Abstract
Calcifying extracellular vesicles (EVs) released from cells within atherosclerotic plaques have received increased attention for their role in mediating vascular calcification, a major predictor of cardiovascular morbidity and mortality. However, little is known about the difference between this pathologic vesicle population and other EVs that contribute to physiological cellular processes. One major challenge that hinders research into these differences is the inability to selectively isolate calcifying EVs from other vesicle populations. In this study, we hypothesized that the formation of mineral within calcifying EVs would increase the density of the vesicles such that they would pellet at a faster rate during ultracentrifugation. We show that after 10 min of ultracentrifugation at 100,000×g, calcifying EVs are depleted from the conditioned media of calcifying coronary artery smooth muscle cells and are enriched in the pelleted portion. We utilized mass spectrometry to establish functional proteomic differences between the calcifying EVs enriched in the 10 min ultracentrifugation compared to other vesicle populations preferentially pelleted by longer ultracentrifugation times. The procedures established in this study will allow us to enrich the vesicle population of interest and perform advanced proteomic analyses to find subtle differences between calcifying EVs and other vesicle populations that may be translated into therapeutic targets for vascular calcification. Finally, we will show that the differences in ultracentrifugation times required to pellet the vesicle populations can also be used to estimate physical differences between the vesicles.
Keywords: calcification, extracellular vesicles, atherosclerosis, ultracentrifugation, isolation
A major challenge in the study of extracellular vesicles (EVs) is the difficulty in isolating populations of interest within the myriad of vesicle types present in biological samples (1–3). Exosomes, microparticles, apoptotic bodies and matrix vesicles are all believed to arise through different cellular mechanisms to perform varying biological functions (1,4,5); however, significant overlap exists in the size and structure of these vesicle subtypes (6,7). Ultracentrifugation is a common technique to isolate all vesicles from samples (3). This isolation is usually followed by common assay techniques to identify changes in RNA or protein expression. However, this non-discriminatory technique is not suitable to identify changes that occur in a vesicle subtype that represents a small portion of the total vesicle population (3). To overcome this problem, specific vesicles of interest are often isolated using an immunoprecipitation-based technique, whereby antibodies against a known antigen on specific vesicles are adsorbed to a larger bead that can be isolated from the solution via magnetic or centrifugation methods (8,9). While this technique can be utilized effectively, it requires a priori knowledge of differential protein expression on the population of interest and therefore is not suitable for finding novel vesicle subpopulations. Further, the expense associated with antibodies and reagents required for immunoprecipitation approaches can be prohibitive for large-scale experiments or those in which many replicates are required.
One specific vesicle subtype of interest to our research is the calcifying EV, also termed as matrix vesicles (10,11). Matrix vesicles have been well-described in bone development, wherein osteoblast-derived vesicles nucleate hydroxyapatite crystals along collagen fibres in the developing bone (12,13). Recently, calcifying EVs derived from macrophages and smooth muscle cells (SMCs) have received increased attention for their role in vascular calcification (10,11,14–17). Matrix vesicles serve as nucleating foci for the formation of microcalcifications within atherosclerotic plaque fibrous caps, which leads to plaque instability, rupture and subsequent myocardial infarction and stroke (18–21). Given the observation that calcifying matrix vesicles have been shown to exhibit an electron dense structure as hydroxyapatite nucleation proceeds (22), we hypothesized that time-dependent ultracentrifugation may be used to enrich calcifying EVs with a greater physical density than other vesicular populations from vascular SMCs. Isolating EVs of interest improves the sensitivity of assays by reducing background from other vesicle subtypes, and herein, we will show that selective enrichment of calcifying EVs enhances the signal-to-noise ratio in various calcification and proteomic assays by reducing the contribution from non-calcifying vesicles. Utilizing this protocol will allow us to better characterize the biophysical and proteomic properties of calcifying EVs and may help identify potential therapeutic targets for vascular calcification.
Methods
Cell culture and conditioned media collection
Human coronary artery (SMCs, PromoCell) were grown to confluence and were cultured in control media consisting of DMEM with 10% (v/v) foetal bovine serum and 1% (v/v) penicillin/streptomycin (control) or a calcifying media consisting of control media supplemented with 10 nM dexamethasone, 100 µM L-ascorbic acid and 10 mM β-glycerophosphate. The media were replaced every 2 days. The SMCs were cultured for at least 14 days, a time point sufficient to induce osteogenic differentiation of the cells in the calcifying media (23). For specified samples, an inhibitor for tissue non-specific alkaline phosphatase (TNAP) was used at a concentration of 1 µM (Calbiochem, #613810). After the prescribed culture period, the culture media were replaced with media containing the same components but lower foetal bovine serum (0.1%). This was done to reduce the noise caused by the presence of vesicles within the serum compared to the vesicles of interest released by the SMCs. After 24 h, the low serum media were collected after centrifugation at 1,000×g for 5 min to remove potential cellular contaminants. The resulting supernatant was then stored at −80°C prior to further processing.
RNA preparation and real-time PCR
Total RNA from the cell culture was isolated using TriZol (Life Technologies). Reverse transcription was performed using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). The mRNA expression was determined by TaqMan-based real-time PCR reactions (Life Technologies). The following TaqMan probes were used: Hs01029144_m1 (human ALPL) and 4326315E (human β-actin). The expression levels were normalized to β-actin. Results were calculated using the ΔΔCt method, and presented as fold increase relative to control.
Alizarin red S staining for calcium deposition
Calcium deposition was assessed by Alizarin red S staining. SMCs were fixed in 10% formalin and stained with 40 mM Alizarin Red S (pH 4.2) for 30 min at room temperature. Excess dye was removed by washing the plates with distilled water. The extent of calcium mineral deposition is proportional to the amount of Alizarin red S stain remaining in the culture well after washing.
Ultracentrifugation isolation of EVs
Samples were thawed on ice, and 50 µl of each sample was collected for nanoparticle tracking analysis (NTA) prior to ultracentrifugation. Of the remaining sample, 1 ml was pipetted into 11×34 mm ultracentrifuge tubes (Beckman Coulter). The tubes were loaded into a fixed angle rotor (Beckman Coulter TLA 120.2) for ultracentrifugation (Beckman Coulter Optima MAX-UP Ultracentrifuge) at 100,000×g at 4°C for 10 min. Following this 10 min ultracentrifugation, 50 µl of the supernatant was collected for NTA. The remaining supernatant was transferred to a new ultracentrifugation tube and the samples were subjected to an additional 20 min of ultracentrifugation at 100,000×g. The collection process, supernatant transfer and ultracentrifugation protocol was repeated for a 30 min ultracentrifugation followed by a final sample collection and ultracentrifugation for 40 min. The pellets from each ultracentrifugation collection time point were washed once by re-suspending the pellets in PBS and using ultracentrifugation at 100,000×g for 40 min to obtain the final pellet for further analyses.
NanoSight NTA
NanoSight NTA (NanoSight LM10, Malvern Instruments Ltd, Malvern, UK) was used to assess vesicle size and concentration within the media samples (24). The samples were diluted 1:10 in PBS prior to injection into the NanoSight chamber. For each sample, 2 NTA videos were collected for 1 min apiece, and fresh media were injected between the 2 recordings. The camera gain was set at a constant value of 13, and the threshold value for vesicle detection was set at 4. The resultant size and concentration output data were averaged to generate the final distribution for each sample.
Alkaline phosphatase activity
To measure TNAP activity, cells or ultracentrifugation pellets were re-suspended in 120 µl of TNAP Assay Buffer (BioVision) and incubated overnight at −30°C. TNAP activity was measured using the BioVision Alkaline Phosphatase Activity Colorimetric Assay kit according to the manufacturer's protocol (BioVision).
EV mineral content
To measure calcium phosphate mineral content within isolated EVs, we modified the fluorescence-based OsteoImage assay (Lonza) used for detection of hydroxyapatite mineral in cell culture. The ultracentrifugation pellets from 1 ml conditioned media were re-suspended and fixed in 10% formalin for 15 min. The vesicles were then washed once in PBS using 100,000×g ultracentrifugation for 40 min. Following this wash, the pellets were re-suspended in 200 µl diluted OsteoImage dye (according to manufacturer protocol) for 30 min at room temperature. The dye was then removed via ultracentrifugation and the vesicles were washed twice in OsteoImage wash buffer using 100,000×g ultracentrifugation for 40 min. After the final wash, the pellet was re-suspended in 100 µl OsteoImage wash buffer and fluorescence intensity was measured using a plate reader. The resultant fluorescence values were normalized to the number of vesicles pelleted by ultracentrifugation (obtained from NanoSight) to account for variations in vesicle numbers in different treatment conditions. Fold changes of these normalized values for calcific vesicles compared to control conditions are reported for each ultracentrifugation time point.
Transmission electron microscopy
Ultracentrifuged vesicle pellets were immersion fixed in 2.5% glutaraldehyde, 2% paraformaldehyde, in 0.1 M cacodylate buffer pH 7.4 [modified Karnovsky's fixative (25)] for at least 1 h at room temperature before being spun down again and embedded in 4% low melting agarose. Pellets were cut to size and placed at 4°C overnight. To continue processing, pellets were washed 3×5 min each in PBS then fixed secondarily in 1% osmium tetroxide in buffer, for 1 h at 4°C. Pellets were then washed 3×5 min in PBS, then 3×5 min in deionized water before being taken through a dehydration series including 30, 50, 70, 95 and 100% ethanol, 10 min each at 4°C. Pellets were then brought to room temperature to continue dehydrating with 100% ethanol (3×10 min each). Infiltration proceeded with 2:1 100% ethanol to LX112 Epon resin for 30 min followed by a 1:1 mixture of these components overnight with the pellets rotated at room temperature. The following day, the sample containers’ caps were left open to evaporate the ethanol, and the remaining resin was removed prior to the addition of 100% freshly made resin. Samples were recapped and rotated overnight. Before embedding the next day, samples were placed in a vacuum oven at 60°C for 2 h. Samples were embedded in flat-bottomed BEEM capsules and cured at 60°C for 48 h. Cured blocks were sectioned using a Leica Ultracut E ultramicrotome with a diamond knife at 80 nm thickness and placed on bare 200 mesh copper grids. Grids were contrast stained with 2% uranyl acetate for 10 min and lead citrate for 5 min. Grids were imaged on a JEOL 1400 TEM equipped with a side mount Gatan Orius SC1000 digital camera.
Mass spectrometry
For mass spectrometry analysis of the pelleted vesicles, the vesicles were lysed in RIPA buffer, and polyacrylamide gel electrophoresis was used to separate the proteins by size. The polyacrylamide gels were stained with coomassie blue to visualize the separated protein bands. The gels were then cut into small strips dictated by the appearance of protein bands that exhibited differences between the treatment groups. Trypsin was used to digest the proteins into peptide sequences prior to mass spectrometry. Peptide samples were analyzed with the high resolution/accuracy LTQ-Orbitrap (Elite model) mass spectrometer fronted with a Nanospray FLEX ion source, and coupled to an Easy-nLC1000 HPLC pump (Thermo Scientific). The peptides were subjected to a dual column set-up: an Acclaim PepMap RSLC C18 trap column, 75 µm×20 mm; and an analytical column packed in-house with 3 µm, 100 Å pore size C18 resin (Bruker) in a 75 µm×200 mm long piece of fused silica capillary. The analytical gradient was run at 350 nl/min from 10 to 30% Solvent B (acetonitrile/0.1% formic acid) for 30 min followed by 5 min of 95% Solvent B. Solvent A was 0.1% formic acid. All reagents were HPLC-grade. The instrument was set at 120 K resolution, and the top 20 precursor ions (within a scan range of 380–2,000 m/z) were subjected to collision-induced dissociation (collision energy 35%) for peptide sequencing (MS/MS). The MS/MS data were queried against the Human UniProt database (downloaded on 27 March 2012) using the SEQUEST search algorithm (26) via the Proteome Discoverer Package (version 1.4, Thermo Scientific) using methionine oxidation as a variable modification, and carbamidomethylation of cysteine residues as a fixed modification. The peptide false discovery rate (FDR) was calculated using Percolator provided by Proteome Discoverer: the FDR was determined based on the number of MS/MS spectral hits when searched against the reverse, decoy Human database (27,28). Peptides were filtered based on a 1% FDR. Proteins used for spectral quantification contained ≥2 unique peptides.
Statistical analysis
All quantitative data are presented as mean values from at least 6 independent experiments (n>6) with error bars representing the standard deviation of the mean. Unless otherwise noted, differences between control and calcifying treatment groups were determined using unpaired t-tests with the statistical significance threshold set as 0.05 (i.e. p<0.05). The mass spectrometry data were generated using the combined data from 2 independent experiments.
Results
Osteogenic reprogramming and calcium mineral deposition
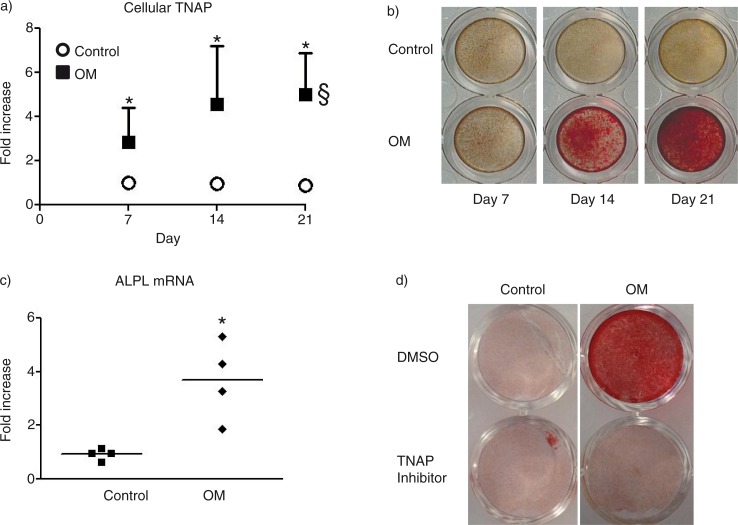
SMCs in calcifying media exhibit a significant 2.8-fold increase in TNAP activity after 7 days in culture that increases to 5-fold after 21 days in culture (Fig. 1a). This elevated TNAP activity is concomitant with an increase in mineralization as shown by Alizarin red S staining (Fig. 1b). The increased TNAP activity was found to correspond to a significant increase in TNAP mRNA expression after 14 days in the calcifying SMC (Fig. 1c). Treating SMCs with 1 µM TNAP inhibitor abrogated the mineralization observed after 21 days in culture (Fig. 1d).
Fig. 1.
Osteogenic transition of vascular SMCs. a) Time-dependent TNAP activity. §=p<0.05 by ANOVA. b) Representative images of Alizarin Red S stained mineralized matrix at day 21. c) ALP mRNA at day 14. d) Representative images of Alizarin Red S stained mineralized matrix at day 21 after treatment with TNAP inhibitor (1 µM). *=p<0.05 control versus OM. n=4 independent experiments. Mean±SD
Ultracentrifugation enrichment of calcifying matrix vesicles
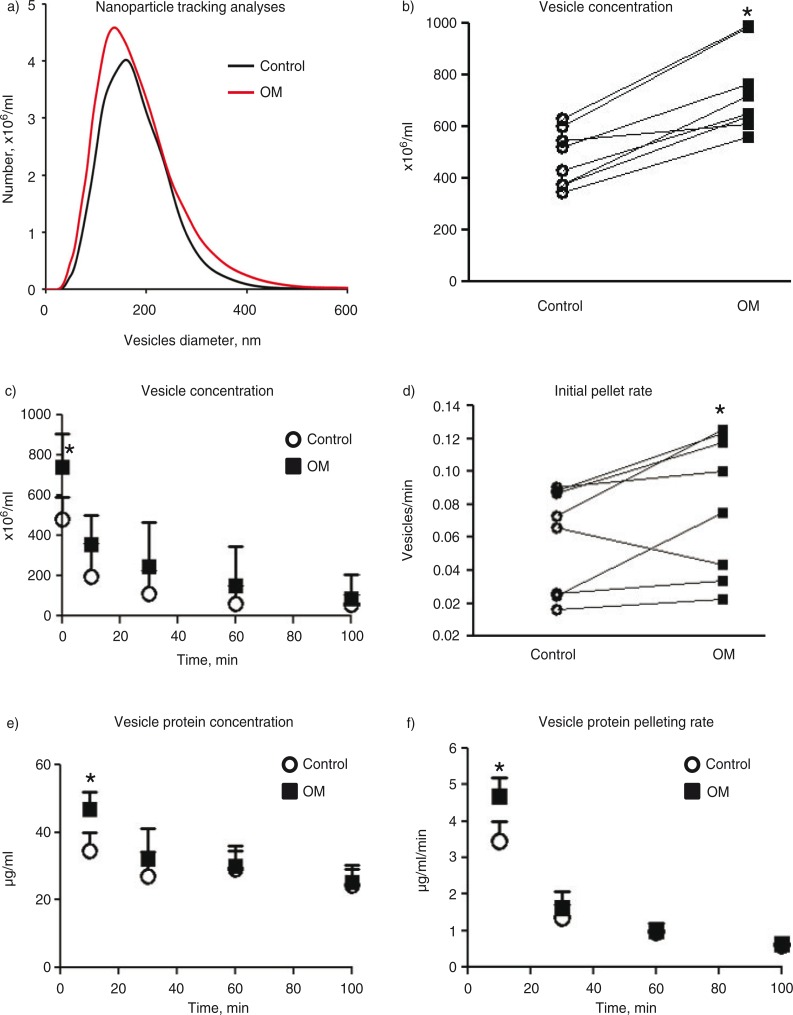
NTAs revealed similar size profiles for EVs released from SMCs cultured in control or calcifying media for greater than 14 days (Fig. 2a). The majority of the measured vesicles for both groups were between 30 and 300 nm in diameter, consistent with previous literature values for exosomes, matrix vesicles and microparticles (1). Vesicles released from calcifying SMCs exhibited a trending but insignificant increase in mean vesicle size (185±17) compared to vesicles from control cultures (175±16 nm, p=0.2); however, the concentration of vesicles measured in the conditioned media from calcifying SMCs was significantly higher compared to control SMCs (Fig. 2b). After 10 min ultracentrifugation at 100,000×g, the vesicle concentration between the 2 treatment groups was no longer different (Fig. 2c). A pair-wise comparison revealed a significant difference in the initial rate (i.e. the slope calculated from the initial vesicle concentration to the vesicle concentration remaining after 10 min) of pelleted vesicles (Fig. 2d) during the first 10 min (calcifying: 39±21 vesicles per minute, control: 28±17 vesicles per minute, p=0.049). No quantifiable difference in concentration was found for all subsequent ultracentrifugation times studied. Pair-wise tests were used for this analysis due to the variability in initial vesicle concentrations from the SMC donors and the desire to measure the rate of vesicles pelleted from the same donor samples over time. Similar to the NTA measurements, BCA protein analyses revealed significantly higher amounts of protein in the 10 min ultracentrifugation pellet of vesicles from calcifying SMCs compared to the protein pelleted from control vesicles (Fig. 2e). Each subsequent centrifugation time after 10 min showed no difference in pelleted protein between control and calcifying samples. Dividing the protein measured in the pellet at each time point by the ultracentrifugation time elapsed since the previous time point yields the rate of protein accumulation in the pellet for each time point considered (Fig. 2f). The rate of pelleted protein per minute was significantly higher in the calcifying samples compared to control samples only over the first 10 min of ultracentrifugation.
Fig. 2.
Ultracentrifugation of extracellular vesicles. a) NanoSight NTA plot of vesicle diameter versus concentration for control and calcifying samples. b) The concentration of vesicles in the conditioned media of control and calcifying samples as measured by NTA. Connecting lines indicate data obtained from the same experiment. c) The concentration of vesicles remaining in the conditioned media samples over the ultracentrifugation time course. d) The rate at which vesicles are pelleted during the first 10 min of ultracentrifugation. Connecting lines indicate data obtained from the same experiment. e) Protein assay of pelleted vesicles over the ultracentrifugation time course. f) The rate of pelleted protein over the ultracentrifugation time course. *=p<0.05. n=8 independent experiments. Mean±SD
Calcification potential of isolated vesicle populations
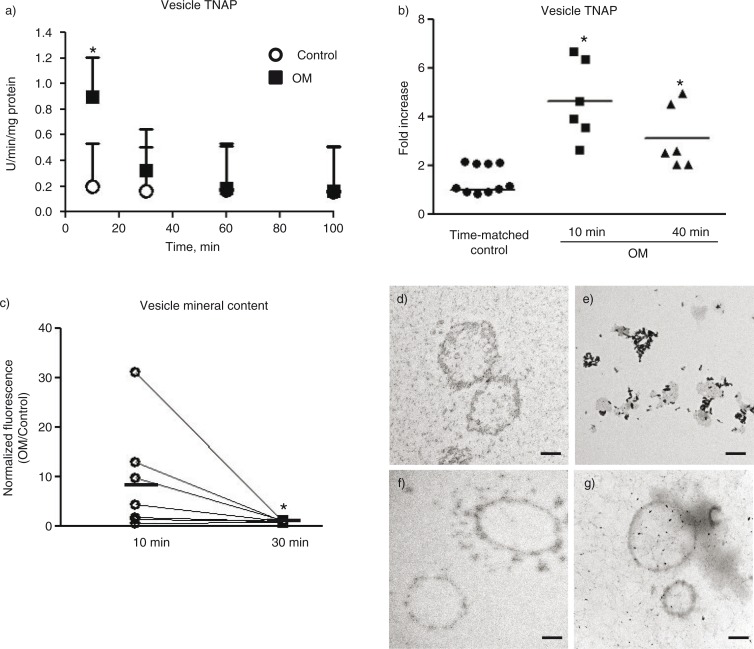
TNAP is a common marker of osteogenic potential and its presence in osteoblast-derived matrix vesicles has been shown to confer calcification potential to the vesicles (22). Therefore, TNAP activity was assayed in the pelleted fraction of each ultracentrifugation time point as a measure of vesicle calcification potential (Fig. 3a). The 10 min ultracentrifugation pellet from osteogenic samples exhibited significantly higher TNAP activity than the pellet from control samples. The pellets in each subsequent time point showed no difference in TNAP activity in osteogenic and control samples. Pelleted vesicles from 10 min ultracentrifugation of calcifying SMC media exhibited a 4.6-fold increase in TNAP activity compared to the control pellet (Fig. 3b). As a comparison of the 10 min ultracentrifugation with commonly used vesicle isolation protocols that use longer ultracentrifugation times, samples were subjected to 100,000×g ultracentrifugation for 40 min (without collecting pellets from earlier time points). Using this protocol, the pelleted portion from osteogenic samples displayed a 3.1-fold increase in TNAP activity compared to the time-matched control samples (Fig. 3b).
Fig. 3.
Calcification potential of pelleted vesicles. a) TNAP activity measured in the pelleted vesicles over the ultracentrifugation time course. b) TNAP activity measured in conditioned media samples after 10 min ultracentrifugation or 40 min ultracentrifugation. Control: time-matched control. Line indicates mean from 6 to 10 independent experiments. c) Normalized fluorescence intensity (per vesicle) and fold increase OM/Control from 7 independent experiments. Line indicates mean. *=p<0.05 by Wilcoxon matched-pairs signed-rank test. d) TEM images of vesicles from control or e) calcifying media pelleted at 10 min ultracentrifugation followed by an additional 20 min ultracentrifugation for the same f) control and g) calcifying samples (30 min total ultracentrifugation). *=p<0.05. n=6–7 independent experiments for A–C. Mean±SD.
To measure the mineral content of the EVs, we utilized OsteoImage-hydroxyapatite-specific fluorescent dye. EVs from SMCs cultured in calcifying media pelleted by 10 min ultracentrifugation at 100,000×g exhibited an 8.9-fold higher fluorescent signal per vesicle than EVs from SMCs cultured in control media (Fig. 3c). The EVs obtained from ultracentrifugation of the 10 min supernatant for an additional 30 min at 100,000×g exhibited significantly less fluorescent signal per vesicle (p=0.03). Only background fluorescence was observed when this assay was performed on media not used in SMC culture. To further assess changes in the isolated vesicle fractions, we utilized TEM to image the contents of the ultracentrifugation pellets. TEM of 10 min pellets revealed vesicular structures in control samples (Fig. 3d). The 10 min ultracentrifugation pellet from calcifying conditioned media exhibit electron dense structures associated with hydroxyapatite crystals (Fig. 3e) that are reminiscent of TEM images previously reported for calcifying matrix vesicles from osteoblasts (29). The 30 min ultracentrifugation pellet for both control (Fig. 3f) and calcifying (Fig. 3g) samples contained vesicular structures closer in appearance to the 10 min pellet from the control samples. The hydroxyapatite-associated structures were not observed in the 30 min ultracentrifugation pellets.
Mass spectrometry profile of calcifying matrix vesicles
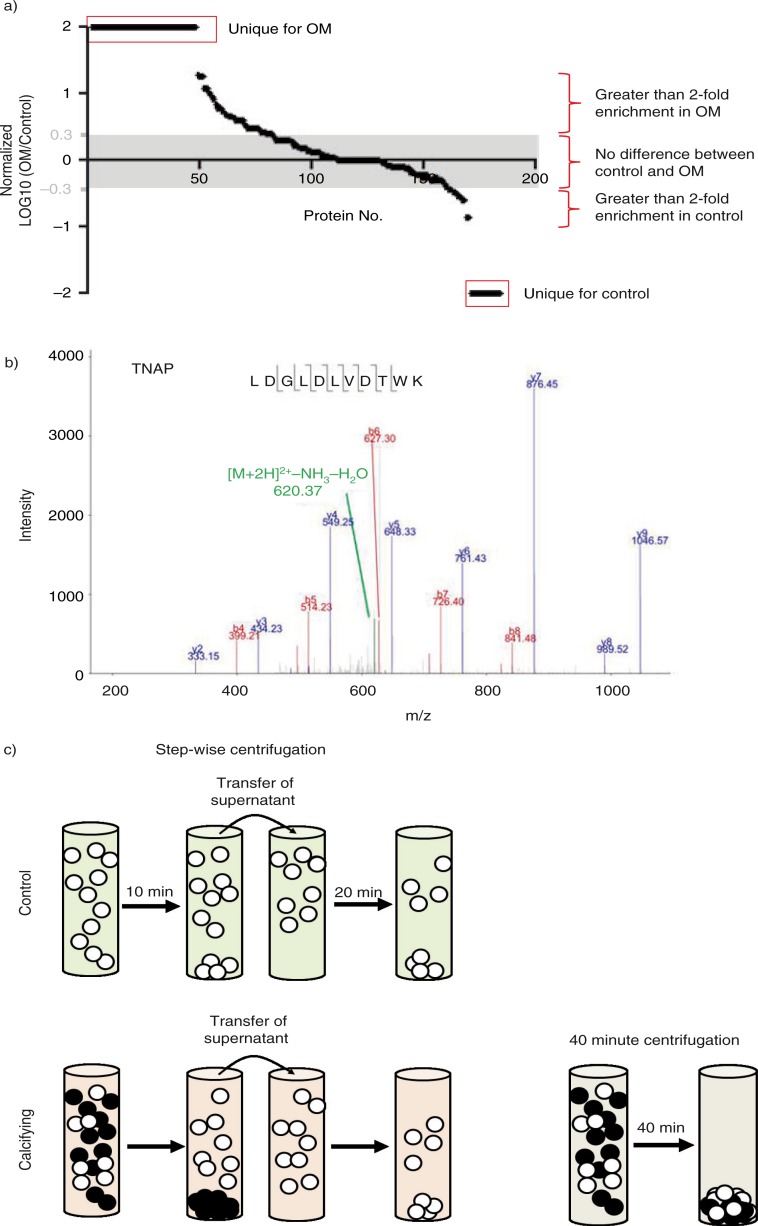
Mass spectrometry was used to perform an unbiased proteomic analysis of control and calcifying media pelleted portions from ultracentrifugation for 10 min, and from the pellets resulting from an additional 20 min ultracentrifugation of the 10 min supernatant (30 total min). A total of 422 proteins (with ≥2 unique peptides) were identified between the 2 conditions. The logarithm was taken of the ratio of the spectral counts identified in the calcifying samples versus control samples. We then plotted these normalized spectral count values from the proteins most enriched in the calcifying samples to those most enriched in the control samples to visualize the distribution of protein enrichment between these 2 treatment groups. After 10 min ultracentrifugation, 48 proteins were found exclusively in the pellets from the calcifying matrix vesicles, whereas only 27 proteins were found exclusively in pellets from the control samples (Fig. 4a). Further, a larger number of proteins were found to be enriched in the 10 min ultracentrifugation pellet of the calcifying samples versus the 10 min ultracentrifugation pellet of the control samples. An example MS2 spectrum of a known calcification marker, TNAP, is shown in Fig. 4b. The logarithmic plot is marked to show proteins with greater than 2-fold enrichment for each treatment group. We used a gene ontology (GO) database to establish the functional annotation of the identified proteins (Tables I and II). Proteins enriched in EVs isolated from control media were found to be associated with binding of extracellular matrix and matrix-associated components (Table I), whereas proteins enriched in calcifying EVs were found to be associated with calcium ion binding and enzymatic functions in addition to matrix and protein binding (Table II). Specifically, our mass spectrometric analyses identified 4 proteins previously shown to be involved in EV-mediated vascular calcification (Table III). In addition, among the proteins detected in the 10 min pellet by mass spectrometry were a variety of transmembrane proteins such as the common plasma membrane protein, sodium/potassium ATPase, indicating that this ultracentrifugation time was sufficient to pellet membrane-derived vesicles.
Fig. 4.
Mass spectrometric profile of pelleted vesicles. a) Log plot of protein peptide spectral counts in calcifying vesicles versus control vesicles after 10 min ultracentrifugation. b) An example MS2 spectrum of a known calcification marker, TNAP, is shown in Fig. 4b. c) Schematic of serial ultracentrifugation protocol to enrich calcifying vesicles versus traditional ultracentrifugation protocol for 40 min.
Table I.
Functional annotation for proteins enriched in control EV
| GO term ID | GO term | Number of genes | p |
|---|---|---|---|
| GO:0005539 | Glycosaminoglycan binding | 3 | 3.17 E − 04 |
| GO:0030247 | Polysaccharide binding | 3 | 4.08 E − 04 |
| GO:0001871 | Pattern binding | 3 | 4.08 E − 04 |
| GO:0030246 | Carbohydrate binding | 3 | 0.001919 |
| GO:0050840 | Extracellular matrix binding | 2 | 0.012363 |
Table II.
Functional annotation for proteins enriched in calcifying EV
| GO term ID | GO term | Number of genes | p |
|---|---|---|---|
| GO:0005509 | Calcium ion binding | 21 | 4.32 E − 09 |
| GO:0005198 | Structural molecule activity | 12 | 1.80 E − 04 |
| GO:0004857 | Enzyme inhibitor activity | 8 | 2.88 E − 04 |
| GO:0030246 | Carbohydrate binding | 7 | 0.006703 |
| GO:0004866 | Endopeptidase inhibitor activity | 6 | 6.29 E − 04 |
| GO:0030414 | Peptidase inhibitor activity | 6 | 8.02 E − 04 |
| GO:0005178 | Integrin binding | 4 | 0.002787 |
| GO:0004867 | Serine-type endopeptidase inhibitor activity | 4 | 0.009633 |
| GO:0051082 | Unfolded protein binding | 4 | 0.017534 |
| GO:0001871 | Pattern binding | 4 | 0.037223 |
| GO:0030247 | Polysaccharide binding | 4 | 0.037223 |
| GO:0001948 | Glycoprotein binding | 3 | 0.013423 |
| GO:0030911 | TPR domain binding | 2 | 0.014259 |
| GO:0030235 | Nitric-oxide synthase regulator activity | 2 | 0.023654 |
Table III.
PSM of known proteins involved in calcification from calcifying EVs
| Donor 1 | Donor 2 | |||
|---|---|---|---|---|
| Protein | 10 min | 30 min | 10 min | 30 min |
| Annexin A2 | 44 | 23 | 53 | 36 |
| Annexin A5 | 6 | 3 | 18 | 12 |
| Annexin A6 | – | – | 8 | 18 |
| S100A9 | 5 | – | – | – |
Discussion
Ultracentrifugation enrichment of calcifying matrix vesicles
Our results show an increase in the mineralization potential of vascular SMCs cultured in calcifying media. Mineral deposition was inhibited by the addition of an inhibitor to TNAP, thus providing the impetus for us to study the subset of calcifying EVs containing this enzyme. In this study we show that the subpopulation of TNAP-positive calcifying EV can be enriched from the total EV population by taking advantage of the increased vesicle density that results from the growth of mineral within the vesicles. Our results indicate that after 10 min of ultracentrifugation at 100,000×g, the dense calcifying vesicles are preferentially pelleted compared to the less dense control vesicle population (Fig. 4c) as assessed by TNAP activity and mineral content. Similar techniques could be used in the future to enrich certain vesicle populations expected to exhibit a physical difference from other vesicle populations.
The purpose of the current study was to take advantage of the physical density change associated with mineralization of calcifying EVs from the larger population of EVs. The method presented in this study allowed us to detect TNAP activity and mineralization in a subset of dense vesicles that were pelleted after only 10 min of ultracentrifugation. Our main goal was to enrich this calcifying subset and reduce the noise caused by other vesicle populations. Therefore, the current study provides a template for calcifying vesicle enrichment but does not address methods of obtaining a larger total number of calcifying vesicles. A viable method to accomplish this goal would be to enhance the calcifying vesicle signal by using collagenase digestion to isolate more calcifying vesicles that become trapped in the extracellular matrix and membrane microvilli during in vitro calcification (30–32). Future studies may combine these 2 methods to both increase the signal due to more matrix vesicles and decrease the noise caused by non-calcifying vesicle populations.
Researchers should be aware that although ultracentrifugation is a non-specific means to isolate vesicles, different ultracentrifugation parameters could produce different results. As we have shown in this study, the calcifying vesicles are completely removed from the conditioned media after 10 min of ultracentrifugation (Fig. 3a). By continuing the ultracentrifugation for longer times, increasing numbers of other vesicle populations are also pelleted. The presence of other vesicle populations in the calcification analyses reduces the signal-to-noise ratio of protein or gene-based assays because the other populations contribute to the total amount of protein or genetic content assayed but do not increase the amount of the calcification measurement. For example, in our study, ultracentrifugation of the conditioned media for 40 min instead of 10 min led to a decrease in the measured amount of TNAP activity per milligram of protein (Fig. 3b). The absolute amount of measured raw TNAP activity did not change between these ultracentrifugation times; however, the accumulation of vesicles with no TNAP activity in the pellet resulted in the observed loss in signal due to an increase in the amount of protein used to normalize the data.
Small but important changes could be missed by not considering the effects of ultracentrifugation speed and time on the vesicle populations that are isolated. In our study, we used 2 independent experiments for our mass spectrometry analyses. These 2 experiments were performed using cultures of primary vascular SMCs obtained from 2 independent donors. In Table III, the donor-to-donor variability can be seen. Many proteins follow similar trends; however, the abundance and behaviour can be slightly different between donors. This is always a challenge when working with primary cells, but we believe it is also a strength, as the results may be more reflective of those obtained from the human population. In order to better assess the variability between donors, technical variation and noise must be minimized. Now that we have a way to enrich our vesicles of interest, we can work to understand the donor variability and the implications on the production and calcification of EVs. We established the current protocol with no a priori assumptions regarding the protein composition of the calcifying EVs; however, we were able to detect many previously established markers of calcifying matrix vesicles (Table III). Once we have enriched our population of interest, we can continue to perform advanced proteomic profiling of calcifying vesicles using targeted mass spectrometry enabled by our spectral library (Fig. 4b) that contains MS fingerprints for all proteins enriched in this vesicle population (Fig. 4a). These targeted proteomics approaches offer increased sensitivity to better characterize subtle differences in protein expression (33) and compare the calcifying matrix vesicles produced by SMCs cultured in the conditions utilized in this study to matrix vesicles from other calcifying cells and those from calcified tissues.
In addition to speed and time, researchers should also consider the effect of media density and viscosity and the length of the ultracentrifugation tubes used to collect the samples. Larger tubes or a more dense or viscous media would require longer or higher speed ultracentrifugation protocols than shorter tubes or less viscous media (see physical density analysis below). By considering all of these parameters, studies may be able to better identify protein changes in vesicle populations that can then be used to establish more sensitive isolation techniques (e.g. immunoprecipitation). Further, by understanding the consequences and theoretical underpinnings of ultracentrifugation, important physical properties of vesicular populations may be ascertained.
Density analysis
The ultracentrifugation methods presented can be used to estimate differences in density between calcifying and non-calcifying EVs. As shown, significantly greater amounts of protein are pelleted after 10 min ultracentrifugation for the calcifying samples compared to the control samples. If the amount of protein per vesicle is approximately constant, the ratio of the protein pelleted rates (Fig. 2f) should match the ratio of the initial pellet rates measured by NTA (Fig. 2d). Taking the ratio of the initial rates of vesicle depletion shown in Fig. 2d gives a value of 1.37, which closely matches the ratio of protein accumulation (Fig. 2e) of 1.35. In other words, the rate at which vesicles are depleted in the calcifying samples over the first 10 min (k calcifying) is 1.35 times higher than the rate at which vesicles are depleted from control samples over the same time (k control):
| 1 |
By considering the forces acting on vesicles during ultracentrifugation, we can show that these results indicate that calcifying vesicles are approximately 35% more dense (i.e. reduced vesicle density, ρ v, compared to the culture media density, ρ s) than control vesicles. At terminal velocity as the vesicles are pelleted, the force produced by ultracentrifugation (F c) is opposed by a buoyancy force (F B) and a drag force on the vesicle (F D).
| 2 |
The F c is found by multiplying the mass of an individual vesicle by the acceleration due to ultracentrifugation.
| 3 |
where r is the radius of an individual vesicle, ρ v is the density of an individual vesicle, ω is the angular velocity of the centrifuge rotor and R is the radius from the edge of the centrifuge arm to the centre of the rotor. The F B is generated by the displacement of the liquid as the vesicle moves through the solution with the ultracentrifugation force.
| 4 |
where ρ s is the density of the culture media solution. At terminal velocity (v t, when the acceleration of the vesicles is zero), the F D experienced by a vesicle is a function of the vesicle radius and the viscosity of the culture media solution (η) such that
| 5 |
As in equation (1), at terminal velocity, the F C is balanced by the F D and F B to give
| 6 |
By rearranging and combining terms, we find a relationship for the density difference between the vesicle and solution as a function of the other parameters.
| 7 |
For the time required to pellet a vesicle (τ) over a certain length (l), since there is no acceleration when the vesicle reaches v t,
| 8 |
The time, τ, required to pellet a vesicle is the reciprocal of the rate at which vesicles are pelleted (k):
| 9 |
Therefore, the original relationship for reduced density becomes
| 10 |
If we reasonably assume that all ultracentrifugation parameters are constant, that the vesicles from each treatment group are similar in size, and that the vesicles travel similar lengths in the tubes, then the ratio of reduced densities simplifies to
| 11 |
The calcifying and control subscripts denote the reduced densities and rates for calcifying and control samples, respectively. As shown in equation (1), from our data this ratio of rates is approximately 1.35, and therefore, the ratio of the reduced densities must also equal 1.35. Using this approach, we can begin to estimate the effects of hydroxyapatite mineralization on the physical properties of the calcifying EVs. Future studies could use spectroscopic techniques to further probe the mineral content within the calcifying vesicles for better analyses of the mass that is added to the vesicles by the growing mineral. The nucleating events associated with calcification remain largely unknown. By better understanding the physical parameters, we can begin to build better models of vesicle interactions and the nucleation of mineral associated with calcification. Further, as shown in this study, we can also take advantage of these physical changes to separate the calcification for enhanced signal-to-noise in our proteomic analyses and ultimately give a clearer fingerprint of calcifying vesicles. These techniques may lead to the identification of novel therapeutic approaches to prevent or alter the nucleation of calcification by EVs.
Acknowledgements
The authors thank Mr. Iwao Yamada and Mr. Brett Pieper for mass spectrometry support.
Footnotes
†These authors contributed equally to the work.
Conflict of interest and funding
This project was supported by a research grant from Kowa Company Ltd. to Dr. Masanori Aikawa. Dr. Masaya Iwashita is an employee of Kowa Company Ltd. Dr. Elena Aikawa is supported by grants from the National Institutes of Health (R01HL114805; R01HL109506).
References
- 1.Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–88. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. J Vis Exp. 2012:e3037. doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Momen-Heravi F, Balaj L, Alian S, Mantel PY, Halleck AE, Trachtenberg AJ, et al. Current methods for the isolation of extracellular vesicles. Biol Chem. 2013;394:1253–62. doi: 10.1515/hsz-2013-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–8. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 6.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126:5553–65. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 7.Bobrie A, Colombo M, Krumeich S, Raposo G, Thery C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012;1:18397. doi: 10.3402/jev.v1i0.18397. doi: http://dx.doi.org/10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. doi: http://dx.doi.org/10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 10.New SE, Aikawa E. Role of extracellular vesicles in de novo mineralization: an additional novel mechanism of cardiovascular calcification. Arterioscler Thromb Vasc Biol. 2013;33:1753–8. doi: 10.1161/ATVBAHA.112.300128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.New SE, Goettsch C, Aikawa M, Marchini JF, Shibasaki M, Yabusaki K, et al. Macrophage-derived matrix vesicles: an alternative novel mechanism for microcalcification in atherosclerotic plaques. Circ Res. 2013;113:72–7. doi: 10.1161/CIRCRESAHA.113.301036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson HC, Garimella R, Tague SE. The role of matrix vesicles in growth plate development and biomineralization. Front Biosci. 2005;10:822–37. doi: 10.2741/1576. [DOI] [PubMed] [Google Scholar]
- 13.Anderson HC. Vesicles associated with calcification in the matrix of epiphyseal cartilage. J Cell Biol. 1969;41:59–72. doi: 10.1083/jcb.41.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapustin AN, Davies JD, Reynolds JL, McNair R, Jones GT, Sidibe A, et al. Calcium regulates key components of vascular smooth muscle cell-derived matrix vesicles to enhance mineralization. Circ Res. 2011;109:e1–12. doi: 10.1161/CIRCRESAHA.110.238808. [DOI] [PubMed] [Google Scholar]
- 15.Bobryshev YV, Killingsworth MC, Lord RS, Grabs AJ. Matrix vesicles in the fibrous cap of atherosclerotic plaque: possible contribution to plaque rupture. J Cell Mol Med. 2008;12:2073–82. doi: 10.1111/j.1582-4934.2008.00230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu HH, Camacho NP. Isolation of calcifiable vesicles from human atherosclerotic aortas. Atherosclerosis. 1999;143:353–62. doi: 10.1016/s0021-9150(98)00322-0. [DOI] [PubMed] [Google Scholar]
- 17.Hsu HH, Camacho NC, Tawfik O, Sun F. Induction of calcification in rabbit aortas by high cholesterol diets: roles of calcifiable vesicles in dystrophic calcification. Atherosclerosis. 2002;161:85–94. doi: 10.1016/s0021-9150(01)00623-2. [DOI] [PubMed] [Google Scholar]
- 18.Maldonado N, Kelly-Arnold A, Cardoso L, Weinbaum S. The explosive growth of small voids in vulnerable cap rupture; cavitation and interfacial debonding. J Biomech. 2013;46:396–401. doi: 10.1016/j.jbiomech.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maldonado N, Kelly-Arnold A, Vengrenyuk Y, Laudier D, Fallon JT, Virmani R, et al. A mechanistic analysis of the role of microcalcifications in atherosclerotic plaque stability: potential implications for plaque rupture. Am J Physiol Heart Circ Physiol. 2012;303:H619–28. doi: 10.1152/ajpheart.00036.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly-Arnold A, Maldonado N, Laudier D, Aikawa E, Cardoso L, Weinbaum S. Revised microcalcification hypothesis for fibrous cap rupture in human coronary arteries. Proc Natl Acad Sci USA. 2013;110:10741–6. doi: 10.1073/pnas.1308814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutcheson JD, Maldonado N, Aikawa E. Small entities with large impact: microcalcifications and atherosclerotic plaque vulnerability. Curr Opin Lipidol. 2014;25:327–32. doi: 10.1097/MOL.0000000000000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson HC. Matrix vesicles and calcification. Curr Rheumatol Rep. 2003;5:222–6. doi: 10.1007/s11926-003-0071-z. [DOI] [PubMed] [Google Scholar]
- 23.Steitz SA, Speer MY, Curinga G, Yang HY, Haynes P, Aebersold R, et al. Smooth muscle cell phenotypic transition associated with calcification: upregulation of Cbfa1 and downregulation of smooth muscle lineage markers. Circ Res. 2001;89:1147–54. doi: 10.1161/hh2401.101070. [DOI] [PubMed] [Google Scholar]
- 24.Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharmaceut Res. 2010;27:796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karnovsky M. A formaldehyde-glutaraldehyde fixative of high osmolarity for use in electron microscopy. J Cell Biol. 1965;27:137A. [Google Scholar]
- 26.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 27.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 28.Kall L, Storey JD, MacCoss MJ, Noble WS. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- 29.Anderson HC, Sipe JB, Hessle L, Dhanyamraju R, Atti E, Camacho NP, et al. Impaired calcification around matrix vesicles of growth plate and bone in alkaline phosphatase-deficient mice. Am J Pathol. 2004;164:841–7. doi: 10.1016/s0002-9440(10)63172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu LN, Yoshimori T, Genge BR, Sauer GR, Kirsch T, Ishikawa Y, et al. Characterization of the nucleational core complex responsible for mineral induction by growth plate cartilage matrix vesicles. J Biol Chem. 1993;268:25084–94. [PubMed] [Google Scholar]
- 31.Balcerzak M, Malinowska A, Thouverey C, Sekrecka A, Dadlez M, Buchet R, et al. Proteome analysis of matrix vesicles isolated from femurs of chicken embryo. Proteomics. 2008;8:192–205. doi: 10.1002/pmic.200700612. [DOI] [PubMed] [Google Scholar]
- 32.Reynolds JL, Joannides AJ, Skepper JN, McNair R, Schurgers LJ, Proudfoot D, et al. Human vascular smooth muscle cells undergo vesicle-mediated calcification in response to changes in extracellular calcium and phosphate concentrations: a potential mechanism for accelerated vascular calcification in ESRD. J Am Soc Nephrol. 2004;15:2857–67. doi: 10.1097/01.ASN.0000141960.01035.28. [DOI] [PubMed] [Google Scholar]
- 33.Lassman ME, McAvoy T, Lee A, Chappell D, Wong O, Zhou H, et al. Practical immunoaffinity-enrichment LC-MS for measuring protein kinetics of low-abundance proteins. Clin Chem. 2014;60:1217–24. doi: 10.1373/clinchem.2014.222455. [DOI] [PMC free article] [PubMed] [Google Scholar]