Abstract
An origin activation checkpoint has recently been discovered in the G1 phase of the mitotic cell cycle, which can be triggered by loss of DNA replication initiation factors such as the Cdc7 kinase. Insufficient levels of Cdc7 activate cell cycle arrest in normal cells, whereas cancer cells appear to lack this checkpoint response, do not arrest, and proceed with an abortive S phase, leading to cell death. The differential response between normal and tumor cells at this checkpoint has led to widespread interest in the development of pharmacological Cdc7 inhibitors as novel anticancer agents. We have used RNAi against Cdc7 in combination with SILAC-based high resolution MS proteomics to investigate the cellular mechanisms underlying the maintenance of the origin activation checkpoint in normal human diploid fibroblasts. Bioinformatics analysis identified clear changes in wide-ranging biological processes including altered cellular energetic flux, moderate stress response, reduced proliferative capacity, and a spatially distributed response across the mitochondria, lysosomes, and the cell surface. These results provide a quantitative overview of the processes involved in maintenance of the arrested state, show that this phenotype involves active rather than passive cellular adaptation, and highlight a diverse set of proteins responsible for cell cycle arrest and ultimately for promotion of cellular survival. We propose that the Cdc7-depleted proteome maintains cellular arrest by initiating a dynamic quiescence-like response and that the complexities of this phenotype will have important implications for the continued development of promising Cdc7-targeted cancer therapies.
Keywords: Quantitative Proteomics, Mass Spectrometry, SILAC, Cdc7, DNA Replication
Introduction
The DNA replication licensing process of the mitotic cell cycle is stringently and precisely regulated to ensure correct duplication of the eukaryotic genome only once per cell cycle.1-3 During early G1 phase, origin “licensing” occurs at thousands of replication origins distributed along human chromosomes and is a prerequisite for the initiation of DNA replication. The origin recognition complex (ORC) binds to DNA at these sites and recruits Cdc6 and Cdt1, which in turn load the minichromosome maintenance replicative helicase (MCM2-7) onto DNA. Together this assembly is termed the pre-replicative complex (pre-RC). Origin activation occurs in late G1 phase and involves phosphorylation of the pre-RC constituents by at least two protein kinases, CDK (cyclin-dependent kinases) and the Cdc7/ASK kinase. This leads to local unwinding of the double-stranded DNA, loading of the replisome (composed of DNA polymerases and auxiliary factors), and progression into S phase for DNA synthesis and chain elongation.1-3 To avoid reduplication of the genome and potentially cancer-causing defects, the DNA replication process is strictly monitored by cell cycle checkpoints and by mechanisms that prevent the formation of pre-RCs during the S, G2, and M phases of the cell cycle.
The highly conserved Cdc7 kinase plays a critical role in the DNA replication initiation pathway by promoting entry into S phase through phosphorylation and activation of the MCM2-7 helicase.4-6 Several other functions have been attributed to the Cdc7 kinase, including cohesion loading onto chromatin for chromosomal segregation in mitosis,7 double-strand break formation during meiotic recombination,8 and activation of the DNA damage repair ATR-Chk1 pathway.9,10 Recently, targeting of Cdc7 with RNAi or small molecule inhibitors has been shown to result in potent cancer-cell-specific killing in a range of tumor types of both epithelial and mesenchymal origin.1,11-13 Cancer cells are thought to establish only limited numbers of replication forks under Cdc7 rate-limiting conditions, causing fork stalling and collapse during an abortive S phase and triggering apoptotic cell death. The specificity of this novel anticancer approach is critically dependent on the fact that untransformed cells avoid lethal S phase progression in the presence of low Cdc7 kinase activity by eliciting a Cdc7 inhibition checkpoint response that arrests cells at the G1/S boundary.11,12,14 Our recent studies in normal human diploid fibroblasts have revealed the molecular architecture and circuitry involved in this enigmatic checkpoint response.15 In arrested cells, FoxO3a drives the activation of ARF leading to loss of Mdm2, p53 stabilization, activation of p21 and mediates p15INK4B upregulation. p53 in turn activates expression of the Wnt/β-catenin signaling antagonist Dkk3, leading to Myc and cyclin D1 down-regulation. The resulting loss of CDK activity inactivates the Rb-E2F pathway and overrides the G1-S transcriptional program leading to a robust G1/S cell cycle arrest.15 Notably, this signaling network includes many common proto-oncogenes and tumor suppressors, which explains why so many tumor types are unable to engage a protective G1/S checkpoint response in response to Cdc7 rate-limiting conditions.
Currently there is intense interest in the development of pharmacological Cdc7 inhibitors as novel anticancer agents.13,16,17 We have recently demonstrated that the cell cycle arrest following activation of the Cdc7-inhibition checkpoint in primary somatic cells is fully reversible on recovery of Cdc7 kinase activity and that cells remain viable while arrested at the G1-S boundary.12 However, an important and unanswered question relates to how primary cells maintain cell viability following activation of the Cdc7-inhibition checkpoint, information that is of crucial importance in understanding how human tissues can potentially avoid toxicity from Cdc7 inhibitors, and thus reinforcing the specificity of such a therapeutic approach. In this paper we apply a global proteomics approach to quantitatively evaluate the cellular-wide response to Cdc7 depletion and to better understand the preservation of the checkpoint phenotype in normal cells. The origin activation checkpoint was triggered by RNAi against Cdc7 in IMR90 fibroblasts and a quantitative SILAC proteomics strategy with high accuracy LC–MS/MS was applied to this experimental system.
We show that the predominant response to Cdc7 depletion is to actively maintain the cells in an arrested state by modulating the cellular metabolic signature, by inducing an oxidative stress response to limit cellular damage, and by suppressing RNA stability and protein translation. Our results suggest a spatially distributed response over different subcellular locations including mitochondria, lysosomes, and the cell surface. In short, Cdc7 depletion triggers a panel of responses that are similar to those evoked during cellular quiescence (G0). Contrary to the widely held assumption that quiescent cells exist in a static and passive phase, we support the opinion that G0 is actually a dynamic and highly active collection of states responsible for reversible cellular arrest.18 In addition to well-defined cellular pathways and networks, our study reveals a novel role for many proteins that have not been previously associated with cell cycle regulation. This global and quantitative proteomics analysis of the Cdc7-depleted cellular environment will provide a valuable reference point for ongoing Cdc7-targeted drug development.
Experimental Section
Cell Culture and SILAC Labeling
IMR90 (ATCC CCL-186), a human primary diploid fibroblast adherent cell line, was obtained from LGC Standards (Middlesex, U.K.) at population doubling (PD) 12. All experiments with IMR90 cells were performed with a PD of less than 22. IMR90 cells were cultured at 37 °C with 5% CO2 in SILAC-specific DMEM media (Invitrogen, Paisley, U.K.), supplemented with 10% dialyzed FBS (Invitrogen), 100 U/mL penicillin and 100 μg/mL streptomycin (Sigma Aldrich, Poole, U.K.) according to manufacturer’s instructions (Invitrogen, Paisley, U.K.). For quantitative SILAC analysis of two labeling states, the cells were divided into two populations, labeled with either “light” l-lysine and l-arginine (Lys0, Arg0) or “heavy” 13C6-lysine and 13C6,15N4-arginine (Lys6, Arg10) (Invitrogen, Paisley, U.K.) and cultured for six passages to achieve full incorporation of the SILAC amino acids.
RNA Interference
Small interfering RNA (siRNA) depletion of Cdc7 was performed on the “light” population, while the “heavy” cells were transfected with a control oligo for three biological replicates. One reverse labeling experiment was performed. Cdc7 expression was inhibited in SILAC-labeled IMR90 cells with custom double-stranded RNA oligonucleotides (Ambion, Warrington, U.K.) at 10 nM concentration for 72 h (5′→3′ sense oligo: GCUCAGCAGGAAAGGUGUUUU; antisense oligo: AACACCUUUCCUGCUGAGCUU). The Cdc7 siRNA used in this study has previously been extensively characterized for transfection efficiency, off-target effects, BrdU incorporation assays and flow cytometry studies.11,15 Nontargeting siRNA was used as the negative control. For the rescue experiment, the full 1725 bp CDC7 cDNA sequence containing four silent, single base pair mutations in the 21 bp Cdc7-siRNA interaction region was inserted into a pCMV6-AC expression vector (OriGene) to fully abolish the siRNA effect.
Cell Population Growth Assessment, Cell Cycle Analysis, and Immunofluorescence
Phase contrast microscopy was performed with an inverted Axiovert 200 M (Carl Zeiss, Welwyn Garden City, U.K.) and Axiovision software. Flow cytometric cell cycle analysis was performed as described previously.19 For detection of BrdU incorporation, cells were pulsed with 100 μM BrdU (2 h), fixed in 3.7% paraformaldehyde (5 min), and permeabilized with 0.2% Triton X-100 (5 min). Coverslips were incubated with 2 N HCl (1 h), washed with PBS, and incubated with primary anti-BrdU antibody (Alexis Biochemicals, Exeter, U.K.), 50 ng/mL propidium iodide, and 50 ng/mL RNase A for 1 h at 37 °C. Slides were mounted in Vectashield mounting medium (Vector Laboratories, Peterborough, U.K.). DNA was visualized with 1.5 μg/mL DAPI. Fluorescence confocal microscopy of random fields of cells was performed on a Leica TCS SP confocal microscope (Leica, Milton Keynes, U.K.). At least 400 cells were scored for BrdU incorporation and quantified as percentages of the total number of cells.
Sample Preparation, Protein Separation, Immunoblotting, and In-Gel Digestion
Crude nuclear extracts and cytoplasmic fractions were prepared by lysing cells in buffer containing 10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, and protease inhibitors. Nuclei were precipitated by centrifuging at 1,000 × g for 5 min at 4 °C, and the cytoplasmic fraction was collected. Nuclear pellets were washed with the same buffer, lysed in modified RIPA buffer (50 mM Tris-HCl pH 7.4, 300 mM NaCl, 1% sodium deoxycholate, 1% NP-40, 1 mM EDTA, 0.1% SDS) for 30 min, sonicated, and centrifuged at 13,000 × g.
SILAC analysis was performed on whole cell extracts, where SILAC-labeled cells were harvested and lysed for 45 min in ice-cold modified RIPA buffer supplemented with protease and phosphatase inhibitors (Roche Diagnostics, Lewes, U.K.). Lysates were sonicated on ice and centrifuged at 14,000 × g, 4 °C for 20 min. Protein concentration was evaluated by the Bio-Rad protein assay kit (Bio-Rad, Hemel Hempstead, U.K.). For Western blot analysis, 60 μg of total protein was loaded in each lane and separated by 4% to 20% SDS-PAGE. Proteins were transferred onto PVDF membranes (Bio-Rad) by semidry electroblotting. Blocking, antibody incubations, and washing steps were performed as described.20 Antibodies used: Cdc7 (MBL International, Woburn, MA), Mcm2 and Orc4 (BD Biosciences, Oxford, U.K.), Mcm2 phospho-Ser-41 and Mcm2 phospho-Ser-53 (Bethyl Laboratories, Montgomery, TX), ASK (Eurogentec, Seraing, Belgium), Mcm7 (Thermo Fisher Scientific, Fremont, CA), NDRG1 (Abcam, Cambridge, U.K.), PCNA (Santa Cruz, CA), ribosomal protein S6, GPX1 and FTH1 (New England Biolabs, Herts, U.K.), and p16 and β-actin (Sigma, Gillingham, U.K.).
For comparative SILAC analysis, “heavy” and “light” whole cell lysates were combined in a 1:1 ratio based on protein concentration (30 μg each) and separated by SDS-PAGE under reducing conditions. Proteins were visualized by silver staining (ProteoSilver Plus, Sigma Aldrich, Poole, U.K.), and equally sized bands were excised from the gel lane and processed with the Progest Investigator (Digilab, Huntingdon, U.K.) using established protocols for reduction and alkylation.21 Finally, gel plugs were rehydrated in 20 μg/mL sequencing grade modified trypsin (Promega, Southampton, U.K.) and incubated overnight at 37 °C. Tryptic peptides were eluted, vacuum-dried, resuspended in 0.1% formic acid, and analyzed by LC–MS/MS.
Mass Spectrometry
LC–MS/MS was performed with an LTQ-Orbitrap Classic mass spectrometer (Thermo Fisher Scientific, U.K.). Peptide samples were loaded using a Surveyor MS Pump and Micro AS autosampler (Thermo Fisher Scientific, U.K.) onto a MiChrom C18 CapTrap for desalting and then introduced into the MS via a nanoelectrospray ion source consisting of a fused silica capillary column (i.d. 100 μm; o.d. 360 μm; length 20 cm; 5 μm C18 Reprosil, Nikkyo Technos Co., Japan). Separation was achieved by a dual gradient, formed of 5–23% Buffer B for 65 min, followed by 23%–40% Buffer B for 30 min, and a step gradient to 60% Buffer B for 5 min (Buffer A = 0.1% formic acid; Buffer B = 100% acetonitrile (ACN), 0.1% formic acid). The pump rate was reduced via a splitter to 0.9 μL/min. Measurements were performed in the positive ion mode. The full scan precursor MS spectra (450–1600 m/z) were acquired in the Orbitrap analyzer with a resolution of r = 60,000. This was followed by data dependent MS/MS fragmentation in centroid mode of the most intense ion from the survey scan using collision induced dissociation (CID) in the linear ion trap (normalized collision energy 35%, activation Q 0.25; electrospray voltage 1.4 kV; capillary temperature 200 °C; isolation width 2.00). This MS/MS scan event was repeated for the top 6 peaks in the MS survey scan. Target ions already selected for MS/MS were dynamically excluded for 40 s. Singly charged ions were excluded from MS/MS analysis. XCalibur software version 2.0.7 (Thermo Fisher Scientific, U.K.) was used for data acquisition.
Protein Identification and Quantification
Raw MS files from all replicate SILAC experiments were uploaded onto the MaxQuant software platform (version 1.0.13.13) for peaklist generation, quantification of SILAC pairs, identification of individual peptides, and assembly into protein groups.22,23 XCalibur raw files were processed by Quant.exe and Identify.exe of the MaxQuant suite, in combination with the Mascot search engine (version 2.2, Matrix Science, U.K.), and searched against a concatenated International Protein Index (IPI) human protein database (version 3.54; containing 75,710 entries, including 262 commonly observed contaminants such as porcine trypsin and some human keratins). This decoy database also contained reversed versions of all sequences and was created with the Sequence Reverser program within MaxQuant. Selected Max-Quant analysis parameters included trypsin enzyme specificity, SILAC doublet measurements of Lys6 and Arg10, 2 missed cleavages, minimum peptide length of 6 amino acids, minimum of 2 peptides (1 of which is unique), top 6 MS/MS peaks per 100 Da, peptide mass tolerance of 7 ppm for precursor ions, and MS/MS tolerance of 0.5 Da. Oxidation of methionine and N-terminal protein acetylation were selected as variable modifications, and cysteine carbamidomethylation was selected as a fixed modification. All proteins were filtered according to a false discovery rate (FDR) of 1% applied at both peptide and protein levels. Proteins were automatically quantified in the MaxQuant software: a minimum of 3 peptide ratio counts from razor and unique peptides were necessary for protein quantification, and the “requantification” option was enabled.22,23 An Experimental Design template was used to specify individual experiments and reverse labeling conditions within the analysis. In the final results files, all Protein Groups with a normalized ratio Significance B score of ≤0.05 were accepted for downstream analysis.
Bioinformatics Analysis
For the visualization and assignment of statistically over-represented functional annotations, Ingenuity Pathway Analysis (IPA; version 8.0),24 STRING (version 8.2),25 and Cytoscape (version 2.6.3)26 software were used. REFSEQ protein identifiers and expression fold changes were uploaded into IPA, and only direct interactions between molecules were selected for consideration by the IPA library. A Fisher’s exact test identified canonical pathways most significant to the data set. The open-source STRING database was used for evaluation of protein–protein interaction partners within the data set.25 A list of all altered proteins were uploaded into STRING using HUGO Gene identifiers as input with a 0.7 Confidence Score cutoff (high confidence). A protein association network was created where molecules are represented as nodes connected via edges that represent the supporting evidence. Cytoscape is an open-source software environment that provides a broad framework for biological data, where molecules are shown as nodes connected by edges that represent data attributes such as protein interactions. Cytoscape can be extended to incorporate many additional plug-in modules such as the BiNGO tool for Gene Ontology (GO) enrichment analysis27 or the MCODE algorithm for identification of densely connected protein clusters based on a graph theoretic clustering algorithm.28 MCODE (version 1.3) was used to identify densely interconnected regions within the STRING protein network. To assess over-representation of GO annotations, HUGO gene identifiers were uploaded into BiNGO (version 2.3) and a hypergeometric test was chosen with the Benjamini Hochberg FDR correction for multiple testing (significance level 0.05). The three GO vocabularies, namely, Biological Processes (GOBP), Cellular Component (GOCC), and Molecular Function (GOMF)29 were searched separately. GO terms are represented as nodes and are linked in a hierarchical layout, where the size of each node is proportional to the number of molecules within the term and nodes are colored by significance of enrichment of each term relative to the whole annotation.
Results
A Subset of Cellular Proteins Is Affected by Suppression of Cdc7 Kinase
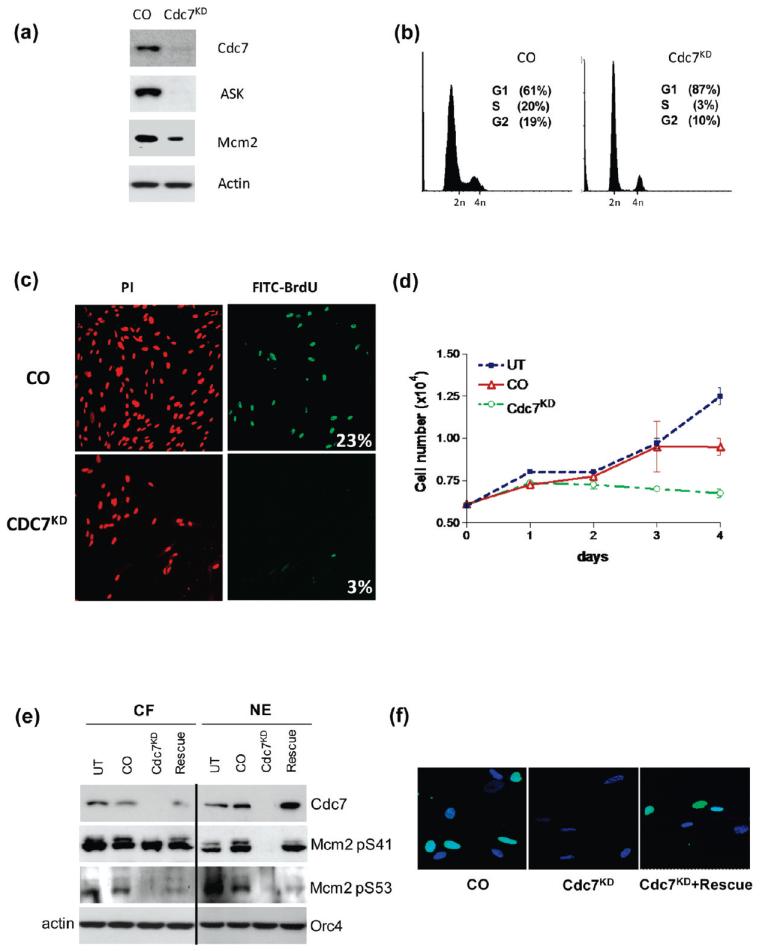
The general experimental approach and tools used in this study are outlined in Figure 1. The specificity of Cdc7 siRNA, the possibility of off-target effects, and rigorous experimental validation of the Cdc7-depleted system have all been previously addressed by our group,12,14,15 and evidence for the efficiency of the Cdc7 knockdown is summarized in Figure 2. Transfection of IMR90 fibroblasts with Cdc7-siRNA (CDC7KD) reduced Cdc7 mRNA levels by 80% relative to control-siRNA (CO) at 72 h, and Cdc7 protein levels were undetectable at this time point (Figure 2a). Protein levels of the Cdc7 regulatory subunit ASK (activator of S-phase kinase) were also substantially reduced. A reduction in total protein level and loss of phosphorylation at Ser-41 and Ser-53 was observed for the major Cdc7 substrate Mcm2 (Figure 2a and e). A shift to hypo-phosphorylated Mcm2 has been previously documented following Cdc7 depletion.11,12 Growth of Cdc7-depleted cells reached a plateau 48 h post-transfection, indicating cessation of cell proliferation (Figure 2d), and the majority of these arrested cells accumulated with G1 DNA content (87%) (Figure 2b). The percentage of BrdU-incorporating cells dropped from 23% in CO cells to 3% in CDC7KD cells (Figure 2c), providing further evidence that Cdc7 depletion led to a G1 arrested state. A rescue experiment was performed by the expression of a CDC7 gene variant containing silent mutations in the siRNA targeting region. This led to a recovery from cell cycle arrest, as demonstrated by re-expression of Cdc7 protein, phosphorylation of Mcm2 at Ser-41 and Ser-53, and increased BrdU incorporation (Figure 2e and f).
Figure 1.
SILAC workflow. A combined strategy involving RNAi against Cdc7, SDS-PAGE protein fractionation, LC–MS/MS data acquisition with a LTQ-Orbitrap (Thermo Fisher Scientific), and downstream bioinformatics for identification, quantification, and functional annotation was applied in this study.
Figure 2.
Cdc7 depletion arrests IMR90 fibroblasts in G1 phase of the cell cycle. (a) Immunoblot analysis for Cdc7, its regulatory subunit ASK, and its substrate Mcm2 in control-siRNA (CO) and Cdc7-siRNA transfected (CDC7KD) IMR90 whole cell extracts (actin loading control). (b) DNA content of CO and CDC7KD cells was assessed by FACS analysis. Percentages of cells found to be in G1, S, and G2 phases of the cell cycle are indicated. (c) CO and CDC7KD cells were stained for BrdU incorporation, and DNA was counterstained with propidium iodide (PI). Percentages of BrdU-incorporating cells are indicated. (d) Cell number was measured at the indicated time points (UT, untreated control). (e) Immunoblot analysis of Cdc7 protein levels and phosphorylation of Mcm2 at Ser-41 and at Ser-53 in cytoplasmic fraction (CF) and nuclear extract (NE) (actin and Orc4 loading controls). “Rescue” indicates IMR90 cells transfected with a CDC7 rescue plasmid. (f) Cells were measured for BrdU incorporation in CO, CDC7KD transfected cells, as well as CDC7KD cells treated with CDC7 rescue plasmid. Light blue indicates BrdU-incorporating cells, and dark blue indicates DAPI-stained nuclei.
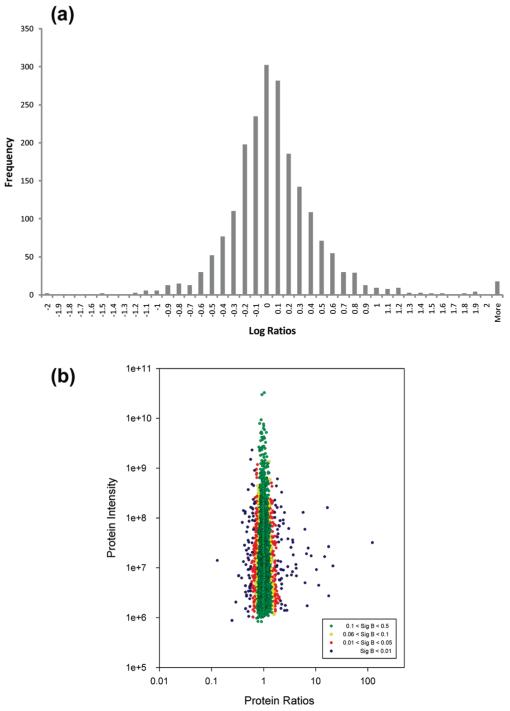
Three biological replicates were performed for SILAC analysis of Cdc7-depleted versus control siRNA transfected cells. A linear regression comparison of protein ratios for each biological replicate demonstrates the reproducibility of the replicates and is shown in Supplementary Figure S1 (Biological Replicate One versus Two: r2 = 0.7385; Biological Replicate One versus Three: r2 = 0.8142; Biological Replicate Two versus Three: r2 = 0.8589). In total, 338 individual SDS-PAGE gel slices from eight technical gel experiments were subjected to MS/MS analysis, resulting in 234,086 SILAC peptide pairs, of which 66% were sequenced (Table 1). Using an FDR of 1% at the protein and peptide level, 2,452 proteins were identified of which 2,045 were assigned quantitative abundance ratios based on a minimum of at least three abundance ratios for razor peptides (Table 1). Plotting the normalized (Cdc7-depleted/control) ratio values for all 2,045 quantified proteins demonstrated that the spread of the entire data set follows a Gaussian distribution and shows that the majority of cellular proteins are unaffected by Cdc7 knockdown (Figure 3a). Protein ratios were grouped into intensity bins of similar magnitude (each bin containing at least 300 proteins) and evaluated as being differentially expressed on the basis of the deviance of outlier ratios from the normalized distribution of all ratios (Significance B).22,23 Plotting the protein intensities versus the protein ratios (Figure 3b) showed that across all intensity ranges, significant ratio outliers (colored by Significance B values) are clearly distinct from the normalized ratio distribution; 315 quantified proteins were found to have a Significance B score of ≤0.05 and were taken to be significantly altered in expression. The SILAC ratios of several of these proteins were verified by Western blotting (Supplementary Figure S2). Supplementary Table S1 contains relevant protein and individual peptide information for each of the 315 differentially expressed proteins. These proteins were further evaluated by downstream applications to detect cellular functional attributes that are affected following impairment of replication origin activation. Supplementary Table S2 contains identification and quantification information relating to the 1,730 unchanged proteins (Significance B > 0.06).
Table 1.
Overview of MS Results Obtained with MaxQuant Softwarea
| SILAC analysis | |
|---|---|
| proteins identifiedb | 2452 |
| proteins quantifiedc | 2045 |
| proteins differentially expressed following Cdc7 depletiond | 315 |
| significantly upregulated proteins | 145 |
| significantly downregulated proteins | 170 |
| number of MS full scans | 2034346 |
| number of MS/MS scan events | 647831 |
| number of MS/MS identified | 246220 |
| peptide sequences identified | 20950 |
| number of SILAC pairs | 234086 |
| number of SILAC pairs sequenced | 154648 (66.06%) |
| number of SILAC pairs identified | 87626 |
Shown are the results obtained by MaxQuant (version 1.0.13.13) analysis of the Cdc7-depleted SILAC data set using defined analysis parameters (outlined in the Experimental Section).
Protein groups identified under MaxQuant search parameters with protein and peptide FDR = 1%.
Protein abundance ratios are calculated as median values using a minimum of 3 quantifiable razor peptides.
Protein ratios with Significance B ≤ 0.05 were accepted as differentially expressed.
Figure 3.
Overview of the distribution of abundances for the entire SILAC data set. (a) The spread of normalized (log) protein abundance ratios expressed as a histogram follows a normal distribution. (b) Normalized protein abundance ratios plotted against protein intensity. Individual proteins are colored by their Significance B (Sig B) value. Green: proteins with no detected change in abundance (1,601 proteins; Sig B) 0.1–0.5). Yellow: proteins with small, nonsignificant changes in abundance (129 proteins; Sig B) 0.06–0.1). Red: proteins with moderately significant abundance changes (181 proteins; Sig B) 0.01–0.05). Blue: proteins with highly significant abundance changes (134 proteins; Sig B < 0.01).
Cdc7 Knockdown Induces a Metabolic and Oxidative-Stress Response
The complete data set of 2,045 quantified proteins was uploaded into IPA, and an expression value cutoff of Significance B ≤ 0.05 was applied, allowing us to distinguish between altered and unaltered proteins in the analysis. Of the 315 proteins with Significance B ≤ 0.05, IPA identified 270 function/pathway eligible molecules, while 45 proteins did not correspond to a functional annotation in the IPA knowledge base. The successful molecules were mapped onto defined canonical pathways and ranked according to significance of association with the data set. The 10 top scoring IPA canonical pathways are listed in Table 2, the mapped proteins are overlaid onto each pathway in Supplementary Figures S3–S12, and details regarding all mapped proteins for each pathway are given in Supplementary Table S3.
Table 2.
IPA Canonical Pathways Affected by Cdc7 Suppressiona
| canonical pathway | BH p-value | no. mapped to pathway/total available for pathway | no. differentially expressed/no. mapped to pathway | differentially expressed molecules |
|---|---|---|---|---|
| Nrf2-mediated oxidative stress response | 4.97 × 10−8 | 68/181 | 18/68 (26.4%) | MGST1, FTL, DNAJC9, DNAJB4, GCLC, DNAJC10, TXNRD1, HMOX1, DNAJC8, ABCC1, DNAJB1, TXN, SQSTM1, DNAJB6, MGST3, EPHX1, FTH1, AKR1B1 |
| synthesis and degradation of ketone bodies | 5.13 × 10−8 | 9/19 | 6/9 (66.6%) | HADHB, ACAA1, ACAT1, HMGCL, HMGCS1, HADHA |
| butanoate metabolism | 1.89 × 10−5 | 25/128 | 8/25 (32%) | HADHB, PPME1, ACAA1, SUCLG2, ACAT1, HMGCL, HMGCS1, HADHA |
| bile acid biosynthesis | 2.29 × 10−4 | 11/97 | 6/11 (54.5%) | ADH5, HADHB, AKR1C1, ACAA1, PTGR1, HADHA |
| valine, leucine, and isoleucine degradation | 2.55 × 10−4 | 25/105 | 7/25 (28%) | HADHB, ACAA1, ACAT1, HMGCL, IVD, HMGCS1, HADHA |
| biosynthesis of steroids | 2.95 × 10−4 | 7/126 | 5/7 (71.4%) | MVD, FDPS, FDFT1, IDI1, LSS |
| granzyme A signaling | 3.06 × 10−4 | 10/18 | 4/10 (40%) | H1F0, HIST1H1C, HIST1H1B, HMGB2 |
| oxidative phosphorylation | 5.47 × 10−4 | 42/156 | 10/42 (23.8%) | ATP6 V1E1, NDUFS1, ATP6 V1C1, NDUFB7, NDUFS2, ATP6 V1G1, ATP6 V1A, ATP6 V1B2, NDUFA8, NDUFS4 |
| glutathione metabolism | 1.14 × 10−3 | 23/92 | 6/23 (26%) | MGST1, PGD, GPX1, GCLC, MGST3, IDH1 |
| propanoate metabolism | 1.74 × 10−3 | 23/128 | 6/23 (26%) | HADHB, ACAA1, SUCLG2, ACAT1, IVD, HADHA |
2,045 quantified proteins were uploaded into Ingenuity Pathway Analysis (IPA) software using REFSEQ identifiers, fold changes, and Significance B p-values. The expression value cut-off was set to 0.05. This resulted in 2,023 molecules being mapped to the Ingenuity knowledge base, of which 270 qualified as function/pathway eligible molecules (molecules that meet the expression value criteria and have at least one functional annotation in the IPA knowledge base). Shown are the top ranking 10 canonical pathways associated with this data set. A Benjamini–Hochberg test was used to test for significance of the over-representation of each pathway (BH p-value). A ratio value is given for the number of proteins identified within the pathway (no. mapped) divided by the total number of molecules for the pathway within the IPA library (total available). Another ratio is given for the number of significantly altered molecules (no. differentially expressed) divided by the number of proteins identified in the pathway (no. mapped). The differentially expressed proteins associated with each pathway are also listed. All proteins mapped to these 10 pathways are detailed in Supplementary Table S3 and proteins are overlaid onto each pathway in Supplementary Figures S3–S12.
“Nrf2-mediated oxidative stress response” was the most significantly affected pathway containing 18 differentially expressed proteins (Table 2). Supplementary Figure S3 clearly demonstrates that they are mainly associated with the “reduction in oxidative damage” and activation of “phase I and II metabolizing enzymes” branches of the pathway. A further 36 unchanged proteins were mapped downstream of Nrf2, indicating that a full-blown stress response was not initiated. The “Glutathione Metabolism” pathway was also identified as significantly altered, containing six up-regulated proteins (26% of molecules mapped to pathway) and provides further evidence for a mild antioxidant stress response (Supplementary Figure S11).
The other top-scoring pathways identified by IPA together indicate altered cellular metabolism, and four major themes are apparent: fatty acid metabolism (“Synthesis and Degradation of Ketone Bodies”, “Butanoate Metabolism”, and “Propanoate Metabolism”), steroid metabolism (“Biosynthesis of Steroids” and “Bile Acid Biosynthesis”), branched chain amino acid metabolism (“Valine, Leucine and Isoleucine Degradation”), and mitochondrial redox metabolism (“Oxidative Phosphorylation”). These metabolic processes are functionally interlinked, and some proteins participate in multiple pathways. For example, the mitochondrial trifunctional enzyme (HADHA and HADHB subunits) and the HMG-CoA synthase (HMGCS1), all of which were found to be up-regulated, are implicated in fatty acid metabolism, ketone body synthesis, and branched-chain amino acid degradation. Ketogenic reactions and the catabolism of branched chain amino acids (valine, leucine, and isoleucine) provide an alternative source of energy during fasting or low glucose conditions. HMG-CoA (synthesized by HMG-CoA synthase) is also a key intermediate in the synthesis of cholesterol, the major precursor for the synthesis of bile acids, steroid hormones, and fat-soluble vitamins. A significant increase in eight enzymes involved in the cholesterol biosynthetic pathway was observed (ACAT1, HMGCS1, MVD, IDI1, FDPS, FDFT1, LSS, and NSDHL). Cdc7 depletion also led to up-regulation of the cholesterol binding proteins NPC1 and NPC2, which facilitate the endosomal/lysosomal transport of cholesterol-containing lipoproteins.30
Cellular-Wide Functional Changes Following Cdc7 Depletion Can Be Categorized by GO Annotation
To identify descriptive GO annotations, the up-regulated (145 proteins) and down-regulated (170 proteins) groups were analyzed separately against each GO ontology. The most significantly enriched annotations for each GO category are presented in Table 3, with the corresponding proteins for each enriched term listed in Supplementary Table S4. An overview of enriched generic GOSlim terms is shown graphically in Figure 4, with the corresponding proteins for each enriched GOSlim term listed in Supplementary Table S5. The pattern of enriched GO terms differs considerably between the up- and down-regulated protein groups. BiNGO analysis for the group of 170 down-regulated proteins strongly suggests that Cdc7 knockdown results in reduced cellular biosynthesis and proliferation. The statistically enriched GOMF and GOBP terms include DNA, RNA, and protein binding; RNA (and mRNA) splicing, processing and metabolism; as well as nucleosome and chromatin assembly. The data set contains 36 down-regulated proteins associated with the GOBP term “nucleobase, nucleoside, nucleotide and nucleic acid metabolic process” (GO:0006139). GO enrichment analysis for the group of 145 up-regulated proteins indicated that catabolic processes, oxidation reduction, and lipid metabolism were the major biological processes enhanced following Cdc7 depletion. The corresponding molecular functions include oxidoreductase activity, catalytic activity, and acetyl-CoA transferase activity. Together, the enriched GO terms clearly demonstrate a reduction in the transcriptional and translational machinery of the cell as well as activation of energy regenerative and metabolic processes during cell cycle arrest triggered by the origin activation checkpoint.
Table 3.
Gene Ontology Term Enrichment for Protein Sets with Increased/Decreased Abundancea
| up-regulated functions |
down-regulated functions |
|||||||
|---|---|---|---|---|---|---|---|---|
| category | GO ID | term | x | Benjamini p-value | GO ID | term | x | Benjamini p-value |
| GO biological | 55114 | oxidation reduction | 28 | 2.46 × 10−10 | 6374 | nuclear mRNA splicing, via spliceosome | 12 | 6.68 × 10−6 |
| process | 44255 | cellular lipid metabolic process | 23 | 1.43 × 10−6 | 375 | RNA splicing, via transesterification reactions | 12 | 6.68 × 10−6 |
| 16096 | isoprenoid metabolic process | 7 | 2.21 × 10−6 | 377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 12 | 6.68 × 10−6 | |
| 42221 | response to chemical stimulus | 23 | 2.54 × 10−6 | 16071 | mRNA metabolic process | 16 | 3.76 × 10−5 | |
| 44248 | cellular catabolic process | 23 | 3.79 × 10−6 | 6397 | mRNA processing | 15 | 3.76 × 10−5 | |
| 6695 | cholesterol biosynthetic process | 6 | 4.60 × 10−6 | 6394 | RNA processing | 19 | 4.07 × 10−5 | |
| 6629 | lipid metabolic process | 24 | 5.26 × 10−6 | 6395 | RNA splicing | 14 | 5.24 × 10−5 | |
| 6091 | generation of precursor metabolites and energy | 14 | 6.93 × 10−6 | 22607 | cellular component assembly | 11 | 6.27 × 10−5 | |
| 9056 | catabolic process | 24 | 1.35 × 10−5 | 6334 | nucleosome assembly | 9 | 7.56 × 10−5 | |
| 16126 | sterol biosynthetic process | 6 | 2.27 × 10−5 | 31497 | chromatin assembly | 9 | 1.59 × 10−4 | |
| GO molecular | 16491 | oxidoreductase activity | 32 | 2.27 × 10−12 | 3723 | RNA binding | 32 | 4.08 × 10−11 |
| function | 3824 | catalytic activity | 81 | 2.39 × 10−9 | 5515 | protein binding | 104 | 9.93 × 10−7 |
| 16616 | oxidoreductase activity, acting on the CH–OH group of donors, NAD or NADP as acceptor | 10 | 1.66 × 10−6 | 8092 | cytoskeletal protein binding | 18 | 2.45 × 10−5 | |
| 9055 | electron carrier activity | 14 | 2.36 × 10−6 | 3779 | actin binding | 15 | 2.45 × 10−5 | |
| 9055 | oxidoreductase activity, acting on CH–OH group of donors | 10 | 2.72 × 10−6 | 5488 | binding | 141 | 4.40 × 10−4 | |
| 50662 | coenzyme binding | 11 | 4.07 × 10−6 | 31072 | heat shock protein binding | 6 | 1.52 × 10−3 | |
| 16651 | oxidoreductase activity, acting on NADH or NADPH | 8 | 7.03 × 10−6 | 43566 | structure-specific DNA binding | 7 | 1.72 × 10−3 | |
| 48037 | cofactor binding | 12 | 1.59 × 10−5 | 3697 | Mol Funct single-stranded DNA binding | 5 | 1.91 × 10−3 | |
| 16408 | C-acyltransferase activity | 4 | 9.49 × 10−5 | 3690 | double-stranded DNA binding | 5 | 8.36 × 10−3 | |
| 3988 | acetyl-CoA C-acyltransferase activity | 3 | 9.49 × 10−5 | 42805 | actinin binding | 2 | 1.84 × 10−2 | |
| GO cellular component | 44444 | cytoplasmic part | 87 | 3.32 × 10−21 | 30530 | heterogeneous nuclear ribonucleoprotein complex | 8 | 1.73 × 10−10 |
| 5737 | cytoplasm | 107 | 5.53 × 10−20 | 44424 | intracellular part | 126 | 1.39 × 10−8 | |
| 323 | lytic vacuole | 18 | 1.55 × 10−12 | 43228 | non-membrane-bounded organelle | 46 | 1.39 × 10−8 | |
| 5764 | lysosome | 18 | 1.55 × 10−12 | 43232 | intracellular non-membrane-bounded organelle | 46 | 1.39 × 10−8 | |
| 5773 | vacuole | 18 | 8.35 × 10−12 | 5622 | intracellular | 128 | 7.16 × 10−8 | |
| 44429 | mitochondrial part | 24 | 3.22 × 10−10 | 30529 | ribonucleoprotein complex | 22 | 5.27 × 10−7 | |
| 5739 | mitochondrion | 30 | 8.71 × 10−10 | 44446 | intracellular organelle part | 64 | 6.01 × 10−7 | |
| 44424 | intracellular part | 114 | 1.39 × 10−8 | 44422 | organelle part | 64 | 6.01 × 10−7 | |
| 31090 | organelle membrane | 34 | 6.42 × 10−7 | 32991 | macromolecular complex | 47 | 1.59 × 10−6 | |
| 5622 | intracellular | 114 | 7.40 × 10−7 | 5737 | cytoplasm | 91 | 1.70 × 10−6 | |
Over-representation of GO annotation terms for the separate sets of up- and down-regulated proteins was computed using the BiNGO plug-in for Cytoscape. Each GO category (GOBP, GOCC, and GOMF) was processed separately. Shown are the top scoring 10 annotations for each category, with corresponding GO ID reference number, number of proteins found to be associated with each term (x), and the Benjamini p-value indicating the significance of the enrichment. All proteins found within each term are listed in Supplementary Table S4.
Figure 4.
GOSlim functional annotation enrichment for up- and down-regulated protein groups. Enrichment of GOSlim annotations was computed using BiNGO for up- and down-regulated protein groups separately (145 up-regulated proteins; 170 down-regulated proteins). Over-represented terms are displayed graphically as hierarchical trees for each ontology (GOBP, GOMF, and GOCC), where each term is shown as a node and parent–daughter terms are connected by edges. The size of the node is proportional to the number of molecules within this group, and the color of the node represents the significance of enrichment (see color scale). To distinguish up- and down-regulated processes, nodes linked by green edges are found only in the up-regulated group, whereas nodes linked by red edges are only found in the down-regulated group. Nodes linked by black edges are common to both groups. All proteins relating to each term and individual term significance scores are listed in Supplementary Table S5.
Protein Association Networks and Interaction Clusters
The STRING database was used to create a protein interaction network from the list of 315 differentially expressed proteins; 175 nodes (proteins) were found to be interconnected by 894 edges representing highly confident protein interactions. The network was imported into Cytoscape and MCODE analysis was used to identify densely connected cluster regions (scores greater than 4.0) (Figure 5). BiNGO GOSlim analysis for each cluster identified the following statistically enriched annotations: Cluster 1 “RNA binding”; Cluster 2 “Proteinaceous extracellular matrix”; Cluster 3 “Ion transport”; Cluster 4 “Respiratory electron transport chain”; Cluster 5 “Lipid metabolic process”; and Cluster 6 “Nucleoplasm” (Figure 5).
Figure 5.
Cdc7 depletion protein–protein network. Network association analysis of altered proteins was created with STRING software and viewed in Cytoscape. All 315 significantly altered proteins were uploaded with an Interaction Confidence Score of ≥0.7, resulting in 175 nodes and 894 edges. Nodes are colored by fold change and are linked by edges that represent types of interaction evidence. The top six MCODE clusters of densely interconnected regions are circled and labeled according to the most enriched GOSlim term for that cluster of proteins. MCODE scores for each cluster, GO term identification number (GO-ID), annotation name, significance of GO term enrichment (corr p-value), and number of molecules within each cluster that match the term (x) are shown in the table below. Further information for each protein node can be found in Supplementary Table S1.
Extracellular Matrix and Calcium Signaling Are Strongly Affected by the Origin Activation Checkpoint
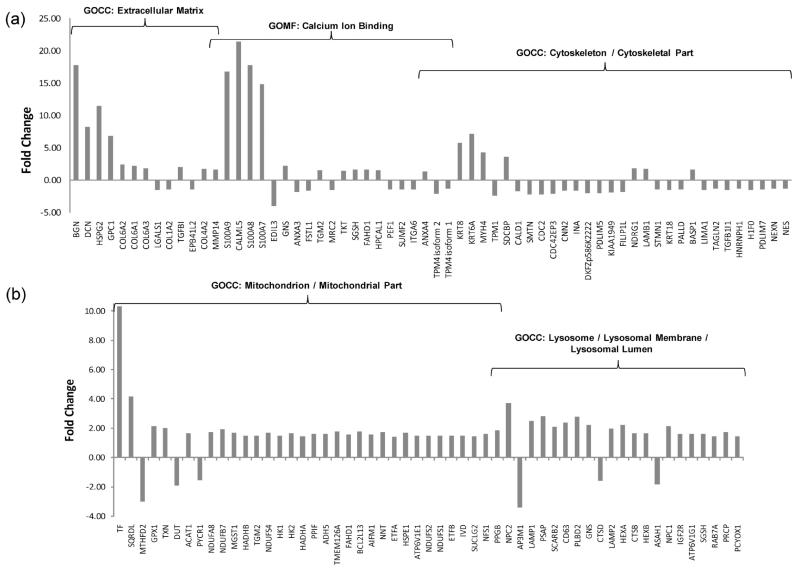
Of the top ranking 15 proteins with the most significantly changed abundance ratios, seven were associated with the extracellular matrix or extracellular region/space (S100A7, BGN, DCN, HSPG2, HMOX1, DCD and GPC1) and four were associated with calcium ion binding (S100A9, CALML5, S100A8 and S100A7). The S100 calcium binding proteins S100A9, S100A8, and S100A7 were up-regulated 16-fold, 17-fold, and 15-fold respectively. In total, 13 significantly altered proteins were located to the extracellular matrix including five collagen isoforms (COL6A2, COL6A1, COL6A3, COL1A2, and COL4A2), and 21 proteins were involved in calcium ion binding (Figure 6a). Coupled with the fact that changes occurred in 32 cytoskeletal proteins (Figure 6a), it appears that modifications in intracellular architecture play a role in the response to checkpoint activation. The present results indicate that very substantial changes occur at the cell surface of Cdc7-suppressed cells involving cell–cell adhesion and communication, suggesting that cell cycle-arrested fibroblasts may exhibit altered cellular exocytosis and may transmit growth-suppressing signals to their surrounding microenvironment and neighboring cells.
Figure 6.
Large abundance changes were associated with extracellular matrix, calcium ion binding, cytoskeleton, mitochondria, and lysosomes. Shown are fold change expression values for proteins associated with the GOCC terms “extracellular matrix”, “cytoskeleton”, “mitochondria”, and “lysosome”, as well as the GOMF term “calcium ion binding”.
Cdc7 Depletion Induces a Spatially Distributed Response Across Different Subcellular Regions
GOCC analysis revealed a spatially distributed response over specific subcellular regions. Depletion of Cdc7 resulted in the altered abundance of 34 mitochondrial proteins primarily involved in oxidative phosphorylation, antioxidant stress response, and fatty acid oxidation (Figure 6b). The data show increased abundance in five subunits of Complex I of the electron transport chain (NDUFA8, NDUFB7, NDUFS4, NDUFS2, and NDUFS1) that functions in the oxidation of NADH and establishes the electrochemical gradient necessary to drive ATP synthesis. Up-regulation of the mitochondrial membrane protein NAD(P) transhydrogenase (NNT) was also found, which is involved in the respiratory chain production of NADPH, as well as up-regulation of the mitochondrial apoptosis-inducing factor (AIF), which in normal cells functions as an NADH oxidase flavoprotein involved in oxidative phosphorylation, redox control, and maintenance of Complex I.31 The mitochondrial hexokinase isoforms HK1 and HK2 bind to the outer mitochondrial membrane and maintain the opening of the mitochondrial pore and were also increased in abundance. These isoforms can redirect newly generated mitochondrial ATP to drive glycolytic pathways, in effect coupling glucose metabolism to oxidative phosphorylation.32-34
This study has identified significant changes in the abundance of 22 lysosomal proteins (Figure 6b), including up-regulation of the lysosomal membrane proteins LAMP1, LAMP2, CD63, and SCARB2/LIMP2. The lysosomal degradation of proteins, nucleic acids, lipids and carbohydrates requires a variety of acid hydrolases that are active within the acidic intralysosomal microenvironment. The acidification of lysosomes and recycling endosomes is established by membrane proton pumps (ATPases), and five subunits of the V-type proton ATPase were up-regulated by Cdc7 depletion (ATP6 V1C1, ATP6 V1A, ATP6 V1G1, ATP6 V1B2, ATP6 V1E1). Several major lysosomal hydrolases and degradative enzymes were found to be altered in abundance, including cathepsins (CTSD, CTSB, CTSA), prosaposin (PSAP), and β-hexosaminidase (HEXA, HEXB). Several lysosomal-associated trafficking molecules were also affected by Cdc7 depletion. RAB7A is important for the fusion of autophagosomes with lysosomes and has recently been found to complex with the novel FYCO1 protein for promotion of microtubule-directed transport of autophagic vesicles.35 Both RAB7A and FYCO1 were significantly up-regulated in abundance. We found down-regulation of three subunits of a major heterotetrameric adapter protein AP-3 (AP3M1, AP3S1, and AP3B1) that participates in receptor-mediated endocytosis. Deficiency of endosome-localized AP-3 is known to result in impaired sorting of the LAMP family of lysosomal membrane proteins from the endosome to the lysosome, resulting in the increased redistribution of LAMPs to the plasma membrane.36 Therefore, the up-regulation of lysosomal membrane glycoproteins presented here could reflect altered endosomal trafficking as a consequence of diminished AP-3 levels.
The heterogeneous nuclear ribonucleoprotein (hnRNP) complex was identified as the most significantly enriched GOCC annotation for the group of down-regulated proteins (Table 3). The hnRNPs are a family of RNA binding proteins that associate with nascent mRNA transcripts in the nucleus and influence mRNA metabolism, stability, splicing, processing, and nucleocytoplasmic transport.37 RNA binding proteins such as hnRNPs maintain genomic stability and link nuclear mRNA transcriptional events with protein translation in the cytoplasm. Eight individual hnRNPs were found to be significantly reduced in expression in our data set (HNRNPA1, HNRNPA0, HNRNPH3, HNRNPA2B1, HNRNPH1, HNRPDL, RALY, and RBMX), as well as five other RNA binding proteins involved in gene regulation (RBM14, RBM3, RBMS2, CIRBP, and KHDRBS1), further supporting the argument for reduced mRNA biogenesis, splicing, and stability in the cell cycle arrested fibroblasts.
Depletion of Cdc7 Leads to Reduced Proliferative Capacity of the Cell
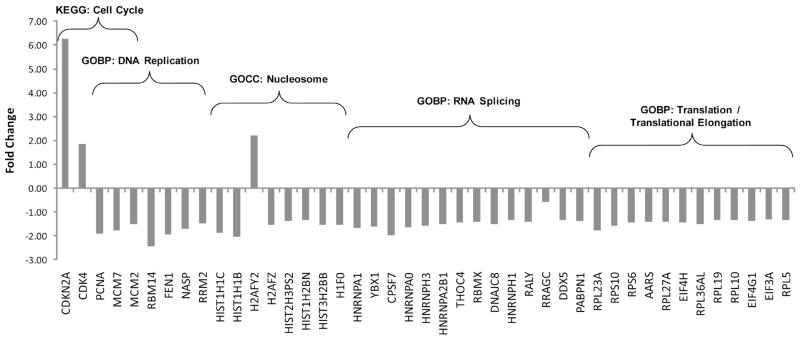
A reduction in key components of the DNA replication machinery was observed following Cdc7 depletion (Figure 7). The suppression of Cdc7 resulted in down-regulation of two subunits of its major substrate, the MCM2-7 helicase (MCM2 and MCM7). Cdc7 depletion also resulted in reduced levels of the proliferating cell nuclear antigen (PCNA), which is a cofactor for DNA polymerases38 and also a reduction in FEN1, an endonuclease involved in synthesis of the lagging DNA strand. Seven histone proteins were decreased in abundance(HIST1H1C, HIST1H1B, H2AFZ, HIST2H3PS2, HIST1H2BN, HIST3H2BB, and H1F0), and one core histone H2AFY2 protein was increased. There was also a reduction in nuclear autoantigenic sperm protein (NASP), which acts as a H1 linker histone chaperone and is involved in nuclear transport of H1 histones and chromatin assembly on newly synthesized DNA.39 Altered abundance of two cyclin-dependent kinases (CDK4 and CDC2/CDK1) was detected, as well as a 6-fold elevation in a cyclin-dependent kinase inhibitor CDKN2A. Together, these results suggest that many elements of the DNA replication machinery are altered when Cdc7 is removed from the cellular environment.
Figure 7.
Cdc7 depletion is associated with reduced cellular proliferation. Shown are fold change expression values for proteins associated with the KEGG pathway term “cell cycle”, the GOCC term “nucleosome”, and the GOBP terms “DNA replication”, “RNA splicing”, and “translation” or “translational elongation”.
Cell cycle arrest can be accompanied by reduced ribosomal biogenesis and mRNA stability.40 Protein translation initiation is a multistage process involving several translation factors that facilitate the recruitment of the ribosome to the mRNA. Eukaryotic initiation factors are known to be inactivated during starvation or stressful conditions to reduce translation of many mRNAs.41 Three eukaryotic translation initiation factors (EIF4H, EIF4G1, and EIF3A) and eight different ribosomal proteins (RPL23A, RPS10, RPS6, RPL27A, RPL36AL, RPL19, RPL10, and RPL5) were decreased in abundance (Figure 7). In addition to the reduction in hnRNPs and the RNA binding proteins mentioned above, several other DNA, RNA, and nucleotide processing molecules were also significantly reduced in abundance following Cdc7 depletion, including YBX1, THOC4, PABPN1, CPSF7, DDX5, and RRM2 (Figure 7).
Discussion
The existence of an origin activation checkpoint that monitors DNA replication initiation has recently been demonstrated in human somatic cells.1,11,12,15 In the study presented here, RNAi against Cdc7 in human fibroblasts was coupled with a high resolution, quantitative proteomics and bioinformatics strategy for evaluation of the global response involved in the maintenance of cell cycle arrest at this checkpoint. In the following, we first discuss the clear evidence that this checkpoint is an actively maintained cellular state, that Cdc7 depletion triggers a multitude of diverse cellular responses at varying subcellular locations, and that perturbation of DNA replication initiation results in reduced proliferative capacity. Subsequently, we compare the Cdc7 depletion-induced cell cycle arrest to G0, noting that the cellular response strongly resembles a quiescence-like state. Finally, we note current limitations on global proteomic measurements and suggest that complementary, directed proteomics experiments aimed at characterizing spatial/temporal features should provide a more complete analysis of the nature of the apparent quiescence-like state induced by Cdc7 depletion.
It is now well accepted that redox-dependent signaling pathways are intrinsically linked to cell cycle progression.42 It is therefore not surprising that key antioxidant molecules were found to be increased in abundance during activation of the Cdc7 checkpoint, suggesting a coordinated response to mild oxidative stress. In particular, the effector molecules of the “Nrf2-mediated oxidative stress response” pathway were highlighted by IPA analysis. Nrf2 is a key cytoprotective transcription factor that undergoes nuclear stabilization in response to elevated levels of oxidative or electrophilic species.43 It induces the transcription of a battery of antioxidant and detoxifying genes containing antioxidant response elements (ARE), which are responsible for eliminating oxidative species and restoring redox homeostasis.43-45 Cdc7 depletion resulted in up-regulation of the two major thiol-containing redox systems, thioredoxin and glutathione, both of which are under the transcriptional regulation of Nrf2. Our results demonstrate a 6-fold increase in abundance of heme oxygenase (HMOX1), which is an ARE-containing sensor of cellular stress and inflammatory insult. A 3-fold increase was found in the iron-sequestering molecule ferritin (FTH1 and FTL) and several xenobiotic metabolizing enzymes were also up-regulated, such as the multidrug-resistance associated protein ABCC1, the biotransformation enzyme epoxide hydrolase (EPHX1), and AKR1C1, a member of the aldo-keto reductase family of soluble NAD(P)H oxidoreductases. We also found elevated levels of sequestosome (SQSTM1), a protein recently shown to exert its pro-survival role by activating a variety of ARE-containing genes.46 However, many proteins downstream of Nrf2, including several classical antioxidant molecules (peroxiredoxins and superoxide dismutases) remained unchanged in abundance. This reinforces the premise that the arrested cells show a stimulus-specific response, are responding to mild levels of stress, and do not require activation of the full complement of stress response proteins. This implicitly suggests that other factors may modulate pathway(s) involving Nrf2 and activation of ARE.
The cytoprotective Nrf2 transcriptional program is also known to be induced during a condition known as stress hormesis, a beneficial cellular preconditioning mechanism whereby exposure to mild stress promotes the expression of antioxidant enzymes and enables the cell to withstand future stresses, such as higher doses of the same stress-inducing agent or general cytotoxic stressors.47,48 It is therefore likely that the antioxidants, detoxifying enzymes, heat-shock chaperones, and growth factor-induced molecules that were found to be altered in Cdc7-depleted cells, actually represent a self-preservation hormetic response to mild stress levels. This observation strongly supports an active role for Cdc7 depletion in stable, viable, and reversible G1 arrest.
Nonproliferating cells held in cell cycle arrest are thought to have lowered metabolic requirements and reduced energy flux. Therefore, it was surprising to discover that Cdc7-depleted cells exhibit increased demand for energy obtained from the breakdown of fats and proteins. Catabolic processes such as lysosomal proteolysis, fatty acid β-oxidation, synthesis/degradation of ketone bodies, and branched-chain amino acid degradation were all over-represented in this analysis. These energy-liberating pathways are most commonly utilized during fasting conditions, and it has long been known that serum deprivation can induce quiescence.18 The Cdc7 -depleted cells, however, are maintained in complete media and receive full nutrients, suggesting that they become more self-reliant in terms of energy generation and less dependent on external energy resources. Additionally, we observed down-regulation of the serum-deprivation response factor (SDPR) and unchanged levels of autophagy-related proteins (ATG3 and ATG4B), all of which would typically be up-regulated during serum starvation.49,50 Therefore, we propose that either the Cdc7-depleted proteome mimics calorie restriction to drive cell cycle arrest or calorie restriction and Cdc7 deprivation share common, but not identical, afferent regulators of quiescence.
In addition to the metabolic, structural, and protective responses implicated in the maintenance of the arrested state, we have also observed changes in a subset of proteins responsible for the characteristic suppression of growth and proliferation during cell cycle arrest. Down-regulation of a range of proteins involved in DNA replication, RNA transcriptional events, and protein translation all support a reduction in protein biosynthetic efficiency and reduced proliferative capacity in Cdc7-depleted cells. Furthermore, proteins specifically involved in amino acid biogenesis were found to be down-regulated. For example, an asparagine biosynthetic enzyme (ASNS) was reduced, as were all three enzymes involved in the three-step serine biosynthesis pathway (PHGDH, PSAT1, and PSPH). We have also observed altered expression of other proteins implicated in growth inhibition and reduced proliferation. The tumor metastasis suppressor protein, N-Myc downstream-regulated gene 1 (NDRG1), was increased in abundance. Although the function of NDRG1 is still unclear, this pleiotropic molecule is known to have tumor-suppressing qualities that inhibit cancer cell growth and metastasis by modulating many cellular processes including differentiation, proliferation, and invasion.51-53 Overexpression of NDRG1 in a prostate cancer model led to a reduction in adhesion, decreased proliferation, and reduced structural components of the ribosome.51 A recent proteomics study revealed that a large number of ribosomal, protein translation, and RNA processing proteins are found within the NDRG1 interactome.54 NDRG1 is a stress-responsive transcriptional target of p5355 and was one of a panel of p53 target genes which were specifically up-regulated by oxidative stress.56 Dong et al. demonstrated that NDRG1 is up-regulated during mimosine-induced cell cycle arrest at late G1 phase and suggest that NDRG1 may be an important factor in G1 arrest.57
It is clear from the present results that impairment of DNA replication origin activation is not limited to processes taking place exclusively in the nucleus. Although subcellular location annotations for proteins are still sparse,58 evidence was obtained that the arrested state is coupled to a diverse range of other processes that are predominantly located in mitochondria, lysosomes, and the cell surface/extracellular region. Not surprisingly, there are also strong indications of changes in protein trafficking. The importin α/β complex mediates the nuclear transportation of many proteins including Cdc7 via the nuclear pore complex59 and the reduced abundance of importin-α-2 (KPNA2) suggests disturbances in nucleocytoplasmic transport. Changes in abundance of transport molecules such as the lysosomal/endosomal trafficking proteins, the Rab GTP-binding proteins (RAB7A, RAB3A, and RAB21), and chaperone proteins (DNAJB6, DNAJB1, DNAJC8, DNAJB4, DNAJC10, and DNAJC9) all imply that dynamic protein trafficking between spatially separated compartments is affected during maintenance of the arrested state.
Together, these results paint a picture of a metabolically active cell utilizing its own resources and energy stores to enhance survival and sustain its existence. It would appear that the origin activation checkpoint at late G1 phase in human fibroblasts is maintained by activation and repression of biological processes that have previously been linked to maintenance of cellular quiescence. In support of this, we have also seen increased abundance of specific molecules that are known to become activated during G0; for example, the small leucine-rich proteoglycan decorin (DCN) was up-regulated 8-fold and is one of a small group of “quiescins” whose expression is elevated during quiescence in human lung fibroblasts60 and myogenic cells.61
To what extent are the changes observed for Cdc7-depleted cells synonymous with quiescence? Quiescence is generally accepted to be a reversible “out-of-cycle” state where cells withdraw from the cell cycle into a nondividing state, in which they survive until mitogenic stimuli persuade them to resume proliferation.62 Despite the fact that the majority of eukaryotic cells exist in this nonproliferating state, the cellular control of quiescence is still poorly understood. The traditional view is that quiescent cells reduce their proliferative potential, lower their metabolic output, and exist in an extended G1 or resting phase outside of the cell cycle. We and others have previously shown that loss of proliferative capacity in G0 is accompanied by down-regulation of replication licensing factors and the licensing repressor geminin.20,63 This led to a new definition of G0 as a “reversible withdrawal from the cell cycle characterized by unlicensed origins and the absence of CDK activity”.64 More recently, there is growing evidence that quiescence is under active transcriptional control and involves a dynamic, coordinated response throughout the cell.
Transcriptomic studies in yeast and mammalian cells have begun to address the role of candidate quiescence factors in the induction and maintenance of the G0 state, although as yet there are no equivalent large-scale proteomic analyses. The fission yeast S. pombe G0 transcriptome has been investigated using nitrogen starvation-induced quiescence.65 A diverse group of gene transcripts were found to be important for maintenance of the G0 state and were implicated in processes such as amine catabolism, oxidoreductase and hydrolase activity, carbohydrate metabolism, autophagy, mitochondrial ATPase function, ubiquitin-dependent proteolysis, cytoskeletal networks, and cell communication. The S. pombe model was also used to identify 33 core genes required for entry into and maintenance of quiescence.66 These genes encoded proteins involved in broad ranging processes such as stress-response, cell cycle kinase activity, chromatin remodelling, RNA splicing, ribosomal biogenesis, translation, lipid and ATP metabolism, protein trafficking, enodosome formation, and vesicle fusion. Many of these genes have human homologues, and it is thought that the genetic profile induced by quiescence may be conserved from yeast to higher eukaryotes.67
The interpretation of yeast G0 as a dynamic state correlates well with studies of human quiescence. Venezia et al. observed that principally quiescent adult hematopoietic stem cells exist in a “state of readiness” to react to environmental changes.68 Two recent studies used gene expression profiling to demonstrate the existence of a transcriptional panel necessary for the regulation of cellular quiescence in human fibroblasts.18,69 Coller et al. identified that fibroblast quiescence is actually a dynamic collection of states with a shared “quiescence programme” of genes required for general enforcement of the G0 phenotype.18 This program included genes important for ensuring the reversibility of the G0 state, intercellular communication, suppression of growth, division, and terminal differentiation, as well as antiapoptotic genes. Liu et al. identified a distinct set of serum deprivation early response genes necessary for induction, rather than maintenance of quiescence, and found that these genes were implicated in immune responses, redox reactions, and extracellular matrix metabolism.69 The various genome-wide studies of cellular quiescence mentioned here are all in agreement that G0 is a highly dynamic and active state regulated across many subcellular regions, and there is increasing evidence that different forms of quiescence are induced by different stimuli.18 Therefore, quiescence should no longer be regarded as a passive, resting state for cells in the absence of mitogenic stimuli and instead should be thought of as an umbrella term for an actively maintained, reversible cellular arrest.
Can proteomics tell us more about the exact nature of the “poised quiescence” state induced by Cdc7 depletion? Generally speaking, the analysis of biological function from large scale transcriptomic and proteomic data sets is still evolving. A number of different approaches, software, and knowledge-bases are now available, each of which tends to have its own strengths and weaknesses. IPA has been extensively used in recent years to analyze large scale “omics” data sets (2781 scientific publications citing the use of IPA, as of June 2010).24 Recently, the combination of STRING and MCODE for identification of enriched clusters within protein networks has been successfully applied in several studies, including a lysine acetylation network70 and a DNA damage response network.71 In the present case, the IPA pathway tool and the BiNGO GO term enrichment analysis identified complementary biological processes affected by Cdc7 depletion. The strategy of applying several different data-mining tools to the analysis seems to be essential at the present stage of development and greatly improved our ability to extract statistically significant and functionally intelligible information from the large-scale proteomic data set. Even so, an interesting feature of the present results was that the IPA/STRING data-mining methods did not classify some of the largest changes in abundance ratios to strongly connected networks and largely missed highly significant changes at the cell surface and extracellular space. Datamining methods such as IPA and STRING emphasize well-documented biology by providing dense networks of previously established knowledge and may neglect more ambiguous proteins and processes. Although this provides a very useful overview, less well-studied biological systems may be poorly detected or defined by these data-mining methods. The existence of “orphan” proteins with large, highly significant changes in abundance may provide priorities for further investigations aimed at defining “new” biological cellular responses.
The observation that global response patterns of the type analyzed here seem to show features and groups of proteins specific to the stimulus opens the way to further unravelling very complex cellular networks. As we have noted elsewhere,58,72 the key to this may be more explicit measurement of the spatio/temporal fluxes of proteins. The existence and preservation of differentiated subcellular structures, as well as the communication links between them, ensure that functional processes are spatially and temporally distributed across many subcellular locations, and this is clearly the case for Cdc7 regulation. The present study is limited temporally in that depletion of Cdc7 by RNAi is a slow process, and some of the subcellular responses observed may represent long-term adaptation of the cells to a “lesion”. This technical limitation makes it difficult to ascertain whether the observed biological changes are a cause or consequence of triggering the origin activation checkpoint. There is still a need for more rapid, dynamic methods of specifically perturbing cellular function, and the recent development of Cdc7 inhibitors will allow modulation of Cdc7 function, so that subsequent dynamic effects can be followed as a function of time. Similarly, there are currently restrictions on the spatial interpretation of the proteomics results. One of the weak points in current database annotations is that subcellular location and function are generally not coupled. All but four of the proteins showing changed abundance ratios were annotated with multiple locations (33 proteins had no associated GOCC term), and many proteins may have functional/regulatory annotations that are probably inconsistent with a single subcellular location.
Global proteomics measurements still have limited proteome coverage that may constrain deciphering of the contributions of different systems to cellular response. For example, we recently identified FOXO3a as a major player for the co-ordination of the origin activation checkpoint.15 The FOXO pathway is known to powerfully tip the balance of the cell toward quiescence,73 and FOXO and Nrf2 are both hormesisinducing, oxidative stress-responsive pathways.47,74 However, the present results do not elucidate to what extent the FOXO and Nrf2 pathways may be parallel, competing, or cooperative pathways and/or the extent to which interpretation of the global proteomics data may be conditioned by previous research priorities encoded in existent databases. With the above-noted spatio/temporal constraints in mind, we have begun complementing the present experiments with further data on nuclear proteins, especially in networks under control by phosphorylation.
Finally, our results should be interpreted in the context of emerging Cdc7-targeted therapies. Targeting Cdc7 has been shown to induce potent cancer-cell killing in a broad range of tumor types as a result of an abortive S phase followed by apoptotic cell death. Here we have shown that primary somatic cells circumvent lethal S phase progression under similar Cdc7 rate-limiting condition by engaging an origin activation checkpoint and undergoing a robust G0/G1 cell cycle arrest. We have shown that viability and maintenance of this arrested state involves modulation of the cellular metabolic signature, initiating a dynamic quiescence-like response, and priming cells for cell cycle re-entry. It now remains to be seen whether Cdc7 inhibitors in healthy tissues, particularly self-renewing tissues characterized by high turnover (e.g., bone marrow and gut mucosa), also induce a protective quiescence-like arrest. If this is the case then we might expect to observe low toxicity with such anticancer agents. The ability to analyze large-scale, systems-wide data sets by proteomics methods is now becoming routine, and we believe that further elucidation of origin activation checkpoint networks will provide ways to go beyond single target “magic bullet” approaches to cancer therapy.
Supplementary Material
Acknowledgment
This study has been supported by Cancer Research U.K. scientific program grant C428/A6263 (KS and GHW) and Wellcome Trust grant 081879/z/06/z (JGZ).
Footnotes
Supporting Information Available: Figure S1: Linear regression comparison of three SILAC biological replicate experiments. Figure S2: Western blot validation of SILAC proteins. Figure S3: NRF2-mediated oxidative stress response pathway. Figure S4: Synthesis and degradation of ketone bodies pathway. Figure S5: Butanoate metabolism pathway. Figure S6: Bile acid biosynthesis pathway. Figure S7: Valine, leucine, and isoleucine degradation pathway. Figure S8: Biosynthesis of steroids pathway. Figure S9: Granzyme A signaling pathway. Figure S10: Oxidative phosphorylation pathway. Figure S11: Glutathione metabolism pathway. Figure S12: Propanoate metabolism pathway. Table S1: Protein information for each of the 315 differentially expressed proteins. Table S2: Protein information for all 1730 proteins that were quantified but found to be unchanged in abundance following Cdc7 depletion. Table S3: Protein names, HUGO gene symbols, fold changes, and p-values for all molecules mapped to each of the 10 top-scoring IPA pathways. Table S4: GO enrichment annotation data for up- and down-regulated protein groups. Table S5: Generic GOSlim enrichment annotation data for up and down regulated protein groups. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Blow JJ, Gillespie PJ. Replication licensing and cancer–a fatal entanglement. Nat. Rev. Cancer. 2008;8(10):799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 2007;41:237–80. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- (4).Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell. 2006;24(1):101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Bousset K, Diffley JF. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12(4):480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jiang W, McDonald D, Hope TJ, Hunter T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 1999;18(20):5703–5713. doi: 10.1093/emboj/18.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008;22(14):1894–1905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell. 2008;135(4):662–678. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- (9).Kim JM, Kakusho N, Yamada M, Kanoh Y, Takemoto N, Masai H. Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene. 2008;27(24):3475–3482. doi: 10.1038/sj.onc.1210994. [DOI] [PubMed] [Google Scholar]
- (10).Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell. 2003;11(1):203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- (11).Montagnoli A, Tenca P, Sola F, Carpani D, Brotherton D, Albanese C, Santocanale C. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res. 2004;64(19):7110–7116. doi: 10.1158/0008-5472.CAN-04-1547. [DOI] [PubMed] [Google Scholar]
- (12).Rodriguez-Acebes S, Proctor I, Loddo M, Wollenschlaeger A, Falzon M, Prevost AT, Sainsbury R, Williams GH, Stoeber K. Targeting DNA replication before it starts: CDC7 as a therapeutic target in p53-mutant breast cancers. Am. J. Pathol. 2010 doi: 10.2353/ajpath.2010.100421. published online ahead of print; doi:, 10.2353/ajpath.2010.100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C, Patton V, Alzani R, Ciavolella A, Sola F, Molinari A, Volpi D, Avanzi N, Fiorentini F, Cattoni M, Healy S, Ballinari D, Pesenti E, Isacchi A, Moll J, Bensimon A, Vanotti E, Santocanale C. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat. Chem. Biol. 2008;4(6):357–365. doi: 10.1038/nchembio.90. [DOI] [PubMed] [Google Scholar]
- (14).Kulkarni AA, Kingsbury SR, Tudzarova S, Hong HK, Loddo M, Rashid M, Rodriguez-Acebes S, Prevost AT, Ledermann JA, Stoeber K, Williams GH. Cdc7 kinase is a predictor of survival and a novel therapeutic target in epithelial ovarian carcinoma. Clin. Cancer Res. 2009;15(7):2417–2425. doi: 10.1158/1078-0432.CCR-08-1276. [DOI] [PubMed] [Google Scholar]
- (15).Tudzarova S, Trotter MWB, Wollenschlaeger A, Mulvey C, Godovac-Zimmermann J, Williams GH, Stoeber K. Molecular architecture of the DNA replication origin activation checkpoint. EMBO J. 2010 doi: 10.1038/emboj.2010.201. published online ahead of print, doi:, 10.1038/emboj.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Swords R, Mahalingam D, O’Dwyer M, Santocanale C, Kelly K, Carew J, Giles F. Cdc7 kinase - a new target for drug development. Eur. J. Cancer. 2010;46(1):33–40. doi: 10.1016/j.ejca.2009.09.020. [DOI] [PubMed] [Google Scholar]
- (17).Ermoli A, Bargiotti A, Brasca MG, Ciavolella A, Colombo N, Fachin G, Isacchi A, Menichincheri M, Molinari A, Montagnoli A, Pillan A, Rainoldi S, Sirtori FR, Sola F, Thieffine S, Tibolla M, Valsasina B, Volpi D, Santocanale C, Vanotti E. Cell division cycle 7 kinase inhibitors: 1H-pyrrolo[2,3-b]pyridines, synthesis and structure–activity relationships. J. Med. Chem. 2009;52(14):4380–4390. doi: 10.1021/jm900248g. [DOI] [PubMed] [Google Scholar]
- (18).Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4(3):e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Eward KL, Obermann EC, Shreeram S, Loddo M, Fanshawe T, Williams C, Jung HI, Prevost AT, Blow JJ, Stoeber K, Williams GH. DNA replication licensing in somatic and germ cells. J. Cell Sci. 2004;117(Pt 24):5875–5886. doi: 10.1242/jcs.01503. [DOI] [PubMed] [Google Scholar]
- (20).Kingsbury SR, Loddo M, Fanshawe T, Obermann EC, Prevost AT, Stoeber K, Williams GH. Repression of DNA replication licensing in quiescence is independent of geminin and may define the cell cycle state of progenitor cells. Exp. Cell Res. 2005;309(1):56–67. doi: 10.1016/j.yexcr.2005.05.027. [DOI] [PubMed] [Google Scholar]
- (21).Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. Ingel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- (22).Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteomewide protein quantification. Nat. Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- (23).Cox J, Matic I, Hilger M, Nagaraj N, Selbach M, Olsen JV, Mann M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009;4(5):698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- (24).Ingenuity Pathway Analysis, Ingenuity Systems. www.ingenuity.com.
- (25).Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess over-representation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- (28).Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. Mechanism of cholesterol transfer from the Niemann-Pick type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J. Biol. Chem. 2006;281(42):31594–31604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- (31).Modjtahedi N, Giordanetto F, Madeo F, Kroemer G. Apoptosis-inducing factor: vital and lethal. Trends Cell Biol. 2006;16(5):264–272. doi: 10.1016/j.tcb.2006.03.008. [DOI] [PubMed] [Google Scholar]
- (32).Robey RB, Hay N. Mitochondrial hexokinases: guardians of the mitochondria. Cell Cycle. 2005;4(5):654–658. doi: 10.4161/cc.4.5.1678. [DOI] [PubMed] [Google Scholar]
- (33).Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J. Exp. Biol. 2003;206(Pt 12):2049–2057. doi: 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- (34).Arora KK, Pedersen PL. Functional significance of mitochondrial bound hexokinase in tumor cell metabolism. J. Biol. Chem. 1988;263(33):17422–17428. [PubMed] [Google Scholar]
- (35).Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 2010;188(2):253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 2004;164(7):1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 2002;3(3):195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- (38).Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129(4):665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- (39).Richardson RT, Alekseev OM, Grossman G, Widgren EE, Thresher R, Wagner EJ, Sullivan KD, Marzluff WF, O’Rand MG. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J. Biol. Chem. 2006;281(30):21526–21534. doi: 10.1074/jbc.M603816200. [DOI] [PubMed] [Google Scholar]
- (40).Ferreira-Cerca S, Hurt E. Cell biology: Arrest by ribosome. Nature. 2009;459(7243):46–47. doi: 10.1038/459046a. [DOI] [PubMed] [Google Scholar]
- (41).Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Burhans WC, Heintz NH. The cell cycle is a redox cycle: linking phase-specific targets to cell fate. Free Radical Biol. Med. 2009;47(9):1282–1293. doi: 10.1016/j.freeradbiomed.2009.05.026. [DOI] [PubMed] [Google Scholar]
- (43).Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009;284(20):13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat. Res. 2008;659(1-2):31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 2004;10(11):549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- (46).Liu Y, Kern JT, Walker JR, Johnson JA, Schultz PG, Luesch H. A genomic screen for activators of the antioxidant response element. Proc. Natl. Acad. Sci. U.S.A. 2007;104(12):5205–5210. doi: 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Son TG, Camandola S, Mattson MP. Hormetic dietary phytochemicals. Neuromol. Med. 2008;10(4):236–246. doi: 10.1007/s12017-008-8037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Lindsay DG. Nutrition, hormetic stress and health. Nutr. Res. Rev. 2005;18(2):249–258. doi: 10.1079/NRR2005110. [DOI] [PubMed] [Google Scholar]
- (49).Gustincich S, Vatta P, Goruppi S, Wolf M, Saccone S, Della Valle G, Baggiolini M, Schneider C. The human serum deprivation response gene (SDPR) maps to 2q32-q33 and codes for a phosphatidylserine-binding protein. Genomics. 1999;57(1):120–129. doi: 10.1006/geno.1998.5733. [DOI] [PubMed] [Google Scholar]
- (50).Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Kovacevic Z, Fu D, Richardson DR. The iron-regulated metastasis suppressor, Ndrg-1: identification of novel molecular targets. Biochim. Biophys. Acta. 2008;1783(10):1981–1992. doi: 10.1016/j.bbamcr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- (52).Bandyopadhyay S, Pai SK, Hirota S, Hosobe S, Takano Y, Saito K, Piquemal D, Commes T, Watabe M, Gross SC, Wang Y, Ran S, Watabe K. Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene. 2004;23(33):5675–5681. doi: 10.1038/sj.onc.1207734. [DOI] [PubMed] [Google Scholar]
- (53).Ellen TP, Ke Q, Zhang P, Costa M. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis. 2008;29(1):2–8. doi: 10.1093/carcin/bgm200. [DOI] [PubMed] [Google Scholar]
- (54).Tu LC, Yan X, Hood L, Lin B. Proteomics analysis of the interactome of N-myc downstream regulated gene 1 and its interactions with the androgen response program in prostate cancer cells. Mol. Cell. Proteomics. 2007;6(4):575–588. doi: 10.1074/mcp.M600249-MCP200. [DOI] [PubMed] [Google Scholar]
- (55).Stein S, Thomas EK, Herzog B, Westfall MD, Rocheleau JV, Jackson RS, 2nd, Wang M, Liang P. NDRG1 is necessary for p53-dependent apoptosis. J. Biol. Chem. 2004;279(47):48930–48940. doi: 10.1074/jbc.M400386200. [DOI] [PubMed] [Google Scholar]
- (56).Han ES, Muller FL, Perez VI, Qi W, Liang H, Xi L, Fu C, Doyle E, Hickey M, Cornell J, Epstein CJ, Roberts LJ, Van Remmen H, Richardson A. The in vivo gene expression signature of oxidative stress. Physiol Genomics. 2008;34(1):112–126. doi: 10.1152/physiolgenomics.00239.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Dong Z, Arnold RJ, Yang Y, Park MH, Hrncirova P, Mechref Y, Novotny MV, Zhang JT. Modulation of differentiation-related gene 1 expression by cell cycle blocker mimosine, revealed by proteomic analysis. Mol. Cell. Proteomics. 2005;4(7):993–1001. doi: 10.1074/mcp.M500044-MCP200. [DOI] [PubMed] [Google Scholar]
- (58).Qattan AT, Mulvey C, Crawford M, Natale DA, Godovac-Zimmermann J. Quantitative organelle proteomics of MCF-7 breast cancer cells reveals multiple subcellular locations for proteins in cellular functional processes. J. Proteome Res. 2010;9(1):495–508. doi: 10.1021/pr9008332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Kim BJ, Lee H. Importin-beta mediates Cdc7 nuclear import by binding to the kinase insert II domain, which can be antagonized by importin-alpha. J. Biol. Chem. 2006;281(17):12041–12049. doi: 10.1074/jbc.M512630200. [DOI] [PubMed] [Google Scholar]
- (60).Coppock D, Kopman C, Gudas J, Cina-Poppe DA. Regulation of the quiescence-induced genes: quiescin Q6, decorin, and ribosomal protein S29. Biochem. Biophys. Res. Commun. 2000;269(2):604–610. doi: 10.1006/bbrc.2000.2324. [DOI] [PubMed] [Google Scholar]
- (61).Nishimura T, Nozu K, Kishioka Y, Wakamatsu J, Hattori A. Decorin expression in quiescent myogenic cells. Biochem. Biophys. Res. Commun. 2008;370(3):383–387. doi: 10.1016/j.bbrc.2008.03.025. [DOI] [PubMed] [Google Scholar]
- (62).Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246(4930):603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- (63).Stoeber K, Tlsty TD, Happerfield L, Thomas GA, Romanov S, Bobrow L, Williams ED, Williams GH. DNA replication licensing and human cell proliferation. J. Cell Sci. 2001;114(Pt 11):2027–2041. doi: 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- (64).Blow JJ, Hodgson B. Replication licensing–defining the proliferative state. Trends Cell Biol. 2002;12(2):72–78. doi: 10.1016/s0962-8924(01)02203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Shimanuki M, Chung SY, Chikashige Y, Kawasaki Y, Uehara L, Tsutsumi C, Hatanaka M, Hiraoka Y, Nagao K, Yanagida M. Two-step, extensive alterations in the transcriptome from G0 arrest to cell division in Schizosaccharomyces pombe. Genes Cells. 2007;12(5):677–692. doi: 10.1111/j.1365-2443.2007.01079.x. [DOI] [PubMed] [Google Scholar]
- (66).Sajiki K, Hatanaka M, Nakamura T, Takeda K, Shimanuki M, Yoshida T, Hanyu Y, Hayashi T, Nakaseko Y, Yanagida M. Genetic control of cellular quiescence in S. pombe. J. Cell Sci. 2009;122(Pt 9):1418–1429. doi: 10.1242/jcs.046466. [DOI] [PubMed] [Google Scholar]
- (67).Yanagida M. Cellular quiescence: are controlling genes conserved. Trends Cell. Biol. 2009;19(12):705–715. doi: 10.1016/j.tcb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- (68).Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, Goodell MA. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2(10):e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Liu H, Adler AS, Segal E, Chang HY. A transcriptional program mediating entry into cellular quiescence. PLoS Genet. 2007;3(6):e91. doi: 10.1371/journal.pgen.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- (71).Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3(104):ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- (72).Godovac-Zimmermann J. Cancer-omics failure: warehouses, magic bullets, space/time and the Life of Brian in a cancer cell. Exp. Rev. Proteomics. 2010;7(3):303–306. doi: 10.1586/epr.10.7. [DOI] [PubMed] [Google Scholar]
- (73).Kops GJ, Medema RH, Glassford J, Essers MA, Dijkers PF, Coffer PJ, Lam EW, Burgering BM. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol. Cell. Biol. 2002;22(7):2025–2036. doi: 10.1128/MCB.22.7.2025-2036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419(6904):316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.