Abstract
The cytoskeleton is a cellular scaffolding system whose functions include maintenance of cellular shape, enabling cellular migration, division, intracellular transport, signaling and membrane organization. In addition, in immune cells, the cytoskeleton is essential for phagocytosis. Following the advances in proteomics technology over the past two decades, cytoskeleton proteome analysis in resting and activated immune cells has emerged as a possible powerful approach to expand our understanding of cytoskeletal composition and function. However, so far there have only been a handful of studies of the cytoskeleton proteome in immune cells. This article considers promising proteomics strategies that could augment our understanding of the role of the cytoskeleton in host-defense mechanisms.
Keywords: actin, cytoskeleton, host defense, immune cell, immune response, mass spectrometry, microtubule, protein, proteomics, purification strategies
Unlike the skeleton in higher organisms, the cellular skeleton is an adaptive and dynamic cellular network of protein polymers involved in cellular function in terms of movement, transport, secretion and shape. It also provides a platform for regional activities such as signaling, bio synthesis and energy production. In immune cells, it regulates a number of cellular functions that are related to the immune response, including migration, extravasation, antigen recognition, phagocytosis and cellular signaling/activation [1].
The eukaryotic cell cytoskeleton consists of three distinct species of filamentous structures: microfilaments, intermediate filaments and microtubules, composed mainly of actin, vimentin/keratin and tubulin, respectively. Although compositionally distinct, there is considerable interaction between these different filamentous networks, especially the more dynamic microfilaments and microtubules. Actin filaments are composed of two intertwined actin fibers and play an important role in maintaining cell morphology, phagocytosis, endocytosis, cell movement, and cell-to-cell and cell-to-matrix attachments. Microtubules are hollow cylindrical structures consisting of polymers of α- and β-tubulin and are mostly involved in intra cellular transport of organ-organelles and the formation of the mitotic spindle. Intermediate filaments are the most stable cytoskeletal structures and serve to maintain cellular shape.
Rapid assembly of actin filaments is the principal driving force behind many forms of cell locomotion and changes in cell shape. Cells can migrate at rates up to approximately 0.5 μm/s [2]. This means that filaments must have a net rate of elongation of approximately 200 monomers per second. Molecular motors move along the actin filaments and microtubules and guide the organization of cellular components. The assembly and disassembly of actin filaments and their organization into higher-order networks is regulated by actin-associated regulatory proteins that, in turn, are controlled by specific signaling pathways [3]. The extensive cytoskeletal fiber network provides a large surface area serving as a platform for binding of proteins that regulate/enable cytoskeletal rearrangement or have unrelated function.
Hundreds of proteins have so far been found to bind to actin [4], and novel actin-binding proteins are constantly being discovered in the post-genomic era. The vast number of ligands with significant affinity for actin strongly suggests that there is probably a large number of binding sites that cover much of the exposed surface of the molecule. The architecture of cytoskeletal networks is controlled by several classes of regulatory cytoskeleton-binding proteins: nucleation-promoting factors (Wiskott-Aldrich syndrome protein [WASP]), which initiate filament formation; capping proteins (tropomodulin, CapZ), which terminate filament growth; polymerases, which promote faster or more sustained filament growth (formin); de polymerizing factors (cofilin), and severing factors, which disassemble filaments (gelsolin); cross linkers (Arp2/3) and stabilizing proteins (L-plastin, ND1-L). The myosin family of motor proteins use F-actin as a track upon which to move. For microtubules, the motors are members of the dynein or kinesin families.
Further understanding of the mechanisms of cytoskeleton dynamics requires an analysis of temporal patterns of cytoskeleton binding proteins (CBPs) association/dissociation. This association of CBP to cytoskeletal fibers is often transient and of low affinity. The purpose of this article is to summarize the available evidence in relation to the immune response obtained by application of proteomics techniques to investigate cytoskeleton composition and dynamics.
The cytoskeleton in immune cells
Immune cells carry out specific effector functions related to the host response to infection and inflammation. Efficient accomplishment of these functions requires a finely regulated cellular cytoskeleton to enable reorganization of the cellular membrane, receptor localization, recruitment of signaling intermediates and changes in the morphology of the cell. All these cytoskeleton-involving processes are necessary for the activation, proliferation, differentiation, secretion, cell–cell interaction and survival of immune cells, and phagocytosis [1,5,6].
Phagocytosis is a major host defense function that is critically dependent on the cytoskeleton. It is defined as a process of elimination of foreign particles larger than 0.5 μm in diameter or host apoptotic cells, by phagocytic leukocytes. It comprises multiple events: particle binding, actin assembly, membrane remodeling, pseudopod extension and phagosome closure. This uptake mechanism is dependent on a complex rearrangement of the actin cytoskeleton [7]. Nascent phagosomes lack the ability to kill pathogens or to degrade the ingested targets. These properties are acquired during the course of phagosomal maturation, a complex sequence of reactions that result in remodeling of the phagosomal membrane and content. Actin assembly is essential for this process [8].
Another immune cell function dependent on the cytoskeleton is signaling from surface receptors. Association of various cell surface receptors with the cytoskeleton has been reported by a number of studies. For example, it was postulated that interaction between EGF receptor (EGFR) and actin is involved in the receptor internalization process and signaling from the EGFR to ERKs[9]. Other surface receptors, such as T-cell receptor [10], the high-affinity receptor for immunoglobulin E (FceR1) [11], B-cell antigen receptor [12] and the tyrosine phosphatase CD45 [13], also associate with the cytoskeleton. Aggregation of the FceR1 stimulates a variety of cellular responses but excessive aggregation inhibits such responses. Actin filaments have been implicated in the inhibitory phenomenon since disruption of filaments enhances the cellular reactions stimulated by the aggregated receptors [11].
An example of the essential role of the cytoskeleton for immune response is Wiskott-Aldrich syndrome. This disease is due to the loss of WASP activity, leading to defects in a broad range of cellular processes and resulting in complex immunodeficiency. WASP is an actin-binding protein and an important regulator of the actin cytoskeleton that is required for a number of immune cell functions, including migration, phagocytosis and immune synapse formation [14]. Similarly, loss of the depolymerizing factor coronin 1a leads to a T-cell immunodeficiency in humans, and inactivation of many other cytoskeletal regulatory proteins leads to immunodeficiencies in mice [6].
From proteins to proteomes
With the availability of whole genome sequences, research attention is shifting from gene sequences and genome content to protein functions and systems biology. Many aspects of protein function are not contained in gene or protein sequences. For example, when, where and to what extent a particular protein is modified in signal transduction is not encoded in DNA, although the potential for modifications and the proteins needed for this activity are genetically encoded. Traditional concepts and techniques employ a reductionist approach, focused on understanding the function of individual proteins. The features by which a protein can be described include expression, localization, interactions, domain structure, modification and activity. Thanks to the combination of developments in new instrumentation, fragmentation methods, availability of completed genome sequences and bioinformatics, there has been a shift from analysis of one protein at the time to more comprehensive proteome analyses. In the past decade, mass spectrometry (MS) has emerged as the dominant technology for in-depth characterization of the protein components of biological systems [15-17]. Owing to its unparalleled ability to acquire high-content, quantitative information about biological samples of high complexity, MS became a major driving force behind proteomics science. Within the past few years MS-driven proteomics has made remarkable progress in generating large-scale datasets for protein modifications, organelle composition, protein–protein interactions and protein profiles in healthy and diseased cells and tissues. Very recently, a Wikipedia-style, online proteomics encyclopedia has been created for sharing datasets, providing a centralized proteomics/MS data repository [18,19,101]. Despite the progress and enthusiasm, full analysis of the highly complex proteome presents a formidable challenge. A variety of general cellular processes such as gene transcription, mRNA processing, export and degradation, protein translation, post-translational modifications (PTMs), subcellular localization and controlled degradation of proteins are fundamental characteristics of cellular function that contribute to the observed complexity. It is this degree of complexity that has led to the increasingly widespread recognition that highly parallel ‘omics’ approaches are needed to unravel the global characteristics of cellular function [20]. Thus, proteomics became a widely accepted and successful approach in biology and biomedicine, with MS as its most important technology. Proteomics has evolved from 2D gel electrophoresis (2DE)/MALDI TOF MS to gel-free liquid chromatography (LC)-MS/MS approaches. Contemporary 1D/2D LC-MS/MS workflows exhibit much higher sensitivity, speed, quantitative dynamic range and ease of use in comparison with gel-based resolving techniques.
Proteomics approaches to analysis of the cytoskeleton in immune cells
Cytoskeletal proteins are often identified in many proteomic studies either as contaminants, owing to their very high abundance, or their specific binding to a large number of other proteins or subcellular structures. Studies of the cytoskeletal proteome can greatly contribute to our understanding of cytoskeleton composition, function, regulation and dynamics. Proteomics methodology offers cytoskeleton researchers a variety of new ways to monitor cytoskeletal function including: identification of novel CBPs; monitoring the time course of protein association/dissociation with/from the cytoskeleton upon cellular activation; and investigating the role of PTMs in cytoskeletal function. Unfortunately, this opportunity has remained largely unused, as so far there have only been a handful of studies of the cytoskeletal proteome in immune cells.
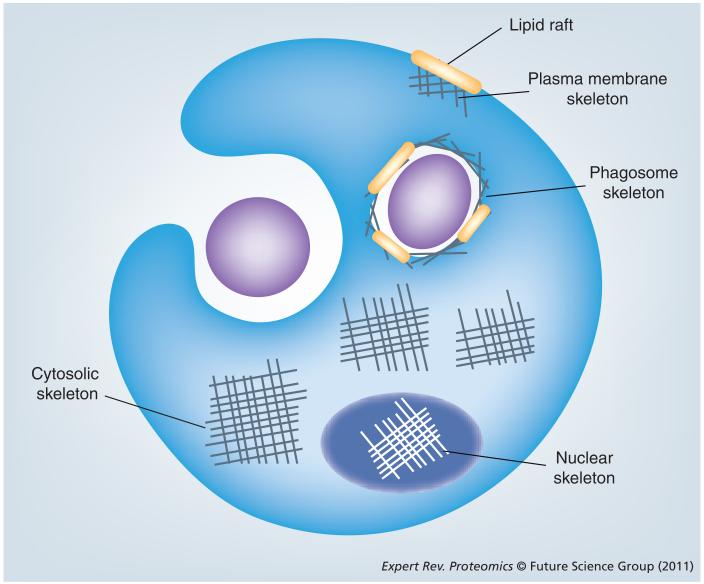
The main cytoskeletal compartments in an immune cell include the plasma membrane, and the cytoplasmic, phagosome and nuclear skeletons (Figure 1). Plasma membranes are organized into functional domains by packing into cholesterol and sphingolipid-enriched liquid-ordered membrane microdomains called ‘lipid rafts’, structures thought to serve as scaffolding platforms for signal transduction [21]. Lipid rafts have been a very popular target of proteomics studies [22]. A specialized part of the cytoskeleton, situated in close proximity to a cell membrane with a protein composition and structure that differs from that of the bulk skeleton in cytosol, is called the plasma membrane or cortical skeleton. This cytoskeleton compartment is essential for signaling from the surface receptors due to its attachment to lipid rafts. Lymphocyte stimulation leads to a lipid-based reorganization of plasma membrane rafts and this dynamic process is controlled by proteins that are linked to the actin fibers [23,24]. Actin function in the membrane skeleton is also required for chemotaxis, cell movement, adhesion and phagocytosis [25]. The dynamics of membrane raft lipids were examined in a recent study of signaling through the plasma membrane IgE receptor FcεRI by use of electrospray ionization MS [26]. Over 100 phospholipid components were quantified in lipid rafts. Stimulation of rat basophil leukemia cells via FcεRI causes a substantial increase in the ratio of polyunsaturated to saturated phospholipids. The use of the actin-modulating agents latrunculin, cytochalasin D and jasplakinolide indicated that the phospholipid composition of lipid rafts is strongly influenced by the actin cytoskeleton [26]. The temporal proteomics profiling of lipid rafts in T cells upon chemokine receptor CCR6 activation has indicated the integration of actin skeleton dynamics [27]. A number of proteins involved in the actin cytoskeleton rearrangement were thereby shown to be actively recruited into lipid rafts following T-cell activation through CCR6 [27]. This is consistent with the biological function of chemokine receptors in leukocyte migration toward sites of inflammation. It is noteworthy that changes in the lipid raft proteome in T cells induced by activation of the chemokine receptor and the T-cell receptor are quite different [28].
Figure 1. The main cytoskeleton compartments in neutrophils/macrophages.
Cytosolic, phagosome, plasma membrane and nuclear skeletons, as indicated. Nuclear actin filaments are schematically represented as white lines, while they are drawn in gray in other cellular compartments. This is to emphasize the different structure of actin fibers in the nucleus.
A proteomics study by Nebl et al. has shown that a subset of plasma membrane skeleton proteins from bovine neutrophils co-isolates with cholesterol-rich, high-density detergent-resistant membrane fragments (DRM-H) [23]. Thereby, a set of 19 major proteins was identified [23], including lipid raft-associated integral membrane proteins stomatin, flotillin 1, flotillin 2 and Lyn kinase. Membrane skeleton DRM-H proteins include fodrin, myosin-IIA, myosin-IG, α-actinin 1, α-actinin 4, vimentin and the F-actin-binding protein supervillin. The authors hypothesized that fodrin, myosin-IIA, myosin-IG and supervillin are components of a mobile, actin-based membrane skeleton that regulates the organization and transport of cholesterol-rich signaling domains at the neutrophil plasma membrane. Taken together, although the actin cytoskeleton is implicated in many lipid raft-mediated signaling processes, little is known about the biochemical basis for actin involvement.
Xu et al. comprehensively investigated the composition of the main cytoskeletal compartments of human neutrophils, including the membrane skeleton [29]. Among the total of 38 proteins identified, mostly cytoskeletal and cell signaling proteins, there were four that were previously not reported to associate with the neutrophil cytoskeleton: ficolin, valosin-containing protein, LCRP8 and major vault protein. LCRP8 is a protein of unknown function, valosin-containing protein may be involved in antigen presentation, while ficolin acts as opsonin, thereby promoting phagocytosis. Major vault protein could be implicated in EGFR signaling and regulation of neutrophil survival [30]. Identification of stomatin and supervillin from the previous study [23] was not confirmed, possibly due to the difference in species, or diverse isolation and separation procedures (1DE vs 2DE). A number of signaling proteins identified in detergent-resistant membranes (Hpast, IQGAP1, Lyn, G protein α and β) support the concept of the involvement of lipid rafts and the underlying cytoskeleton in cellular signaling. This study also identified 72 proteins in the cytosolic skeleton and 27 in the detergent-insoluble phagosome skeleton [29]. The most remarkable finding was that of a number of metabolic enzymes not previously reported to associate with the cytoskeleton: phosphoglycerate mutase (glycolysis), transketolase, phosphogluconate dehydrogenase (pentose phosphate pathway), N-acetylglucosamine kinase (glutamate and amino sugar metabolism), glucosidase (glycoprotein processing) and aldehyde dehydrogenase (oxidation of aldehydes). Somewhat surprisingly, metabolic enzymes were the most represented functional group (20.3%) apart from the cytoskeleton and associated proteins (46.8%). This finding is in agreement with the comprehensive proteomic study of microtubule-associated proteins (MAPs) by Patel et al., indicating that the highest share of proteins (31.3%) was involved in various metabolic pathways, while the second most abundant group consisted of proteins involved in gene synthesis/protein expression (29.8%) [31]. The study identified a total of 409 MAPs in murine RAW264.7 macrophage cells by use of a proteomic approach. These results are consistent with reports that numerous ‘soluble’ enzymes can be associated with the cytoskeleton [32]. It has been previously reported that interactions of glycolytic enzymes with the cytoskeleton are of relatively low affinities and transient [33], with the role of regulating metabolism through localized enrichment of the enzymes and resulting changes in enzymatic activity [33]. Such association of metabolic enzymes with cytoskeletal fibers is consistent with the reports showing that saponin permeabilization of the plasma membrane did not result in significant loss of cellular contents, even over an extended period of time, and that permeabilized cells retain almost normal protein biosynthetic capacity [34]. This suggests that diffusion within the cell is limited by the attachment of proteins to the cytoskeleton. Consistently, disruption of the actin cytoskeleton resulted in a loss of function and rapid diffusion of a number of cytosolic proteins, suggesting that actin fibers were largely responsible for the retention of functional organization.
Macrophages rapidly modulate their microtubule cytoskeleton to perform acute immune functions. Activated macrophages increase in size, and their phagocytic, secretory and migratory capacities increase within hours of stimulation [35,36]. Patel et al. examined the effect of macrophage activation on MAPs by proteomic profiling before and after IFN-γ/lipopolysaccharide treatment [31]. A number of proteins showed altered abundances between control and treated samples, of which 53 had greater than a threefold enhanced microtubule binding, while MAP release also occurred simultaneously [31]. This study has increased our understanding of how the cytoskeleton proteome is modulated in response to macrophage activation. The augmented microtubule-association of the chaperone protein heat-shock protein (HSP)90β following stimulation by IFN-γ/lipopolysaccharide was demonstrated for the first time, and the data are suggestive of a critical role for HSP90β in microtubule stability [31]. Furthermore, this report provided a good example of the use of bioinformatic tools to interpret large sets of proteomics data. It was thereby found that the set of MAPs interact with one another at a significantly higher frequency than a random set of proteins.
Meng and Wilkins have utilized proteomics to study the cytoskeleton within the unmanipulated natural killer-like cell line YTS [37] and identified a total of 126 different proteins. Again, a sizeable fraction of identified proteins (8%) were classified as energy metabolism enzymes, suggesting a role of cytoskeleton in the targeting and organization of cellular metabolism by providing areas of localized synthesis. Similarly to the previous study of macrophage cytoskeletal preparations [31], large numbers of proteins (10%) were classified as DNA-binding or transcriptional elements. These findings are consistent with the recently identified nuclear function for the actin cytoskeleton in the organization of chromatin and gene expression [38]. The only aforementioned study not identifying any DNA-binding proteins was that of Xu et al. where nuclei were removed prior to the preparation of subcellular fractions [29]. Therefore, identification of DNA-binding proteins in cytoskeleton preparations might be due to the use of total cellular lysates. Alternatively, increasing numbers of proteins are known to have multiple subcellular locations, such as glycolytic enzymes in the nucleus [39,40], and there is increasing evidence that such multiple locations are important to cellular function [41]. Thus far, analysis of the nuclear cytoskeleton in immune cells has not been approached by proteomics. Comparison of the nuclear cytoskeleton proteome in untreated and activated immune cells would be an interesting new avenue in cytoskeleton research as the structure of nuclear actin fibers remains poorly defined (discussed in more detail within the ‘Future directions’ section).
Cytoskeleton purification strategies & proteomics
Several different strategies have been used in previous proteomics-based studies of the cytoskeleton. The neutrophil detergent-resistant membrane cytoskeleton preparations by Xu et al. [29] and Nebl et al. [23] were based on the fact that cytoskeleton-associated proteins constitute a major part of detergent-insoluble pellets, as interactions between cytoskeletal proteins provide resistance against disruption by nonionic detergents. The low-speed pellet fraction, obtained by sedimentation of cell lysates at 2000-5000 g, represents the membrane skeleton [10,13]. Low-speed supernatants further sedimented at over 200,000 g are referred to as the high-speed pellet. This is the cytoplasmic actin cytoskeleton [13]. Apart from cosedimentation, CBPs can also be affinity purified by use of a tubulin affinity column [42] or by a procedure involving the stabilization of microtubules by Taxol and subsequent release of CBPs from Taxol-stabilized microtubules with a high-ionic-strength homogenization buffer [43]. The advantage of the latter approach is that cytoskeletal proteins are removed before MS analysis, thus avoiding the possibility of highly abundant cytoskeletal proteins masking the lower abundant MAPs. A novel technique developed by Meng et al. for the enrichment of cytoskeleton from natural killer cells, uses magnetic Dynal® beads to capture fibers and associated proteins [37]. The mechanism of cytoskeletal capture by Dynal beads is unknown. A possible explanation could involve the physical entrapment of the beads within the actin fiber network.
One of the best-studied functions of immune cells in which cytoskeletal elements play a key role is phagocytosis. The function of the phagosome skeleton is to give structure to a phagosome and to facilitate fusion between phagosomes and late endosomes [44]. A detergent-insoluble phagosome fraction yields cytoskeletal fibers associated with phagosomes [29]. Unlike most approaches relying on isolation of organelles based on their intrinsic density, isolation of latex bead-containing phagosomes is facilitated by the low buoyant density of the phagocytized latex. Thus, phagosomes are floated into a region of the sucrose gradient where other cellular organelles are not detected. Previous morphological and biochemical analysis of latex bead phagosome preparations indicated the virtual absence of contamination by mitochondria, Golgi vesicles, endosomes and plasma membrane [45].
There is currently a great need for methodological studies comparing the available cytoskeleton-enrichment procedures in order to establish a method providing highest recovery of cytoskeletal proteins with minimal contamination. A degree of cytoskeleton enrichment should be established in each study by determining the ratio of actin, tubulin or other relevant cytoskeletal proteins in the enriched preparation versus total cellular lysates.
So far the only criterion used for evaluation of the afore-mentioned cytoskeleton isolation procedures have been the percentage of cytoskeletal proteins among the total number of identified proteins. According to this measure, the detergent-resistant cytosol and the Dynal bead preparations achieved an almost identical share of 46% cytoskeletal proteins, while microtubule cosedimentation accomplished only 13.5%. The low incidence of accepted cytoskeletal proteins obtained with microtubule co sedimentation probably reflects the removal of microtubule fibers prior to the MS identification or, alternatively, the inclusion of a large number of contaminating proteins. However, in view of the enormous number of proteins that transiently associate with the cytoskeleton, simply using the percentage of ‘accepted’ cytoskeletal proteins among the total number of identified proteins is not an ideal criterion for the evaluation of the purity of cytoskeletal preparations.
Taken together, better enrichment procedures and/or alternative methods for validating cytoskeletal preparations are both of crucial importance for further characterization of the cytoskeletal proteome. Cytoskeletal proteomics can profit from extensive experience gained in proteomics analyses of other subcellular organelles [19,41,46-51]. For example, while insoluble proteins continue to be more difficult to analyze, many proteomics analyses now use shotgun proteomics methods in which total protein extracts are proteolysed to peptides prior to the MS analyses [52]. This greatly improves the ability to detect and quantify less soluble proteins and is certainly applicable to cytoskeletal proteins. Furthermore, some of the purification methods used with other organelles [19,41] may be applicable to cytoskeletal proteins. The same is true for methods that validate organellar proteins as bona fide constituents of the organelle. Especially relevant are proteomics methods using quantitative correlations between large sets of different proteins to validate whole groups of organellar proteins, thereby avoiding many of the uncertainties involved in trying to devise procedures that achieve ‘100% purity’ of the target organelle [41].
Essential controls for proteomics studies of cytoskeleton & proteomics data validation
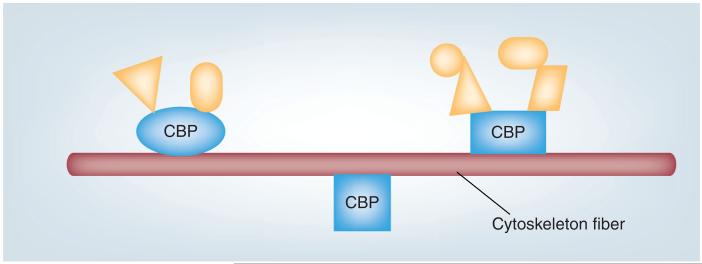
Controls employing a cytoskeleton preparation from cells pretreated with cytoskeleton-disrupting agent such as cytochalasin D are essential for evaluating the specificity of cytoskeleton-enrichment procedures. Furthermore, a control sample representing total cellular proteins is essential in order to establish an enrichment factor for each CBP and cytoskeletal protein. Proteomic studies of the immune cell cytoskeleton are likely to result in an increasing number of identified CBPs and it is important to validate these newly observed interactions by use of alternative methods whenever possible. Immunohistochemistry and/or double-high-resolution fluorescence imaging of cytoskeleton fibers and CBP can provide additional valuable information on their association in living cells. Western blotting can provide a quantitative estimate of the CBP fraction that is bound to cytoskeleton fibers versus the freely soluble unbound protein and can also be used for validation of the proteomics data [29]. Furthermore, it should be investigated whether CBP association with cytoskeleton fibers is due to a direct or indirect interaction (Figure 2). This can be delineated by use of pure cytoskeletal fibers [53], together with a purified or recombinant individual CBP. Dissection of protein–protein interactions can be performed with high sensitivity and reliability by use of our modified affinity pull-down approach [54], or antibody-based pull-down from cross-linked cellular protein (disuccinimidyl suberate), in order to conserve proximity information. Furthermore, any CBP–cytoskeleton interaction may be validated by its disruption through CBP site-directed mutagenesis [55].
Figure 2. Cytoskeleton-binding proteins.
CBPs attach to cytoskeleton fibers directly (large proteins) or indirectly (CBP-interacting proteins; smaller proteins). CBPs and CBP-interacting proteins associate with cytoskeleton fibers in order to direct their subcellular localization, as a concentrating mechanism at specific locations within the cell or to mediate cytoskeletal rearrangement/stabilization. The interactions of CBPs and CBP-interacting proteins are often transient.
CBP: Cytoskeleton-binding protein.
Functional relevance of newly identified CBPs for cytoskeleton function
Techniques such as RNA interference suppression and disruption of CBP–cytoskeleton interaction by site-directed mutagenesis should be carried out in order to investigate the significance of newly identified CBPs for cytoskeleton function and defense functions of immune cells. The function of the cytoskeleton following targeted perturbations of CBPs can be assessed in detail by the use of quantitative speckle fluorescence microscopy [56], or the assay of cellular functions that are known to be critically dependent on the cytoskeleton.
Future directions for proteomics approaches to cytoskeleton analyses
Proteomics methods continue to develop very rapidly. Further improvements in protein identification have been achieved with new advances in MS instruments. The hybrid linear quadrupole ion trap-Orbitrap™ combines a linear ion trap with an Orbitrap mass analyzer, a novel mass analyzer that provides the high mass accuracy of fourier transform MS with the lower cost and ease of operation of an ion trap instrument. The result is an instrument capable of high mass accuracy, high resolution, large ion capacity and large dynamic range [57]. Both the throughput and sensitivity of Orbitrap instruments are still being improved, as are the high-performance LC methods used for resolution of peptides. These developments will strongly enhance the proportion of the total cellular proteome that can be analyzed.
Proteomics is increasingly moving to quantitative measurements of protein abundance rather than simple descriptive detection of the presence of proteins in any given biological sample [58,59]. This opens the door to investigation of quantitative responses of cells to functional stimuli and also provides new ways to characterize the spatio/temporal behavior of cellular proteins, including their presence in specific subcellular locations. At the same time, proteomics methods for detection of PTMs continue to develop strongly [60]. Protein phosphorylation is one of the most important PTMs in cytoskeleton regulation and has been studied intensively. The use of proteomics approaches in analysis of CBP and cytoskeletal protein phosphorylation should be intensified, keeping in mind that the capability of phosphopeptide identification has recently been further enhanced by a new fragmentation technique. Electron transfer dissociation has been developed for use in MS/MS to preserve labile PTMs in peptides during fragmentation [61]. Quantitative phosphoproteomics has already been used extensively to dissect pathways associated with T-cell receptor signaling [62-64], but has not yet been applied to studies of the immune cell cytoskeletal phosphoproteome.
Actin has been identified as a potential candidate for SUMOylation [65], although the nucleus does not show any phalloidin staining of actin fibers and it is known that the majority of SUMOylated proteins are found in the nucleus. The absence of classical actin filaments within the nucleus, as determined by phalloidin staining, has raised questions regarding the form and function of nuclear actin. Nuclear actin SUMOylation [66] raises the intriguing possibility that this PTM also regulates the nuclear structure of actin. SUMOylation could interfere with the formation of classical actin filaments by supporting the formation of unconventional actin structures, such as the antiparallel lower dimer, an arrangement possibly adopted by nuclear actin. This assumption is based on the lower dimer-specific antibody staining of nuclear actin but not cytoplasmic actin [67]. Further studies are needed to establish the role of SUMOylation in the formation of nucleus-specific actin structures in immune cells.
Overall, the application of proteomics to the study of the immune cell cytoskeleton has so far been underexploited. All of the cytoskeleton proteomic studies mentioned previously except for that of Patel et al. [43] were descriptive, and the four different compartments of the cytoskeleton (cytosolic, plasma membrane, phagosome and nuclear) have not been systematically exploited to reduce the complexity of samples for proteomics analyses. This is particularly evident for the understudied immune cell nuclear cytoskeleton, where investigations of nuclear response to immune challenge would be particularly interesting. Cytoskeleton proteomics analyses of PTMs such as phosphorylation, acetylation, methylation and proteolysis are also still relatively sparse and there has been limited use of proteomics methods for the detection of direct protein–protein interactions. We are awaiting a potential boom in new information on the cytoskeleton, and future proteomics studies should particularly aspire to identify spatio/temporal changes in cytoskeletal protein abundance, location and form (both transcriptional and post-translational) following various treatments of immune cells.
Expert commentary
Despite its importance in immune defense, the cytoskeleton in immune cells has not been a very popular target for proteomics studies, with only a few publications thus far. A variety of immune cell types were used, as well as almost as many methods for enriching the cytoskeleton. The main purpose was to define the protein composition of the cytoskeleton and associated proteins. Only one of the studies examined how the protein composition of MAPs changes upon cell stimulation. The variety of models and MS technologies used makes comparisons between the various studies challenging. The expected major cytoskeletal proteins were found consistently in all studies and so were proteins not previously known to associate with the cytoskeleton. By identifying novel CBPs, proteomic approaches can lead to the generation of hypotheses to direct further investigations aimed at improving our understanding of cytoskeleton-dependent immune defense functions.
Five-year view
Protein quantitation strategies based on MS/MS sequencing and high-throughput shotgun proteomics [52,68,69] have transformed MS from a merely descriptive tool to a tool for quantitative measurement of dynamic changes in protein abundance, form and location. Although quantitative proteomics methods based on MS/MS sequencing suffer from limited coverage of lower-abundance proteins, this restriction appears to be largely due to insufficient peptide resolution rather than the sensitivity of current mass spectrometers. Taken together, new sample preparation methods [70-72], ultra-high-pressure LC [73], anion and cation mixed-bed ion exchange techniques [74] and increased sensitivity of MS spectrometers [57], can all be expected to result in major improvements in protein coverage of biological samples. The quantitative proteomic approaches can give exquisitely detailed information on cytoskeleton dynamics and the exact transcriptional and post-translational form of proteins, and should be combined with high-resolution light microscopy to investigate cytoskeleton rearrangements during processes relevant for functions of immune cells such as membrane signaling, phagocytosis, migration, adhesion and degranulation.
Aiming for proteomic analysis of different subcellular cytoskeleton compartments, especially the thus far entirely overlooked nuclear skeleton proteome, is a valid strategy to obtain improved detection of lower-abundance proteins due to the reduced sample complexity.
Proteomics experiments usually do not distinguish among transcriptional, translational, post-translational or catabolic regulation of protein abundance. On the other hand, gene-expression analysis by cDNA microarrays does not distinguish between transcriptional regulation, mRNA stabilization and translational regulation. Combined transcriptomics and proteomics approaches can therefore give new insight into mechanisms for modulation of protein abundance following cell activation [75]. Temporal profiling of immune cells should also include metabolomics - the comprehensive analysis of the small molecule biological sample composition. Since metabolites are often the indirect products of gene expression, this approach complements insights obtained by transcriptomics and proteomics. Consequently, combined transcriptomics, metabolomics, proteomics and bioinformatics approaches could link gene expression and protein production to metabolic consequences during immune cell activation. Such a complete systems biology approach is likely to become common practice in the future.
Single-cell analysis has been beyond the capability of ‘omics’ technology. This is rapidly changing with the recent examples of single-cell genomics, transcriptomics, proteomics and metabolomics, owing to emerging technologies that range from micro/nanofluidics to microfabricated interfaces for MS to third- and fourth-generation automated DNA sequencers [76]. Single-cell analysis is the new frontier in omics, and single-cell omics has the potential to transform systems biology through new discoveries derived from cellular heterogeneity and communication.
The impact of mathematics on biology has so far been very modest; however, the interpretation of large amounts of raw data obtained from integrated high-throughput transcriptomics, metabolomics, proteomics and physiological approaches will present an increasing challenge and requires further development of bioinformatics tools and mathematical models in immunology [77,78]. Models must be constructed, analyzed, simulated, validated and verified.
We are much better at taking cells apart than putting them together. An in vitro reconstitution of biological processes from component molecules presents a powerful but difficult approach to studying functional organization in biology and defining the necessary and sufficient conditions for a cytoskeletal process. Complex cytoskeletal structures observed in cells can be reconstituted in vitro from purified protein components [79]. New methods will be needed in order to also allow for the reconstruction of integral membrane proteins and metabolic processes [80].
As ever-increasing amounts of new data become available through the use of ‘omics’ approaches, more flexibility will be needed to allow necessary revisions of the existing conceptual models.
Key issues.
The cytoskeleton is involved in all main functions of immune cells related to the response to infection.
Studies of the cytoskeletal proteome can vastly contribute to our understanding of cytoskeleton composition, function, regulation and dynamics.
The opportunity to use proteomics approaches in cytoskeleton research has remained largely underexploited.
Further research of the mechanisms of cytoskeleton dynamics requires quantitative proteomic analysis of temporal patterns of cytoskeleton-binding proteins association/dissociation, rather than simple descriptive detection of the presence of proteins.
Newly identified cytoskeleton-binding proteins by the use of proteomics methodology should be validated with other approaches such as Western blotting and double-fluorescence imaging.
Improved cytoskeleton purification strategies will facilitate the acquisition and interpretation of proteomics data.
There is a need for proteomic analysis of subcellular cytoskeleton compartments in immune cells, especially the thus far entirely overlooked nuclear skeleton proteome.
Available methodological strategies should be increasingly used in order to improve the detection of low-abundance proteins.
Proteomics, transcriptomics, metabolomics and bioinformatics approaches need to be progressively combined in cytoskeleton research.
In vitro reconstitution of cytoskeleton processes would aid in the validation of proteomics data.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with financial interests in or financial conflicts with the subject matter or material discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Kustermans G, El Mjiyad N, Horion J, Jacobs N, Piette J, Legrand-Poels S. Actin cytoskeleton differentially modulates NF-κB-mediated IL-8 expression in myelomonocytic cells. Biochem. Pharmacol. 2008;76(10):1214–1228. doi: 10.1016/j.bcp.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 2.Selve N, Wegner A. Rate of treadmilling of actin filaments in vitro. J. Mol. Biol. 1986;187(4):627–631. doi: 10.1016/0022-2836(86)90341-4. [DOI] [PubMed] [Google Scholar]
- 3.Disanza A, Steffen A, Hertzog M, Frittoli E, Rottner K, Scita G. Actin polymerization machinery: the finish line of signaling networks, the starting point of cellular movement. Cell Mol. Life Sci. 2005;62(9):955–970. doi: 10.1007/s00018-004-4472-6. [DOI] [PubMed] [Google Scholar]
- 4.dos Remedios CG, Chhabra D, Kekic M, et al. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 2003;83(2):433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 5.Marin-Esteban V, Charron D, Gelin C, Mooney N. Chemotherapeutic agents targeting the tubulin cytoskeleton modify LPS-induced cytokine secretion by dendritic cells and increase antigen presentation. J. Immunother. 2010;33(4):364–370. doi: 10.1097/CJI.0b013e3181cd1094. [DOI] [PubMed] [Google Scholar]
- 6.Wickramarachchi DC, Theofilopoulos AN, Kono DH. Immune pathology associated with altered actin cytoskeleton regulation. Autoimmunity. 2010;43(1):64–75. doi: 10.3109/08916930903374634. [• Shows that perturbations in the actin cytoskeleton affect the organization of plasma membrane domains.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liebl D, Griffiths G. Transient assembly of F-actin by phagosomes delays phagosome fusion with lysosomes in cargo-overloaded macrophages. J. Cell Sci. 2009;122(Pt 16):2935–2945. doi: 10.1242/jcs.048355. [DOI] [PubMed] [Google Scholar]
- 8.Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453(7192):241–245. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]
- 9.Song W, Xuan H, Lin Q. Epidermal growth factor induces changes of interaction between epidermal growth factor receptor and actin in intact cells. Acta Biochim. Biophys. Sin. (Shanghai) 2008;40(8):754–760. [PubMed] [Google Scholar]
- 10.Caplan S, Baniyash M. Normal T cells express two T cell antigen receptor populations, one of which is linked to the cytoskeleton via zeta chain and displays a unique activation-dependent phosphorylation pattern. J. Biol. Chem. 1996;271(34):20705–20712. doi: 10.1074/jbc.271.34.20705. [DOI] [PubMed] [Google Scholar]
- 11.Gimborn K, Lessmann E, Kuppig S, Krystal G, Huber M. SHIP downregulates FcεR1-induced degranulation at supraoptimal IgE or antigen levels. J. Immunol. 2005;174(1):507–516. doi: 10.4049/jimmunol.174.1.507. [DOI] [PubMed] [Google Scholar]
- 12.Onabajo OO, Seeley MK, Kale A, et al. Actin-binding protein 1 regulates B cell receptor-mediated antigen processing and presentation in response to B cell receptor activation. J. Immunol. 2008;180(10):6685–6695. doi: 10.4049/jimmunol.180.10.6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stie J, Jesaitis AJ. Reorganization of the human neutrophil plasma membrane is associated with functional priming: implications for neutrophil preparations. J. Leukoc. Biol. 2007;81(3):672–685. doi: 10.1189/jlb.0806513. [DOI] [PubMed] [Google Scholar]
- 14.Thrasher AJ, Burns SO. WASP: a key immunological multitasker. Nat. Rev. Immunol. 2010;10(3):182–192. doi: 10.1038/nri2724. [DOI] [PubMed] [Google Scholar]
- 15.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [•• Quantitative lipid raft proteome analysis in T cells upon plasma membrane receptor activation. Active recruitment of cytoskeleton-associated proteins into lipid rafts was revealed upon receptor activation.] [DOI] [PubMed] [Google Scholar]
- 16.Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422(6928):193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 17.Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450(7172):991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 18.Mathivanan S, Ahmed M, Ahn NG, et al. Human Proteinpedia enables sharing of human protein data. Nat.Biotechnol. 2008;26(2):164–167. doi: 10.1038/nbt0208-164. [DOI] [PubMed] [Google Scholar]
- 19.Foster LJ, de Hoog CL, Zhang Y, Xie X, Mootha VK, Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125(1):187–199. doi: 10.1016/j.cell.2006.03.022. [•• Quantitative analysis of microtubule-associated proteins upon macrophage stimulation.] [DOI] [PubMed] [Google Scholar]
- 20.Godovac-Zimmermann J, Kleiner O, Brown LR, Drukier AK. Perspectives in spicing up proteomics with splicing. Proteomics. 2005;5(3):699–709. doi: 10.1002/pmic.200401051. [DOI] [PubMed] [Google Scholar]
- 21.Razzaq TM, Ozegbe P, Jury EC, Sembi P, Blackwell NM, Kabouridis PS. Regulation of T-cell receptor signalling by membrane microdomains. Immunology. 2004;113(4):413–426. doi: 10.1111/j.1365-2567.2004.01998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster LJ. Lessons learned from lipid raft proteomics. Expert Rev. Proteomics. 2008;5(4):541–543. doi: 10.1586/14789450.5.4.541. [DOI] [PubMed] [Google Scholar]
- 23.Nebl T, Pestonjamasp KN, Leszyk JD, Crowley JL, Oh SW, Luna EJ. Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J. Biol. Chem. 2002;277(45):43399–43409. doi: 10.1074/jbc.M205386200. [DOI] [PubMed] [Google Scholar]
- 24.Viola A, Gupta N. Tether and trap: regulation of membrane-raft dynamics by actin-binding proteins. Nat. Rev. Immunol. 2007;7(11):889–896. doi: 10.1038/nri2193. [DOI] [PubMed] [Google Scholar]
- 25.Jones GE. Cellular signaling in macrophage migration and chemotaxis. J. Leukoc. Biol. 2000;68(5):593–602. [PubMed] [Google Scholar]
- 26.Han X, Smith NL, Sil D, Holowka DA, McLafferty FW, Baird BA. IgE receptor-mediated alteration of membrane-cytoskeleton interactions revealed by mass spectrometric analysis of detergent-resistant membranes. Biochemistry. 2009;48(27):6540–6550. doi: 10.1021/bi900181w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin SL, Chien CW, Han CL, et al. Temporal proteomics profiling of lipid rafts in CCR6-activated T cells reveals the integration of actin cytoskeleton dynamics. J. Proteome Res. 2010;9(1):283–297. doi: 10.1021/pr9006156. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi M, Katagiri T, Kosako H, Iida N, Hattori S. Global analysis of dynamic changes in lipid raft proteins during T-cell activation. Electrophoresis. 2007;28(12):2035–2043. doi: 10.1002/elps.200600675. [DOI] [PubMed] [Google Scholar]
- 29.Xu P, Crawford M, Way M, Godovac-Zimmermann J, Segal AW, Radulovic M. Subproteome analysis of the neutrophil cytoskeleton. Proteomics. 2009;9(7):2037–2049. doi: 10.1002/pmic.200800674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolli S, Zito CI, Mossink MH, Wiemer EA, Bennett AM. The major vault protein is a novel substrate for the tyrosine phosphatase SHP-2 and scaffold protein in epidermal growth factor signaling. J. Biol. Chem. 2004;279(28):29374–29385. doi: 10.1074/jbc.M313955200. [DOI] [PubMed] [Google Scholar]
- 31.Patel PC, Fisher KH, Yang EC, Deane CM, Harrison RE. Proteomic analysis of microtubule-associated proteins during macrophage activation. Mol. Cell Proteomics. 2009;8(11):2500–2514. doi: 10.1074/mcp.M900190-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clegg J, Kell D, Knull H, Welch GR, Wilson J. Macromolecular interactions: tracing the roots. Trends Biochem. Sci. 2001;26(2):91. doi: 10.1016/s0968-0004(00)01739-4. [DOI] [PubMed] [Google Scholar]
- 33.Knull HR, Walsh JL. Association of glycolytic enzymes with the cytoskeleton. Curr. Top. Cell Regul. 1992;33:15–30. doi: 10.1016/b978-0-12-152833-1.50007-1. [DOI] [PubMed] [Google Scholar]
- 34.Hudder A, Nathanson L, Deutscher MP. Organization of mammalian cytoplasm. Mol. Cell Biol. 2003;23(24):9318–9326. doi: 10.1128/MCB.23.24.9318-9326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nacife VP, Soeiro Mde N, Gomes RN, D’Avila H, Castro-Faria Neto HC, Meirelles Mde N. Morphological and biochemical characterization of macrophages activated by carrageenan and lipopolysaccharide in vivo. Cell Struct. Funct. 2004;29(2):27–34. doi: 10.1247/csf.29.27. [DOI] [PubMed] [Google Scholar]
- 36.Hermann P, Rubio M, Nakajima T, Delespesse G, Sarfati M. IFN-α priming of human monocytes differentially regulates Gram-positive and Gram-negative bacteria-induced IL-10 release and selectively enhances IL-12p70, CD80, and MHC class I expression. J. Immunol. 1998;161(4):2011–2018. [•• Focuses on methods to perform subcellular fractionation.] [PubMed] [Google Scholar]
- 37.Meng X, Wilkins JA. Compositional characterization of the cytoskeleton of NK-like cells. J. Proteome Res. 2005;4(6):2081–2087. doi: 10.1021/pr0502121. [DOI] [PubMed] [Google Scholar]
- 38.Pederson T. As functional nuclear actin comes into view, is it globular, filamentous, or both? J. Cell Biol. 2008;180(6):1061–1064. doi: 10.1083/jcb.200709082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheng WY, Wang TC. Proteomic analysis of the differential protein expression reveals nuclear GAPDH in activated T lymphocytes. PLoS One. 2009;4(7):e6322. doi: 10.1371/journal.pone.0006322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shakib K, Norman JT, Fine LG, Brown LR, Godovac-Zimmermann J. Proteomics profiling of nuclear proteins for kidney fibroblasts suggests hypoxia, meiosis, and cancer may meet in the nucleus. Proteomics. 2005;5(11):2819–2838. doi: 10.1002/pmic.200401108. [DOI] [PubMed] [Google Scholar]
- 41.Qattan AT, Mulvey C, Crawford M, Natale DA, Godovac-Zimmermann J. Quantitative organelle proteomics of MCF-7 breast cancer cells reveals multiple subcellular locations for proteins in cellular functional processes. J. Proteome Res. 2010;9(1):495–508. doi: 10.1021/pr9008332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chuong SD, Good AG, Taylor GJ, Freeman MC, Moorhead GB, Muench DG. Large-scale identification of tubulin-binding proteins provides insight on subcellular trafficking, metabolic channeling, and signaling in plant cells. Mol. Cell Proteomics. 2004;3(10):970–983. doi: 10.1074/mcp.M400053-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Vallee RB. A taxol-dependent procedure for the isolation of microtubules and microtubule-associated proteins (MAPs) J. Cell Biol. 1982;92(2):435–442. doi: 10.1083/jcb.92.2.435. [•• Describes the quantitative proteomics approach enabling precise predictions of the placement of individual phosphorylation events within a signaling pathway.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kjeken R, Egeberg M, Habermann A, et al. Fusion between phagosomes, early and late endosomes: a role for actin in fusion between late, but not early endocytic organelles. Mol. Biol. Cell. 2004;15(1):345–358. doi: 10.1091/mbc.E03-05-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li N, Mak A, Richards DP, et al. Monocyte lipid rafts contain proteins implicated in vesicular trafficking and phagosome formation. Proteomics. 2003;3(4):536–548. doi: 10.1002/pmic.200390067. [DOI] [PubMed] [Google Scholar]
- 46.Wiederhold E, Veenhoff LM, Poolman B, Slotboom DJ. Proteomics of Saccharomyces cerevisiae organelles. Mol. Cell Proteomics. 2010;9(3):431–445. doi: 10.1074/mcp.R900002-MCP200. [• Presents a model for the actin-small ubiquitin-related modifier post-translational modification (SUMO) complex and show that SUMOylation is required for the nuclear localization of actin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan W, Aebersold R, Raines EW. Evolution of organelle-associated protein profiling. J. Proteomics. 2009;72(1):4–11. doi: 10.1016/j.jprot.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michelsen U, von Hagen J. Isolation of subcellular organelles and structures. Methods Enzymol. 2009;463:305–328. doi: 10.1016/S0076-6879(09)63019-6. [DOI] [PubMed] [Google Scholar]
- 49.Newberg J, Hua J, Murphy RF. Location proteomics: systematic determination of protein subcellular location. Methods Mol. Biol. 2009;500:313–332. doi: 10.1007/978-1-59745-525-1_11. [DOI] [PubMed] [Google Scholar]
- 50.Ploscher M, Granvogl B, Reisinger V, Masanek A, Eichacker LA. Organelle proteomics. Methods Mol. Biol. 2009;519:65–82. doi: 10.1007/978-1-59745-281-6_5. [DOI] [PubMed] [Google Scholar]
- 51.Gauthier DJ, Lazure C. Complementary methods to assist subcellular fractionation in organellar proteomics. Expert Rev. Proteomics. 2008;5(4):603–617. doi: 10.1586/14789450.5.4.603. [DOI] [PubMed] [Google Scholar]
- 52.Lu B, Motoyama A, Ruse C, Venable J, Yates JR., 3rd Improving protein identification sensitivity by combining MS and MS/MS information for shotgun proteomics using LTQ-Orbitrap high mass accuracy data. Anal. Chem. 2008;80(6):2018–2025. doi: 10.1021/ac701697w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chuong SD, Mullen RT, Muench DG. Identification of a rice RNA- and microtubule-binding protein as the multifunctional protein, a peroxisomal enzyme involved in the β-oxidation of fatty acids. J. Biol. Chem. 2002;277(4):2419–2429. doi: 10.1074/jbc.M109510200. [DOI] [PubMed] [Google Scholar]
- 54.Radulovic M, Crane E, Crawford M, Godovac-Zimmermann J, Yu VP. CKS proteins protect mitochondrial genome integrity by interacting with mitochondrial single-stranded DNA-binding protein. Mol. Cell Proteomics. 2010;9(1):145–152. doi: 10.1074/mcp.M900078-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sohn J, Parks JM, Buhrman G, et al. Experimental validation of the docking orientation of Cdc25 with its Cdk2-CycA protein substrate. Biochemistry. 2005;44(50):16563–16573. doi: 10.1021/bi0516879. [DOI] [PubMed] [Google Scholar]
- 56.Danuser G, Waterman-Storer CM. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct. 2006;35:361–387. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- 57.Olsen JV, Schwartz JC, Griep-Raming J, et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell Proteomics. 2009;8(12):2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan S, Aebersold R, Chen R, et al. Mass spectrometry based targeted protein quantification: methods and applications. J. Proteome Res. 2009;8(2):787–797. doi: 10.1021/pr800538n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Godovac-Zimmermann J. Cancer-omics failure: warehouses, magic bullets, space/time and The Life of Brian in cancer cells. Expert Rev. Proteomics. 2010;7(3):303–306. doi: 10.1586/epr.10.7. [DOI] [PubMed] [Google Scholar]
- 60.Mann K, Poustka AJ, Mann M. Phosphoproteomes of Strongylocentrotus purpuratus shell and tooth matrix: identification of a major acidic sea urchin tooth phosphoprotein, phosphodontin. Proteome Sci. 2010;8(1):6. doi: 10.1186/1477-5956-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chi A, Huttenhower C, Geer LY, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl Acad. Sci. USA. 2007;104(7):2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen V, Cao L, Lin JT, et al. A new approach for quantitative phosphoproteomic dissection of signaling pathways applied to T cell receptor activation. Mol. Cell Proteomics. 2009;8(11):2418–2431. doi: 10.1074/mcp.M800307-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mayya V, Lundgren DH, Hwang SI, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein–protein interactions. Sci. Signal. 2009;2(84):ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 64.Grant MM, Scheel-Toellner D, Griffiths HR. Contributions to our understanding of T cell physiology through unveiling the T cell proteome. Clin. Exp. Immunol. 2007;149(1):9–15. doi: 10.1111/j.1365-2249.2007.03395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell Proteomics. 2005;4(1):56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hofmann WA, Arduini A, Nicol SM, et al. SUMOylation of nuclear actin. J. Cell Biol. 2009;186(2):193–200. doi: 10.1083/jcb.200905016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jockusch BM, Schoenenberger CA, Stetefeld J, Aebi U. Tracking down the different forms of nuclear actin. Trends Cell Biol. 2006;16(8):391–396. doi: 10.1016/j.tcb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 68.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 69.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17(10):994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 70.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 71.Hubner NC, Ren S, Mann M. Peptide separation with immobilized pI strips is an attractive alternative to in-gel protein digestion for proteome analysis. Proteomics. 2008;8(23-24):4862–4872. doi: 10.1002/pmic.200800351. [DOI] [PubMed] [Google Scholar]
- 72.Mulvey C, Thur B, Crawford M, Godovac-Zimmermann J. How many proteins are missed in quatitative proteomics experiments based on current MS/MS sequencing methods? Proteomics. 2010:61–63. doi: 10.4137/PRI.S5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Livesay EA, Tang K, Taylor BK, et al. Fully automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal. Chem. 2008;80(1):294–302. doi: 10.1021/ac701727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Motoyama A, Xu T, Ruse CI, Wohlschlegel JA, Yates JR., 3rd Anion and cation mixed-bed ion exchange for enhanced multidimensional separations of peptides and phosphopeptides. Anal. Chem. 2007;79(10):3623–3634. doi: 10.1021/ac062292d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fessler MB, Malcolm KC, Duncan MW, Worthen GS. A genomic and proteomic analysis of activation of the human neutrophil by lipopolysaccharide and its mediation by p38 mitogen-activated protein kinase. J. Biol. Chem. 2002;277(35):31291–31302. doi: 10.1074/jbc.M200755200. [DOI] [PubMed] [Google Scholar]
- 76.Wang D, Bodovitz S. Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol. 2010;28(6):281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Louzoun Y. The evolution of mathematical immunology. Immunol. Rev. 2007;216:9–20. doi: 10.1111/j.1600-065X.2006.00495.x. [DOI] [PubMed] [Google Scholar]
- 78.Bellomo N, Forni G. Complex multicellular systems and immune competition: new paradigms looking for a mathematical theory. Curr. Top. Dev. Biol. 2008;81:485–502. doi: 10.1016/S0070-2153(07)81017-9. [DOI] [PubMed] [Google Scholar]
- 79.Bieling P, Laan L, Schek H, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450(7172):1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 80.Liu Y, Sanoff HK, Cho H, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4(4):e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 101.Proteomics encyclopeida. www.humanproteinpedia.org.