Abstract
The smallest gene HBx of Hepatitis B virus (HBV) is recognized as an important viral oncogene (V-oncogene) in the hepatocarcinogenesis. Our previous work demonstrated that RMP is a cellular oncogene (C-oncogene) required for the proliferation of hepatocellular carcinoma (HCC) cells. Here we presented the collaboration between V-oncogene HBx and C-oncogene RMP in the development of HCC. The coexpression of HBx and RMP resulted in the cooperative effect of antiapoptosis and proliferation of HCC cells. In vivo, overexpression of RMP accelerated the growth of HBx-induced xenograft tumors in nude mice and vice versa HBx promoted the growth of RMP-driven xenograft tumors. Although HBx didn't regulate the expression of RMP, HBx and RMP interact with each other and collocalized in the cytoplasm of HCC cells. HBx and RMP collaboratively inhibited the expression of apoptotic factors and promoted the expression of antiapoptotic factors. This finding suggests that HBV may induce, or at least partially contributes to the carcinogenesis of HCC, through its V-oncoprotein HBx interacting with the C-oncoprotein RMP.
Keywords: HBx, viral oncogene
Introduction
A leading cause for the development of hepatocellular carcinoma (HCC) is chronic infection with Hepatitis B Virus (HBV). More than half of HCC cases worldwide are due to chronic HBV infection. HBV infection-related liver diseases remain a major global health problem 1.
Within the 3.2 kilobases (Kb) HBV genome, there are four overlapping genes encoding the viral core protein (capsid), surface proteins (envelope), reverse transcriptase, and X protein (HBx).
HBx is a major regulator encoded by HBV genome, which plays an important role in the development of HBV-associated HCC. Chronic HBV infection develops into HCC through several factors, including the integration of viral DNA into the genome of the host cell, the chronic inflammation due to the immune response of the host to the HBV infection. As a typical viral oncogene (V-oncogene) of HBV, HBx is predominantly nuclear at low expression levels and mostly cytoplasmic at high expression levels. Some of cytosolic HBx are localized in the outer mitochondrial membrane 2-4.
RNAs are synthesized by RNA polymerases (RNAPs). In eukaryotic cells, different kinds of RNAs are transcribed by RNAP I, II and III in the nuclear genome 5. RNAP I, II and III are composed of 14, 12 and 17 subunits respectively, among which RPB5, RPB6 and RPB8 are commonly shared by all three RNAPs 6. HBx has been identified to associate with RPB5 subunit of RNAP II and regulate transcription activity 7. Further, from HepG2 cell line, we identified RPB5-mediating protein (RMP) with RPB5 as a probe 8. We found that the general transcriptional factor TFIIF was associated with RNAP II through the interaction between RAP30 and RPB5 subunits. In yeast, the biosynthesis and assembly of eukaryotic RNA polymerase required both RMP and RPB5. Deletion of RMP led to abnormally substantial accumulation of all three RNAPs of I, II and III in the cytoplasm 5.
There are multiple variants of RMP, including the unconventional prefolding RPB5 interactor (URI). Recently, both Dr. Krek group and ours showed that RMP exhibited the typical activity of a cellular oncogene (C-oncogene) during the carcinogenesis 9,10. URI, which was overexpressed in tissues and cell lines of human ovarian carcinomas, was required for the survival of ovarian cancer cells 9. The molecular mechanism analysis revealed that URI associated and inactivated PP1γ (phosphatase 1 gamma), which enhanced the S6K1 survival signaling 11. The release of PP1γ by the phosphorylation of URI results in apoptosis and cell death. The research of our group found that RMP is actively involved in the proliferation of HCC cells 10. RMP presents an antiapoptotic role, which in turn supports the proliferation and growth of HCC cells. Depletion of RMP induced apoptosis of HCC cells. RMP is also required for development and growth of HCC xenograft tumors in vivo. Interestingly, RMP has been shown to play distinct roles in normal hepatic cells and HCC cells 12.
Despite of many studies conducted toward HBV and HBx, the mechanism of how HBx induces the carcinogenesis of HCC remains largely obscure. As both HBx and RMP associate with RNAP by interacting with RPB5 and demonstrate the oncogenic property, we investigated the relationship between HBx and RMP, the two oncogenes in the carcinogenesis of HCC.
In this article, we identified the collaboration between HBx and RMP. The two oncogenes cooperatively stimulated the proliferation of HCC cells, demonstrating an antiapoptotic effect on the HCC cells, which contributed to hepaticcarcinogenesis.
Materials and Methods
Plasmids, Cell Lines and Reagents
The plasmid pFlagCMV4 was from Sigma. pGPU6-Neo was from GenePharma Co. (Shanghai, China). pCDNA3.1/Neo/RMP and pEx-5/GFP/Neo/HBx were constructed by and purchased from GenePharma Co. (Shanghai, China). The plasmids pGPU6/Neo-SCR and pGPU6/Neo-RMPi for RMP depletion were constructed as described previously 10. HepG2 cell line stably expressing HBx 13,14 was a gift from Dr. Xiaodong Zhang in Nankai University. All other cell lines used in this article were obtained from our own laboratory of the Department of Cell Biology at Soochow University. Cells were cultured in DMEM medium (Gibco-BRL, Shanghai, China) supplemented with 10% Fetal Bovine Serum (FBS) (Sino-American Biotechnology Co, Shanghai, China). The Annexin-V-FITC/PI apoptosis detection kit was from BD Biosciences (Shanghai, China). Methyl-thiazolyl-tetrazolium (MTT) was from Sigma (Shanghai, China). Cell Counting Kit-8 (CCK-8) was purchased from Dojindo Laboratories (Shanghai, China).
Antibodies against Bax, Bad, Bcl-2, p53, AFP and β-actin were purchased from Boster Company (Wuhan, China). The RMP antibody was a product from AppTec Pharmaceutical (Suzhou, China). Antibody against HBx was purchased from Santa Cruz Biotechnology, Inc (Shanghai, China).
Cell Culture, Transfection, Western Blot Analysis and Immunoprecipitation
A total of 2×105cells were seeded into each well of a 24-well plate, 24 hrs prior to transfection. Cell numbers were chosen to finally reach 90% confluency at the time of transfection. These cells were transfected with lipofectamine 2000 (Invitrogen, Shanghai, China) according to the manufacturer's protocol. LF2000/DNA complexes were allowed to form in OptiMEM with final concentration of 10 μg/ml LF2000. Selection of stably transfected cells were conducted as previously described 10,15.
For immunoprecipitation, the supernatants of cell lysates were collected after centrifugation and immunoprecipitated with 20 μl of 50% anti-FLAG M2 or HBx resin, followed by rotation for 2 hrs at 4°C. Then the resin was washed 4 times with buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl) (TBS). The bound proteins were eluted, fractionated by 12.5% SDS-PAGE, transferred onto PVDF membranes, and subjected to Western blot analysis with the antibody. The proteins were visualized by enhanced chemiluminescence (ECL), according to the manufacturer's instructions (Amersham Pharmacia Biotech) 10,15.
Cell Viability Assay
Cell proliferation assay by Cell Counting Kit-8 (CCK-8) was conducted as following. Cells were seeded into a 96-well plate (5×104 cells/cell) and cultured in 100 μl of DMEM supplemented with 10% FBS. At the indicated time points, medium was exchanged for 110 μl of DMEM with CCK-8 reagent (10 μl CCK-8 and 100 μl DMEM), and the cells were incubated for 2 hrs. Absorbance was measured at a wavelength of 450 nm for each well, with the reference wavelength set at 600 nm. An increase or decrease in absorbance values at 450 nm in the experimental wells relative to the initial value indicated cell growth or death, respectively. Cell growth was monitored every 24 hrs over 5 days. All experiments were independently repeated at least three times.
MTT assays were carried out as described previously 10,15. Briefly, cells were cultured 96-well plate for 24 hrs, and 30 μl of MTT (5 mg/ml) was added to each well. Crystals were formed and then dissolved by adding 300 μl isopropanol acidified with HCl (0.04 N) containing 10% Triton X-100. Finally, 200 μl of the blue formazan mixtures was transferred to 96-well plates. Microplate Reader (Model 550, BIO-RAD, Shanghai, China) was used to read the plates at 570 nm.
Analysis of Apoptosis and cell cycle by Flow Cytometry
Cells were exposed to ionising radiation (IR) using a 60Co γ-irradiator at a dose of 2 or 6 Gy. Cells radiated or unradiated were then stained with Annexin V-FITC and propidium iodide (PI) and analyzed for apoptosis according to the manufacturer's instructions (BD Biosciences, Shanghai, China). Briefly, cells were washed with buffer for 2 times, and re-suspended in 400 μl of Dulbecco's phosphate-buffered saline (PBS). Then, 100 μl of cell suspension was incubated with 10 μl of PI (50 μg/ml) and 5 μl Annexin V-FITC for 15 minutes at room temperature (RT) in the dark, followed by the analysis with flow cytometry. Cells that stained positive for only Annexin V-FITC were in the early stage of apoptosis, whereas cells that stained positive for both Annexin V-FITC and PI were in the stage of late apoptosis or primary necrosis.
Tumor Formation Assay in Nude Mice
Tumor formation assay was carried out as described previously 10, 12. Xenograft tumors developed in the nude mice after inoculation were subjected to treatment by vectors of RMP overexpression, RMP depletion or HBx. Briefly, a suspension (25 μl/mouse) of Lipofectamine 2000 (20 μg) was mixed with expression plasmids, empty vector (10 μg) or PBS in a final concentration of 100 μg /ml and incubated for 20 min to allow them to complex. Then the complex was delivered by multiple intratumor injection every other day for 2 weeks. At the end of the fourth week, tumors were dissociated and evaluated. The animal operations and procedures were approved by the Committee on the Use of Live Animals in Teaching and Research of Soochow University.
Reverse transcription and Quantitative real time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Shanghai, China). Thermoscript RT system (Invitrogen, Shanghai, China) was applied for the reaction of reverse-transcription. The real time PCR was performed in triplicate in a total volume of 20 μl containing 10 μl SsoFastEvaGreensupermix (Bio-Rad) with SYBR Green, 2 μl cDNA, 2 μl each of the primers, and 6 μl RNase-free water. The PCR program was 94°C for 2 min, followed by 40 cycles at 94°C for 15 s, 60°C for 15 s, and 72°C for 30 s. qRT-PCR assay was run on a Mini OpticonTM Real-time PCR instrument. Each reaction contains 3 technical replicates for qRT-PCR analysis. Primers for RMP were 5'- TCC GAA TAA ATA CTG GAA AG -3' and 5'-AAG GCT CTG TAA ATG TCT GC -3'. Primers for Bax were 5'- TTT TGC TTC AGG GTT TCA TC -3' and 5'- GAC ACT CGC TCA GCT TCT TG -3'. Primers for Bcl-2 were 5'- GGT GGG AGG GAG GAA GAA -3' and 5'- CGC AGA GGC ATC ACA TCG -3'. Primers for GAPDH were 5'-GAC CTG ACC TGC CGT CTA-3' and 5'- AGG AGT GGG TGT CGC TGT -3'. Primers for HBx were 5'-ACCGACCTTGAGGCCTACTT-3' and 5'-GCTTGGCAGAGGTGAAAAAG-3'.
Immunohistochemistry Detection
Tissues of nude mice were subjected to formalin fixation, paraffin embedding and sectioning for immunohistochemistry assays as described previously 16. The slides were first blocked in PBST containing 3% BSA (PBST-BSA) at 37°C for 30 minutes and incubated overnight at 4°C with mouse monoclonal antibodies (1:500 in PBST-BSA). Slides were then washed 3 times in PBST (10 minutes each time) and incubated for 2 hrs with the secondary antibody (anti-mouse HRP, 1:200) in PBST at room temperature. Slides were then washed 3 times in PBST and mounted with 3 μl of Vectashield for further analysis. Finally sections were examined at high power (x 400) under a standard light microscope. Cell staining was regarded as positive if nuclear was homogeneous stained or 10% or more of cytoplasm was heterogeneously stained.
Immunofluorescence and confocal microscopy
HepG2 cells were grown on glass coverslips, washed once in PBS, and fixed with 2% formaldehyde in PBS for 30 min at room temperature (RT). Cells were then permeabilized for 5 min with 100% cold methanol, and dried at -25°C. GFP-fused proteins were detected after being counterstained with 0.0005% Evans Blue. For immunostaining, coverslips were blocked in PBS with 1.5% BSA at RT for 1 hr and incubated with primary antibodies in a humidifying chamber at RT for 1 hr (1:300 monoclonal antibody). The cells were washed 5 times with PBS and incubated at RT for 1 hr with secondary antibodies (1:30 Cy3-labled goat anti-rabbit IgG and FITC-labled goat anti-mouse IgG). Subsequently, the cells were washed 5 times in PBS and mounted with Vectashield mounting medium. A confocal laser scanning microscope (TCS-SP2, Leica Co., Ltd) was applied to acquire the immunofluorescent images.
Statistical analysis
Values are expressed as the mean +/- standard error (SE). Student's t-test was used to determine the significance of the difference between compared groups. P< 0.05 indicated significant differences.
Results
HBx and RMP cooperatively promoted the proliferation of HCC cells
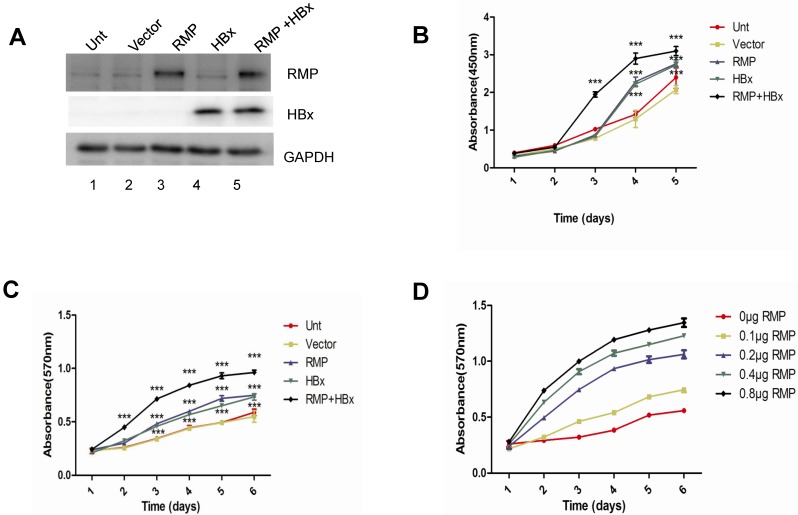
As both HBx and RMP associate with RPB5 of RNA polymerase II and exhibit oncogenic property in the carcinogenesis of HCC, we examined the combined effect of HBx and RMP on the proliferation of HCC cells. HepG2 cells were transfected with expression vectors of HBx or RMP alone, or both (RMP+HBx). The expression of RMP and HBx were determined by Western blot analysis. As shown in Fig. 1A, the protein level of RMP was elevated in cells transfected with RMP vector, while HBx expressed only in the cells transfected with HBx vector (lane 4). The expression of both HBx and RMP was enhanced in the cells transfected with both vectors of HBx and RMP (RMP+HBx, lane 5) (Fig. 1A). Then we examined the effect of RMP and HBx on the growth of HCC cells. Cell Counting Kit-8 (CCK-8) was used as described in the Material and Methods. As shown in the Fig. 1B, the cells expressing either HBx or RMP grew faster than the control cells untransfected or transfected with vector alone. Interestingly the coexpression of both RMP and HBx resulted in stronger promotion of cell proliferation than either RMP or HBx expression alone, indicating a cooperative effect between RMP and HBx on the growth of HCC cells. To confirm the results, we examined the cells with an alternative method of MTT which was extensively applied to test proliferation 10,15. The results also showed that RMP and HBx enhanced the growth of HCC cells with a cooperative effect (Fig. 1C), which is consistent with the results by CCK-8 (Fig. 1B). We further examined if RMP and HBx cooperated to promote the cell growth in a dose dependent manner. HepG2 cells stably expressing HBx 13,14 was co-transfected with increasing amounts of RMP expression vector. As shown in Fig. 1D, increasing amount of RMP exhibited stronger cooperative promotion of the growth of HCC cells expressing HBx, indicating an interaction and cooperation between HBx and RMP.
Fig 1.
HBx and RMP cooperatively promote the proliferation of HCC cells. (A). HepG2 cells were transfected with vectors of RMP, HBx or both vectors of RMP and HBx. The untransfected (Unt) cells or cells transfected with empty vector (Vector) were applied as controls. Cell lysates were extracted for the Western blot analysis with the antibodies against RMP, HBx or GAPDH. (B) and (C). HBx and RMP cooperatively promote the proliferation of HCC cells. HepG2 cells were transfected with vectors as indicated. CCK8 (B) and MTT (C) assays were carried out on the days indicated for the determination of cell growth as described in Materials and Methods. (D). Dose-dependent effect of HBx and RMP on the growth of HCC cells. HepG2 cells stably expressing HBx were transiently transfected with increasing amount of RMP expression vectors as indicated. MTT assay was carried out on the days indicated for the determination of cell growth. Each measurement was made in triplicate. Student's t-test, ***, p<0.001, relative to controls.
The cooperative antiapoptotic effect of HBx and RMP on HCC cells
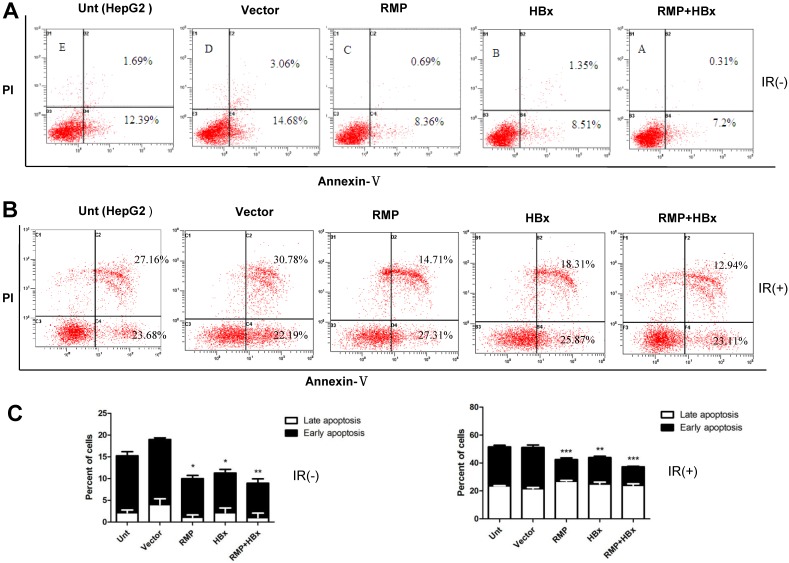
As apoptosis is an important factor affecting cell proliferation, we examined the co-effect of HBx and RMP on the apoptosis of HCC cells. Cells transfected with various vectors were subjected to apoptosis analysis by flow cytometry. As shown in Fig. 2A, there was decreased population of both early and late apoptotic cells in HepG2 overexpressing either HBx or RMP, compared with cells untransfected or transfected with empty vector. Apparently, HepG2 cells co-expressing both HBx and RMP demonstrated the least apoptotic cells among all five groups of transfected and untransfected cells (Fig. 2A and 2C, left panel). To confirm the results, the effect of HBx and RMP on the induced apoptosis was also carried out by using 60Co irradiation (Fig. 2B). More than half population of untreated HCC cells (Unt) or cells treated with empty vector (vector) were in early and late apoptotic phase after treatment of radiation. In contrast, the late apoptotic cells were significantly decreased in the HCC cells expressing either RMP or HBx. The least late apoptotic cells were observed in the HCC cells expressing both RMP and HBx, although early apoptosis remained constant in all the transfected or untreated cells (Fig. 2B and 2C, right panel). These results suggest that RMP and HBx exert a cooperatively antiapoptotic effect on the HCC cells.
Fig 2.
The cooperative effect of HBx and RMP in the antiapoptosis of HCC cells. (A). HepG2 cells were transfected with expression vectors as indicated. Two days later, cells were harvested and subjected to apoptosis analysis by flow cytometry. The percentage of apoptotic cells were scored and depicted graphically in (C, left panel). (B). HepG2 cells were transfected with expression vectors as indicated. One day later, cells were irradiated at 6 Gy and cultured for additional 2 days. Finally cells were harvested and subjected to apoptosis analysis by flow cytometry. The percentage of apoptotic cells were scored and depicted graphically in (C, right panel). Student's t test, *, p<0.05; **, p<0.01; ***, p<0.001, relative to controls.
RMP promoted the growth of HCC xenograft tumors induced by HBx
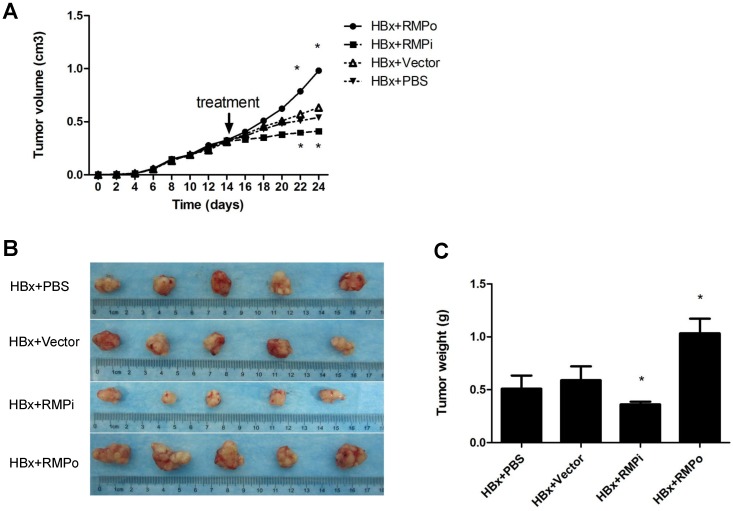
We also examined the effect of RMP on the xenograft tumors developed from HBx-expressing HepG2 cells. Mice were inoculated with HepG2 cells which stably expressed HBx as detected by Western blot (Fig.1A). Tumors were developed to considerable size 2 weeks later. Then the mice were treated by intratumor injection with RMP expression plasmid (RMPo), RMP depletion plasmid (RMPi), empty vector or PBS, respectively. As shown in Fig. 3A, the overexpression of RMP accelerated HBx-induced growth of HCC xenograft tumors (HBx+RMPo), while the depletion of RMP retarded the growth of xenograft tumors (HBx+RMPi), compared with the control tumors treated with PBS or empty vector. As results, RMPo enlarged the HCC xenogroft tumors, while RMPi dramatically reduced the size of the tumors (Fig. 3B and 3C), suggesting a collaboration between HBx and RMP in promoting the growth of HCC tumors.
Fig 3.
RMP promotes the growth of HCC xenograft tumors induced by HBx. (A). Nude mice were inoculated subcutaneously with HepG2 cells stably transfected with HBx expression vector for tumor formation. Two weeks after inoculation, tumors in the nude mice were injected every other day with indicated vectors, or PBS as a control. Tumor size was measured everyday after inoculation and expressed as the mean volume of tumors from five mice, which was depicted graphically. (B). HCC xenograft tumors were dissected at 24thday after inoculation and photographs of tumors from each group are shown. Quantitation of mean tumor weight is shown in (C). Student's t test, *, p<0.05, relative to controls.
HBx promoted the growth of HCC xenograft tumors induced by RMP
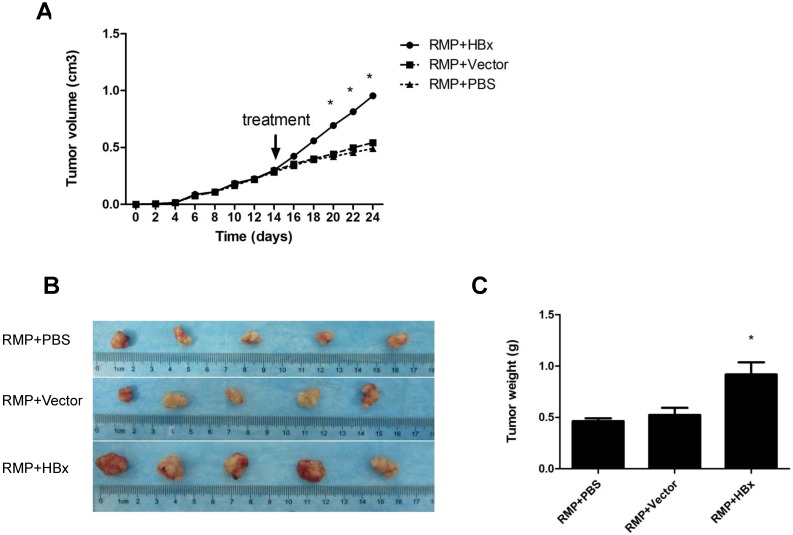
To further confirm the cooperation of RMP and HBx on the growth of HCC tumor, we examined the effect of HBx on the growth of HCC xenograft driven by RMP overexpression. Previously we established xenograft tumors with SMMC-7721 cells stably overexpressing RMP (RMPo) which were verified by Western blot 10, 12. Two weeks after inoculation with RMP-expressing 7721 cells, the developed tumors were administrated with HBx vector, empty vector or PBS, respectively, as indicated. As shown in Fig. 4A, the xenograft tumors grew much faster after the treatment with HBx vector (RMP+HBx), compared with the control tumors treated with PBS or empty vector (Vector). As results, the expression of HBx promoted the RMP-induced tumors, resulting in the biggest tumors among all 3 groups (Fig. 4B and 4C).
Fig 4.
HBx promotes the growth of HCC xenograft tumors induced by RMP. (A). Nude mice were inoculated subcutaneously with SMMC-7721 cells stably transfected with RMP expression vectors for tumor formation. Two weeks after inoculation, tumors in the nude mice were injected every other day with indicated vectors, or PBS as a control. Tumor size was measured everyday after inoculation and expressed as the mean volume of tumors from five mice, which was depicted graphically. (B). Tumors were dissected at 24th day after inoculation and photographs of tumors from each group are shown. Quantitation of mean tumor weight is shown in (C). Student's t test, *, p<0.05, relative to controls.
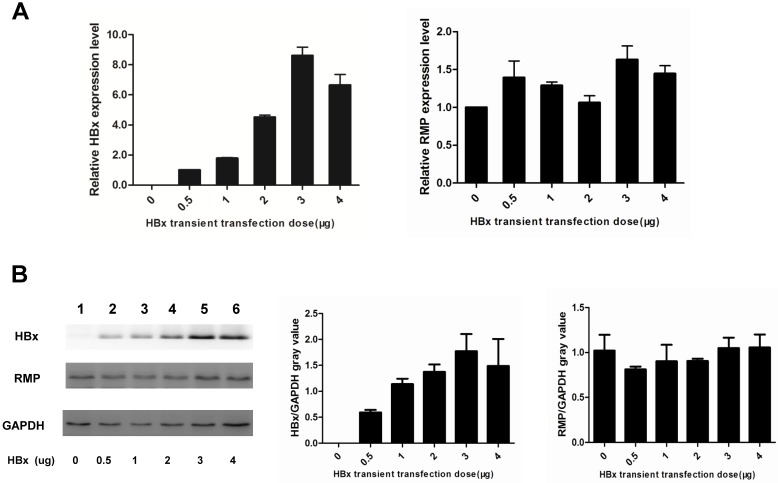
HBx did not regulate the expression of RMP in HCC cells
As HBx has been shown to cooperate with oncogenes by upregulating their expression 13,14,17, we examined if HBx regulated the expression of endogenous oncogene RMP in HCC cells. As shown in Fig. 5A, the transcription of RMP mRNA was not affected by transfection with increasing amount of HBx expression vectors, as detected by qRT-PCR. Then we further examined whether HBx regulated translation, stability or degradation of RMP protein. As shown in Fig.5B, the expression of RMP protein was neither affected by the transfection with increasing amount of HBx expression vectors, demonstrating that HBx did not regulate the expression of RMP at either transcription or translation levels. These results suggest that the cooperation between HBx and RMP is not mediated by the regulation of RMP expression, but by the possible interaction between HBx and RMP.
Fig 5.
HBx does not regulate the expression of RMP in HCC cells. (A). HepG2 cells were transfected with increasing amounts of HBx expression vector as indicated. qRT-PCR was performed for the quantative determination of the expression of HBx (left panel) and endogenous RMP (right panel). (B). HepG2 cells were transfected with increasing amounts of HBx expression vector as indicated. Cell lysates were extracted for the Western blot analysis (left panel) with antibodies against HBx, RMP or GAPDH as indicated. The protein expression of HBx (middle panel) and RMP (right panel) was quantitated against GAPDH. Results are reported as means ±S.D. of three independent experiments.
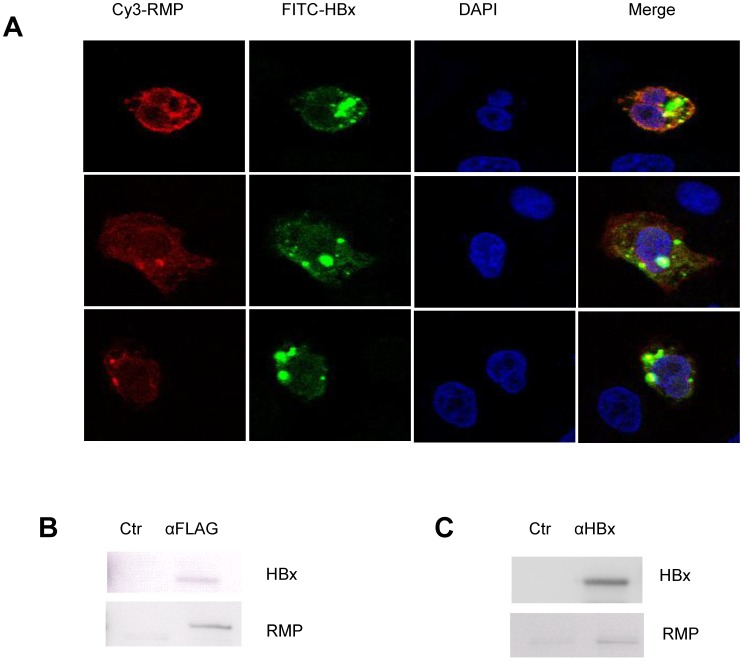
HBx interacted with RMP in HCC cells
As no evidence showed that HBx could regulate the expression of RMP, we wondered if there was an interaction between HBx and RMP. At first, we examined the localization and colocalization of HBx and RMP in HCC cells. The immunofluorescent staining was performed with HepG2 cells transfected with both RMP and HBx vectors. As shown in Fig. 6A, both RMP and HBx were localized primarily in the cytoplasm and less in the nucleus of HCC cells. The merged image demonstrated that RMP and HBx colocalized in the cytoplasm of HepG2 cells.
Fig 6.
HBx interacts with RMP in HCC cells. (A). HepG2 cells were cotransfected with HBx and Flag-RMP expression vectors. Cells were fixed and immunostained with antibodies against RMP conjugated with Cy3 and HBx antibody conjugated with FITC. The stained cells were analyzed and photographed under a confocal laser scanning microscope. (B). Flag-RMP expression vectors were cotransfected into HepG2 cells stably expressing HBx. Cell lysates were extracted and immunoprecipitation was performed with antibody against Flag. The immunoprecipitates were fractionated in 12.5% SDS-PAGE and subjected to Western blot analysis with antibodies against HBx and RMP as indicated. Preimmune IgG was applied as control of immunoprecipiation (Ctr). (C). Lysates were extracted from HepG2 cells stably expressing HBx. Immunoprecipitation was performed with antibody against HBx or with the preimmue IgG as a control (Ctr). The immunoprecipitates were fractionated in 12.5% SDS-PAGE and subjected to Western blot analysis with antibodies against HBx and RMP as indicated.
Then immuoprecipitation was also performed to determine the molecular interaction between HBx and RMP. The results were shown in Fig. 6B, HCC cells were cotransfected with vectors encoding HBx and Flag-RMP. The exogenously expressed Flag-RMP was immunoprecipitated by Flag antibody. The Western blot analysis showed that HBx was coimmunoprecipitaed with Flag-RMP in contrast to the control immunoprecipitation by IgG (Fig. 6B).
We further examined if the HBx could interact with the endogenous RMP. The lysates of HepG2 cells stably transfected with HBx was incubated with an antibody against HBx. The immunoprecipitates were examined by Western blot analysis with both HBx and RMP antibodies. As shown in Fig. 6C, the endogenous RMP was immunoprecipitated together with HBx, indicating that HBx was also associated with endogenous RMP which may be within a complex of RNA polymerase II.
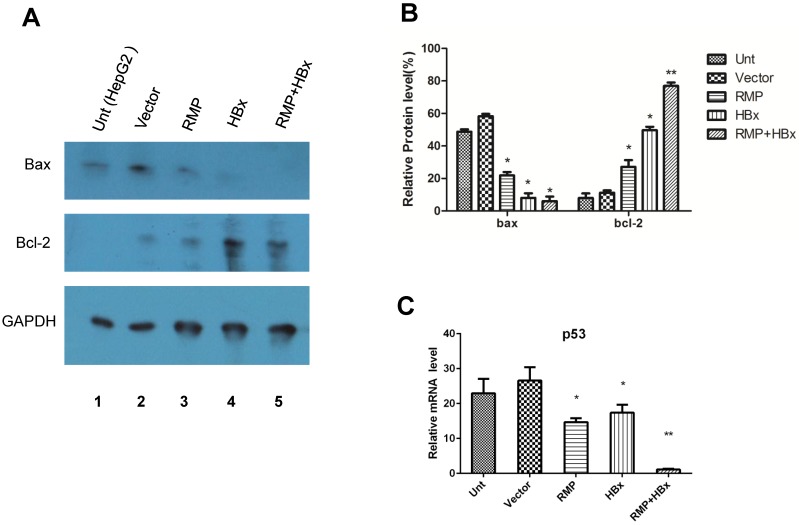
HBx and RMP collaboratively regulated the apoptotic genes
To investigate the mechanism of the collaboration between HBx and RMP on HCC growth, we examined the effect of HBx or RMP on the expression of apoptotic genes. As shown in Fig. 7A, both RMP and HBx suppressed the expression of proapoptotic factor Bax. The co-expression of RMP and HBx demonstrated even stronger inhibition on Bax expression than either HBx or RMP alone (Fig. 7A and 7B). In contrast, the expression of antiapoptotic factor Bcl-2 was increased by RMP or HBx. Co-transfection of RMP and HBx expression vectors resulted in stronger expression of Bcl-2 than either HBx or RMP alone (Fig.7A and 7B).
Fig 7.
HBx and RMP collaborately regulate apoptotic genes. (A). HepG2 cells (lane 1) were transfected with empty vector (lane 2), expression vectors for RMP (lane 3), HBx (lane 4), or both RMP and HBx (lane 5). Protein was extracted and Western blot analysis was carried out with antibodies against Bax, Bcl-2 and GAPDH which was applied for the normalization. The relative expression of Bax and Bcl-2 against GAPDH were graphically depicted in (B). (C). HepG2 cells were transfected with vectors as indicated. The mRNA was extracted and quantative RT-PCR was carried out for the determination of expression of p53. The expression of GAPDH was also determined for the normalization of expression of above factors. Student's t test, *, p<0.05; **, p<0.01, relative to controls.
The collaborative effect of RMP and HBx was also observed on the expression of p53 and caspase-3, which was detected by qRT-PCR (Fig. 7C). The expression of p53 was inhibited by either RMP or HBx to some extents. And the cotransfection of HBx and RMP vectors significantly suppressed p53 expression (Fig. 7C).
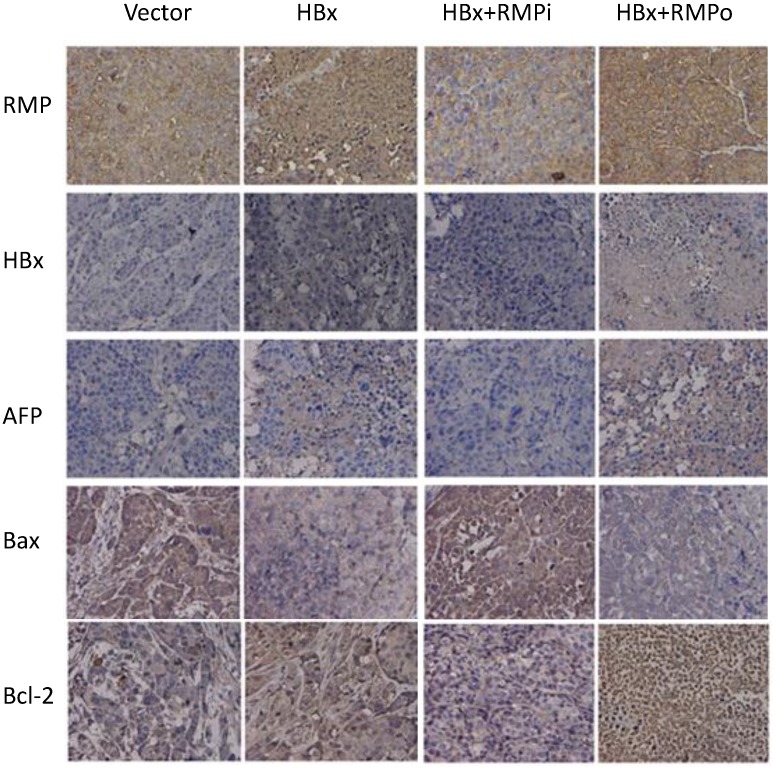
To confirm the effect of RMP and HBx on the expression of apoptotic genes, we further examined the expression of apoptotic genes in the HCC xenograft tumors induced by RMP or HBx. As shown in Fig. 8, RMP expression increased when the xenograft tumors were treated with RMP expression vector (RMPo) and decreased when tumors were treated with RMP interference vector (RMPi). HBx was expressed in the xenograft tumors derived from HepG2 cells which stably expressed the HBx. The expression of AFP was increased by expression of both HBx and RMP. The expression of proapoptotic factor Bax was also decreased by HBx or the coexpression of HBx and RMP (HBx+RMPo), while increased by the depletion of RMP (HBx+RMPi) (Fig. 8). In contrast, the expression of antiapoptotic factor Bcl-2 was increased by HBx alone or coexpression of HBx and RMP (HBx+RMPo), but decreased by the depletion of RMP (HBx+RMPi).
Fig 8.
Immunohistochemical analysis of expression of apoptotic factors in mouse xenograft tumors. HepG2 cells stably expressing HBx were inoculated subcutaneously into the dorsal flanks of nude mince. Two weeks later, the established tumors were subjected to intratumor injection with vectors as indicated on the top. Tumors were sectioned and immunohistochemical examination was carried out with antibodies as indicated to the left.
Discussion
RMP/URI has been shown to play multiple roles in transcription regulation 8,18,19, nutrient-related signaling pathways 20 and maintenance of genomic stability 21. Recent studies identified that RMP/URI acts as an oncogene which is involved in the carcinogenesis of both HCC and ovarian cancer 9,10.
RMP has multiple binding partners which might contribute to the complexity of RMP function. RMP binds with a multiprotein complex, including SKP2, PFD2, PFD4, RPB5 of the core subunit of RNAP II, the ATPases TIP48 and TIP49, which is within chromatin-remodeling complexes. RMP was found to be copurified with paf1 complex 16, which is involved in the histone methylation and cell cycle control. RMP interacts with DNA methyltransferase I (DNMT1)-associating protein (DMAP1), and is implicated in gene silencing 22. The association of yeast homologue of RMP (Bud27p) with the histone demethylase domain of Gis 1p' jmjC 5,23, suggests that RMP might be involved in the epigenetic regulation.
HBx encoded by the HBV genome is crucial for the carcinogenesis of HCC. In the culture system, not only the transcription of viral genes, but also that of host genes, are activated by HBx. Similar to RMP, HBx is located mainly in cytosolic. Some cytosolic HBx are localized in mitochondria 1-3, 24-26, where RMP was located 11. Our results demonstrated the colocalization of RMP and HBx in the cytosol. In the cultured cells, HBx is localized in the outer mitochondrial membrane, which is consistent with the reported interaction of HBx with voltage-dependent anion channel (VDAC) spanning the outer mitochondrial membrane 2-4.
Although HBx does not bind directly to DNA, it may regulate gene expression via protein-protein interaction. Actually HBx has been shown to be involved in transcription 27, signaling pathways, protein degradation, cell-cycle control 28,29, and carcinogenesis 30. Molecular evidence indicates that HBx is an oncoprotein. However, the detailed mechanism of HBx-mediated hepatocarcinogenesis with its partners remains unclear.
In this article, we report that HBx and RMP cooperatively stimulate the proliferation and suppress the apoptosis of HCC cells. Our studies show that HBx and RMP were colocalized in the HCC cells and could be co-immunoprecipitated. Consistent with the presence of physical interaction, functional collaboration was observed between HBx and RMP as well. The cell growth assay demonstrated that coexpression of both HBx and RMP resulted in a cooperative effect in the promotion of the HCC cell growth. A similar collaboration was observed on the apoptosis of HCC cells. Coexpression of HBx and RMP cooperatively suppressed the apoptosis of HCC. The collaboration between HBx and RMP was also observed in vivo by the nude mice experiments.
Apparently, apoptotic factors are the targets of both RMP and HBx. Our results demonstrated that RMP or HBx alone suppressed the expression of proapoptotic factor such as Bax, while at the same time enhancing the expression of antiapoptotic factors, such as Bcl-2. This is consistent with the reported result that Bcl-2 was elevated by the expression of HBx in a microarray 31. Coexpression of both RMP and HBx cooperatively regulates the expression of apoptotic factors.
Although much more clinical data is required to confirm the possible collaboration between HBx and RMP in the carcinogenesis of HCC, the immunohistochemical examination in HCC tissues demonstrated that enhanced expression of RMP and HBx was accompanied by reduced expression of proapoptotic factors, together with elevated expression of antiapoptotic factors.
There are several possible lines of mechanism for the cooperation between HBx and RMP, as common targets or pathways were identified to be shared by both HBx and RMP.
RMP has been found to function in the mTOR survival signaling. PP1γ is able to suppress the cell survival by inhibiting S6 kinase (S6K1) and BAD phosphorylation. RMP binds with PP1γ which is inhibited in the coupled state. S6K1 phosphorylates the RMP, resulting in the release of PP1γ from the inactive state 11.
Interestingly, HBx has also been involved in the mTOR signaling. Overexpression of HBx in HCC cells upregulated mammalian target of rapamycin (mTOR) downstream effector S6 kinase (S6K1), and increased as well the cell proliferation and vascular endothelial growth factor (VEGF) production. HBx transgenic mouse model demonstrated that HBx-modulated mTOR/S6K1 signaling was associated with liver tumorigenesis 32.
Both HBx and RMP function in the DNA damage repair. HBx associates with the DNA damage-binding (DDB) protein, which binds to the breakends of damaged DNA 33. In C.elegans, RMP is required for the DNA stability and genome integrity. Depletion of RMP resulted in DNA damage and germline apoptosis in a p53-dependent manner 19.
Dr. Murakami group identified DNA methytransferase 1 (DNMT1)-associating protein (DMAP1) as one of the RMP-interacting partners. DMAP1 facilitated the nuclear localization of RMP and the corepressor activity of RMP in a dose-dependent manner 22. Interestingly, HBx has also been demonstrated to upregulate the expression of DNMT1 34.
A similar cooperation was observed between two oncogenes of HBx and AIB1 (amplified in breast cancer 1) in the hepatocarcinogenesis of HCC 32. HBx was found to inhibit the ubiquitination and degradation of AIB1, therefore stabilize AIB1 protein and enhance its expression. Interestingly, recent studies show that HBx also upregulates oncogene expression of Rab18 and Lin28A/Lin28B and deregulates the proliferation of hepatoma cells 13,14. However in our study, HBx did not affect the expression of endogenous RMP. Instead, the cooperation relies on the interaction between HBx and RMP, as verified by the inmmunoprecipitation and colocalizaion in HCC cells.
This finding may be of important significance, which may explain or partially explain the hepatocarcinogenesis by the infection of HBV. After HBV infection, HBx expresses and targets to the RPB5 of RNAP II, which in turn associated with the endogenous cellular oncoprotein RMP. The physical vicinity and interaction, between endogenous cellular oncoprotein RMP and HBV oncoprotein HBx leads to functional collaboration which inhibits the apoptosis of HCC cells, and contributes to the carcinogenesis of HCC. If this model is true, disruption of connections between HBx and RMP may be a potential way for the prevention and treatment of HCC.
Acknowledgments
This study was partly supported by Nature Science Foundation of China (81172347, 81272301), Special Research Found for the Doctoral Program of Higher Education (20123201110011), A project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions and the Graduate Innovation Project of Jiangsu Province (CXZZ12-0834 and CXZZ13-0828).
References
- 1.Rawat S, Clippinger AJ, Bouchard MJ. Modulation of apoptotic signaling by the hepatitis B virus X protein. Viruses. 2012;4:2945–72. doi: 10.3390/v4112945. doi:10.3390/v4112945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clippinger AJ, Bouchard MJ. Hepatitis B virus HBx protein localizes to mitochondria in primary rat hepatocytes and modulates mitochondrial membrane potential. Journal of virology. 2008;82:6798–811. doi: 10.1128/JVI.00154-08. doi:10.1128/JVI.00154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahmani Z, Huh KW, Lasher R, Siddiqui A. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. Journal of virology. 2000;74:2840–6. doi: 10.1128/jvi.74.6.2840-2846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoshan-Barmatz V, Israelson A, Brdiczka D, Sheu SS. The voltage-dependent anion channel (VDAC): function in intracellular signalling, cell life and cell death. Current pharmaceutical design. 2006;12:2249–70. doi: 10.2174/138161206777585111. [DOI] [PubMed] [Google Scholar]
- 5.Miron-Garcia MC, Garrido-Godino AI, Garcia-Molinero V, Hernandez-Torres F, Rodriguez-Navarro S, Navarro F. The prefoldin bud27 mediates the assembly of the eukaryotic RNA polymerases in an rpb5-dependent manner. PLoS genetics. 2013;9:e1003297.. doi: 10.1371/journal.pgen.1003297. doi:10.1371/journal.pgen.1003297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woychik NA, Liao SM, Kolodziej PA, Young RA. Subunits shared by eukaryotic nuclear RNA polymerases. Genes & development. 1990;4:313–23. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]
- 7.Cheong JH, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. The EMBO journal. 1995;14:143–50. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorjsuren D, Lin Y, Wei W, Yamashita T, Nomura T, Hayashi N. et al. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Molecular and cellular biology. 1998;18:7546–55. doi: 10.1128/mcb.18.12.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theurillat JP, Metzler SC, Henzi N, Djouder N, Helbling M, Zimmermann AK. et al. URI is an oncogene amplified in ovarian cancer cells and is required for their survival. Cancer cell. 2011;19:317–32. doi: 10.1016/j.ccr.2011.01.019. doi:10.1016/j.ccr.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 10.Yang H, Gu J, Zheng Q, Li M, Lian X, Miao J. et al. RPB5-mediating protein is required for the proliferation of hepatocellular carcinoma cells. The Journal of biological chemistry. 2011;286:11865–74. doi: 10.1074/jbc.M110.136929. doi:10.1074/jbc.M110.136929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R. et al. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Molecular cell. 2007;28:28–40. doi: 10.1016/j.molcel.2007.08.010. doi:10.1016/j.molcel.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Yang S, Wang H, Guo Y, Chen S, Zhang MY, Shen J. et al. RMP plays distinct roles in the proliferation of hepatocellular carcinoma cells and normal hepatic cells. International journal of biological sciences. 2013;9:637–48. doi: 10.7150/ijbs.6439. doi:10.7150/ijbs.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You X, Liu F, Zhang T, Li Y, Ye L, Zhang X. Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis. 2013;34:1644–52. doi: 10.1093/carcin/bgt089. doi:10.1093/carcin/bgt089. [DOI] [PubMed] [Google Scholar]
- 14.You X, Liu F, Zhang T, Lv N, Liu Q, Shan C. et al. Hepatitis B virus X protein upregulates Lin28A/Lin28B through Sp-1/c-Myc to enhance the proliferation of hepatoma cells. Oncogene. 2013 doi: 10.1038/onc.2012.618. doi:10.1038/onc.2012.618. [DOI] [PubMed] [Google Scholar]
- 15.Gu J, Sun D, Zheng Q, Wang X, Yang H, Miao J. et al. Human Elongator complex is involved in cell cycle and suppresses cell growth in 293T human embryonic kidney cells. Acta biochimica et biophysica Sinica. 2009;41:831–8. doi: 10.1093/abbs/gmp072. [DOI] [PubMed] [Google Scholar]
- 16.Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D. et al. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Molecular and cellular biology. 2005;25:5052–60. doi: 10.1128/MCB.25.12.5052-5060.2005. doi:10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Tong Z, Li T, Chen Q, Zhuo L, Li W. et al. Hepatitis B virus X protein stabilizes amplified in breast cancer 1 protein and cooperates with it to promote human hepatocellular carcinoma cell invasiveness. Hepatology. 2012;56:1015–24. doi: 10.1002/hep.25751. doi:10.1002/hep.25751. [DOI] [PubMed] [Google Scholar]
- 18.Wei W, Dorjsuren D, Lin Y, Qin W, Nomura T, Hayashi N. et al. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. The Journal of biological chemistry. 2001;276:12266–73. doi: 10.1074/jbc.M009634200. doi:10.1074/jbc.M009634200. [DOI] [PubMed] [Google Scholar]
- 19.Mita P, Savas JN, Djouder N, Yates JR 3rd, Ha S, Ruoff R. et al. Regulation of androgen receptor-mediated transcription by RPB5 binding protein URI/RMP. Molecular and cellular biology. 2011;31:3639–52. doi: 10.1128/MCB.05429-11. doi:10.1128/MCB.05429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M. et al. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302:1208–12. doi: 10.1126/science.1088401. doi:10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- 21.Parusel CT, Kritikou EA, Hengartner MO, Krek W, Gotta M. URI-1 is required for DNA stability in C. elegans. Development. 2006;133:621–9. doi: 10.1242/dev.02235. doi:10.1242/dev.02235. [DOI] [PubMed] [Google Scholar]
- 22.Delgermaa L, Hayashi N, Dorjsuren D, Nomura T, Thuy le TT, Murakami S. Subcellular localization of RPB5-mediating protein and its putative functional partner. Molecular and cellular biology. 2004;24:8556–66. doi: 10.1128/MCB.24.19.8556-8566.2004. doi:10.1128/MCB.24.19.8556-8566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tronnersjo S, Hanefalk C, Balciunas D, Hu GZ, Nordberg N, Muren E. et al. The jmjN and jmjC domains of the yeast zinc finger protein Gis1 interact with 19 proteins involved in transcription, sumoylation and DNA repair. Molecular genetics and genomics: MGG. 2007;277:57–70. doi: 10.1007/s00438-006-0171-3. doi:10.1007/s00438-006-0171-3. [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Kim HY, Lee S, Kim SW, Sohn S, Kim K. et al. Hepatitis B virus x protein induces perinuclear mitochondrial clustering in microtubule- and Dynein-dependent manners. Journal of virology. 2007;81:1714–26. doi: 10.1128/JVI.01863-06. doi:10.1128/JVI.01863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huh KW, Siddiqui A. Characterization of the mitochondrial association of hepatitis B virus X protein, HBx. Mitochondrion. 2002;1:349–59. doi: 10.1016/s1567-7249(01)00040-x. [DOI] [PubMed] [Google Scholar]
- 26.Shirakata Y, Koike K. Hepatitis B virus X protein induces cell death by causing loss of mitochondrial membrane potential. The Journal of biological chemistry. 2003;278:22071–8. doi: 10.1074/jbc.M301606200. doi:10.1074/jbc.M301606200. [DOI] [PubMed] [Google Scholar]
- 27.Lee SG, Rho HM. Transcriptional repression of the human p53 gene by hepatitis B viral X protein. Oncogene. 2000;19:468–71. doi: 10.1038/sj.onc.1203312. doi:10.1038/sj.onc.1203312. [DOI] [PubMed] [Google Scholar]
- 28.Jia L, Wang XW, Harris CC. Hepatitis B virus X protein inhibits nucleotide excision repair. International journal of cancer Journal international du cancer. 1999;80:875–9. doi: 10.1002/(sici)1097-0215(19990315)80:6<875::aid-ijc13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 29.Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. Journal of virology. 2004;78:12725–34. doi: 10.1128/JVI.78.23.12725-12734.2004. doi:10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cougot D, Neuveut C, Buendia MA. HBV induced carcinogenesis. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2005;34(Suppl 1):S75–8. doi: 10.1016/s1386-6532(05)80014-9. [DOI] [PubMed] [Google Scholar]
- 31.Zhang WY, Xu FQ, Shan CL, Xiang R, Ye LH, Zhang XD. Gene expression profiles of human liver cells mediated by hepatitis B virus X protein. Acta pharmacologica Sinica. 2009;30:424–34. doi: 10.1038/aps.2009.22. doi:10.1038/aps.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yen CJ, Lin YJ, Yen CS, Tsai HW, Tsai TF, Chang KY. et al. Hepatitis B virus X protein upregulates mTOR signaling through IKKbeta to increase cell proliferation and VEGF production in hepatocellular carcinoma. PloS one. 2012;7:e41931.. doi: 10.1371/journal.pone.0041931. doi:10.1371/journal.pone.0041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee TH, Elledge SJ, Butel JS. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. Journal of virology. 1995;69:1107–14. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng DL, Zhang L, Cheng N, Xu X, Deng Q, Teng XM. et al. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. Journal of hepatology. 2009;50:377–87. doi: 10.1016/j.jhep.2008.10.019. doi:10.1016/j.jhep.2008.10.019. [DOI] [PubMed] [Google Scholar]