Abstract
Signaling pathways and small RNAs direct diverse cellular events, but few examples are known of defined signaling pathways directly regulating small RNA biogenesis. We show that ERK phosphorylates Dicer on two conserved residues in its RNAse IIIb and dsRNA-binding domain, and phosphorylation of these residues is necessary and sufficient to trigger Dicer’s nuclear translocation in worms, mice, and human cells. Phosphorylation of Dicer on either site inhibits Dicer function in the female germ line and dampens small RNA repertoire. Our data demonstrate that ERK phosphorylates and inhibits Dicer during meiosis I for oogenesis to proceed normally in C. elegans and that this inhibition is released before fertilization for embryogenesis to proceed normally. The conserved Dicer residues, their phosphorylation by ERK, and the consequences of the resulting modifications implicate an ERK-Dicer nexus as a fundamental component of the oocyte-to-embryo transition and an underlying mechanism coupling extracellular cues to small RNA production.
INTRODUCTION
Organismal development requires precise control of signaling networks and molecular pathways. The RAS-ERK signaling pathway and the Dicer-dependent small RNA biogenesis pathway are two evolutionarily conserved pathways that regulate diverse cell biological and developmental events in higher eukaryotes/metazoans (Murchison et al., 2007; Sundaram, 2006). This work defines a direct regulatory link between the ERK signaling pathway and Dicer and the specific biological outcomes they may mediate.
The RTK-RAS-ERK pathway transduces extracellular signals into specific cellular responses through a conserved kinase cascade that results in the phosphorylation and activation of extracellular-signal regulated kinase (ERK), which is the terminal effector kinase of this pathway (Chang and Karin, 2001). ERK is a conserved proline-directed serine/threonine kinase that phosphorylates its substrates in order to direct many cellular and developmental processes (Arur et al., 2009; Chang and Karin, 2001; Welker et al., 2011).
The Dicer-dependent small RNA biogenesis pathway generates miRNAs and siRNAs that regulate a vast array of developmental and cellular processes via their ability to inhibit the translation and / or stability of target mRNAs (Denli et al., 2004; Grishok et al., 2001; Ketting et al., 2001; Tijsterman and Plasterk, 2004). Biogenesis of miRNAs and siRNAs occurs via a series of processing steps that result in the production of their mature forms by the RNAse III enzyme Dicer (Denli et al., 2004; Fire et al., 1998; Lee et al., 2002; Zhang et al., 2007). Dicer has been predominantly observed to carry out this function in the cytoplasm (Tijsterman and Plasterk, 2004), but in some contexts Dicer localizes to the nucleus (Barbato et al., 2007; Barraud et al., 2011; Emmerth et al., 2010; Sinkkonen et al., 2010). The mechanisms or pathways that trigger Dicer’s nuclear localization in vivo remain to be elucidated.
Developing oocytes and early-stage embryos are transcriptionally quiescent in mouse, zebrafish, and C. elegans (Su et al., 2007; Xia et al., 2012; Zuccotti et al., 2011); thus post-transcriptional and translational regulation of gene expression is crucial for oocyte development and the oocyte-to-embryo transition. Both the RAS-ERK pathway and Dicer have been shown to regulate key steps in oogenesis. For example, during mouse oogenesis active ERK regulates meiotic maturation (Verlhac et al., 1993; Verlhac et al., 1996; Verlhac et al., 2000); during C. elegans germ line development active ERK regulates various steps of meiotic I progression and oocyte maturation (Arur et al., 2009; Hubbard and Greenstein, 2000; Lee et al., 2007; Lopez et al., 2013; Verlhac et al., 1993; Verlhac et al., 1996; Verlhac et al., 2000). In both C. elegans and mouse oocytes, active ERK is rapidly de-phosphorylated and inactivated just before fertilization, but the functional significance of this inactivation is not known.
Similar to ERK expression during oogenesis, Dicer expression during oogenesis is well conserved from C. elegans to humans (Flemr et al., 2013; Knight and Bass, 2001; Murchison et al., 2007). Systemic loss of Dicer in worms and mammals has drastic effects on oogenesis: in worms, it blocks meiotic maturation (Knight and Bass, 2001); in mice, it blocks meiotic maturation at meiosis II (Flemr et al., 2013; Murchison et al., 2007). But, loss of dicer function specifically in mouse oocytes does not lead to a change in miRNA levels, suggesting that dicer mediates its effect on oocyte development either through siRNAs (Flemr et al., 2013) or in a small RNA independent manner. Additionally, specific loss of maternal inheritance of Dicer from growing oocytes results in failure in oocyte progression and subsequent failure in embryonic progression; a similar phenotype is evidenced upon abrogation of miRNA activity (via specific deletion in Dgcr8) from oocytes, demonstrating that maternally inherited Dicer activity and miRNAs are essential for zygotic development (Su et al., 2007; Tang et al., 2007). These studies demonstrate that while Dicer activity may be dispensible in the oocytes during meiosis I and oocyte development, it is essential for embryonic development. With a major role of Dicer in generation of the small RNA repertoire, these data suggest a switch in the activity of small RNA biogenesis pathway, coinciding with the oocyte-to-embryo transition.
In a prior study, using a functional genomic approach we identified Dicer as an MPK-1/ERK substrate during C. elegans germ line development (Arur et al., 2009). Here, we show that MPK-1 phosphorylates DCR-1 in its RNAse IIIb and dsRNA-binding domains during most of oogenesis, but that DCR-1 is rapidly dephosphorylated just before fertilization. Phosphorylation of either domain is necessary and sufficient for DCR-1’s nuclear localization and inhibits standard DCR-1 functions. Phosphorylation of DCR-1 on the dsRNA-binding domain confers on it the ability to negatively regulate MPK-1 ERK activation in the loop region of the germ line. The inhibition of DCR-1 function via phosphorylation during oogenesis and reactivation (via dephosphorylation) prior to fertilization is essential for oocyte-to-embryo transition. Thus our study provides a cellular and molecular mechanism for how the ERK regulates the oocyte-to-embryo transition via the regulation of the Dicer. As we find that Dicer is phosphorylated on these residues in mice and humans, our work suggests that the ERK-Dicer nexus may function in many cellular and developmental contexts.
RESULTS
Erk phosphorylates Dicer on two conserved phosphorylation sites
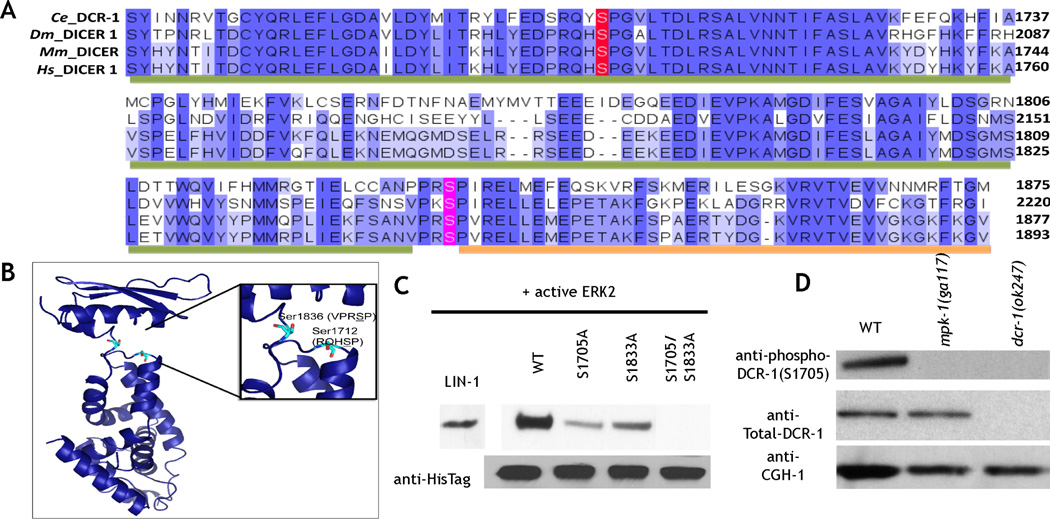
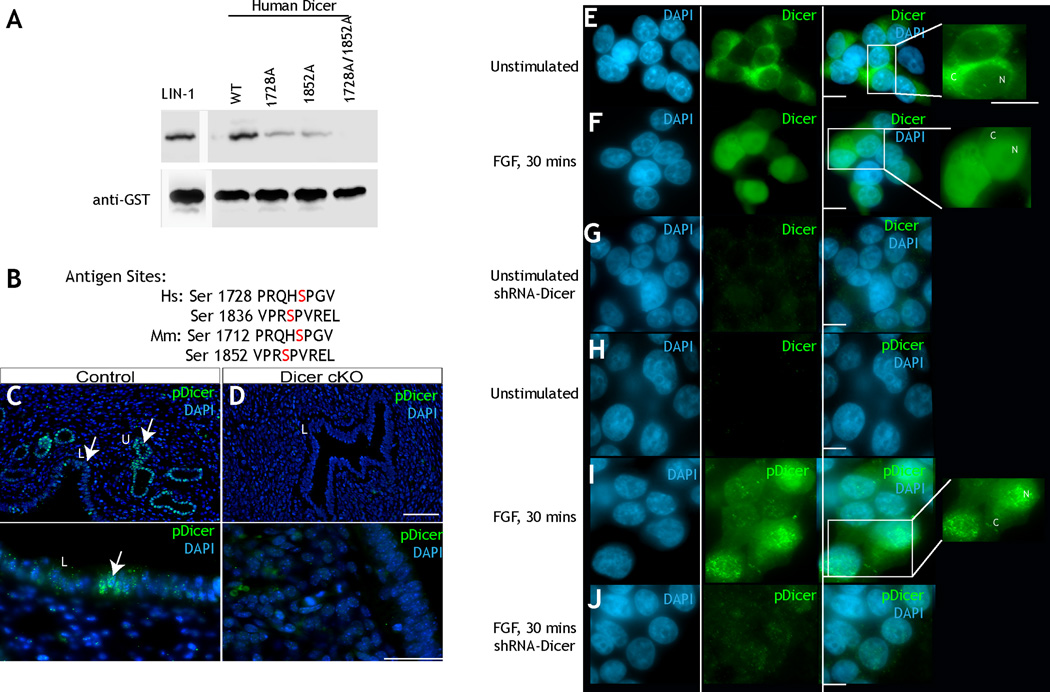
Our prior study identified Dicer as a potential evolutionarily conserved substrate of ERK (Arur et al., 2009). C. elegans Dicer (DCR-1) contains two evolutionarily conserved potential ERK phosphorylation sites at Serine 1705 and Serine 1833 (Fig. 1A–B), within DCR-1’s RNAse IIIb domain and dsRNA binding domain, respectively (Fig. 1A–B). To determine if ERK phosphorylates these sites, we performed in vitro kinase assays with active murine ERK2 on wild-type recombinant C-terminal fragments of DCR-1 as well as those in which Ser 1705 and Ser 1833 are mutated singly or doubly to alanine (Supplement). ERK robustly phosphorylates wild-type DCR-1; mutation of either Ser 1705 or Ser 1833 reduced ERK-dependent phosphorylation of DCR-1; mutation of both Ser 1705 and Ser 1833 abrogated ERK-dependent phosphorylation of DCR-1 (Fig. 1C). Thus, ERK phosphorylates DCR-1 on Ser 1705 and Ser 1833 in vitro (Fig. 1C).
Figure 1. C. elegans DCR-1 is phosphorylated by ERK on evolutionarily conserved residues.
A. Multiple sequence alignment of Dicer C-terminus showing the conserved serine residues in C. elegans (Ce) (1668–1875), D. melanogaster (Dm) (2018–2220), M. musculus (Mm) (1675–1877), and Homo sapiens (Hs) (1691–1893). Residues are colored based on percentage identity, and the amino acids corresponding to the RiboC domain (Ribonuclease III C terminal domain, also called RNAse IIIb domain) and the DSRM motif (double stranded RNA binding motif) domains are indicated with green and orange bars, respectively. B. Structure of RNaseIIIb and dsRNA binding domains of mouse Dicer generated from PDB ID 3C4B, highlighting residues Ser1712 and Ser 1836, analogous to worm residues Ser 1705 and Ser 1833. The figure was generated using pymol. C. In vitro ERK2 kinase assays of 6X His tagged C. elegans DCR-1 C terminal (1464–1910) protein. LIN-1: positive control. Mutation of either Ser 1705 or 1833 to alanine reduced phosphorylation; mutation of both residues to alanine abrogated phosphorylation. Anti-His tag western: loading control. D. Western blot analysis of whole worm lysates from indicated genotypes. The phospho (Ser 1705)-DCR-1 antibody (top) detects a single band at about 220 kDa; this band is absent from mpk-1(0) and dcr-1(0) animals. Total DCR-1 (middle) detects a band in both wild-type and mpk-1(0) animals but is lost from dcr-1(0) animals. CGH-1 a germ line enriched helicase (bottom) is the loading control.
To determine whether DCR-1 is phosphorylated on Ser 1705 and Ser 1833 in vivo, we generated rabbit polyclonal antibodies specific to the phosphorylated forms of each epitope (Experimental Procedures). Western blot analysis of worm lysates from wild-type, dcr-1(ok247) (termed dcr-1(0) since they lack dcr-1 mRNA), and mpk-1 (ga117) (termed mpk-1(0) since they lack mpk-1 mRNA (Church et al., 1995)) mutants reveals that each antibody recognizes a single band of 220 kDa in wild-type extracts but not in dcr-1(0) or mpk-1(0) extracts (Fig. 1D). An antibody specific to total DCR-1 also recognizes a single band at 220 kDa that is absent from the dcr-1(0) worm extracts, but present in mpk-1(0) worm extracts. Thus, DCR-1 is phosphorylated in an mpk-1 dependent manner on Ser 1705 and Ser 1833 in C. elegans (Fig. 1D).
Phospho-DCR-1 localizes to the nucleus during oogenesis
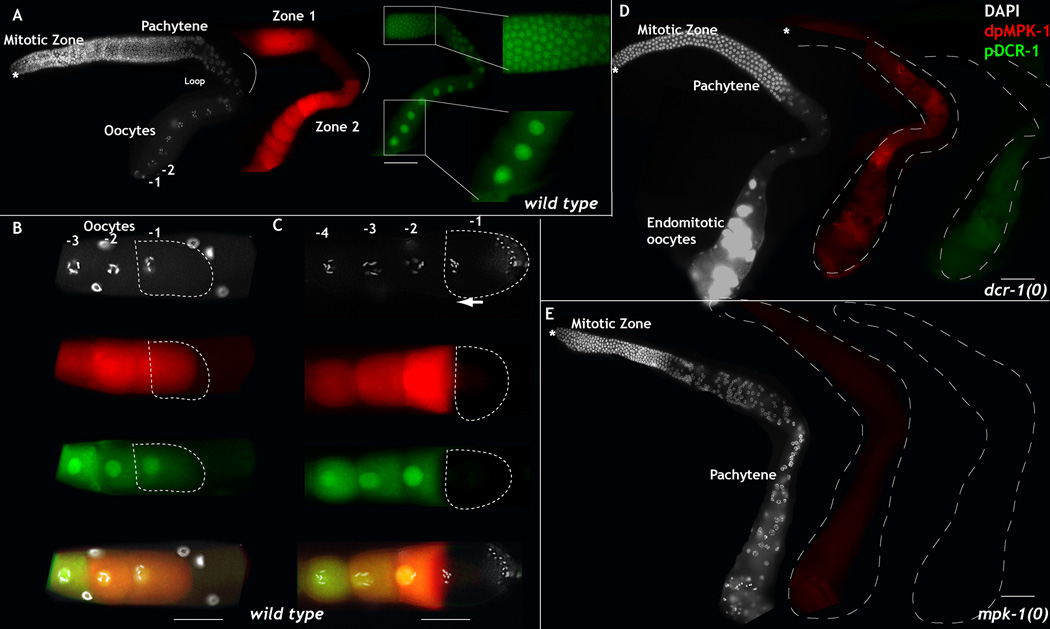
Using each phospho-DCR-1 specific antibody, we followed the in vivo dynamics of DCR-1 phosphorylation in the C. elegans germ line and observed identical staining patterns with both antibodies (not shown). We thus combined the two antibodies and used them to detect phospho-DCR-1 localization in the germ line. We find that phospho-DCR-1 accumulates in a pattern similar to that of active MPK-1: like active MPK-1, phospho-DCR-1 first becomes detectable in the mid-pachytene stage (zone 1 of MPK-1 activation, Fig. 2A); unlike active MPK-1, which is transiently downregulated in the loop region (Fig. 2A), phospho-DCR-1 persists in the loop region and continues to be phosphorylated in growing oocytes, where MPK-1 is reactivated (Fig. 2A). Both active MPK-1 and phospho-DCR-1 are dramatically downregulated in the terminal oocyte, which is mature and ready to ovulate and undergo fertilization (Fig. 2C), but in the immature oocyte, both active MPK-1 and phospho-DCR-1 are visible, and phospho-DCR-1 is nuclear (Fig. 2B). Thus, DCR-1 is phosphorylated during most of oogenesis, but rapidly dephosphorylated in the terminal oocyte shortly before fertilization.
Figure 2. Phosphorylated Dicer is nuclear in C. elegans germ line and dependent on MPK-1 activation.
A-E: Dissected adult hermaphroditic C. elegans germ lines oriented from left to right with the distal tip cell (asterisk) on the left and oocytes on the right, stained for DAPI (to visualize DNA, white), dpMPK-1 (red), and phospho-DCR-1 (S1705 and S1833, green). A. Wild-type germ line exhibits two zones of dpMPK-1 activation zones 1 and 2 in mid-pachytene and oocytes respectively. The mitotic zone and loop region do not show MPK-1 activation. Phosphorylated DCR-1 localizes to the nucleus from mid-pachytene stage continuously through the loop region and oocytes. −1, −2 marks the linear birth order of the growing oocytes. −1 being the oldest. B-C. Three sibling oocytes in a linear birth order −1, −2, −3 are illustrated. B. In an immature diakinetic oocytes, −1 oocyte nucleus is centrally located, dpMPK-1 levels are high and DCR-1 is phosphorylated and nuclear. C. In a mature oocyte, the nuclear envelope breaks down, DNA condenses and the nucleus migrates to the anterior (arrow), MPK-1 and DCR-1 are no longer phosphorylated. D-E. Phospho-DCR-1 at Ser1705 and Ser 1833 is dependent on the presence of dcr-1 (D) and mpk-1 (E). Scale bar: 20 µm
DCR-1 is known to localize and function in the cytoplasm to generate small RNAs (Morse et al., 2002), but we find that phospho-DCR-1 localizes largely to the nucleus (Fig. 2A–C). The nuclear signal for phospho-DCR-1 is specific to DCR-1 as complete loss of dcr-1 abrogates phospho-DCR-1 staining (Fig. 2D). Phosphorylation of DCR-1 is also dependent on MPK-1 activity in vivo since loss of mpk-1 function causes loss of phospho-DCR-1 staining (Fig. 2E), while total DCR-1 is still present as seen in Western Blot analysis (Fig. 1D). Thus, we conclude that DCR-1 is phosphorylated in the C. elegans germ line in an MPK-1 dependent manner and phosphorylation of DCR-1 correlates with its nuclear translocation.
Phosphorylation of DCR-1 triggers its nuclear localization
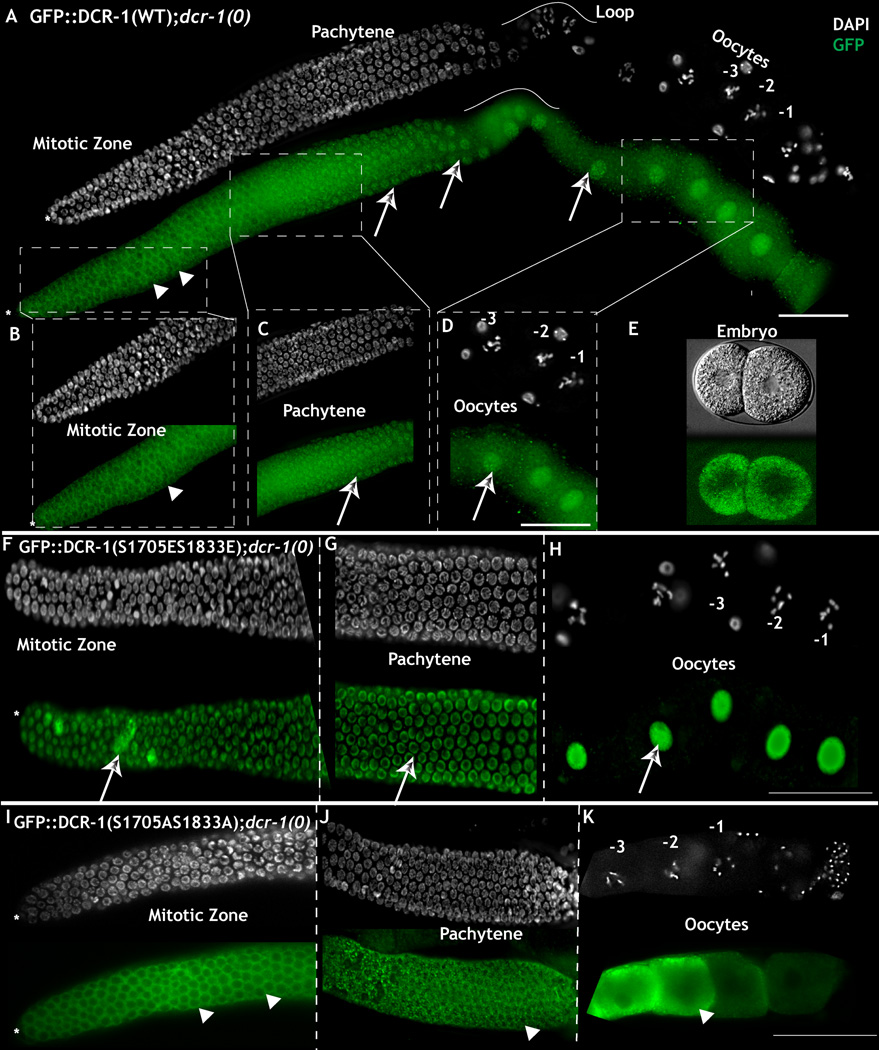
To test whether phosphorylation of DCR-1 is necessary and sufficient to target DCR-1 to the nucleus, we generated seven N-terminal GFP-tagged DCR-1 transgenes: one wild-type version and six others in which we mutated Ser 1705, Ser 1833, or both to glutamic acid (E) to mimic the phosphorylated state of these residues or to alanine (A) to mimic the non-phosphorylatable state of these residues. All transgenes contain the endogenous DCR-1 promoter and 3’ UTR. When introduced into a dcr-1(0) background, the wild-type DCR-1 transgene construct fully rescues the dcr-1(0) phenotype of endomitotic oocytes and displays the expected subcellular localization pattern in the germ line: cytoplasmic in those regions of the germ line that lack active MPK-1 (except for the loop region; Fig. 3A–B), nuclear in those regions of the germ line that possess active MPK-1 and in the loop region (Fig 3A, C-D). In dcr-1(0) worms, GFP expression of the dually phosphomimetic DCR-1S1705E/S1833E transgene localizes to the nucleus throughout the germ line, even in those regions that lack MPK-1 activity (Fig. 3F-3H). In contrast, GFP expression of the dually non-phosphorylatable DCR-1S1705A/S1833A transgene localizes to the cytoplasm throughout the germ line, even in those regions marked by high levels of active MPK-1 (Fig. 3I-3K). These results suggest that phosphorylation of DCR-1 is necessary and sufficient for the localized nuclear targeting of DCR-1 observed in the C. elegans germ line.
Figure 3. Phosphorylation of Dicer is necessary and sufficient for nuclear translocation.
A. Dissected adult hermaphroditic C. elegans germ line bearing the DCR-1WT transgene completely rescues the dcr-1(0) germ line phenotype. GFP staining (to visualize GFP::DCR-1, green) exhibits cytoplasmic localization in mitotic cells (arrow heads) and cytoplasm and nuclear staining in pachytene, loop and oocytes (arrows). B: Magnified view of the mitotic zone. DCR-1WT protein exhibits cytoplasmic accumulation (arrow head). C-D: DCR-1WT protein exhibits nuclear (arrows) and cytoplasmic staining in pachytene (C) and oocytes (D). E: Embryos produced from DCR-1WT; dcr-1(0) animals exhibit cytoplasmic accumulation. Live GFP image F-H: DCR-1S1705E/S1833E protein localizes primarily to the nucleus in the mitotic region (arrow, F), pachytene (arrows, G) and oocytes (H). I-K: DCR-1S1705A/S1833A protein localizes primarily to the cytoplasm in the mitotic zone (arrow heads, I), cytoplasm and P granules (arrow heads, J) and oocytes (arrow heads, K). Note animals shown in I-K were assayed at 6hr past mid-L4, earlier than those assayed in panel A-H, because expression of this DCR-1 transgene leads to multiple meiotic defects later in oogenesis (see text). Scale bar: 20 µm. (See also Figure S1 and S3)
In dcr-1(0) worms, we note that GFP expression of DCR-1 that carries singly mutated phospho-mimetic residue at either S1705E or S1833E displays a largely nuclear localization throughout the germ line (Fig. S1D–F and J–L). In contrast GFP expression of DCR-1 that carries singly non-phosphorylatable S1705A or S1833A transgene displays cytoplasmic localization in the distal region where MPK-1 is not active (Fig. S1 A – C and G–I). Thus, phospho-mimetic modification of either site is sufficient to induce DCR-1 to localize to the nucleus.
Completion of meiosis I during oogenesis requires somatic but not germ line DCR-1 function
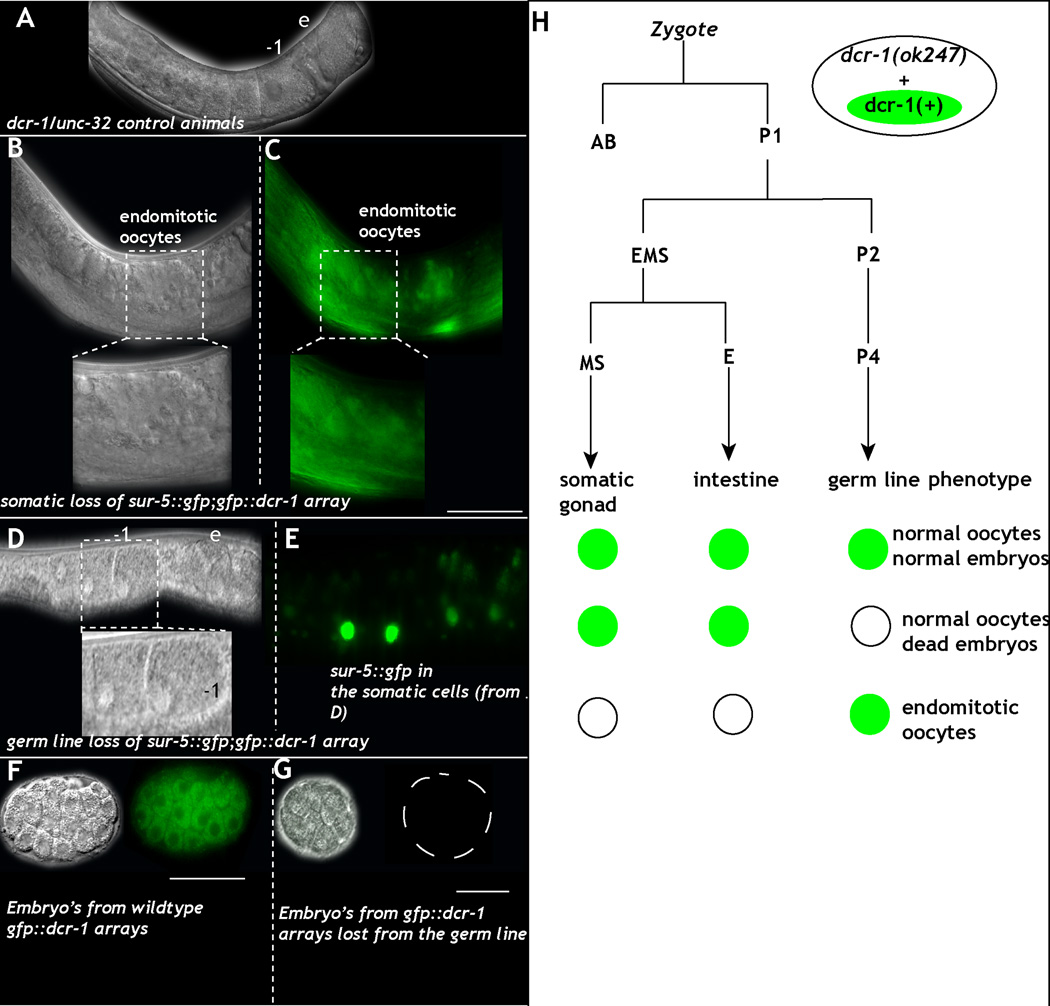
Worms homozygous for dcr-1(0) are able to complete early somatic development, likely due to perdurance of the maternal dcr-1 activity. Adult homozygous dcr-1(0) animals are sterile and exhibit defects in oocyte maturation and ovulation (Knight and Bass, 2001), with the oocytes undergoing endomitosis due to failed ovulation. Ovulation in C. elegans is tightly linked to the somatic gonad that communicates with the germ line (Govindan et al., 2009; Knight and Bass, 2001). Thus, we performed a genetic mosaic analysis to determine whether the ovulation defects were due to an autonomous function of DCR-1 in the germ line or a non-autonomous function of DCR-1 in the somatic gonad.
To produce dcr-1 genetic mosaics for phenotypic analysis, we used a standard transgene-loss assay (Yochem et al., 1998), where transgene carrying cells are marked with GFP. In dcr-1(0)/unc-32 animals, we generated extrachromosomal arrays containing transgenes with gfp::dcr-1 and sur-5::gfp. The sur-5::gfp allows us to follow the presence/absence of the array (and thus dcr-1 function) in the soma (by direct observation) and germ line (by inheritance) to identify worms that either lose the array in the germ line (P lineage) or the soma (MS lineage) during embryonic division (Fig. 4H). Worms that lost the gfp::dcr-1; sur-5::gfp extrachromosomal array from the soma but not the germ line display severe endomitotic oocyte phenotypes, similar to that observed in dcr-1(0) null worms (Fig. 4B–C compare to Fig. 2D). In contrast, worms that retain gfp::dcr-1; sur-5::gfp in the soma, but lose it from the germ line exhibit normal germ line development (Fig. 4D–E). Germ lines from these animals produce embryos that begin to divide, but arrest and die shortly thereafter (Fig. 4G). Thus, DCR-1 acts in a non-autonomous manner in the soma to regulate oocyte maturation and ovulation. Germ line DCR-1 product appears dispensable for oocyte maturation, contributing instead to an essential process (or processes) in early embryonic development. We further tested this by mating dcr-1(0) mosaic animals that retained gfp::dcr-1; sur-5::gfp dcr-1(0) in the soma but not germ line with wild type males (4 distinct mosaic animals were mated independently). Germ lines from these mated animals produced embryos that died at various stages of early cell divisions, much like the unmated animals. These mosaic data indicate an essential role for maternal-germline-encoded DCR-1 in early embryonic development.
Figure 4. Loss of dcr-1 from the germ line results in normal germ line development but dead embryos.
A: DIC images of control dcr-1(0)/unc-32 heterozygous animals reveal linear row of oocytes (-1, per birth order) and formation of embryos (e). B-C: DIC (B) image of the germ line from an animal that lost the sur-5::gfp;gfp::dcr-1 array in the soma. The germ line produces endomitotic oocytes (inset- magnification of endomitotic oocyte). (C) GFP image of B. D-E: DIC (D) image of the germ line from animal that lost the sur-5::gfp;gfp::dcr-1 array in the germ line but retains it in the soma (magnified in inset. −1, −2 marks oocytes). E: GFP image of D. Green exhibits sur-5::gfp in the somatic cells. (F-G): Embryos obtained from animals that retain both germ line and somatic array (F) exhibit normal divisions and polarization of the embryo. G: Embryos obtained from animals that lose the array from the germ line die at various stages before gastrulation. H: Schema of the mosaic analysis performed using the dcr-1(+) extrachromosomal array marked with sur-5::gfp. Animals were individually scored for presence of the array in the germ line (presence in progeny), cells of the somatic gonad (gonadal sheath and distal tip cell), and the intestine. Location of array loss within the lineage was inferred by the absence of GFP in all scored tissues descending from a given cell. Green circles indicate the presence of an array, while open circles indicate its absence. Loss of the dcr-1 array in the germ line lineage results in presence of normal appearing germ line but dead embryos. Loss of dcr-1 array from the somatic gonad results in endomitotic oocytes and sterile animals. Scale bar: 10µm. (See also Figure S2, Table S3).
In our analysis, we observed one other potential role for dcr-1 in the germ line. Animals in which dcr-1 function was depleted preferentially in the germ line (via the use of RNAi in an rrf-1 null mutant background (Sijen et al., 2001) that disrupts somatic RNAi; Fig. S2A–B) displayed ectopic germ line dpMPK-1 levels while retaining normal oocyte development (Fig. S2A–B). These observations indicate that while MPK-1 phosphorylates DCR-1, DCR-1 also appears to inhibit MPK-1 activity in the loop region of the germ line.
Phosphomimetic DCR-1 transgenes allow germ line progression and fertilization but fail to support subsequent embryonic development
Each identified phosphorylation site resides within a key functional domain of DCR-1: Ser 1705 within the RNAseIIIb domain, and Ser 1833 within the dsRNA-binding domain. To assess the effect of phosphorylation of each residue on DCR-1 function, we assayed the ability of the phosphomimetic transgenes - DCR-1S1705E, DCR-1S1833E and DCR-1S1705E/S1833E- to rescue the dcr-1(0) null phenotype, relative to DCR-1WT transgene. We generated all transgenes via two independent methods (see Supplement); all DCR-1 transgenes expressed GFP::DCR-1 expression at least 80% of the corresponding wild-type transgene, and the levels of expression do not correlate with the severity of the phenotypes (Fig. S3, Tables S1 and S2).
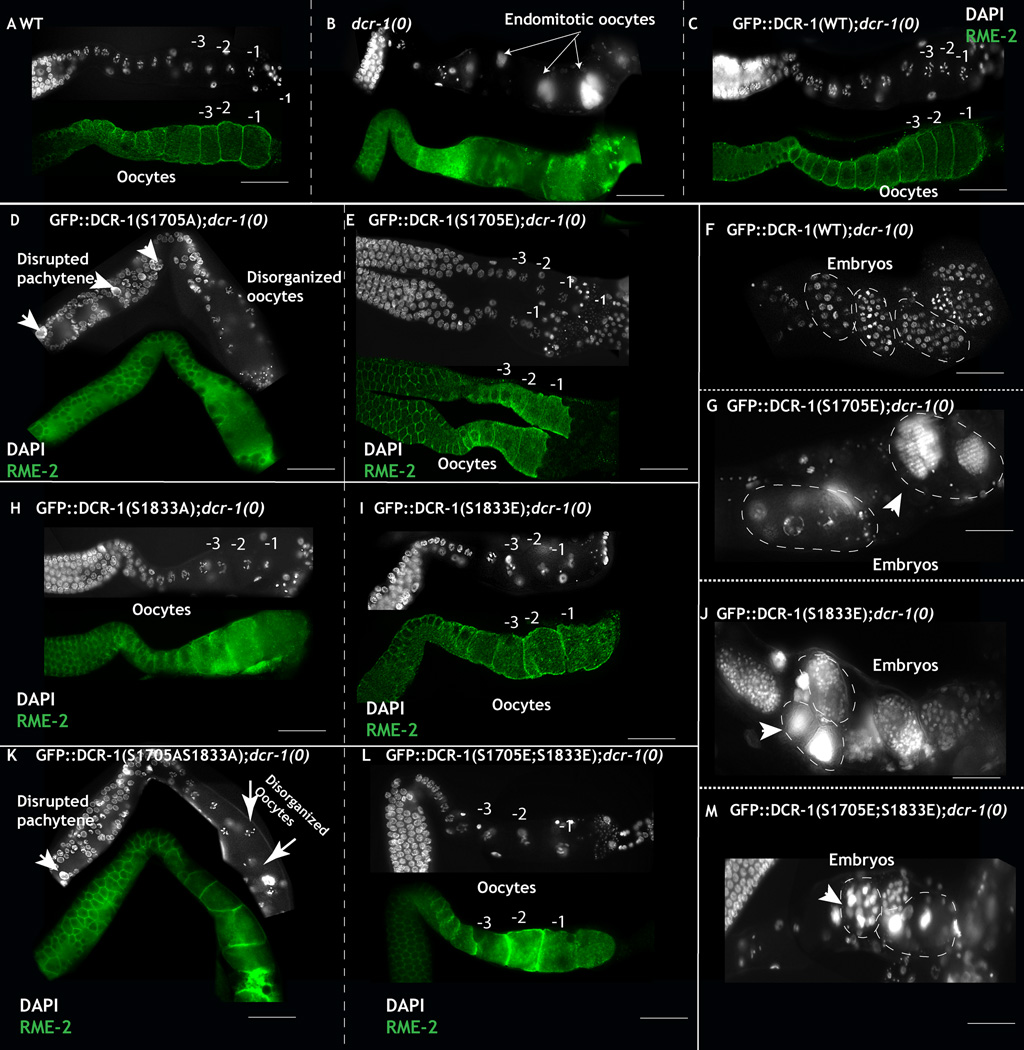
Expression of either the phosphomimetic DCR-1S1705E, or DCR-1S1833E, or DCR-1S1705E/S1833E rescued the ovulation defects of the dcr-1(0) mutants (Table S1 and S2), suggesting that these phosphomimetic mutants rescue DCR-1’s somatic function. The transgenes also restored active MPK-1 levels to its wild-type pattern in the germ line (Fig. S2C–D, Table S1 and S2) and gave rise to germ lines that appeared morphologically wild-type and contained wild-type appearing oocytes (Fig 5E, I and L). These oocytes underwent fertilization and produced embryos that died during embryogenesis in the uterus (Fig. 5 G, J, M compare to F, wild-type embryos, Table S1 and S2), a phenotype reminiscent of worms that lack dcr-1 function specifically in the germ line (see above). Analysis of these embryos revealed defects such as endoreduplicated chromosomes and failure in the early embryonic cleavage divisions (Fig. 5G, J, M). Thus, the DCR-1 phospho-mimetic transgenes are sufficient to rescue the somatic function of DCR-1, but they cannot rescue the requirement of DCR-1 function for early embryogenesis, leading to a model, in which phosphorylation of DCR-1 suppresses DCR-1 activity in the germ line while allowing DCR-1 to be rapidly activated by dephosphorylation in anticipation of its requirement during early embryonic development.
Figure 5. Phosphomimetic DCR-1 transgenes restore oocyte development and result in inviable embryos.
A: Dissected germ lines from adult wild-type hermaphrodites animals contain a linear row of oocytes marked with the oocyte specific marker RME-2 (green) and exhibit normal chromosomal morphology. −1 marks the oldest oocyte in the linear germ line. B: Systemic loss of dcr-1 function results in failure in oocyte ovulation and maturation; instead oocytes undergo endomitotic divisions and the animals are sterile. Arrows point to the endomitotic DNA. C: Expression of DCR-1WT transgene in the dcr-1(0) background rescues the ovulation defects, the animals are fertile and produce viable embryos. D: Expression of DCR-1S1705A transgene in dcr-1(0) animals rescues ovulation defects, instead the germ lines display defects in pachytene progression, polyploid pachytene nuclei (arrow heads), disorganization of pachytene cell membranes and disorganized oocyte development. E: Expression of DCR-1S1705E transgene in dcr-1(0) animals rescues the ovulation defects and oocytes appear morphologically wild-type. F: Embryos born of wild-type mothers develop into multicellularity inside the uterus as shown. Each DAPI dot exhibits one nucleus. G: Embryos born from DCR-1S1705E transgene in dcr-1(0) animals exhibit failure to divide normally, and chromosomes that endoreduplicate. Arrowhead: endoreduplicated chromosomes (arrow heads). These embryos are inviable. H: Expression of DCR-1S1833A transgene in dcr-1(0) animals rescues ovulation defects with morphologically wild-type oocytes, additionally, the germ lines exhibit ectopic MPK-1 activation (Fig. S2). I: Expression of DCR-1S1833E transgene in dcr-1(0) animals rescues the ovulation defects and yields morphologically wild-type oocytes. J: Embryos born from the DCR-1S1833E;dcr-1(0) mothers are inviable and exhibit endoreduplicated chromosomes (arrow heads). K: Expression of DCR-1S1705/S1833A transgene in dcr-1(0) animals rescues the ovulation defects, instead the germ lines exhibit polyploid pachytene nuclei and disorganized oocytes (arrows). L: Expression of DCR-1S1705E/S1833 transgene in dcr-1(0) animals rescues ovulation defects and yields morphologically wild-type oocytes. L: Embryos born from the DCR-1S1705E/S1833E;dcr-1(0) mothers are inviable and display chromosome segregation defects and endoreduplicated chromosomes (arrow heads). (See also Fig. S3, Table S1)
To directly assess whether the phospho-mimetic DCR-1 transgenes functioned in the germ line or the soma, we performed a mosaic analysis using the three DCR-1 phosphomimetic transgenes, as we had for wild-type DCR-1 (above). As observed for the wild-type DCR-1 transgene, the presence of the phospho-mimetic transgene in the soma, but not the germ line, rescued the oocyte ovulation and maturation defects observed in dcr1-(0) worms and produced a normal linear row of morphologically normal oocytes that gave rise to dead embryos (Table S3). Conversely, the absence of the DCR-1 transgene from the soma led to the canonical dcr-1 oocyte ovulation and maturation phenotypes. We also performed similar genetic mosaic studies in parallel with the non-phosphorylatable DCR-1 transgenes, and found their presence in the soma but not the germ line rescued the oocyte ovulation and maturation defects observed in dcr-1(0) worms. The apparent absolute requirement for dcr-1 function in the soma to promote normal oocyte ovulation and maturation precludes the ability to assess the effect of expressing the phospho-mimetic or non-phosphorylatable DCR-1 transgene specifically in the germ line. Nonetheless, these findings suggest that the germ line phenotypes obtained from these DCR-1 transgenes in dcr-1 null worms result specifically from expression of the transgene in the germ line.
Non-phosphorylatable DCR-1 transgenes unveil distinct roles of S1705 and S1833 phosphorylations in MPK regulation and oocyte development
Expression of the non-phosphorylatable DCR-1 transgenes- DCR-1S1705A, DCR-1S1833A and DCR-1S1705A/S1833A in the dcr-1(0) animals resulted in rescue of the endomitotic phenotype of the dcr-1(0) worms, suggesting that these DCR-1 transgenes exhibit normal DCR-1 function in the soma. Analysis of these animals, however, revealed distinct effects of DCR-1 phosphorylation at each site (Table S1 and S2), which we detail below individually.
Phenotypes obtained from non-phosphorylatable DCR-1S1833A transgene in dcr-1(0) null worms
Expression of DCR-1S1833A transgene in the dcr-1(0) animals gave rise to morphologically wild-type germ lines that produced normal oocytes in 87% (52/60) of germ lines (Fig. 5H, Table S1 and S2). Upon fertilization, these oocytes yielded overtly normal and viable embryos that developed normally, and produced self-fertile adults. The remaining 13% of worms displayed a small multiple oocyte phenotype and were sterile (Table S1 and S2), similar to that observed when the RAS-MPK-1/ERK pathway is elevated in the germ line (Lee et al., 2007). Both classes of worms, those that produced normal linear row of oocytes (87%, (Fig. S2D)) and those that produced small multiple oocytes (13%), exhibited elevated levels of active MPK-1 in the loop region. Thus, phosphorylation of Ser 1833 is necessary to inhibit MPK-1 activation in the loop region of the germ line, indicating that DCR-1 functions in a negative feedback loop with dpMPK-1 in the germ line.
Phenotypes obtained from non-phosphorylatable DCR-1S1705A transgene in dcr-1(0) null worms
In dcr-1(0) homozygous mutant worms, expression of the non-phosphorylatable S1705A DCR-1 transgene led to severe defects in the pachytene region and developing oocytes, the regions of the germ line where phospho-DCR-1 is normally detectable (see Fig. 2A, zone 1, and zone 2). In the pachytene region, we observed large polyploid pachytene cells, disruption of pachytene cell membrane, defects in pachytene progression, and disruption of oocyte development and organization (Fig. 2A, Fig. 5D, Table S1 and S2). The animals were sterile. The DCR-1S1705A phenotypes are semi-dominant as we observed similar phenotypes, albeit at reduced frequencies, in wild-type worms that carried the DCR-1S1705A transgene (Table S1 and S2). Our data suggests that blocking phosphorylation at Ser 1705 likely generates an over-active DCR-1 that is incompatible with meiotic progression (Table S1). These data suggest that phosphorylation of DCR-1 on Ser 1705 in the RNAse IIIb domain during pachytene and in developing oocytes is necessary for suppression of DCR-1 activity and normal progression of meiotic prophase and normal oogenesis. Expression of DCR-1S1705A/S1833A transgene in the dcr-1(0) background resulted in an additive phenotype (Fig. 5K, Table S1 and S2): germ lines displayed large polyploid pachytene cells, disrupted pachytene cell membranes and disorganized oocytes (Fig. 5J, Table S1 and S2). Thus, phosphorylation of DCR-1 is essential for oogenesis to proceed normally.
Modification at Ser 1705 and Ser 1833 impacts the ability of DCR-1 to generate small RNAs
To investigate the molecular implications of phosphorylation of DCR-1S1705 and DCR-1S1833 modification on DCR-1 in vivo, we performed deep sequencing analysis of small RNA (miRNA, siRNA, and other RNAs in the range roughly between 20 and 30 nt) from populations carrying the various DCR-1 transgenes in an otherwise dcr-1(0) genetic background. Since homozygous dcr-1(0) animals are sterile and worms homozygous for most of the phospho-site transgenes are either sterile or give rise to dead embryos, we obtained each DCR-1 transgene-carrying homozygous population from the heterozygous parents. Homozygote animals (identified by selection against a balancer chromosome) were synchronized at the L4 / adult molt, with sequencing analysis performed at 6 hours, 12 hours and 24 hours past L4 / adult molt stage. As controls, parallel sequencing analysis was carried out on equivalently obtained populations from N2 (wild-type), dcr-1(ok247null), and drsh-1(ok369). Sequencing was performed independently on both sets of transgenes, those obtained from Biolistic transformation (Praitis et al., 2001) as well as those obtained from MosSci mediated integration (Frokjaer-Jensen et al., 2008).
We first analyzed the differences in miRNA abundance between the dcr-1(0) and wild-type animals. Surprisingly, we found only modest differences in bulk miRNA levels between wild-type and dcr-1(0) animals (Fig. S4). miRNA levels were reproducibly lower in dcr-1(0) animals, but only by <2 fold when expressed in terms of total small RNA, with some noise in the system likely due to variable levels of tRNA and rRNA degradation products. Examining specific mature miRNA species, we evidenced only modest differences in relative abundance in wild-type versus DCR-depleted populations, with a notable exception of the embryo-specific miR35-miR42 cluster (Fig. S5, dark blue), for which the lack of embryonic development in the absence of DCR-1 activity might explain the very low accumulation.
Although the observed effects of zygotic DCR-1 depletion on miRNA accumulation were of limited magnitude, we note that consistent results have been reported by several groups (Grishok et al., 2001; Knight and Bass, 2001). A strong and persistent maternally-derived contribution of dcr-1(+) protein from the mothers of the animals being analyzed appears likely to explain the persistence of miRNA populations in dcr-1(0) animals derived from heterozygous mothers. These data are consistent with observations that demonstrate the sufficiency of maternal DCR-1 contributions to rescue to adulthood an otherwise early-embryonic lethal phenotype in dcr-1(0) null homozygotes ((Grishok et al., 2001) J. Maniar and A. Fire, unpublished data, this work). On a conceptual level, depletion of a processing enzyme that isn't rate-limiting in development will only begin to impact overall development at the point where either the enzyme is sufficiently dilute to be rate limiting.
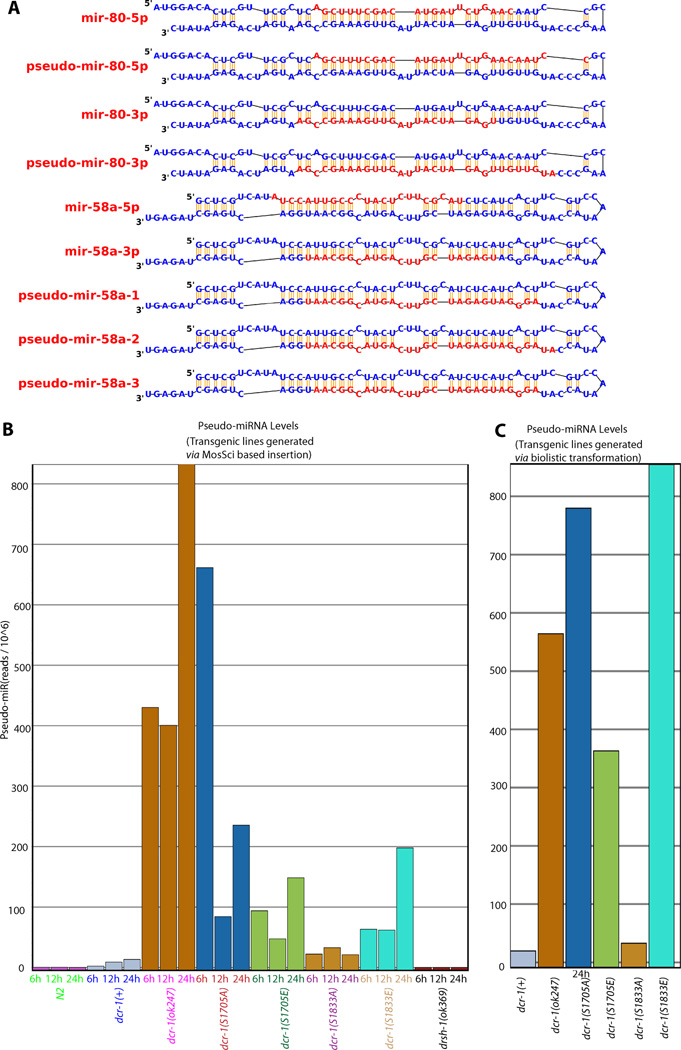
During the course of the miRNA analysis, we noticed some RNA species that appeared at higher levels in libraries derived from dcr-1(0) animals but were absent in libraries from wild-type (N2) control animals. These species carry 2–5nt extensions on either the 5’ or the 3’ end of standard miRNAs (Fig 6A). As a working model, we propose that these "pseudo-miRNAs" are derived from dcr-1-independent processing of pre-miRNAs that accumulate under conditions of dcr-1 depletion. We note that appearance of these RNAs was fully dependent on DRSH-1 (Drosha) (Fig. 6C), consistent with derivation from a standard miRNA precursor either by DRSH-1 or by other means (biological or chemical). Since the indicated pseudo-miRNAs were only observed in conditions of dcr-1 depletion, we can use their incidence as an alternative (and positive) metric for DCR-1 depletion. We chose the most abundant pseudo-miRNAs (which derive from the precursors to two abundant miRNAs, miR-58 and miR-80, Fig. S5) to test this model. We note that miR-58 and miR-80 remained abundant small RNA species in the DCR-1 phospho site-mutant transgene libraries, consistent with the remarkable (albeit incomplete) persistence of the maternal DCR-1 activity.
Figure 6. Alternative miRNA cleavage products that appear under conditions of DCR-1 depletion.
A: Schematic of miR58 and miR80 precursors with the standard and DCR-1-depletion dependent "pseudo" miRNAs shown. Note that these five miRNA-related molecules were chosen based on the following criteria: (i) miRNA-precursor inclusion, (ii) >10-fold relative enrichment in all three dcr-1(ok247) time points relative to N2 controls, and (iii) >99% confidence in the >10-fold enrichment. Numerous additional products meeting somewhat less stringent fold-enrichment criteria were also evident, but only these five species were included in the counts in Panels B and C). Secondary structures shown were redrawn from miR-base models (Griffiths-Jones et al., 2008). B: Abundance of pseudo-miRNAs in the set of strains derived by Mos-Sci integration. C: Abundance of pseudo miRNAs in the set of strains derived by Biolistic transformation. (See also Figure S4–S6)
Analysis of the accumulation of pseudo-miRNAs in each of the strains (wild-type and phospho-mutants), revealed some pseudo-miRNAs in each of the rescued backgrounds with both phosphomimetics accumulating pseudo-miRNAs at levels substantially above the wild-type rescue (Fig. 6B–C). These data are consistent with a model in which phosphorylation of DCR-1 at either Ser 1705 or Ser 1833 can impair DCR-1’s dicing activity. Among the non-phosphorylatable DCR-1 transgenes, we observe substantial pseudo-miRNA accumulation in two independent DCR-1S1705A transgenes, while DCR-1S1833A accumulates pseudo-miRNA populations only slightly more than a DCR-1WT transgene. These data are consistent with a possible role for Ser 1705 affecting DCR-1 activity, either in cycling through phosphorylation or in contributing to the folding, stability, or activity of the enzyme.
We next assayed for the 26G class of endo siRNAs, for which a substantial fraction have been shown to be germ line and DCR-1 dependent (Han et al., 2009; Kamminga et al., 2012). These endo siRNAs can be divided into Class I or spermatogenic class and Class II or the oogenic and the embryogenic class, based on observed incidence in the individual tissues (Han et al., 2009; Kamminga et al., 2012). DCR-1 is known to have a key role in 26G accumulation, as all classes are absent in animals homozygous for a viable (but conditionally sterile) mutant in the DCR-1 helicase domain (Welker et al., 2010). Additional components, including ERI-1 (a putative RNAse) are also required for all classes of 26G accumulation, while other components are required specifically for the maternal/oocyte or sperm classes (Han et al., 2009). There is a window of time during which both Class I and Class II 26G RNAs are detected (late L4 larvae and young adults), with depletion of Class I 26G RNAs as the animals get older.
By assigning each gene a count of 26G RNAs in the background of diverse Dicer mutations, we can track the efficacy of DCR-1 in contributing to the production of Class I and Class II 26G RNAs. By this criterion, we see that the DCR-1WT transgene rescued dcr-1(0) for 26G RNAs of both Class I and II (Fig. S6B). Phospho-mimetic transgenes DCR-1S1705E or DCR-1S1833E produced 26G RNA profiles that mimic dcr-1(0) (Fig. S6D, S6F compare to S6A). These results are consistent with models in which the phosphorylation of DCR-1 reduces its ability to produce 26G RNAs. Non-phosphorylatable DCR-1S1833A transgene mimicked the DCR-1WT transgene and rescued both the Class I and Class II 26G RNAs in an otherwise dcr-1(0) background (Fig. S6E), while non-phosphorylatable DCR-1S1705A resulted in a loss of 26G RNAs (Fig. S6C). These 26G RNA profiles were specific to dcr-1(0) and modifications of DCR-1, since depletion of drsh-1 did not impact their accumulation (Fig. S6F). Interestingly, while the germ line phenotypes of the DCR-1S1705A protein suggested that the DCR-1 unphosphorylated at S1705 was over active, the RNA profiles demonstrate that the population of 26G RNAs are not produced. These data together suggest that the inability to phosphorylate DCR-1 at S1705 may result in a protein that functions as a neomorph, rather than a hypermorph.
Together, these data provide a biochemical validation of the mutant rescue assays and demonstrate that phosphorylated residues in DCR-1 impact the ability of the protein to contribute to the production of a normal small RNA repertoire.
Phosphorylation of Dicer is evolutionarily conserved
The serine residues phosphorylated by ERK in C. elegans DCR-1 are conserved in the mouse and human forms of DCR-1 (Fig. 1A). Thus, we used in vitro kinase assays to determine if active ERK phosphorylates the homologous amino acids, Ser 1728 and Ser 1852, on human Dicer (Fig. 1A–B). As observed for C. elegans DCR-1, mutation of either serine to alanine reduced ERK-dependent phosphorylation of DCR-1, and mutation of both sites abrogated phosphorylation (Fig. 7A). To determine whether Dicer is phosphorylated on these amino acids in human cells, we followed the localization of total and phospho-Dicer in HEK293T cells (human embryonic kidney 293T). In these cells, under basal serum starved conditions, total Dicer localizes to the cytoplasm and phospho-Dicer is undetectable (Fig. 7E, 7H). Upon stimulation with fibroblast growth factor (FGF), a portion of total Dicer translocates to the nucleus and phospho-Dicer, as detected by our antibody is found in the nucleus (Fig. 7F, 7I). Treatment of stimulated and unstimulated 293T cells with shRNA specific for Dicer resulted in loss of staining of both total and phospho-Dicer, demonstrating specificity of the antibodies to Dicer (Fig. 7G, 7J). Additionally, inhibition of ERK activation via treatment of FGF stimulated HEK293T cells with MEK inhibitor UO126 reduces phosphorylated Dicer accumulation as assayed by western blot analysis (Fig. S7). This suggests that phosphorylation of Dicer is dependent on ERK activity in human HEK 293T cells. Thus, ERK phosphorylation of Dicer can trigger its nuclear localization in worms and humans. Consistent with Dicer being phosphorylated on these residues in higher metazoans, we also found that Dicer is phosphorylated and localizes to the nucleus in mouse uterine cells (Fig. 7C–D). Thus, we conclude ERK mediated phosphorylation of Dicer and its regulation of Dicer’s subcellular localization is evolutionarily conserved.
Figure 7. Phosphorylation of Dicer is evolutionarily conserved.
A: Kinase assay using murine active ERK2 (NEB) on a recombinant form of C terminal truncated wild-type human Dicer protein and versions of Dicer where Ser 1712 and/or Ser 1836 are mutated to alanine. Loss of either Ser 1712 or Ser 1836 reduces phosphorylation of Dicer; mutation of both sites to alanine abrogates phosphorylation. B: Sequence of the sites that are phosphorylated in human Dicer and recognized by the worm phospho-DCR-1 antibody. C: Wild-type mouse uterus stained with DAPI (blue) and phospho-Dicer (S1705 and S1833, Green). The luminal epithelium is labeled with the L, and uterine epithelium with U. Phospho-Dicer accumulates in the nuclei of luminal epithelium and uterine epithelium. Image is at a 20× magnification. D: Dicer was specifically inactivated in Müllerian duct mesenchyme-derived tissues of the reproductive tract of the female mouse, using an Amhr2-Cre allele ((Parker et al., 2008). This resulted in reduction / absence of the uterine ducts (Panel B). Compared to panel A, we did not see any accumulation of phospho-Dicer in the luminal epithelium (L) of Dicer conditional knockout cells. Image is at a 20× magnification. E: Unstimulated 293T cells show total Dicer in the cytoplasm. Stimulation (F) with FGF for 30 minutes results in nuclear translocation of total Dicer (N, inset). G: treatment with short RNA hairpin for dicer removes Dicer staining. H: Unstimulated cells do not show staining for phospho-Dicer. H-I: Upon stimulation with FGF phospho-Dicer is evidenced in the nuclei. J: Treatment with short hairpin for Dicer abrogates phospho-Dicer staining. (See also Figure S7)
DISCUSSION
Our work reports the impact of ERK-mediated phosphorylation of Dicer on the regulation and function of Dicer during the oocyte-to-embryo transition in C. elegans. Below we discuss the regulation of Dicer localization and function by phosphorylation, how this regulation influences oogenesis and the oocyte-to-embryo transition, and the conserved nature of the ERK-Dicer regulatory nexus.
Phosphorylation of Dicer triggers its nuclear localization and alters its function
In its canonical function, Dicer, a core RNAse III enzyme for small RNA biogenesis (Andl et al., 2006; Gent et al., 2010; Murchison and Hannon, 2004; Zhang et al., 2007), acts in the cytoplasm to generate miRNAs and siRNAs into their mature form. Dicer, however, has been found to localize to the nucleus in some contexts. For example, in fission yeast, a C-terminally-tagged version of Dicer localizes to perinuclear foci; this nuclear localization of Dicer depends on its dsRNA binding domain (Barraud et al., 2011; Emmerth et al., 2010), the protein domain within which one of our identified phosphorylation sites resides (Ser 1833 in worms). The mechanism by which Dicer translocates to the nucleus in yeast, is unclear as fusion of Dicer’s C-terminal end to GFP is not sufficient to direct GFP to the nucleus. Thus, the C-terminal end of Dicer does not behave as a simple nuclear localization signal. Dicer was also found to localize to chromosomes during mitosis in vertebrate cultured cells (Sinkkonen et al., 2010). The localization of Dicer to chromosomes in this context was specific to rDNA loci, but did not correlate with processing of pre-rRNA or the methylation status of these loci. Thus, while Dicer was known to localize to the nucleus the mechanisms that target it to the nucleus remained unknown.
Our work shows that ERK-mediated phosphorylation of the RNAse III and dsRNA-binding domains of the C-terminal region of DCR-1 is necessary and sufficient to direct DCR-1 to the nucleus the C. elegans germ line. These phosphorylation sites are conserved from worms to humans, and we show in human cell lines that nuclear translocation of Dicer correlates with its phosphorylation. Thus, we suggest that phosphorylation of Dicer on the two identified residues is a general, evolutionarily conserved mechanism that targets Dicer to localize to the nucleus.
Dicer function during oocyte development and oocyte-to-embryo transition
In mice and C. elegans, loss of Dicer function in oocytes has been reported to cause infertility (Flemr et al., 2013; Knight and Bass, 2001; Murchison et al., 2007). But, in both cases, the process of oogenesis proceeds apparently normally until just before fertilization (or after oocyte maturation). In mouse oocytes that lack dicer function, no detectable defects were observed in growing oocytes, the number of oocytes or the process of first polar body extrusion; defects in spindle organization and chromosome segregation were however observed after oocytes began to mature. In C. elegans, our genetic mosaic studies indicate that dcr-1 function in the germ line is dispensable for oogenesis, but essential for embryogenesis (Fig. 4): worms that lack dcr-1 function in the germ line produce overtly normal oocytes, but once fertilized and forced to continue to develop in the absence of DCR-1, these oocytes yield inviable embryos. Our functional analysis also reveals that DCR-1 and MPK-1 ERK function in a feedback loop, to restrict the activation of MPK-1 at the loop region.
As shown previously (Knight and Bass, 2001), we find that systemic loss of dcr-1 function in worms produces endomitotic oocytes, with our genetic mosaic studies indicating that this phenotype arises from the loss of dcr-1 from the soma, not the germ line (Fig. 4). Thus, even though DCR-1 is expressed at the RNA and protein level throughout the germ line, it does not appear to function to promote oocyte development.
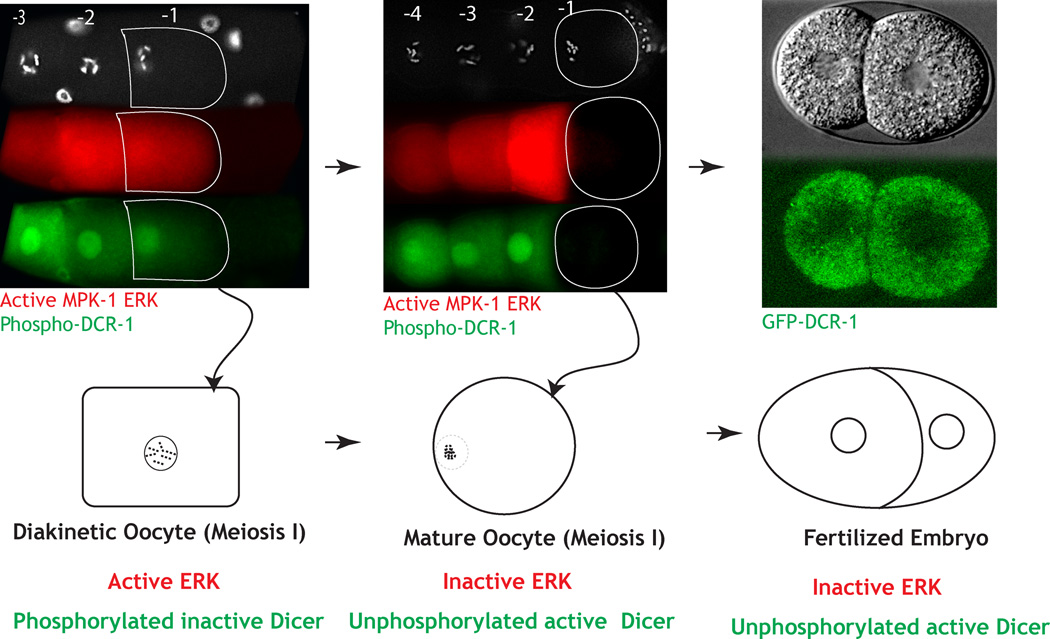
Our work herein leads to the mechanistic model (Fig. 8) on how the ERK-Dicer regulatory nexus could regulate the oocyte-to-embryo transition. For this model, MPK-1 is active in the mid-pachytene stage of oogenesis (zone 1; Fig. 2A), briefly downregulated in the loop region, reactivated in maturing oocytes (zone 2; Fig. 2A), and then downregulated in the maturing oocyte (-1) just before fertilization. Within zones 1 and 2, active MPK-1 promotes pachytene progression and oocyte development and maturation, respectively. MPK-1 drives these oogenic processes at least in part through its phosphorylation and inhibition of Dicer function within developing oocytes. Thus, until the very end of oogenesis Dicer function is inhibited. Then just before oocytes are to be fertilized, a switch is flipped, and within oocytes MPK-1 activity is downregulated, followed by the rapid dephosphorylation of Dicer. Dephosphorylation of Dicer then renders it functional in anticipation of fertilization and embryogenesis. Thus, for oogenesis to occur normally, Dicer function must be inhibited, and for embryogenesis to occur normally, Dicer function must be reinitiated by dephosphorylation. The presence of active ERK during oogenesis drives Dicer phosphorylation; its absence coupled with a phosphatase leads to dephosphorylation of Dicer. The precipitous switch from active to inactive ERK that occurs in the terminal oocyte just before fertilization signals the exact timing of the oocyte-to-embryo transition and the likely reprogramming of small RNA production.
Figure 8. Model: Phosphorylation of Dicer in oocytes aids oocyte-to-embryo transition.
During diakinesis (A, B), the oldest oocyte (-1 in A, B) contains active ERK and phosphorylated Dicer (green, inactive). As this oocyte exits diakinesis and matures (-1 in panel C, D), it turns off active ERK and Dicer is de-phosphorylated (presumably active). Unphosphorylated Dicer is functionally active to promote normal embryogenesis. Thus phosphorylation and dephosphorylation of Dicer appears to be necessary for the reprogramming of an oocyte to an embryo.
The ERK-Dicer nexus: a regulatory switch across species?
Our work in C. elegans suggests that ERK through its ability to phosphorylate Dicer acts as a switch to alter small RNA biogenesis during oogenesis or during the oocyte-embryo transition. We also find that Dicer is phosphorylated in mouse tissue and in human and mouse cell lines, with the phosphorylation and nuclear localization of Dicer in human and mouse cell lines dependent on FGF and the RAS-RAF-MEK-ERK cascade. Thus, Dicer function and with it small RNA biogenesis is under the direct control of ERK (and perhaps other proline-directed serine/threonine kinases) in mammals. We hypothesize that the ERK-Dicer regulatory nexus is a conserved regulatory mechanism that exerts switch-like control over Dicer function and small RNA generation in diverse cell and tissue types within most if not all metazoans.
As Dicer has been show to possess tumor suppressor function (Wang et al., 2011), the mis- or de-regulation of this switch, as likely occurs under oncogenic activation of the RAS-ERK pathway, may directly contribute to the progression and pathology of cancer. However, due to the tumor heterogeneity, specific loss of Dicer activity in certain cell types has been difficult to assess. Our identification of the Dicer-depletion-dependent miRNA precursors as an in vivo biomarker for Dicer activity may be a very useful tool to assess Dicer activity spatially or temporally in Dicer depleted tumor cells.
EXPERIMENTAL PROCEDURES
Strains and reagents used
Standard C. elegans culture conditions at 20°C were used unless noted. Strains used in the study were: I: rrf-1(pk1417); III: dcr-1(ok247) III/hT2GFP I; II: EG4322 (ttTi5605; unc-119(ed3)III. dcr-1(ok247)/unc-32 III. Strains generated in the study: vizIs5 (GFP::DCR-1(WT);dcr-1(ok247)); vizIs11 (GFP::DCR-1(S1833A);dcr-1(ok247)); vizIs24 (GFP::DCR-1(S1833E);dcr-1(ok247); vizIs8 (GFP::DCR-1(S1705E);dcr-1(ok247); vizIs25 (GFP::DCR-1(S1705A);dcr-1(ok247). These strains were generated via biolistic transformation (Merritt et al., 2008). Strains generated via MosSci mediated insertion (Frokjaer-Jensen et al., 2008) vizSi1(GFP::DCR-1(WT)II;unc-119dcr-1(ok247))III; vizSi2 (GFP::DCR-1(S1705E)II;unc-119dcr-1(ok247)III; vizSi3(GFP::DCR-1(S1705A)II;unc-119dcr-1(ok247)III; vizSi4 (GFP::DCR-1(S1833A)II;unc-119dcr-1(ok247))III; vizSi5 (GFP::DCR-1(S1833E)II;unc-119dcr-1(ok247)III; visSi6 (GFP::DCR-1(S1705E, S1833E)II;unc-119dcr-1(ok247)III; vizSi7 (GFP::DCR-1(S1705AS1833A);unc-119dcr-1(ok247)III.
Generation and purification of phospho-Dicer antibody
To generate pDCR-1 antibodies, the phospho-peptides RQY*SPGVL, and PR*SPIREL were synthesized and HPLC fractionated to >95% purity (Sigma, MO.), as described earlier(Arur et al., 2009). Sera were generated in rabbits via Yenzyme Inc, CA. Each phospho-purified DCR-1 antibody was purified 4 times to obtain the desired specificity. Specificity at each step of purification was assayed as described (Arur et al., 2009).
Transgenic constructs and generation of transgenic lines
The GFP::DCR-1 construct contains 3.75 kb of the 5’ promoter and UTR region and 3 Kb of the 3’ UTR region of the dcr-1 gene with GFP inserted immediately before the translational start. dcr-1 promoter and 3’UTR genomic sequences were amplified from Bristol N2 genomic DNA and inserted into either the pMMO16 vector or the pCFJ150 vector for biolistic transformation or MosSci based transformation respectively. S1705A/S1833A and the S1705E/S1833E mutations were generated via Dpn1 mediated site directed mutagenesis as described earlier (Arur et al., 2009). All constructs were sequenced verified. Unless mentioned, MosSci lines are presented in the figures. Both biolistic transformation lines as well as the MosSci lines produced a similar range of phenotypes. For MosSci injections, ttTi5605; unc-119(ed3) was crossed into dcr-1(ok247) and unc-119(ed3)dcr-1(ok247) recombinants were selected. Injected lines were assayed for non-unc phenotypes and dcr-1(ok247) rescue. The expression of each integrated transgene was then assessed in the dcr-1(0) background by anti-GFP immunostaining analysis.
Mosaic analysis
Genetic mosaic analysis for dcr-1 was performed using a rescuing dcr-1(+) extrachromosomal array, vizEx001 [dcr-1(+) sur-5::gfp] carrying the cell autonomous sur-5::gfp pMM01(Yochem et al., 1998). dcr-1(ok247) animals bearing vizEx001 array were used for the analysis. Young adult hermaphrodites were examined on a Zeiss Axioskop using DIC and fluorescence microscopy with a 100X Plan-Apochromat (NA 1.4) objective lens to identify array loss. To determine the point of array loss in animals exhibiting mosaic expression of the sur-5::gfp marker, the following cells were examined: DTC, gonadal sheath, spermatheca, body wall muscles, intestine, and the germ line (the presence of GFP-positive progeny could only be scored if at least one gonad arm produced fertilizable embryos). To identify germ line-loss mosaics, L4 hermaphrodites were cultured individually. Animals producing embryos of GFP-negative progeny were further examined by fluorescence microscopy. Dead embryos produced by dcr-1(ok247) germ line mosaics were assayed by dissection of mothers and analysis of the embryos.
Supplementary Material
ACKOWLEDGEMENTS
The authors thank Drs. Tim Schedl, David Greenstein and Andy Golden for extensive and helpful discussions and reagents. SA thanks Dr. David Greenstein for extensive guidance on the mosaic analysis. We thank Dr. Jill Schumacher and members of the Arur Lab for comments on the manuscript. The following funding agencies supported this work: NSF-IOS-0744261, NIH NS036570 (JBS), NIH GM37706 (AZF), NIH T32CA009299 (GG) while in Dr. Richard Behringer’s lab, and NIH GM98200, Center for Genetics and Genomics, and Institutional Research Grant (CCSG) (SA).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA ACCESS
The small RNA profiling data have been deposited at the NCBI Bioproject database Accession Number PRJNA265871, and Sequence Read Archive Data Base Accession Number SRP049421.
AUTHOR CONTRIBUTIONS
MD, TF, KSM, GG, AK, AZF, SA performed experiments. MD, TF, BL, AK, JL, AZF, JBS and SA analyzed the data. SA conceived the study and SA, AZF and JBS wrote the manuscript.
REFERENCES
- Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S, Ohmachi M, Nayak S, Hayes M, Miranda A, Hay A, Golden A, Schedl T. Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc Natl Acad Sci U S A. 2009;106:4776–4781. doi: 10.1073/pnas.0812285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato C, Ciotti MT, Serafino A, Calissano P, Cogoni C. Dicer expression and localization in post-mitotic neurons. Brain Res. 2007;1175:17–27. doi: 10.1016/j.brainres.2007.07.088. [DOI] [PubMed] [Google Scholar]
- Barraud P, Emmerth S, Shimada Y, Hotz HR, Allain FH, Buhler M. An extended dsRBD with a novel zinc-binding motif mediates nuclear retention of fission yeast Dicer. The EMBO journal. 2011;30:4223–4235. doi: 10.1038/emboj.2011.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Church DL, Guan KL, Lambie EJ. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development. 1995;121:2525–2535. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Buhler M. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Developmental cell. 2010;18:102–113. doi: 10.1016/j.devcel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan JA, Nadarajan S, Kim S, Starich TA, Greenstein D. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development. 2009;136:2211–2221. doi: 10.1242/dev.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic acids research. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Gu W, Claycomb JM, Batista PJ, Mello CC, Conte D. Cloning Argonaute-associated small RNAs from Caenorhabditis elegans. Methods Mol Biol. 2011;725:251–280. doi: 10.1007/978-1-61779-046-1_17. [DOI] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJ, Greenstein D. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn. 2000;218:2–22. doi: 10.1002/(SICI)1097-0177(200005)218:1<2::AID-DVDY2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 2012;8:e1002702. doi: 10.1371/journal.pgen.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Ohmachi M, Arur S, Nayak S, Francis R, Church D, Lambie E, Schedl T. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics. 2007;177:2039–2062. doi: 10.1534/genetics.107.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AL, 3rd, Chen J, Joo HJ, Drake M, Shidate M, Kseib C, Arur S. DAF-2 and ERK couple nutrient availability to meiotic progression during Caenorhabditis elegans oogenesis. Developmental cell. 2013;27:227–240. doi: 10.1016/j.devcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3’ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Current opinion in cell biology. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GS, Maity TS, Bass BL. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J Mol Biol. 2008;384:967–979. doi: 10.1016/j.jmb.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Filipowicz W, Svoboda P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PloS one. 2010;5:e12175. doi: 10.1371/journal.pone.0012175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, Eppig JJ. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302:104–117. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Current biology : CB. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram MV. RTK/Ras/MAPK signaling. WormBook. 2006:1–19. doi: 10.1895/wormbook.1.80.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Plasterk RH. Dicers at RISC; the mechanism of RNAi. Cell. 2004;117:1–3. doi: 10.1016/s0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- Verlhac MH, de Pennart H, Maro B, Cobb MH, Clarke HJ. MAP kinase becomes stably activated at metaphase and is associated with microtubule-organizing centers during meiotic maturation of mouse oocytes. Developmental biology. 1993;158:330–340. doi: 10.1006/dbio.1993.1192. [DOI] [PubMed] [Google Scholar]
- Verlhac MH, Kubiak JZ, Weber M, Geraud G, Colledge WH, Evans MJ, Maro B. Mos is required for MAP kinase activation and is involved in microtubule organization during meiotic maturation in the mouse. Development. 1996;122:815–822. doi: 10.1242/dev.122.3.815. [DOI] [PubMed] [Google Scholar]
- Verlhac MH, Lefebvre C, Kubiak JZ, Umbhauer M, Rassinier P, Colledge W, Maro B. Mos activates MAP kinase in mouse oocytes through two opposite pathways. The EMBO journal. 2000;19:6065–6074. doi: 10.1093/emboj/19.22.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Czech B, Crunk A, Wallace A, Mitreva M, Hannon GJ, Davis RE. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome research. 2011;21:1462–1477. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Molecular cell. 2011;41:589–599. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Pavelec DM, Nix DA, Duchaine TF, Kennedy S, Bass BL. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA. 2010;16:893–903. doi: 10.1261/rna.2122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, He H, Wang Y, Liu M, Zhou T, Lin M, Zhou Z, Huo R, Zhou Q, Sha J. PCBP1 is required for maintenance of the transcriptionally silent state in fully grown mouse oocytes. Cell Cycle. 2012;11:2833–2842. doi: 10.4161/cc.21169. [DOI] [PubMed] [Google Scholar]
- Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR, 3rd, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccotti M, Bellone M, Longo F, Redi CA, Garagna S. Fully-mature antral mouse oocytes are transcriptionally silent but their heterochromatin maintains a transcriptional permissive histone acetylation profile. Journal of assisted reproduction and genetics. 2011;28:1193–1196. doi: 10.1007/s10815-011-9562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.