Abstract
The dengue virus (DENV) circulates between humans and mosquitoes and requires no other mammals or birds for its maintenance in nature. The virus is well-adapted to humans, as reflected by high-level viraemia in patients. To investigate its high adaptability, the DENV induction of host type-I interferon (IFN) was assessed in vitro in human-derived HeLa cells and compared with that induced by the Japanese encephalitis virus (JEV), a closely related arbovirus that generally exhibits low viraemia in humans. A sustained viral spread with a poor IFN induction was observed in the DENV-infected cells, whereas the JEV infection resulted in a self-limiting and abortive infection with a high IFN induction. There was no difference between DENV and JEV double-stranded RNA (dsRNA) as IFN inducers. Instead, the dsRNA was poorly exposed in the cytosol as late as 48 h post-infection (p.i.), despite the high level of DENV replication in the infected cells. In contrast, the JEV-derived dsRNA appeared in the cytosol as early as 24 h p.i. Our results provided evidence for the first time in DENV, that concealing dsRNA in the intracellular membrane diminishes the effect of the host defence mechanism, a strategy that differs from an active suppression of IFN activity.
The dengue virus (DENV), a member of the family Flaviviridae, is the causative agent of dengue, which is a major mosquito-borne disease in tropical and subtropical regions of the world. A recent study estimates 390 million dengue infections annually, of which 96 million have apparent manifestations (at any level of disease severity)1. There are four different serotypes of the DENV, all of which cause a spectrum of diseases ranging from a mild, non-specific febrile syndrome called classical dengue fever (DF), to the severe forms of the disease, dengue haemorrhagic fever (DHF) and dengue shock syndrome (DSS)2. The cells of the mononuclear phagocyte lineage are considered to be the primary target of DENV3,4. The presence of this virus in cells in organs, such as the Kupffer cells and endothelial cells in the liver and the vascular endothelial cells in the lung, has been reported5. However, the mechanism of cellular tropism and the viral kinetics are not clarified yet. Additionally, an understanding of the role of the host immune responses in disease evolution is still wanting. Thus far, little progress has been made on the development of animal models that will help elucidate the pathogenesis or contribute to the development of antiviral therapy. Despite the great need, antiviral drugs and effective vaccines are unfortunately not yet available for dengue.
Like other flaviviruses, the DENV circulates between vertebrate animals and mosquitoes, mainly Aedes aegypti. Although the virus is maintained through a sylvatic cycle in non-human primates and an urban cycle in humans, the former is not necessary, a characteristic that differentiates it from the yellow fever virus, the prototype flavivirus6,7. In both mild and severe cases of dengue, patients present with a high-level viraemia 4 to 5 days before defervescence8, making humans a viral reservoir, or a natural amplifier of the virus. In contrast to this, patients infected with the Japanese encephalitis virus (JEV) hardly exhibit viraemia. The JEV circulates between swine, wild birds and Culex mosquitoes, with accidental infection of humans, a dead-end host because of the low-level viraemia9,10.
The host innate immune response mediated by type-I interferon (IFN-α/β) and its subsequent signaling pathway is one of the most important frontline, non-specific defences against the early phase of virus infections11. The type-I IFN induction is triggered by viral components called pathogen-associated molecular patterns (PAMPs), mainly virus-derived double-stranded RNA (dsRNA) in the case of flavivirus infection12. Retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) are cytosolic pattern recognition receptors (PRRs) that sense virus-derived or synthetic dsRNA. RIG-I and MDA5 positively regulate IFNα/β expression through the IFN regulatory factor (IRF)-3 and nuclear factor-kappa B (NF-κB). The secreted type-I IFNs attach to the interferon-α/β receptors (IFNAR) 1 and 2 to activate the Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling pathway. STAT1 and STAT2 combine with IRF-9 to form a heterotrimeric transcription factor complex known as ISGF3 and induce the expression of hundreds of IFN-stimulated genes (ISGs), which perform various antiviral activities13.
The flaviviruses have developed several mechanisms to escape from or counterattack the type-I IFN pathway. The hepatitis C virus non-structural protein 3/4A (NS3/4A) is known to suppress the RIG-I/Cardif-induced IFN response14. The tick-borne encephalitis virus (TBEV) and JEV have been reported to conceal their dsRNA in intracellular membrane vesicles to hide from the cytosolic PRRs, thereby delaying the type-I IFN induction15,16,17. The type-I IFN regulation by DENV non-structural proteins NS2B/318,19, NS4A/B20,21, and NS522 has been previously described.
This study aims to investigate further the innate immune response to a DENV infection by evaluating the IFN induction in a human-derived cell line and correlating it with the viral replication and spread. Here, we provided evidence about the DENV strategy of concealing its double-stranded RNA in the intracellular membrane to induce a poor IFN induction, thereby allowing the viral replication to proceed. This finding could contribute to understanding why the DENV, in comparison with the JEV, replicate efficiently in a human host.
Results
The DENV and JEV exhibit differential dissemination in human-derived HeLa cells
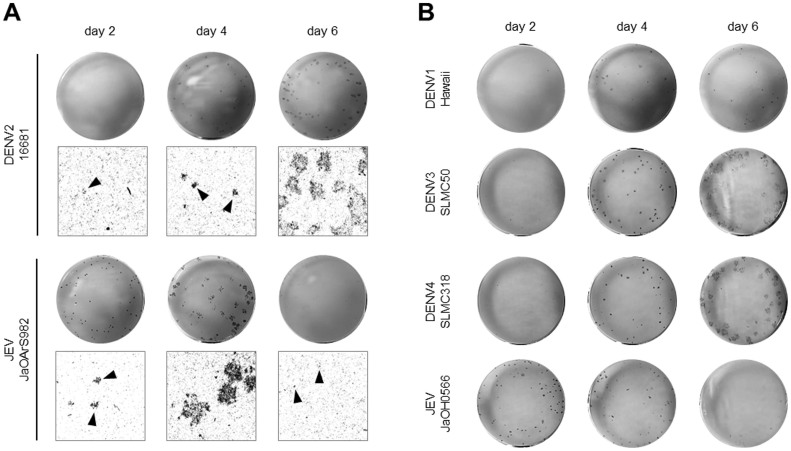
The dengue virus (DENV) dissemination was evaluated in human-derived HeLa cells. The Japanese encephalitis virus (JEV), a flavivirus more adapted to swine and birds than humans, was included for comparison. The HeLa cells were infected with DENV2 strain 16681 or JEV strain JaOArS982. The foci of the infected cells were detected by immunostaining every 2 days until day 6 post-infection (p.i.), the time after which cells started dying. At 4 days p.i., visible foci of DENV-infected cells were observed and they continued to enlarge until 6 days p.i., whereas, the foci of JEV-infected cells developed from 2 to 4 days p.i., but they almost disappeared by 6 days p.i (Fig. 1A). This abortive focus formation in the JEV-infected primate cells was demonstrated in a previous study17. To confirm this observation, the viral spread of other DENV serotypes and JEV strains was evaluated in HeLa cells. Patient-derived DENV3 (strain SLMC-50) and DENV4 (strain SLMC-318) produced a foci pattern similar to DENV2 strain 16681 (Fig. 1B). At 4 days p.i., only tissue culture-adapted DENV1 (strain Hawaii)-infected cells showed visible foci, the size of which remained unchanged until 6 days p.i. At 2 days p.i., JEV strain JaOH0566 (isolated from human brain) produced foci, which disappeared by 6 days p.i., as was observed for JEV strain JaOArS982 (Fig. 1B).
Figure 1. DENV and JEV focus formation in HeLa cells.
(A) HeLa cells were infected with DENV2 strain 16681 or JEV strain JaOArS982 at 40 focus forming units (FFU)/well. At the indicated days p.i., the foci were detected by immunostaining. The upper figures show macroscopic images, and the lower figures show 100× magnified images. The black arrowheads indicate the foci. (B) HeLa cells were infected with DENV1 strain Hawaii, DENV3 strain SLMC-50, DENV4 strain SLMC-318, or JEV strain JaOH0566 at 40 FFU/well. At the indicated days p.i., foci were detected by immunostaining.
An anti-IFN cocktail does not enhance DENV infection
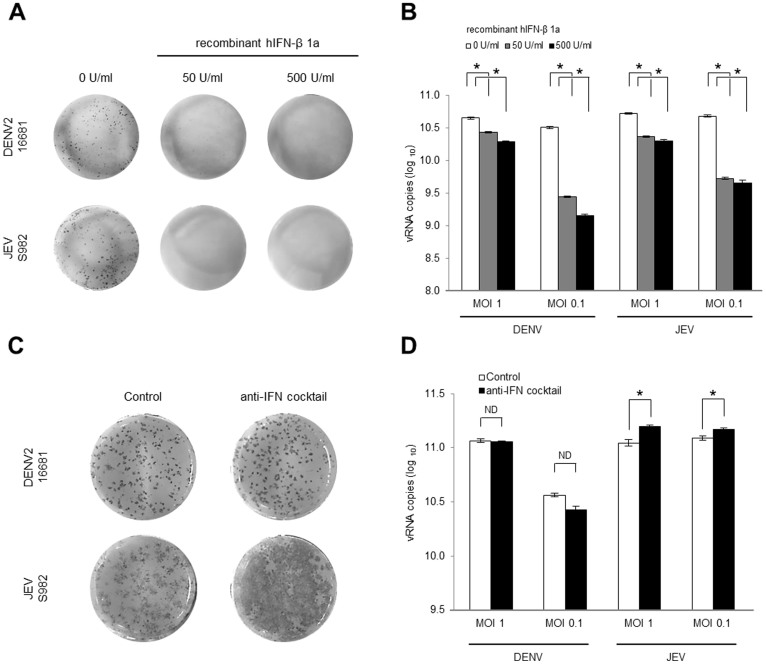
IFN produced by cells in response to an infection can regulate viral growth and spread. To confirm the effect of type-I IFN on the DENV and the JEV, DENV- or JEV-infected cells were treated with extracellular human recombinant-IFN-β1a at 12 h p.i. The IFN treatment inhibited the focus formation and growth of the DENV and the JEV (Fig. 2A and B), demonstrating the similar sensitivity of both viruses to IFN. A previous study demonstrated that JEV dissemination in primate cell lines is restricted by type-I IFN17. To clarify its role in virus dissemination and growth, type-I IFN was blocked using an anti-IFN antibody cocktail (anti-human IFN-β and anti-human IFN-α/βR2). The anti-IFN treatment had no effect on the DENV focus formation or viral growth (Fig. 2C and D), whereas the number and size of foci produced by the JEV were increased (Fig. 2C). The JEV growth in culture fluid was significantly increased with anti-IFN treatment (Fig. 2D). These results indicate that the JEV infection induces a type-I IFN response that restricts focus formation in HeLa cells. On the other hand, the lack of response to the anti-IFN treatment in the DENV-infected cells suggests an absence of or poor type-I IFN induction.
Figure 2. Effect of IFN-β on DENV and JEV focus formation in HeLa cells.
HeLa cells were infected with DENV2 strain 16681 or JEV strain JaOArS982. At 12 h p.i., the culture medium was replaced with 0 to 500 U/ml of the human recombinant-IFN-β1a in MEM. At 4 days p.i., foci were detected by immunostaining (A) and the vRNA in the culture fluid was quantified by RT-qPCR (B). The HeLa cells were treated with anti-IFN cocktail consisting of 2,000 neutralization U/ml of the anti-human IFN-β Ab and 20 μg/ml of the anti-human IFN-α/βR2 (CD118) 1 h before the viral infection. The concentration of the anti-IFN cocktail was maintained after the viral infection. At 4 days p.i. the foci were detected by immunostaining (C), and the vRNA in the culture fluid was quantified by RT-qPCR (D). The mean ± SE of the vRNA copy number was obtained from two independent duplicate experiments. The values between two groups were tested by Welch test analysis. The asterisks indicate a statistical significance at p < 0.01, and ND indicates no significant difference.
IFN is weakly induced in the DENV-infected cells
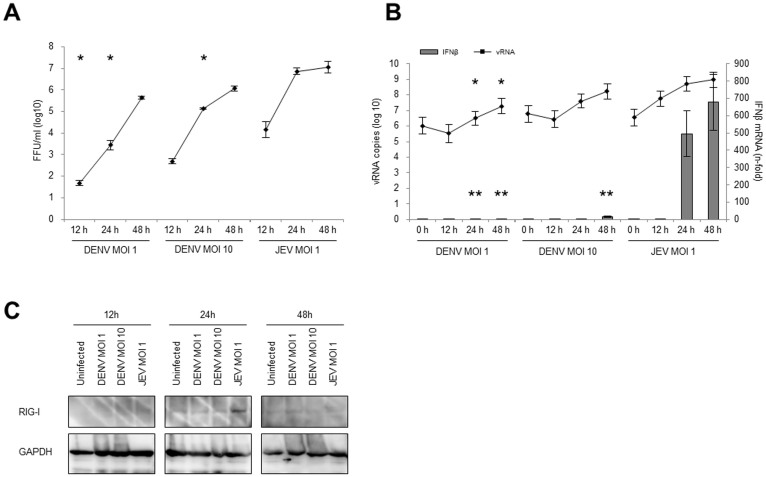
To quantify the type-I IFN induction during viral infection, IFN-β mRNA was measured by quantitative reverse transcription PCR (RT-qPCR). The extracellular virus from supernatants and intracellular viral RNA (vRNA) were also quantified by focus forming assay and RT-qPCR, respectively, to assess the relationship between IFN-β induction and virus replication. At MOI 1 and 10, the extracellular levels of the DENV increased logarithmically from 12 to 48 h p.i. with a maximum titre of 5.65 ± 0.05 (log10) FFU/ml and 6.06 ± 0.09 (log10) FFU/ml, respectively. In comparison, the extracellular levels of the JEV increased rapidly from 12 to 24 h, after which it started to plateau and reached a maximum titre of 7.06 ± 0.22 (log10) FFU/ml at 48 h p.i (Fig. 3A). High levels of intracellular vRNA were measured, which peaked at 7.29 ± 0.50 (log10) DENV RNA copies/μg of total RNA at MOI 1, 8.22 ± 0.50 (log10) DENV RNA copies/μg of total RNA at MOI 10, and 8.97 ± 0.48 (log10) JEV RNA copies/μg of total RNA at MOI 1 by 48 h p.i. (Fig. 3B and Supplemental Table S2). However, the IFN-β expression in the DENV-infected cells was extremely low, which was 1.93-fold (MOI 1) and 15.74-fold (MOI 10) higher than that in the uninfected cells. In contrast, the JEV-infected cells (MOI 1) showed highest level of IFN-β induction (678.85-fold higher than uninfected cells) at 48 h p.i (Fig. 3B and Supplemental table S2).
Figure 3. Virus growth and innate immune response against DENV and JEV infection.
HeLa cells were infected with DENV2 strain 16681 at MOI 1 or 10 or JEV strain JaOArS982 at MOI 1. At the indicated time points, the culture fluid and the cells were harvested. (A) The virus titers in the culture fluid were determined by FFA. (B) The vRNA and the IFN-β gene expression in the DENV- and the JEV-infected cells that were quantified by RT-qPCR. The absolute vRNA copy numbers and the relative-fold increase in the IFN-β mRNA levels were normalized with GAPDH mRNA levels. The mean ± SE of the vRNA copy number was obtained from three independent duplicate experiments. The values between the DENV- (MOI 1) and JEV- (MOI 1) infected cells or the DENV- (MOI 10) and the JEV- (MOI 1) infected cells were tested by Welch test analysis. The single asterisks indicate a significant difference of vRNA (* p < 0.01) and double asterisks indicate a significant difference of IFN-β mRNA (** p < 0.01). (C) Representative cropped blots of RIG-I and GAPDH in DENV- and JEV-infected cells are shown. The infected cells were harvested and the target proteins were detected by the immunoblotting assay. GAPDH was used as an internal control. All the samples were derived from the same experiment and blotting was processed in parallel. The full-length blotting images are presented in Supplemental Fig. S4.
In flavivirus infections, the dsRNA is detected by cytosolic PRRs and induces a type-I IFN response. The JEV dsRNA is recognized by RIG-I, while DENV dsRNA is recognized by both RIG-I and MDA523,24. To confirm the upregulation of PRRs in both viral infections, RIG-I and MDA5 were examined with immunoblotting. Unexpectedly, MDA5 level was below detection in this study (data not shown). The DENV did not induce RIG-I expression at any time point, though RIG-I was clearly up-regulated in the JEV-infected cells at 24 h p.i. (Fig. 3C). In conclusion, the DENV-infected cells seems to lack the IFN-β induction activity, even though the cells were infected with DENV at a high MOI and showed a high level of viral RNA.
The exogenous DENV dsRNA induced a high level of IFN-β expression in DENV-infected cells
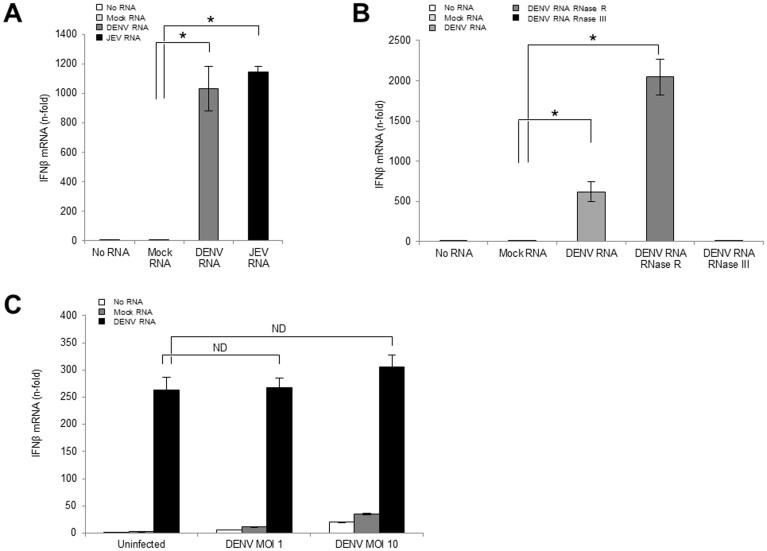
The viral dsRNA, produced in cells after flavivirus infection, is a major type-I IFN inducer12. Our data on the low IFN-β induction in DENV-infected cells implied an inability of the DENV dsRNA as IFN-β inducer. To evaluate the difference between IFN-β induction activity of the DENV-derived dsRNA and the JEV-derived dsRNA, the viral dsRNA was transfected into the cells. The dsRNA was prepared by using a hot phenol chloroform extraction to keep its double strand structure as described in methods. Unexpectedly, both the DENV and the JEV dsRNAs induced a similar high level of IFN-β at 6 h post transfection of uninfected cells (Fig. 4A). The data showed that the DENV and JEV dsRNAs were similar as IFN-inducers. Moreover, to confirm that viral dsRNA, but not single-stranded RNA (ssRNA) induces IFN-β, the DENV dsRNA was treated prior to cell transfection with RNase R for ssRNA digestion or RNase III for dsRNA digestion. The RNase III-digested DENV RNA lost the IFN-β induction activity (Fig. 4B), showing that viral dsRNA is important for IFN-β induction. After RNase digestion, the amount of total RNA was measured, and 500 ng/well of RNA was transfected into cells. As a result, the ratio of dsRNA in the RNase R-treated sample increased, and this may explain why RNase R-treated RNA demonstrated a higher IFN-β induction than the non-treated RNA (Fig. 4B).
Figure 4. Effect of viral dsRNA on the IFN activation pathway.
(A) One microgram/well of the mock RNA, the DENV RNA, or the JEV RNA was transfected into uninfected cells. (B) Five hundred nanogram/well of the non-treated, RNase R-treated or the RNase III-treated DENV RNA was transfected into uninfected cells. (C) The cells were infected with DENV2 strain 16681 at MOI 1 or 10 prior to the IFN-β stimulating assay. After 48 h p.i., 500 ng/well of the mock RNA or the DENV RNA were transfected into the cells with the uninfected control. Six hours after the RNA transfection, the cells were harvested, and the IFN-β and the GAPDH mRNA levels were quantified by RT-qPCR. The relative-fold increase of the IFN-β mRNA levels was normalized with the GAPDH mRNA levels. The mean ± SE of the IFN-β mRNA levels were obtained from two independent duplicate experiments. The values between two groups were tested by Welch test analysis. The asterisks indicate statistical significance at p < 0.01, and ND indicate no significant difference.
To evaluate the IFN-β activation pathway in DENV-infected cells, mock RNA or the DENV dsRNA was transfected into uninfected or DENV-infected cells. Results showed that the uninfected and the DENV-infected cells expressed almost equal levels of IFN-β after stimulation by DENV dsRNA (Fig. 4C). Similar to the DENV dsRNA, the JEV dsRNA showed IFN-β induction in the uninfected, the DENV- and the JEV-infected cells (Supplemental Fig. S1). The JEV-infected cells with the exogenous inducer JEV dsRNA produced an extremely high level of IFN-β, which was likely due to a cumulative effect of the exogenous inducer and the viral infection itself (Supplemental Fig. S1). By 48 h p.i., 79.03% ± 2.08% and 78.02% ± 2.54% of the cells were infected with DENV at MOI 1 and JEV at MOI 1, respectively (Fig. 5B). It indicates that IFN-β induced by exogenous dsRNA is not derived from cells not yet infected by the virus within the culture but from DENV- or JEV-infected cells. These results indicated that the DENV dsRNA can induce IFN-β. The results in Fig. 3B where poor IFN-β induction in DENV-infected cells was shown could mean that the IFN-β induction pathway was not well activated.
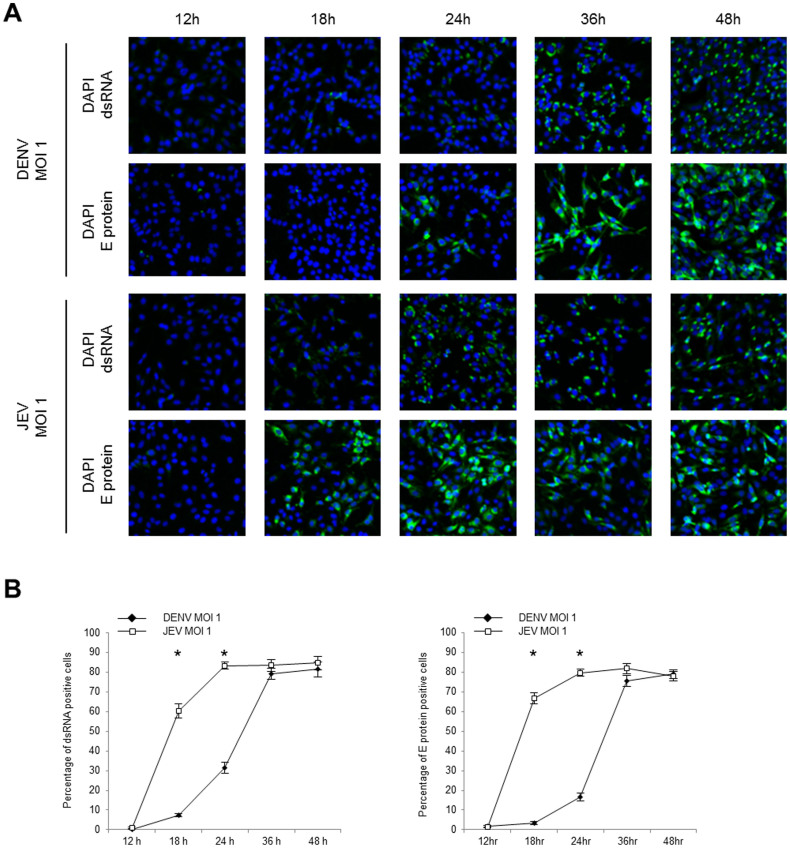
Figure 5. dsRNA and E protein expression in DENV- and JEV-infected cells.
HeLa cells were infected with DENV2 strain 16681 or JEV strain JaOArS982 at MOI 1 in chamber slides. At the indicated time points, the infected cells were fixed and permeabilized by 1% NP-40. (A) The viral dsRNA and E protein were visualized by using the immunofluorescence assay. The nuclei were stained with DAPI. (B) The percentages of the dsRNA and the E protein expression were obtained by calculating the mean percentage of positively stained cells in 3 different fields (241.1 ± 10.8 total cells/field). The percentages of positively stained cells were arcsine transformed prior to the Welch test analysis. The statistically significant differences between DENV- and JEV-infected cells are indicated (* p < 0.01). The error bars indicate standard error of the means.
The DENV dsRNA is predominantly absent in the cytosol
Due to the pivotal role of dsRNA in the IFN induction, its expression was evaluated. By using an immunofluorescence assay, the viral dsRNA was detected in most of the DENV- and JEV-infected cells at 36 h and 24 h p.i., respectively (Fig. 5A). Both infections caused a maximum of approximately 80% of total cells expressing dsRNA (Fig. 5B). The analysis of the viral E protein revealed similar kinetics and percentages of dsRNA expressing cells, confirming the above results.
The localization of dsRNA is important for recognition by PRRs. Intracellular localization of dsRNA was determined through differential permeabilization methods followed by immunofluorescence staining17. Nonidet P-40 (NP-40) is known to permeabilize all cellular membrane structures such as the plasma membrane and endoplasmic reticulum (ER) membrane. On the other hand, digitonin, a mild non-ionic detergent, permeabilizes membranes enriched with cholesterol, such as the plasma membrane, but it preserves intracellular membranes with low levels of cholesterol, such as the ER membrane25,26.
Both of the permeabilization methods were initially evaluated with a cytosol marker, MEK1/2, and an intracellular membrane marker, calregulin, which localizes in the ER lumen. Both markers were positively stained when the cells were permeabilized with 1% NP-40. Digitonin permeabilization (0.5 mM) enabled only the staining of MEK1/2 and not calregulin (Supplemental Fig. S2). As a negative control, the permeabilization buffer without digitonin did not result in the staining of the organelle markers (Supplemental Fig. S2).
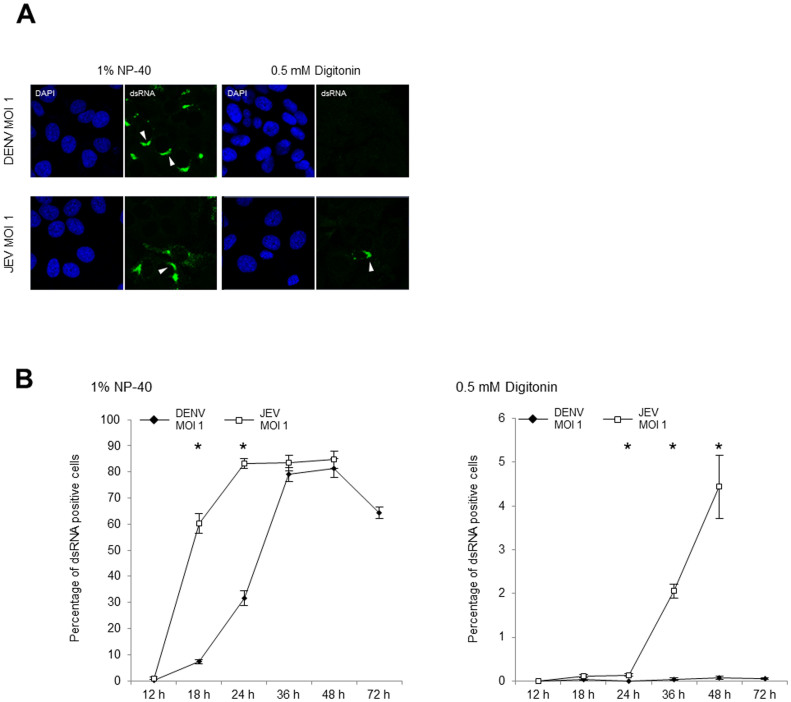
The Digitonin permeabilization of the DENV-infected cells resulted in negative staining for dsRNA in the cytosol (Fig. 6A and B) until 72 h p.i. although more than 60% of the cells had DENV dsRNA in the cells (Fig. 6B). On the other hand, the cytosolic dsRNA was detected in the JEV-infected cells from 24 h p.i., with an increasing number of positively stained cells until 48 h p.i (Fig. 6A, B, and Supplemental Fig. S3 for the low magnification images for each time p.i.). The experiment was continued until the cells started dying (72 h p.i. for DENV and 48 h p.i. for JEV). The DENV dsRNA was predominantly absent in the cytosol up to 72 h p.i., and the absence of cytosolic dsRNA was correlated with an extremely lower IFN-β induction in the DENV-infected cells.
Figure 6. Immunofluorescence detection of the cytosolic DENV and JEV dsRNA.
(A) The localization of the dsRNA in DENV2 strain 16681 (MOI 1) and JEV strain JaOArS982 (MOI 1) infected cells were observed at 24 h p.i. The infected cells were permeabilized with 1% NP-40 or 0.5 mM of digitonin and stained for dsRNA using the immunofluorescence assay. The white arrowheads indicate the viral dsRNA. (B) The percentage of cells positively stained for dsRNA with 1% NP-40 or 0.5 mM digitonin permeabilization was calculated in 3 different fields (241.1 ± 10.8 total cells/field for NP-40, and 1241.4 ± 37.4 total cells/field for digitonin permeabilization). The percentages of positively stained cells were arcsine transformed prior to the Welch test analysis. The statistically significant differences between the DENV- and JEV-infected cells are indicated (* p < 0.01). The error bars indicate standard error of the means. The experiment was continued until the cells started dying (72 h p.i. for DENV and 48 h p.i. for JEV).
Discussion
Our results demonstrated a sustained replication and dissemination of the DENV in human-derived HeLa cells due to the lack of the type-I IFN induction, in contrast to the self-limiting and abortive replication of the JEV as an effect of the high-level type-I IFN induction (Fig. 1, 2 and 3). Several experiments on the exogenous viral dsRNA transfection clarified there was no difference between the DENV- and the JEV-derived dsRNAs as an IFN-β inducer (Fig. 4A, Supplemental Fig. S1). Moreover, we showed that the DENV-infected cells could induce high level of IFN-β when a certain amount of DENV dsRNA was introduced into the infected cells (Fig. 4C). Our data clarified the cytosolic absence of the DENV dsRNA (Fig. 6), the primary PAMPs of flaviviruses, in comparison with cytosolic presence of the JEV dsRNA coupled with IFN-β induction (Fig. 3B and 6). In the human-derived cell line HEK-293 and the non-human primates cell line LLC-MK2, the infection with the DENV also exhibited a poor IFN-β induction up to 24 h p.i. despite a high level of the viral RNA in the cells, and the dsRNA was absent from the cytosol in the LLC-MK2 cells (data not shown). To evaluate the cytosolic dsRNA, RT-qPCR coupled with RNase III and RNase R treatment was done, but significant results were not observed due to the possible low amount of dsRNA in the cytosol. A previous study showed a delayed type-I IFN induction in JEV-infected porcine cells coupled with a delayed cytosolic exposure of the viral dsRNA, which was concealed in intracellular membranes at an early phase of infection. The delay contributed to the efficient JEV dissemination in the cells derived from porcine, one of the amplifiers of JEV in nature17. Here we showed a poor IFN-β induction coupled with a poor cytosolic exposure of the DENV dsRNA in HeLa cells suggesting a dsRNA concealing strategy of the DENV, similar to that of JEV, for efficient viral replication and dissemination in human cells.
Recently, electron tomography studies have revealed the detailed morphology of the DENV replication machinery in intracellular membrane vesicles27,28. During replication, flaviviruses modify the structure of the ER convoluted membranes (also called paracrystalline arrays) and the smooth membrane structures (SMS)29,30. The SMS consist of vesicle packets (Vp), virus induced vesicles (Ve), and tubular structures (T). In Ve, the viral dsRNA and several non-structural (NS) proteins such as NS1, NS2B/3, and NS5 accumulate, likely with host proteins to assemble the replication complex (RC) for viral RNA synthesis31,32. The lumen of the Ve connects to the cytosol through an approximately 11.2 nm pore28. According to Överby and Weber's model of a TBEV infection, the dsRNA leaking from the small pore at the later phase of the infection is recognized by the cytosolic PRRs33. Additionally, Miorin et al. demonstrated the free movement of newly synthesized TBEV RNA within a limited perinuclear region34. These studies suggest that flavivirus RNA can move into a specific cytoplasmic area. However, the crucial factor that triggers the cytosolic dsRNA exposure or determines the vRNA localization has not yet been identified. Obviously, the total copy number of dsRNA is an important factor, but this does not adequately explain the phenomenon, because both the DENV- and the JEV-infected cells reached similarly high levels of viral RNA and dsRNA expression (Fig. 3B, 5, and Supplemental Table S2) yet have differential localization of dsRNA (Fig. 6 and Supplemental Fig. S3). It has also been shown before, that the timing of the JEV dsRNA exposure to the cytosol differs between human cells and porcine cells, despite identical kinetics of viral RNA replication in the two cell lines17. To date, several host factors involved in the flavivirus RC have been reported35,36. Those factors that have a function in nucleic acid transport or membrane remodelling would be ideal candidates for future investigations.
Some DENV proteins have been reported to regulate the type-I IFN response18,19,20,21,22,37. The NS2B/3 protein inhibits the IFN activation through a disruption of MITA, a membrane protein involved in IFN induction that is triggered by viral RNA and dsDNA18,19. The IFN signaling pathway can also be inhibited by co-expression of the DENV NS2A, NS4A and 4B proteins20,21. Moreover, NS4A and 4B co-expression resulted in enhanced inhibition of the IFN-sensitive response element20,21. Jones et al. reported that DENV NS5 mediates the degradation of STAT222,37. In our study, the exogenous DENV dsRNA induced a similar level of IFN-β in the DENV-infected cells as the uninfected cells (Fig. 4C). It suggested that the IFN activation pathway was intact in the infected cells. One reason for the absence of active inhibition of the IFN-β induction by DENV in this study could be that the effect of dsRNA stimulation (500 ng/well) was stronger than the effect of anti-IFN activity by DENV non-structural protein. To investigate the balance between the IFN induction owing to the presence of dsRNA and the anti-IFN activity owing to DENV NS2B/3 protein, the IFN-β induction should be evaluated at lower concentration of the exogenous dsRNA inducer in future study. It is possible that both active inhibition and limited access to dsRNA might account for the poor IFN-β response. In the case of the TBEV infection, the dsRNA-concealing mechanisms can work especially at early phase of infection (up to 12 to 16 h p.i.)15. In a temporary immune evasion strategy, the dsRNA concealing may play a key role in delaying the IFN expression and the consequent stimulation of the ISGs until the IFN-modulating proteins of the DENV are sufficiently expressed in the late phases of the infection, at least up to 24 h p.i.
The DENV is one of the flaviviruses that circulates between humans and mosquitoes, and requires no other mammals and birds for its maintenance in nature. The good adaptability of the DENV to humans should be studied from various perspectives, such as the vectors, host cell receptors, and immune systems responses. Several research groups have demonstrated the anti-type-I IFN activities of the DENV block both the IFN induction and signaling pathways. In this study, we demonstrated a dsRNA-concealing mechanism of the DENV may contribute to the evasion of IFN response. The combination of anti-type-I IFN induction, signaling, and dsRNA concealing may allow DENV to replicate more effectively in human cells. These findings may contribute to the better understanding of the DENV infection in humans.
Methods
Cells and viruses
Human epithelial HeLa and mosquito C6/36 E2 cells were maintained in a minimum essential medium (MEM) supplemented with 10% foetal calf serum (FCS) and 0.2 mM nonessential amino acids. The HeLa cells were grown at 37°C with 5% CO2, and the C6/36 E2 cells were grown at 28°C without CO2. A tissue culture-adapted dengue virus serotype1 (DENV1) strain Hawaii, infectious clone-derived DENV2 strain 16681 (GenBank accession no. U87411)38, and patient-derived DENV3 strain SLMC-50 and DENV4 strain SLMC-318 from the Philippines were used for this study. For comparison, two Japanese encephalitis virus (JEV) strains, a mosquito-derived JaOArS982 (infectious clone)39 and a patient-derived JaOH0566 were used. Viruses were propagated in the C6/36 E2 cells to generate working stocks.
Focus forming assay (FFA) and virus titration
Focus formation and virus titration were performed in the HeLa cells. The HeLa cells were seeded on 24- or 96-well plates until a 80 to 90% confluency. The virus stocks were diluted ten-fold in the MEM supplemented with 2% of FCS and 0.2 mM nonessential amino acids. After the removal of the growth medium, the virus dilutions were inoculated to the cells, and the plates were incubated at 37°C, 5% CO2. After 90 min of virus adsorption, the MEM containing 2% FCS and 1.25% methylcellulose was overlaid on the cells. In another study, the HeLa cells were treated with an anti-IFN-β cocktail of 2,000 neutralization U/ml of anti-human IFN-β (PBL Interferon Source) and 20 μg/ml of anti-human IFN-α/βR2 (CD118) (PBL Interferon Source) 1 h before the viral inoculation, after which the same concentration of anti-IFN-β cocktail was maintained until focus staining was performed as previously described17. In a separate study, the HeLa cells were treated with either 50 or 500 U/ml of human recombinant-IFN-β1a (PBL Interferon Source) at 12 h post infection (p.i.).
At the indicated time points, the infected cells were fixed with 4% paraformaldehyde (PFA). The fixed cells were permeabilized by 1% nonidet P-40 (NP-40) in phosphate buffered saline (PBS) followed by blocking with Blockace (DS Pharma). For the primary antibody (Ab), the mouse monoclonal Ab against the flavivirus-specific envelope (E) glycoprotein, 12D11/7E8 was used to detect both the DENV- and the JEV-infected cells40. Five hundred times diluted goat anti-mouse IgG + M Ab conjugated with horse radish peroxidase (American Qualex) was used as a secondary Ab, and the infected cells were visualized with 0.8 mg/ml of 3,3′-Diaminobenzidine, tetrahydrochloride (DAB). For virus titration, the DENV- and JEV-infected cells were immunostained at 4 and 3 days p.i., respectively.
Real-time quantitative reverse transcription PCR
The intracellular host mRNA and viral RNA (vRNA) and the vRNA in culture fluid were quantified by using quantitative reverse transcription (RT-qPCR). To quantify the vRNA, DENV and JEV consensus primers were designed for the NS5 coding gene. The primer sets for the IFN-β mRNA and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were described in previous studies17,41.The total RNA was harvested from the cells by using the RNeasy Mini Kit (Qiagen). One microgram of RNA in a total volume of 20 μl was reverse transcribed with Superscript III (Invitrogen) in the presence of the Oligo(dT)12–18 primer (Invitrogen) and NS5 gene specific reverse primer to make complementary DNA for cellular mRNA and vRNA, respectively. In another study, the vRNA in culture fluid was extracted using the QIAamp Viral RNA Mini Kit (Qiagen). Eleven microliters of the extracted RNA in a total volume of 20 μl was reverse transcribed with Superscript III in the presence of NS5 gene specific reverse primer to make complementary DNA. Three microliters of the product was used for SYBR green real-time PCR (Applied Biosystems). The primer sets used in this study are shown in Supplemental Table S1. The number of copies of the cellular mRNA and vRNA were calculated by absolute quantification based on plasmid standards. Both the copy numbers of vRNA and IFN-β mRNA were normalized to that of the GAPDH mRNA. The level of the IFN-β mRNA was expressed as a fold increase over the uninfected cells.
Immunoblotting assay
The HeLa cells were cultured in 6-well plates, and the cells were infected with the DENV or the JEV. At the indicated time p.i., cells were washed in PBS and collected using trypsin-EDTA. The collected cells were lysed by incubating them on ice with 8 M urea, 2% 3-[(3-Cholamidopropyl)dimethylammonio]-propanesulfonate (CHAPS), and 0.2% of sodium dodecyl sulfate (SDS) in PBS for 30 min. A four-fold dilution of acetone was added to the cell lysate, and incubated at −80°C for 1 h to allow for precipitation. After centrifugation at 15,000 rpm for 15 min, the pellet was dissolved in loading buffer (0.25 M trishydroxymethylaminomethane-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.01% bromophenol blue, and 5% 2-mercaptoethanol). After heat denaturation at 100°C for 3 min, the samples and the Precision Plus Protein Standards (Bio Rad) were loaded in a 5 to 20% SDS-polyacrylamide gel electrophoresis (Atto) and an electrophoresis was run for 90 min at 20 mA/gel. The proteins were transferred to a polyvinylidene difluoride membrane by semi-dry blotting for 2 h at 100 mA/membrane followed by blocking with Blockace. For the primary antibodies, anti-retinoic acid-inducible gene-I (RIG-I), anti-melanoma differentiation-associated gene 5, and anti-GAPDH antibodies (Santa Cruz Biotechnology) were used at 200 times dilution in Blockace. To detect the primary antibodies, an anti-rabbit IgG (American Qualex) and an anti-goat IgG (Santa Cruz Biotechnology) conjugated with horse radish peroxidase were used at 5,000 times dilution in Blockace as secondary antibodies. The protein bands were visualized by using Luminata Forte Western HRP substrate (Millipore) and detected by LAS-4000 mini-luminescent-image analyser (Fujifilm).
IFN-β stimulating assay
To prepare the double-stranded RNA (dsRNA), the HeLa cells were cultured in a 75 cm2 flask, and the cells were infected with the DENV or the JEV at MOI 1. At 48 h p.i., infected cells were harvested and the total RNA was extracted using a hot acidic phenol: chloroform (5: 1) preparation (Sigma) to maintain the structure of the viral dsRNA. The extracted RNA was treated with 6.8 Kunitz units of DNase (Qiagen), and the aliquots were kept at −80°C until use. The mock RNA was prepared from the uninfected HeLa cells as a negative control. For the RNase digestion, 10 μg of the extracted RNA was digested by 20 U of RNase R (Epicentre Biotechnologies) in the reaction buffer or 1 U of RNase III (Epicentre Biotechnologies) in the reaction buffer (0.33 mM trishydroxymethylaminomethane-acetate, pH 7.5, 66 mM potassium acetate, 10 mM magnesium acetate and 0.5 mM dithiothreitol). After the RNase treatment, the RNA was extracted using acidic phenol: chloroform (5: 1), and the RNA concentration was evaluated for the IFN stimulation assay.
For the IFN stimulation, the HeLa cells were cultured in a 12 well plate, and the cells were infected with the DENV or the JEV at the indicated MOI 48 h prior to stimulation, with uninfected control. One or 0.5 μg of RNA was mixed with 1 μl of Plus Reagent (Invitrogen) in 95 μl of Opti-MEM (Gibco). After 5 min, 3 μl of Lipofectamine LTX (Invitrogen) was added to the mixture, incubated for 30 min at room temperature, and transfected into cells. After 6 h of transfection, all of the cells were harvested and the total RNA was extracted. The IFN-β and GAPDH mRNA were quantified by RT-pPCR as described above.
Immunofluorescence assay
The HeLa cells were seeded in an 8-well Millicell EZ slide (Millipore) and infected with the DENV or the JEV. The cells were fixed by incubating them on ice with 4% PFA containing 4% sucrose for 20 min, followed by a wash with HEPES buffer (20 mM HEPES, pH 7.3, 110 mM potassium acetate, 5 mM sodium acetate, 2 mM MgCl2, 1 mM EGTA, 2 mM dithiothreitol)42. To permeabilize only the plasma membrane, the weak detergent digitonin (Sigma) was used. The fixed cells were incubated on ice with 0 to 500 mM of digitonin in HEPES buffer for 8 min. To permeabilize all of the cellular membranes, the cells were incubated with 1% NP-40 in PBS for 15 min at room temperature. Anti-MEK1/2 and anti-calregulin antibodies (Santa Cruz Biotechnology) were used to stain the cytosolic and intracellular membrane markers at 100 and 50 times dilution in Blockace, respectively. To visualize the viral dsRNA and E protein, mouse IgG2a K1 monoclonal Ab (20 μg/ml in Blockace) (English & Scientific Consulting Kft.), and 12D11/7E8 monoclonal Ab were used. The primary antibodies were detected by Alexa Fluor 488 or 594 conjugated secondary antibodies (Invitrogen) at 500 times dilution in Blockace. The stained cells were sealed with ImmunoSelect Antifading Mounting Medium (Dianova GmbH) which contains 4′, 6-Diamidino-2-phenylindole (DAPI). The images were captured using the LSM 780 confocal laser scanning microscope (Carl Zeiss). The cell number in a field was counted using ImageJ software, an open-source image processing program. The percentage of cells positively stained for dsRNA and E protein were calculated in 3 different fields.
Author Contributions
K.M., L.A.E.-M. and L.U. designed the experiments. L.U. performed the experiments and analysed the data and L.A.E.-M. provided technical support on the experiments. Y.T., K.O., D.H., F.Y. and T.N. provided comments and technical advice. K.M., L.A.E.-M. and C.C.B. helped in manuscript writing. All of the authors discussed the results and commented on the manuscripts.
Supplementary Material
Supplemental information
Acknowledgments
We would like to thank Mya Myat Ngwe Tun, Guillermo Posadas-Herrera, and Koutarou Aoki from the Department of Virology, Institute of Tropical Medicine (NEKKEN), Nagasaki University, Nagasaki, Japan for their support; Shuzo Urata, Youhei Kurosaki, and Jiro Yasuda from the Department of Emerging Infectious Disease, Institute of Tropical Medicine (NEKKEN), Nagasaki University, Nagasaki, Japan, for providing technical advice; and Richard M. Kinney from the Center for Disease Control and Prevention, Division of Vector-borne Diseases, Fort Collins, CO, USA, for kindly donating the DENV2 strain 16681 infectious clone. This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, the Global COE program, MEXT, Japan, the Japan Initiative for Global Network on Infectious Diseases (J-GRID), MEXT, Japan, and a Grant-in-Aid for Scientific Research from the Ministry of Health, Labour, and Welfare, Japan.
References
- Bhatt S. et al. The global distribution and burden of dengue. Nature 496, 504–507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamarapravati N. et al. World Health Organization technical report series. Viral Haemorrhagic Fevers. Ch. 2, 13–24 (World Health Organization, Geneva, 1985). [PubMed] [Google Scholar]
- Guabiraba R. & Ryffel B. Dengue virus infection: current concepts in immune mechanisms and lessons from murine models. Immunology 141, 143–156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K., Kyle J. L. & Harris E. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J. Virol. 80, 11418–11431 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessie K., Fong M. Y., Devi S., Lam S. K. & Wong K. T. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J. Infect. Dis. 189, 1411–1418 (2004). [DOI] [PubMed] [Google Scholar]
- Wang E. et al. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 74, 3227–3234 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. N., Knipe D. M. & Howley P. M. Fields Virology 5th edition Ch. 34, 1172–1185 (Lippincott Williams & Wilkins, Philadelphia, 2007). [Google Scholar]
- Vaughn D. W. et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 181, 2–9 (2000). [DOI] [PubMed] [Google Scholar]
- Misra U. K. & Kalita J. Overview: Japanese encephalitis. Prog. Neurobiol. 91, 108–120 (2010). [DOI] [PubMed] [Google Scholar]
- van den Hurk A. F., Ritchie S. A. & Mackenzie J. S. Ecology and geographical expansion of Japanese encephalitis virus. Annu. Rev. Entomol. 54, 17–35 (2009). [DOI] [PubMed] [Google Scholar]
- Randall R. E. & Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89, 1–47 (2008). [DOI] [PubMed] [Google Scholar]
- Munoz-Jordan J. L. & Fredericksen B. L. How flaviviruses activate and suppress the interferon response. Viruses 2, 676–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au-Yeung N., Mandhana R. & Horvath C. M. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT 2, e23931 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaka M. et al. Hepatitis C virus non-structural proteins responsible for suppression of the RIG-I/Cardif-induced interferon response. J. Gen. Virol. 88, 3323–3333 (2007). [DOI] [PubMed] [Google Scholar]
- Miorin L., Albornoz A., Baba M. M., D'Agaro P. & Marcello A. Formation of membrane-defined compartments by tick-borne encephalitis virus contributes to the early delay in interferon signaling. Virus Res. 163, 660–666 (2012). [DOI] [PubMed] [Google Scholar]
- Overby A. K., Popov V. L., Niedrig M. & Weber F. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J. Virol. 84, 8470–8483 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada-Murao L. A. & Morita K. Delayed cytosolic exposure of Japanese encephalitis virus double-stranded RNA impedes interferon activation and enhances viral dissemination in porcine cells. J. Virol. 85, 6736–6749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre S. et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 8, e1002934 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. Y. et al. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 8, e1002780 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan J. L. et al. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79, 8004–8013 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan J. L., Sanchez-Burgos G. G., Laurent-Rolle M. & Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 100, 14333–14338 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J., Laurent-Rolle M., Shi P. Y. & Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83, 5408–5418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y. M. et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82, 335–345 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006). [DOI] [PubMed] [Google Scholar]
- Plutner H., Davidson H. W., Saraste J. & Balch W. E. Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J. Cell Biol. 119, 1097–1116 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Xiao N. & DeFranco D. B. Use of digitonin-permeabilized cells in studies of steroid receptor subnuclear trafficking. Methods 19, 403–409 (1999). [DOI] [PubMed] [Google Scholar]
- Welsch S. et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5, 365–375 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junjhon J. et al. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 88, 4687–4697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. & Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6, 363–374 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary K. & Blair C. D. Sequential events in the morphogenesis of japanese Encephalitis virus. J. Ultrastruct. Res. 72, 123–129 (1980). [DOI] [PubMed] [Google Scholar]
- Westaway E. G., Mackenzie J. M., Kenney M. T., Jones M. K. & Khromykh A. A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71, 6650–6661 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati M. et al. Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Antiviral Res. 87, 125–148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overby A. K. & Weber F. Hiding from intracellular pattern recognition receptors, a passive strategy of flavivirus immune evasion. Virulence 2, 238–240 (2011). [DOI] [PubMed] [Google Scholar]
- Miorin L. et al. Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J. Virol. 87, 6469–6481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Z., Yuan Z., Rice C. M. & MacDonald M. R. Flavivirus replication complex assembly revealed by DNAJC14 functional mapping. J. Virol. 86, 11815–11832 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino B., Nougairede A., Wurtz N., Gould E. & de Lamballerie X. Role of host cell factors in flavivirus infection: Implications for pathogenesis and development of antiviral drugs. Antiviral Res. 87, 281–294 (2010). [DOI] [PubMed] [Google Scholar]
- Jones M. et al. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 79, 5414–5420 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney R. M. et al. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 230, 300–308 (1997). [DOI] [PubMed] [Google Scholar]
- Takamatsu Y. et al. NS1' protein expression facilitates production of Japanese encephalitis virus in avian cells and embryonated chicken eggs. J. Gen. Virol. 95, 373–383 (2014). [DOI] [PubMed] [Google Scholar]
- Kinoshita H. et al. Isolation and characterization of two phenotypically distinct dengue type-2 virus isolates from the same dengue hemorrhagic Fever patient. Jpn. J. Infect. Dis. 62, 343–350 (2009). [PubMed] [Google Scholar]
- Moue M. et al. Toll-like receptor 4 and cytokine expression involved in functional immune response in an originally established porcine intestinal epitheliocyte cell line. Biochim. Biophys. Acta 1780, 134–144 (2008). [DOI] [PubMed] [Google Scholar]
- Wilson G. L., Dean B. S., Wang G. & Dean D. A. Nuclear import of plasmid DNA in digitonin-permeabilized cells requires both cytoplasmic factors and specific DNA sequences. J. Biol. Chem. 274, 22025–22032 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental information