Abstract
Background
Children with high-grade glioma, including diffuse intrinsic pontine glioma (DIPG), have a poor prognosis despite multimodal therapy. Identifying novel therapeutic targets is critical to improve their outcome. We evaluated prognostic roles of telomere maintenance mechanisms in children with HGG, including DIPG.
Methods
A multi-institutional retrospective study was conducted involving 50 flash-frozen HGG (35 non-brainstem; 15 DIPG) tumors from 45 children (30 non-brainstem; 15 DIPG). Telomerase activity, expression of hTERT mRNA (encoding telomerase catalytic component) and TERC (telomerase RNA template) and alternative lengthening of telomeres (ALT) mechanism were assayed. Cox Proportional Hazard regression analyses assessed association of clinical and pathological variables, TERC and hTERT levels, telomerase activity, and ALT use with progression-free or overall survival (OS).
Results
High TERC and hTERT expression was detected in 13/28 non-brainstem HGG samples as compared to non-neoplastic controls. High TERC and hTERT expression was identified in 13/15 and 11/15 DIPG samples, respectively, compared to controls. Evidence of ALT was noted in 3/11 DIPG and 10/19 non-brainstem HGG specimens. ALT and telomerase use were identified in 4/19 non-brainstem HGG and 2/11 DIPG specimens. In multivariable analyses, increased TERC and hTERT levels were associated with worse OS in patients with non-brainstem HGG, after controlling for tumor grade or resection extent.
Conclusions
Children with HGG and DIPG, have increased hTERT and TERC expression. In children with non-brainstem HGG, increased TERC and hTERT expression levels are associated with a worse OS, making telomerase a promising potential therapeutic target in pediatric HGG.
Keywords: Pediatric, high-grade glioma, telomerase
INTRODUCTION
The outcome for children with high-grade gliomas (HGG) remains poor despite multimodal therapy with surgery, radiation, and conventional chemotherapy. Molecularly targeted agents may offer new therapeutic options for this devastating disease. One potential target is telomerase, a ribonucleoprotein complex that includes a catalytic subunit encoded by the hTERT gene and an RNA template (TERC) [1–3]. Telomerase elongates telomeric DNA by adding hexanucleotide repeats to compensate for progressive DNA loss with each cell division.[1,4]
Telomerase activity is detectable in 85–95% of cancer types [5–7]. By contrast, normal somatic tissues, including brain, do not demonstrate telomerase activity [5,7]. hTERT mRNA expression correlates tightly with telomerase activity and is generally restricted to tumor cells [8–12]. Although TERC RNA is present in most normal cells, TERC expression is upregulated during tumorigenesis [13,14].
Telomerase activity is highly variable in adult anaplastic astrocytomas (10–100%) and glioblastoma multiforme (GBM) (26–100%) [15–20]. Higher telomerase activity levels are associated with shorter overall survival (OS) for adult GBM [18,19]. A subset of the telomerase-negative tumors maintains telomere length by an alternative lengthening of telomeres (ALT) mechanism involving DNA homologous recombination [21,22]. One study reported that 11% and 44% of adult and pediatric GBM, respectively, demonstrated evidence of ALT [23]. Adults with ALT-positive GBM were reported to have longer survival times [24,25]. ATRX and DAXX are members of a chromatin-remodeling complex that deposits histone H3.3 onto chromatin and telomeres [26–28]. It has recently been shown that mutations in H3F3A, encoding H3.3, and chromatin remodeling genes ATRX and DAXX drive pediatric glioma [29]. The presence of H3F3A/ATRX/TP53 mutations was found to be strongly associated with ALT [29]. Three point mutations were identified which led to single amino acid substitutions at two sites in the H3.3 tail, H3.3K27M and H3.3G34V/R [29–31]. Furthermore, 60–71% of DIPGs harbored the H3.3K27M mutation, while none had H3.3G34R/V mutations [31]. H3.3K27M mutation defines a clinically and biologically distinct subgroup of DIPG and is associated with shorter survival [31].
Little data exist regarding the role of telomere maintenance mechanisms and correlation to outcome in children with HGG or DIPG. To the best of our knowledge, this is the first report to describe telomerase expression and its prognostic significance in children with HGG, including DIPG.
MATERIALS AND METHODS
We conducted a retrospective study of children with Grade III (anaplastic astrocytomas) or IV gliomas (GBM), including DIPG, diagnosed between 1980 and 2011 who had flash-frozen tumor tissue available at Cincinnati Children’s Hospital Medical Center (CCHMC; Cincinnati, OH), Children’s Memorial Hospital (Chicago, IL), New York Medical Center (New York, NY), Nationwide Children’s Hospital (Columbus, OH), Henry Ford Hospital (Detroit, MI), or the Brain Tumor Tissue Bank (Ontario, Canada). Institutional review board approval was obtained at CCHMC and at participating institutions (as applicable). Clinical information obtained included histopathology, extent of resection, primary tumor location, and time from diagnosis to progression/relapse, death, or last follow-up. Fifty HGG specimens from 45 patients were acquired, including 15 DIPGs and 35 non-brainstem HGGs. Four non-brainstem HGG patients provided multiple tumor samples at different times in their disease courses.
Tissue Processing and Analysis
All tumors were centrally reviewed for histological confirmation by an experienced neuropathologist (LM). Study controls included matched non-tumoral flash-frozen cortical brain specimens from autopsies of six DIPG patients on this study (median age, 5.1 years; range, 2.8–12.5 years), five flash-frozen non-tumoral cortical brain specimens from patients (age range, 1.8–18 years) who had undergone epilepsy surgery, and commercial RNA derived from parietal cortex of a 26-year-old man (Agilent, Santa Clara, CA), pooled pons from 20 people (age range, 18–54 years; Clontech, Mountain View, CA) and total brain from an 18-year-old man (Clontech, Mountain View, CA). All tissue specimens were stored at −80°C until processed for analysis.
Three measures of telomerase expression, (i) hTERT mRNA and (ii) TERC RNA levels by quantitative real-time polymerase chain reaction (qRT-PCR), and (iii) telomerase enzyme activity by telomeric repeat amplification protocol (TRAP) assay, were performed on tumor and control specimens. Although the TRAP assay provides a direct assessment of telomerase enzymatic activity, it may be subject to false negative results if telomerase is inactivated by time or heat during specimen processing. hTERT mRNA levels have been shown to correlate well with telomerase activity [8–10]. Telomeric restriction fragment (TRF) analysis by Southern blot was used to evaluate ALT activity [32]. Molecular assays were performed by investigators blinded to patients’ outcome.
hTERT and TERC Expression
Total RNA was extracted by RNAzol reagent (Molecular Research Center, Cincinnati, OH), and one μg RNA was converted to cDNA using First Strand cDNA Synthesis kit (USB, Cleveland, Ohio). hTERT mRNA expression analysis was performed by qRT-PCR using a primer/probe set that spans exon junctions 3–4 and therefore detects all gene splice forms (Hs00972650_m1; Applied Biosystems, Carlsbad, CA). TERC RNA was performed similarly by qRT-PCR using commercially available primers and probes (Hs03297287_s1; Applied Biosystems, Carlsbad, CA). mRNA levels of housekeeping genes glyceraldehyde phosphate dehydrogenase and TATA-box binding protein using commercially available primer/probe sets (Hs03929097_g1 and Hs00427621_m1, Applied Biosystems, Carlsbad, CA) were used as normalizers. The mean hTERT and TERC levels from normal brain specimens were used as the reference level to which the tumor specimen levels were compared. qRT-PCR assays were performed for each sample at least twice in triplicate. Relative expression (expressed as relative quantification, RQ) levels of hTERT mRNA and TERC RNA levels were compared to the mean expression level from non-neoplastic control brain samples using the comparative Ct method [33]. RQ values >1 defined increased expression levels as compared to the mean values for control non-neoplastic specimens.
Telomerase Enzyme Activity
Telomerase enzyme activity was assessed in tumor and control non-tumoral samples using the TRAPeze Telomerase Detection Kit (Millipore, Billerica, MA) as previously described [34]. Recombinant RNasin ribonuclease inhibitor was added to the CHAPS lysis buffer before the extraction to a concentration of 200 units/ml lysis buffer. Testing was performed using 400 ng total protein per tumor/control specimen. Tumors were considered to have telomerase activity if a characteristic laddering pattern of telomere amplification products was detected by gel electrophoresis. Lysates without detectable enzymatic activity were reanalyzed at higher protein concentrations to detect low levels of telomerase activity. HeLa cell line (30 ng total protein) and CHAPS buffer were used as positive and negative controls, respectively.
ALT Use
DNA was extracted from tumor specimens using the Gentra Puregene kit (Qiagen, Dusseldorf, Germany). Assessment of telomere restriction fragment (TRF) lengths by Southern blot was performed on tumor specimens using the TeloTAGGG Telomere Length Assay Kit (Roche Diagnostics, Indianapolis, IN). For all samples, 1 μg of total genomic DNA was digested and separated by gel electrophoresis. Samples were considered positive for ALT if they exhibited heterogeneous telomere lengths, including high molecular weight (>21 kb) telomeres. The osteosarcoma Saos-2 and histiocytic lymphoma U937 cell lines were used as positive and negative controls, respectively, for ALT.
H3F3A Gene Sequencing
Histone 3.3 gene H3F3A was partially sequenced as described by Wu et al [30]. Briefly, a region of interest in H3F3A gene was sequenced by direct sequencing of PCR amplified products using the primers forward: 5′-GATTTTGGGTAGACGTAATCTTCA-3′, reverse: 5′-TTTCCTGTTACTCATCTTTTTGTT-3′ and a high fidelity DNA polymerase Optimase (Transgenomic, Omaha, NE). Available matched non-neoplastic samples were also sequenced when available. The Sanger sequencing was performed on both strands using the above forward and reverse primers.
Statistical Analyses
TERC expression levels were analyzed in base-2 log-transformed scale to provide hazard ratios (HR) in terms of doubling of qRT-PCR levels. hTERT is a low-copy-number transcript, and no amplification may be reported in qRT-PCR analysis with the maximum of 50 cycles. A constant of 1 was added to all hTERT expression RQ values to enable analysis in base2 log-transformed scale since the original data contained some values that were 0. TRAP and ALT variables were categorized as positive or negative, as described above. The following clinical and demographic variables were reviewed: age at diagnosis, pathological grade (WHO grade III vs. IV), tumor location (DIPG/brainstem vs. non-brainstem), specimen disease timing (initial vs. relapse), and extent of surgical resection (total vs. partial).
Associations of biological markers and clinical and demographic variables with progression-free survival (PFS) and OS were evaluated using Cox Proportional Hazard (PH) models and log-rank tests as appropriate. Due to sample size limitations, multivariable models were limited to 2 variables. Four patients had multiple tumor specimens (n=9 specimens) from different time points, and the samples were analyzed for temporal changes in telomerase maintenance mechanisms descriptively. For the outcome analysis, assay results from the earliest specimen were used for patients who had multiple tumor samples available. PFS was defined as time from diagnosis to first disease progression or death and to last follow-up visit for patients without progression or death. OS was defined as time from diagnosis to death or last follow-up visit.
Comparison of hTERT and TERC expression values between DIPG and non-brainstem HGG specimens and between relapsed and newly-diagnosed non-brainstem HGG specimens was performed using the Kruskal-Wallis test. Results with p-values <0.05 were reported as statistically significant without any correction for multiplicity.
RESULTS
Patient Characteristics
Fifty tumor samples from 45 patients were included. Table 1 summarizes the clinical characteristics of 35 non-brainstem HGG tumors from 30 patients and of 15 DIPG tumors from 15 patients. Two non-brainstem samples were secondary HGG that developed after low-grade astrocytoma or medulloblastoma.
Table 1.
Subject Characteristics
| Characteristic of Subjects with Non-brainstem HGG | Results |
|---|---|
| No. of patients | 30 |
| No. of tumor specimens | 35 |
| Median age | 13.1 years |
| Age range | 0.2–22.2 years |
| Male:female | 18:12 |
| Pathologic grade (III:IV) | 8:22 |
| Specimen timing (diagnosis:relapse) | 21:11 |
| Characteristic of Subjects with DIPG | |
| No. of patients | 15 |
| No. of tumor specimens | 15 |
| Median age | 5.1 years |
| Age range | 2.7–12.5 years |
| Male:female | 3:12 |
| Pathologic grade (III:IV) | 3:12 |
| Specimen timing (diagnosis:autopsy) | 1:14 |
HGG, high-grade glioma
DIPG, diffuse intrinsic pontine glioma
hTERT and TERC expression
Table 2 summarizes clinical and demographic information and results from telomere maintenance assays, including hTERT and TERC expression analysis. Among 15 DIPG tumor specimens, 86.7% and 73.3% had increased TERC RNA and hTERT mRNA expression, respectively, compared to the mean normal controls. Of 28 non-brainstem HGG specimens analyzed, 46.4% had increased TERC RNA and hTERT mRNA expression levels compared to normal controls. Median TERC values were significantly (3.4-fold) higher in children with DIPG compared to those with non-brainstem HGG (Kruskal-Willis, p=0.001) (Table 3). Median hTERT values trended towards being significantly higher (12-fold) in the DIPG cohort compared to the non-brainstem HGG cohort (Kruskal-Willis, p=0.056). When hTERT and TERC values were compared between the non-brainstem HGG specimens from diagnosis (n=22) vs. relapse (n=6), no statistically significant differences were noted.
Table 2.
Summary of Clinical Characteristics and Telomere Maintenance Assay Results
| ID | Gender | Age (years) | Tumor | Pathology | Specimen Timing | Resection | Telomerase activity by TRAP | ALT | TERT (RQ) | TERC (RQ) | Time to Death or Last Follow-Up (years) | Deceased |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 8.8 | DIPG | GBM | A | N/A | − | + | 5.38 | 8.56 | 0.84 | Y |
| 2 | F | 12.1 | DIPG | AA | D | Partial | N/A | N/A | 52.29 | 1.46 | 2.26 | Y |
| 3 | F | 22.2 | HGG | GBM | D | Partial | − | + | 0 | 0.31 | 1.53 | N |
| 4 | F | 12 | DIPG | AA | A | N/A | − | + | 0.5 | 5.03 | 1.21 | Y |
| 5 | F | 10.2 | HGG | AA | D | Partial | − | + | 0.11 | 0.23 | 11.19 | N |
| 6 | F | 17.9 | HGG | GBM | R | Partial | N/A | N/A | 0.49 | 1.58 | 3.47 | Y |
| 7 | M | 17.9 | HGG | AA | D | Partial | + | N/A | 80.21 | 1.4 | 1.18 | Y |
| 8 | F | 2.5 | HGG | AA | D | Total | − | N/A | 0 | 4.031 | 5.36 | N |
| 9# | M | 17.9 | HGG | AA | D | Partial | − | + | 0 | 0.84 | 2.98 | Y |
| 10# | M | 17.9 | HGG | GBM | R | Partial | + | + | 4.058 | 0.32 | 2.98 | Y |
| 11 | M | 5.9 | HGG | GBM | D | Partial | N/A | N/A | 0.44 | 1.7 | 2 | N |
| 12 | M | 13.8 | HGG | AA | D | Total | N/A | N/A | 0.36 | 1.18 | 0.27 | N |
| 13 | M | 12.1 | HGG | GBM | D | Total | N/A | N/A | 0 | 0.43 | 3.23 | N |
| 14 | M | 3.7 | HGG | AA | D | Partial | − | N/A | 0.14 | 0.21 | 0.26 | N |
| 15* | M | 12.1 | HGG | GBM | R | Total | N/A | N/A | 57.91 | 5.29 | 3.23 | N |
| 16* | M | 12.1 | HGG | GBM | R | Total | N/A | N/A | 71.41 | 1.73 | 3.23 | N |
| 17^ | F | 0.02 | HGG | GBM | R | Partial | − | N/A | N/A | N/A | 21.38 | N |
| 21 | F | 5.1 | DIPG | GBM | A | N/A | − | N/A | 3.58 | 5.22 | 1.16 | Y |
| 22 | F | 3.2 | DIPG | AA | A | N/A | N/A | N/A | 5.44 | 26.016 | 0.56 | Y |
| 23 | M | 5.4 | DIPG | GBM | A | N/A | + | − | 6.37 | 2.47 | 0.68 | Y |
| 24 | F | 12.5 | DIPG | GBM | A | N/A | + | − | 4.83 | 6.4 | 0.81 | Y |
| 25 | F | 12.1 | DIPG | GBM | A | N/A | N/A | N/A | 0.8 | 2.38 | 0.78 | Y |
| 26 | M | 2.8 | DIPG | GBM | A | N/A | − | − | 1.68 | 2.45 | 2.64 | Y |
| 27 | F | 3.4 | DIPG | GBM | A | N/A | − | − | 0.12 | 0.53 | 0.42 | Y |
| 36 | F | 4.7 | DIPG | GBM | A | N/A | − | − | 9.027 | 2.65 | 1.58 | Y |
| 37 | F | 4.8 | DIPG | GBM | A | N/A | − | − | 3.92 | 5.82 | 1.25 | Y |
| 45 | F | 9.6 | DIPG | GBM | A | N/A | − | + | 15.22 | 5.92 | 0.64 | Y |
| 46 | F | 4.2 | DIPG | GBM | A | N/A | − | − | 0.14 | 0.84 | 0.81 | Y |
| 47 | F | 4.6 | DIPG | GBM | A | N/A | + | − | 7.12 | 2.85 | 1.95 | Y |
| 28 | F | 6.9 | HGG | GBM | R | Total | + | − | 5.31 | 1.056 | 3.58 | Y |
| 29 | F | 20 | HGG | GBM | R | Partial | − | + | 0.33 | 0.18 | 0.83 | N |
| 30 | M | 6 | HGG | GBM | D | Partial | − | N/A | 8.45 | 19.36 | 0.08 | Y |
| 31 | F | 15.8 | HGG | AA | D | Partial | − | − | 0.3 | 1.44 | 2.25 | N |
| 32 | F | 13.8 | HGG | GBM | D | Total | + | − | 499.71 | 0.2 | 1 | Y |
| 33 | M | 15.2 | HGG | GBM | D | Partial | + | − | 2.99 | 3.21 | 1.42 | Y |
| 34 | F | 7.4 | HGG | AA | D | Partial | − | + | 0 | 3.13 | 1.58 | Y |
#,^,*, +: serial specimens from same patient; HGG, non-brainstem high-grade glioma; DIPG, diffuse intrinsic pontine glioma; GBM, glioblastoma multiforme; AA, anaplastic astrocytoma; RQ, relative quantification; Specimen timing—A, autopsy; D, diagnosis; R, relapse
Table 3.
hTERT and TERC Expression by Tumor Location
| Variable | Tumor | n | Mean | SD | RQ Median | Range |
|---|---|---|---|---|---|---|
| hTERT | DIPG | 15 | 7.76 | 12.95 | 4.83 | 0.12–52.3 |
| Non-brainstem HGG | 28 | 23.3 | 94.6 | 0.4 | 0–499 | |
| TERC | DIPG | 15 | 5.24 | 6.19 | 2.85 | 0.53–26.01 |
| Non-brainstem HGG | 28 | 1.97 | 3.79 | 0.84 | 0.15–19.34 |
HGG, high-grade glioma; DIPG, diffuse intrinsic pontine glioma; RQ, relative quantification; SD, standard deviation
Telomerase Activity by TRAP
Thirty-eight tumor (26 non-brainstem HGG; 12 DIPG) and five non-neoplastic matched brain specimens were analyzed for telomerase activity by TRAP (Supplemental Figure 1; Table 2). Twenty-five percent (3/12) of DIPG specimens and 42% (11/26) of non-brainstem HGG samples had telomerase activity, compared to none of the non-neoplastic control specimens. Ninety-two percent (11/12) of TRAP-positive non-brainstem HGG samples were Grade IV tumors. The lower level of telomerase-positive DIPG specimens may be secondary to technical issues related to time to processing of autopsy specimens and resulting inactivation of any telomerase enzyme present in tumor cells, especially in light of the lower correlation of TRAP results with hTERT mRNA levels in specimens obtained by autopsy versus surgery.
ALT Use
Telomere length of 30 (19 non-brainstem HGG; 11 DIPG) specimens was analyzed by Southern blot (Supplemental Figure 2; Table 2). Twenty-seven percent (3/11) of DIPG and 53% (10/19) non-brainstem HGG specimens showed evidence of ALT use. Interestingly, 21% (4/19) of non-brainstem HGG and 18% (2/11) of DIPG specimens used both ALT and telomerase. Specifically, 6/13 (46%) ALT-positive (non-brainstem and DIPG) tumors also showed evidence of telomerase use. Finally, 3/19 (16%) non-brainstem HGG and 2/11 (18%) DIPG tumors exhibited no telomere maintenance mechanism (i.e., demonstrated no telomerase activity on TRAP, no evidence of ALT on TRF analysis and had hTERT mRNA RQ ≤1).
H3F3A Gene Sequencing
Partial sequencing of H3F3A gene, which encodes histone 3 variant H3.3, was performed on 12 DIPG and 23 non-brainstem HGG specimens. Six (50%) DIPG specimens demonstrated a K27M mutation, but none had G34V/G34R mutations. Three (13%) K27M mutations, one (4.5%) G34V mutation and one (4.5%) G34R mutation were detected in non-brainstem HGG specimens. Patients with K27M mutations tended to be younger (7.4–11.6 years). Both tumor specimens with G34R/G34V mutations used ALT to maintain telomere length and were from young adult patients (20–22 years). When considering the DIPG specimens only, 3/6 of tumors with K27M mutations used ALT and 0/5 wild-type H3.3 tumors used ALT.
Association Between Telomere Maintenance Mechanism and Patient Outcome
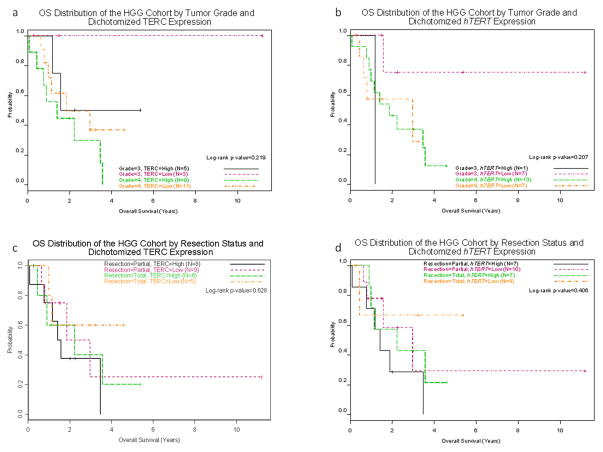
Overall median survival times for patients with DIPG and non-brainstem HGG were consistent with published literature. For the DIPG cohort, median PFS and OS were 0.63 years (range, 0.13–1.25 years) and 0.84 years (0.42–2.64 years), respectively. For the non-brainstem HGG cohort, median PFS and OS were 0.83 years (0.08–3.67 years) and 2.25 years (0.08–11.19 years), respectively. Kaplan-Meier plots display OS distributions for non-brainstem cohort stratified by hTERT level or TERC level and extent of resection or tumor grade (Figure 1a–d). Note that conversion of hTERT and TERC into dichotomous variables by splitting them into high and low levels at their median value in the HGG cohort for the Kaplan-Meier analysis lead to reduction in power and thus loss of statistical significance seen with the Cox analysis where these markers were treated as continuous variables. For the DIPG cohort, telomerase activity, ALT use, hTERT and TERC values were not associated with either PFS or OS in neither single- nor multivariable models (Supplementary Figure 3).
Figure 1.
Figure 1a–d. Overall survival distributions by Kaplan Meier analysis of non-brainstem HGG cohort stratified hTERT expression and by resection status or tumor grade. hTERT and TERC expression were dichotomized into high (> median value) and low (< median value) groups for these plots only. The Cox model results are based on the continuous version of this variable.
In the non-brainstem HGG cohort, extent of resection (gross-total vs. subtotal), age at diagnosis, and tumor grade (III vs. IV) were considered using single-variable models. Higher tumor grade was associated with a worse PFS that approached statistical significance (log-rank; P=0.072) in the single-variable models; no other variable demonstrated statistically significant association with PFS.
Among patients with non-brainstem HGG, 24 experienced progression and 18 had died at the time of analysis; therefore, our multivariable models could only accommodate two variables simultaneously. Given the well-established role of resection status and tumor grade in determining outcome, we chose to fit two-variable models that included either resection status or tumor grade and one telomere maintenance variable. Age at diagnosis was also considered but was not associated with PFS or OS in our cohort. In a Cox model incorporating hTERT and extent of resection, increased hTERT expression level was associated with worse OS (P=0.021) in children with non-brainstem HGG. Similarly, when modeled simultaneously with tumor grade, increased TERC expression level was associated with worse OS (P=0.016). Our data also contained some evidence for an association between higher TERC expression and worse OS based on a Cox model also accounting for extent of resection (P=0.079). No such associations with PFS were noted. Neither ALT use nor telomerase activity was associated with PFS or OS in single-variable or multiple-variable models for the non-brainstem HGG cohort.
DISCUSSION
This study is the first to describe telomerase expression and its prognostic significance in children with HGG, including DIPG. We report that children with HGG, particularly DIPG, have increased hTERT and TERC levels compared to normal controls. More importantly, increased hTERT mRNA and TERC RNA expression are associated with worse OS in children with non-brainstem HGG. These findings are similar to reports in other pediatric cancers, including neuroblastoma, ependymoma and Wilms tumor [35–37]. Neither telomerase activity nor ALT use was associated with PFS or OS in our non-brainstem HGG cohort, which may be due to lack of power to detect differences in these categorical variables. Similarly, no statistically significant association between patient outcome and telomere maintenance variables, including expression levels of the telomerase components, telomerase activity and ALT use, were identified in the DIPG cohort, which may have been because of the smaller sample size and short survival times typical of children with DIPG.
In the current study, 46% of HGG specimens (56% of non-brainstem HGG; 25% of DIPG tumors) used ALT to maintain telomere lengths, which builds on a prior study that included pediatric GBM tumors [23]. The prevalence and pattern of somatic H3F3A mutations in our cohort are generally consistent with prior studies [29,31]. Specifically, G34V/G34R mutations were not found in our DIPG cases. Our finding of K27M and G34V mutations in 13% and 9%, respectively, of non-brainstem HGG specimens is consistent with prior publications.[30] Our K27M mutation prevalence in DIPG of 50% (95% CI: 21.1%–78.9%) is comparable to the reported prevalence [30,31]. Although previous studies have identified the connection between ATRX/DAXX mutations, G34R/G34V mutations and ALT use, they have not fully evaluated any potential link between K27M mutations and ALT use. Our findings that all 3 DIPG patients with ALT use also had K27M mutations, suggests that the acquisition of a K27M mutation may be necessary but not sufficient for the induction of ALT use in DIPG. Further investigation of this relationship between K27M mutation and ALT use in a larger DIPG cohort is needed.
A novel observation in this study is that 21% of non-brainstem HGG and 18% of DIPG specimens showed both telomerase expression and ALT use. It is unclear whether these tumors are composed of separate cell populations (e.g., neural stem cell and tumor bulk populations) that utilize different telomere maintenance mechanisms or one cell population that uses both mechanisms. Further studies are required to fully understand this finding and its potential impact on response to therapies, especially in light of a prior publication that has suggested that the use of ALT in human glioma stem cells confers radiation resistance [38]. Finally, 16% of non-brainstem HGG and 18% of DIPG samples demonstrated no telomere maintenance mechanism (i.e., no telomerase activity, increased hTERT mRNA levels or ALT use), compared to 40% of adult GBM [39].
Normal somatic cells without telomerase activity have been shown to constitutively express TERC but not hTERT. However, TERC is known to be upregulated during tumorigenesis [14,40]. Our study analyzed several serial tumor samples from patients at different disease time points for evidence of known telomere maintenance mechanisms. Our data from a small subset of patients suggest that telomerase activity and hTERT mRNA levels, but not TERC levels, may be upregulated over the disease course, possibly in response to therapeutic interventions (data not shown), which is consistent with other publications that have noted that prior therapies can affect telomerase activity in other cell types [41–43]. Although our non-brainstem HGG cohort included diagnostic and relapsed tumor specimens from patients who had undergone radiotherapy and chemotherapy, a comparison of TERC or hTERT expression levels from diagnosis versus relapse specimens did not reveal statistically significant differences, albeit in a small, unpaired cohort.
Our results suggest that telomerase inhibition may be a promising therapeutic approach for children with HGG, including those with DIPG. Imetelstat (Geron, Menlo Park, CA) is a telomerase inhibitor that binds to the template region of TERC has demonstrated in vitro and in vivo activity in GBM [40, 41] and has undergone Phase I-II trials in adults with advanced solid tumors [44]. Since telomerase is active in a variety of pediatric cancers, imetelstat may be an effective drug for pediatric malignancies [45–48,36,49–52]. The Children’s Oncology Group has recently completed a Phase I trial of imetelstat in children with relapsed solid tumors. Elucidation of the telomere maintenance mechanisms used by pediatric HGG tumors is important in determining subpopulations of children with HGG who are most likely to benefit from treatment with telomerase inhibitors.
Supplementary Material
Representative telomerase enzyme activity by Telomere Repeat Amplification Protocol (TRAP). Tumor samples in lanes 3, 4, and 8 are DIPG; remains samples are non-brainstem HGG. Tumor samples in lanes 1–7 and 12 are telomerase-positive. Tumor samples in lanes 8–11 show no telomerase activity. Lane 13, positive control (PC). Lane 14, negative control (NC). Lane 15, PCR amplification control. IC, PCR internal control.
Representative telomeric restriction fragment (TRF) analysis by Southern blot. Tumor samples in lanes 1, 6, 9, and 10 are DIPG; remains samples are non-brainstem HGG. Samples in lanes 2–8 demonstrate Alternative Lengthening of Telomeres (ALT) activity. Tumor samples in lanes 9–12 are ALT-negative. Lane 14, positive control (PC) for ALT (osteosarcoma cell line Saos-2). Lane 15, negative control (NC) for ALT (histiocytic lymphoma U937 cell line). Ladder band sizes in lanes 1 and 16 labeled on far right in kilobase pairs (Kbp).
Overall survival distributions by Kaplan Meier analysis of DIPG cohort stratified hTERT expression, TERC expression, TRAP activity or ALT use.
Table 4.
Multivariable Cox Proportional Hazards Model Results for Non-Brainstem HGG Assessing Association with OS
| Variable | HR | P value |
|---|---|---|
| Extent of resection | 2.1 | 0.18 |
| hTERT | 1.3 | 0.021 |
| Extent of resection | 1.6 | 0.41 |
| TRAP | 2.2 | 0.14 |
| Extent of resection | 1.4 | 0.55 |
| TERC | 1.4 | 0.079 |
| Extent of resection | 1.0 | 1.0 |
| ALT use | 0.5 | 0.3 |
| Tumor grade | 2.6 | 0.21 |
| hTERT | 1.2 | 0.11 |
| Tumor grade | 2.9 | 0.17 |
| TRAP | 1.4 | 0.49 |
| Tumor grade | 5.0 | 0.042 |
| TERC | 1.6 | 0.016 |
| Tumor grade | 3.7 | 0.21 |
| ALT use | 0.6 | 0.46 |
HGG, high-grade glioma; OS, overall survival
Acknowledgments
We thank Kelly Verel, Nicole Reinholdt, Adrianne Gerkhardt, Marcela White, and Dr. Rachel Brody for their hard work on sample and data collection. We thank Rebecca Turner for her assistance with sample collection and regulatory support. We thank Dr. Mi-Ok Kim and Chunyan Liu for their statistical input. Rachid Drissi was supported by an Institutional and Translational Science Award, NIH/NCRR Grant number UL1RR026314. Kathleen Dorris was supported by grant National Institute of Environmental Health Sciences T32 ES010957. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Some tumor samples were obtained from patients treated on a Cincinnati Children’s Hospital Medical Center clinical trial that was supported by Genentech.
Footnotes
Conflict of Interest: None.
Some of these data were previously presented at a poster session at the 2011 Society of Neuro-Oncology Annual Meeting.
The authors declare that they have no conflict of interest.
References
- 1.Blackburn EH. Structure and function of telomeres. Nature. 1991;350 (6319):569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al. The RNA component of human telomerase. Science. 1995;269 (5228):1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 3.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17 (4):498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 4.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11 (5):1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266 (5193):2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 6.Broccoli D, Young JW, de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc Natl Acad Sci U S A. 1995;92 (20):9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33 (5):787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 8.Takakura M, Kyo S, Kanaya T, Tanaka M, Inoue M. Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res. 1998;58 (7):1558–1561. [PubMed] [Google Scholar]
- 9.Wu A, Ichihashi M, Ueda M. Correlation of the expression of human telomerase subunits with telomerase activity in normal skin and skin tumors. Cancer. 1999;86 (10):2038–2044. doi: 10.1002/(sici)1097-0142(19991115)86:10<2038::aid-cncr22>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 10.Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59 (3):551–557. [PubMed] [Google Scholar]
- 11.Poole JC, Andrews LG, Tollefsbol TO. Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT) Gene. 2001;269 (1–2):1–12. doi: 10.1016/s0378-1119(01)00440-1. [DOI] [PubMed] [Google Scholar]
- 12.Ducrest AL, Amacker M, Mathieu YD, Cuthbert AP, Trott DA, Newbold RF, Nabholz M, Lingner J. Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res. 2001;61 (20):7594–7602. [PubMed] [Google Scholar]
- 13.Avilion AA, Piatyszek MA, Gupta J, Shay JW, Bacchetti S, Greider CW. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996;56 (3):645–650. [PubMed] [Google Scholar]
- 14.Blasco MA, Rizen M, Greider CW, Hanahan D. Differential regulation of telomerase activity and telomerase RNA during multi-stage tumorigenesis. Nat Genet. 1996;12 (2):200–204. doi: 10.1038/ng0296-200. [DOI] [PubMed] [Google Scholar]
- 15.Langford LA, Piatyszek MA, Xu R, Schold SC, Jr, Shay JW. Telomerase activity in human brain tumours. Lancet. 1995;346 (8985):1267–1268. doi: 10.1016/s0140-6736(95)91865-5. [DOI] [PubMed] [Google Scholar]
- 16.Chong EY, Lam PY, Poon WS, Ng HK. Telomerase expression in gliomas including the nonastrocytic tumors. Hum Pathol. 1998;29 (6):599–603. doi: 10.1016/s0046-8177(98)80009-9. [DOI] [PubMed] [Google Scholar]
- 17.Le S, Zhu JJ, Anthony DC, Greider CW, Black PM. Telomerase activity in human gliomas. Neurosurgery. 1998;42 (5):1120–1124. doi: 10.1097/00006123-199805000-00099. discussion 1124–1125. [DOI] [PubMed] [Google Scholar]
- 18.Huang F, Kanno H, Yamamoto I, Lin Y, Kubota Y. Correlation of clinical features and telomerase activity in human gliomas. J Neurooncol. 1999;43 (2):137–142. doi: 10.1023/a:1006258817785. [DOI] [PubMed] [Google Scholar]
- 19.Boldrini L, Pistolesi S, Gisfredi S, Ursino S, Ali G, Pieracci N, Basolo F, Parenti G, Fontanini G. Telomerase activity and hTERT mRNA expression in glial tumors. Int J Oncol. 2006;28 (6):1555–1560. doi: 10.3892/ijo.28.6.1555. [DOI] [PubMed] [Google Scholar]
- 20.Maes L, Van Neste L, Van Damme K, Kalala JP, De Ridder L, Bekaert S, Cornelissen M. Relation between telomerase activity, hTERT and telomere length for intracranial tumours. Oncol Rep. 2007;18 (6):1571–1576. doi: 10.3892/or.18.6.1571. [DOI] [PubMed] [Google Scholar]
- 21.Bryan TM, Marusic L, Bacchetti S, Namba M, Reddel RR. The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum Mol Genet. 1997;6 (6):921–926. doi: 10.1093/hmg/6.6.921. [DOI] [PubMed] [Google Scholar]
- 22.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21 (4):598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 23.Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, Netto GJ, Epstein JI, Lotan TL, Westra WH, Shih Ie M, Iacobuzio-Donahue CA, Maitra A, Li QK, Eberhart CG, Taube JM, Rakheja D, Kurman RJ, Wu TC, Roden RB, Argani P, De Marzo AM, Terracciano L, Torbenson M, Meeker AK. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179 (4):1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald KL, McDonnell J, Muntoni A, Henson JD, Hegi ME, von Deimling A, Wheeler HR, Cook RJ, Biggs MT, Little NS, Robinson BG, Reddel RR, Royds JA. Presence of alternative lengthening of telomeres mechanism in patients with glioblastoma identifies a less aggressive tumor type with longer survival. J Neuropathol Exp Neurol. 2010;69 (7):729–736. doi: 10.1097/NEN.0b013e3181e576cf. [DOI] [PubMed] [Google Scholar]
- 25.Hakin-Smith V, Jellinek DA, Levy D, Carroll T, Teo M, Timperley WR, McKay MJ, Reddel RR, Royds JA. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361 (9360):836–838. doi: 10.1016/s0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- 26.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140 (5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24 (12):1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107 (32):14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482 (7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 30.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Baker SJ. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44 (3):251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, Bartels U, Albrecht S, Schwartzentruber J, Letourneau L, Bourgey M, Bourque G, Montpetit A, Bourret G, Lepage P, Fleming A, Lichter P, Kool M, von Deimling A, Sturm D, Korshunov A, Faury D, Jones DT, Majewski J, Pfister SM, Jabado N, Hawkins C. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124 (3):439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura M, Stone RC, Hunt SC, Skurnick J, Lu X, Cao X, Harley CB, Aviv A. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat Protoc. 2010;5 (9):1596–1607. doi: 10.1038/nprot.2010.124. [DOI] [PubMed] [Google Scholar]
- 33.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3 (6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 34.Holt SENJ, Wright WE, Shay JW. Comparison of the telomeric repeat amplification protocol (TRAP) to the new TRAPeze Telomerase Detection Kit. Methods in Cell Science. 1996;18 (3):237–248. [Google Scholar]
- 35.Maitra A, Yashima K, Rathi A, Timmons CF, Rogers BB, Shay JW, Gazdar AF. The RNA component of telomerase as a marker of biologic potential and clinical outcome in childhood neuroblastic tumors. Cancer. 1999;85 (3):741–749. [PubMed] [Google Scholar]
- 36.Dome JS, Bockhold CA, Li SM, Baker SD, Green DM, Perlman EJ, Hill DA, Breslow NE. High telomerase RNA expression level is an adverse prognostic factor for favorable-histology Wilms’ tumor. J Clin Oncol. 2005;23 (36):9138–9145. doi: 10.1200/JCO.2005.00.562. [DOI] [PubMed] [Google Scholar]
- 37.Choi LM, Kim NW, Zuo JJ, Gerbing R, Stram D, Lukens JN, Matthay KK, Seeger RC, Reynolds CP. Telomerase activity by TRAP assay and telomerase RNA (hTR) expression are predictive of outcome in neuroblastoma. Medical and pediatric oncology. 2000;35 (6):647–650. doi: 10.1002/1096-911x(20001201)35:6<647::aid-mpo35>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 38.Silvestre DC, Pineda JR, Hoffschir F, Studler JM, Mouthon MA, Pflumio F, Junier MP, Chneiweiss H, Boussin FD. Alternative lengthening of telomeres in human glioma stem cells. Stem cells. 2011;29 (3):440–451. doi: 10.1002/stem.600. [DOI] [PubMed] [Google Scholar]
- 39.Royds JA, Al Nadaf S, Wiles AK, Chen YJ, Ahn A, Shaw A, Bowie S, Lam F, Baguley BC, Braithwaite AW, MacFarlane MR, Hung NA, Slatter TL. The CDKN2A G500 allele is more frequent in GBM patients with no defined telomere maintenance mechanism tumors and is associated with poorer survival. PLoS One. 2011;6 (10):e26737. doi: 10.1371/journal.pone.0026737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol Cell Biol. 1999;19 (6):3989–3997. doi: 10.1128/mcb.19.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natarajan M, Mohan S, Konopinski R, Otto RA, Herman TS. Induced telomerase activity in primary aortic endothelial cells by low-LET gamma-radiation is mediated through NF-kappaB activation. The British journal of radiology. 2008;81 (969):711–720. doi: 10.1259/bjr/57867919. [DOI] [PubMed] [Google Scholar]
- 42.Ram R, Uziel O, Eldan O, Fenig E, Beery E, Lichtenberg S, Nordenberg Y, Lahav M. Ionizing radiation up-regulates telomerase activity in cancer cell lines by post-translational mechanism via ras/phosphatidylinositol 3-kinase/Akt pathway. Clin Cancer Res. 2009;15 (3):914–923. doi: 10.1158/1078-0432.CCR-08-0792. [DOI] [PubMed] [Google Scholar]
- 43.Akiyama M, Ozaki K, Kawano T, Yamada O, Kawauchi K, Ida H, Yamada H. Telomerase activation as a repair response to radiation-induced DNA damage in Y79 retinoblastoma cells. Cancer letters. 2013;340 (1):82–87. doi: 10.1016/j.canlet.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Asai A, Oshima Y, Yamamoto Y, Uochi TA, Kusaka H, Akinaga S, Yamashita Y, Pongracz K, Pruzan R, Wunder E, Piatyszek M, Li S, Chin AC, Harley CB, Gryaznov S. A novel telomerase template antagonist (GRN163) as a potential anticancer agent. Cancer Res. 2003;63 (14):3931–3939. [PubMed] [Google Scholar]
- 45.Tabori U, Ma J, Carter M, Zielenska M, Rutka J, Bouffet E, Bartels U, Malkin D, Hawkins C. Human telomere reverse transcriptase expression predicts progression and survival in pediatric intracranial ependymoma. J Clin Oncol. 2006;24 (10):1522–1528. doi: 10.1200/JCO.2005.04.2127. [DOI] [PubMed] [Google Scholar]
- 46.Hiyama E, Yamaoka H, Matsunaga T, Hayashi Y, Ando H, Suita S, Horie H, Kaneko M, Sasaki F, Hashizume K, Nakagawara A, Ohnuma N, Yokoyama T. High expression of telomerase is an independent prognostic indicator of poor outcome in hepatoblastoma. Br J Cancer. 2004;91 (5):972–979. doi: 10.1038/sj.bjc.6602054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poremba C, Hero B, Heine B, Scheel C, Schaefer KL, Christiansen H, Berthold F, Kneif S, Stein H, Juergens H, Boecker W, Dockhorn-Dworniczak B. Telomerase is a strong indicator for assessing the proneness to progression in neuroblastomas. Medical and pediatric oncology. 2000;35 (6):651–655. doi: 10.1002/1096-911x(20001201)35:6<651::aid-mpo36>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 48.Dome JS, Chung S, Bergemann T, Umbricht CB, Saji M, Carey LA, Grundy PE, Perlman EJ, Breslow NE, Sukumar S. High telomerase reverse transcriptase (hTERT) messenger RNA level correlates with tumor recurrence in patients with favorable histology Wilms’ tumor. Cancer Res. 1999;59 (17):4301–4307. [PubMed] [Google Scholar]
- 49.Poremba C, Scheel C, Hero B, Christiansen H, Schaefer KL, Nakayama J, Berthold F, Juergens H, Boecker W, Dockhorn-Dworniczak B. Telomerase activity and telomerase subunits gene expression patterns in neuroblastoma: a molecular and immunohistochemical study establishing prognostic tools for fresh-frozen and paraffin-embedded tissues. J Clin Oncol. 2000;18 (13):2582–2592. doi: 10.1200/JCO.2000.18.13.2582. [DOI] [PubMed] [Google Scholar]
- 50.Tabori U, Dome JS. Telomere biology of pediatric cancer. Cancer Invest. 2007;25 (3):197–208. doi: 10.1080/07357900701208683. [DOI] [PubMed] [Google Scholar]
- 51.Didiano D, Shalaby T, Lang D, Grotzer MA. Telomere maintenance in childhood primitive neuroectodermal brain tumors. Neuro Oncol. 2004;6 (1):1–8. doi: 10.1215/S1152851703000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanders RP, Drissi R, Billups CA, Daw NC, Valentine MB, Dome JS. Telomerase expression predicts unfavorable outcome in osteosarcoma. J Clin Oncol. 2004;22 (18):3790–3797. doi: 10.1200/JCO.2004.03.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative telomerase enzyme activity by Telomere Repeat Amplification Protocol (TRAP). Tumor samples in lanes 3, 4, and 8 are DIPG; remains samples are non-brainstem HGG. Tumor samples in lanes 1–7 and 12 are telomerase-positive. Tumor samples in lanes 8–11 show no telomerase activity. Lane 13, positive control (PC). Lane 14, negative control (NC). Lane 15, PCR amplification control. IC, PCR internal control.
Representative telomeric restriction fragment (TRF) analysis by Southern blot. Tumor samples in lanes 1, 6, 9, and 10 are DIPG; remains samples are non-brainstem HGG. Samples in lanes 2–8 demonstrate Alternative Lengthening of Telomeres (ALT) activity. Tumor samples in lanes 9–12 are ALT-negative. Lane 14, positive control (PC) for ALT (osteosarcoma cell line Saos-2). Lane 15, negative control (NC) for ALT (histiocytic lymphoma U937 cell line). Ladder band sizes in lanes 1 and 16 labeled on far right in kilobase pairs (Kbp).
Overall survival distributions by Kaplan Meier analysis of DIPG cohort stratified hTERT expression, TERC expression, TRAP activity or ALT use.