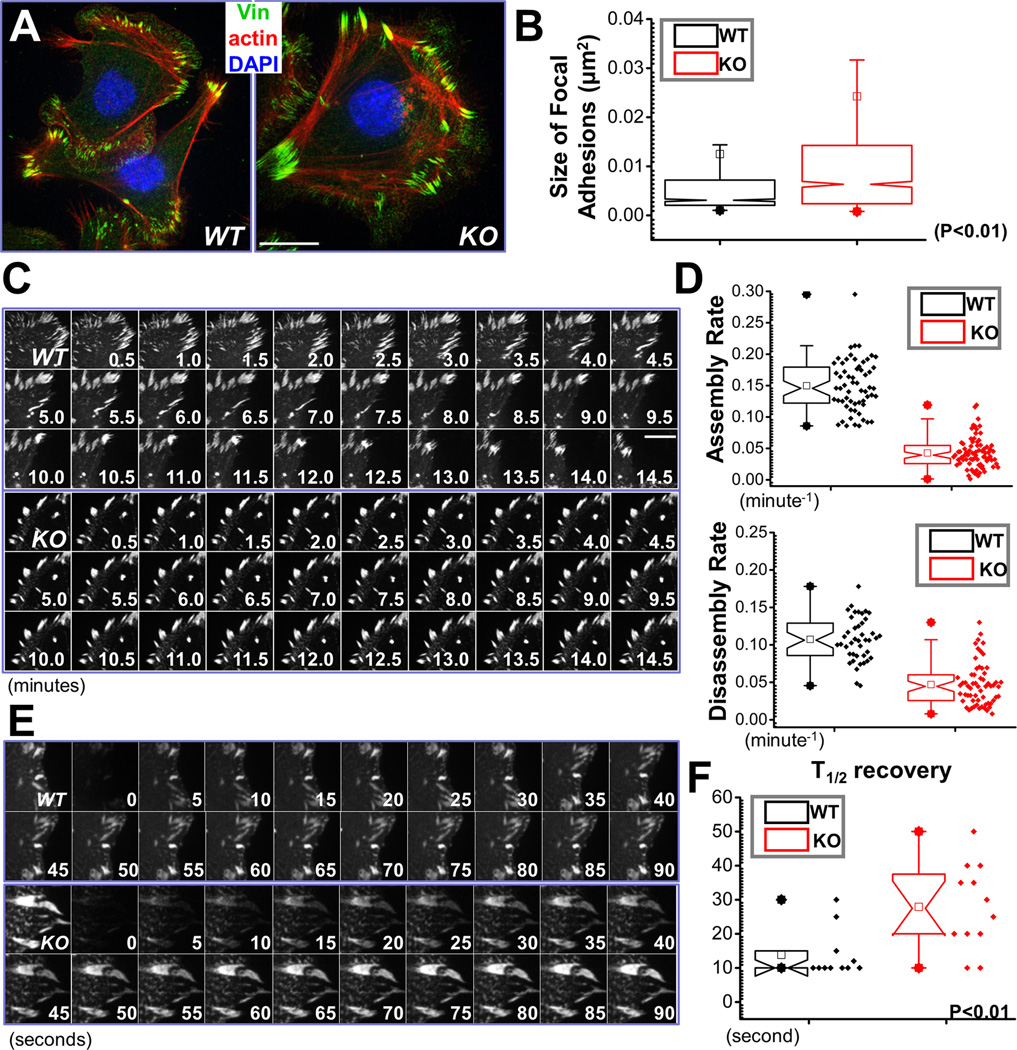
Figure 3. MAP4K4 regulates FA dynamics.
(A) Immunolabeling of WT and MAP4K4 null cells for F-actin (red), nuclei (DAPI; blue), and FA marker vinculin (Vin; green). Scale bar = 20 µm. (B) Box and whisker plot indicating the size distribution of FAs in WT and KO cells. N>300, and P<0.01. (C) Representative time-lapse images (montages) of DsRed-Zyxin expressing keratinocytes. Note formation and dissolution of FAs in WT cells and very static FAs in KO cells. Scale bar = 10 µm. (D) Box and whisker plots revealing slow assembly and disassembly rates of FAs in MAP4K4 KO cells relative to their WT counterparts. N>50 for each group, and P<0.01. (E) Fluorescence recovery after photobleaching (FRAP) was used to visualize reduced dynamics of FAs in MAP4K4 KO vs WT cells. Representative time-lapse images (montage) of FAs are shown. Scale bar = 5 µm. (F) Box-and-whisker diagram quantifying the differences in half-time (T1/2) of FRAP between WT and KO cells. N>10, and P<0.01.
We further employed confocal videomicroscopy to trace and examine the behavior of individual FA (Wu et al., 2008). To visualize FAs, we transfected cells with plasmids encoding DsRed-Zyxin, a fluorescently labeled FA marker protein (Wu et al., 2008). Representative examples of the perturbations in FAs dynamics arising from MAP4K4-deficiency are shown in complete form in Supplementary Video 1, and in montages in Figure 3C. During the interval of observation, FAs in MAP4K4 null cells were often static, while many FAs in WT keratinocytes underwent continual bouts of formation, maturation and disassembly. Quantification of the kinetics of individual FAs revealed dramatic decrease in both the assembly and disassembly rates of FAs in KO cells (Fig. 3D).The defects in FA dynamics were further substantiated by fluorescence recovery after photobleaching (FRAP) experiments (Fig. 3E and supplementary Fig. 3). By comparison with GFP-Paxillin transfected WT cells, we found that deletion of MAP4K4 resulted in a substantial increase in half-times of fluorescence recovery after bleaching (Fig. 3F). Together, these data provide compelling evidence that MAP4K4 regulates cell migration by promoting FA dynamics.
Identification of MAP4K4 Binding Partners
MT targeting promotes FA turnover (Ezratty et al., 2005; Kaverina et al., 1999; Rodriguez et al., 2003; Wu et al., 2008). The phenotype of MAP4K4 deletion is highly analogous to what we observed in ACF7 KO cells (Wu et al., 2008). However, unlike ACF7 null cells, MAP4K4 KO cells exhibit bundled MT filaments that associate with peripheral FAs (Fig. 4A). Confocal videomicroscopy further indicates comparable MT targeting frequency between WT and MAP4K4 KO cells (Supplementary Fig. 4A). Additionally, MT plus end dynamics are largely unaltered in MAP4K4 KO cells (Fig. 4B). These results suggest that MAP4K4 does not regulate MT dynamics or FA targeting, but instead more likely acts downstream of MTs to facilitate FA turnover.
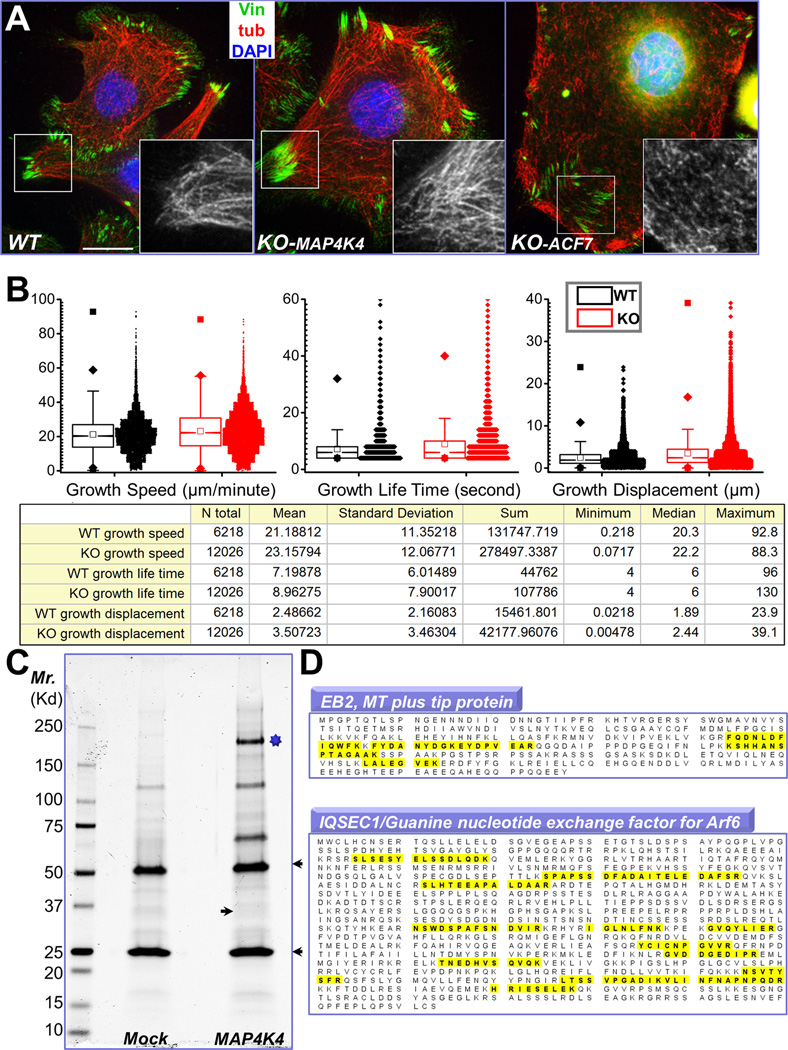
Figure 4. Identification of MAP4K4 binding partners by tandem affinity purification.
(A) Immunofluorescence for MTs (red), FAs (vinculin, green), and nuclei (DAPI, blue) shows altered MT organization in ACF7 KO cells but not in MAP4K4 deficient cells. Boxed areas are magnified as insets, where only MT staining is shown. Note bundled MT filaments merge toward peripheral FAs in WT and MAP4K4 KO cells. Scale bar = 20 µm. (B) Box and whisker plots for EB1 plus end dynamics in WT and MAP4K4 KO cells. Descriptive statistics of all the results, including mean and standard deviation, were shown in the table below. (C) MAP4K4 and its associated proteins were isolated by tandem affinity purification and resolved by SDS-PAGE. IgG heavy chain and light chain are marked by arrowheads. The putative band for MAP4K4 is marked by star. Putative band for EB2 is marked by an arrow. (D) Both EB2 and IQSEC1 were found as MAP4K4 binding proteins. Identified peptides for each protein were highlighted (yellow).
Rho family small GTPases are known to play a pivotal in regulating FA dynamics and cell migration (Jaffe and Hall, 2005). However, biochemical pull-down assays revealed comparable overall levels of the GTP-bound state of RhoA, Rac1, and Cdc42 in WT and MAP4K4 KO cells (Supplementary Fig. 4B–D). Talin associates with integrin cytoplasmic tail and is involved in mechanotransduction and activation of integrin (Calderwood et al., 2013). Proteolysis of talin by calpain has been shown to be critical for FA dynamics (Franco et al., 2004). However, our immunoblot assays demonstrate similar level of talin and cleaved talin in WT and MAP4K4 null cells (Supplementary Fig. 4E). Together, these findings suggest that MAP4K4 regulates FA dynamics through a distinct mechanism other than activation of Rho GTPases or talin proteolysis.
To probe deeply into the role of MAP4K4 in FA dynamics, we performed tandem affinity purification to identify its binding partners. Purified MAP4K4 binding proteins were resolved by electrophoresis and examined by silver staining (Fig. 4C). Interestingly, our analysis with LC-MS/MS identified two MAP4K4 interacting proteins with potential relevance in cytoskeletal dynamics and FA turnover, EB2 and IQSEC1 (Fig. 4D).
MAP4K4 Interaction with EB2
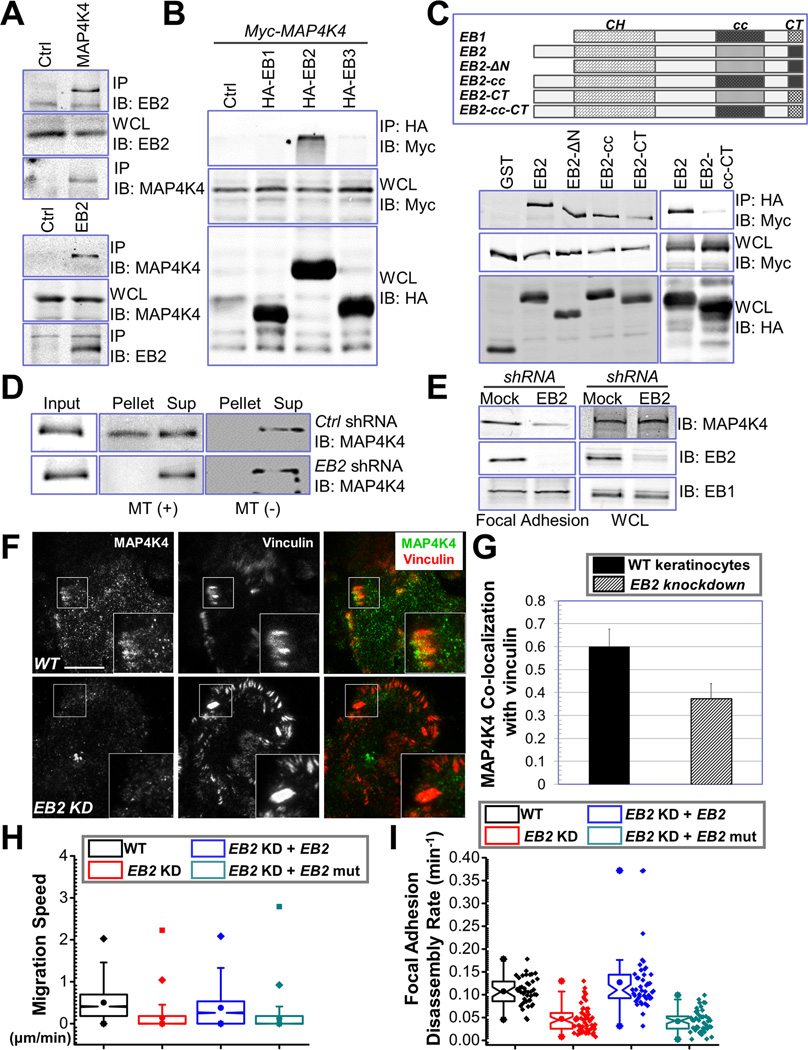
To verify MAP4K4 interaction with EB2, we carried out endogenous coimmunoprecipitation assays. Consistent with MS results, we detected significant amount of EB2 in α-MAP4K4 immunoprecipitates and MAP4K4 in α-EB2 precipitates (Fig. 5A).
Figure 5. EB2 recruits MAP4K4 to MTs and promotes FA dynamics.
(A) Keratinocyte lysate was immunoprecipitated with α-EB2 or α-MAP4K4 antibodies. Immunoprecipitate (IP) as well as whole cell lysate (WCL, 10 µg total protein) was subjected to immunoblot with different antibodies as indicated. (B) HEK293T cells were transfected with different plasmids as indicated. Cell lysates were immunoprecipitated with α-HA antibody and immunoblotted with different antibodies as indicated. For WCL, 10 µg total proteins were used. Note only EB2 specifically pulls down MAP4K4. (C) Schematic representation of EB1, EB2, and various EB2 mutants used for coimmunoprecipitation assays (top panel). CH: calponin homology domain; cc: coiled-coil domain; CT: acidic C terminus. Association of EB2 or EB2 mutants with MAP4K4 was determined by coimmunoprecipitation as described above (lower panel). For WCL, 10 µg total proteins were used. (D) Control or EB2 knockdown cells were transfected with plasmid encoding MAP4K4. MT binding was examined by co-sedimentation assay. Pellet and supernatant as well as an aliquot of pre-cleared cell lysate (input) were immunoblotted to assess the level of MAP4K4. (E) Presence of MAP4K4, EB2, and EB1 in isolated FAs or whole cell lysate (WCL, 20 µg) from control or EB2-shRNA treated cells was determined by immunoblots. (F–G) WT keratinocytes and EB2 knockdown (KD) cells were subjected to immunofluorescence staining and examined by TIRF microscopy (F). Co-localization between MAP4K4 and vinculin was determined by Pearson correlation coefficient and quantified (G). Scale bar = 20 µm. N=10, P<0.01. (H–I) Quantifications of migration velocities and FA disassembly for control, EB2-knockdown (KD) cells, and cells rescued with WT EB2 or EB2 mutant (EB2-cc-CT). Note that only WT EB2 restored FA dynamics and rescued the speed of migration. For cell motility assay (I), N= 30 cells × 120 time points for each group. P<0.01 between WT and EB2 KD; EB2 KD and EB2 KD + EB2; and EB2 KD + EB2 and EB2 KD + EB2 mutant. For FA disassembly (J), N>40 for each group. P<0.01 between WT and EB2 KD; EB2 KD and EB2 KD + EB2; and EB2 KD + EB2 and EB2 KD + EB2 mutant.
Mammalian cells including keratinocytes express three EB proteins that share substantial sequence homology (Komarova et al., 2009). To test the potential interaction with other EBs, we exogenously expressed EB1, EB2, and EB3 together with MAP4K4 in cells. Co-immunoprecipitation showed that MAP4K4 specifically associates with only EB2, but not EB1 or EB3 (Fig. 5B). This specificity is consistent with our SILAC analysis of FAs, which shows comparable fold changes of EB2 and MAP4K4 in FAs upon disruption of MTs (Fig. 1B). Sequence alignment of three EB family members indicates three unique regions in EB2, the N terminus, the acidic C terminus (CT) and an internal region close to the coiled coil domain (cc) (Fig. 5C). To examine their contribution to binding with MAP4K4, we introduced deletion or swapping mutations to EB2 by either removing the N terminus or replacing the unique regions in EB2 with corresponding sequences from EB1 (Fig. 5C). We then carried out co-immunoprecipitation to determine their respective binding affinity to MAP4K4. Our results show that, whereas the N terminus of EB2 is dispensable for MAP4K4 interaction, EB1 swapping mutation at the cc domain or CT diminished association with MAP4K4 at different degree (Fig. 5C). Interaction with MAP4K4 was barely detectible when we used the EB2 mutant containing swapping mutations at both cc domain and CT (Fig. 5C), suggesting that MAP4K4 associates with EB2 at multiple locations.
MAP4K4 associates with MTs (Fig. 1C) but does not possess any recognizable MT binding motif. As a MT binding protein, EB2 could function as an adaptor protein to recruit MAP4K4 to MTs. To test this hypothesis, we prepared lentiviral vector encoding EB2 shRNA sequence as reported (Supplementary Fig. 5A) (Komarova et al., 2009; Komarova et al., 2005) and performed MT pull down assay for MAP4K4. Interestingly, depletion of endogenous EB2 greatly reduces MT binding of MAP4K4 (Fig. 5D). Additionally, knockdown of EB2 leads to a significant decrease of MAP4K4 level in FAs (Fig. 5E–G). These results strongly suggest that EB2 acts as an essential adaptor, permitting MT-delivery of MAP4K4 to FAs.
Consistent with our hypothesis, knockdown of EB2 in mouse primary keratinocytes led to severe defects in cell motility (Fig. 5H). Additionally, FAs in EB2-depleted cells were stabilized, as determined by confocal videomicroscopy (Fig. 5I). Exogenous expression of an EB2 mutant with multiple silent mutations within the shRNA recognition sequence restored cell motility and FA dynamics in knockdown cells (Fig. 5H and I). By contrast, expression of MAP4K4 non-binding mutant of EB2 (cc-CT mutant, Fig. 5C) failed to rescue the defects (Fig. 5H and I), indicating an essential role for MAP4K4 interaction in this process. The cc-CT mutation in EB2 does not alter its MT binding affinity (Supplementary Fig. 5B), or change its ability to track MT plus ends (Supplementary video 2).
MAP4K4 Activates Arf6 through its Interaction with IQSEC1
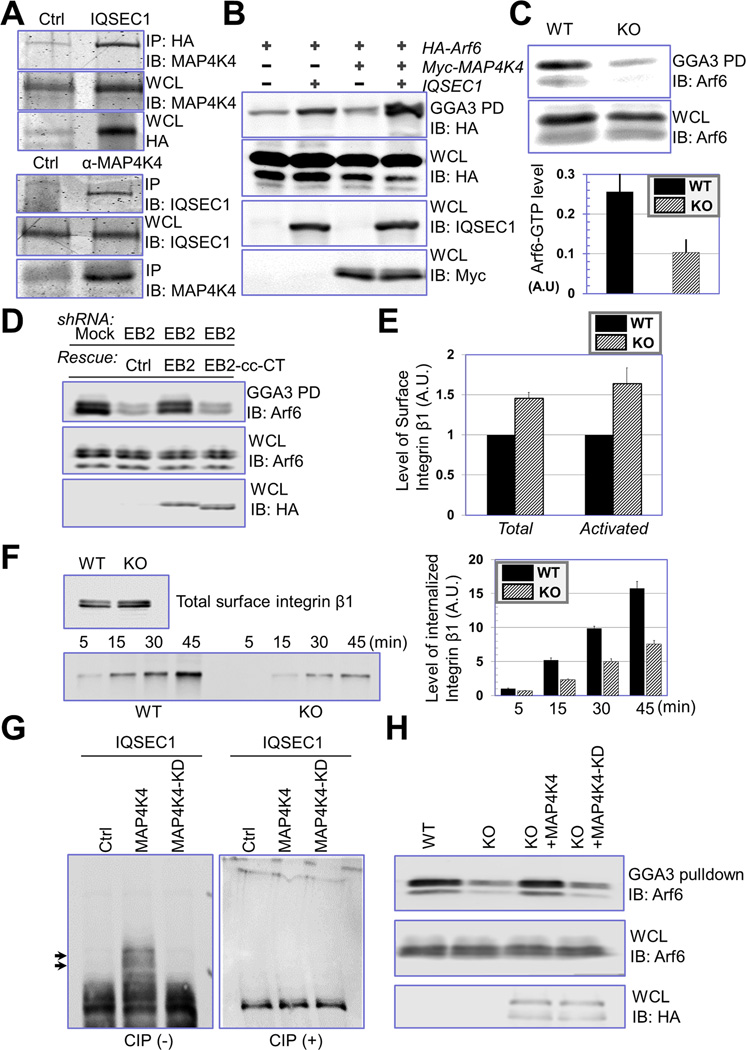
Our MS analysis identified IQSEC1 as a MAP4K4 binding protein (Fig. 4C–D), and presence of the N terminal peptide of IQSEC1 suggests that MAP4K4 associates with the longer splicing isoform of IQSEC1 (Dunphy et al., 2006). Coimmunoprecipitation assay confirmed interaction between IQSEC1 and MAP4K4 (Fig. 6A).
Figure 6. MAP4K4 promotes IQSEC1 and Arf6 activity.
(A) Interaction between IQSEC1 and MAP4K4 was confirmed by coimmunoprecipitation. Cells were transfected with plasmids encoding Myc-tagged MAP4K4 with or without HA-tagged IQSEC1. Cell lysate was immunoprecipitated with α- HA and blotted with different antibodies as indicated (top panel). To verify interaction of endogenous proteins, lysate of WT keratinocytes were immunoprecipitated with control or α-MAP4K4 IgG, and blot with α-IQSEC1 antibody (bottom panel). An aliquot of whole cell lysate (20 µg) were examined by immunoblot as well. (B) HEK293T cells were transfected with HA-tagged Arf6 together with MAP4K4 or IQSEC1 in different combinations as indicated. Level of GTP-bound Arf6 was determined by GGA3 pull down and immunoblot with α-HA antibody. An aliquot of WCL (10 µg) was immunoblotted with different antibodies as indicated to verify comparable expression level of different genes. (C) Level of endogenous Arf6-GTP was determined by GGA3 pull down coupled with α-Arf6 immunoblot (top panel). Quantification from densitometry analysis shows significant decrease of Arf6 activity upon loss of MAP4K4 (lower panel). For WCL, 20 µg total proteins were used. N=4, and P<0.05. (D) Level of Arf6-GTP was determined by GGA3 pull down coupled with α-Arf6 immunoblot in control cells, EB2 knockdown cells, and EB2 knockdown cells rescued with either WT EB2 or EB2 cc-CT mutant (HA tagged). For WCL, 20 µg total proteins were used. (E) Surface level of β1-integrin (pan β1-integrin or activated β1-integrin) was determined by flow cytometry. N=3, P<0.01 for both total integrin and activated integrin. (F) Internalization of β1-integrin in WT and MAP4K4 KO cells was determined by reversible biotinylation. Biotinylated proteins were isolated by streptavidin agarose and subjected to immunoblot with anti-β1-integrin antibody. Relative level of internalized integrin was determined by densitometry and quantified (right panel). N=3, and P<0.05 for each time point. (G) IQSEC1 was isolated from transfected cells by immunoprecipitation and phosphorylation of IQSEC1 was determined by electrophoresis with Phos-tag acrylamide. Note upper shifted bands (arrows) that represent hyperphosphorylated proteins are only present in cells co-transfected with IQSEC1 and WT MAP4K4. CIP: calf intestinal phosphatase. (H) Arf6-GTP level was determined by GGA3 pull down for WT, MAP4K4 KO cells, and KO cells rescued with WT MAP4K4 or MAP4K4 KD mutant (HA tagged). For WCL, 20 µg of total proteins were used.
Mounting evidence suggests that IQSEC1, an Arf6 GEF protein, regulates cell adhesion to matrix via Arf6 (Dunphy et al., 2006; Hiroi et al., 2006; Morishige et al., 2008; Someya et al., 2010). We first examined the potential role of MAP4K4 in activation of Arf6. To this end, we exogenously expressed IQSEC1 with or without MAP4K4 and performed GGA3 pull down assay to determine the level of GTP-bound Arf6 (D'Souza-Schorey and Chavrier, 2006). Expression of IQSEC1 (the longer isoform as BRAG2b) alone only modestly enhanced the Arf6 activity, whereas co-expression of both IQSEC1 and MAP4K4 led to a significant increase in GTP-Arf6 level (Fig. 6B). Consistently, loss of MAP4K4 in keratinocytes significantly reduced endogenous Arf6-GTP level as expected (Fig. 6C). To test whether MAP4K4 binding with MTs is involved in Arf6 activation, we carried out GGA3 pull-down analysis in EB2 knockdown cells. As expected, depletion of endogenous EB2 leads to dramatic reduction of GTP-bound Arf6 level (Fig. 6D). Expression of WT EB2 but not EB2 mutant deficient for MAP4K4 interaction (cc-CT mutant of EB2) can restore Arf6 activation, strongly suggesting that MAP4K4 association with EB2 is essential for Arf6 activation in cells (Fig. 6D).
IQSEC1 and Arf6 control cell adhesion by regulation of integrin internalization and intracellular trafficking (D'Souza-Schorey and Chavrier, 2006; Dunphy et al., 2006). To determine the role of MAP4K4 in this process, we first quantitated the surface expression of endogenous β1 integrin by flow cytometry. Our results indicate that KO of MAP4K4 in skin keratinocytes leads to an ~50% increase in surface level of β1 integrin (total or active β1 integrin) relative to WT control cells (Fig. 6E and supplementary Fig. 6A). Immunoblotting of cell lysates demonstrated that overall expression of β1 integrin was not affected by loss of MAP4K4 (data not shown). As reported in other cell lines (Dunphy et al., 2006; Ezratty et al., 2005), β1 integrin in keratinocytes is undergoing active endocytosis and intracellular trafficking. Internalized β1 integrin can be detected in intracellular vesicles and co-localize with various endosomal markers, such as Rab5 (Supplementary Fig. 6B–C). To quantitatively assess the rate of β1 integrin internalization, we carried out reversible biotinylation assay (Wu et al., 2005). Our results reveal that endocytosis of β1 integrin in MAP4K4 null cells is significantly reduced (Fig. 6F). Together, our results provide compelling evidence that MAP4K4 promotes FA dynamics by regulating IQSEC1/Arf6 pathway and controlling endocytosis of integrin. Recent study suggested that IQSEC1 might regulate α5β1 integrin endocytosis via activation of Arf5 (Moravec et al., 2012). However, our biochemical pull-down suggests that level of GTP-bound Arf5 is not significantly affected by deletion of MAP4K4 (Supplementary Fig. 6D).
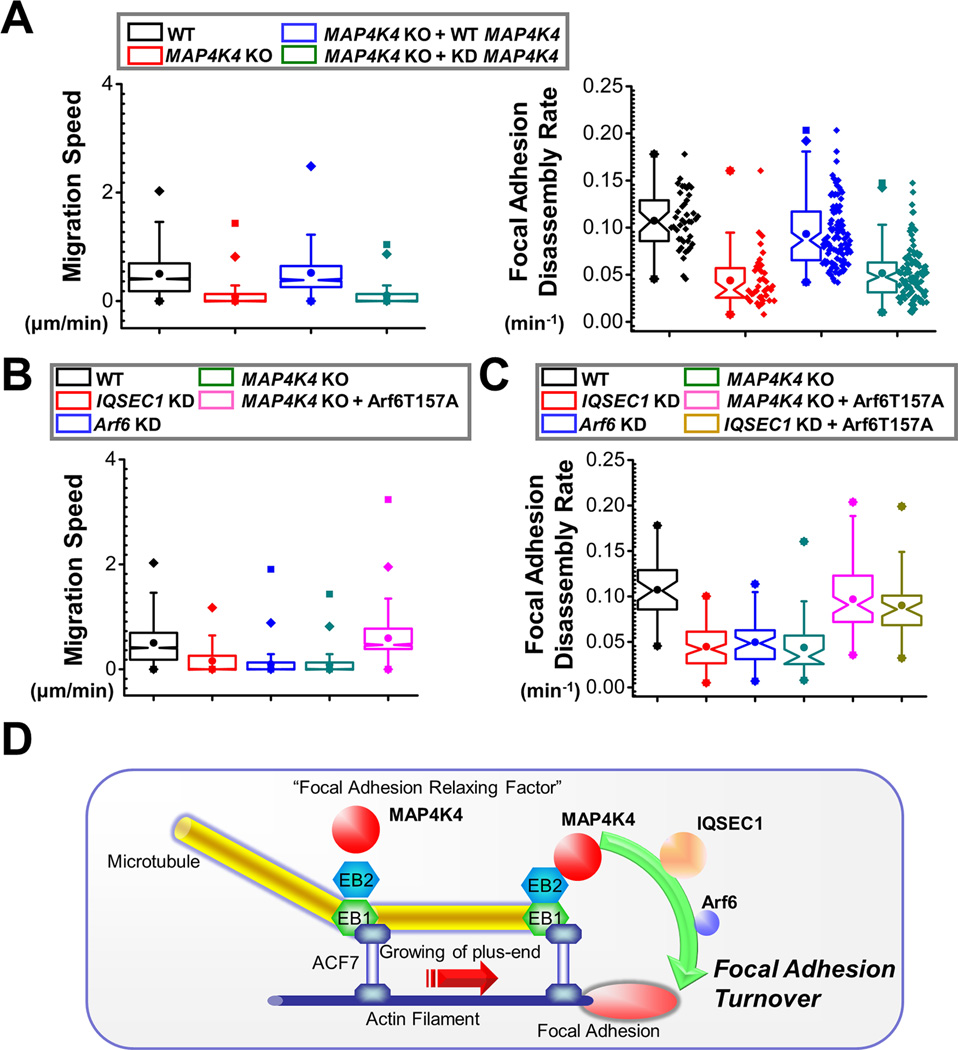
Activity of GEF proteins can be regulated by post-translational modifications, such as phosphorylation. Co-expression of IQSEC1 with WT MAP4K4 but not kinase defective (KD) mutant of MAP4K4 leads to significant phosphorylation of IQSEC1 in vivo, which is sensitive to treatment of alkaline phosphatase (Fig. 6G). To examine the potential relevance in IQSEC1 activity, we re-expressed WT or KD mutant of MAP4K4 in MAP4K4 deficient cells. Pull-down results indicate that WT but not KD mutant of MAP4K4 can rescue Arf6 activity in MAP4K4 KO cells, suggesting that MAP4K4 might regulate Arf6 activation via protein phosphorylation (Fig. 6H). When expressed in MAP4K4 KO cells, WT but not KD mutant of MAP4K4 can enhance FA disassembly and cell movement, suggesting that the kinase activity of MAP4K4 is essential for its role in these processes (Fig. 7A).
Figure 7. MAP4K4 and IQSEC1 interaction regulates FA dynamics and cell motility.
(A) Cell motility and FA dynamics were analysed for WT, MAP4K4 KO cells, and KO cells rescued with WT MAP4K4 or MAP4K4 KD mutant. For cell motility assay (left panel), N= 30 cells × 120 time points for each group. P<0.01 between WT and MAP4K4 KO, MAP4K4 KO and MAP4K4 KO + WT MAP4K4, and MAP4K4 KO + WT MAP4K4 and MAP4K4 KO + KD MAP4K4. For FA disassembly (right panel), N>40 for each group. P<0.01 between between WT and MAP4K4 KO, MAP4K4 KO and MAP4K4 KO + WT MAP4K4, and MAP4K4 KO + WT MAP4K4 and MAP4K4 KO + KD MAP4K4. (B, C) Quantifications of migration velocities (B) and FA disassembly rate (C) for control, IQSEC1-knockdown cells, Arf6-knockdown cells, MAP4K4 KO cells, and cells rescued with Arf6 T157A. For cell motility assay, N= 30 cells × 120 time points for each group. P<0.01 between WT and IQSEC1 KD; WT and Arf6 KD, WT and MAP4K4 KO, and MAP4K4 KO and MAP4K4 KO + Arf6 T157A. For FA disassembly, N>40 for each group. P<0.01 between between WT and IQSEC1 KD; WT and Arf6 KD, WT and MAP4K4 KO, MAP4K4 KO and MAP4K4 KO + Arf6 T157A, and IQSEC1 KD and IQSEC1 KD + Arf6 T157A. (D) A working model summarizing the role of MAP4K4 in FA turnover and cell migration. We posit that MT dynamics are coordinated by cytoskeletal crosslinkers, such as ACF7 that guides MT growth toward FAs. MT interacting protein EB2 can bind and deliver MAP4K4 to FAs, where MAP4K4 can subsequently activate IQSEC1 and Arf6, leading to FA turnover and efficient cell movement.
To determine the relevance of IQSEC1/Arf6 pathway in migration of keratinocytes, we prepared shRNAs for both genes (Supplementary Fig. 7). Consistent with reports from other cell lines (Dunphy et al., 2006; Morishige et al., 2008; Someya et al., 2001), depletion of endogenous IQSEC1 or Arf6 in mouse keratinocytes markedly stabilized FAs and impaired cell motility (Fig. 7B and C). To determine whether Arf6 acts downstream to MAP4K4 in these processes, we transfected MAP4K4 KO cells with plasmid encoding Arf6 T157A, the fast cycling mutant of Arf6 (Santy, 2002). Relative to mock transfection, expression of Arf6 T157A significantly rescued cell motility and FA disassembly defects in MAP4K4 null cells (Fig. 7B and C). In addition, expression of Arf6 T157A restored FA dynamics in cells depleted with IQSEC1 (Fig. 7C). Taken together, our results delineate a signaling cascade whereby MT dynamics regulate FA turnover through MAP4K4 interaction with IQSEC1 and subsequent activation of Arf6 (Fig. 7D).
DISCUSSION
Accumulating evidence suggests that FAs can serve as the “hotspot” for crosstalk between MT and actin networks (Rodriguez et al., 2003). Small’s group has shown that MTs can specifically grow towards FAs, a process likely guided by underlying F-actin filaments (Kaverina et al., 1999; Krylyshkina et al., 2002). Targeting of MTs toward FAs promotes dissolution of FAs at the cell periphery, a feature in line with the long-standing observation that cells treated with MT depolymerizing drugs contain enlarged and stabilized FAs (Bershadsky et al., 1996; Ezratty et al., 2005; Wu et al., 2008). Further evidence has implicated dynamin and clathrin in this process, strongly suggesting that FA dissolution is mediated by the endocytic pathway (Ezratty et al., 2009; Ezratty et al., 2005). However, the molecular mechanisms by which MT targeting contributes to FA dynamics have not been established. It has been speculated that MTs can serve as macromolecular tracks to deliver an unknown “disassembly factor” to facilitate FA turnover (Krylyshkina et al., 2002). But the nature of this “disassembly factor” and how this “disassembly factor” can activate endocytic machinery to promote FA dissolution remain unclear.
Our study presents compelling evidence that MAP4K4 acts as a MT-dependent “disassembly factor” to promote FA dynamics. Our model suggests that MAP4K4 associates with MT via its interaction with EB2. Upon targeting to FAs via MTs, MAP4K4 can bind and phosphorylate IQSEC1, which in turn activates Arf6 and endocytosis, leading to turnover of FAs. A schematic outlining this model is presented in Figure 7D. Consistent with this model, ablation of MAP4K4 in skin epidermal cells impairs wound healing in vivo. Furthermore, isolated MAP4K4 null cells display diminished Arf6 activity, stabilized FAs, and aberrant cell motility in vitro. Our results illuminate a hitherto unrecognized signaling cascade underlying FA dynamics and cell migration.
Interestingly, our study suggests that loss of MAP4K4 in primary skin keratinocytes inhibits both FA assembly and disassembly. This finding is consistent with our previous report on ACF7, whose deletion abolishes targeting of MT plus ends to FAs and inhibits both FA assembly and disassembly (Wu et al., 2008). In addition, treatment with nocodazole leads to stabilization of FAs, and assembly and disassembly rate of FAs are decreased similarly (Supplementary Fig. 1A) (Wu et al., 2008). Together, these results argue that in addition to FA dissolution, MT dynamics and targeting of MTs to FAs may play an equally important role in FA assembly. The underlying molecular mechanisms deserve further investigation.
Our study has also elucidated the role of EB2 in MT functionality. Growing MT ends accumulate a special group of MT interacting proteins, often referred to as MT plus-end tracking proteins (Akhmanova and Steinmetz, 2008). Mammalian cells also express three EB proteins, EB1, EB2 and EB3, which are encoded by separate genes. It has been reported that EB1 and EB3 together play a major role in mammalian cell to control MT dynamics by promoting growth and suppressing catastrophe of MTs, whereas EB2 is dispensable in these processes (Komarova et al., 2009; Komarova et al., 2005). In addition, unlike EB1 and EB3, enrichment of EB2 at MT plus ends is less prominent (Komarova et al., 2009). Our study unravels a specific and essential role of EB2 in mediating the crosstalk between MTs and FAs. Inactivation of EB2 in keratinocytes leads to aberrant FA dynamics and decreased motility, at least in part through its specific interaction with MAP4K4. Future studies will be needed to fully understand how the three EB proteins act in different cellular processes in vivo. We favor the hypothesis that unlike EB1 and EB3, EB2 might not be directly involved in regulation of MT dynamic instability. Instead, via its unique sequence, EB2 may function as an adaptor molecule to recruit signaling molecules to MTs, which is required for various cellular processes, including cell motility.
Under different conditions, turnover of FAs can be mediated by clathrin-dependent endocytosis or macropinocytosis (Ezratty et al., 2009; Gu et al., 2011). The role of Arf6 in endocytic membrane trafficking is well established, but it also likely regulates macropinocytosis (Schweitzer et al., 2011). Further investigation will be required to determine whether MAP4K4-induced Arf6 activity promotes FA turnover via clathrin-dependent endocytosis or macropinocytosis, or both. Nevertheless, identification of the interaction between MAP4K4 and IQSEC1 provides an intriguing link between Arf6 signaling and MT network, fulfilling the gap in our understanding of MT-induced turnover of FA.
In closing, our findings provide critical insights into the mechanics of MT-induced FA turnover and now pave the way for probing more deeply into the intricate signaling network orchestrating the crosstalk between F-actin and MT cytoskeletal networks and localized sites of membrane-associated activity.
EXPERIMENTAL PROCEDURES
SILAC (Stable Isotope Labeling) and FA fractionation
HaCaT cells were labeled with either “heavy” or “light” isotopic lysine using a SILAC protein quantification kit (Thermo scientific, Rockford, IL, USA) according to manufacturer’s instruction. Briefly, cells were grown in DMEM:F12 medium supplemented with 10% dialyzed fetal bovine serum and either the “heavy” form of L-Lysine-2HCl (4, 4, 5, 5-D4) or “light” L-Lysine for more than six generations to achieve more than 98% of labeling efficiency (Ong et al., 2003).
To isolate FA proteins, subconfluent HaCaT cells were treated with nocodazole (13 µm) or vehicle control for 30 minutes. Fractionation was carried out essentially as described (Kuo et al., 2011). Briefly, cells were hypotonically shocked with triethanolamine-containing solution. Cell bodies were removed by hydrodynamic force with PBS repetitively. FA proteins were then collected and denatured by scraping and sonication. Samples were analyzed with LC-MS/MS afterwards.
Generation of MAP4K4 cKO Mice
The MAP4K4 flox strain was kindly provided by Dr. Min Xie at University of Texas Southwestern Medical Center and Prof. Michael Schneider at Imperial College London. The breeding stock of MAP4K4 flox animals were provided by Dr. Edward Skolnik at New York University. MAP4K4 cKO animals were generated by breeding MAP4K4fl/fl mice to K14-Cre transgenic mice. All mice used in this study were bred and maintained at the ARC (animal resource center) of University of Chicago in accordance with institutional guidelines.
TIRF microscopy
TIRF imaging was done using Leica GSD/TIRFM Ground State Depletion Superresolution Microscope. Solid state laser light (405nm, 488nm, 532nm, 642nm) was focused at the back focal plane of a 160× NA 1.43 objective. Signals were recorded by Andor iXon3 897 high speed EMCCD camera. For TIRF imaging, the stained cells were mounted in an open chamber filled with 1× PBS.
Pearson's coefficient for co-localization was calculated using NIH ImageJ software.
FRAP and FA Assembly/Disassembly Measurements
Kinetics of FA assembly and disassembly were performed essentially as previously described (Wu et al., 2008). Keratinocytes were plated on fibronectin-coated dishes and transfected with plasmid encoding DsRed-Zyxin. Time series of images were acquired on a 3i Marianas Yokogawa-type spinning-disc confocal microscope equipped with a 100× α-plane (1.45 oil) lens and an EM charge-coupled device camera. The rate constants for FA assembly and disassembly were obtained by calculating the slope of relative fluorescence intensity increases or decreases of individual FA on a semilogarithmic scale against time.
FRAP assays were performed essentially as described (Wu et al., 2008). Briefly, cells were plated on fibronectin-coated 3.5-cm dishes and transfected with plasmid encoding GFP-paxillin. Three or two prebleach events were performed followed by a 1-s bleach event. Fluorescence recovery was recorded for 140 seconds after photobleaching events, and data from these photokinetic experiments were analyzed using DeltaVision software.
Flow cytometry
Flow cytometry analysis of surface integrin was carried out essentially as described (Wu et al., 2008).
Statistical analysis
Statistical analysis was performed using Excel or OriginLab software. Box plots are used to describe the entire population without assumptions on the statistical distribution. A student t test was used to assess the statistical significance (P value) of differences between two experimental conditions.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to Dr. Min Xie at University of Texas Southwestern Medical Center and Prof. Michael Schneider at Imperial College London for sharing their MAP4K4 floxed mice in advance of publication, and to Dr. Edward Skolnik at New York University for providing us the breeding stocks. We thank Dr. Yulia Komarova at University of Illinois at Chicago for providing the EB2 construct. We thank Dr. Martha Vaughan at National Institutes of Health for providing the IQSEC1 construct. We thank Dr. Elaine Fuchs at the Rockefeller University for sharing reagents. We thank Dr. Don Wolfgeher at University of Chicago Proteomics Core lab and Dr. Vytas Bindokas and Dr. Chrstine Labno at University of Chicago Light Microscopy Core and Dr. Kathryn Fox at University of Chicago for their excellent technical assistance. The animal studies were carried out in the ALAAC-accredited animal research facility at the University of Chicago. This work was supported by a grant R01-AR063630 from the National Institutes of Health, the Research Scholar Grant (RSG-13-198-01) from the American Cancer Society, and the V scholar award from V foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nature reviews Molecular cell biology. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Bershadsky A, Chausovsky A, Becker E, Lyubimova A, Geiger B. Involvement of microtubules in the control of adhesion-dependent signal transduction. Current biology : CB. 1996;6:1279–1289. doi: 10.1016/s0960-9822(02)70714-8. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annual review of cell and developmental biology. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA, Campbell ID, Critchley DR. Talins and kindlins: partners in integrin-mediated adhesion. Nature reviews Molecular cell biology. 2013;14:503–517. doi: 10.1038/nrm3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JO, Li H, Lundquist EA. The MIG-15 NIK kinase acts cell-autonomously in neuroblast polarization and migration in C. elegans. Developmental biology. 2008;324:245–257. doi: 10.1016/j.ydbio.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nature reviews Molecular cell biology. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends in cell biology. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Dunphy JL, Moravec R, Ly K, Lasell TK, Melancon P, Casanova JE. The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Current biology : CB. 2006;16:315–320. doi: 10.1016/j.cub.2005.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty EJ, Bertaux C, Marcantonio EE, Gundersen GG. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. The Journal of cell biology. 2009;187:733–747. doi: 10.1083/jcb.200904054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nature cell biology. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Rodgers MA, Perrin BJ, Han J, Bennin DA, Critchley DR, Huttenlocher A. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nature cell biology. 2004;6:977–983. doi: 10.1038/ncb1175. [DOI] [PubMed] [Google Scholar]
- Goldspink DA, Gadsby JR, Bellett G, Keynton J, Tyrrell BJ, Lund EK, Powell PP, Thomas P, Mogensen MM. The microtubule end-binding protein EB2 is a central regulator of microtubule reorganisation in apico-basal epithelial differentiation. Journal of cell science. 2013;126:4000–4014. doi: 10.1242/jcs.129759. [DOI] [PubMed] [Google Scholar]
- Gu Z, Noss EH, Hsu VW, Brenner MB. Integrins traffic rapidly via circular dorsal ruffles and macropinocytosis during stimulated cell migration. The Journal of cell biology. 2011;193:61–70. doi: 10.1083/jcb.201007003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi T, Someya A, Thompson W, Moss J, Vaughan M. GEP100/BRAG2: activator of ADP-ribosylation factor 6 for regulation of cell adhesion and actin cytoskeleton via E-cadherin and alpha-catenin. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10672–10677. doi: 10.1073/pnas.0604091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annual review of cell and developmental biology. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Kaverina I, Krylyshkina O, Small JV. Microtubule targeting of substrate contacts promotes their relaxation and dissociation. The Journal of cell biology. 1999;146:1033–1044. doi: 10.1083/jcb.146.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, et al. Mammalian end binding proteins control persistent microtubule growth. The Journal of cell biology. 2009;184:691–706. doi: 10.1083/jcb.200807179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y, Lansbergen G, Galjart N, Grosveld F, Borisy GG, Akhmanova A. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Molecular biology of the cell. 2005;16:5334–5345. doi: 10.1091/mbc.E05-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylyshkina O, Kaverina I, Kranewitter W, Steffen W, Alonso MC, Cross RA, Small JV. Modulation of substrate adhesion dynamics via microtubule targeting requires kinesin-1. The Journal of cell biology. 2002;156:349–359. doi: 10.1083/jcb.200105051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo JC, Han X, Hsiao CT, Yates JR, 3rd, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nature cell biology. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Lewellyn L, Cetera M, Horne-Badovinac S. Misshapen decreases integrin levels to promote epithelial motility and planar polarity in Drosophila. The Journal of cell biology. 2013;200:721–729. doi: 10.1083/jcb.201209129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravec R, Conger KK, D'Souza R, Allison AB, Casanova JE. BRAG2/GEP100/IQSec1 interacts with clathrin and regulates alpha5beta1 integrin endocytosis through activation of ADP ribosylation factor 5 (Arf5) The Journal of biological chemistry. 2012;287:31138–31147. doi: 10.1074/jbc.M112.383117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishige M, Hashimoto S, Ogawa E, Toda Y, Kotani H, Hirose M, Wei S, Hashimoto A, Yamada A, Yano H, et al. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nature cell biology. 2008;10:85–92. doi: 10.1038/ncb1672. [DOI] [PubMed] [Google Scholar]
- Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nature cell biology. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Santy LC. Characterization of a fast cycling ADP-ribosylation factor 6 mutant. The Journal of biological chemistry. 2002;277:40185–40188. doi: 10.1074/jbc.C200481200. [DOI] [PubMed] [Google Scholar]
- Schweitzer JK, Sedgwick AE, D'Souza-Schorey C. ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis. Seminars in cell & developmental biology. 2011;22:39–47. doi: 10.1016/j.semcdb.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya A, Moss J, Nagaoka I. The guanine nucleotide exchange protein for ADP-ribosylation factor 6, ARF-GEP100/BRAG2, regulates phagocytosis of monocytic phagocytes in an ARF6-dependent process. The Journal of biological chemistry. 2010;285:30698–30707. doi: 10.1074/jbc.M110.107458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya A, Sata M, Takeda K, Pacheco-Rodriguez G, Ferrans VJ, Moss J, Vaughan M. ARF-GEP(100), a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2413–2418. doi: 10.1073/pnas.051634798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YC, Treisman JE, Skolnik EY. The Drosophila Ste20-related kinase misshapen is required for embryonic dorsal closure and acts through a JNK MAPK module on an evolutionarily conserved signaling pathway. Genes & development. 1998;12:2371–2380. doi: 10.1101/gad.12.15.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Gan B, Yoo Y, Guan JL. FAK-mediated src phosphorylation of endophilin A2 inhibits endocytosis of MT1-MMP and promotes ECM degradation. Dev Cell. 2005;9:185–196. doi: 10.1016/j.devcel.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Wu X, Kodama A, Fuchs E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell. 2008;135:137–148. doi: 10.1016/j.cell.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Shen QT, Oristian DS, Lu CP, Zheng Q, Wang HW, Fuchs E. Skin stem cells orchestrate directional migration by regulating microtubule-ACF7 connections through GSK3beta. Cell. 2011;144:341–352. doi: 10.1016/j.cell.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wang X, Li Z, Gotoh N, Chapman D, Skolnik EY. Mesodermal patterning defect in mice lacking the Ste20 NCK interacting kinase (NIK) Development. 2001;128:1559–1572. doi: 10.1242/dev.128.9.1559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.