Abstract
Background
Extracellular vesicles (EVs) are small nanometre-sized vesicles that are circulating in blood. They are released by multiple cells, including tumour cells. We hypothesized that circulating EVs contain protein kinases that may be assessed as biomarkers during treatment with tyrosine kinase inhibitors.
Methods
EVs released by U87 glioma cells, H3255 and H1650 non-small-cell lung cancer (NSCLC) cells were profiled by tandem mass spectrometry. Total AKT/protein kinase B and extracellular signal regulated kinase 1/2 (ERK1/2) levels as well as their relative phosphorylation were measured by western blot in isogenic U87 cells with or without mutant epidermal growth factor receptor (EGFRvIII) and their corresponding EVs. To assess biomarker potential, plasma samples from 24 healthy volunteers and 42 patients with cancer were used.
Results
In total, 130 different protein kinases were found to be released in EVs including multiple drug targets, such as mammalian target of rapamycin (mTOR), AKT, ERK1/2, AXL and EGFR. Overexpression of EGFRvIII in U87 cells results in increased phosphorylation of EGFR, AKT and ERK1/2 in cells and EVs, whereas a decreased phosphorylation was noted upon treatment with the EGFR inhibitor erlotinib. EV samples derived from patients with cancer contained significantly more protein (p=0.0067) compared to healthy donors. Phosphorylation of AKT and ERK1/2 in plasma EVs from both healthy donors and patients with cancer was relatively low compared to levels in cancer cells. Preliminary analysis of total AKT and ERK1/2 levels in plasma EVs from patients with NSCLC before and after sorafenib/metformin treatment (n=12) shows a significant decrease in AKT levels among patients with a favourable treatment response (p<0.005).
Conclusion
Phosphorylation of protein kinases in EVs reflects their phosphorylation in tumour cells. Total AKT protein levels may allow monitoring of kinase inhibitor responses in patients with cancer.
Keywords: biomarker, cancer, kinase inhibitor, signalling, plasma
Extracellular vesicles (EVs) are nanometre-sized vesicles released by cells into body fluids such as blood and urine. They can be formed in the cytoplasm by inward budding of endosomal membranes and subsequent fusion with the plasma membrane (exosomes), or they can be shed directly from the plasma membrane (microvesicles) (1–3). Importantly, EVs can be isolated from blood, cerebrospinal fluid (CSF) and urine. Because they contain tumour-derived nucleic acids and proteins, including kinases such as AKT, EGFR (epidermal growth factor receptor) and MET (4–6), EVs provide a potential biosource for diagnostics (7,8).
Kinases are important targets for treatment in patients with cancer (9). However, not all patients respond favourably to kinase inhibitors and most patients acquire drug resistance. Therefore, biomarkers that aid in prediction and real-time monitoring of treatment responses in patients could be of great value (10). The (re-)activation of protein kinases in tumour cells promotes autophosphorylation and has been associated with drug resistance during treatment (11–13). A blood-based test providing information on tumour kinase phosphorylation could allow for longitudinal monitoring of drug resistance and personalized treatment strategies. Here we hypothesized that phosphorylation of protein kinases in EVs could serve as a predictive biomarker during treatment with tyrosine kinase inhibitors.
Materials and methods
Cells and compounds
H1650 (NCI-H1650; ATCC) and H3255 (NCI-H3255; ATCC) non-small-cell lung cancer (NSCLC) cells were maintained in RPMI-1640 (Lonza Biowhittaker, Verviers, Belgium). The glioma cell lines U87 (U87MG; ATCC) and U87-EGFRvIII (dr. Web Cavenee, Ludwig Cancer Inst., USA) were cultured in DMEM (Lonza Biowhittaker). All cell culture media were supplemented with 10% foetal bovine serum (FBS), 100 U/ml penicillin and 0.1 mg/ml streptomycin, referred to as complete DMEM. EV-depleted medium was obtained by 2 h ultracentrifugation of serum at 100,000×g before use as a medium supplement. Drug treatments were performed with erlotinib (LC Laboratories, Woburn, MA, USA) in DMEM containing EV-depleted serum. Cell viability was assessed by 2 h incubation with MTT 3-(4,5-dimethylthiazolyl-2)-2,5-diophenyl tetrazolium bromide (Sigma-Aldrich, Zwijndrecht, The Netherlands).
Isolation of EVs
EVs were isolated from plasma or conditioned media as described previously (14). Supernatants from cells, cultured for 32 h in serum-free or with 10% EV-depleted serum, were centrifuged at 500×g for 10 min to remove cells. After another centrifugation step at 2,000×g for 15 min, the samples were concentrated by retention and collection using 100 kDa cut-off filters (Amicon Ultra-15, Millipore, Amsterdam, The Netherlands) and stored at −20°C. The precleared plasma samples were diluted 10-fold in PBS before ultracentrifugation. Centrifugation at 10,000×g for 45 min was applied, before EVs were pelleted by ultracentrifugation at 100,000×g for 90 min, using a SW32TI rotor (Beckmann Instruments, Woerden, The Netherlands). The EV pellet was treated with proteases (0.5 g/L, Trypsin, Lonza Biowhittaker) or erlotinib (10 µM) at 37°C for 1 h when indicated. Subsequently, EVs were washed in PBS followed by centrifugation at 100,000×g for 90 min. All sequential centrifugation steps were performed at 4°C. Pellets were solubilized in M-PER mammalian protein extraction reagent, supplemented with phosphatase and protease inhibitor cocktails (Pierce, Thermo Scientific, Breda, The Netherlands).
LC-MS/MS analysis
EVs were isolated from 2 independent cell cultures (10× T175) for each cell line and isolated as described above. Full sample yield (corresponding to 10–15 µg of protein) was loaded on NuPAGE precast gradient gels, and proteins were separated at 125 V. After electrophoresis, gels were fixed in 50% ethanol containing 3% phosphoric acid and stained with Coomassie R-250 and washed in Milli-Q water (Supplementary Fig. 1). Gel lanes were cut in 5 slices and each slice was processed by in-gel-digestion according to the protocol previously published by Shevchenko (15). Briefly, samples were treated with dithiothreitol (10 mM) for 1 h at 56°C and iodoacetamide (50 mM) for 45 min at RT. The slices were dried and incubated overnight with trypsin (6.25 ng/µL). Peptides were extracted first in 1% formic acid and then in 50% ACN/5% formic acid. Subsequently, peptides were separated by an Ultimate 3,000 nanoLC-MS/MS system (Dionex LC-Packings, Amsterdam, The Netherlands). After injection, peptides were trapped on a 10 mm×100 µm ID trap column and separated at 300 nl/min in a 10–40% buffer gradient (80% ACN+0.05% formic acid) in 60 min. Peptides were measured on-line after nanospray ionization, using a Q Exactive mass spectrometer (ThermoFisher, Bremen, Germany). Intact masses were measured at resolution 70,000 (at m/z 200) in the orbitrap. MS/MS spectra were acquired at resolution 17,500 (at m/z 200) in the orbitrap using an AGC target value of 2×105 charges and an underfill ratio of 0.1%.
Protein identification and quantification
MS/MS spectra were searched against the Uniprot human reference proteome FASTA file (release Sept 2012; 147,951 entries) using MaxQuant 1.3.0.5 (default settings) (16). Cysteine carboxamidomethylation was treated as fixed modification and methionine oxidation and N-terminal acetylation as variable modifications. Peptide and protein identifications were filtered at an FDR of 1% using the decoy database strategy. Proteins were quantified by spectral counting (17,18), that is, the sum of all MS/MS spectra for each identified protein. These counts were normalized by dividing the spectral counts per protein by the sum of all counts per sample and multiplying by the average sum across all samples (18).
Immunohistochemistry and western blot analysis
For immunohistochemistry, cells were fixed in 2% paraformaldehyde in PBS for 15 min. Permeabilization was performed with ice-cold methanol (10 min), followed by 1 h incubation with 5% bovine serum albumin (BSA). Staining with primary antibodies against EGFR (Cell Signaling #2232, 1:200) or phospho-Y (4G10, Millipore, USA, 1:200) was performed for 1–2 h at room temperature. Protein content of samples was determined with the Micro BCA protein assay (Pierce, Thermo Scientific, The Netherlands). For western blotting, equal protein amounts or equal volumes (Fig. 1d) were used. Electrophoresis was performed on precast 4–12% SDS polyacrylamide gels (NuPAGE®, Life Technologies, Bleiswijk, The Netherlands) and transferred to PVDF membranes for immunoblotting of proteins. Membranes were probed with antibodies against phospho-EGFR-y1068 (Cell Signaling, Leiden, The Netherlands #2236s), EGFR (CS #2232), phospho-AKT-s473 (CS #4051s), AKT (CS #9272), phospho-ERK1/2-t202/204 (CS #9101s), ERK1/2 (CS #9107s), α-tubulin (CS #2144s), Alix (CS #2171s), CD63 (BD Bioscience, Etten-Leur, The Netherlands 556,019), cytochrome-C (BD Bioscience 556,433) and GAPDH (CS #2118). IRDye infrared dye (LI-COR Biosciences, Bad Homburg, Germany) labelled secondary antibodies were used for detection. Membranes were scanned using an Odyssey infrared imaging system and accompanying software (version 3.0, Licor Biosciences). Signal intensity was quantified using Image J imaging analysis software (NIH, Bethesda, MD).
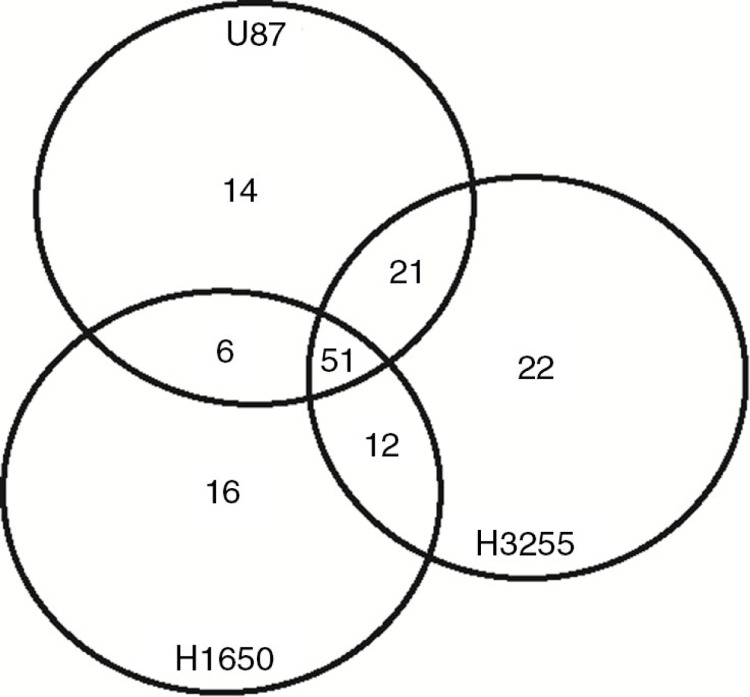
Fig. 1.
Venn diagram of protein kinases detected in EVs from 3 different cell lines. All protein kinases were identified by a cross search using protein IDs of the human kinome.
Study approval and plasma collection
All patients and/or their guardians gave written and oral consent prior to providing blood for isolation of circulating EVs. The protocol was approved by the Dutch Competent Authority, Centrale Commissie Mensgebonden Onderzoek (CCMO) and The Ethics Committee, Medisch Etische Toesting Commissie (METC) VUmc (protocol number: NL38229.029.11). We collected 12 ml of EDTA blood (in 2× BD vacutainer #368661) – yielding 3–6.5 ml of plasma – from patients with various types of cancer (n=32) and healthy donors (n=24). Samples were centrifuged twice at 2,000×g for 10 min to remove cells and frozen at −80°C until further processing.
Statistical analysis
Statistical analysis was performed using the 2-tailed (un)paired Student's t-test embedded in Graphpad Prism software (version 5.0 for Windows, GraphPad software, CA, USA). For the survival analyses the log-rank test was applied. All p-values <0.05 were considered significant.
Results
Protein kinases released in EVs from cancer cell lines
We first investigated the variety and abundance of protein kinases in EVs from different cell lines. EVs were isolated from conditioned media of 2 NSCLC cell lines (H1650 and H3255) and U87 glioma cells, lysed and subjected to tandem mass spectrometry. In total, 3,773 different proteins were identified in at least 2 independent samples. Marker proteins for exosomes such as TSG101, CD63, FLOT1 and Alix were abundantly present in the samples (see Supplementary Table I for full proteome results). We next searched for protein kinases in the data set by overlaying all protein IDs with publicly available protein kinase IDs that were previously published (19). Kinases were found from all 7 designated kinase groups (tyrosine kinase, tyrosine kinase like, AGC, serine/threonine (STE), CMCG, casein kinase 1, calcium and calmodulin–dependent kinases), but also several kinases from atypical classes (RGC, other) were present. EVs from U87 cells contained 92 different kinases, while EVs from H1650 and H3255 NSCLC cells yielded, respectively, 85 and 106 protein kinases (Fig. 1). This analysis shows that the majority of the kinases in EVs are present in EVs from multiple cell lines.
As most clinically approved drugs, target kinases from the tyrosine kinase group, we focused on this subset of the kinome. Multiple kinases from the tyrosine kinase group were found, with comparable representation (32 from a total of 130 kinases) as in the human genome (90 from 518), suggesting rather random inclusion of protein kinases during release of EVs. Both receptor tyrosine kinases (EGFR, ERBB2, EPHA2, AXL, MET, PDGFR-B) as well as non-receptor tyrosine kinases were present (SRC, JAK1, FAK, YES1) in EVs (Table I). In addition multiple cell cycle kinases (CDK2, PLK1 and AURKB) and well-known drug targets were identified such as mTOR, EGFR and MET. For quantitation of protein levels in the different samples, average spectral counts are shown. EGFR and EPHA2 were among the most prominent receptor tyrosine kinases in EVs, while JAK1 and YES1 non-receptor tyrosine kinases were released with particular high levels. Wild-type EGFR was abundantly present in both H3255 and U87 EVs. H3255 EVs uniquely showed high levels of ERBB2, while only U87 EVs contained PDGFRB. Also downstream mediators of these receptors were detected. For example, AKT was found in EVs from U87 and H3255 cells, while ERK1/2 was present in all samples.
Table I.
Overview of average spectral counts for the tyrosine kinases detected in extracellular vesicle samples from different cell lines
| Kinase | H1650 | H3255 | U87 |
|---|---|---|---|
| AXL | 1 | 1 | 11 |
| BLK | 0 | 0 | 5 |
| CSK | 3 | 5 | 5 |
| DDR1 | 0 | 17 | 0 |
| DDR2 | 0 | 2 | 25 |
| EGFR | 20 | 587 | 137 |
| EPHA1 | 0 | 7 | 0 |
| EPHA2 | 46 | 61 | 66 |
| EPHA4 | 0 | 9 | 1 |
| EPHA5 | 0 | 0 | 1 |
| EPHB2 | 2 | 30 | 67 |
| EPHB3 | 0 | 19 | 0 |
| EPHB4 | 2 | 98 | 1 |
| ERBB2 | 0 | 126 | 0 |
| ERBB3 | 0 | 10 | 0 |
| FRK | 0 | 6 | 0 |
| FYN | 0 | 10 | 18 |
| IGF1R | 0 | 8 | 5 |
| INSR | 0 | 9 | 5 |
| JAK1 | 2 | 26 | 63 |
| JAK2 | 0 | 0 | 1 |
| LYN | 2 | 49 | 16 |
| MERTK | 0 | 4 | 0 |
| MET | 2 | 22 | 21 |
| MST1R | 1 | 14 | 0 |
| PDGRFB | 0 | 0 | 12 |
| PTK2 | 4 | 2 | 1 |
| SRC | 2 | 4 | 20 |
| TYK2 | 0 | 15 | 19 |
| YES1 | 6 | 51 | 45 |
| AKT1a | 0 | 14 | 3 |
| AKT2a | 0 | 4 | 2 |
| MTORa | 16 | 16 | 3 |
| ERK1 (MAPK3)a | 2 | 4 | 6 |
| ERK2 (MAPK1)a | 7 | 27 | 22 |
Not tyrosine kinase.
Protein kinase phosphorylation in cells and EVs in vitro
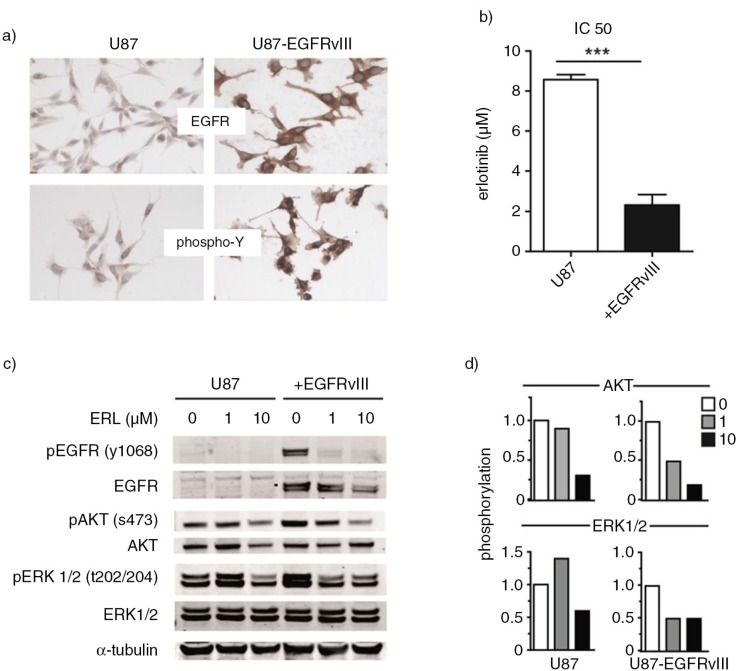
We next investigated phosphorylation of protein kinases in tumour cells and their EVs. Different receptor tyrosine kinases were found to engage similar signalling components, ultimately influencing a relatively small number of downstream kinases (20,21). We reasoned that, in particular, the protein kinases AKT and ERK1/2 could be valuable biomarkers as they reside downstream of multiple oncogenic receptors that are currently being targeted by drugs. AKT and ERK1/2 could therefore provide information on activity of multiple receptor protein kinases. To study differential phosphorylation, an isogenic human cell line model was used, consisting of U87 glioma cells with and without overexpression of the constitutively active mutant kinase EGFR (EGFRvIII) (Fig. 2a). Introduction of EGFRvIII in these cells enhances the proliferation rate and results in increased sensitivity of these cells to the EGFR inhibitor erlotinib (Fig. 2b). IC50 values as measured by the MTT cell viability assay were 8.5 µM for the parental cells and 2.3 µM for the EGFRvIII cells (p<0.001). Impact of EGFRvIII on phosphorylation of downstream targets was measured by western blot. Increased phosphorylation of AKT and ERK1/2 was found in cells expressing EGFRvIII in comparison to U87 parental cells (Fig. 2c). To visualize the impact of erlotinib treatment on phosphorylation of AKT and ERK1/2, quantification of the respective phospho-staining intensity was performed (Fig. 2d). Treatment of EGFRvIII cells with low concentrations of erlotinib (1 µM) reduced phosphorylation of both AKT and ERK1/2. In U87 parental cells in contrast, reductions in phosphorylation were only noted at a 10-fold higher concentration of erlotinib. No effect was found on EGFR, AKT and ERK1/2 protein expression levels (Fig. 2c).
Fig. 2.
U87 cells with or without overexpression of the constitutively active EGFRvIII receptor were used as model cell line. First, these cells were cultured on slides for immunostaining. Stainings were performed with EGFR and phospho-Y antibodies. (a) Sensitivity of cell lines to the EGFR inhibitor erlotinib was determined in the MTT cell viability assay. (b) Subsequently, U87 and U87-EGFRvIII cells were treated with increasing concentrations of erlotinib or DMSO vehicle. Phosphorylation of EGFR, AKT and ERK1/2 was analyzed by western blot. (c) Intensity of AKT and ERK1/2 phospho-stainings was quantified by Image J. Relative phosphorylation during erlotinib treatment is shown. (d) To assess statistical significance, the 2-tailed unpaired Student's t-test was applied. ***p≤0.001.
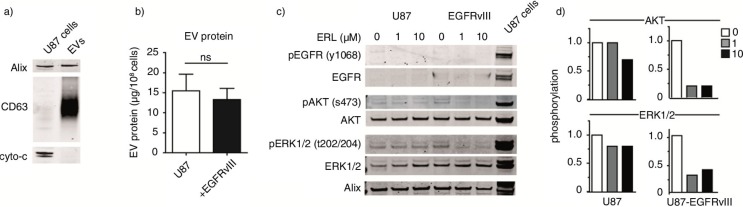
To further subtype the EVs released by U87 cells in vitro, relative abundance of the exosome markers Alix, CD63 and apoptotic marker cytochrome-C was measured (Fig. 3a). Increased levels of CD63 and Alix were observed, while cytochrome-C was not detected in EV samples. These results suggest a significant enrichment of exosomes in the EV pool. To determine whether EGFRvIII expression influences EV production, protein yields from isolations were measured. Total protein yield as measured by the Micro BCA protein assay was normalized to cell counts. No significant differences were observed in the yield from the isolations of U87 cells or EGFRvIII-containing cells (15.5 and 13.3 µg, respectively; Fig. 3b). To investigate whether EVs could provide a read-out of oncogenic signalling in cancer cells, EVs were isolated from cells treated with erlotinib and subjected to western blot. Total AKT and ERK1/2 protein levels were readily detected by western blot, while EGFR western blot analysis of EV samples appeared less efficient. Phosphorylation as well as total protein levels of protein kinases was lower in EVs than in U87-EGFRvIII cells, used as positive control for immunostaining. On the other hand, phosphorylation of EGFR, AKT and ERK1/2 kinases was elevated in EVs from EGFRvIII cells in comparison to EVs from parental U87 cells (Fig. 3c). Similar to what was found in cells, treatment of the erlotinib-sensitive U87-EGFRvIII cells with 1 µM erlotinib for 32 h resulted in a decrease in phosphorylation of AKT and ERK1/2 in EVs. However, in the EVs from the erlotinib-resistant parental U87 cells only a modest reduction in phosphorylation was observed at a 10-fold higher erlotinib dose (Fig. 3c and d). Altogether these results indicate that the phosphorylation profiles of AKT and ERK1/2 kinases in EVs may reflect the intracellular phosphorylation status during treatment with erlotinib.
Fig. 3.
EVs were isolated from U87 cells. To further characterize EVs, relative abundance of exosome markers Alix, CD63 and the apoptotic marker cytochrome-C was analyzed by western blot. (a) EV protein yield from cells with or without EGFRvIII was determined by Micro BCA protein assay. Results were normalized to cell counts as measured by hemacytometer. (b) U87 cells with or without EGFRvIII were treated with erlotinib and EVs were isolated. Phosphorylation of EGFR and downstream mediators AKT and ERK1/2 in EVs was determined by western blot. U87 cells overexpressing EGFRvIII were used as positive control for immunostaining. (c) Intensity of phosphostainings was quantified by Image J. Relative phosphorylation during treatment with erlotinib is shown. (d) To assess statistical significance, the 2-tailed unpaired Student's t-test was applied. ns=non-significant.
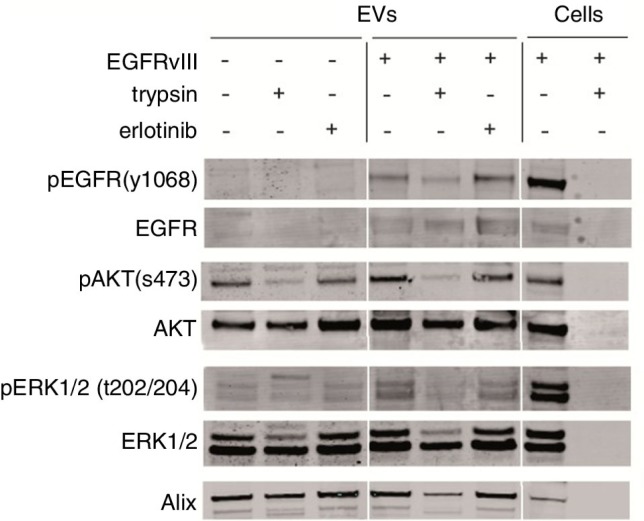
In order to determine whether the double-layered membranes of EVs protect their content from proteases, EVs were incubated with proteases, erlotinib and PBS-control. Protease treatment caused small changes in kinase protein levels in EVs. In whole cells lysates in contrast, complete digestion was observed after 1 h of treatment (Fig. 4). Simultaneously, a reduction of the Alix levels was found in the protease treated samples. These results suggest that structure and/or stability of EVs may be affected by proteases. Short-term exposure of the EVs to high concentrations of erlotinib (10 µM) had no effect on either phosphorylation or total protein levels in the EVs (Fig. 4). Altogether we conclude that AKT and ERK1/2 protein kinases reside within EVs, which protects them from potential detrimental influences of enzymes in the circulation.
Fig. 4.
EVs were isolated from culture medium of an equal number of U87 cells with or without EGFRvIII. EV pellets were treated with proteases (trypsin, 0.5 g/L) or erlotinib (10 µM) for 1 h at 37°C. Subsequently, EVs were washed in PBS and lysed. Western blot analysis was performed using equal volumes to determine protein levels and their phosphorylation. U87 cells overexpressing EGFRvIII were used as positive control for immunostaining.
Protein kinase phosphorylation in blood derived EVs
We next investigated whether AKT and ERK1/2 kinase phosphorylation may be used as biomarker in patients with cancer. Since limited information is available on the production of EVs by different cancer cells, a cohort of 42 patients was recruited with a variety a cancer types (Table II). Three patients with oesophageal cancer, 3 patients with breast cancer, 21 patients with NSCLC and 4 patients with glioma were included among others (Table II). Thirty-three patients (76%) had metastatic disease, while 9 patients (21%) had either glioma or localized disease. At the time, the first blood samples were obtained none of the patients received active systemic treatment.
Table II.
Patient characteristics
| Sample type | Mean age (±SD) | Gender (% male) | Metastatic disease (%) |
|---|---|---|---|
| Healthy donors (n=22) | 49.5 (9.9) | 12 (55) | – |
| Non-small-cell lung cancer (NSCLC) (n=21) | 57.9 (12.3) | 11 (52) | 20 (95) |
| Glioma (n=4) | 68.8 (4.9) | 2 (50) | 0 (0) |
| Breast cancer (n=3) | 52 (8.2) | 0 (0) | 2 (67) |
| Oesophageal cancer (n=3) | 55 (1) | 2 (67) | 2 (67) |
| Colorectal cancer (n=2) | 67.5 (14.8) | 2 (100) | 2 (100) |
| Cholangio cancer (n=2) | 62.5 (13.4) | 1 (50) | 1 (50) |
| Endometrial cancer (n=1) | 63 | 0 (0) | 1 (100) |
| Pancreatic cancer (n=1) | 64 | 0 (0) | 0 (0) |
| Renal cell cancer (n=1) | 52 | 1 (100) | 1 (100) |
| Melanoma (n=1) | 61 | 0 (0) | 1 (100) |
| Small-cell lung cancer (SCLC) (n=1) | 77 | 0 (0) | 1 (100) |
| Ewing sarcoma (n=1) | 49 | 1 (100) | 1 (100) |
| Cancer of unknown primary (n=1) | 71 | 1 (100) | 1 (100) |
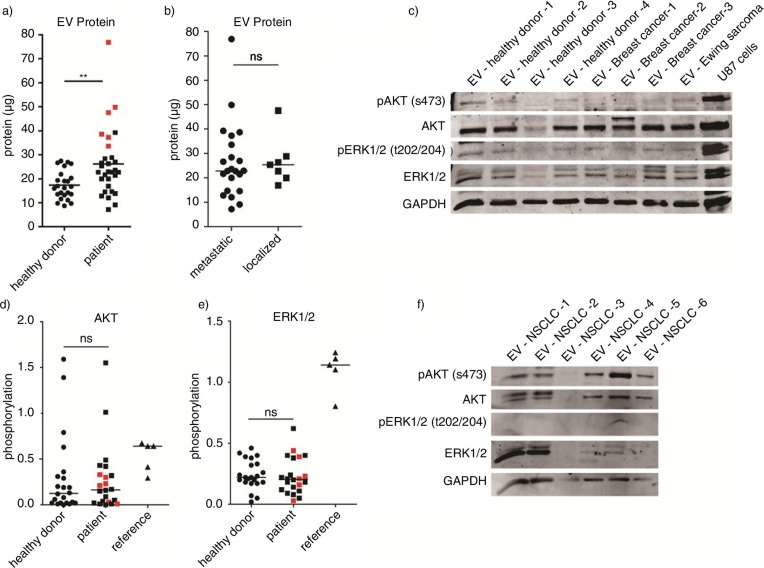
First, EV protein levels in blood of healthy volunteers (n=22) and the patients with cancer were measured. Plasma samples were diluted in PBS, and EVs were isolated. EV protein yield from each sample was measured using the Micro BCA protein assay (Fig. 5a). EV samples derived from patients with cancer contained significantly more protein (p=0.0067) compared to healthy donors. In 6 patients, increased EV protein levels were observed. Two of these patients had NSCLC, while the others had CRC, SCLC, oesophageal cancer and RCC. Five out of six patients were diagnosed with metastatic disease. No significant difference was detected in the amount of EV protein between patients with metastatic disease and patients with localized disease (p=0.78) (Fig. 5b).
Fig. 5.
Plasma was collected from 32 patients with cancer and 28 healthy donors. Total protein yield from each isolation was measured by the Micro BCA protein assay. (a–b) Phosphorylation of AKT and ERK1/2 was measured by western blot using equal protein amounts from 21 patients and 22 healthy donors. U87 cells overexpressing EGFRvIII were used as positive control for immunostaining. (c) Signal intensities were quantified by Image J imaging analysis software and phosphorylation was measured by calculating the relative signal intensity from phosphorylated and total protein levels. Subgroup analysis was performed with patients having high EV protein levels as shown in red. (a, d–e) To determine variability in total EV kinase levels in plasma, maximum protein amounts were loaded from a subgroup of patients with NSCLC. (f) Statistical significance was tested using the 2-tailed paired Student's t-test. ns=not significant, p≤0.05, **p≤0.01, ***p≤0.001.
Subsequently, AKT and ERK1/2 protein levels and their phosphorylation status in circulating EVs in patients and healthy donors were determined. Western blot analysis was performed using equal protein amounts from 21 randomly selected patients and 22 healthy donors. Total AKT protein was readily detected in all EV samples, whereas total ERK1/2 levels showed more variation between samples (Fig. 5c, d and Supplementary Fig. 2). Alix staining was not detectable in EV samples isolated from plasma, whereas GAPDH staining was high (Supplementary Fig. 1). Subsequently, phosphorylation of AKT and ERK1/2 was measured in EV samples from both patients as well as healthy volunteers. As compared to phosphorylation levels of these proteins in cancer cells, phosphorylation of AKT and ERK1/2 was relatively low (Fig. 5c–e). Semi-quantification of phosphorylation was performed by calculating the relative signal intensity from phosphorylated and total protein levels. No significant differences were observed in the phosphorylation status of AKT and ERK1/2 between patients with cancer and healthy volunteers, p=0.78 and p=0.76, respectively (Fig. 5d and e). Additionally, the subgroup of patients with high protein levels did not demonstrate increased AKT (p=0.56) or ERK1/2 phosphorylation (p=0.98) as compared to the healthy donors (Fig. 5d and e). To determine variability in total EV kinase levels in plasma, maximum protein amounts were loaded from a subgroup of patients with NSCLC. Substantial variation in total AKT and ERK1/2 levels were noted in EVs from these patients (Fig. 5f).
Total AKT levels in EVs correlate with response to sorafenib/metformin treatment
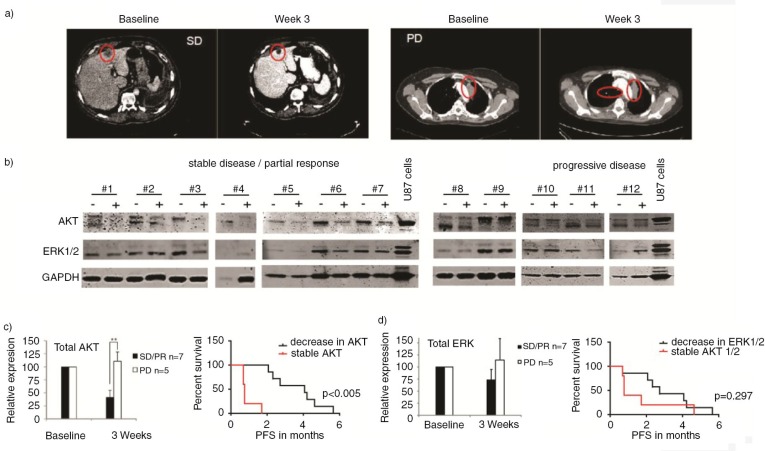
A subgroup of 12 patients with KRAS mutated NSCLC (n=12) were included in an on-going single arm phase II study (more detailed information can be found in Supplementary Table II). Based on the KRAS mutation status, patients were selected for treatment with sorafenib and metformin. Sorafenib is a small molecule tyrosine kinase inhibitor, which inhibits angiogenesis in tumours through blockade of VEGFR2/3 and PDGF receptors. In addition, it is known to have affinity for CRAF and BRAF, both mediators downstream of KRAS. Metformin, on the other hand, reduces systemic glucose and insulin levels, thereby interfering with insulin/insulin-like growth factor receptor (IGFR1) signalling in cancer cells. Since AKT and ERK1/2 act downstream of IGFR1 and RAS/RAF, respectively, EV protein levels were investigated as potential predictive biomarkers. Consistent with previous results both before and during treatment, phosphorylation in EVs was low (data not shown). Before and 3 weeks after onset of treatment, CT scans and blood plasma samples were collected. Treatment with sorafenib/metformin resulted in partial response/stable disease or progressive disease (PD) at 3 weeks according to RECIST criteria (Fig. 6a). Median progression free survival (PFS) of the favourable response group was 4.1 months (95% CI 0.6–7.5), while the resistant group responded for only 0.8 months (95% CI 0.7–0.8). Overall survival of the favourable response group was 6.5 months (95% CI 3.1–9.9), while the patients with PD survived for only 2.1 months (95% CI 0.3–3.9). Plasma EVs were isolated and samples were subjected to western blot analysis of AKT and ERK1/2 protein levels (Fig. 6b). Protein levels at baseline and during treatment were quantified and correlated to the treatment response (Fig. 6c). AKT was detected in plasma EV samples of all patients (n=12). No reduction in the AKT and ERK1/2 protein levels in EVs were observed during treatment with sorafenib/metformin in patients with PD at 3 weeks. However, in patients with partial response/stable disease (n=7), significant reductions were observed in AKT protein levels in plasma EVs (p<0.005, Fig. 6b and c). Similarly, ERK1/2 levels in EVs decreased during treatment in patients with a favourable treatment response; however, this difference did not reach significance (p=0.297, Fig. 6d). Overall we conclude that substantial variation is observed in AKT and ERK1/2 protein levels in EVs particularly during treatment with sorafenib/metformin. AKT protein levels in EVs may serve as potential predictive biomarker for the response of patients with NSCLC to sorafenib and metformin.
Fig. 6.
Representative CT images of patient #7 at baseline and with stable disease (SD) at 3 weeks on sorafenib/metformin and of patient #9 at baseline and with progressive disease (PD) at 3 weeks on sorafenib/metformin. (a) Western blot analysis was performed of plasma EVs of NSCLC patients (n=12). (b) Total AKT and ERK1/2 levels in EVs, 3 weeks after start of treatment, are shown. Patients with PD and SD or partial response (PR) on sorafenib/metformin treatment are grouped. (b) Samples are paired pre- and on-treatment samples depicted with – and +, respectively. Signal intensities were quantified by Image J imaging analysis software. (c) Kaplan Meier curves were generated for the treatment response (PFS) of patients with stable and decreased AKT and ERK1/2 levels in EVs. (c–d) To assess statistical significance, the 2-tailed unpaired t-test was applied. *p<0.001, **p<0.0001. The log-rank test was used for statistical analysis of survival analyses.
Discussion
We found that EVs contain multiple protein kinases and that their phosphorylation corresponds to the profile in tumour cells in vitro. In addition, preliminary evaluation of EV associated AKT protein levels in patients reveals a correlation with response to sorafenib/metformin treatment.
Presence of multiple tumour-derived drug targets in EVs suggests potential as biomarker platform. Several techniques have been developed to predict and monitor effects of kinase inhibitors. Mutational analysis of genes encoding kinase inhibitor targets, such as EGFR and BRAF in tumour tissue biopsies, proved to be a powerful tool to predict treatment response in patients with NSCLC and melanoma (22,23). Tumour mutations are also readily detectable in several blood compartments such as plasma, EVs and blood platelets (8,24) (25), allowing for non-invasive diagnostics. Important advantage of blood-based biomarkers is that they are easily available and also allow longitudinal monitoring during treatment. Mutations in DNA or RNA are stable and tumour specific, but biological effects may be hampered by, for example methylation or RNA interference. Protein levels and protein phosphorylation, in contrast, directly correlate with biological effects in cells, yielding powerful biomarkers (26). Circulating peptide and protein signatures were also shown to be predictive for treatment response (27–30).
Previous studies indicated that upon treatment with small molecule kinase inhibitors, multiple modulations of phosphorylation occur in tumours of patients (31,32). For example, in tumour biopsies from patients with gastric cancer, a reduction in AKT phosphorylation was found, with an interesting association with apoptosis (33). Another study, investigating EVs as potential biomarker platform, indicated that analysis of protein levels in EVs allows real-time monitoring of treatment effects induced in tumours in vivo (7). Here, we show that proteins in EVs are protected against external influences and also that post-translational modifications such as phosphorylation are conserved. Moreover, our in vitro data suggest that circulating EVs may provide a read-out of oncogenic signalling in tumour cells. However, despite very high phosphorylation in cancer cells, we found relatively low phosphorylation of AKT and ERK1/2 in plasma EVs. Since phosphorylation was low, but fairly detectable in cell culture samples, it is unlikely that phosphorylation is lost during the vesicle isolation. Levels in healthy donors suggest that AKT and ERK1/2 studies may suffer from a high background of vesicles released by other cells. Therefore, it should be evaluated whether more tumour-specific proteins may give lower background and therefore are more attractive as a biomarker. Because of the role of AKT and ERK1/2 in signalling of multiple receptor tyrosine kinases, phosphorylation of these proteins in cells as well as their EVs may still be of great interest. Although we found that phosphorylation of AKT and ERK1/2 in EVs is low, it remains to be investigated whether this event has any value as a biomarker. For such a study, it is important to develop a more sensitive and specific method for quantitative measurements of phosphorylated proteins in EVs. These may be based on, for example, ELISA, µNMR, peptide array or mass spectrometry techniques (7,34).
Current EV isolation methods typically yield relatively low protein levels, which limits protein biomarker studies. For clinical applications, the EV studies may benefit from a simple and robust isolation strategy that also allows specific isolation of tumour-derived EVs from blood. Flow cytometry methods have been described, but may be particularly beneficial for the larger vesicles (e.g. apoptotic bodies) (35). Capture-based isolation procedures are convenient but use surface markers to specifically bind subsets of EVs (exosomes, microvesicles), and this may also bias and limit biomarker studies. We did not detect Alix in our plasma EV analysis, while in our in vitro analysis a pronounced exosome signature was noted in the EV samples with abundant Alix, TSG101 and CD63. To the best of our knowledge, it is currently unknown which subtype of EVs contains the most clinically useful biomarkers. Future biomarker research may therefore also benefit from further subtyping of the different EVs. Combinations of antibodies against EV subtype markers (e.g. CD63, cytochrome-C) with antibodies raised against epitopes exclusively present on mutant proteins (36) may help in isolation of tumour-specific EVs.
We found increased EV protein amounts in patients with cancer compared to healthy volunteers. In our sorafenib/metformin analysis, high AKT levels during treatment were associated with treatment failure and poor survival. MET protein levels in EVs were previously found to be correlated with overall survival of patients with melanoma (5). These results suggest that high kinase levels in EVs are a poor prognostic feature in patients with cancer. It would be of interest to investigate the value of EV counts as well as EV protein amounts both at baseline as well as during treatment, in conjunction with protein kinase levels as biomarker. The biomarker analysis also showed downmodulation of total AKT and to some extent ERK1/2 protein levels during treatment with sorafenib/metformin. Unfortunately, no tumour tissue samples were available for comparative analyses between tumour tissue and EV levels. Whether these results can be related to direct biological effects of the treatment on tumour cells or reflect a mere systemic response remains enigmatic. However, this analysis warrants further investigation and proves potential of protein kinase levels as biomarker in patients with cancer.
Supplementary Material
Acknowledgements
We kindly thank C. Mietus, M. Lavaei, Dr. J.C. Reijneveld, Dr. D.M. Pegtel for technical input and fruitful discussions and Dr. X.O. Breakefield for continuous support.
Footnotes
Both authors contributed equally.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
References
- 1.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 2.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3:321–30. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 3.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Di Vizio D, Kim J, Hager MH, Morello M, Yang W, Lafargue CJ, et al. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009;69:5601–9. doi: 10.1158/0008-5472.CAN-08-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–91. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–24. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 7.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835–40. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDermott U, Settleman J. Personalized cancer therapy with selective kinase inhibitors: an emerging paradigm in medical oncology. J Clin Oncol. 2009;27:5650–9. doi: 10.1200/JCO.2009.22.9054. [DOI] [PubMed] [Google Scholar]
- 10.Bernards R. It's diagnostics, stupid. Cell. 2010;141:13–17. doi: 10.1016/j.cell.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366:207–15. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dave B, Migliaccio I, Gutierrez MC, Wu MF, Chamness GC, Wong H, et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J Clin Oncol. 2011;29:166–73. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 14.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107:6328–33. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 16.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 18.Pham TV, Piersma SR, Warmoes M, Jimenez CR. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics. 2010;26:363–9. doi: 10.1093/bioinformatics/btp677. [DOI] [PubMed] [Google Scholar]
- 19.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 20.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen JY, Lin JR, Cimprich KA, Meyer T. A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision. Mol Cell. 2012;45(2):196–209. doi: 10.1016/j.molcel.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 23.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson RJ, Balaj L, Hulleman E, van Rijn S, Pegtel DM, Walraven M. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118(13):3680–3. doi: 10.1182/blood-2011-03-344408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–8. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 27.Taguchi F, Solomon B, Gregorc V, Roder H, Gray R, Kasahara K, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst. 2007;99:838–46. doi: 10.1093/jnci/djk195. [DOI] [PubMed] [Google Scholar]
- 28.Deprimo SE, Huang X, Blackstein ME, Garrett CR, Harmon CS, Schoffski P, et al. Circulating levels of soluble KIT serve as a biomarker for clinical outcome in gastrointestinal stromal tumor patients receiving sunitinib following imatinib failure. Clin Cancer Res. 2009;15:5869–77. doi: 10.1158/1078-0432.CCR-08-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran HT, Liu Y, Zurita AJ, Lin Y, Baker-Neblett KL, Martin AM, et al. Prognostic or predictive plasma cytokines and angiogenic factors for patients treated with pazopanib for metastatic renal-cell cancer: a retrospective analysis of phase 2 and phase 3 trials. Lancet Oncol. 2012;13:827–37. doi: 10.1016/S1470-2045(12)70241-3. [DOI] [PubMed] [Google Scholar]
- 30.Taguchi A, Politi K, Pitteri SJ, Lockwood WW, Faca VM, Kelly-Spratt K, et al. Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell. 2011;20:289–99. doi: 10.1016/j.ccr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guix M, Granja Nde M, Meszoely I, Adkins TB, Wieman BM, Frierson KE, et al. Short preoperative treatment with erlotinib inhibits tumor cell proliferation in hormone receptor-positive breast cancers. J Clin Oncol. 2008;26:897–906. doi: 10.1200/JCO.2007.13.5939. [DOI] [PubMed] [Google Scholar]
- 32.Demetri GD, Heinrich MC, Fletcher JA, Fletcher CD, Van den Abbeele AD, Corless CL, et al. Molecular target modulation, imaging, and clinical evaluation of gastrointestinal stromal tumor patients treated with sunitinib malate after imatinib failure. Clin Cancer Res. 2009;15:5902–9. doi: 10.1158/1078-0432.CCR-09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rojo F, Tabernero J, Albanell J, Van Cutsem E, Ohtsu A, Doi T, et al. Pharmacodynamic studies of gefitinib in tumor biopsy specimens from patients with advanced gastric carcinoma. J Clin Oncol. 2006;24:4309–16. doi: 10.1200/JCO.2005.04.2424. [DOI] [PubMed] [Google Scholar]
- 34.Piersma SR, Labots M, Verheul HM, Jimenez CR. Strategies for kinome profiling in cancer and potential clinical applications: chemical proteomics and array-based methods. Anal Bioanal Chem. 2010;397:3163–71. doi: 10.1007/s00216-010-3784-7. [DOI] [PubMed] [Google Scholar]
- 35.Yuana Y, Bertina RM, Osanto S. Pre-analytical and analytical issues in the analysis of blood microparticles. Thromb Haemost. 2011;105:396–408. doi: 10.1160/TH10-09-0595. [DOI] [PubMed] [Google Scholar]
- 36.Wilmott JS, Menzies AM, Haydu LE, Capper D, Preusser M, Zhang YE, et al. BRAF(V600E) protein expression and outcome from BRAF inhibitor treatment in BRAF(V600E) metastatic melanoma. Br J Cancer. 2013;108:924–31. doi: 10.1038/bjc.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.