Abstract
Concurrent proteomics analysis of the nuclei and mitochondria of MCF7 breast cancer cells identified 985 proteins (40% of all detected proteins) present in both organelles. Numerous proteins from all five complexes involved in oxidative phosphorylation (e.g., NDUFA5, NDUFB10, NDUFS1, NDUF2, SDHA, UQRB, UQRC2, UQCRH, COX5A, COX5B, MT-CO2, ATP5A1, ATP5B, ATP5H, etc.), from the TCA-cycle (DLST, IDH2, IDH3A, OGDH, SUCLAG2, etc.), and from glycolysis (ALDOA, ENO1, FBP1, GPI, PGK1, TALDO1, etc.) were distributed to both the nucleus and mitochondria. In contrast, proteins involved in nuclear/mitochondrial RNA processing/translation and Ras/Rab signaling showed different partitioning patterns. The identity of the OxPhos, TCA-cycle, and glycolysis proteins distributed to both the nucleus and mitochondria provides evidence for spatio-functional integration of these processes over the two different subcellular organelles. We suggest that there are unrecognized aspects of functional coordination between the nucleus and mitochondria, that integration of core functional processes via wide subcellular distribution of constituent proteins is a common characteristic of cells, and that subcellular spatial integration of function may be a vital aspect of cancer.
Keywords: quantitative proteomics, mass spectrometry, breast cancer, MCF7 cells, subcellular organelles, subcellular location, nucleus, mitochondria, TCA cycle, oxidative phosphorylation, glycolysis
1. INTRODUCTION
Two overall characteristics of cellular function that are clearly involved in the wide diversity of cancers include high degrees of genetic instability and major changes in cellular energy metabolism. The genetic instability, which is associated with the cell nucleus, allows cells to escape from a variety of normal restrictions on proliferation,1 whereas the changes in energy metabolism, which involve mitochondria, seem to be associated with the need for production of new cellular components in proliferating cells. There are suggestions that cancer might even be primarily a metabolic disease2-11 and increasing evidence that the aerobic glycolysis and/or glutaminolysis characteristic of many cancer cells mainly reflects the needs of proliferating cells for production of new cellular components.4
Mitochondria are involved in a wide spectrum of disease5,12 and closely connected to cancer through energy and proliferation requirements. Mitochondria are highly dynamic organelles that vary with cell/tissue type,13-15 undergo fission-fusion processes,16-21 retain connections to the endoplasmic reticulum22,23 and participate in the integration of cellular signaling processes, including between the nucleus and mitochondria.24 The mitochondrial genome encodes only 13 proteins that are part of the oxidative-phosphorylation complexes I, III, IV and V of the mitochondrial inner membrane.25 Since most mitochondrial proteins are nuclear encoded, mechanisms for the biogenesis of mitochondria, the coordination of mitochondrial biogenesis with the cell cycle and the establishment/maintenance of the mitochondrial proteome have been intensively investigated.26-29 Several mechanisms for protein trafficking involving mitochondria are known. These include classical import via mitochondrial import systems that involve mitochondrial import sequences, participation of chaperones, protein folding/unfolding steps and the direction of proteins to specific mitochondrial compartments such as the inner membrane, outer membrane, intermembrane space and matrix,30 but other diverse mechanisms also operate for many proteins.31 Mitochondria show spatial juxtaposition with the endoplasmic reticulum that seems to be important in lipid metabolism and trafficking, Ca2+ homeostasis, apoptosis and possibly other functions22,23 and there are some indications that nonclassical pathways for direction of proteins from the endoplasmic reticulum to mitochondria may exist.32 A variety of nuclear receptors such as the estrogen, PPARγ2, glucocorticoid, and c-ErbAα1 (thyroid hormone) receptors as well as transcription factors such as AP1, CREB, NF-κB, Stat3 and p53 can translocate to mitochondria where they seem to be involved in transcriptional, DNA repair and apoptotic functions.33-36 Recently an export system that transfers proteins from mitochondria to peroxisomes via vesicular trafficking has also been identified.37 These dynamic properties of mitochondria mean that the mitochondrial proteome is not static, but varies with cell type and state.
Recent work has re-emphasized that many cellular proteins show multiple subcellular locations. We recently used proteomics methods to show that at least 50% and perhaps as much as much as 75% of proteins detected in MCF7 cells show multiple locations involving significant proportions of each protein.38 In yeast, recent studies indicate that at least 30% of mitochondrial proteins also have other subcellular locations.39 In the context of nuclear import/export, there has long been evidence that dynamic relocation of proteins to and from the nucleus is crucial to cellular function and there is increasing evidence for changes in nuclear transport following perturbations such as oxidative stress.40,41 Other clear evidence that dynamic spatial redistribution of proteins to many cellular locations is coupled to cellular function has recently been obtained by a genome-wide screen showing that over 200 yeast proteins (4–5% of the genome) are spatially translocated as a consequence of hypoxia, with much of the protein translocation occurring prior to transcriptional changes.42
These features suggest that to understand the energetic/integrative functions of mitochondria and the nature of their role in cancer and other diseases, it will be necessary to study the dynamic features of the mitochondrial proteome. It will be essential to also look at concurrent changes in the nuclear proteome. As a preparatory step for dynamic response studies, the present work establishes that for nuclei and mitochondria purified by sucrose gradient fractionation approximately 1000 proteins (40% of all detected proteins) can be detected and monitored concurrently in both mitochondria and the nucleus in MCF7 breast cancer cells with current proteomics methods. We present extensive evidence that the observed partitioning of these proteins between the nucleus and mitochondria represents real functional distributions and is not an artifact of the subcellular fractionation methods. Analysis of the distribution between the nucleus and mitochondria for specific subgroups of proteins involved in oxidative phosphorylation, the tricarboxylic acid cycle, RNA processing/translation, glycolysis and Ras-related signaling suggests that dispersion of component proteins over many subcellular locations is common and that different cellular functions use different patterns of subcellular spatial dispersion. The identity of many of these proteins suggests that spatial distribution of numerous proteins over multiple sites is critical to function, and that there are unrecognized aspects of the functional coordination between mitochondria and nuclei that need further investigation. We suggest that extensive subcellular dispersion of the constituent proteins of core cellular functions is a fundamental characteristic of cells that may be consistent with requirements for robustness in complex systems and vital to cancer.
2. MATERIAL AND METHODS
2.1. Culturing, Harvesting and Breakage of Cells
The mammary epithelial adenocarcinoma breast cancer cell line MCF7 was purchased from the ATCC, HTB-22, Manassas, VA and cultured at 37 °C with 5% CO2 in Dulbecco’s Modified Eagle Media DMEM/F-12 (Gibco, Invitrogen, Paisley, U.K.) with 2 mM l-Glutamine, 15 mM HEPES, 10% defined fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco, Invitrogen, Paisley, U.K.) and 50 μg/mL gentamicin (Gibco, Invitrogen, Paisley, U.K.).
Cells were washed three times within tissue culture flasks with cold phosphate buffer saline (PBS) buffer in order to remove most of the FBS, followed by harvesting using a cold plastic cell scraper. Approximately 107 cells obtained per 75 cm2 flask were suspended in a cold hypotonic osmotic shock buffer consisting of 10 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCL (pH 7.4) by vortex mixing and left to swell on ice for 10 min. The buffer exchange was started by a 10 min 1000× g spin at 4 °C, followed by pellet resuspension in 3 mL isotonic 0.3 M sucrose. Another spin at 4 °C for 10 min at 1000× g was done in order to obtain cells in three volumes of isotonic sucrose breaking buffer containing 300 mM sucrose, 1 mM EDTA, Heparin 5 U/mL, 10 mM HEPES, 5 mM MgCl2, pH 7.4. Cells were broken gently by liquid shear in a tight-fitting glass Dounce homogenizer (0.05–0.08 mm clearance). Phase contrast microscopy was used to ensure that >95% of cells were broken.
2.2. Organelle Preparation
The isolation of nuclei and mitochondria was based on the procedure of Wang et al.43 The main modifications included additional washes of nuclei and mitochondria as described below. Nuclei were spun down at 800× g for 10 min at 4 °C to produce a crude nuclear pellet, while the supernatant was kept for isolation of the mitochondrial and cytoplasmic fractions. All the procedures were performed at 4 °C with fresh protease and phosphatase inhibitor cocktail supplements (Roche Diagnostics, Mannheim, Germany). The validity of the subcellular fractionation was assessed by Western blotting as described below.
2.3. Preparation of Nuclear Proteins.38,44,45
To isolate the nuclei with an intact nuclear membrane while simultaneously reducing cellular debris, the nuclear pellet was suspended in a hypotonic buffer containing 0.1% Triton-X100 and 2 mM EDTA, passed through a syringe needle (22 gauge) and spun at 3000 rpm for 5 min in order to sediment intact nuclei. This treatment was repeated twice to maximize removal of cell debris.45 The final nuclear pellet was resuspended by vortex mixing in ice-cold hypotonic buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 5 mM MgCl2, 2 mM EDTA, 1 mM DTT, 0.1% Triton X-100) supplemented with freshly dissolved protease and phosphatase inhibitor cocktails and incubated for 15 min at 4 °C on a rotating platform. Nuclei were spun down and extracted with four volumes of high salt breaking buffer containing 20 mM HEPES, pH 7.9, 700 mM NaCl, 1 mM EDTA, 1.5 mM MgCl2 and 10% glycerol, for 2 h on a rotating platform at 4 °C.44 The extract was centrifuged for 10 min at 4 °C at 300× g to remove any residual cell debris. The supernatant was subjected to acetone precipitation using 4 volumes of 80% ice-cold acetone, left for a minimum of 1h at −20 °C and then spun at 13 000 rpm for 30 min at 4 °C to recover nuclear proteins. The pellet was air-dried for 5 min to eliminate any acetone residue and then resuspended in a 1× protein solubilization buffer (10 mM PIPES pH 7.3, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholic acid).38
2.4. Preparation of Mitochondrial Proteins.43
The supernatant above the crude nuclear pellet obtained as described above was transferred to a clean tube and centrifuged for 10 min at 7000× g at 4 °C to pellet crude mitochondria. The pellet was retained and the remaining supernatant was centrifuged again at 14 000× g for 45 min at 4 °C (using a TLA-100.4 rotor, Optima TLX 120 Bench top Ultracentrifuge Beckman Coulter, Beckman, MN), and the pellet was retained. The supernatant was used for the preparation of the cytosolic fraction and the two pellets were combined as a crude mitochondrial fraction. The pellet was then washed with MSHE buffer containing 210 mM Mannitol, 70 mM sucrose, 1 mM EGTA, 2 mM EDTA, 5 mM HEPES, pH 7.4, 10 mM Tris-HCL pH 7.4, and resuspended in 2 mL of sucrose solution (0.25 M sucrose, 0.15 mM MgCl2, 10 mM Tris-HCl pH 6.7). The mitochondrial suspension was then laid on the top of a discontinuous sucrose gradient (0.43–1.46M) and further processed according to our established protocol.38 Briefly, centrifugation of the sucrose gradient was performed for 18 h at 14 440× g in a swing-bucket rotor (TST41 rotor, Optima LE-80K centrifuge, Beckman, MN). Mitochondria formed a band at ~1.16 to 1.3 M sucrose. The refractive index (Rf) and the molarity were estimated by use of a refractometer (Sun Instruments, Torrence, CA). The gradient fractions were then diluted 1:1 with Dilution Buffer (1 mM EDTA, Heparin 5 U/mL, 10 mM, HEPES, 5 mM MgCl2, pH 7.4) and spun for 60 min at 22 000× g in a TLA-100.4 fixed rotor (TLX Ultracentrifuge, Beckman, Chaska, MN) in order to pellet the mitochondria from the sucrose suspension. This pellet was further subjected to fractionation and mass spectrometric analysis as described below.
2.5. Preparation of Cytoplasmic Proteins.38
The supernatant retained from the second pelleting of mitochondria was diluted with an equal volume of dilution buffer (1 mM EDTA, Heparin 5U/mL, 10 mM HEPES, 5 mM MgCl2, pH 7.4) and centrifuged for 60 min at 22 000× g. The clarified cytosol fraction was then subjected to acetone precipitation as described above and dissolved in protein solubilization buffer.
2.6. SDS-PAGE Gel Fractionation and Processing
Protein concentrations were determined using a BioRad detergent compatible (DC) protein assay. Thirty micrograms of protein of the nuclear, mitochondrial and cytosolic fractions was separated by 10% (w/v) SDS-PAGE with a Mini-Protean III system (BioRad, Herts, U.K.) and the proteins were visualized by silver staining (ProteoSilver Plus kit, Sigma-Aldrich, Poole, Dorset, U.K.). The gel was cut into 36 horizontal slices and each slice further cut into 1 × 1 mm2 pieces. Destaining was done using 100 mM sodium thiosulphate and 30 mM potassium ferricyanide. Samples were reduced by 10 mM dithiothreitol DTT and alkylated with 100 mM iodoacetamide by use of the ProGest Investigator Instrument (DigiLab, Genomics Solutions, Cambs, U.K.) according to the established protocol.38 Subsequently, 200 ng of sequencing grade modified trypsin (Promega, Hamps, U.K.) in 50 mM ammonium bicarbonate was added to dry gel pieces and the mixture incubated overnight at 37 °C as described previously.46 Tryptic peptides were eluted from the gel, concentrated and dissolved in 0.1% formic acid for LC–MS/MS analysis.
2.7. Prefractionation of Proteins by Isoelectric Focusing
An Agilent 3100 OFFGEL fractionator was used with a 24 well setup and 24 cm long 3–10 NL pH gradient IPG strips (Agilent Technologies, U.K.) according to the protocol of the supplier and Geiser et al.47 The nucleus and the mitochondrial samples were focused with a maximum current of 50 μA and maximum voltage of 8000 V until 64 kVh was reached. The runs took approximately 40 h with the temperature set to 20 °C. After IEF, groups of three contiguous fractions (volume between 100–150 μL) were combined and acetone precipitated as described above, dissolved in 6 M urea, reduced, alkylated, trypsinized, cleaned up by use of StageTips C1848,49 (ThermoFisher Scientific, U.K.), dried and dissolved in 0.1% formic acid for LC–MS/MS analysis.
2.8. Tests of Fractionation Reproducibility by Western Blotting
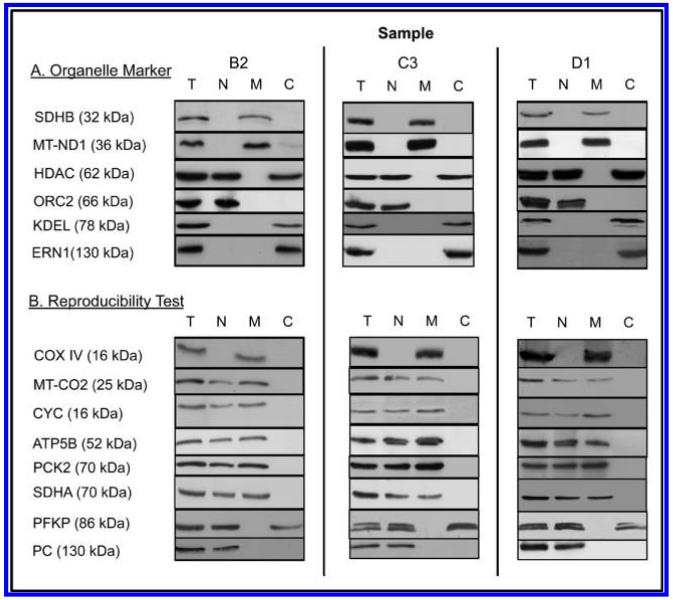
To check reproducibility of the fractionation procedures, a set of cyclic permuted samples was prepared using the methods described above. MCF7 cells were harvested and processed as noted above in detail. Cells were divided into four aliquots: A, B, C, and D, all of which were individually homogenized. Homogenate “A” was used for Western blotting as the total cell lysate, while the homogenates B-D were processed to give cytosol, nucleus and unbroken cell pellet. Each fraction was then divided into three aliquots to give nine samples: B-1, B-2, B-3; C-1, C-2, C-3; D-1, D-2, D-3, each of which had 2 fractions: supernatant and nucleus/unbroken cells. Each of the nine fractions was then individually processed to produce nine nuclear preparations (B-1n to D-3n), nine mitochondrial preparations (B-1m to D-3m) and nine cytoplasmic samples (B-1c to D-3c). For the triplicate Western blotting experiments, the cyclic permuted sets of samples {B1n, C2n, D3n}, {B2n, C3n, D1n} and {B3n, C1n, D2n}, and the corresponding mitochondrial and cytoplasm sets, were prepared. The first of these triplicate sets was used for testing and optimization of individual antibodies. The second was used for the experiments shown in Figure 7 (see text) and the third was held in reserve.
Figure 7.
Western blotting of a cyclic permuted triplicate preparation (see Material and Methods) of a cellular total lysate (T) and of nuclear (N), mitochondrial (M) and cytoplasmic (C) subcellular preparations. Sample loadings and dilutions of primary and secondary antibodies were adjusted to verify presence/absence of the proteins in the different fractions, that is, relative spot intensities between different samples are not indicative of relative abundance in different fractions.
Samples for Western blotting were diluted with the SDS-Sample buffer (4× solution: 250 mM Tris-HCL pH 6.8, 30% glycerol, 6% SDS, 0.02% bromophenol blue and 300 mM DTT), vortexed and heated for 3–5 min at 95 °C. Each sample was electrophoretically separated by 10–12% (w/v) SDS-PAGE with a Mini-Protean III system (BioRad, Herts, U.K.), using standard techniques according to Laemmli.50 Gels were electro-transferred to nitrocellulose membranes with a semidry transfer apparatus (BioRad, Herts, U.K.). The nitrocellulose membranes were blocked with 5% BSA or milk-TBS-Tween buffer (TBST: Tris-buffer saline, 0.1% Tween20) for a minimum of 2 h at 4 °C under slow agitation on an orbital shaker and followed by a rinse for 5 s in TBST. The dilution of primary antibodies in TBST was optimized according to the manufacturer’s advice and incubation performed overnight with slow agitation at 4 °C. The membranes were washed and then agitated three times with 100 mL TBST for 10 min to remove residual primary antibody and finally incubated for 1 h at room temperature with the secondary antibodies under slow agitation on an orbital shaker. The dilution of the secondary antibodies was optimized in TBST according to the results. The membranes were washed three times with TBST for 10 min each while being agitated and finally for 5 min with TBS. The following primary antibodies were used: anti-SDHA, anti-HDAC, and anti-IRE1α (ERN1) (catalogue numbers, 5839S, 2062S, and 3294S Cell Signaling, New England BioLabs, U.K.), anti-KDEL (ADI-SPA-827, Enzo Life Science, U.K.), anti-PCK2 (TA302563, OriGene Technologies, Rockville, MD), anti-COXIV, anti-ATP5B, anti-MT-CO2, anti-PFKP, anti-MT-ND1, anti-PCB, anti-SDHB, anti-CYC1 and anti-ORC2 (ab14744, ab14730 ab3298, ab119796, ab74257, ab113020, ab84622, ab128337, ab68348, Abcam, U.K.). Peroxidase-conjugated secondary antibodies were obtained from Cell Signaling (7074, New England BioLabs, U.K.). The chemiluminescence reagents ECL Plus and Hyper Film were from GE Healthcare (Bucks, U.K.).
2.9. LC–MS/MS Mass Spectrometric Analysis and Database Searching
According to our established protocol as described previously,38 peptide samples were analyzed with a LTQ-Orbitrap-XL MS (Thermo Fisher Scientific, Surrey, U.K.), using a capillary column, NTCC-360/100-5-153 (100 μm I.D, 5 μm C18 particles, 15 cm length, Nikkyo Technos CO, Tokyo, Japan) and a nanoelectrospray ion source at a flow rate reduced via a splitter of 0.9 μL/min. The mobile phase comprised 0.1% formic acid (Solvent A) and 100% acetonitrile with 0.1% formic acid (solvent B). The gradient ranged from 5–23% B in 65 min followed by 23–40% B in 30 min with a flow of 350 μL/min from the Thermo SURVEYOR MS Pump Plus. The peptide solutions were desalted using a Michrom C18 Captrap and eluted to the LTQ-Orbitrap MS with a linear gradient of 5–60% buffer B. The range 450 to 1600 m/z of the full scan FTMS was acquired after screening in the MS positive ion mode with a resolution of r = 60 000.
A data dependent MS/MS fragmentation was done for the most intense ion from the survey scan using the following parameters for collision induced dissociation (CID): normalized collision energy 35%, activation Q 0.25; electrospray voltage 1.4 kV; capillary temperature 200 °C: isolation width 2.00. The targeted ions were dynamically excluded for 40s and the scan was repeated for the top 3 peaks of the same MS/MS fragmentation.
2.10. MS Data Interpretation
Peptides and proteins were identified using the ProteinProphet and PeptideProphet algorithms with Scaffold software (Scaffold v 2.06.02, Proteome Software Inc. Portland, OR) with the MASCOT algorithm (Matrix Science, v2.2.04). Each MS/MS spectrum was searched against the human protein sequence database (IPI v3.87, file ipi.HUMAN.v3.87.fasta.gz) downloaded from the European Bioinformatics Institute server. The MASCOT searches were run using the following parameters: cysteine carbamidomethylation modifications and methionine oxidation, 2 missed cleavages were allowed, precursor error tolerance at <20 ppm; MS/MS fragment tolerance set to 0.2 Da and charge set to +2; full trypsin specificity (N and C-terminal also applied). The tryptic peptide matches had confidence values of ≥95%. Only proteins with two or more unique peptides matching were considered positively identified.
The initial mapping of peptides to proteins gave 6022 protein sequences (IPI numbers). For each of the four samples (nucleus, SDS-PAGE fractionation; nucleus, pI fractionation; mitochondrion, SDS-PAGE fractionation; mitochondrion, pI fractionation), the protein IPI sequence groups compatible with the MS data were labeled according to the underlying gene using the IPI cross reference table (ipi.HUMAN.xres.gz, v3.87). In addition to the IPI numbers, original accession numbers from the IPI cross-reference table are given in Supplementary Tables 1 and 2 for all identified proteins. In subsequent analysis, consensus protein sequence groups across the samples were determined for the three sample combinations (N-SDS, N-pI), (M-SDS, M-pI) and (N-SDS, N-pI, M-SDS, M-pI) by using all identified unique peptides that were assigned to the same gene to requery the full IPI human protein sequence data set. In these remappings, only protein sequences that contained all identified peptides from the underlying gene were accepted, giving a total of 5015 protein sequences. In all cases the resulting consensus protein sequence groups were independent, that is, none of the final protein sequence groups have any common protein sequence. In a few cases (13, see text), there was no protein sequence that contained all peptides—for this small number of cases, two independent protein sequence groups were established by manual inspection of the mapping of the peptides to compatible protein sequences.
2.11. Subcellular Location Annotation
Gene Ontology Cellular Component (GO CC) annotations for each experimental IPI sequence were downloaded from the GOA FTP site at the European Bioinformatics Institute (http://www.ebi.ac.uk/FTP/) using a GO slim with the controlled vocabulary: GO:0005576, extracellular region; GO:0005634, nucleus; GO:0005737, cytoplasm; GO:0005739, mitochondrion; GO:0005768, endosome; GO:0005773, vacuole; GO:0005783, endoplasmic reticulum; GO:0005794, Golgi apparatus; GO:0005829, cytosol; GO:0005886, plasma membrane. For our nuclear and mitochondrial fractions, only a handful of the IPI sequences had annotations to the endosome or vacuole and these annotations were not used in further analysis. For each protein sequence group (see Supporting Information Table 2), the GO CC annotations given are the union of the annotations for the individual sequences in the protein sequence group.
3. RESULTS
On the basis of work previously published by us38 and others,51,52 we used subfractionation of cellular organelles by sucrose gradient centrifugation. The protein samples from nuclear (N) and mitochondrial (M) fractions of sucrose gradient subcellular fractionations of MCF7 cells were further fractionated by SDS gel electrophoresis or pI prior to trypsin treatment and MS analyses. Each of the four MS data sets (N-SDS, N-pI, M-SDS, M-pI) was processed individually to identify protein sequence groups compatible with the observed peptides in the sample. This gave a total of 6022 different protein sequences grouped in 2700 different protein sequence groups over the four samples (Supporting Information Table 1). The protein sequence groups corresponded to 2384 genes, the difference arising because the observation of different numbers/identities of peptides from the same gene in the four samples led to slightly different groups of protein sequences from different samples for 514 genes. For each gene, the sets of unique peptides observed in the individual samples were combined and the full human IPI sequence set was requeried with the combined peptides to search for consensus protein sequence groups consistent with all four samples. This gave 5012 protein sequences in 2397 different protein sequence groups, of which 1163 (48.5%) corresponded to a single protein sequence among the 91,464 nonredundant protein sequences in the Human IPI data set (Supporting Information Table 2). For 13 genes two different, independent sets of protein sequence groups from the same gene were required to interpret the detected peptides. Thus, for these 13 genes, direct evidence for the existence of (at least) two different protein isoforms was obtained from the sets of detected peptides (see below).
3.1. Separation of the Nuclear and Mitochondrial Fractions
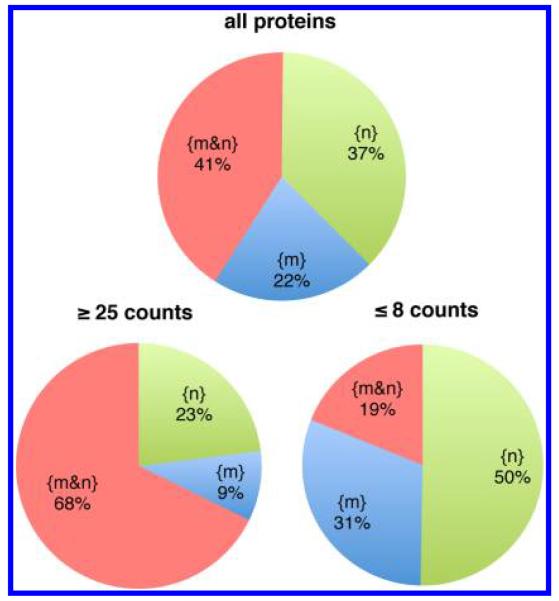
Figure 1 shows the distribution of the detected proteins between the nucleus and mitochondria. Over all 2397 protein sequence groups, 985 (41%) of the proteins were found in both locations. We have used the data sets in three ways to examine the separation between the nuclear and mitochondrial preparations and the influence of the sampling properties of the MS spectral counting detection on the observed distribution. First, by looking at high abundance proteins. For 195 proteins, with more than 25 sequenced peptides (counts) in the nucleus, in the mitochondria or in both, 68% of proteins are observed in both locations. There are 63 very abundant proteins that are seen in only one of the two locations. These proteins have large numbers of spectral counts and many different sequenced peptides, as shown in Table 1 for selected proteins. The large numbers of peptides/spectral counts observed for these high abundance proteins only in the nucleus or only in the mitochondria indicate that any direct cross contamination of the two organelles is very small. To avoid confusion, we use the following terminology from this point: {M} and {N} denote the sets of proteins identified in the experimental sucrose gradient fractions for mitochondria and nucleus respectively. Over all proteins in both fractions, the subsets {m}, {n} and {m&n} denote the sets of proteins detected only in mitochondria, only in the nucleus or in both respectively, that is, {m&n} is the intersection of {M} and {N}. If used without brackets, M or N denotes subcellular locations.
Figure 1.
Partitioning of the sets of proteins detected in the nuclear fraction {N} (1880 proteins) and in the mitochondrial fraction {M} (1502 proteins) between sets of proteins detected only in the nucleus {n}, only in mitochondria {m} or in both organelles {m&n}. (Top) All proteins. (Lower left) High abundance proteins with ≥25 counts in {M}, in {N} or in both. (Lower right) Lower abundance proteins with ≤8 counts in {M}, in {N} or in both.
Table 1. Partitioning of High Abundance Proteins Between the Nucleus and Mitochondria.
| nucleus |
mitochondria |
|||||
|---|---|---|---|---|---|---|
| gene | protein name | location | counts | peptides | counts | peptides |
| Selected high abundance proteins seen only in mitochondria | ||||||
| CAT | Catalase | M | 0 | 0 | 48 | 18 |
| ESYT1 | Isoform 1 of Extended synaptogamin-1 | M | 0 | 0 | 51 | 22 |
| ITGB1 | Isoform Beta-1A of Integrin beta-1 | M | 0 | 0 | 52 | 18 |
| ITGB5 | Integrin beta-5 | M | 0 | 0 | 26 | 13 |
| LMAN1 | Protein ERGIC-53 | M | 0 | 0 | 29 | 10 |
| PGRMC2 | membrane-associated progesterone receptor component 2 | M | 0 | 0 | 26 | 6 |
| POR | NADPH--cytochrome P450 reductase | M | 0 | 0 | 61 | 26 |
| RHOA | Transforming protein RhoA | M | 0 | 0 | 47 | 8 |
| SLC1A5 | Neutral amino acid transporter B(0) | M | 0 | 0 | 33 | 9 |
| SLC3A2 | Isoform 1 of 4F2 cell-surface antigen heavy chain | M | 0 | 0 | 176 | 24 |
| sum over 18 high abundance proteins | M | 0 | 0 | 847 | 236 | |
| Selected high abundance proteins seen only in the nucleus | ||||||
| CDC5L | Cell division cycle 5-like protein | N | 40 | 22 | 0 | 0 |
| FBL | rRNA 2′-O-methyltransferase fibrillarin | N | 83 | 13 | 0 | 0 |
| HDAC1 | Histone deacetylase 1 | N | 38 | 9 | 0 | 0 |
| KDM1A | Isoform 1 of Lysine-specific histone demethylase 1A | N | 26 | 12 | 0 | 0 |
| LRRK2 | Leucine-rich repeat serine/threonine-protein kinase 2 | N | 63 | 8 | 0 | 0 |
| NOP58 | Nucleolar protein 58 | N | 65 | 18 | 0 | 0 |
| PRKDC | DNA-dependent protein kinase catalytic subunit | N | 98 | 64 | 0 | 0 |
| PRPF31 | Isoform 1 of U4/U6 small nuclear ribonucleoprotein Prp31 | N | 27 | 12 | 0 | 0 |
| RCC2 | Protein RCC2 | N | 131 | 23 | 0 | 0 |
| WTAP | Isoform 1 of Pre-mRNA-splicing regulator WTAP | N | 33 | 16 | 0 | 0 |
| sum over 45 high abundance proteins | N | 2369 | 714 | 0 | 0 | |
A second test for possible cross contamination of the nuclear and mitochondrial fractions involved looking at cellular functions that differ between these two organelles. We used proteins involved in RNA processing and protein translation for this comparison since the 13 mitochondria-transcribed proteins (see below) have different subcellular machinery that shows considerable resemblance to prokaryotic organisms.53 For example, the RNase exosome complex, which exists in both the nucleus and the cytoplasm, is involved in many types of RNA processing associated with nuclear-transcribed proteins, including mRNA decay.54,55 The nine core proteins of this complex were observed exclusively in the nuclear fraction (Supporting Information Table 3). For the 13 proteins that are transcribed in mitochondria, the (nuclear encoded) protein cofactors used by mitochondria in transcription and in RNA processing have been less completely characterized, but involve proteins such as TSFM, TACO1, SLIRP and PUS1,53 which we observed exclusively in the mitochondrial fraction (Supporting Information Table 3). A large majority of the proteins of the large mitochondrial ribosome subunit were also detected exclusively in the mitochondrial fraction (Supporting Information Table 3). Some exceptions were observed, which is probably related to previous evidence that some mitochondrial ribosomal proteins have other functions, especially in apoptosis and cell cycle regulation, including in the nucleus.53 Overall the distribution observed for proteins involved in RNA processing and translation is again indicative of very little direct cross contamination between the two organelles.
The third characterization of the separation of mitochondria and nuclei used the proteins for which the peptide data directly gave evidence of multiple isoforms from the same gene. Different patterns of isoform distribution between the two organelles were found for different proteins (Table 2). For example, from the CUX1 gene, the homeobox cut-like 1 protein was observed in the nucleus, whereas the substantially smaller CASP protein was observed predominantly in mitochondria (Table 2). In the present context, the proteins syntaxin-16 (STX16), prolyl 4-hydroxylase subunit alpha-1 (P4HA1) and heterogeneous ribonucleoprotein Q (SYNCRIP) showed peptides consistent with distribution of specific isoforms to one or the other of the organelles (Table 2). The substantial numbers of peptides/counts observed for these proteins, especially heterogeneous ribonucleoprotein Q, is another strong indication for clean separation of the nucleus and mitochondria.
Table 2. Partitioning of Protein Isoforms between the Nucleus and Mitochondriaa.
| nucleus |
mitochondria |
|||||
|---|---|---|---|---|---|---|
| gene | descriptive protein name | location | peptides | counts | peptides | counts |
| STX16 | Isoform D of Syntaxin-16 | N | 6 | 7 | 0 | 0 |
| STX16 | Isoform B of Syntaxin-16 | M | 0 | 0 | 8 | 13 |
| P4HA1 | Isoforms 2,3 of Prolyl 4-hydroxylase subunit alpha-1 | N | 7 | 7 | 0 | 0 |
| P4HA1 | Isoform 1 of Prolyl 4-hydroxylase subunit alpha-1 | M | 0 | 0 | 11 | 20 |
| SYNCRIP | Isoform 1 of Heterogeneous nuclear ribonucleoprotein Q | N | 21 | 184 | 0 | 0 |
| SYNCRIP | Isoforms 3,5 of Heterogeneous nuclear ribonucleoprotein Q | M | 0 | 0 | 13 | 46 |
| CCDC6 | CCDC6 protein (Fragment) | N | 5 | 6 | 0 | 0 |
| CCDC6 | Coiled-coil domain-containing protein 6 | N | 5 | 6 | 0 | 0 |
| CNBP | Isoform 1 of Cellular nucleic acid-binding protein | M+N | 4 | 6 | 3 | 4 |
| CNBP | Isoform 5 of Cellular nucleic acid-binding protein | N | 4 | 4 | 0 | 0 |
| CUX1 | Protein CASP | M+N | 2 | 2 | 9 | 9 |
| CUX1 | Homeobox protein cut-like 1 | N | 12 | 14 | 0 | 0 |
| HDLBP | Vigilin | M+N | 16 | 28 | 14 | 16 |
| HDLBP | cDNA FLJ56889, moderately similar to Vigilin | N | 3 | 3 | 0 | 0 |
| HLA-A | HLA class I histocompatibility antigen, Cw-8 alpha chain | M | 0 | 0 | 3 | 3 |
| HLA-A | HLA class I histocompatibility antigen, A-2 alpha chain | M+N | 5 | 8 | 10 | 27 |
| HNRNPC | Isoform C2, Heterogeneous nuclear RNPs C1/C2 | M+N | 19 | 210 | 13 | 40 |
| HNRNPC | Isoform C1, Heterogeneous nuclear RNPs C1/C2 | M+N | 11 | 28 | 6 | 7 |
| RBM14/4 | RNA-binding protein 14 | M+N | 15 | 54 | 6 | 9 |
| RBM14/4 | RNA-binding protein 4 | M+N | 5 | 9 | 5 | 5 |
| SERBP1 | SERPINE1 mRNA binding protein 1 | N | 11 | 14 | 0 | 0 |
| SERBP1 | Plasminogen activator inhibitor 1 RNA-binding protein | M+N | 2 | 4 | 8 | 15 |
| TMPO | Lamina-associated polypeptide 2, isoforms beta/gamma | M+N | 7 | 15 | 4 | 8 |
| TMPO | Lamina-associated polypeptide 2, isoform alpha | M+N | 19 | 47 | 11 | 11 |
| TPM1 | Isoforms 1,4 of Tropomyosin alpha-1 chain | M+N | 14 | 36 | 6 | 6 |
| TPM1 | Isoform 5 of Tropomyosin alpha-1 chain | N | 2 | 2 | 0 | 0 |
Details of the peptides observed in the nucleus and in mitochondria, genes, protein sequence groups and protein names are given in Supporting Information Table 2. In a few cases the protein sequence group includes more than one closely related sequence (isoform).
Overall, these three tests provide very strong evidence that the approximately 40% of detected proteins observed in both the nucleus and mitochondria are not an artifact of incomplete separation of the two organelles. The data in Figure 1 for lower abundance proteins (≤8 counts) also suggests that the sampling properties of the MS spectral counting may result in some of the lowest abundance proteins being detected only in one organelle despite being present in both, that is, the level of ~40% probably represents a lower limit on the proportion of proteins present in both organelles.
3.2. Partitioning of Oxidative Phosphorylation Proteins
We have analyzed in some detail the distribution of four sets of proteins that are closely associated with cellular energy metabolism and with the role of mitochondria as integrators of both cellular energy and cellular signaling processes: oxidative-phosphorylation (OxPhos) proteins, tricarboxylic-acid-cycle (TCA) proteins, glycolysis (GLS) proteins and Ras and Ras-related Rab (RAS) proteins. Two of these (OxPhos and TCA) are core mitochondrial functions, one is a largely cytosolic process (GLS) and one involves more distributed, membrane-related, signaling processes (RAS). In keeping with their functional characteristics, these groups of proteins show different patterns with regard to their partitioning between mitochondria and the nucleus (see below).
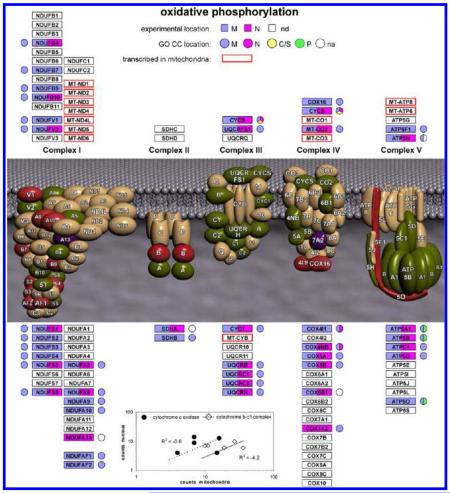
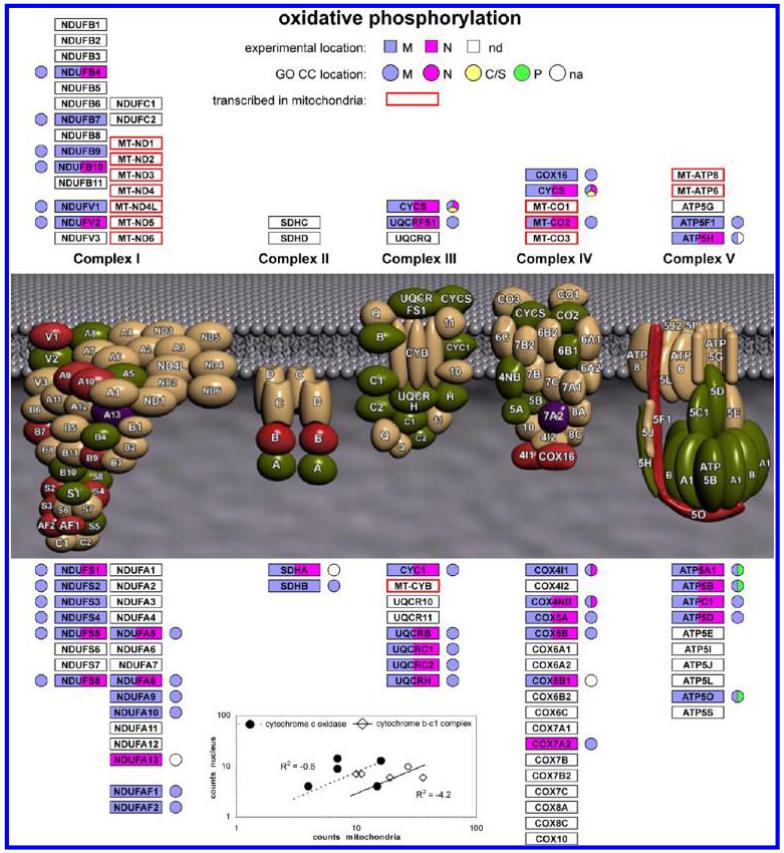
We begin with the analysis of the OxPhos proteins (Figure 2). Overall, 45 proteins were detected from complexes 1–5, including an appreciable proportion of proteins from each complex. Most of the 13 proteins transcribed in mitochondria, which are parts of complexes 1, 3, 4 and 5, are intrinsic membrane proteins and only the MT-CO2 subunit of complex 4 (cytochrome c oxidase) was detected in the present experiments, in both the nucleus and mitochondria. In fact, only 15 of the 45 proteins were detected exclusively in {m}, with 27 in {m&n}, and 3, with small numbers of counts (Supporting Information Table 2), only in {n}. For the proteins in {m&n}, there was no correlation between the number of counts in {M} and in {N} as shown in the inset to Figure 2 for proteins from complex 3 (Cytochrome b-c1 complex) and complex 4. This indicates that proteins that are components of a particular mitochondrial complex do not partition to the nucleus as that complex and may have other roles in the nucleus. The sampling properties of spectral counting do not seem to be dominant in these observations. For example, from complex 5 (ATP synthase) the delta subunit (ATP5D) was observed with 28 counts in {n} and 6 counts in {m}. Figure 2 also shows the GO Cellular Component (GO CC) annotation for these proteins. Although most of the protein sequence groups included at least one sequence with annotation, one (ATP5H) only had annotation for a sequence not found by the MS data and three protein sequence groups (NDUFA13, SDHA, COX6B1) had no sequence with GO CC annotation at the gene level. For two of these (NDUFA13, SDHA), the consensus sequence from the MS data corresponded to a sequence apparently so far only observed at the cDNA level (Supporting Information Table 2). Overall, the OxPhos proteins seem to resemble the pattern seen for mitochondrial ribosomal proteins, that is, a proportion of the proteins in complexes 1–5 were also observed in the nucleus. Although less information is currently available, some OxPhos proteins might have other functions in the nucleus, similar to some mitochondrial ribosomal proteins. It should also be noted that although oxidative phosphorylation is a core mitochondrial process, some of the components are known to be distributed to other subcellular locations, for example, the presence of ATP synthase and other OxPhos components in the plasma membrane and probably elsewhere in the cell.56-59
Figure 2.
Summary of experimental MS data and GO CC annotation terms for proteins in oxidative phosphorylation complexes 1–5. nd denotes proteins that were not detected. na denotes proteins for which there was no GO CC annotation for the protein sequence groups detected by MS. In some cases other protein sequences from the same gene that were excluded by the MS data had GO CC annotation, which is shown in the figure. (Inset) Correlation between counts in mitochondria and counts in the nucleus for proteins in the {m&n} data set for complex 3 (Cytochrome b-c1 complex) and complex 4 (cytochrome c oxidase).
3.3. Partitioning of Tri-Carboxylic-Acid Cycle Proteins
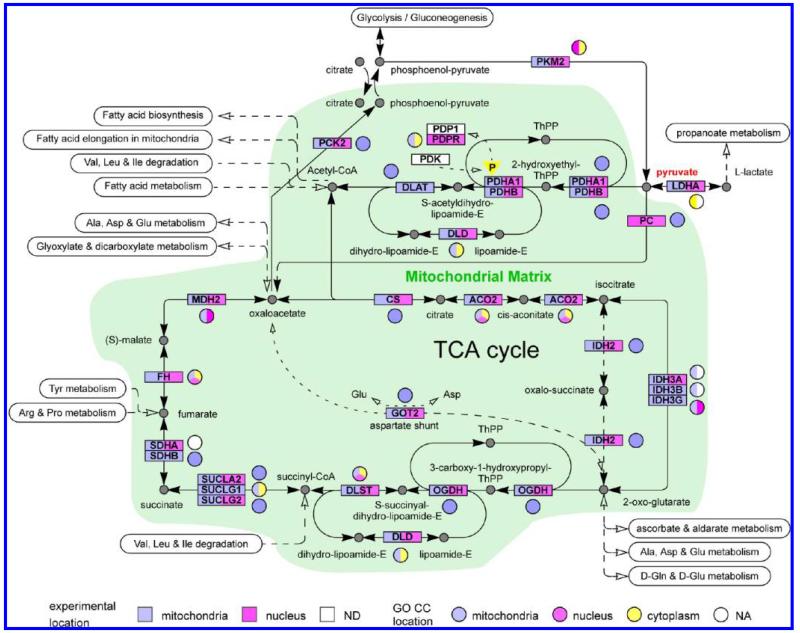
The TCA proteins show a different pattern of distribution between the mitochondria and nucleus, as summarized in Figure 3. Almost all of the proteins in the TCA-cycle, the pyruvate dehydrogenase system and the interface to the last steps of glycolysis were detected in the present experiments (Figure 3). The only exceptions were the pyruvate dehydrogenase kinase (PDK) and the pyruvate dehydrogenase phosphatase, where only the regulatory subunit (PDPR), but not the catalytic subunit (PDP) was detected. These enzymes act to regulate the activity of the pyruvate dehydrogenase complex by phosphorylation (down regulation)/dephosphorylation.60 Several notable characteristics of the present data are summarized in Figure 3, which also shows the current GO CC annotations: (a) although 5 proteins from the TCA-cycle have previously been annotated to the nucleus, in MCF7 cells 18 of the 25 proteins shown are observed in both N and M; (b) for 3 genes we observed protein sequences different from previously annotated sequences (IDH3A, IDH3B, LDHA) with different subcellular distributions from the previously annotated sequences; (c) for 1 gene (SDHA) we observed a consensus sequence that corresponds to a sequence that is apparently so far only described at the cDNA level; (d) For 5 sequence groups (DLST, IDH2, MDH2, PCK2, PKM2, Supporting Information Table 2), the MS data was consistent with a unique, identical protein sequence being present in both N and M; (e) for 13 protein sequence groups the MS data could not rigorously exclude different protein isoforms in N and M, although for 4 other proteins (GOT2, IDH3A, SDHA, SUCLG2) the union of peptides observed in both sites was consistent with a single protein sequence (Supporting Information Table 2); (f) 5 protein sequence groups (DLAT, IDH3B, IDH3G, SHDB, SUCLG1) were detected solely in mitochondria and 2 protein sequence groups (PC, PDPR) detected solely in the nucleus. It should also be noted that although the TCA-cycle is a core mitochondrion process, many of these proteins either are known to also be cytosolic (Figure 3) or to have cytosolic counterparts (MDH1, IDH1, GOT1, PCK1), of which MDH1, IDH1 and GOT1 were observed in both M and N in the present experiments (Supporting Information Table 2).
Figure 3.
Summary of experimental MS data and GO CC annotation terms for proteins in the TCA-cycle and in the interface to glycolysis/gluconeogenesis. ND denotes proteins that were not detected. NA denotes proteins for which there was no GO CC annotation for the protein sequence groups detected by MS. In some cases, other protein sequences from the same gene that were excluded by the MS data had GO CC annotation, which is shown in the figure.
From a functional viewpoint, both branches of the TCA cycle seem to be “short-circuited” in the nucleus by the absence of a few critical proteins (the pathway involving IDH2 is thought to be a minor contributor61) again suggesting that these proteins have different roles in the nucleus and in mitochondria. Furthermore, if PC/PDPR (6/2 peptides and 6/2 counts in the nucleus respectively, Supporting Information Table 2) are not present or strongly reduced in the mitochondria of MCF7 cells, this would be expected to have major effects on the metabolic flux of pyruvate through acetyl-CoA, oxaloacetate or lactate (Figure 3). Interestingly, both PKM2 and some isoforms of LADH (Supporting Information Table 2) were also detected in both M and N (Figure 3). We further address the reproducibility of these results below and consider their possible functional implications in the discussion.
3.4. Qualitative Description of Annotation Data and Overall Protein Distribution
The availability of concurrent proteomics data for the nucleus and mitochondria, together with the demonstration that these organelles are cleanly separated by the sucrose gradient fractionation, facilitates the analysis of protein sets involved in functional processes that primarily operate in one or the other of the organelles (OxPhos, TCA cycle) or of protein sets involving functional systems that strongly differ between the two organelles (RNA processing/translation). To analyze how other functional processes such as glycolysis or Ras-related signaling might involve the nucleus and mitochondria, it is necessary to confront the issue of possible contamination of the experimental nuclear and mitochondrial fractions with other subcellular components such as the cytosol or other membranous structures such as the endoplasmic reticulum or the plasma membrane. We address this in this section and the following two sections. In this section we use a simple enumeration of the Gene Ontology Cellular Component (GO CC) annotation terms for the proteins observed in the nucleus and mitochondrion to show the distribution of the annotation data and to obtain a qualitative overview of the experimental data. In the following section, we use a more complete hierarchical analysis that takes directly into account that many proteins have GO CC annotations to multiple subcellular locations. This is used to directly test for contamination of the experimental nuclear and mitochondrial fractions by the plasma membrane, Golgi apparatus and endoplasmic reticulum. The subsequent section provides experimental evidence using Western blotting that directly measures possible interorganelle contamination and provides evidence for reproducibility of the proteomics results for selected proteins of functional interest.
To characterize database information on subcellular location we mapped the GO CC annotations to a hierarchy of high-level annotation terms {nucleus (N), cytoplasm (C), plasma membrane (P), extracellular region (X)} and low-level annotation terms {mitochondrion (M), cytosol (S), endoplasmic reticulum (R), Golgi apparatus (G)} as previously described.38 M, S, R and G are all daughters of cytoplasm in the GO ontology and the cytoplasm annotation was therefore used only when no low level annotation was available. Overall, 36% of the proteins in the full GO CC human set had annotations to more than one subcellular location with this classification scheme. Of the 5015 protein sequences identified as compatible with the MS results, 2057 sequences had GO CC annotation. At the gene level, 1835 of the 2397 protein sequence groups (76.6%) included at least one protein sequence with GO CC annotation (Supporting Information Table 2). Of the 562 protein sequence groups without annotation, 267 were protein sequence groups with no annotation for the gene and for 295 protein sequence groups there was at least one protein sequence with GO CC annotation for the corresponding gene, but the sequence was not found in the protein sequence groups determined in the present study.
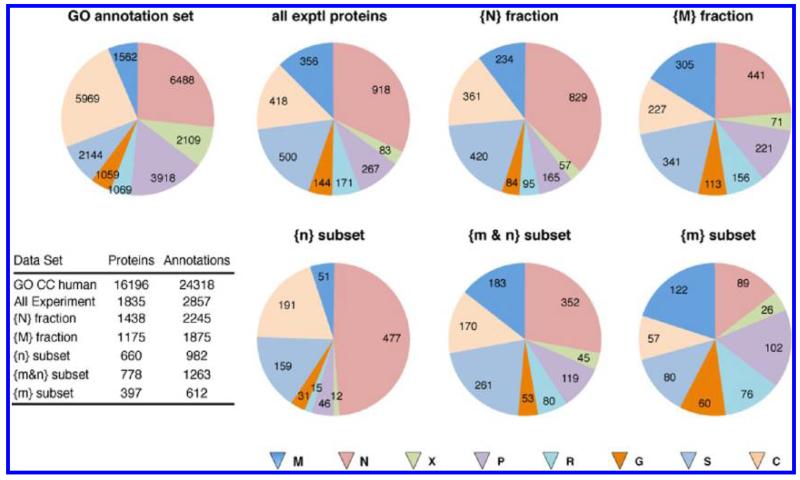
Figure 4 shows the distributions obtained with simple enumeration of the GO CC terms for the full GO human data set and for the full set of proteins observed experimentally. An initial, qualitative characterization of the experimental data with respect to previously determined GO CC annotations, to distribution between the nucleus and mitochondria, and to possible contamination is obtained by examining six subsets of the experimental protein data (Figure 4; note that across all six different experimental data subsets, 72–78% of observed proteins have annotation). For example, (a) nuclear and mitochondrial proteins are enriched in the sucrose gradient fractions compared to the complete human GO CC set (see all experimental proteins); (b) the nuclear {N} and mitochondrial {M} fractions show the expected differential enrichment of the respective protein annotation sets between the two subcellular organelles; (c) the proportion of C annotations is always fairly small, that is, for many proteins there is a low-level M, S, R, or G annotation; (d) all protein subsets show a substantial number of annotations other than N or M; (e) all protein subsets show an appreciable, but relatively constant proportion of proteins annotated as cytosol; (f) the proportion of annotations to potential membranous contaminants (P, R, G) varies more strongly, with the smallest proportion in the {n} subset, a moderate proportion in the {m&n} subset and the largest proportion in the {m} subset. Qualitatively these results support the conclusion that the fractionation has been successful with respect to M/N separation and enrichment, but emphasize that there are appreciable numbers of proteins with multiple subcellular locations and many proteins common to both organelles. It is not clear from this simple analysis whether the distribution of proteins annotated with C, S, P, R, or G represents multiple locations for individual proteins or is influenced by contamination of the nuclear and mitochondrial fractions with other cellular components. This is a crucial question for the use of sucrose gradient fractionation together with MS-based proteomics and is therefore addressed in detail in the next two sections.
Figure 4.
Enumeration of the GO cell component annotations for the different sets of proteins. The numbers of proteins with the indicated annotation are given on the pie slices. The pie slices are sized as a proportion of all annotations in each data set. Total numbers of proteins and annotations for each data set are given in the inset table. Multiple annotations to the same location were counted only once.
3.5. Hierarchical Analysis of Subcellular Distribution with Explicit Allowance for Multiple Subcellular Locations
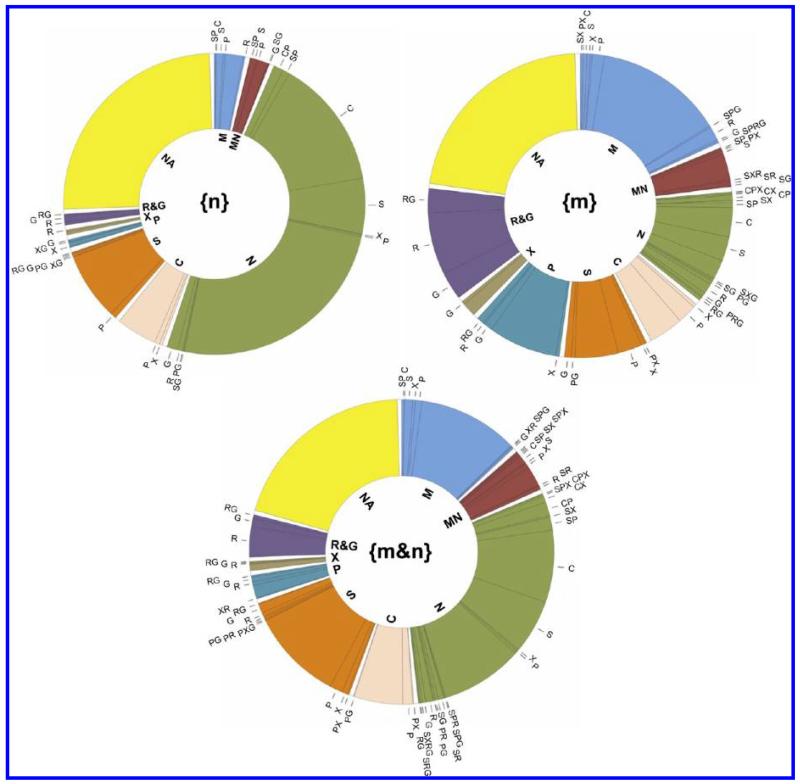
Because a substantial proportion of the experimentally detected proteins with GO CC annotations are annotated to multiple subcellular locations (44%), it is necessary to use a hierarchical classification that allows for more than the 8 categories used in the previous section. For example, for the 1438 annotated proteins identified in the experimental {N} fraction, there were 86 different combinations of the 8 simple types of GO CC annotations. Assuming that the GO CC annotations are not wildly inappropriate for application to MCF7 cells, for example, because the GO CC data tabulates many different cell types under many different experimental conditions, this already suggests that the experimental data is considerably influenced by proteins that have multiple subcellular locations. To allow for this complexity, to retain the simpler overview of only 8 major subcellular locations, and to concentrate on the nucleus and mitochondria, we used the following classification hierarchy: (a) all proteins were first classified into the annotation categories: M-[C,S,P,X,R,G], M&N-[C,S,P,X,R,G], N-[C,S,P,X,R,G], and other-1, that is, those proteins with M and/or N annotations were subdivided into subsets M, N or M&N, but carried tags for the other locations; (b) the other-1 proteins, with no N or M annotations, were subsequently classified into the categories: C-[P,X,R,G], S-[P,X,R,G], other-2; (c) the other-2 proteins were classified into the categories P, X, R&G and NA, where NA denotes that no annotation was found for the protein and R&G denotes an annotation of R, G or, R and G. Many of the R&G proteins were annotated with both endoplasmic reticulum and Golgi apparatus and there were relatively few proteins in this category. The intermediate classification C or S was used to keep the ambiguity of the C annotation explicit (it could be combinations of M, S, R and G). Several important characteristics of the data are evident from the resulting distributions (Figure 5). First, for all three data subsets, a substantial proportion of the proteins (22–26%) have no subcellular location annotation. Second, a substantial proportion of the proteins with annotation are annotated to multiple subcellular locations (42–47%). Third, even though many proteins have annotations to multiple locations, there are many proteins seen in the nucleus and mitochondria that have not previously been annotated to these locations (see below). These results are consistent with our previous conclusion that a majority of proteins have multiple subcellular locations and that current annotations underestimate the multiplicity of locations,38 as well as with increasing evidence that such multiple locations are subject to dynamic changes (see Discussion).
Figure 5.
Hierarchical distribution of GO CC annotation data for the proteins detected only in mitochondria {m} (517 proteins), only in the nucleus {n} (895 proteins) or in both {m&n} (985 proteins) using the hierarchical classification described in the text. The color code corresponds to the hierarchy level given in the center of the doughnut. The annotation terms are: M, mitochondria; MN, mitochondria and nucleus; N, nucleus; C, cytoplasm; S, cytosol; P, plasma membrane; X, extracellular region; R&G, endoplasmic reticulum and/or Golgi apparatus; NA, no annotation.
This type of hierarchical classification can be turned around to analyze whether the nucleus and mitochondria are systematically contaminated with plasma membrane, endoplasmic reticulum or Golgi apparatus proteins. For example, it can be tested whether proteins seen in the subset {m&n} and annotated with plasma membrane arise mainly from promiscuous contamination in the sucrose gradient of both organelles with plasma membrane. In this case the relative abundance of the {m&n} proteins in the {M} and {N} fractions should be correlated and reflect the degree of contamination of each organelle. Conversely, if plasma membrane proteins are among the proteins observed in the {n} and {m} subsets of proteins, these are necessarily different proteins and are evidence against promiscuous contamination of both organelles.
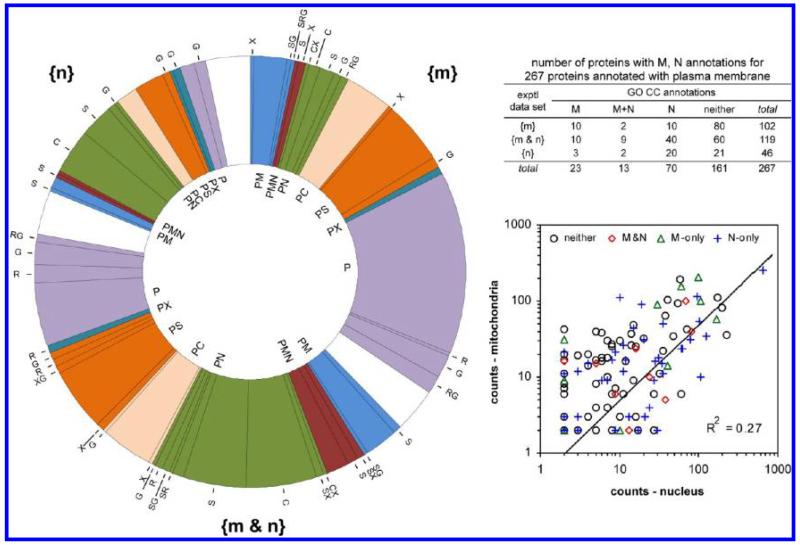
Over all experimental data, 267 of the 1835 proteins with GO CC annotations had an annotation to plasma membrane. The complete distribution of the P annotations in hierarchical form is shown in Figure 6. Based on the GO CC annotations, 40 of 517 {m}, 14 of 985 {m&n} and 4 of 895 {n} proteins are currently annotated solely to plasma membrane, but not to M, N or other locations. The remaining 209 proteins with P annotations have additional annotations to multiple locations, but only 106 of them to M and/or N. Over the three experimental data subsets {m}, {m&n} and {n}, the distribution of the numbers of proteins and their GO CC annotations is shown in the inset to Figure 6. The existence of 102 proteins (515 peptides, 1163 counts) solely in the {m} subset and 46 proteins (186 peptides, 291 counts) solely in the {n} subset is evidence against adventitious cocontamination of both the nucleus and mitochondria. For the 119 proteins in the {m&n} subset with annotation to plasma membrane, there was only low correlation in the abundances observed for the individual proteins in the nuclear and mitochondrial gradient fractions (Figure 6). These results suggest that the plasma membrane proteins partition individually to the nucleus and/or mitochondria and do not partition as a coherent group of proteins. These results are strong evidence against promiscuous contamination of the mitochondrial and nuclear fractions by plasma membrane.
Figure 6.
Evaluation of possible contamination of the nuclear and mitochondrial sucrose gradient fractions by plasma membrane. (Left) Hierarchical distribution of the GO CC annotations for the experimental subsets {m}, {n} and {m&n} for 267 proteins annotated to plasma membrane. The color code corresponds to the upper level annotations shown inside the doughnut. (Inset table) Distribution of the GO CC M and N annotations over the experimental subsets {m}, {n} and {m&n}. (Right) For the 119 proteins in the {m&n} experimental data subset, the correlation between the number of counts in the {M} and {N} gradient fractions. The GO CC annotation for each protein is indicated by M (only), N (only), M+N, or neither. The correlation coefficient R2 was calculated with linear data for all 119 proteins. The data are shown on a log scale to disperse the points.
Further evidence against promiscuous contamination can be obtained by considering groups of proteins with related functional characteristics. There is increasing evidence that mitochondria participate in the integration of cellular signaling processes, apparently through the participation of the mitochondrial outer membrane. Examination of the distribution of Ras and Ras-related Rab proteins between the nucleus and mitochondria provides evidence against adventitious contamination of mitochondria and the nucleus by plasma membrane and evidence for mitochondrial participation in signaling integration. Of the 19 Ras and Ras-related Rab proteins detected (Supporting Information Table 4), 17 were detected only in mitochondria, one (Rab-10) was detected only in the nucleus, and one (NRas) was detected in both locations, albeit with predominance in mitochondria. As shown in Supporting Information Table 4, many of these Ras and Rab proteins are presently annotated to plasma membrane. Several Rab proteins already have identified functions in mitochondria, for example, Rab2062 and Rab32.63 In yeast 12 GTP-binding proteins, mostly of the Ras family, were identified in the mitochondrial outer membrane.64 This is good evidence that any adventitious contamination of the nuclear and mitochondrial fractions by plasma membrane is very small and does not obscure real functional differences in subcellular protein distribution between the two organelles, and is also consistent with the idea that mitochondria are intimately involved in signal integration in cells.
We have carried out similar analyses for proteins with GO CC annotations of endoplasmic reticulum and Golgi apparatus, with similar results (Table 3). In all three cases the correlation between nuclear and mitochondrial counts for proteins in the {m&n} subset is small (Table 3). For all three membranous components P, R and G, the number of proteins observed in the {m} and {n} subsets is appreciable. Furthermore, for each of the P, R and G membranous cellular components, there are appreciable numbers of more abundant proteins that are observed exclusively in {m} or {n} and provide good evidence against adventitious contamination of the {M} and {N} gradient fractions (Table 3). Overall these results indicate that there is little promiscuous contamination of the M and N gradient fractions by plasma membrane, endoplasmic reticulum or the Golgi apparatus.
Table 3. Correlation of GO CC Subcellular Location Annotations P, R, G and S with Nucleus (N) and Mitochondrion (M) Annotations and Experimental Data.
| GO CC annotation plasma membrane (P) number of proteins with N and/or M annotation |
||||||
|---|---|---|---|---|---|---|
| data set | M | M+N | N | neither | sum | R2 {m&n}a |
| {m} | 10 | 2 | 10 | 80 | 102 | |
| {m&n} | 10 | 9 | 40 | 60 | 119 | 0.27 |
| {n} | 3 | 2 | 20 | 21 | 46 | |
| sum | 23 | 13 | 70 | 161 | 267 | |
| selected{m} and {n} proteins |
||||||
|---|---|---|---|---|---|---|
| data set | gene | {N} peptides | {N} counts | {M} peptides | {M} counts | |
| {n} | SRP72 | 20 | 31 | 0 | 0 | |
| LRRK2 | 8 | 63 | 0 | 0 | ||
| ENAH | 13 | 23 | 0 | 0 | ||
| {m} | CAT | 0 | 0 | 18 | 48 | |
| RHOA | 0 | 0 | 8 | 47 | ||
| LAMP2 | 0 | 0 | 5 | 36 | ||
| GO CC annotation endoplasmic reticulum (R) number of proteins with N and/or M annotation |
||||||
|---|---|---|---|---|---|---|
| data set | M | M+N | N | neither | sum | R2 {m&n}a |
| {m} | 9 | 3 | 9 | 55 | 76 | |
| {m&n} | 1 | 5 | 21 | 53 | 80 | 0.15 |
| {n} | 2 | 0 | 5 | 8 | 15 | |
| sum | 12 | 8 | 35 | 116 | 171 | |
| selected {m} and {n} proteins |
||||||
|---|---|---|---|---|---|---|
| data set | gene | {N} peptides | {N} counts | {M} peptides | {M} counts | |
| {n} | PDCD6 | 7 | 11 | 0 | 0 | |
| P4HA1 | 7 | 7 | 0 | 0 | ||
| RRS1 | 9 | 18 | 0 | 0 | ||
| {m} | POR | 0 | 0 | 26 | 61 | |
| TMX1 | 0 | 0 | 11 | 40 | ||
| LMAN1 | 0 | 0 | 10 | 29 | ||
| GO CC annotation Golgi apparatus (G) number of proteins with N and/or M annotation |
||||||
|---|---|---|---|---|---|---|
| data set | M | M+N | N | neither | sum | R2 {m&n}a |
| {m} | 3 | 1 | 9 | 47 | 60 | |
| {m&n} | 3 | 0 | 13 | 37 | 53 | −0.08 |
| {n} | 0 | 2 | 13 | 16 | 31 | |
| sum | 6 | 3 | 35 | 100 | 144 | |
| selected {m} and {n} proteins |
||||||
|---|---|---|---|---|---|---|
| data set | gene | {N} peptides | {N} counts | {M} peptides | {M} counts | |
| {n} | AFGF1 | 6 | 9 | 0 | 0 | |
| COPA | 18 | 21 | 0 | 0 | ||
| GTPBP4 | 11 | 23 | 0 | 0 | ||
| {m} | LMAN1 | 0 | 0 | 10 | 29 | |
| LPCAT1 | 0 | 0 | 7 | 17 | ||
| SLC1A5 | 0 | 0 | 9 | 33 | ||
| GO CC annotation Cytosol (S) number of proteins with N and/or M annotation |
||||||
|---|---|---|---|---|---|---|
| data set | M | M+N | N | neither | sum | R2 {m&n}a |
| {m} | 5 | 7 | 20 | 48 | 80 | |
| {m&n} | 12 | 15 | 91 | 143 | 261 | 0.56 |
| {n} | 8 | 8 | 64 | 79 | 159 | |
| sum | 25 | 30 | 175 | 270 | 500 | |
| selected {m} and {n} proteins |
||||||
|---|---|---|---|---|---|---|
| data set | gene | {N} peptides | {N} counts | {M} peptides | {M} counts | |
| {n} | DDX6 | 20 | 60 | 0 | 0 | |
| SNRPB | 6 | 87 | 0 | 0 | ||
| TRIM25 | 12 | 30 | 0 | 0 | ||
| {m} | BAG6 | 0 | 0 | 10 | 14 | |
| GGH | 0 | 0 | 7 | 23 | ||
| PPT1 | 0 | 0 | 8 | 32 | ||
R2{m&n} is the correlation between the number of nuclear counts and the number of mitochondrial counts for proteins in subset {m&n}.
The endoplasmic reticulum results provide a particularly useful test since spatial juxtaposition between mitochondria and the MAM (mitochondria-associated membrane) region of the endoplasmic reticulum apparently may involve up to 20% of the surface area of mitochondria, is important in both lipid metabolism and trafficking, Ca2+ homeostasis, apoptosis and may have additional roles.22,23 The proteins involved in this contact region are still a subject of investigation and appear to be subject to dynamic changes.23,65 Our {M} fraction contains some proteins that have been suggested to be part of the MAM (ACAT1, ATP2A2, ATP2A3, CALR, CANX, CYB5R1, CYB5R3, CYP1A1, ERLIN2, ERP44, HSPA5, HSPA9, PDIA3, TMX1), but not others (DGAT2, ERLIN1, HMOX1, ITPR1, MFN1, MFN2, PACS2, PEMT, PTDSS1, PTDSS2, RYR1). Many of these proteins have other known functions outside the MAM, but this suggests that there could be partial capture of endoplasmic reticulum proteins in this specialized region from the mitochondrial side even though there is not general contamination of the mitochondria with endoplasmic reticulum. This may actually be an advantage for functional studies (see discussion).
In summary, strong evidence was obtained that the nucleus and mitochondria are cleanly distinguished in the sucrose gradient fractions. Similarly, the observed distribution of the proteins speaks against the possibility that the majority of the proteins observed in both the nucleus and mitochondria ({m&n} subset) correspond to adventitious contamination of both organelles by other membranous cellular components such as the plasma membrane, endoplasmic reticulum or the Golgi apparatus. Collectively the above results suggest that even though many of the detected proteins have other known locations/functions, the observation of the majority of these proteins in the nuclear and mitochondrial fractions of the sucrose gradient fractionation reflects real functional distributions rather than artifacts of the fractionation method (see discussion).
3.6. Experimental Western Blotting Tests for Contamination and Reproducibility
Although the MS data provides much more information than can be obtained by monitoring a small number of proteins with antibodies, we have performed direct Western blotting (WB) experiments for three reasons: (1) to provide a comparison with more conventional tests of the purity of the nuclear and mitochondrial fractions and to directly test for contamination; (2) to test the reproducibility of the fractionations and verify the reproducibility of the nucleus/mitochondrial protein distributions for selected, functionally interesting proteins; and, (3) to begin to establish a set of proteins/antibodies that can be used as routine controls of the cell breakage and sucrose gradient fractionation processes prior to investment of large amounts of mass spectrometer time.
As shown in Figure 7, there are four outstanding characteristics of the results. (1) The sample preparation procedures are highly reproducible. For all 14 proteins identical presence/absence of the proteins in different subcellular locations was obtained in multiple preparations. (2) The proteins chosen as potential organelle markers (mitochondria: SDHB, MT-ND1; nucleus: ORC2; Golgi apparatus: KDEL; endoplasmic reticulum: ERN1) show presence of the proteins only in the expected subcellular fractions. HDAC is partitioned between the nucleus and cytoplasm, but not mitochondria, as previously described.66,67 (3) The subcellular distributions for all proteins detected by MS agree with the Western blot results (the organelle markers KDEL, ERN1 and MT-ND1 were not observed by MS). (4) Many proteins that are associated functionally with specific subcellular locations in fact show multiple locations. For example, although SDHB, MT-ND1 and COX4 are observed solely in the mitochondrial preparation, other “mitochondrial” proteins (MT-CO2, CYC1, ATP5B, PCK2, SDHA) are observed in both the nucleus and mitochondria. Similarly, the “cytoplasmic” glycolytic protein PFKP was observed in both the cytoplasm and nucleus, but not in mitochondria, in exact agreement with the MS results. Finally, although usually described as a mitochondrial protein, PC was detected only in the nucleus in MCF7 cells, again in agreement with the MS results.
Overall, the WB experiments confirm the reproducibility of the nuclear and mitochondrial preparations, show little cross contamination between mitochondria and the nucleus and are consistent with little contamination of the nuclear and mitochondrial preparations with plasma membrane, endoplasmic reticulum or the Golgi apparatus. Furthermore, the WB experiments confirm the presence of selected, interesting proteins that are “unexpected” in the nucleus and/or mitochondria according to the dominant functional roles presently ascribed to these proteins.
3.7. Dealing with Cytosolic Proteins
Operatively, cytosolic proteins provide the greatest challenge to experimental verification of functions at multiple subcellular locations. Compared to N, M, P, R and G proteins, cytosolic proteins are less readily sequestered as a group and all of these membrane-bounded cellular organelles are bathed in the cytosol. In keeping with this, cytosolic proteins show the greatest correlation between the number of counts in the nucleus and mitochondria for proteins in the subset {m&n} (Table 3). However, numerous examples are already known of cytosolic proteins that carry out functional roles at the surfaces of these organelles. We have therefore consciously chosen not to treat the nuclear and mitochondrial preparations to stringently remove cytosolic proteins and propose to detect their functional relevance by other approaches (see discussion). In this section we analyze the data for proteins with GO CC annotation of cytosol to elucidate the nature of the distributions that might need to be analyzed. In the next section we show the results recorded for proteins involved in glycolysis as an example for cytosolic proteins involved in energy metabolism.
Of the 1835 proteins with GO CC annotation, 418 were annotated to cytoplasm and 500 were annotated to cytosol. Because of the ambiguity of the cytoplasm annotation, in the following we use only the 500 proteins annotated with cytosol. Of these, 80 were observed in the {m} subset, 261 in the {m&n} subset and 159 in the {n} subset (Table 3); 194 were annotated purely to cytosol and 306 were annotated to 34 other combinations of the 8 major subcellular locations used in our classification hierarchy (Figure 5). Cytosolic proteins in the {m&n} subset show a higher, but still moderate, level of correlation between nuclear and mitochondrial counts than was observed for proteins annotated with plasma membrane, endoplasmic reticulum or the Golgi apparatus (Table 3). The degree of correlation was not influenced by removing proteins with lower abundance (≤8 counts) in M and/or N. Two limiting mechanisms that could lead to high levels of correlation would seem to be: (a) there are cytosolic proteins that promiscuously bind to all membranous surfaces in proportion to their cytosolic concentration, or (b) there are relatively stable protein complexes in the cytosol that partition as complexes between the nucleus and mitochondria. Examination of a series of protein groups known to be involved in complexes (Table 4) suggests that the constituents of cytosolic protein complexes partition to varying degrees between mitochondria and the nucleus. The strong divergence in the values of counts{M}/{N} shown in Table 4 suggests that partitioning of cytosolic proteins to mitochondria and the nucleus is not adventitious and may have functional importance. For example, mechanisms are known that locate mRNAs for some mitochondrial proteins to the mitochondrial outer membrane surface,68,69 and some mitochondrial proteins seem to be at least partially cotranslationally inserted into mitochondria.70
Table 4. Distribution of Selected Cytosolic Protein Groups between Mitochondria and the Nucleus.
| nucleus |
mitochondria |
||||||
|---|---|---|---|---|---|---|---|
| protein group (no. of proteins) or proteina | data set | peptides | counts | peptides | counts | counts M/N | R2 {m&n}b |
| 60S ribosomal proteins (24) | {m&n} | 173 | 1303 | 108 | 278 | 0.21 | −0.06 |
| 60S ribosomal protein L3 | {n} | 15 | 63 | 0 | 0 | ||
| 60S ribosomal protein L5 | {n} | 13 | 68 | 0 | 0 | ||
| 60S ribosomal protein L6 | {n} | 9 | 70 | 0 | 0 | ||
| 40S ribosomal proteins (22) | {m&n} | 183 | 595 | 52 | 141 | 0.24 | 0.25 |
| 40S ribosomal protein S8 | {n} | 8 | 94 | 0 | 0 | ||
| Eukaryotic translation initiation factors (10) | {m&n} | 92 | 187 | 38 | 44 | 0.23 | 0.38 |
| Proteasome (10) | {m&n} | 41 | 62 | 55 | 85 | 1.4 | 0.01 |
| T-complex protein 1 (7) | {m&n} | 45 | 68 | 90 | 152 | 2.2 | 0.10 |
| T-complex protein 1 subunit delta | {m} | 0 | 0 | 8 | 12 | ||
| Coatomer (6) | {m&n} | 86 | 210 | 20 | 32 | 0.15 | 0.57 |
| Coatomer subunit alpha | {n} | 18 | 21 | 0 | 0 | ||
| F-actin-capping protein (3) | {m&n} | 33 | 104 | 20 | 37 | 0.36 | 0.94 |
Number of proteins detected experimentally in the {m&n} data subset.
Correlation between the number of counts in {N} and {M} for proteins in the {m&n} data subset.
More detailed examination of the distribution between the nucleus and mitochondria of individual proteins from cytosolic protein groups suggests that some partition as well-defined complexes. For example, in the {m&n} subset the F-actin capping protein (Table 4) shows strong correlation between nuclear and mitochondrial counts, with the slope of the linear correlation close to the ratio of total counts in each location. For other complexes such as coatomers and ribosomes that involve larger numbers of proteins, the correlations are smaller and for these groups, some constituent proteins with high abundance in only one of the two locations were also observed (Table 4). These features might reflect compositional differences between nuclear-associated and mitochondrial-associated complexes, that some proteins have additional functions and hence different abundance in one of the locations, or that the proteins mainly partition individually rather than as complexes. Given the limitations of sampling with spectral counting (Figure 1), this has not been further pursued with the present data.
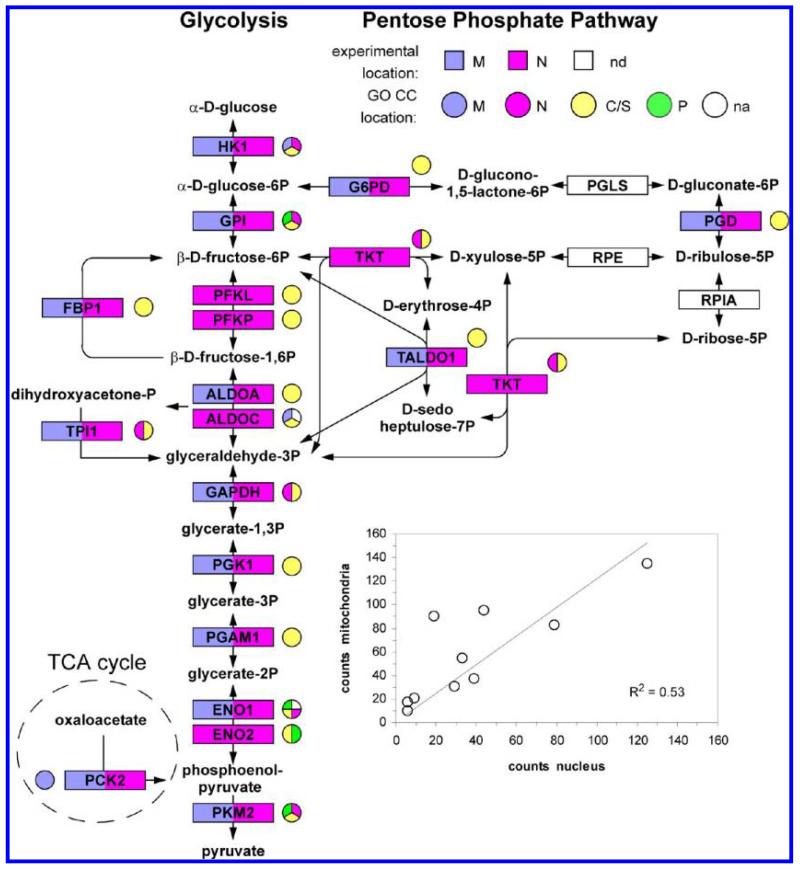
3.8. Proteins Involved in Glycolysis
Glycolysis provides an example of the distribution of proteins involved in a cytosolic process over the nucleus and mitochondrion. Among the glycolytic enzymes that were detected, all 14 were observed in the nucleus and 10 were also detected in mitochondria (Figure 8). For the 7 proteins involved in the pentose phosphate pathway and its interface to glycolysis, 4 of the 7 proteins were detected, all 4 were observed in the nucleus and 3 were also observed in mitochondria. Two protein sequence groups (ALDOC, ENO1) corresponded to sequences that did not have previous GO CC annotation. For 5 proteins (ENO1, FBP1, PFKP, PKM2 and TALDO1), the MS data was consistent with a single protein sequence in the indicated sites; for 4 proteins (GAPDH, GPI, PGAM1, PGK1) the consensus sequence was consistent with a single sequence, but the MS data did not rigorously exclude different sequences in M and N; for the remaining 9 proteins, the MS data was consistent with multiple sequences from the gene (Supporting Information Table 2). The GO CC annotations for the 14 glycolytic proteins show that 6 have previously been annotated to the nucleus, 2 to mitochondria and 4 to the plasma membrane. For the 10 glycolysis proteins that were observed here in both the nucleus and mitochondria, there was a very moderate degree of correlation between the counts in {M} and {N} (inset to Figure 8).
Figure 8.
Summary of the experimental data and GO CC annotation terms for glycolysis and pentose phosphate pathway proteins. nd denotes proteins that were not detected by MS. na denotes proteins for which there was no GO CC annotation for the protein sequence group detected in the MS experiments. In some cases other protein sequences from the same gene that were excluded by the MS data had GO CC annotation, which is shown on the figure. (Inset) Correlation between the number of counts in the nucleus and counts in mitochondria for the ten glycolysis enzymes detected in both locations.
There is increasing evidence for important functional roles of glycolytic proteins in the nucleus that are consistent with these results. Seven years ago we demonstrated that in kidney cells the nuclear abundances of G6PD, TPI1. GAPDH, PGAM1, PKM2 and LADH were changed by hypoxia.71 In recent years participation of ENO1, FBP1, GAPDH, PKM2 and LADH in transcription in the nucleus has been demonstrated by others (Supporting Information Table 5). Very recently, connections to the cell cycle have been established for nuclear FBP172 and PGAM1 in the nucleus of cancer cells has been observed.73 This is also the case for fructose-2,6-biphosphatase 3 (PFKFB3)74 which, although not a direct participant in the glycolysis enzymatic cascade, has major effects on glycolysis through the allosteric regulator fructose 2,6-bisphosphate that apparently include variations in cellular energy source (glutamine vs glucose) as a function of the cell cycle in cancer cells.74,75 There are scattered experimental reports of further functions of these and other glycolytic enzymes in the nucleus, mitochondria and elsewhere in cells (Supporting Information Table 5). Furthermore, GPI, HK1 and TKT have been detected in the nucleus in large scale antibody screening of subcellular protein locations76 (Supporting Information Table 5). Most of the glycolytic enzymes also show direct, physical interactions with multiple proteins that have been annotated previously to the nucleus (Supporting Information Table 5). While direct investigation of possible nuclear functions for other individual glycolytic enzymes detected in the present experiments is now desirable, we conclude that there is already a substantial body of corroborating evidence showing that a majority of glycolytic enzymes can be present and have important functions in the nucleus (see discussion). Apart from hexose kinases HK1 and HK2,77-79 the presence of glycolytic enzymes in mitochondria has been less researched, but we note evidence that in cancer cells glycolytic enzymes may form large cytosolic complexes73 and that association of large protein complexes that include glycolytic enzymes with yeast and plant mitochondria have been reported,80-82 that membrane association of glycolytic enzymes has been found in pancreatic cancer cells,83 and that many glycolytic enzymes have been reported to concentrate in the MAM contacts between mitochondria and the endoplasmic reticulum after viral infection.84 Overall, there are very substantial indications that even for nominally cytosolic glycolytic proteins the present methods reflect functionally important subcellular protein distributions.
3.9. New Subcellular Annotation Data
The present results suggest that increasing the proportion of proteins covered and increasing the coverage of alternative locations for individual proteins in subcellular location databases will be important for deeper understanding of cellular function. There seem to be some endemic bottlenecks to the generation and inclusion of such data in public databases (see discussion). With this in mind, we have prepared a set of “secure” new annotations to mitochondria and the nucleus from the present experimental data (Supporting Information Table 6). These secure assignments correspond to sequences where a unique protein sequence among the 91 564 protein sequences of the IPI human data set was identified. An overview of the secure assignments that could be made is shown in Table 5.
Table 5. Comparison of Experimental Locations with GO CC.
| GO CC annotation founda | ||||||
|---|---|---|---|---|---|---|
| GO CC annotation |
||||||
| data set uniqueb | M | M+N | N | otherc | NAd | sum |
| {m} | 50 | 10 | 17 | 86 | 0 | 163 |
| {m&n} | 61 | 19 | 140 | 111 | 0 | 331 |
| {n} | 13 | 10 | 200 | 55 | 0 | 278 |
| GO CC annotation |
||||||
|---|---|---|---|---|---|---|
| data set multipleb | M | M+N | N | otherc | NAd | sum |
| {m} | 49 | 13 | 49 | 123 | 0 | 234 |
| {m&n} | 10 | l | 7 | 17 | 0 | 35 |
| {m&n} | 0 | 5 | 25 | 21 | 0 | 51 |
| {m&n} | 64 | 23 | 132 | 142 | 0 | 361 |
| {n} | 18 | 10 | 257 | 97 | 0 | 382 |
| sum | 265 | 91 | 827 | 652 | 0 | 1835 |
| GO CC annotation not founde | ||||||
|---|---|---|---|---|---|---|
| GO CC annotation |
||||||
| data set uniqueb | M | M+N | N | otherc | NAd | sum |
| {m} | 8 | 0 | 1 | 13 | 31 | 53 |
| {m&n} | 8 | 1 | 26 | 15 | 34 | 84 |
| {n} | 0 | 0 | 51 | 12 | 49 | 112 |
| GO CC annotation |
||||||
|---|---|---|---|---|---|---|
| data set multipleb | M | M+N | N | otherc | NAd | sum |
| {m} | 3 | 1 | 6 | 20 | 37 | 67 |
| {m&n} | 5 | 1 | 5 | 4 | 5 | 20 |
| {m&n} | 3 | 0 | 11 | 3 | 6 | 23 |
| {m&n} | 3 | 3 | 20 | 21 | 20 | 67 |
| {n} | 2 | 2 | 34 | 13 | 85 | 136 |
| sum | 32 | 8 | 154 | 101 | 267 | 562 |
| sum all proteins | 297 | 99 | 981 | 753 | 267 | 2397 |
Proteins (1835) for which at least one protein in the MS protein sequence group had a GO CC annotation.
Proteins for which there was a unique MS protein sequence are indicated by bold. Proteins for which the MS protein sequence group included multiple sequences are indicated by italics.
Proteins which had GO CC annotations to other locations, but not to M and/or N.
Proteins that had no GO CC annotation at the gene level.
Proteins (562) for which there was no sequence in the MS protein sequence group with GO CC annotation. For 267 of these proteins, there was no GO CC annotation at the gene level; 295 of these proteins had a GO CC annotation for a sequence from the same gene that was not contained in the MS protein sequence group.
Among the 1835 protein sequence groups with GO CC annotation, 772 could be assigned to unique protein sequences and locations in the data sets {m}, {m&n} and {n}. Of these 269 agreed with the previously annotated partitioning between M and N (shaded diagonal in Table 5), 251 corresponded to additional annotations to M/N for proteins already annotated with one of the two (off-diagonal portion of Table 5) and 252 corresponded to annotation to M and/or N for proteins previously annotated to other locations (column “other” in Table 5). For the 1063 proteins where the MS data corresponded to protein sequence groups with multiple sequences, at least one of which had GO CC annotation, 335 corresponded to the previous annotation, 328 corresponded to additional annotation to M/N, and 400 corresponded to M and/or N annotations for proteins previously annotated to other subcellular locations. For these 1063 proteins it was not possible to assign a unique protein sequence to the subcellular locations, but the information at the gene level is useful for biological interpretations.
For the 562 protein sequence groups without GO CC annotation, 267 corresponded to genes for which no GO CC annotation was available at the gene level, and 114 of these could be assigned to unique protein sequences; 295 protein sequence groups corresponded to a gene that had a protein sequence with GO CC annotation, but the annotated protein sequence was not contained in our MS protein sequence groups. One-hundred thirty-five of these could be assigned to a unique protein sequence different from that previously annotated. The same partitioning between M and N as for the annotated sequence was shown by only 98 of the 295 sequence groups. These 295 sequence groups suggest there may be quite extensive partitioning of different protein isoforms from the same gene to different subcellular locations similar to that shown in Table 2, although with the present data incomplete annotation of the previously annotated sequences cannot be excluded. Over all data, the secure annotations include assignment of 686 unique protein sequences to M and 879 unique protein sequences to N (Supporting Information Table 6).
4. DISCUSSION
There is increasing evidence that distribution of proteins to multiple subcellular locations is crucial to cellular function.38-42,70,85-87 In the following we discuss three topics: (a) the use of sucrose gradient fractionation in MS-based proteomics to obtain data about subcellular spatial distribution of proteins, (b) how the observed distributions might be related to MCF7 cells as cancer cells, and (c) the implications of our results for how such spatial distributions may relate to overall control of cellular function.
4.1. Sucrose Gradient Fractionation and Subcellular Protein Distributions
Sucrose and other forms of gradient fractionation have long been used to separate subcellular components, but the degree to which “pure” organelles and suborganellar fractions can be prepared without contamination or loss of important components has been uncertain. By analyzing in detail the distribution of different classes of proteins between mitochondria and the nucleus, we were able to directly test whether contamination obscures real, potentially functionally important features of the experimental subcellular protein distributions. On the basis of the results, we suggest that the previous uncertainties about sucrose gradient fractionation, largely engendered by the diversity of proteins observed in individual fractions/subcellular sites, seem to have arisen mainly due to an underestimation of the multiplicity of subcellular locations shown by many proteins.38 Independent of our results, collected results from many research groups are gradually verifying that a large proportion of cellular proteins have multiple subcellular locations and that many proteins have different functional roles in different locations.
The dispersion of proteins over the nucleus and mitochondria observed here is increasingly evident in other studies and it is notable that an increasing number of seemingly well-known proteins have different functional roles in the two sites and translocate between the two organelles. For example, in addition to its well-known nuclear roles, p53 is also present in mitochondria where it may have roles in both apoptosis and DNA repair.35 Similarly, telomerase has been found to have antioxidative stress roles in mitochondria.92 Prohibitin (PHB) translocates from mitochondria to the nucleus under oxidative stress and seems have a dual function as an antiapoptotic molecule that maintains mitochondrial structure and as a transcriptional activator in the nucleus.93 Prohibitin may also have roles in metabolic switching between glycolysis and fatty acid oxidation, roles as a nuclear transcriptional regulator in interaction with p53, E2F, and Rbn, and roles in regulation of cell cycle transit (see refs in 93). It is known to locate in the nuclear matrix in osteosarcoma MG-63 cancer cells94 and in MCF7 cells we observed both PHB and PHB2 in the {m&n} protein set (Supporting Information Table 2). The functional roles of these proteins were first elucidated in the nucleus and only subsequently did alternative functions in mitochondria become apparent. The present results suggest that there may be many proteins with well-known mitochondrial functions that have other roles in the nucleus (and vice versa).
Our results suggest that, when used with care, fractionation by sucrose gradient centrifugation is sufficiently reproducible and reliable in reflecting real subcellular distributions of proteins that it should be a vital tool for using high-throughput MS-based proteomics to analyze static and dynamic aspects of the subcellular locations of proteins and their involvement in cellular function. The determination of sets of proteins common to two sites such as {m&n} helps in resolving ambiguities, but these kinds of system-wide analyses may not offer certainty for single proteins and are perhaps best viewed as a screen to identify “interesting” proteins whose presence and functional role in specific locations can then be confirmed by targeted analyses. The almost 1000 proteins that we identified as being present in both the nucleus and mitochondria of MCF7 cells suggests that the set of proteins common to different subcellular sites may often be large and that complementary high throughput approaches that detect dynamic changes in the abundance or form of proteins in different sites as a response to cellular stimulation may provide the most direct way to select the most interesting proteins associated with particular cellular functions.38 For example, using methods similar to those described here with augmentation by SILAC labeling, we have recently been able to show in IMH90 cells that engagement of the “origin activation checkpoint” for DNA translation initiation that is associated with cell cycle arrest88,89 involves shuttling between the nucleus and cytoplasm of at least 50 proteins, including a few of the mitochondrial proteins identified in the nucleus in the present study of MCF7 cells (Mulvey et al., in preparation). Such dynamic studies are likely to be particularly important for dealing with cytosolic proteins.
There is also increasing evidence for interorganelle contact regions, for example, of endoplasmic reticulum with both the plasma membrane90 and mitochondria.22,23 It is likely that future studies will first use overall organelle separations where, for example, mitochondrial preparations may capture some proteins from interorganelle interfaces such as the MAM region, and then use finer suborganelle separations that attempt to capture contact regions such as the MAM.84,91 Dynamic measurements will be advantageous in both cases, first to screen for involvement of specialized contact regions in specific functional processes and then to more deeply characterize such involvement.
4.2. Potential Connections to Cancer
MCF7 cells are closely related to breast cancer, but belong to the group of cancer cells that may still obtain a majority of their energetic requirements from oxidative phosphorylation.95 With regard to the joint involvement of the nucleus and mitochondria in cancer, one of the most intriguing results of the present work is that many proteins involved in oxidative phosphorylation and in the TCA cycle are partitioned between the nucleus and mitochondria. Some of these proteins have been implicated in cancer and other diseases, but there is currently little information about their functional roles in the nucleus.
The present experiments detected novel features of the interface between the TCA-cycle and glycolysis. In MCF7 cells, pyruvate carboxylase (PC) seems to be at least predominantly located in the nucleus, the mitochondrial form of phosphoenolpyruvatecarboxykinase (PCK2) partitions to the nucleus, and both PKM2 and LADH partition to both the nucleus and mitochondria. These proteins are in a critical location relative to glycolysis, the pyruvate dehydrogenase system, the TCA-cycle and the lactate production characteristic of many cancer cells (Figure 3). We note that PKM2 is known to participate in transcription in the nucleus in systems involving both HIF1α98 and Oct4,99 that LADH is known to participate in transcriptional complexes in the nucleus100 and that we have previously detected the interaction of LADH with the mitochondrial single strand RNA binding protein.44 These observations are suggestive of major changes in the flux through pyruvate, acetyl-CoA and oxaloacetate (Figure 3) that may be coupled to other roles of these proteins in the nucleus and mitochondria. Many other TCA-cycle proteins are also observed in the nucleus (Figure 3), mutations in other TCA-cycle proteins are known to underlie certain types of cancers101,102 and in yeast at least three TCA-cycle proteins translocate under hypoxia.42 This suggests that the TCA-cycle proteins may also have the many-faceted spatial/functional connections to the nucleus that are emerging for glycolysis proteins.
There is currently little information about nuclear functions for the 30 OxPhos proteins that we detected in the nucleus. Particularly interesting is the unusual isoform detected for SDHA, a protein that is connected to various kinds of cancer and other diseases,101,102 that has previously been observed in the nucleus102 and that in yeast is one of seven OxPhos proteins that showed changes in subcellular location under hypoxia.42 Other OxPhos proteins that we detected have previously been observed not only in in the matrix/inner membrane, but also in the mitochondrial outer membrane where some of them may be present as unprocessed precursor proteins.64 A surprise was to detect MT-CO2 (COX2) from the cytochrome c oxidase complex in the nucleus (Figures 2 and 7, Supporting Information Table 2)—this seems to be the first report of a mitochondria-encoded protein in the nucleus. The presence in the plasma membrane of ATP-synthase56-59 and of other OxPhos proteins elsewhere in cells,57,59 which were detected with a wide variety of methods including imaging, indicates that OxPhos proteins can have locations and functional roles outside mitochondria. There is clearly much work ahead in further validating and elucidating the nuclear roles of OxPhos proteins.
Although aerobic glycolysis is now widely regarded as a characteristic of proliferating/cancer cells and the role of hypoxia-inducible-factor (HIF) has been intensively studied, very complex networks that may vary with different cancers are involved.105 The presence of numerous glycolytic enzymes in the nucleus now appears likely to be an integral part of metabolic changes in proliferating cells and in many types of cancer cells. For a few glycolytic enzymes, especially hexokinase,77-79 nuclear/mitochondrial distribution is known to be important to cancer metabolism. The present results suggest further investigation of the functional roles of glycolytic enzymes in mitochondria and the nucleus is needed, especially in the light of indications that they concentrate in the MAM region of mitochondria84 and that mitochondrial restructuring and bioenergetic plasticity are also important for stem cell differentiation/regression.96 Both PKM297,98 and PFKFB374 have recently been proposed as “master” proteins for controlling the metabolic changes involved in hypoxia, proliferation and cancer, but a substantial body of other literature and the present results suggest that much more complicated networks with feedback loops involving many glycolysis, TCA-cycle, OxPhos and signaling proteins may be involved. Furthermore hypoxia causes dynamic changes in protein subcellular location involving functions that go far beyond changes in glycolysis and include TCA-cycle, OxPhos and chromatin remodeling proteins among many others.42,71 Several years ago we demonstrated that numerous glycolytic enzymes translocate to the nucleus under hypoxia71 - the present results suggest that similar dynamic subcellular distributions may be operative for other functional systems such as oxidative phosphorylation and the TCA-cycle and may contribute to cancer and other diseases.
4.3. The Influence of Subcellular Protein Distributions on Cellular Function
The outstanding general characteristic of the cellular functions that we analyzed is the distribution of proteins over multiple subcellular locations: two functions involve numerous proteins that are distributed over at least the nucleus and mitochondria (OxPhos, TCA cycle); one includes proteins distributed over at least the nucleus, mitochondria and cytosol (glycolysis); one (Ras/Rab signaling) appears to involve at least plasma membrane/mitochondria partitioning; and, only one (RNA processing/translation) seemed to be more specifically associated with either the nucleus or mitochondria, presumably as a characteristic of differences between prokaryotic and eukaryotic cells. We believe that it is time to start thinking about cellular function in terms of processes that involve wide distribution of constituent proteins over multiple subcellular locations as a means of integrating different cellular functions. The present study delineates a large number of enzymes, kinases, phosphatases, etc. that are distributed between the nucleus and mitochondria and whose activity in the “wrong” location might depend on the concentrations of substrates, cofactors and other proteins in that “wrong” location. It is clear that many proteins are widely distributed over diverse subcellular locations (Figure 5), and as we suggested previously,38 the most important proteins for overall control of cellular function may be those that distribute most widely and are able to undergo dynamic changes in abundance/form/activity at different subcellular locations in response to cellular environment. The methods presented here begin to offer ways that appear to be reproducible and reliable to identify such proteins.
As more and more examples of proteins with multiple subcellular locations/functions are encountered, related questions arise about whether changes in protein abundance and/or form in a specific subcellular site involve input to the site (transcription/translation), removal from the site (degradation), whether pre-existing proteins undergo spatial translocations to or from the site, and how these changes are coupled to post-translational modifications. A critical point that often remains obscure is whether exactly the same protein is being observed in the different sites. Furthermore, information regarding protein functions at different sites obtained by conventional biological studies may be difficult to incorporate into databases because the protein identifications are often based only on antibodies and may not distinguish among the multiple protein sequences from the same gene detected by genomics studies. Examples that sequence isoforms are critical would be PKM2, where alteration of a single internal peptide gives an isoform critical to the glycolytic pathway in proliferating cells that is closely connected to hypoxia and cancer98 or the glycolysis enzyme ENO1, where an alternate site for initiation of translation gives the nuclear Myc promoter-binding protein-1 (MBP-1) that is connected to cell death, cancer and senescence.103,104 Furthermore, the spatial/functional dispersion of proteins can be connected to a plethora of different post-translational modifications, as exemplified by the wide-ranging, different functionalities of GAPDH.85,86 Complementation of full-proteome scans using bottom-up proteomics with top-down proteomics methods that could rapidly and routinely fully characterize the transcriptional and post-translational features of a few tens of selected proteins seems to be a remaining task for proteomics. It would be desirable that conventional biological experiments, are routinely accompanied by verification by MS-based proteomics of the exact structures for the crucial proteins.
A related characteristic that needs further evaluation is the quantitative amount of proteins in different subcellular locations. We note, for example, that direct visualization methods such as fluorescence labeling have often not reported glycolytic enzymes in the nucleus in high throughput experiments that try to screen the cellular location of large numbers of proteins. On the other hand, targeted analyses do detect them and their activities in the nucleus. This should remind us that cells are highly nonlinear systems and that functions such as regulation of transcription, with potentially high amplification factors, may only involve trace, catalytic proportions of the total cellular protein. This may also result in strong asymmetries in other kinds of experiments. For example, in interactome databases glycolytic enzymes are often found as direct interaction partners of specific nuclear proteins if the nuclear proteins were used as the baits, but detection of these interactions seems to be drowned out by cytosolic interactions if glycolytic enzymes are used as baits with whole cell lysates. Where cellular functions with high amplification factors are involved, experimental methods with high dynamic range for direct detection or methods which isolate subcellular regions may be required. We suspect that factors of this nature may underlie why we detect many proteins in both the nucleus and mitochondria whose presence in those sites has been verified by information from targeted experiments, but is not reported with some other experimental strategies.
In conclusion, the present data demonstrates that sucrose gradient fractionation seems to be a valuable, reproducible method to screen subcellular protein distributions and that large numbers of proteins are partitioned between the nucleus and mitochondria. It includes a large number of proteins in “wrong” locations relative to their presently defined roles in specific cellular functions. Several interesting entry points for further investigation of the metabolic changes exhibited by MCF7 breast cancer cells have been identified. The partitioning of many proteins to both the nucleus and mitochondria also provides a very rich backdrop for studies of the dynamic responses of both the nucleus and mitochondria of MCF7 cells to breast-cancer-related stimulations, which are currently in progress. More generally, we have grown increasingly accustomed to the idea that the dogma of one gene gives one mRNA gives one protein is inadequate. Now it seems necessary to also re-examine the corollary that one protein gives one subcellular location gives one function.38,70,87 In doing so we might remember that cells seem to have a high degree of plasticity and that, for example, in protists there can be major differences in the organization of subcellular location/function.106 We might also take note of complex systems theory and begin to consider whether the functional diversity, partial overlap with other processes, highly diverse networks and spatial distribution characteristics may be a reflection of the need for robustness in complex systems.107
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The Wellcome Trust grant to J.G.Z. and a King Faisal Foundation scholarship to A.Q.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supplementary Table 1. Assignments of MS data to peptides and proteins for the nucleus-SDS, nucleus-pI, mitochondria-SDS and mitochondria-pI subcellular preparations and fractionations. Supplementary Table 2. Summary of the protein sequence groups across all four samples (N-SDS, N-pI, M-SDS, M-pI) and GO CC (cell component) annotations. Supplementary Table 3. Partitioning of RNA processing and translation proteins between nucleus and mitochondria. Supplementary Table 4. Distribution between nucleus and mitochondria for Ras and Ras-related Rab proteins. Supplementary Table 5. Literature evidence for alternate spatio/functional distribution of glycolytic enzymes. Supplementary Table 6. Assignment of unique protein sequences to mitochondria (M) or the nucleus (N). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- (2).Seyfried TN, Shelton LM. Cancer as a metabolic disease. Nutr. Metab. (London) 2010;7:1–22. doi: 10.1186/1743-7075-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hainaut P, Plymoth A. Cancer as a metabolic disease. Curr. Opin. Oncol. 2012;24(1):56–57. doi: 10.1097/CCO.0b013e32834e388a. [DOI] [PubMed] [Google Scholar]
- (4).Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Duchen MR, Szabadkai G. Roles of mitochondria in human disease. Essays Biochem. 2010;47:115–137. doi: 10.1042/bse0470115. [DOI] [PubMed] [Google Scholar]
- (6).Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discovery. 2010;9(6):447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- (7).Bellance N, Lestienne P, Rossignol R. Mitochondria: from bioenergetics to the metabolic regulation of carcinogenesis. Front. Biosci. 2009;14:4015–4034. doi: 10.2741/3509. [DOI] [PubMed] [Google Scholar]
- (8).Ferreira LM. Cancer metabolism: the Warburg effect today. Exp. Mol. Pathol. 2010;89(3):372–380. doi: 10.1016/j.yexmp.2010.08.006. [DOI] [PubMed] [Google Scholar]
- (9).Locasale JW, Cantley LC. Altered metabolism in cancer. BMC Biol. 2010;8:1–3. doi: 10.1186/1741-7007-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13(6):472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- (11).Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- (12).Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Krivakova P, Cervinkova Z, Lotkova H, Kucera O, Rousar T. [Mitochondria and their role in cell metabolism] Acta Med. (Hradec Kralove) Suppl. 2005;48(2):57–67. [PubMed] [Google Scholar]
- (15).Campello S, Scorrano L. Mitochondrial shape changes: orchestrating cell pathophysiology. EMBO Rep. 2010;11(9):678–684. doi: 10.1038/embor.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Westermann B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell. Biol. 2010;11(12):872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- (17).Chan DC. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- (18).Berman SB, Pineda FJ, Hardwick JM. Mitochondrial fission and fusion dynamics: the long and short of it. Cell Death Differ. 2008;15(7):1147–1152. doi: 10.1038/cdd.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Perfettini JL, Roumier T, Kroemer G. Mitochondrial fusion and fission in the control of apoptosis. Trends Cell Biol. 2005;15(4):179–183. doi: 10.1016/j.tcb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- (20).Grandemange S, Herzig S, Martinou JC. Mitochondrial dynamics and cancer. Semin. Cancer Biol. 2009;19(1):50–56. doi: 10.1016/j.semcancer.2008.12.001. [DOI] [PubMed] [Google Scholar]
- (21).Karbowski M, Youle RJ. Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 2003;10(8):870–880. doi: 10.1038/sj.cdd.4401260. [DOI] [PubMed] [Google Scholar]
- (22).de Brito OM, Scorrano L. An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J. 2010;29(16):2715–2723. doi: 10.1038/emboj.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Fujimoto M, Hayashi T. New insights into the role of mitochondria-associated endoplasmic reticulum membrane. Int. Rev. Cell Mol. Biol. 2011;292:73–117. doi: 10.1016/B978-0-12-386033-0.00002-5. [DOI] [PubMed] [Google Scholar]
- (24).Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annu. Rev. Biochem. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- (25).Chen JQ, Yager JD, Russo J. Regulation of mitochondrial respiratory chain structure and function by estrogens/estrogen receptors and potential physiological/pathophysiological implications. Biochim. Biophys. Acta. 2005;1746(1):1–17. doi: 10.1016/j.bbamcr.2005.08.001. [DOI] [PubMed] [Google Scholar]
- (26).Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- (27).Baker BM, Haynes CM. Mitochondrial protein quality control during biogenesis and aging. Trends Biochem. Sci. 2011;36(5):254–261. doi: 10.1016/j.tibs.2011.01.004. [DOI] [PubMed] [Google Scholar]
- (28).Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 2011;1813(7):1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Eichner LJ, Giguere V. Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion. 2011;11(4):544–552. doi: 10.1016/j.mito.2011.03.121. [DOI] [PubMed] [Google Scholar]
- (30).Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- (31).Becker T, Bottinger L, Pfanner N. Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem. Sci. 2012;37(3):85–91. doi: 10.1016/j.tibs.2011.11.004. [DOI] [PubMed] [Google Scholar]
- (32).Wang G, Deschenes RJ. Plasma membrane localization of Ras requires class C Vps proteins and functional mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26(8):3243–3255. doi: 10.1128/MCB.26.8.3243-3255.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Shen L, Sun X, Fu Z, Yang G, Li J, Yao L. The fundamental role of the p53 pathway in tumor metabolism and its implication in tumor therapy. Clin. Cancer Res. 2012;18(6):1561–1567. doi: 10.1158/1078-0432.CCR-11-3040. [DOI] [PubMed] [Google Scholar]
- (34).Chen JQ, Cammarata PR, Baines CP, Yager JD. Regulation of mitochondrial respiratory chain biogenesis by estrogens/estrogen receptors and physiological, pathological and pharmacological implications. Biochim. Biophys. Acta. 2009;1793(10):1540–1570. doi: 10.1016/j.bbamcr.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Leigh-Brown S, Enriquez JA, Odom DT. Nuclear transcription factors in mammalian mitochondria. Genome Biol. 2010;11(7):1–9. doi: 10.1186/gb-2010-11-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lee J, Sharma S, Kim J, Ferrante RJ, Ryu H. Mitochondrial nuclear receptors and transcription factors: who’s minding the cell? J. Neurosci. Res. 2008;86(5):961–971. doi: 10.1002/jnr.21564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Braschi E, Goyon V, Zunino R, Mohanty A, Xu L, McBride HM. Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Curr. Biol. 2010;20(14):1310–1315. doi: 10.1016/j.cub.2010.05.066. [DOI] [PubMed] [Google Scholar]
- (38).Qattan AT, Mulvey C, Crawford M, Natale DA, Godovac-Zimmermann J. Quantitative organelle proteomics of MCF-7 breast cancer cells reveals multiple subcellular locations for proteins in cellular functional processes. J. Proteome Res. 2010;9(1):495–508. doi: 10.1021/pr9008332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ben-Menachem R, Tal M, Shadur T, Pines O. A third of the yeast mitochondrial proteome is dual localized: a question of evolution. Proteomics. 2011;11(23):4468–4476. doi: 10.1002/pmic.201100199. [DOI] [PubMed] [Google Scholar]
- (40).Patel VP, Chu CT. Nuclear transport, oxidative stress, and neurodegeneration. Int. J. Clin. Exp. Pathol. 2011;4(3):215–229. [PMC free article] [PubMed] [Google Scholar]
- (41).Kodiha M, Stochaj U. Nuclear transport: a switch for the oxidative stress-signaling circuit? J. Signal Transduct. 2012;2012:1–18. doi: 10.1155/2012/208650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Henke RM, Dastidar RG, Shah A, Cadinu D, Yao X, Hooda J, Zhang L. Hypoxia elicits broad and systematic changes in protein subcellular localization. Am. J. Physiol. Cell Physiol. 2011;301(4):C913–928. doi: 10.1152/ajpcell.00481.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wang J, Bai L, Li J, Sun C, Zhao J, Cui C, Han K, Liu Y, Zhuo X, Wang T, Liu P, Fan F, Guan Y, Ma A. Proteomic analysis of mitochondria reveals a metabolic switch from fatty acid oxidation to glycolysis in the failing heart. Sci. China C: Life Sci. 2009;52(11):1003–1010. doi: 10.1007/s11427-009-0140-2. [DOI] [PubMed] [Google Scholar]
- (44).Radulovic M, Crane E, Crawford M, Godovac-Zimmermann J, Yu VP. CKS proteins protect mitochondrial genome integrity by interacting with mitochondrial single-stranded DNA-binding protein. Mol. Cell. Proteomics. 2010;9(1):145–152. doi: 10.1074/mcp.M900078-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Clawson GA. Nuclei, Mitochondria, Lysosomes and Peroxisomes. In: Reid E, editor. Cancer Cell Organelles. Wiley; Chichester: 1982. pp. 188–201. [Google Scholar]
- (46).Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- (47).Geiser L, Dayon L, Vaezzadeh AR, Hochstrasser DF. Shotgun proteomics: a relative quantitative approach using Off-Gel electrophoresis and LC-MS/MS. Methods Mol. Biol. 2011;681:459–472. doi: 10.1007/978-1-60761-913-0_27. [DOI] [PubMed] [Google Scholar]
- (48).Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2(8):1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- (49).Hubner NC, Ren S, Mann M. Peptide separation with immobilized pI strips is an attractive alternative to in-gel protein digestion for proteome analysis. Proteomics. 2008;8(23-24):4862–4872. doi: 10.1002/pmic.200800351. [DOI] [PubMed] [Google Scholar]
- (50).Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- (51).Foster LJ, de Hoog CL, Zhang Y, Xie X, Mootha VK, Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125(1):187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- (52).Taylor SW, Warnock DE, Glenn GM, Zhang B, Fahy E, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. An alternative strategy to determine the mitochondrial proteome using sucrose gradient fractionation and 1D PAGE on highly purified human heart mitochondria. J. Proteome Res. 2002;1(5):451–458. doi: 10.1021/pr025533g. [DOI] [PubMed] [Google Scholar]
- (53).Shutt TE, Shadel GS. A compendium of human mitochondrial gene expression machinery with links to disease. Environ. Mol. Mutagen. 2010;51(5):360–379. doi: 10.1002/em.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Lykke-Andersen S, Tomecki R, Jensen TH, Dziembowski A. The eukaryotic RNA exosome: same scaffold but variable catalytic subunits. RNA Biol. 2011;8(1):61–66. doi: 10.4161/rna.8.1.14237. [DOI] [PubMed] [Google Scholar]
- (55).Schaeffer D, van Hoof A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc. Natl. Acad. Sci. U.S.A. 2011;108(6):2366–2371. doi: 10.1073/pnas.1013180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Moser TL, Kenan DJ, Ashley TA, Roy JA, Goodman MD, Misra UK, Cheek DJ, Pizzo SV. Endothelial cell surface F1-F0 ATP synthase is active in ATP synthesis and is inhibited by Angiostatin. Proc. Natl. Acad. Sci. U.S.A. 2001;98(12):6656–6661. doi: 10.1073/pnas.131067798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Yonally SK, Capaldi RA. The F(1)F(0) ATP synthase and mitochondrial respiratory chain complexes are present on the plasma membrane of an osteosarcoma cell line: An immunocytochemical study. Mitochondrion. 2006;6(6):305–314. doi: 10.1016/j.mito.2006.10.001. [DOI] [PubMed] [Google Scholar]
- (58).Ravera S, Panfoli I, Calzia D, Aluigi MG, Bianchini P, Diaspro A, Mancardi G, Morelli A. Evidence for aerobic ATP synthesis in isolated myelin vesicles. Int. J. Biochem. Cell Biol. 2009;41(7):1581–1591. doi: 10.1016/j.biocel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- (59).Panfoli I, Ravera S, Bruschi M, Candiano G, Morelli A. Proteomics unravels the exportability of mitochondrial respiratory chains. Expert Rev. Proteomics. 2011;8(2):231–239. doi: 10.1586/epr.11.1. [DOI] [PubMed] [Google Scholar]
- (60).Sugden MC, Holness MJ. Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am. J. Physiol. Endocrinol. Metab. 2003;284(5):E855–862. doi: 10.1152/ajpendo.00526.2002. [DOI] [PubMed] [Google Scholar]
- (61).Hartong DT, Dange M, McGee TL, Berson EL, Dryja TP, Colman RF. Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nat. Genet. 2008;40(10):1230–1234. doi: 10.1038/ng.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Hackenbeck T, Huber R, Schietke R, Knaup KX, Monti J, Wu X, Klanke B, Frey B, Gaipl U, Wullich B, Ferbus D, Goubin G, Warnecke C, Eckardt KU, Wiesener MS. The GTPase RAB20 is a HIF target with mitochondrial localization mediating apoptosis in hypoxia. Biochim. Biophys. Acta. 2011;1813(1):1–13. doi: 10.1016/j.bbamcr.2010.10.019. [DOI] [PubMed] [Google Scholar]
- (63).Bui M, Gilady SY, Fitzsimmons RE, Benson MD, Lynes EM, Gesson K, Alto NM, Strack S, Scott JD, Simmen T. Rab32 modulates apoptosis onset and mitochondria-associated membrane (MAM) properties. J. Biol. Chem. 2010;285(41):31590–31602. doi: 10.1074/jbc.M110.101584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Zahedi RP, Sickmann A, Boehm AM, Winkler C, Zufall N, Schonfisch B, Guiard B, Pfanner N, Meisinger C. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell. 2006;17(3):1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Lynes EM, Bui M, Yap MC, Benson MD, Schneider B, Ellgaard L, Berthiaume LG, Simmen T. Palmitoylated TMX and calnexin target to the mitochondria-associated membrane. EMBO J. 2012;31(2):457–470. doi: 10.1038/emboj.2011.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Kim JY, Casaccia P. HDAC1 in axonal degeneration: A matter of subcellular localization. Cell Cycle. 2010;9(18):3680–3684. doi: 10.4161/cc.9.18.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Pacheco M, Nielsen TO. Histone deacetylase 1 and 2 in mesenchymal tumors. Mod. Pathol. 2012;25(2):222–230. doi: 10.1038/modpathol.2011.157. [DOI] [PubMed] [Google Scholar]
- (68).Matsumoto S, Uchiumi T, Saito T, Yagi M, Takazaki S, Kanki T, Kang D. Localization of mRNAs encoding human mitochondrial oxidative phosphorylation proteins. Mitochondrion. 2012;12(3):391–398. doi: 10.1016/j.mito.2012.02.004. [DOI] [PubMed] [Google Scholar]
- (69).Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One. 2008;3(6):1–12. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Yogev O, Pines O. Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim. Biophys. Acta. 2011;1808(3):1012–1020. doi: 10.1016/j.bbamem.2010.07.004. [DOI] [PubMed] [Google Scholar]
- (71).Shakib K, Norman JT, Fine LG, Brown LR, Godovac-Zimmermann J. Proteomics profiling of nuclear proteins for kidney fibroblasts suggests hypoxia, meiosis, and cancer may meet in the nucleus. Proteomics. 2005;5(11):2819–2838. doi: 10.1002/pmic.200401108. [DOI] [PubMed] [Google Scholar]
- (72).Mamczur P, Sok AJ, Rzechonek A, Rakus D. Cell cycle-dependent expression and subcellular localization of fructose 1,6-bisphosphatase. Histochem. Cell Biol. 2012;137(1):121–136. doi: 10.1007/s00418-011-0884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Kowalski W, Nocon D, Gamian A, Kolodziej J, Rakus D. Association of C-terminal region of phosphoglycerate mutase with glycolytic complex regulates energy production in cancer cells. J. Cell Physiol. 2012;227(6):2613–2621. doi: 10.1002/jcp.22998. [DOI] [PubMed] [Google Scholar]
- (74).Colombo SL, Palacios-Callender M, Frakich N, Carcamo S, Kovacs I, Tudzarova S, Moncada S. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108(52):21069–21074. doi: 10.1073/pnas.1117500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Yalcin A, Clem BF, Simmons A, Lane A, Nelson K, Clem AL, Brock E, Siow D, Wattenberg B, Telang S, Chesney J. Nuclear targeting of 6-phosphofructo-2-kinase (PFKFB3) increases proliferation via cyclin-dependent kinases. J. Biol. Chem. 2009;284(36):24223–24232. doi: 10.1074/jbc.M109.016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Barbe L, Lundberg E, Oksvold P, Stenius A, Lewin E, Bjorling E, Asplund A, Ponten F, Brismar H, Uhlen M, Andersson-Svahn H. Toward a confocal subcellular atlas of the human proteome. Mol. Cell. Proteomics. 2008;7(3):499–508. doi: 10.1074/mcp.M700325-MCP200. [DOI] [PubMed] [Google Scholar]
- (77).Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J. Exp. Biol. 2003;206(Pt 12):2049–2057. doi: 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- (78).Neary CL, Pastorino JG. Nucleocytoplasmic shuttling of hexokinase II in a cancer cell. Biochem. Biophys. Res. Commun. 2010;394(4):1075–1081. doi: 10.1016/j.bbrc.2010.03.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).John S, Weiss JN, Ribalet B. Subcellular localization of hexokinases I and II directs the metabolic fate of glucose. PLoS One. 2011;6(3):1–13. doi: 10.1371/journal.pone.0017674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Brandina I, Graham J, Lemaitre-Guillier C, Entelis N, Krasheninnikov I, Sweetlove L, Tarassov I, Martin RP. Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim. Biophys. Acta. 2006;1757(9-10):1217–1228. doi: 10.1016/j.bbabio.2006.07.001. [DOI] [PubMed] [Google Scholar]
- (81).Giege P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell. 2003;15(9):2140–2151. doi: 10.1105/tpc.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Graham JW, Williams TC, Morgan M, Fernie AR, Ratcliffe RG, Sweetlove LJ. Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell. 2007;19(11):3723–3738. doi: 10.1105/tpc.107.053371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Liu X, Zhang M, Go VL, Hu S. Membrane proteomic analysis of pancreatic cancer cells. J. Biomed. Sci. 2010;17:1–13. doi: 10.1186/1423-0127-17-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Zhang A, Williamson CD, Wong DS, Bullough MD, Brown KJ, Hathout Y, Colberg-Poley AM. Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Mol. Cell. Proteomics. 2011;10(10):1–16. doi: 10.1074/mcp.M111.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal. 2011;23(2):317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Sirover MA. Subcellular dynamics of multifunctional protein regulation: Mechanisms of GAPDH intracellular translocation. J. Cell. Biochem. 2012:2193–2200. doi: 10.1002/jcb.24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Yogev O, Naamati A, Pines O. Fumarase: a paradigm of dual targeting and dual localized functions. FEBS J. 2011;278(22):4230–4242. doi: 10.1111/j.1742-4658.2011.08359.x. [DOI] [PubMed] [Google Scholar]
- (88).Tudzarova S, Trotter MW, Wollenschlaeger A, Mulvey C, Godovac-Zimmermann J, Williams GH, Stoeber K. Molecular architecture of the DNA replication origin activation checkpoint. EMBO J. 2010;29(19):3381–3394. doi: 10.1038/emboj.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Mulvey C, Tudzarova S, Crawford M, Williams GH, Stoeber K, Godovac-Zimmermann J. Quantitative proteomics reveals a "poised quiescence" cellular state after triggering the DNA replication origin activation checkpoint. J. Proteome Res. 2010;9(10):5445–5460. doi: 10.1021/pr100678k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Carrasco S, Meyer T. STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu. Rev. Biochem. 2011;80:973–1000. doi: 10.1146/annurev-biochem-061609-165311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 2011;108(35):14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Ahmed S, Passos JF, Birket MJ, Beckmann T, Brings S, Peters H, Birch-Machin MA, von Zglinicki T, Saretzki G. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J. Cell Sci. 2008;121(Pt 7):1046–1053. doi: 10.1242/jcs.019372. [DOI] [PubMed] [Google Scholar]
- (93).Sripathi SR, He W, Atkinson CL, Smith JJ, Liu Z, Elledge BM, Jahng WJ. Mitochondrial-nuclear communication by prohibitin shuttling under oxidative stress. Biochemistry. 2011;50(39):8342–8351. doi: 10.1021/bi2008933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Shi SL, Li QF, Liu QR, Xu DH, Tang J, Liang Y, Zhao ZL, Yang LM. Nuclear matrix protein, prohibitin, was down-regulated and translocated from nucleus to cytoplasm during the differentiation of osteosarcoma MG-63 cells induced by ginsenoside Rg1, cinnamic acid, and tanshinone IIA (RCT) J. Cell. Biochem. 2009;108(4):926–934. doi: 10.1002/jcb.22324. [DOI] [PubMed] [Google Scholar]
- (95).Guppy M, Leedman P, Zu X, Russell V. Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem. J. 2002;364(Pt 1):309–315. doi: 10.1042/bj3640309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Folmes CD, Nelson TJ, Terzic A. Energy metabolism in nuclear reprogramming. Biomark. Med. 2011;5(6):715–729. doi: 10.2217/bmm.11.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, Cantley LC. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329(5998):1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Luo W, Semenza GL. Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget. 2011;2(7):551–556. doi: 10.18632/oncotarget.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Lee J, Kim HK, Han YM, Kim J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int. J. Biochem. Cell Biol. 2008;40(5):1043–1054. doi: 10.1016/j.biocel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- (100).Zheng L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114(2):255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- (101).Briere JJ, Favier J, Gimenez-Roqueplo AP, Rustin P. Tricarboxylic acid cycle dysfunction as a cause of human diseases and tumor formation. Am. J. Physiol. Cell Physiol. 2006;291(6):C1114–1120. doi: 10.1152/ajpcell.00216.2006. [DOI] [PubMed] [Google Scholar]
- (102).Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim. Biophys. Acta. 2011;1807(11):1432–1443. doi: 10.1016/j.bbabio.2011.07.003. [DOI] [PubMed] [Google Scholar]
- (103).Ghosh AK, Kanda T, Steele R, Ray RB. Knockdown of MBP-1 in human foreskin fibroblasts induces p53-p21 dependent senescence. PLoS One. 2008;3(10):1–7. doi: 10.1371/journal.pone.0003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Lo Presti M, Ferro A, Contino F, Mazzarella C, Sbacchi S, Roz E, Lupo C, Perconti G, Giallongo A, Migliorini P, Marrazzo A, Feo S. Myc promoter-binding protein-1 (MBP-1) is a novel potential prognostic marker in invasive ductal breast carcinoma. PLoS One. 2010;5(9):1–10. doi: 10.1371/journal.pone.0012961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Shaw RJ, Cantley LC. Decoding key nodes in the metabolism of cancer cells: sugar & spice and all things nice. F1000 Biol. Rep. 2012;4:1–7. doi: 10.3410/B4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Ginger ML, McFadden GI, Michels PA. Rewiring and regulation of cross-compartmentalized metabolism in protists. Philos. Trans. R. Soc. Lond., B: Biol. Sci. 2010;365(1541):831–845. doi: 10.1098/rstb.2009.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Whitacre JM, Bender A. Networked buffering: a basic mechanism for distributed robustness in complex adaptive systems. Theor. Biol. Med. Model. 2010;7:1–20. doi: 10.1186/1742-4682-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.