Abstract
Self-healing (SH) poly(lactic-co-glycolic acid) (PLGA) microspheres are a unique class of functional biomaterials capable of microencapsulating process-sensitive proteins by simple mixing and heating the drug-free polymer in aqueous protein solution. Drug-free SH microspheres of PLGA 50/50 with percolating pore networks of varying porosity (ε = 0.49–73) encapsulate increasing lysozyme (~1–10% w/w) with increasing ε, with typically ~20–25% pores estimated assessible to entry by the enzyme from the external solution. Release kinetics of lysozyme under physiological conditions is continuous over > 2 weeks and most strongly influenced by ε and protein loading before reaching a lag phase until 28 days at the study completion. Recovered enzyme after release is typically predominantly monomeric and active. Formulations containing acid-neutralizing MgCO3 at >4.3% exhibit >97% monomeric and active protein after the release with full mass balance recovery. Hence, control of SH polymer ε is a key parameter to development of this new class of biomaterials.

Keywords: biodegradable polymers, healing, microencapsulation, porosity, proteins
1. Introduction
Biodegradable polymers are now used in more than a dozen marketed controlled-release drug products ranging from both hydrophilic and hydrophobic small molecules to small water soluble peptides.[1,2] After the development of the commercial 1-month polymer depot for human growth hormone, the Nutropin Depot (Alkermes/Genentech),[3] and its subsequent removal from the market,[4] injectable polymer depots are currently limited to molecules without significant tertiary structure (i.e., occuring in proteins). Key challenges to expand polymer depot formulations have involved: (a) protein instability,[5–10] (b) difficulties associated with use of organic solvents,[11] (c) manufacturing costs,[4] (d) difficulties to control the release kinetics (particularly the initial burst),[12–14] and large needle sizes.[15]
To address a number of the aforementioned challenges in polymer depot development, we have recently developed an entirely new encapsulation method based on polymer healing,[16,17] which shifts the encapsulation paradigm for large molecules in biodegradable polymers (e.g., poly(lactic-co-glycolic acids (PLGAs)) from an organic solvent-based process to that occurring entirely in an aqueous solution.[18,19] The basic concept designed to protect process sensitive agents (e.g., proteins and DNA[9]) involves first creating a drug-free polymer platform (e.g., microspheres), which contains an interconnecting pore network. The porous microspheres are then submerged in an aqueous solution containing the biomacromolecule desired for encapsulation at temperatures (T) typically below the polymer glass transition temperature (Tg) of the polymer.[17,20] Under mild agitation to minimize inter-particle healing (i.e., aggregation), the T is raised > Tg to initiate intra-polymer healing of the pores. Polymer surface pores seal off from the external solution initiating self-healing encapsulation and those pores within the polymer become steadily more spherical.[17] As the technique relies on the natural tendency of polymers to self-heal defects above their Tg, we have termed this process “self-healing microencapsulation.” This process can occur whether the polymer is hydrated or dry, follows Williams-Landel-Ferry time temperature superposition behavior, and is strongly influenced by the pore size and polymer end-capping.[19]
We have devised two types of self-healing encapsulation processes to date—that is, passive loading during which no attempt is made to increase partitioning of the biomacromolecule to be loaded from the aqueous solution into the polymer, and active loading during which there is an additional driving force to cause partitioning into the polymer to encapsulate with high efficiency. The latter technique is akin to remote loading of doxorubicin in DOXIL stealth liposomes by ammonium precipitation of the drug as it diffuses into the empty liposome.[17] The feasibility of the encapsulation method was demonstrated by showing the potential for high drug loading,[18] high encapsulation efficiency,[18,21,22] low initial burst, [18] excellent protein stability,[18,21,22] the ability to terminally sterilize the pre-formed polymer before loading,[21] and controlled release of biomacromolecules in vitro and in vivo.[18,21,22] However, there are several important parameters that require careful examination in order to better understand both the potential and limitations of self-healing encapsulation.
Therefore, herein we first examined the influence of various pore-forming excipients on the loading of bovine serum albumin (BSA) and the related initial release and stability of the protein. Then, we examined the influence of a key parameter controlling the self-healing polymer performance—porosity (ε)—on the passive self-healing of a model enzyme, lysozyme. Porosity of pre-formed PLGA microspheres was manipulated by incorporating into the polymer various levels of a pore-forming agent (MgCO3),[14,23] varying the inner-water phase content during double-emulsion solvent-evaporation,[24–26] and adjusting concentration of carrier solvent to form the microparticle.[24,27,28] The resulting microspheres were throroughly evaluated for their ε, morphology, self-healing encapsulation characteristics, and enzyme stability and release behavior. This data below describes in full detail the ε effects expanding from our initial disclosure of the significant effect of ε on SH loading of proteins.[18] The pore structure of the polymer will affect both passive and active loading strategies by creating the pathway for both entry and distribution of the encapsulating agent as well as template for polymer healing. The encapsulation, morphological, stability, and release behavior below provides previously unknown and important insight as to how porosity can be manipulated by varying microsphere formulation parameters and how this affects novel microspheres formed by self-healing.
2. Experimental Section
2.1. Materials
PLGA with an inherent viscosity (i.v.) = 0.57 dL/g (50:50, PLGA DL LOW IV, Lot No. W3066-603, lauryl ester end group, 51 kD) was purchased from Lakeshore Biomaterials, (Birmingham, AL), formerly Alkermes. α, α-Trehalose dihydrate was purchased from Pfanstiehl (Waukegon, IL) and polyvinyl alcohol) (9–10 kDa, 80% mol hydrolyzed) was purchased from Sigma Aldrich (St. Louis, MO). Magnesium carbonate (MgCO3), Bovine serum albumin (BSA), fraction V, and lysozyme (from chicken egg white) were purchased from Sigma Aldrich (St. Louis, MO). Coomassie Plus Protein Reagent was purchased from Pierce (Thermo Fisher Scientific, Rockford, IL). All other common salts, reagents, and solvents unless otherwise specified were purchased from Sigma Aldrich (St. Louis, MO).
2.2. Methods
2.2.1. Conjugating BSA to a pH-Insensitive Fluorescent Coumarin
1.2 g of BSA was dissolved in 40 ml of 0.2 M sodium bicarbonate pH 4.5 and to this was added 2 ml of 10 mg/ml 7-methoxycoumarin-3-carbonyl azide in DMSO while stirring. The solution was stirring continuously at room temperature in darkness for 90 min. To quench the reaction, 4 ml of 1.5 M hydroxylamine hydrochloride was added and then the solution was extensively dialyzed using a 25,000 MWCO membrane against dd H2O at 4°C.
2.2.2. Formulation of Self-Healing PLGA Microspheres with Varying ε for BSA Loading
For blank particles to be loaded with BSA-Coumarin, an emulsion was first created by adding 150 ul of a porosigen in 1X PBS aqeuous solution (300 mg BSA, 269 mg of dextran, 55 mg of dextran 300 mg Kollidon PVP, 50 mg Kollidon PBP, 500 mg sucrose, 250 mg sucrose, 20 mg PEG MW 17,5000, 10 mg PEG MW 17,5000, 50 mg Gelatin (Type A), or 50 mg Gelatin (Type B) in 1g PBS) to 700 mg PLGA (50:50, 0.19 dL/g) that had been dissolved in 1 ml of CH2Cl2 in a small glass test tube. The aqeuous/polymer solutions were immediately homogenized in an ice water bath at 10,000 rpm for 1.0 minutes creating the first emulsion. 2 ml of 5% PVA (9–10kDa, 80% hydrolyzed) was then added to the first emulsion and the mixture vortexed at a high setting for 15 seconds, creating the second emulsion and the resulting solution was injected into 100 ml of 0.5% PVA (9–10kDa, 80% hydrolyzed) solution under continuous stirring. Microspheres were stirred 3h at room temperature, and collected using sieves (Newark Wire Cloth Company, Newark, NJ) to separate by size and washed thoroughly with dd H2O to remove residual PVA, porosigen, and solvent. The particles were sieved to 20–45 μm and 45–90 μm fractions and immediately freeze dried. The 45–90 μm size range was used for BSA-coumarin encapsulation.
2.2.3. Formulation of Self-Healing PLGA Microspheres with Varying ε for Lysozyme Loading
For each of the three sets of variating formulations (MgCO3 content, inner water phase volume, polymer concentration), an emulsion was first created using trehalose dehydrate solution (500 mg in 1g PBS, pH 7.4) that was added to PLGA (50:50, 0.57 dL/g) that had been dissolved in 1 ml of CH2Cl2 in a 5–6 ml syringe. Accordingly, for varying MgCO3 content, 200 μl trehalose dehydrate solution was added to 320 mg PLGA with 0, 4.8, 14.4, or 39.5 mg MgCO3 suspended in the polymer solution before emulsification. For varying inner water phase volume, 25, 100, 200, or 350 μl trehalose dehydrate solution was added to 320 mg PLGA, and for varying polymer concentration, 200 μl trehalose dehydrate solution was added to 200, 260, 320, or 400 mg PLGA with 4.8 mg MgCO3 suspended in the polymer solution before emulsification. The aqeuous/polymer solutions were immediately homogenized in an ice water bath at 17,000 rpm for 1.0 minutes creating the first emulsion. 2 ml of 5% PVA (9–10kDa, 80% hydrolyzed) was then added to the first emulsion and the mixture homogenized at 6,000 rpm for 25 seconds (20 seconds for varying MgCO3 content particles), creating the second emulsion and the resulting solution was injected into 100 ml of 0.5% PVA (9–10kDa, 80% hydrolyzed) solution under continuous stirring. Microspheres were stirred 3h at room temperature, and collected with sieves (Newark Wire Cloth Company, Newark, NJ) to separate by size and washed thoroughly with dd H2O to remove residual PVA, sugar, salt, and solvent. The particles were sieved to 20–63 μm and 63–90 μm fractions immediately freeze dried. The 20–63 μm size range was used for this study, as this is encompasses a common size distribution used in commercial products and is large enough to avoid phagocytosis but small enough to be injected through small size needles.
2.2.4. Microencapsulation of BSA-Coumarin in Self-Healing PLGA Microspheres
Approximately 1 ml of 205 mg/ml 4 °C BSA-coumarin in PBS was added to approximately 100 mg of blank particles and the microsphere/protein solutions were incubated at 4 °C for 44 h on a rocking platform, and then transferred to a 43 °C incubator on a slow speed rotary shaker for 22 h. Microspheres were removed and washed thoroughly with dd H2O, centrifuging at 3,200 rpm for 5 min to collect the microspheres after each of 10 washes, and then freeze dried. Healing times require typically 12–48 hours due to the slow process of polymer chain reptation into the pores during healing. The times can be reduced by increasing temperature, adding polymer plasticizers, or decreasing polymer molecular weight.[17,18]
2.2.5. Microencapsulation of Lysozyme in Self-Healing PLGA Microspheres
Approximately 1 ml of 250 mg/ml 4 °C lysozyme solution was added to approximately 80 mg of blank particles and the microsphere/protein solutions were incubated at 4 °C for 72 h on a rocking platform, and then transferred to a 43 °C incubator on a slow speed rotary shaker for 46 h. Microspheres were removed and washed thoroughly with dd H2O, centrifuging at 3,800 rpm for 5 min to collect the microspheres after each of 10 washes, and then freeze dried.
2.2.6. Determination of Microsphere Loading, Release Kinetics, and Residual Protein
For analysis of loading and residual protein after the release study, approximately 4 mg of dry microspheres were dissolved in approximately 1.5 ml of acetone, and the protein insoluble in the solvent was sedimented by centrifugation at 13,200 rpm for 10–15 min, and the supernatant removed. This extraction was repeated three times, and the residual acetone was removed via evaporation. The protein sediment was dissolved in PBST for analysis.
For the release study, 1.0 ml of PBST (PBS + 0.02% Tween 80), pH 7.4 was added to approximately 4–10 mg of microspheres and incubated at 37 °C. Release medium was removed and assayed for protein content at each time point and replaced by fresh media.
For determination of soluble lysozyme content in the release media and in the initial loading assays, a Coomassie protein assay was run, using Coomassie Plus Protein Reagent (Thermo Scientific Pierce) and measuring the absorbance at 595 nm. For assay of soluble lysozyme monomer remaining in the particle, soluble BSA-Coumarin loading, soluble BSA-coumarin in the release media, and soluble BSA-coumarin remaining in the particle, the reconstituted protein after polymer removal above or protein in the release media was analyzed by using SE-HPLC (Tosoh Biosciences TSKgel G3000SWxl) using a guard column (Shodex Protein KW-G), with a mobile phase of 0.05 M potassium phosphate, 0.2M NaCl, pH 7.0 at an isocratic flow rate of 0.9 ml/min. The absorbance at 215 and 280 nm was measured for lysozyme, and the fluorescence detection at 480 nm emission (384 nm excitation) for BSA-Coumarin.
Insoluble lysozyme was recovered after removing all soluble lysozyme by centrifugation and washing with 1X PBST. The collected water-insoluble protein was then dissolved in 6 M urea, 1 mM EDTA, 10 mM DL-dithiothreitol (Cleland’s Reagent) (DTT), and after brief vortexing and assaying the protein using Coomassie Plus Protein Reagent as above. Standards were analyzed in the same denaturing and reducing solution.
2.2.7. Determination of the ε of Self-Healing Microspheres
Porosity measurements on blank particles were made by Porous Materials, Inc. (Ithaca, NY) using an AMP-60K-A-1 mercury porosimeter, generating pore volume versus pressure data. The pore volume was reported as amount of volume per gram (cc/g). Total microparticle volume was calculated as the sum of the pore volume and the polymer volume, where the polymer density (1.25 g/cc, provided by manufacturer) and sample weight of the porosimetry sample were used to calculate the pore volume. Porosity was calculated as the quotient of pore volume to total microparticle volume. Pressure associated with microsphere packing and surface wetting, before mercury intrusion into the pores had taken place, was not calculated into the final pore volume. An example plot generated by the instrument to determine the pore intrusion volume is shown in Figure S1.
2.2.8. Activity of Lysozyme
Lysozyme was extracted from the microspheres and dissolved in PBST, pH 7.4, at approximately 8.5 (± 1) μg/ml. At the same time, standard solutions were dissolved in the same buffer at the same fixed concentration. For analysis, 0.15 ml of soluble protein solution was combined with 0.15 ml of 1.5 mg/ml Micrococcus lysodeikticus in 1X PBS, pH 7.4 and the absorbance at 450 nm was monitored every 30 s for a period of 5 min. The activity was calculated using the decrease in absorbance for the linear portion (between 0.5 and 3.0 min) assuming one unit of enzyme activity will reduce the ΔA450nm by 0.001/min. Specific activity is defined in units of activity per mg of protein and is given as % of the specific activity of the native, standard lysozyme. The actual amount of soluble monomer lysozyme in the solution was determined via SE-HPLC and was used for the specific activity calculations.
2.2.9. Scanning Electron Microscopy
Surface images of microspheres were taken after a brief gold coating (60s) using a Hitachi S3200N scanning electron microscope at voltages ranging from 5 to 10 kV.
3. Results and Discussion
3.1. Determining the effects of excipient loading on self-healing loading, and initial release and stability of BSA
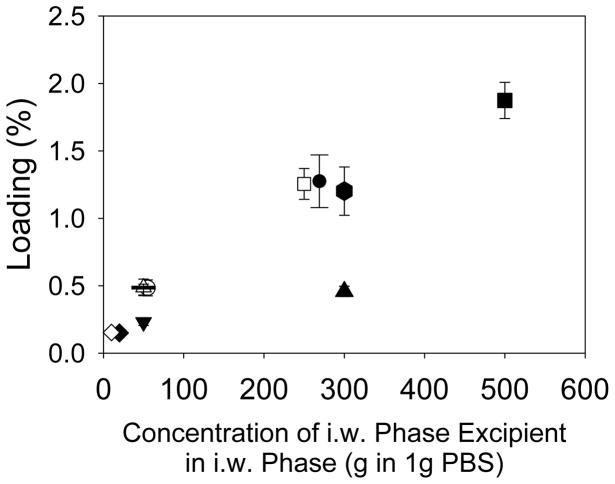
Pores form naturally in PLGA microspheres prepared by in-liquid hardening processes (e.g., water-in-oil-in-water emulsion-solvent evaporation) owing to the carrier organic solvent employed to dissolve the polymer and the aqueous inner water phase commonly used to form the primary emulsion. During drying in liquid, the organic solvent is removed with simultaneous polymer phase shrinkage and/or water exchange, and upon final drying, any water and organic solvent remaining is removed to all but residual levels creating the pores within the dry polymer. In control studies, we first used the inherent ε of the polymer to attempt to load protein by healing, but no significant protein could enter the polymer for encapsulation (data not shown). Then, we added various water-soluble excipients into the polymer microspheres (including a protein, as is normally encapsulated by the water-in-oil-in-water emulsion method) at varying levels in hopes of creating a percolating pore network, and recorded the loading of coumarin-BSA by self-healing encapsulation. As shown in Figure 1, irrespective of several excipients types, we observed the general trend that more BSA-coumarin could be loaded as the concentration of excipient in the inner water phase was increased. A possible explanation could be that increased excipient leads to increasing space (i.e., porosity) in the polymer matrix for protein entry.
Figure 1.
The effect of the concentration of the i.w. phase porosigen on % loading of coumarin-labelled BSA. All formulation parameters were identical except for the identity and concentration of the i.w. phase porosigen in 1 g PBS, pH 7.4, which was either 270 mg dextran (●), 55 mg dextran (○), 300 mg Kollidon (PVP)(▲), 50 mg Kollidon (PVP) (△), 500 mg sucrose (■), 250 mg sucrose (□), 20 mg PEG (◆), 10 mg PEG (◇), 50 mg gelatin type A (▼), 50 mg gelatin type B (—), and 300 mg BSA (⬢).
After a short 7 day release study, the remaining BSA-coumarin was recovered and analyzed (Figure 2). Of the four formulations with high loading (i.e. loading above 0.75% w/w), only that which used BSA as the porosigen had near complete recovery of the loaded BSA-coumarin. This result suggests that the BSA may have helped to stabilize the BSA-Coumarin by helping buffer the acidic solution pH or through providing additional interface-competing molecules, as has been reported previously.[23,28] For the other formulations of higher drug load (sucrose and dextrose formulations), a significant amount (up to 40%) of the protein was not recovered after 7d of release incubation, suggestive of insoluble protein aggregation.
Figure 2.
Mass balance of recovered BSA-coumarin after loading and subsequent 7d release. Dark bars represent the protein released in the first 7d of release, while the grey bars represent the protein recovered after digestion of the remaining particles. All protein released/recovered is normalized by the amount of protein loaded. Blank particles were prepared using 150 ul of i.w. phase (above) in 1 g PLGA (50:50, i.v. = 0.19 dL/g) in 1 ml CH2C12, with all particles between 20 and 90 μm. N=3.
3.2. Creating Self-Healing PLGA Microspheres with Interconnected Pore Networks of Varying ε
As we saw above with the affect of adjusting pore-forming content, which no doubt affects polymer ε, we next sought to systemically study the influence of ε on encapsulation and microsphere performance. In addition, as BSA has a significant tendency toward aggregation during release, we employed another model protein, lysozyme, to more readily evaluate the influence of this parameter. Finally, as we found earlier,[18] it is easier to obtain higher drug loading with higher molecular weight PLGAs, we employed a medium molecular weight PLGA of 51 kDa in place of the lower molecular weight PLGA.
To create porous microspheres for self-healing encapsulation, a percolating pore-network is necessary for the entering biomacromolecule to reach deep within the polymer for release[18] and pores must not become too large (< 3 μm have found to work well)[18] otherwise full healing time becomes prohibitively long.[17] Percolation during microparticle formation is achieved with incorporation of a simple osmotic agent (e.g., a sugar such as trehalose) or a pore-former such as MgCO3, the latter of which reacts with acid in the polymer (e.g., residual acids) to create osmotic salts and/or gaseous CO2.
3.2.1. Effect of Formulation Variables on Self-Healing Microsphere Morphology
Self-healing PLGA microspheres were prepared by varying base content, aqueous inner water phase volume, and polymer solution concentration, as shown in Figure 3, and Figure S2 and Figure S3. All four self-healing formulations that incorporated differing amounts of pore-forming MgCO3 (0, 1.5, 4.3, 11.0% w/w) had a surprisingly similar surface morphology (Figure 3A–D) (enlarged version available as Figures S4A–D), although the amount of surface pores appeared to increase with the amount of base encapsulated and also those surface pores visibly decreased slightly in size with increasing base content.
Figure 3.

Effect of base content on pre-loaded and lysozyme-loaded microparticle morphology. Four PLGA formulations with differing theoretical loading of base were created and loading in lysozyme solution was conducted at 4°C for 72h before healing at 42°C for 46h. SEM images were taken before (A–D) and after closing (E–H). Formulations used trehalose in PBS (pH 7.4) in the inner water phase with varying amounts of theoretical loading of MgCO3 (w/w): A,E) 0%, B,F) 1.5%, C,G) 4.3%, and D,H) 11.0%. White scale bars = 10 μm. See Fig. S4 for enlarged version.
Similarly, the surface morphology of four blank formulations prepared with differing amounts of inner water phase (25, 100, 200, and 350 μl of 500 mg trehalose dehydrate in 1 g PBS, pH 7.4) all had a similar appearance to one another (Figure S2A–D) except for the formulation prepared with the lowest i.w. phase (25 μl), which appeared less porous (Figure S2A). The four blank formulations that were created using differing concentrations of PLGA in the organic phase (200, 260, 320, and 400 mg PLGA in 1 ml CH2Cl2) had similar surface morphologies, but the pore size slightly decreased with increasing polymer concentration (Figure S3A–D).
3.2.2. Effect of Formulation Variables on Mercury Intrusion Volume and ε of Self-Healing Microspheres
The ε of the dry self-healing microspheres was calculated from the intrusion volume, which was determined by mercury porosimetry (Figure S1). A very large quantity of polymer sample was consumed with the testing by the vendor at significant cost, and thus only one test was run and there is no statistical variance. Porosity was calculated as the percentage of pore volume to the entire volume in the microspheres.
The intrusion volume per gram showed a similar trend as the MgCO3 content: generally increasing with increasing base content, although a slight decrease was seen with the highest base content. This intrusion volume per gram was 1.20 (0.0% MgCO3), 1.44 (1.5% MgCO3), 1.76 (4.3% MgCO3), and 1.57 cc/g (11% MgCO3). As seen in Table 1, these measurements were 60.0% (0.0% MgCO3), 64.3% (1.5% MgCO3), 68.8% (4.3% MgCO3), and 66.2% (11% MgCO3).
Table 1.
Effect of formation variables on self-healing microsphere propertiesa
| Method of adjusting porosity in Formulation | Variable in formulation | Porosity of self-healing microspheres (%) | 28-day lysozyme release (%) | Monomeric enzyme remaining (%) | Insoluble enzyme remaining (%) | Mass balance (%)b |
|---|---|---|---|---|---|---|
| Varying Base Content | 0% MgCO3 | 60.0 | 30.4 ± 0.7 | 57 ± 2 | 4 ± 1 | 92 ± 2 |
| 1.5% MgCO3 | 64.3 | 41.3 ± 0.5 | 47 ± 1 | 3 ± 1 | 91 ± 1 | |
| 4.3% MgCO3 | 68.8 | 51 ± 4 | 46 ± 1 | 3 ± 1 | 99 ± 4 | |
| 11% MgCO3 | 66.2 | 57 ± 4 | 51 ± 1 | 1.4 ± 0.1 | 110 ± 4 | |
| Varying Inner Water Phase Volume | 25 μL | 49.5 | 30 ± 3 | 76 ± 4 | 11 ± 2 | 117 ± 5 |
| 100 μL | 60.6 | 25.9 ± 0.3 | 64 ± 3 | 6 ± 1 | 96 ± 3 | |
| 200 μL | 64.0 | 33 ± 2 | 62 ± 2 | 5 ± 1 | 100 ± 3 | |
| 350 μL | 59.7 | 24.5 ± 0.6 | 55 ± 1 | 5 ± 1 | 85 ± 1 | |
| Varying Polymer Concentration | 200 mg/mL | 65.2 | 34.4 ± 0.3 | 55 ± 2 | 4 ± 1 | 93 ± 2 |
| 260 mg/mL | 61.0 | 20.6 ± 0.8 | 74 ± 2 | 6 ± 1 | 100 ± 2 | |
| 320 mg/mL | 72.7 | 46.6 ± 0.5 | 42 ± 2 | 6 ± 3 | 95 ± 3 | |
| 400 mg/mL | 64.6 | 57 ± 2 | 30 ± 2 | 3 ± 1 | 89 ± 2 |
All data reported as mean ± SEM (n=3); formulation conditions listed in Experimental Methods;
Mass balance = 28-day release + monomeric remaining + insoluble remaining; no soluble aggregate in residual samples.
Similarly, the intrusion volume per gram generally increased with increasing inner water phase volume, though a slight decrease was seen in the formulation with the highest volume level. The intrusion volume per gram was 0.78 (25 μl), 1.23 (100 μl), 1.43 (200 μl), and 1.18 cc/g (350 μl), corresponding to the following ε values: 49.5% (25 μl), 60.6% (100 μl), 64.0% (200 μl), and 59.7% (350 μl).
Thus, as the both excipient level increased and the volume of inner water phase volume increased, the porosity seemed to increase until some critical value was reached. At that point, the porosity decreased. One possible explanation for this was that too much aqueous phase was present at some point prior to the microparticle hardening, allowing the porosigen to be released out of the microparticle quicker than for the other formulations. Thus, by the end of the hardening phase, such particles may actually have had less water uptake due to a lower osmotic gradient between the microparticle and the outer water phase, leading to a less porous particle. Similarly, the higher amount of inner aqeous phase may have provided a greater surface area for the organic phase pockets to migrate to, increasing the rate of phase separation and hardening, and this decreasing the overall porosity.
With changes in polymer concentration, the intrusion volume per gram showed no distinct trend. The intrusion volumes were 1.50 (200 mg), 1.25 (260 mg), 2.13 (320 mg), and 1.46 cc/g (400 mg), corresponding to porosities of 65.2% (200 mg), 61.0% (260 mg), 72.7% (320 mg), and 64.6% (400 mg). A complicating factor in the latter data is that the amount of base suspended in the polymer (instead of base loading) was fixed as polymer concentration was increased. Opposing factors (base content, initial organic solvent level, and degree of particle shrinkage) appeared to be responsible for the absence of an expected or observed trend.
3.3. Self-Healing Encapsulation of Lysozyme
The above self-healing microspheres were submerged in 250 mg/mL aqueous lysozyme at 4 °C for 72 h to allow the protein to enter the polymer pore-network before healing the pores at 43 °C for 48 h. The effects of various ε-varying self-healing formulations on microsphere morphology, loading, release, and stability of lysozyme are described below.
3.3.1. Morphology of Self-Healed Microspheres
After the pore closing/encapsulation step, all microspheres with varying levels of MgCO3 had very similar fully healed morphologies, with the highest base content (11% w/w) having a slightly rougher surface (Figure 3E–H). Similarly, healed microspheres prepared from varying inner water phase volume also healed well with indistinguishable surfaces (Figure S2E–H). For the microspheres prepared with varying polymer concentration (Figure S3E–H), the number of visible pores on the surface after healing appeared to increase with polymer concentration as well, suggesting that there was incomplete pore closing with high polymer concentration (400 mg in 1 ml CH2Cl2) formulation (Figure S3H).
3.3.2. Effect of Formulation Parameters and Polymer ε on Loading of Lysozyme
As reported before,[18] increasing ε at constant polymer concentration from ~50–70% shows a steady, apparently somewhat linear, increase in protein loading. Addition of values from different polymer concentrations to the loading vs. ε plot (Figure 4) also supports the positive correlation between the two variables, although linearity may occur only over a limited range. The highest ε value (> 70%) showed a decrease in expected loading. Additional data would be necessary to confirm this observation, although certainly at some level of ε the loading is logically expected to decrease (i.e., no loading is possible as ε approaches unity). The increase in loading with ε up to a point is easily rationalized based on the expectation that more available pores in the polymer matrix available for loading will allow more external protein solution to enter the polymer before healing. Interesting was the x-intercept of ~47% in Figure 4, which is expected to have some relationship with the lower percolation threshold of the microspheres,[28] was significantly higher than the expected percolation threshold values (e.g., < 35%). This result also suggests that if formulations were prepared with even lower ε that these data would not be expected to fall on the best fit line.
Figure 4.
Effects of pre-loaded PLGA microparticle formulation parameters on subsequent lysozyme loading (% w/w) via self-healing encapsulation. Twelve total formulations were prepared, investigating the effect on loading from varying amounts of theoretical MgCO3 content, volumes of inner water phase solution, or polymer concentration: Self-healing formulations were created with A) differing levels of MgCO3 suspended in the organic phase with 200 μL inner water phase volume and 320 mg/mL polymer concentration, B) differing volumes of inner water phase volume with no MgCO3 and 320 mg/mL polymer concentration and C) differing amounts of PLGA (50:50, i.v. = 0.57 dL/g) in 1 ml CH2Cl2 with suspended 4.8 mg MgCO3 and 200 μL inner water phase volume. Open symbols are replotted from Reinhold et al.[1] for completeness.
The loading as a function of various pore-determining variables is also displayed in Figure 5. The most significant changes were recorded for varying MgCO3 and inner water phase levels (Figure 5A–B) affecting the loading by as much as ~8-fold changes in protein loading. It is important to point out that the drops in loading for formulations prepared with the highest level of base and inner water phase content, 11% and 350 μL, respectively, were also associated with similar drops in polymer ε. This resulted in maximal loading and ε for the formulations prepared with the 4.3% MgCO3 and 200 μL levels. It should be noted that 200 μL/mL has long-been known as the rough upper limit on phase volume ratio before significant loss in encapsulation efficiency by the conventional W/O/W method.[29]
Figure 5.

Enzyme loading as a function of porosity for SH-microspheres prepared with varying formulation parameters. Triangles represent varying theoretical base content microspheres (differing amounts of MgCO3 in the organic phase: 0%, 1.5%, 4.3%, and 11.0%,), circles represent varying inner water phase volume microspheres (differing volumes of inner water phase solution (trehalose in PBS, pH 7.4): 25 μl, 100 μl, 200 μl, and 350 μl), and squares represent varying polymer concentration microspheres (differening amounts of PLGA (50:50, i.v. = 0.57 dL/g) in 1 ml CH2Cl2: 200 mg, 260 mg, 320 mg, and 400 mg).
3.3.3. Effect of Formulation Parameters and Polymer ε Estimated Fraction of Pores Utilized for Self-Healing Loading
By assuming equivalent partitioning of the protein between external aqueous loading solution and the internal polymer pores, a fraction of pores utilized for self-healing loading was estimated, as shown in Figure 6. Very interesting was the insensitivity of formulation variables on the fraction of pores utilized (mostly 20–25%) and most of the pores (> 65%) apparently were still available for loading after healing. Thus, in most of the formulations, up to 75% of the available pore volume appears to go unutilized. As the same loading for self-healing particles has been observed for vastly different size loading molecules (i.e. 2 million Da dextran versus 10 kDA dextran),[18] it is believed that this unused porosity is not due to size limitations of the pore network, but because of some other phenomenon. An alternative explanation is that most of the pores were reached by the protein and, because of large internal polymer surface and increase in entropy of pore water, a decrease in protein partitioning into the pore volume (i.e., partition coefficient < 1) occurred. Further experimental and theoretical analysis will be necessary to resolve whether these two possibilities or some combination of the two are dominant. Two formulations clearly different from the rest were microspheres prepared from very low inner water phase (~6% pores utilized) and high polymer concentration (30%). The low fraction of pores for the low inner water phase volume microspheres could have resulted from the proximity to the low percolation threshold and/or the reduced level of trehalose used to create the percolating pore network. The high polymer concentration used in the formulation was expected to cause more rapid hardening of the polymer, causing an artificially high ε and entrapping a larger fraction of the osmotically active trehalose.
Figure 6.

Estimates of pore volume utilized for microspheres prepared with varying formulation parameters. The fraction of pore volume utilized for self-healing encapsulation was estimated by self-healing microsphere porosity (measured by mercury porosimetry) and assuming concentration of lysozyme in pores for encapsulation equaled external solution concentration (i.e., unity partitioning). Four SM-formulations were created with A) differing levels of MgCO3 suspended in the organic phase with 200 μL inner water phase volume and 320 mg/mL polymer concentration, B) differing volumes of inner water phase volume with no MgCO3 and 320 mg/mL polymer concentration and C) differing amounts of PLGA (50:50, i.v. = 0.57 dL/g) in 1 ml CH2Cl2 with suspended 4.8 mg MgCO3 and 200 μL inner water phase volume.
3.4. Release Kinetics of PLGA-Encapsulated Lysozyme
The release rate of the particles (Figure 7) directly correlated with the amount of base used to create the blank particles (Figure 7A). As seen in Table 1, the amount released from the microspheres after 28 days was (% ± SEM) 30 ± 1% (0% MgCO3), 41 ± 1% (1.5% MgCO3), 51 ± 4% (4.3% MgCO3), and 57 ± 4% (11.0% MgCO3). Increases in base content resulted in increased burst release over the first couple of days. All formulations released continuously over about 14 days and then reached a lag period of very low or no release. Mostly soluble protein remained in the polymer at 28 days (see below). For changes in inner water phase volume, the 28-d release of these formulations all released similarly, between 24 and 34% of lysozyme by Day 28 (Figure 7B). The amount released from the microspheres after 28 days was (% ± SEM) 29.7 ± 0.3% (25 μl), 25.9 ± 0.3% (100 μl), 33 ± 2% (200 μl), and 24.5 ± 0.6% (350 μl). For formulations with varying polymer concentration, like the resulting polymer ε of the self-healing microspheres (see above), the 28-d release showed no immediately obvious trend (Figure 7C). The highest release rate was from the 400 mg-PLGA formulation, while the slowest and least released over 28d was the 260 mg-PLGA formulation. The amount released from the microspheres after 28 days was (% ± SEM) 34.4 ± 0.3% (200 mg), 20.6 ± 0.9% (260 mg), 46.6 ± 0.5% (320 mg), and 57 ± 2% (400 mg). As increased ε and loading is expected to increase release rate, the dip in the rate of the 260 mg-polymer formulation could have resulted from its low ε (61%) with similar loading to the 200 mg and 320 mg formulations. By contrast, the 400 mg-polymer formulation, despite having lower ε (65%) than the 320 mg formulation (73%), had a significantly higher lysozyme loading (~10% vs. ~8%). It should be noted that the microparticles displayed a lag phase, as has been reported previously,[23] which for these formulations went past the final release point tested. Complete release of the encapsulated protein from the biodegradable microparticle is expected to take place after the 28d test point.
Figure 7.

Controlled release of lysozyme from self-healing microspheres from microspheres created using differing formulation parameters during blank PLGA microspheres preparation. Formulations were created with A) differing levels of MgCO3 in the organic phase: 0%(△), 1.5% (●), 4.3% (◆), and 11.0% (■), B) differing volumes of inner water phase solution (trehalose in PBS, pH 7.4): 25 μl (●), 100 μl (△), 200 μl (◆), and 350 μl (□), or C) using a constant base amount (4.8 mg) but differing amounts of PLGA (50:50, i.v. = 0.57 dL/g) in 1 ml CH2Cl2: 200 mg (●), 260 mg (△), 320 mg (◆) and 400 mg □). Data in (A) with dashed lines are replotted from Reinhold et al. [1] for completeness.
3.5. Recovery and Stability of Lysozyme After Release
In addition to the evaluation of release kinetics, the encapsulated lysozyme was recovered by extraction at the end of the evaluated 28-day release period to determine the physical stability and activity of the lysozyme remaining in the polymer. The amount of protein remaining, including soluble monomer and insoluble aggregates, was quantified, as displayed in Table 1. No soluble aggregate was observed in the samples by SEC-HPLC. Thus, monomeric content listed in the table also refers to soluble protein content. As seen in the table, generally essentially all the protein was recovered between released, remaining soluble and remaining insoluble enzyme as the mass balance was close to 100% in most cases. In addition, most of the protein was still soluble monomeric protein at the end of the 28 day incubation. As expected, the least insoluble protein and most complete recovery was observed in the samples with the highest MgCO3 content (4.3 and 11%), which is consistent with our past experience with this enzyme in this type of PLGA with this base in cylindrical implants.[30] Moreover, lysozyme is a relatively stable protein in PLGA, with very little aggregation tendency, compared with albumin.[30] The one exception was the formulation with the very phase volume ratio (25 μL/mL), which exhibited 11% insoluble aggregate remaining in the polymer at the end of the release period. This is likely because of the much lower loading of this specimen, which results in lower buffering by the protein and should a constant level of protein be damaged at an interface, then a correspondingly higher fraction of protein would be damaged in this instance.
Similarly, in most cases >90% of the remaining soluble protein after 28 days of release was enzymatically active, as shown in Figure 8. The important exception was the formulation with the very phase volume ratio (25 μL/mL). Similar to the elevated aggregation in this sample (Table 1), a much lower activity in the remaining protein was observed. It is noted that in the samples with the higher level of base loaded ((4.3 and 11%), >97% of monomeric protein and full enzymatic activity was recovered after 28 days release. This data strongly supports the conclusion that the self-healing method is very mild in terms of protecting proteins during encapsulation, as has been reported.[18,21,22] We note that MgCO3 and related basic excipients (Mg(OH)2 and ZnCO3) have been studied extensively for development of biodegradable polymer delivery systems. Heller et al. pioneered the use of Mg(OH)2 in poly(ortho esters) for stabilizing the ortho ester bond for long-term controlled release,[31] many later formulations of which were evaluated in the clinic. Similarly ZnCO3 was incorporated in the commercial Nutropin Depot.[3] We have also monitored histology at the injection site of PLGA containing MgCO3 and found negligible increases in inflammation.[32] Therefore, we continue to incorporate this class of excipients in PLGAs to control both microclimate pH[23] and increase release of protein.[14]
Figure 8.

Specific activity of lysozyme remaining in various microparticle formulations after 28 d release. Formulations of blank particles were created with A) differing levels of MgCO3 in the organic phase: 0%, 1.5%, 4.3%, and 11.0%, B) differing volumes of inner water phase solution (trehalose in PBS, pH 7.4): 25 μl, 100 μl, 200 μl, and 350 μl, or C) using differing amounts of PLGA (50:50, i.v. = 0.57 dL/g) in 1 ml CH2Cl2: 200 mg, 260 mg, 320 mg, and 400 mg. [% Specific activity was calculated as a percentage of the specific activity of fresh, unencapsulated lysozyme.]
4. Conclusions
In closing, pore-forming excipient loading and ε are clearly key parameters that requires monitoring during encapsulation by the novel self-healing method. Porosity values in the neighborhood of ~60–70% appeared to work well for loading high levels of protein by the passive encapsulation method and a variety of pore forming substances could be used to successfully prepared SH-microspheres for elevated loading, as demonstrated with BSA studies. Release and stability of a model protein, lysozyme, was excellent in certain formulations (e.g., with 4.3% MgCO3) for over 2 weeks, although improvements will be needed to accomplish continuous release beyond this time point and release the entire protein. Future studies will also need to be focused on mapping the pore network of protein as it enters the SH microspheres, to reach a higher fraction of pores in the polymer, and to determine methods for active loading of pharmaceutical proteins to reduce losses of high-cost human recombinant proteins, commonly found in pharmaceutical products.
Supplementary Material
Figure S1. A representative pore volume result obtained using mercury porosimetry. Pressure was required to spread and wet the microspheres before penetration into pore network. Here, a pressure of 10.4 bars (1.4 μm) was required for wetting and spreading. Therefore the size of pores measured was 1.4 μm or smaller.
Figure S2. Effect of inner water phase volume on blank SM-microsphere particle porosity and loaded particle morphology. Four PLGA formulations were created and loading/pore closing in lysozyme solution was conducted at 4°C for 72h and 42°C for 46h. SEM images were taken before (AD) and after closing (E–H). Formulations used trehalose in PBS (pH 7.4) in the inner water phase with differing volumes: A,E) 25 μl, B,F) 100 μl, C,G) 200 μl, and D,H) 350 μl.
Figure S3. Effect of polymer concentration on blank SM-microsphere particle porosity and loaded particle morphology. Four PLGA formulations were created and loading/pore closing in lysozyme solution was conducted at 4°C for 72h and 42°C for 46h. SEM images were taken before (AD) and after closing (E–H). Formulations used a constant base amount (4.8 mg) and differing amounts of PLGA (50:50, i.v. = 0.57 dL/g) in 1 ml CH2Cl2: A,E) 200 mg, B,F) 260 mg, C,G) 320 mg, and D,H) 400 mg.
Figure S4 (Enlarged version of Fig. 3). Effect of base content on preloaded and lysozyme-loaded microparticle morphology. Four PLGA formulations with differing theoretical loading of base were created and loading in lysozyme solution was conducted at 4°C for 72h before healing at 42°C for 46h. SEM images were taken before (AD) and after closing (E–H). Formulations used trehalose in PBS (pH 7.4) in the inner water phase with varying amounts of theoretical loading of MgCO3 (w/w): A,E) 0%, B,F) 1.5%, C,G) 4.3%, and D,H) 11.0%.
Acknowledgments
This research was supported in part by NIH HL 68345 and EB 08873. Special thanks to Morgan Giles for aid with manuscript formatting.
Footnotes
Supporting Information is available from the Wiley Online Library
Contributor Information
Dr. Samuel E. Reinhold, Email: reinhold@umich.edu, Upsher-Smith Laboratories, Inc., 6701 Evenstad Drive, Maple Grove, MN 55369 USA
Dr. Steven P. Schwendeman, Email: schwende@umich.edu, Ara G. Paul Professor and Chair, Department of Pharmaceutical Sciences and the Biointerfaces Institute, University of Michigan, North Campus Research Complex, 2800 Plymouth Rd, Ann Arbor, MI 48109 USA
References
- 1.Wischke C, Schwendeman SP. Internation Journal of Pharmaceutics. 2008;364:298. doi: 10.1016/j.ijpharm.2008.04.042. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Chan HF, Leong KW. Advacned Drug Delivery Reviews. 2012;65:104. doi: 10.1016/j.addr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson OL, Cleland JL, Lee HJ, Charnis M, Duenas E, Jaworowicz W, Shepard D, Shahzamani A, Jones AJ, Putney SD. Nature Medicine. 2012;2:795. doi: 10.1038/nm0796-795. [DOI] [PubMed] [Google Scholar]
- 4. [accessed: September 2013];Genentech Press Releases. www.gene.com/media/press-releases/7447/2004-06-01/genentech-and-alkermes-announce-decision.
- 5.Schwendeman SP, Cardamone M, Brandon MR, Klibanov A, Langer R. In: Microparticulate Systems for the Delivery of Proteins and Vaccines. Cohen S, Bernstein H, editors. Ch 1 Marcel Dekker; New York: 1996. [Google Scholar]
- 6.Kissel T, Koneberg R. In: Microparticulate Systems for the Delivery of Proteins and Vaccines. Cohen S, Bernstein H, editors. Ch 2 Marcel Dekker; New York: 1996. [Google Scholar]
- 7.Putney SD, Burke PA. Nature Biotechnology. 1998;16:153. doi: 10.1038/nbt0298-153. [DOI] [PubMed] [Google Scholar]
- 8.van de Weert M, Hennink WE, Jiskoot W. Pharmaceutical Research. 2000;10:1159. doi: 10.1023/a:1026498209874. [DOI] [PubMed] [Google Scholar]
- 9.Schwendeman SP. Critical Reviews in Therapeutic Drug Carrier Systems. 2002;19:73. doi: 10.1615/critrevtherdrugcarriersyst.v19.i1.20. [DOI] [PubMed] [Google Scholar]
- 10.van der Walle CF, Sharma G, Ravi Kumar M. Expert Opinions on Drug Delivery. 2009;2:177. doi: 10.1517/17425240802680169. [DOI] [PubMed] [Google Scholar]
- 11.Whitaker MJ, Davies OR, Serhatkulu G, Stolnik-Trenkic S, Howdle SM, Shakesheff KM. Journal of Controlled Release. 2005;101:85. doi: 10.1016/j.jconrel.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Allison SD. Expert Opinions on Drug Delivery. 2008;5:615. doi: 10.1517/17425247.5.6.615. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Wang BM, Schwendeman SP. Journal of Controlled Release. 2002;82:289. doi: 10.1016/s0168-3659(02)00137-2. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein H, Zhang Y, Khan AM, Tracy MA Alkermes. 5 656 297. US Patent. 1997
- 15.Ravivarapu HB, Moyer KL, Dunn RL. Journal of Pharmaceutical Sciences. 2000;89:732. doi: 10.1002/(SICI)1520-6017(200006)89:6<732::AID-JPS4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 16.Wool RP. Soft Matter. 2008;4:400. doi: 10.1039/b711716g. [DOI] [PubMed] [Google Scholar]
- 17.Mazzara JM, Balagna MA, Thouless MD, Schwendeman SP. Journal of Controlled Release. 2013;171:172. doi: 10.1016/j.jconrel.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 18.Reinhold SE, Desai KGH, Zhang L, Olsen KF, Schwendeman SP. Angewandte Chemie International Edition. 2012;51:10803. doi: 10.1002/anie.201206387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwendeman SP, Kang J, Reinhold S University of Michigan. 8 017 155. US Patent. 2011
- 20.Blasi P, D’Souza SS, Selmin F, DeLuca PP. Journal of Controlled Release. 2005;108:1. doi: 10.1016/j.jconrel.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Desai KGH, Kadous S, Gupta RK, Schwendeman SP. Pharmaceutical Research. 2013;30:1768. doi: 10.1007/s11095-013-1019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desai KGH, Schwendeman SP. Journal of Controlled Release. 2013;165:62. doi: 10.1016/j.jconrel.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu G, Mallery SR, Schwendeman SP. Nature Biotechnology. 2000;18:52. doi: 10.1038/71916. [DOI] [PubMed] [Google Scholar]
- 24.Schlicher EJAM, Postma NS, Zuidema J, Talsma H, Hennink WE. International Journal of Pharmaceutics. 1997;153:235. [Google Scholar]
- 25.Ghaderi R, Sturesson C, Carlfors J. International Journal of Pharmaceutics. 1996;141:205. [Google Scholar]
- 26.Crotts G, Park TG. Journal of Controlled Release. 1995;35:91. [Google Scholar]
- 27.Li WI, Anderson KW, Mehta RC, Deluca PP. Journal of Controlled Release. 1995;37:199. [Google Scholar]
- 28.Ehtezazi T, Washington C. Journal of Controlled Release. 2000;68:361. doi: 10.1016/s0168-3659(00)00270-4. [DOI] [PubMed] [Google Scholar]
- 29.Cohen S, Yoshioka T, Lucarelli M, Hwang LH, Langer R. Pharmaceutical Research. 1991;8:713. doi: 10.1023/a:1015841715384. [DOI] [PubMed] [Google Scholar]
- 30.Gaozhong Zhu. PhD thesis. Ohio State University; 1999. Stabilization and Controlled Release of Proteins Encapsulated in Poly(lactide-co-glycolide) Delivery Systems. [Google Scholar]
- 31.Heller J. Biomaterials. 1990;9:659. doi: 10.1016/0142-9612(90)90024-k. [DOI] [PubMed] [Google Scholar]
- 32.Cui C, Stevens VC, Schwendeman SP. Vaccine. 2007;25:500. doi: 10.1016/j.vaccine.2006.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A representative pore volume result obtained using mercury porosimetry. Pressure was required to spread and wet the microspheres before penetration into pore network. Here, a pressure of 10.4 bars (1.4 μm) was required for wetting and spreading. Therefore the size of pores measured was 1.4 μm or smaller.
Figure S2. Effect of inner water phase volume on blank SM-microsphere particle porosity and loaded particle morphology. Four PLGA formulations were created and loading/pore closing in lysozyme solution was conducted at 4°C for 72h and 42°C for 46h. SEM images were taken before (AD) and after closing (E–H). Formulations used trehalose in PBS (pH 7.4) in the inner water phase with differing volumes: A,E) 25 μl, B,F) 100 μl, C,G) 200 μl, and D,H) 350 μl.
Figure S3. Effect of polymer concentration on blank SM-microsphere particle porosity and loaded particle morphology. Four PLGA formulations were created and loading/pore closing in lysozyme solution was conducted at 4°C for 72h and 42°C for 46h. SEM images were taken before (AD) and after closing (E–H). Formulations used a constant base amount (4.8 mg) and differing amounts of PLGA (50:50, i.v. = 0.57 dL/g) in 1 ml CH2Cl2: A,E) 200 mg, B,F) 260 mg, C,G) 320 mg, and D,H) 400 mg.
Figure S4 (Enlarged version of Fig. 3). Effect of base content on preloaded and lysozyme-loaded microparticle morphology. Four PLGA formulations with differing theoretical loading of base were created and loading in lysozyme solution was conducted at 4°C for 72h before healing at 42°C for 46h. SEM images were taken before (AD) and after closing (E–H). Formulations used trehalose in PBS (pH 7.4) in the inner water phase with varying amounts of theoretical loading of MgCO3 (w/w): A,E) 0%, B,F) 1.5%, C,G) 4.3%, and D,H) 11.0%.