Abstract
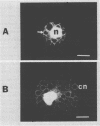
The kinetic complexity of chloroplast DNA isolated from the chromophytic alga Olisthodiscus luteus has been determined. Using optical reassociation techniques, it was shown that the plastid DNA of this alga reacted as a single component with a second order rate constant of 4.1 molar−1 and second−1 (Cot½ 0.24 molar second) under conditions equivalent to 180 millimolar Na+ and 60°C. Given the 92 × 105 dalton complexity calculated for this chloroplast genome, an Olisthodiscus cell contains 650 plastome copies. Although this complement remains constant throughout the growth cycle of the organism, the ploidy level of an individual chloroplast shows significant plasticity and is dependent upon the number of chloroplasts present per cell. Experiments with the DNA fluorochrome Hoechst dye 33258 (bisbenzimide) demonstrate that plastids isolated from all phases of cell growth each possess a ring-shaped nucleoid containing detectable DNA. Olisthodiscus chloroplast DNA showed no sequence mismatch when thermal denaturation profiles of reassociated chloroplast DNA were examined, thus all plastome copies are essentially identical. Finally, reassociation studies demonstrated that no foldback (short inverted repeat) sequences were present in the plastid genome although significant hairpin loop structures were observed in control nuclear DNA samples.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldrich J., Cattolico R. A. Isolation and Characterization of Chloroplast DNA from the Marine Chromophyte, Olisthodiscus luteus: Electron Microscopic Visualization of Isomeric Molecular Forms. Plant Physiol. 1981 Sep;68(3):641–647. doi: 10.1104/pp.68.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook J. R., Kolodner R., Bogorad L. Zea mays chloroplast ribosomal RNA genes are part of a 22,000 base pair inverted repeat. Cell. 1977 Aug;11(4):739–749. doi: 10.1016/0092-8674(77)90288-4. [DOI] [PubMed] [Google Scholar]
- Bendich A. J., Anderson R. S. Characterization of families of repeated DNA sequences from four vascular plants. Biochemistry. 1977 Oct 18;16(21):4655–4663. doi: 10.1021/bi00640a020. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Cattolico R. A., Boothroyd J. C., Gibbs S. P. Synchronous Growth and Plastid Replication in the Naturally Wall-less Alga Olisthodiscus luteus. Plant Physiol. 1976 Apr;57(4):497–503. doi: 10.1104/pp.57.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelm B. K., Hoben P. J., Hallick R. B. Cellular content of chloroplast DNA and chloroplast ribosomal RNA genes in Euglena gracilis during chloroplast development. Biochemistry. 1977 Feb 22;16(4):782–786. doi: 10.1021/bi00623a033. [DOI] [PubMed] [Google Scholar]
- Christiansen C., Christiansen G., Bak A. L. The influence of glucosylation on the renaturation rate of T4 phage DNA. Biochem Biophys Res Commun. 1973 Jun 19;52(4):1426–1433. doi: 10.1016/0006-291x(73)90660-8. [DOI] [PubMed] [Google Scholar]
- Coleman A. W. Use of the fluorochrome 4'6-diamidino-2-phenylindole in genetic and developmental studies of chloroplast DNA. J Cell Biol. 1979 Jul;82(1):299–305. doi: 10.1083/jcb.82.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovin T. M. Recognition mechanisms of DNA-specific enzymes. Annu Rev Biochem. 1976;45:889–920. doi: 10.1146/annurev.bi.45.070176.004325. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Calvo J. M. Alternative secondary structures of leader RNAs and the regulation of the trp, phe, his, thr, and leu operons. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6186–6190. doi: 10.1073/pnas.76.12.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittel M. D., Black C. H., Sandine W. E., Fraser D. K. Use of normal probability paper in determining thermal melting values of deoxyribonucleic acid. Can J Microbiol. 1968 Mar;14(3):239–245. doi: 10.1139/m68-040. [DOI] [PubMed] [Google Scholar]
- Pellegrini M. Three-dimensional reconstruction of organelles in Euglena gracilis Z. I. Qualitative and quantitative changes of chloroplasts and mitochondrial reticulum in synchronous photoautotrophic culture. J Cell Sci. 1980 Jun;43:137–166. doi: 10.1242/jcs.43.1.137. [DOI] [PubMed] [Google Scholar]
- Rawson J. R., Boerma C. Influence of growth conditions upon the number of chloroplast DNA molecules in Euglena gracilis. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2401–2404. doi: 10.1073/pnas.73.7.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J. D. Restriction fragments from Chlamydomonas chloroplast DNA. Methods Enzymol. 1980;65(1):785–795. doi: 10.1016/s0076-6879(80)65073-3. [DOI] [PubMed] [Google Scholar]
- Siu C. H., Chiang K. S., Swift H. Characterization of cytoplasmic and nuclear genomes in the colorless alga Polytoma. V. Molecular structure and heterogenity of leucoplast DNA. J Mol Biol. 1975 Oct 25;98(2):369–391. doi: 10.1016/s0022-2836(75)80125-2. [DOI] [PubMed] [Google Scholar]
- Walbot V. The dimorphic chloroplasts of the C4 plant Panicum maximum contain identical genomes. Cell. 1977 Aug;11(4):729–737. doi: 10.1016/0092-8674(77)90287-2. [DOI] [PubMed] [Google Scholar]