Summary
Circadian clocks play a major role in orchestrating daily physiology, and their disruption can evoke metabolic diseases such as fatty liver and obesity. To study the role of circadian clocks in lipid homeostasis, we performed an extensive lipidomic analysis of liver tissues from wild type and clock-disrupted mice, fed either ad libitum or night fed. To our surprise, a similar fraction of lipids (~17%) oscillated in both mouse strains, most notably triglycerides, but with completely different phases. Moreover, several master lipid regulators (e.g. PPARα) and enzymes involved in triglyceride metabolism retained their circadian expression in clock-disrupted mice. Nighttime restricted feeding shifted the phase of triglyceride accumulation and resulted in ~50% decrease in hepatic triglyceride levels in wild type mice. Our findings suggest that circadian clocks and feeding time dictate the phase and levels of hepatic triglyceride accumulation, however oscillations in triglycerides can persist in the absence of a functional clock.
Introduction
Circadian clocks play a principal role in coordinating our daily physiology and metabolism. Animal studies and epidemiological evidence suggest that disturbance of circadian rhythms through environmental and genetic effects can lead to metabolic diseases such as hyperlipidemia, fatty liver and obesity (Asher and Schibler, 2011; Bass, 2012; Froy, 2010; Green et al., 2008). These observations highlight the central role of circadian regulation in lipid homeostasis. Dyslipidemia and obesity are associated with high morbidity and mortality rates, hence elucidating the mechanism involved in temporal regulation of lipids are of great interest.
The mammalian circadian clock comprise of a master pacemaker, located in the brain that synchronizes subsidiary peripheral oscillators, present in virtually all cells of the body. The master clock is entrained by daily light-dark cycles, whereas feeding time appears to be the dominant timing cue (Zeitgeber) for peripheral clocks. Both the master and peripheral clocks tick in a self-sustained and cell-autonomous fashion. The currently held molecular model for the generation of circadian rhythms is based on interlocked negative transcription feedback loops that drive daily oscillations of core clock and clock-controlled genes (Brown et al., 2012; Dibner et al., 2010; Feng and Lazar, 2012). Briefly, BMAL1/CLOCK drive the expression of Period (i.e. Per1, Per2 and Per3) and Cryptochrome (i.e. Cry1 and Cry2) genes. In turn, PER and CRY proteins accumulate and repress the transcription of their own genes. An additional essential feedback loop, involves the orphan nuclear receptors of the REV-ERB and ROR families. BMAL1 activates Rev-erb transcription, which in turn suppresses Bmal1 expression (Bugge et al., 2012; Cho et al., 2012; Preitner et al., 2002; Solt et al., 2012).
Extensive transcriptome profiling performed throughout the day in liver and additional peripheral organs has demonstrated the pervasive circadian control of physiology and metabolism (Akhtar et al., 2002; McCarthy et al., 2007; Panda et al., 2002; Storch et al., 2002; Vollmers et al., 2009). These studies have revealed that a substantial fraction (~10%) of all liver mRNAs are expressed in a rhythmic fashion, many of them play a role in metabolic processes, including cholesterol and lipid metabolism. Several enzymes participating in lipid biosynthesis and catabolism are expressed in a daily manner (e.g. cytochrome P450s, HMGCoA reductase, Lipin), (Panda et al., 2002). Additional studies have shown diurnal regulation of triglyceride and cholesterol levels in plasma (Hussain and Pan, 2009). Consequently, various genetic mouse models for disrupted clock exhibit impaired lipid metabolism. Clock mutant and Bmal1 knockout mice develop hyperlipidemia, and hepatic steatosis (Shimba et al., 2011; Turek et al., 2005). PER2-deficient mice display altered lipid metabolism (Grimaldi et al., 2010) and ablation of REV-ERBs can lead to hepatic steatosis (Bugge et al., 2012).
Hitherto, circadian research in mammals has largely been focused on dissecting transcription expression profiles of core clock and output genes including genes involved in metabolism and cellular homeostasis. Though these gene expression profiles have hinted that many metabolic pathways and their products are oscillating during the day, direct measurements of metabolites throughout the day are still in their infancy. Ueda and colleagues have quantified the spectra of hundreds of metabolites throughout the day, both in mouse and human plasma samples (Kasukawa et al., 2012; Minami et al., 2009). They successfully established a metabolite timetable method to determine internal body time using these profiles. These studies have primarily focused on measuring internal body time by blood metabolomics and to a lesser extent aimed at identifying these metabolites, their metabolic pathways and circadian clocks dependency. Recent studies performed with human (i.e. blood plasma and saliva) and mouse samples (i.e. liver) identified and measured the levels of about 300 named metabolites throughout the day (Dallmann et al., 2012; Eckel-Mahan et al., 2012). These analyses screened for a variety of primary metabolites (e.g. amino acids, carbohydrates, nucleotides and lipids). Remarkably, a large fraction of oscillating metabolites identified by these recent reports were lipids.
In this study, we extensively examined the circadian changes in lipid abundance in mouse liver and dissected its clock/feeding dependency. To this aim, we performed a temporal and quantitative lipidomic analysis of livers from wild type (WT) and clock-disrupted mice (i.e. Per1/2 null mice), fed either ad libitum or exclusively during the night. We found that a similar fraction of lipids (~17%) oscillated in both mouse strains, most notably triglycerides (TAG), but with completely different phases. Moreover, several master lipid regulators and enzymes involved in TAG metabolism retained their circadian expression in clock-disrupted mice. Feeding-time had a prominent effect on the phase and levels of hepatic TAG in both WT and Per1/2 null mice. Remarkably, upon nighttime restricted feeding, WT mice exhibited a sharp decrease (~50%) in hepatic TAG levels. Our findings suggest that circadian clocks and feeding time dictate the phase and levels of hepatic TAG accumulation, however oscillations in TAG can persist in the absence of a functional clock.
Results
Mouse liver lipidome
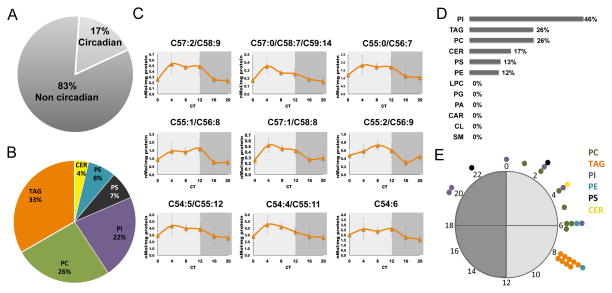
To obtain a temporal depiction of the circadian changes in lipid abundance in mouse liver we performed a wide lipidomic analysis. WT mice were fed ad libitum and housed under a 12 h light dark regimen for several consecutive days. Throughout the last day of the experiment, animals were maintained in constant darkness and were sacrificed every 4 h. Livers were harvested and lipids were identified and quantified by shotgun lipidomics (Han et al., 2012). Altogether 159 lipids were measured (Table S1A). These include different triglycerides (TAG), phospholipids (i.e. phosphatidylinositol (PI), phosphatidylcholine (PC), lysophosphatidylcholine (LPC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidic acid (PA) and phosphatidylglycerol (PG)), sphingolipids (i.e. ceramide (CER) and sphingomyelin (SM)), cardiolipin (CL) and acyl carnitine (CAR). A nonparametric algorithm, JTK_CYCLE (Hughes et al., 2010), was used to identify lipids that display circadian rhythmicity. Out of the 159 lipids, 27 (~17%) were identified as oscillating with P value <0.05 (Figure 1A, Figure S1 and Table S1B). Among the 27 oscillating lipids, the majority consisted of TAG species (~33%), (Figures 1B and 1C). When dissected based on their types, it appeared that oscillating lipid species were highly abundant among PI (46%), and enriched among TAG (26%) and PC (26%), (Figure 1D). Analysis of the phase distribution of the different oscillating lipids revealed that the vast majority of them peaked during the subjective light phase. Namely, 23 out of the 27 reached their zenith levels between Circadian Time (CT) 1 to CT8 (Figure 1E). Hence, we concluded that oscillating lipids mostly consist of TAG species and are predominantly temporally gated to reach their zenith levels during the subjective light phase.
Figure 1. Analysis of mouse liver lipidome.
A., The percentage of lipids that were found to exhibit a circadian pattern of accumulation in livers of WT mice based on JTK_CYCLE analysis (6 time points, n=4 for each, P value <0.05). Out of the 159 measured and identified, 27 lipids exhibited circadian pattern of accumulation. B., Oscillating lipids species distributed according to their types. C., Accumulation profiles of oscillating TAG presented as mean +/− STDEV. D., The percentage of oscillating lipid species within each lipid type. E., Day-time distribution of peak phases of oscillating lipids. TAG in orange, PC in green, PI in purple, PS in black, PE in light blue, and CER in yellow. Circadian Time (CT), CT0 is the time the light used to be turned on and CT12 is the time light used to be turned off. Dark gray represents the subjective night and light gray the subjective day. See also Figure S1.
Circadian expression of enzymes participating in hepatic TAG metabolism
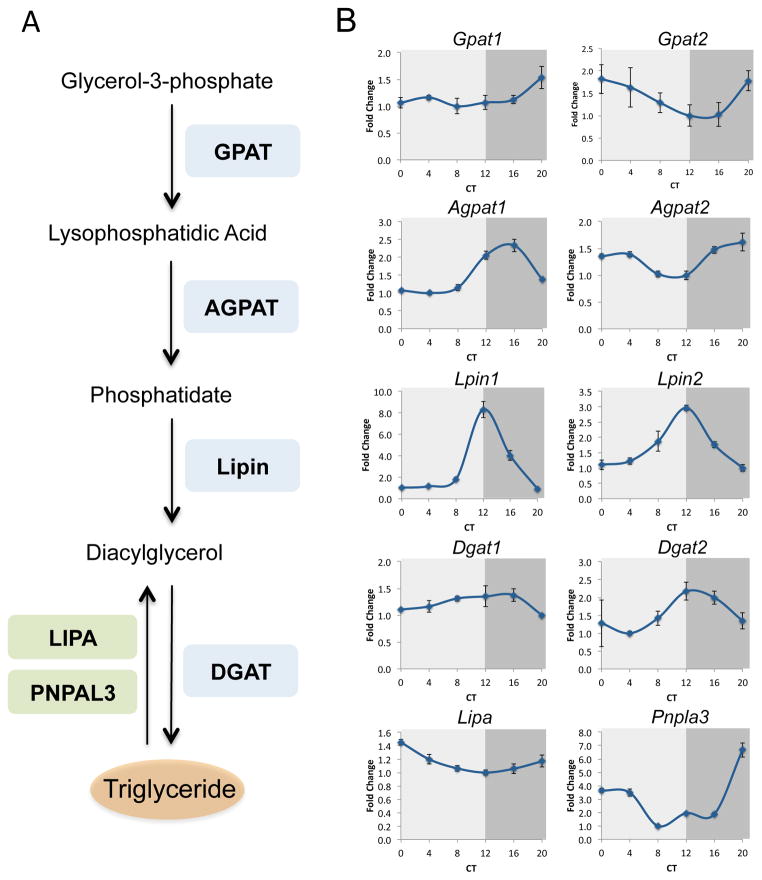
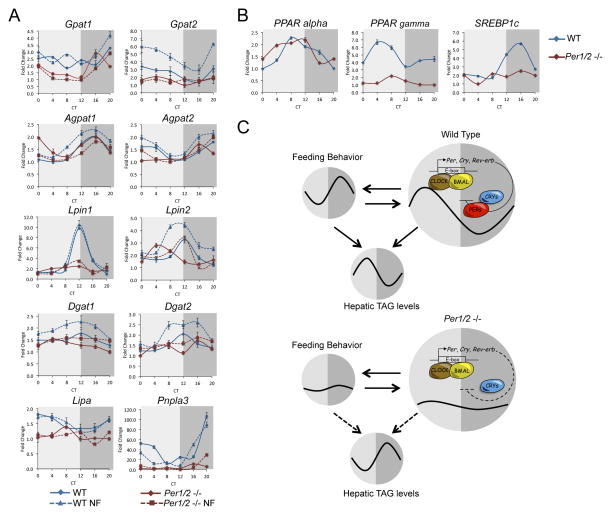
Hepatocytes harbor the ability to synthesize, store and catabolize TAG. A prominent fraction (i.e. 33%) of oscillating lipids consisted of TAG species (Figure 1B). Strikingly, all oscillating TAG peaked around CT8 and dipped around CT20 with amplitude of about 2 fold (Figure 1C and 1E), suggesting that their accumulation in the liver is timely controlled. To corroborate these findings, we examined the circadian expression profile of multiple enzymes that participate in TAG metabolism in WT mice. The glycerol-3-phosphate pathway is the predominant biosynthesis pathway for TAG in the liver and consists of several successive steps (Figure 2A), (Takeuchi and Reue, 2009). The first key and rate-limiting step is the acylation of glycerol 3-phosphate by glycerol-3-phosphate acyltransferase (GPAT) enzymes to synthesize lysophosphatidic acid (LPA). Subsequently, an additional fatty acid is transferred to LPA by the family of 1-acylglycerol-3-phosphate acyltransferase (AGPAT) enzymes to produce phosphatidate (PA). Diacylglycerol (DAG) is synthesized from PA by the Lipin family of proteins, through their phosphatidate phosphatase-1 (PAP) enzymatic activity. Finally, DAG is converted to TAG through the action of diacylglycerol acyltransferase (DGAT) enzymes.
Figure 2. Circadian regulation of hepatocytes triglyceride metabolism.
A., Schematic depiction of the triglyceride biosynthesis pathway (glycerol-3-phosphate pathway) and triglyceride catabolism. Glycerol-3-phosphate acyltransferase (GPAT), 1-acylglycerol-3-phosphate acyltransferase (AGPAT), diacylglycerol acyltransferase (DGAT), Patatin-like phospholipase domain containing 3 (PNPLA3), and Lysosomal acid lipase (LIPA). B., WT mice were sacrificed under constant darkness at 4 h intervals throughout the day. Total RNA was prepared from liver and mRNA expression levels were determined by quantitative real-time PCR and presented as fold change relative to the lowest value. Data are presented as mean +/− STDEV, with a mix of 4 animals per time point. Circadian Time (CT).
WT mice were sacrificed under constant darkness at 4 h intervals throughout the day, livers were harvested and RNA was prepared. The expression level of the different enzymes was quantified by real-time PCR. Remarkably, we identified circadian accumulation pattern for multiple enzymes in the glycerol-3-phosphate pathway, covering all subsequent steps involved in TAG biosynthesis (Figure 2B). While Gpat1 mRNA levels were relatively constant throughout the day, Gpat2 exhibited shallow circadian oscillations with zenith levels around CT0 and nadir levels around CT12. Next, both Agpat1 and Agpat2 transcripts accumulated in a circadian manner, albeit with a phase difference, the former peaked at CT16 while the latter at CT0. Subsequently, the mRNA expression profile of both Lpin1 and Lpin2 was circadian with zenith levels at CT12 and nadir levels at CT0. Finally, the transcript levels of Dgat2 but not Dgat1 were oscillating with a similar phase as Lpin1 and Lpin2. These findings demonstrated that the glycerol-3-phosphate pathway is expressed in a circadian manner.
Hepatic TAG catabolism occurs through the activity of multiple lipases, releasing one fatty acid at a time, producing diacylglycerols, and eventually glycerol (Figure 2A), (Quiroga and Lehner, 2012). To examine the circadian regulation of TAG catabolism in the liver, we analyzed the expression levels of two major haptic lipases, the Patatin-like phospholipase domain containing 3 (Pnpla3) and the Lysosomal acid lipase (Lipa), (Quiroga and Lehner, 2012). The mRNA expression levels of both Pnpla3 and to a much lesser extent Lipa were circadian, with nadir levels around CT12 (Figure 2B). Taken together, our data evinced that multiple hepatic enzymes that participate in TAG homeostasis oscillate in a circadian manner. Conceivably, these enzymes function in concert to generate the above-described circadian oscillation of TAG in the liver.
Liver lipidome analysis of Per 1/2 null mice
To address the clock-dependency of the above-illustrated circadian profile of lipid accumulation in mouse liver, we performed a lipidomic analysis on liver samples obtained from Per1/2 −/− mice. Mice lacking both PER1 and PER2 exhibit arrhythmic locomotor activity under constant darkness and circadian expression of core clock and clock-controlled genes is largely diminished (Zheng et al., 2001). Consistently, expression levels of core clock genes (i.e. Bmal1, Clock, Cry, and Rev-erbα) were relatively constant throughout the day and as expected, Per1 and Per2 mRNA levels were undetectable in these mice (Figure S2).
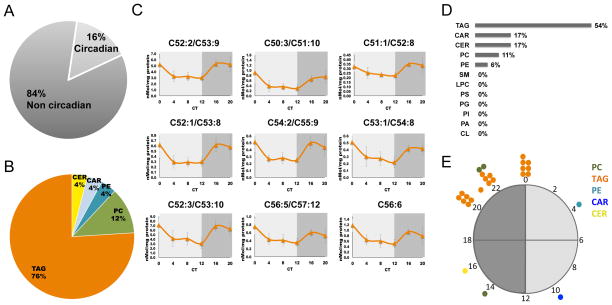
Per1/2 null mice were fed ad libitum, and sacrificed under constant darkness, at 4 h intervals throughout the day. Livers were harvested and lipids were identified and quantified by shotgun lipidomics (Table S2A). Analysis based on the JTK_CYCLE algorithm evinced that out of the 159 classified and quantified lipids, 25 (~16%) were identified as oscillating with P value <0.05 (Figure 3A, Figure S3 and Table S2B). The preponderance of them consisted of TAG (~76%), (Figures 3B, 3C and Figure S3). Moreover, when dissected based on their types, oscillating lipid species were highly enriched among the TAG (~54%), (Figure 3D). Phase distribution analysis revealed a mirror image to that obtained for WT samples (compare Figure 1E with Figure 3E). Thus, all oscillating TAG reached their peak levels during the late night, more specifically between CT20 to CT0 (Figures 3C, 3E and Figure S3). The observed oscillations in lipids accumulation, most notably TAG in the absence of PER1/2, suggest that their circadian accumulation might be clock-independent.
Figure 3. Liver lipidome analysis of Per 1/2 null mice.
A., The percentage of lipids that were found to exhibit a circadian pattern of accumulation in livers of PER1/2 −/− mice based on JTK_CYCLE analysis (6 time points, n=4 for each, P value <0.05). Out of the 159 measured and identified lipids, 25 lipids exhibited circadian pattern of accumulation. B., Oscillating lipids species distributed according to their types. C., Accumulation profiles of representative oscillating TAG presented as mean +/− STDEV. Circadian Time (CT). D., The percentage of the oscillating lipid species within each lipid type. E., Day-time distribution of peak phases of oscillating lipids. TAG in orange, PC in green, PE in light blue, CAR in blue, and CER in yellow. See also Figure S2 and Figure S3.
The effect of feeding-time on circadian TAG accumulation in the liver
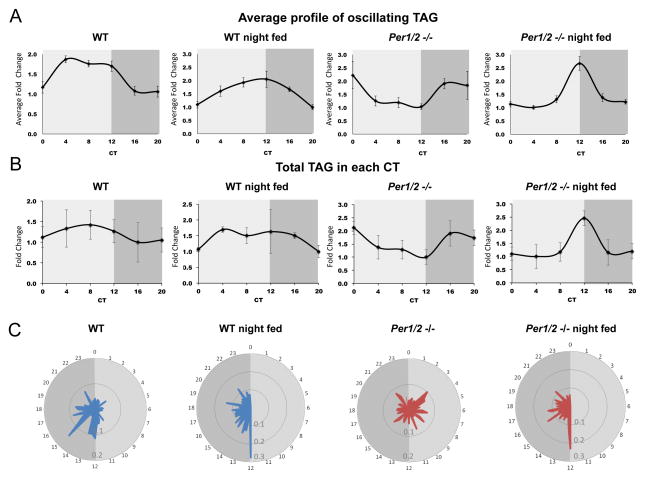
Food is a major source for lipids in general and triglycerides in particular. To examine the direct effect of food ingestion on the accumulation pattern of TAG in the liver, we performed a limited lipidomic analysis, this time measuring only TAG species in WT and Per1/2 null mice, fed exclusively during the night, namely between CT12 and CT0 (Table S3A and Table S4A). JTK_CYCLE based analysis showed that out of 35 measured TAG species, 16 were oscillating in WT and 18 in Per1/2 null mice (~ 46 and ~51% respectively), (P value <0.05), (Figure S4 and Tables S3B and S4B). All oscillating TAG species peaked around CT12 in both mouse strains (Figure S4). However, while WT mice exhibited a gradual buildup and decline of TAG with zenith levels at CT12, Per1/2 −/− mice displayed a prominent and sharp peak in TAG accumulation at CT12 (Figure 4A and Figure S4). The surge in TAG accumulation in night fed animals was probably due to lack of food ingestion prior to CT12 as mice were sacrificed shortly before food was provided. The prominent phase shift in TAG accumulation from around CT20 in Per1/2 −/− mice, and CT8 in WT mice fed ad libitum, to CT12 upon night feeding implied that feeding-fasting cycles can shape the phase of TAG accumulation in mouse liver.
Figure 4. Analysis of feeding and clock-dependency of circadian hepatic TAG accumulation.
A., Profile of oscillating TAG identified based on JTK_CYCLE analysis for WT and Per1/2 −/− mice fed either ad libitum or exclusively during the night. Data are presented as mean +/− STDEV of fold induction for all oscillating TAG relative to the lowest point for each one. B., Accumulation profiles of total TAG levels in livers of WT and Per1/2 −/− mice fed either ad libitum or exclusively during the night. Data are presented as mean +/− STDEV of fold induction for total TAG levels relative to the lowest point. The corresponding P values based on JTK_CYCLE analysis were 0.31, 0.03, 0.02, 0.02, respectively. C., A radar plot presenting the circadian food consumption of WT and Per1/2 −/− mice fed either ad libitum or exclusively during the night. The time of day is indicated in hours, and y-axis shows the food consumption in grams. See also Figures S4, S5 and S7, and Table S6.
Analysis of feeding and clock-dependency of circadian hepatic TAG accumulation
Hitherto, we obtained a temporal depiction of circadian hepatic TAG accumulation in WT and Per1/2 −/− mice, fed either ad libitum or exclusively during the night (Figure 4A). Altogether, we identified circadian oscillations in 9, 16, 19 and 18 out of 35 quantified TAG species, which peaked at ~ZT8, ~ZT12, ~ZT20 and ~ZT12, in WT fed ad libitum, WT night fed, Per1/2 −/− fed ad libitum and Per1/2 −/− night fed mice, respectively. By mass this corresponded to 22%, 41%, 61% and 53% of total TAG liver mass, respectively. Hence, these changes were also reflected to some extent in circadian oscillations of total hepatic TAG levels, particularly in the case of WT night fed, Per1/2 −/− fed ad libitum and Per1/2 −/− night fed mice (P value <0.05 based on the JTK_CYCLE analysis), (Figure 4B).
The ability of feeding-time to shift the phase of TAG accumulation raised the question whether the striking difference in the phase of TAG oscillations in WT versus Per1/2 −/− mice fed ad libitum might be the outcome of different daily eating habits of the two genotypes. Therefore, we temporally monitored the food intake of WT and Per1/2 null mice. WT mice ingested ~70% of their total daily food intake during the subjective dark phase, and ~30% during the subjective light phase. Whereas Per1/2 −/− mice largely differed in their eating habits and ingested relatively equal amounts of food throughout the day (Figure 4C and Figure S5). Altogether, both WT and Per1/2 −/− consumed a similar amount of food throughout the day (Figure 6C). Thus, though Per1/2 null mice differed in their eating habits from WT mice, their eating behavior could not fully explain the circadian accumulation pattern of TAG in these mice, as they consumed food constantly throughout the day.
Figure 6. The effect of feeding time and circadian clocks on total TAG levels in the liver.
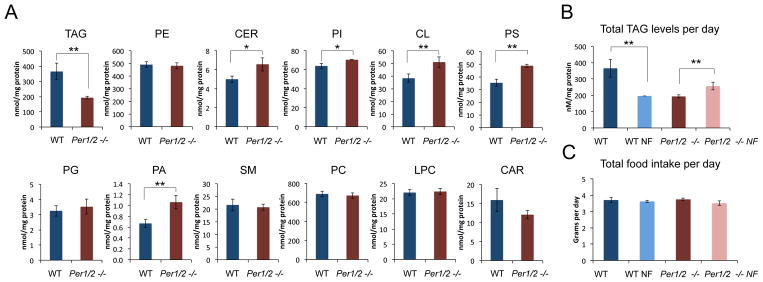
A., Comparison of the total hepatic levels of the different lipid types, quantified throughout the day, between WT and Per1/2 −/− mice fed ad libitum. B., Comparison of the total hepatic TAG levels, quantified throughout the day in WT fed ad libitum, WT night fed, Per1/2 −/− fed ad libitum and Per1/2 −/− night fed mice. C., Comparison of the total daily food intake of WT fed ad libitum, WT night fed, Per1/2 −/− fed ad libitum and Per1/2 −/− night fed mice. The data are presented on a bar graph (mean +/− SEM, n=4 for the lipid analysis and n=8 for the food consumption). Night Fed (NF). P values < 0.05 are marked in * and < 0.01 in **. See also Figure S6.
Dissection of oscillating TAG species based on feeding time and circadian clocks
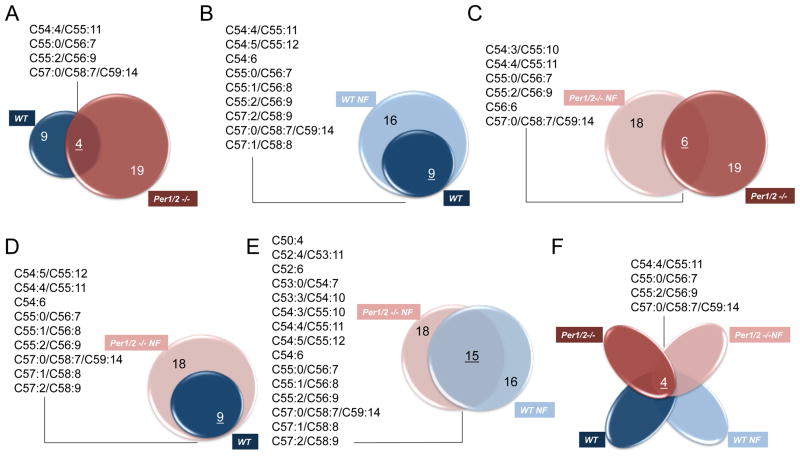
To uncover principles that shape TAG landscape in the liver, namely identify which TAG species are primarily clock driven and which are mainly regulated by food intake, we dissected the overlap between the different TAG species that oscillated under the 4 different experimental setup (i.e. WT fed ad libitum, WT night fed, Per1/2 −/− fed ad libitum, Per1/2 −/− night fed), (Figure 5). There was a relatively small commonality between the oscillating TAG species in WT mice fed ad libitum and Per1/2 −/− mice fed ad libitum (4 out of 9 and 19, respectively), (Figure 5A). Upon nighttime feeding, the number of oscillating TAG species in WT mice increased from 9 to 16. All 9 species that oscillated under ad libitum regimen continued to oscillate in night fed animals (Figure 5B). In sharp contrast, there was a little overlap between the oscillating TAG species under ad libitum and nighttime feeding in Per1/2 −/− mice. Only 6 species overlapped out of the 19 that oscillated in ad libitum and 18 in night fed animals (Figure 5C). Remarkably, we observed an extensive overlap between the TAG species that oscillate in WT night fed and Per1/2 −/− night fed animals, (15 out of 16 and 18, respectively), (Figure 5E). These findings suggest that there is a distinct population of oscillating TAG species that respond to feeding-fasting cycles independently of PER1/2 (Figure S6A). Strikingly, the TAG species that oscillated in Per1/2 −/− fed ad libitum largely differed from the one that appeared to be food driven (Figure S6B). This in line with our findings that Per1/2 −/− mice fed ad libitum eat constantly throughout the day and hence feeding fasting cycles do not determine their circadian TAG accumulation.
Figure 5. Dissection of oscillating TAG species based on feeding time and circadian clocks.
Overlap analysis between the specific TAG species that oscillated under the different experimental setup. A., WT fed ad libitum and Per1/2 −/− fed ad libitum. B., WT night fed and WT fed ad libitum. C., Per1/2 −/− night fed and Per1/2 −/− fed ad libitum. D., WT fed ad libitum and Per1/2 −/− night fed. E., WT night fed and Per1/2 −/− night fed. F., WT fed ad libitum, WT night fed, Per1/2 −/− fed ad libitum and Per1/2 −/− night fed. For each scheme, the list of overlapping TAG species is detailed. Night Fed (NF).
To further characterize these different TAG populations, we plotted them according to the number of carbons. TAG consist of a glycerol backbone to which three molecules of fatty acids are attached. Hence the carbon number provides insight regarding the fatty acid composition of the different TAG. In WT mice fed ad libitum the oscillating TAG comprised of 54 and higher number of carbons, indicating that they primarily contain long-chain fatty acids (C14–20), whereas in Per1/2 −/− mice fed ad libitum a large fraction included also TAG with carbon number below 54, suggesting that they also contain medium-chain fatty acid (C8–12), (Figure S6C). Under night fed condition both mice strains exhibited a very similar distribution (Figure S6D), which actually reflect the strong overlap between the oscillating species in these two groups (Figure 5E). Overall, these analyses suggested that the oscillating TAG species in Per1/2 −/− fed ad libitum largely differ from the TAG identified in all other tested conditions (i.e. WT fed at ad libitum, WT night fed, and Per1/2 −/− night fed). Taken together with our finding that Per1/2 −/− ingest food constantly throughput the day, it is unlikely that the observed TAG oscillations in Per1/2 −/− are food driven.
The effect of feeding-time and circadian clocks on total TAG levels in the liver
Pervious experiments performed with Per2 single knockout mice have demonstrated that their daily food intake is identical to WT mice but their total TAG plasma levels are drastically reduced (~50%), (Grimaldi et al., 2010). In line with these observations, quantification of the total hepatic TAG levels throughout the day revealed a sharp decrease (~50%) in hepatic TAG content of Per1/2 −/− mice compared with WT mice (Figure 6A). The level of none of the other measured lipid types was decreased in Per1/2 null mice compared with WT mice. The levels of PE, PG, SM, PC, LPC, and CAR did not differ between the two mouse strains. Whereas CER, PI, CL, PS and PA levels in Per1/2 null mice were elevated (Figure 6A). Interestingly, 2 weeks of nighttime feeding drastically reduced the total hepatic TAG content in WT mice, (~50% reduction), (Figure 6B). By contrast, nighttime feeding of Per1/2 −/− mice resulted in a significant increase of about 25% in hepatic TAG content (Figure 6B). This indicated that time restricted feeding drastically affects the TAG levels in the liver. These differences were evident in almost every single TAG species that was quantified in our lipidomic analysis (Table S5). It should be noted that under all 4 tested conditions (i.e. WT mice fed ad libitum or night fed, Per1/2 mice −/− fed ad libitum or night fed), we did not observe any significant differences in the total daily food intake of the mice (Figure 6C), indicating that the observed changes in hepatic TAG levels were not due to differences in their food consumption. These findings implied that both feeding and clock-dependent mechanisms not only dictate the phase of hepatic TAG accumulation, but also play a prominent role in determining the total hepatic TAG levels throughout the day.
Feeding and clock driven expression of enzymes and master regulators of hepatic TAG metabolism
To further corroborate these findings and dissect both the clock and/or feeding dependency we determined the expression profile of enzymes participating in TAG homeostasis in WT and Per1/2 null mice, fed either ad libitum or exclusively during the night (Figure 7A). Comparison under ad libitum feeding conditions demonstrated that the expression profile of several enzymes in TAG metabolism, notably Gpat1, Gpat2, Agpat2, Lpin1, Lpin2, Dgat2, Lipa and Pnpla3 differ between the two genotypes. Both Lpin1 and Gpat2 mRNA levels oscillated in WT mice and were relatively low and constant in Per1/2 null mice, indicating that they are expressed in a clock-dependent manner. Remarkably, several enzymes exhibited circadian oscillations in Per1/2 null mice (i.e. Gpat1, Agpat1, Agpat2, Lpin2, Lipa and Pnpla3). Notably, although Pnpla3 mRNA levels were sharply reduced in Per1/2 null mice compared to WT mice, Pnpla3 continued to oscillate in Per1/2 −/− mice with ~5-fold amplitude. The expression profile of Gpat1, Agpat2, Lpin2, Lipa and Pnpla3 was phase shifted in Per1/2 −/− mice. For example, Agpat2 and Pnpla3 mRNA levels accumulated around CT0 in WT mice and around CT16 in Per1/2 −/− mice. Similarly, Lpin2 accumulated about 8 h earlier in Per1/2 −/− mice (i.e. ~CT4 compared to ~CT12). The observed oscillations in the absence of PER1/2 suggest that the circadian accumulation of these enzymes might be clock-independent.
Figure 7. Circadian expression of enzymes participating in TAG homeostasis and master lipid regulators in WT and Per1/2 null mice.
A., WT and Per1/2 −/− mice, fed either ad libitum or exclusively during the night were sacrificed under constant darkness, at 4 h intervals throughout the day. Total RNA was prepared from liver and mRNA expression levels of enzymes participating in TAG metabolism were determined by quantitative real-time PCR and presented as fold change relative to the lowest value. B., Circadian mRNA expression profiles of master lipid regulators in WT and Per1/2 −/− mice fed ad libitum. Data are presented as mean +/− STDEV, with a mix of 4 animals per time point. Dark grey represent the subjective night and light gray the subjective day. C., A schematic model depicting the intricate circadian regulation of hepatic TAG levels by circadian clocks and feeding behavior. Night Fed (NF). Circadian Time (CT).
To examine the direct effect of feeding time, we compared the expression pattern in mice fed either ad libitum or exclusively during the night. The circadian mRNA expression pattern of Lpin1 and Gpat2 persisted under both feeding conditions suggesting that their accumulation is primarily clock-dependent and less affected by feeding time. Altogether, in WT mice, nighttime feeding had very little effect on the phase of oscillating enzymes. By contrast, in Per1/2 −/− the expression phase of Gpat1, Agpat2, Lpin2, Lipa, and Pnpla3 was altered upon nighttime feeding, pointing out that their circadian expression responds to changes in feeding time. Overall, our analysis evinced that the circadian expression of several hepatic enzymes participating in TAG metabolism is clock-dependent whereas other enzymes retain their circadian expression in the absence of a functional clock and respond to feeding.
The circadian oscillations in expression of enzymes participating in TAG homeostasis in Per1/2 −/− mice, prompted us to examine the expression profile of master regulators of lipid metabolism in these mice. The nuclear receptors, Peroxisome Proliferator-Activated Receptor alpha (PPARα), and gamma (PPARγ) that promote the expression of genes involved in fatty acids uptake, utilization, and catabolism, are expressed in a circadian manner (Yang et al., 2006). Indeed, both PPARα, and PPARγ mRNA levels oscillated in livers of WT mice; the former peaked around CT8 and the later around CT4 (Figure 7B). Remarkably, the circadian expression pattern of PPARα persisted in Per1/2 null mice with a similar magnitude and amplitude. A 4-fold reduction in the mRNA levels of PPARγ was observed in Per1/2 null mice. Yet, the PPARγ transcript levels retained their circadian expression and peaked around CT8 (~2-fold induction). Hence, both PPARα, and PPARγ cycled in the absence of PER1/2 proteins. The Sterol Regulatory Element-Binding Protein (SREBP1), a transcription factor that regulates the expression of genes involved in cholesterol and lipid metabolism, accumulates in a daily manner (Le Martelot et al., 2009). As expected, in WT mice, SREBP1c mRNA levels oscillated in a circadian manner with zenith levels at ~CT16 and nadir levels at ~CT4. By contrast, the expression levels of SREBP1c in Per1/2 null mice were relatively low and constant throughout the day (Figure 7B). The expression levels of other central regulators of lipid homeostasis such as Hepatocyte Nuclear Factor 4 (HNF4), Liver X Receptors alpha (LXRα) and beta (LXRβ), were relatively similar and constant throughout the day in both mouse strains (Data not shown).
Thus, we concluded that though the core clock circuitry is non-oscillating in the absence of PER1/2 (Figure S2), we do observe circadian oscillations in the expression of several master lipid regulators and enzymes that participate in TAG homeostasis.
Discussion
We provide a comprehensive temporal and quantitative analysis of lipid accumulation in livers of WT and clock-disrupted mice, fed either ad libitum or exclusively during the night. In view of the wide involvement of lipids in various key cellular functions (e.g. energy storage and provision, membrane composition, and signal transduction), our finding that ~17% of all measured lipids in mouse liver exhibit circadian pattern of accumulation have wide ramifications. Remarkably, our liver lipidomic analysis showed that numerous TAG species accumulate in a circadian manner, all reaching their peak levels around CT8 (Figure 1). In this conjuncture, expression analysis of enzymes participating in TAG metabolism in the liver, revealed a circadian accumulation pattern for multiple enzymes covering all subsequent steps involved in TAG biosynthesis (Figure 2).
As aforementioned, circadian transcriptome analyses have been highly valuable in identifying core clock and output genes including many genes involved in various metabolic pathways. Nevertheless, since the majority of cellular processes are tightly regulated at multiple levels beyond transcription (e.g. translation rate, post translational modification, protein stability) these approaches are limited and provide only a partial depiction. This is particularly pertinent for metabolic pathways that often rely on intricate cascade of enzymatic reactions and are modulated by enzymes activity and substrate availability. Hence, measurements of the absolute levels of metabolites are essential. Indeed, multiple enzymes involved in TAG metabolism in the liver were expressed in a circadian manner (Figure 2), yet they exhibited a wide range of expression phases. Thus, solely based on the mRNA expression data it would have been difficult to predict whether the final products (i.e. TAG) would accumulate in a circadian manner and identify their actual phase. Thus, our analysis clearly showed that in contrast to the disparate expression of enzymes involved in TAG homeostasis, all oscillating TAG peak around CT8 in WT mice fed ad libitum (Figure 1). A plausible explanation for that is that post-transcriptional control of these enzymes in a temporal manner might serve as a determinant of TAG synthesis. Several examples in the literature highlight the role of post-transcriptional control of enzymes in TAG biosynthesis. Both, AMP-activated protein kinase (AMPK), a sensor for cellular energy levels, and casein kinase have been implicated in the phosphorylation and regulation of GPAT enzymatic activity (Coleman and Lee, 2004). Notably, AMPK and casein kinase are significant components within the core clock circuitry (Dibner et al., 2010). Phosphorylation of Lipin1, on multiple serine and threonine residues, in response to various stimuli (e.g. insulin and amino acids) regulates its subcellular localization and its PAP enzymatic activity (Harris et al., 2007; Peterson et al., 2011). Lastly, posttranscriptional control via translation regulation has been also linked to DGAT accumulation (Coleman and Lee, 2004).
Previous genome wide analyses of the temporal DNA binding of BMAL1 and additional core clock proteins using chromatin immunoprecipitation combined with deep sequencing showed that their binding sites are enriched among genes involved in lipid metabolism, notably triglyceride biosynthesis (Cho et al., 2012; Koike et al., 2012; Rey et al., 2011). Binding sites for BMAL1, CLOCK, PER1/2, and CRY1/2 were identified for multiple enzymes in the glycerol-3-phosphate pathway. Specifically, BMAL1, CLOCK, PER1, and CRY1/2 were found to bind the same intergenic loci, upstream to the Lpin1 transcription-starting site, albeit with a different phase. Similarly, BMAL1, CLOCK, PER1/2 and CRY1/2 were present on the same DNA region, upstream to the Gpat2 transcription-starting site, though during different times of the day (~ZT4 for BMAL/CLOCK and ~ZT16 for PER/CRY). Hence, it is conceivable that both Lpin1 and Gpat2 are direct targets of BMAL/CLOCK, PER/CRY transcriptional regulation, as the prominent mRNA circadian expression of both Lpin1 and Gpat2 was strongly dampened in clock-disrupted mice (Figure 7).
Interestingly, we found that a similar fraction of lipids (~17%) were oscillating in both WT and Per1/2 null mice fed ad libitum, most notably TAG. However, they largely differed in their accumulation phase and composition. These observations are intriguing as mice lacking both PER1 and PER2 are arrhythmic under constant darkness and their circadian expression of core clock genes is largely abolished (Figure S2), (Zheng et al., 2001). This raises the question; what are the molecular mechanisms that drive the circadian oscillations in TAG accumulation in the absence of a functional clock. We do show that feeding-fasting cycles can strongly shape the phase of TAG accumulation in mouse liver and that Per1/2 −/− mice differ in their eating habits compared to WT mice. However, their feeding behavior cannot fully explain the oscillations of TAG in these mice, as they consume equal amount of food throughout the day, and hence food ingestion cannot serve as a timing cue. Moreover, overlap analyses of the oscillating TAG species, identified in WT and Per1/2 −/− mice, fed ad libitum or exclusively during the night, point out that the TAG species that oscillated in Per1/2 −/− mice fed ad libitum, largely differed from the oscillating TAG species found in night fed Per1/2 −/− mice and WT mice (Figure 5). Conceivably, the TAG species that oscillated in Per1/2 −/− fed ad libitum, comprise of a distinct population that is not driven by feeding-fasting cycles.
Further studies are required to identify the molecular mechanisms that drive the circadian accumulation of TAG in the absence of a functional clock. It should be noted that while the expression of core clock genes is relatively constant throughout the day in Per1/2 null mice (Figure S2) and these mice exhibit arrhythmic locomotor activity and arrhythmic feeding behavior under constant darkness (Figure 4C), (Zheng et al., 2001). Our analyses do demonstrate the persistence of circadian oscillation in mRNA expression of several enzymes that participate in TAG metabolism and master lipid regulators such as PPARs (Figure 7). Hence, this suggests that different circadian outputs (e.g. locomotor activity, feeding behavior TAG accumulation, mRNA expression) are selectively affected in the absence of PER1/2; some do persist while others dampen.
Recently, it has been shown that time-restricted feeding for more than 3 months can prevent obesity, hepatic steatosis, and metabolic syndrome in mice fed high fat diet (Hatori et al., 2012; Sherman et al., 2012). Time-restricted feeding primarily protected mice from the adverse effects of a high fed diet but several metabolic parameters were also significantly improved upon time restricted feeding of standard diet, among them hepatic steatosis (Hatori et al., 2012; Sherman et al., 2012). In this conjuncture, our analysis evinced that time restricted feeding of regular chow diet in WT mice for a short time period as 2 weeks results in a dramatic decrease (~50%) in total TAG levels in the liver, whereas the total food intake was unchanged. Elevated plasma and hepatic TAG levels are associated with hepatic steatosis and liver malfunction. The striking effect of nighttime feeding on hepatic TAG levels is of obvious clinical importance, as current pharmacological interventions are less efficient and often associated with various adverse side effects (McKenney and Sica, 2007).
In summary, our study evinces that both circadian clocks and feeding-fasting cycles play a prominent role in the regulation of circadian TAG accumulation and total TAG levels in the liver. Nevertheless, in the absence of a functional clock (i.e. Per1/2 −/− mice) and thus lack of feeding rhythms, circadian oscillations in hepatic TAG levels do persist, albeit with a complete different phase. This suggests that additional mechanism play a role in their circadian accumulation (Figure 7C).
Experimental Procedures
Animals
All animal experiments and procedures were conducted in conformity with the Institutional Animal Care and Use Committee (IACUC) guidelines. For liver lipids and mRNA profiling, we analyzed three months old males that were obtained from WT and Per1/2 −/− mouse colonies derived from the original background previously described (Zheng et al., 2001). Mice were kept under 12 h light/dark regimen for 2 weeks and fed either ad libitum or exclusively during the dark phase. Throughout the last day mice were kept under constant darkness and were sacrificed at four hours intervals around the clock. Livers were harvested, rinsed in PBS, and rapidly frozen in liquid nitrogen. CT0 corresponded to the time light used to be turned on and CT12 to the time lights used to be turned off in the animal facility.
Shotgun lipidomic analysis
Mice were sacrificed by cervical dislocation; livers were harvested and rinsed with PBS. A ventro lateral section, which corresponds to the right lobe, was cut and instantly frozen in liquid nitrogen. Liver wafers were pulverized into a fine powder by a stainless steel Bio-pulverizer (12 wells, capacity 10–100 mg per well, BioSpec Products, Bartlesville, OK) at the temperature of liquid Nitrogen. Tissue fine powders (~ 15 mg) were weighed from each liver sample and homogenized in PBS by using 2.0-ml cryogenic vials (Corning Life Sciences, Tewksbury, MA). Protein levels in the homogenates were quantified using a bicinchoninic acid protein assay kit (Thermo Scientific, Rockford, IL) with bovine serum albumin as standards. All determined lipid levels were normalized to the protein content of individual samples.
Individual homogenate of the liver samples was accurately transferred into a disposable glass culture test tube. An internal standard mixture for quantitation of all reported lipid classes were added prior to lipid extraction (see Supplemental Experimental Procedures). Lipids were extracted by methyl-tert-butyl ether (Matyash et al., 2008). Each lipid extract was re suspended in a volume of 100 μl of CHCl3/MeOH (1:1, v/v) per mg of protein, flushed with N2, capped, and stored at −20 °C. The whole process of the lipid extraction was performed in a lab with an ambient temperature of 22.7±0.2 °C and conducted in parallel with the controls.
For ESI direct infusion analysis, lipid extract was further diluted to a final concentration of ~500 fmol/μL by CHCl3/MeOH/isopropanol (1/2/4, v/v/v) with or without 0.02% (v/v) LiOH-saturated MeOH solution, and the mass spectrometric analysis was performed on a QqQ mass spectrometer (Thermo TSQ VANTAGE, San Jose, CA) equipped with an automated nano spray device (TriVersa NanoMate, Advion Bioscience Ltd., Ithaca, NY) and operated with Xcalibur software (Han et al., 2008). Identification and quantification of the different lipid molecular species were performed using an automated software program (Yang et al., 2009).
RNA analysis by real-time quantitative PCR
RNA extraction and transcript quantification by real-time PCR technology was performed as previously described (Asher et al., 2010). Normalization was performed relative to geometrical mean of 3 house keeping genes: Tbp, Hprt, and Gapdh. Primers and probes are listed under Supplementary experimental procedure.
Statistics
Data represent mean ± SEM of 4 animals per time point. Rhythmicity of lipids was assessed with the nonparametric test, JTK_CYCLE, previously described for the analysis of rhythmic transcripts and metabolites (Hughes et al., 2010). A window of 24 h was used for the determination of circadian periodicity, and a P value of <0.05 was considered as statistically significant. Lipid profiles were crosschecked by visual inspection and false positive were excluded.
Measurements of mice daily food consumption
The daily food consumption of mice was monitored using the Phenomaster metabolic cages (TSE Systems). Measurements of food intake were performed at 15 min resolution throughout the day.
Supplementary Material
Highlights.
Lipidomic analysis reveals circadian oscillations in hepatic triglyceride levels.
Circadian oscillations in hepatic triglycerides persist in clock-disrupted mice.
PPARs and triglyceride enzymes are circadianly expressed in clock-disrupted mice.
Feeding-time regulates the phase and levels of hepatic triglyceride accumulation.
Acknowledgments
We thank U. Albrecht for the Per1/2 double knockout mice, S. Anpilov for his help with the JTK_CYCLE analysis, A. Orbach for his technical assistance with the mouse feeders. The work performed in the laboratory of G.A. was supported by the Israel Science Foundation (ISF), the Abish-Frenkel Foundation, the HFSP Career Development Award (HFSP CDA00014/2012), and the European Research Council (ERC-2011 METACYCLES 310320). The work conducted in the laboratory of X.H. was supported by National Institute on Aging Grant R01 AG31675 and intramural institutional research funds. L.R.N received a postdoctoral fellowship from the Feinberg graduate school, Weizmann Institute of Science.
References
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- Brown SA, Kowalska E, Dallmann R. (Re)inventing the circadian feedback loop. Dev Cell. 2012;22:477–487. doi: 10.1016/j.devcel.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109:2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A. 2012;109:5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell. 2012;47:158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O. Metabolism and circadian rhythms--implications for obesity. Endocr Rev. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, Granneman JG, Piomelli D, Leff T, Sassone-Corsi P. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang K, Gross RW. Microfluidics-based electrospray ionization enhances the intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: development of an automated high-throughput platform for shotgun lipidomics. Rapid Commun Mass Spectrom. 2008;22:2115–2124. doi: 10.1002/rcm.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang K, Gross RW. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TE, Huffman TA, Chi A, Shabanowitz J, Hunt DF, Kumar A, Lawrence JC., Jr Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J Biol Chem. 2007;282:277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, Panda S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MM, Pan X. Clock genes, intestinal transport and plasma lipid homeostasis. Trends Endocrinol Metab. 2009;20:177–185. doi: 10.1016/j.tem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasukawa T, Sugimoto M, Hida A, Minami Y, Mori M, Honma S, Honma K, Mishima K, Soga T, Ueda HR. Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci U S A. 2012;109:15036–15041. doi: 10.1073/pnas.1207768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Lo Sasso G, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney JM, Sica D. Role of prescription omega-3 fatty acids in the treatment of hypertriglyceridemia. Pharmacotherapy. 2007;27:715–728. doi: 10.1592/phco.27.5.715. [DOI] [PubMed] [Google Scholar]
- Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, Ikeda S, Yasui A, van der Horst GT, Soga T, Ueda HR. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Quiroga AD, Lehner R. Liver triacylglycerol lipases. Biochim Biophys Acta. 2012;1821:762–769. doi: 10.1016/j.bbalip.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman H, Genzer Y, Cohen R, Chapnik N, Madar Z, Froy O. Timed high-fat diet resets circadian metabolism and prevents obesity. Faseb J. 2012;26:3493–3502. doi: 10.1096/fj.12-208868. [DOI] [PubMed] [Google Scholar]
- Shimba S, Ogawa T, Hitosugi S, Ichihashi Y, Nakadaira Y, Kobayashi M, Tezuka M, Kosuge Y, Ishige K, Ito Y, Komiyama K, Okamatsu-Ogura Y, Kimura K, Saito M. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One. 2011;6:e25231. doi: 10.1371/journal.pone.0025231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, Yoo SH, Takahashi JS, Butler AA, Kamenecka TM, Burris TP. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Reue K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am J Physiol Endocrinol Metab. 2009;296:E1195–1209. doi: 10.1152/ajpendo.90958.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Cheng H, Gross RW, Han X. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal Chem. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.