Summary
The LXR-regulated E3 ubiquitin ligase IDOL controls LDLR receptor stability independent of SREBP and PCSK9, but its relevance to plasma lipid levels is unknown. Here we demonstrate that the effects of the LXR–IDOL axis are both tissue- and species-specific. In mice, LXR agonist induces Idol transcript levels in peripheral tissues but not in liver, and does not change plasma LDL levels. Accordingly, Idol-deficient mice exhibit elevated LDLR protein levels in peripheral tissues but not in the liver. By contrast, LXR activation in cynomolgus monkeys induces hepatic IDOL expression, reduces LDLR protein levels, and raises plasma LDL levels. Knockdown of IDOL in monkeys with an antisense oligonucleotide blunts the effect of LXR agonist on LDL levels. These results implicate IDOL as a modulator of plasma lipid levels in primates and support further investigation into IDOL inhibition as a potential strategy for LDL lowering in humans.
Introduction
Coronary heart disease is the leading cause of death in the industrialized world, and elevated plasma cholesterol levels are one of its major causes. Despite the efficacy of statin drugs in lowering LDL cholesterol levels and decreasing CVD risk, additional therapeutics strategies are needed. Sterol homeostasis in animals is governed through the coordinate actions of two sterol-sensing transcription factors, the Liver X Receptors (LXRs) and the Sterol Regulatory Element Binding Proteins (SREBPs)(Brown and Goldstein, 1997; Peet et al., 1998).
The LXRs control the expression of a battery of genes involved in the uptake, transport, efflux and excretion of cholesterol in a tissue-specific manner. In addition, in the setting of cellular cholesterol excess, LXRs limit the uptake of cellular cholesterol through transcriptional induction of the gene encoding the E3 ubiquitin ligase, the inducible degrader of the LDLR (IDOL) (Zelcer et al., 2009). IDOL binds directly to the cytoplasmic tail of the LDLR and promotes its ubiquitination by the UBE2D1/E1 complex (Calkin et al., 2011; Zhang et al., 2011). This ubiquitinated LDLR then enters the MVB protein-sorting pathway and is shuttled to the lysosome for degradation (Scotti et al., 2011; Sorrentino et al., 2013b).
Previous work showed the acute hepatic overexpression of Idol in mice reduces LDLR protein levels and increases plasma cholesterol levels (Zelcer et al., 2009). Furthermore, targeted deletion of IDOL in cultured mouse cells leads to increased LDLR protein levels and increased LDL uptake, and this increase is additive with the effect of statin treatment (Scotti et al., 2011). However, treatment of mice with an LXR agonist does not raise plasma LDL levels (Grefhorst et al., 2002), raising the question of the physiological impact of the LXR-Idol pathway in the liver. The effect of loss of IDOL function on plasma cholesterol levels is currently unknown. Thus, it is unclear whether IDOL might be a suitable target for a new class of lipid-lowering drugs.
Here we demonstrate that the effects of the LXR-IDOL pathway on LDLR protein levels are both tissue and species-specific. In mice, hepatic LDLR protein is relatively resistant to LXR agonists or loss of IDOL activity. By contrast, LXR agonists raise plasma LDL cholesterol levels in primates through an IDOL-dependent mechanism. These observations support further investigation of the LXR–IDOL pathway as a potential therapeutic target and suggest that IDOL inhibition may mitigate undesirable effects of synthetic LXR agonists on plasma LDL cholesterol levels in humans.
Results
Tissue-specific regulation of LDLR protein by the LXR-IDOL pathway in mice
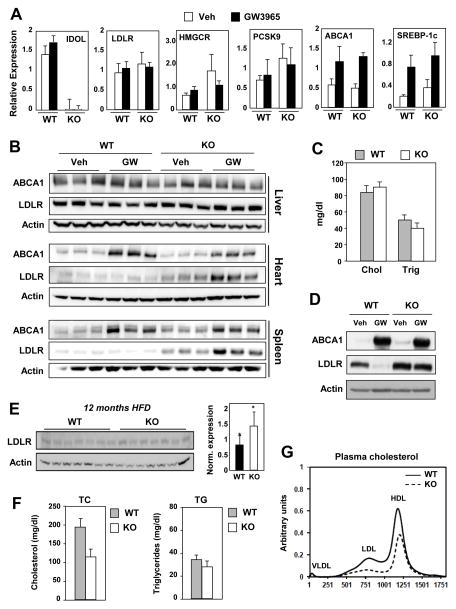
Previous work demonstrated that mouse embryonic stem cells lacking IDOL expression had increased basal LDLR protein expression and were refractory to the degradation effects of LXR agonists (Scotti et al., 2013; Scotti et al., 2011). To determine the consequences of loss of IDOL expression for cholesterol homeostasis in vivo, we generated global IDOL-deficient mice. Analysis of mRNA expression in Idol–/– mice confirmed an absence of Idol transcripts and showed no significant change in the expression of a battery of other genes linked to cholesterol metabolism, including LDLR, PCSK9, and HMGCR (Figure 1A). We observed no difference in total or LDL cholesterol levels in the plasma between chow-fed wild-type (WT) and Idol–/– mice (Figure 1C and S1A). Also, we did not detect any significant changes in plasma apolipoprotein B levels in Idol–/– mice (data not shown). Treatment of Idol–/– mice with synthetic LXR agonist did not change hepatic LDLR protein or plasma LDL levels (Figure 1B and S1B). Feeding a high-cholesterol western diet for 12 months uncovered a subtle but reproducible increase in hepatic LDLR protein levels in Idol–/– mice (Figure 1E). This was associated with a trend towards reduced plasma cholesterol levels that did not reach statistical significance (Figure 1F,G).
Figure 1. IDOL controls peripheral LDLR expression in mice.
WT and Idol-null mice were treated with the LXR agonist GW3965 at 40 mg/kg for 3 days (n = 3–4/group). (A) Hepatic gene expression was quantified by real-time PCR and normalized to 36B4. (B) Protein lysates from liver, heart, and spleen were analyzed by immunoblotting with antibodies against ABCA1, LDLR, and β-Actin. (n = 3–4 mice/treatment) (C) Total serum cholesterol and triglyceride levels for chow-fed animals. (D) Immunoblot analysis of protein extracts from thioglycollate-elicited peritoneal macrophages isolated from WT and Idol-null mice and treated with GW3965 (1 μM). Immunoblot shown is representative of three independent experiments. (E) WT and Idol-null mice were fed western diet for 12 months. Protein lysates from liver were analyzed by immunoblotting and quantified using ImageJ (n=7 mice/genotype). Significance was determined by student t-test (* p < 0.05). (F) Total serum cholesterol and triglyceride levels for the animals shown in E. (G) Plasma lipoprotein cholesterol distribution for the animals shown in E. Error bars represent SEM.
In contrast to the liver findings, the basal levels of LDLR protein were much higher in heart, spleen, and macrophages of Idol–/– mice than in those of WT mice (Figure 1B,D). We have also observed increased LDLR expression in a number of other peripheral tissues from Idol–/– mice (data not shown). Furthermore, LXR agonist treatment reduced LDLR protein levels in heart, spleen, and macrophages of WT but not Idol–/– mice. This effect correlated with induction of protein levels of ABCA1, an established LXR target.
Activity of the hepatic LXR-IDOL Pathway is Species-Specific
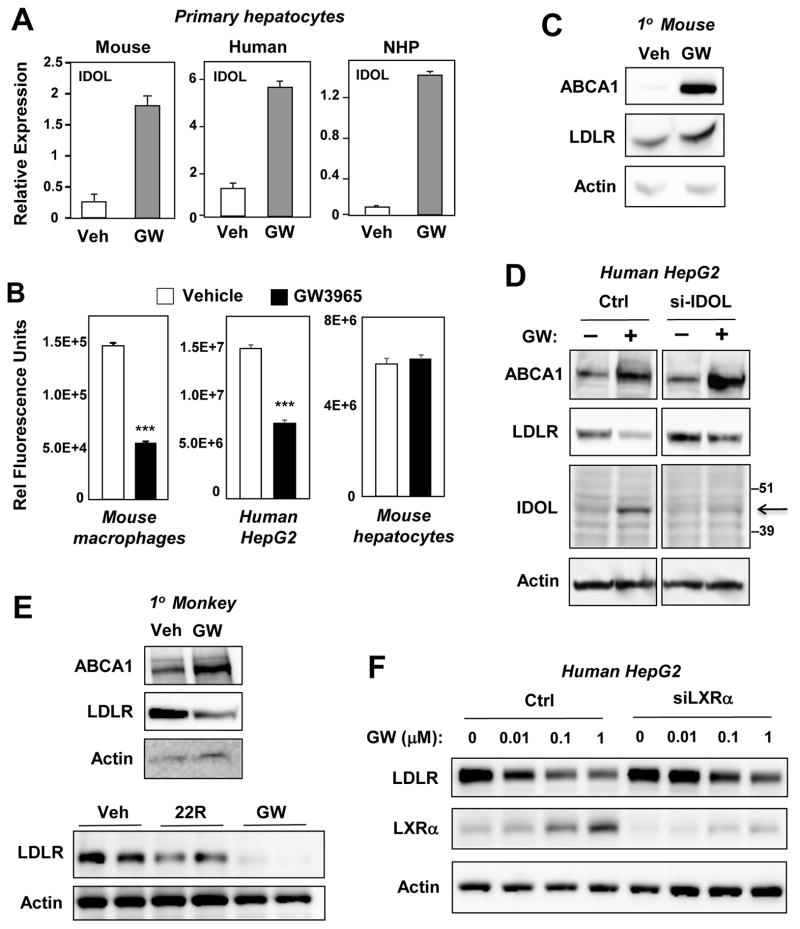
Species-specific differences in lipid metabolism are common. In the case of LXR, several differences between mouse and human gene regulation have been described. For example, Cyp7A1 is a target of LXRs in mice but not humans, and the CETP gene is an LXR target that is not present in mice (Honzumi et al., 2010; Luo and Tall, 2000; Repa et al., 2000). To address the possibility of a species-specific regulation of IDOL, we compared the activity of the LXR–IDOL pathway in mice, humans, and cynomolgus monkeys. An LXR agonist induced IDOL mRNA expression in sterol-starved hepatocytes from all species (Figure 2A). To examine the functional consequences of this observation, LDL uptake studies were performed. LXR agonist treatment blocked LDL uptake in mouse macrophages and human HepG2 hepatoma cells, but not in mouse hepatocytes (Figure 2B and S2A,B,C). Consistent with these differences in lipid uptake, synthetic and sterol LXR agonists caused degradation of the LDLR only in monkey hepatocytes and human hepatoma cells (Figure 2C,D,E and S2D). Interestingly, we were able to detect endogenous IDOL protein in human but not mouse hepatocytes using a validated monoclonal antibody that recognizes both mouse and human proteins (Scotti et al., 2013) (Fig. 2D and data not shown). Importantly, knockdown of IDOL with siRNA reduced IDOL protein and LDLR degradation as expected in HepG2 cells (Figure 2D). These data demonstrate that the effects of the LXR–IDOL pathway on the LDLR are both tissue- and species-specific.
Figure 2. Species-specific regulation of hepatic LDLR protein expression by the LXR–IDOL pathway.
Primary hepatocytes from mice, humans, and nonhuman primates were serum-starved and treated with GW3965 (1 μM) for 24 h. (A) Regulation of IDOL mRNA expression by LXR agonists. (B) Quantification of cellular uptake of DiI-LDL uptake in the presence or absence of GW3965. Cell-associated fluorescence was measured with a Typhoon Instrument. n=3. ***P< 0.001. (C,D) Immunoblot analysis of protein lysates from primary mouse hepatocytes (C) or human HepG2 cells (D) treated with GW3965 (1 μM) for 24 h. (E) Immunoblotting of protein lysates from cynomolgus monkey hepatocytes treated with GW3965 (1 μM) or 22R-hydroxycholesterol (2.5 μM) for 24 h. (F) Immunoblot analysis of protein lysates from control or LXRα-siRNA-transfected HepG2 cells treated for 24 h with increasing doses of GW3965. Data are representative of three independent experiments. Error bars represent SEM.
One important difference between mouse and primate cells is the ability of LXRs to induce expression of the LXRα gene in response to ligand activation in primates but not mice (Laffitte et al., 2001). We hypothesized that this species-specific feed-forward mechanism might allow certain LXR target genes to be more responsive to LXR pathway activation in humans and nonhuman primates compared to mice. In support of this hypothesis, blocking the autoregulation of LXRα expression in HepG2 cells with siRNAs blunted the ability of LXR agonists to induce the degradation of the LDLR (Fig. 2F).
The LXR-IDOL axis is active in nonhuman primates
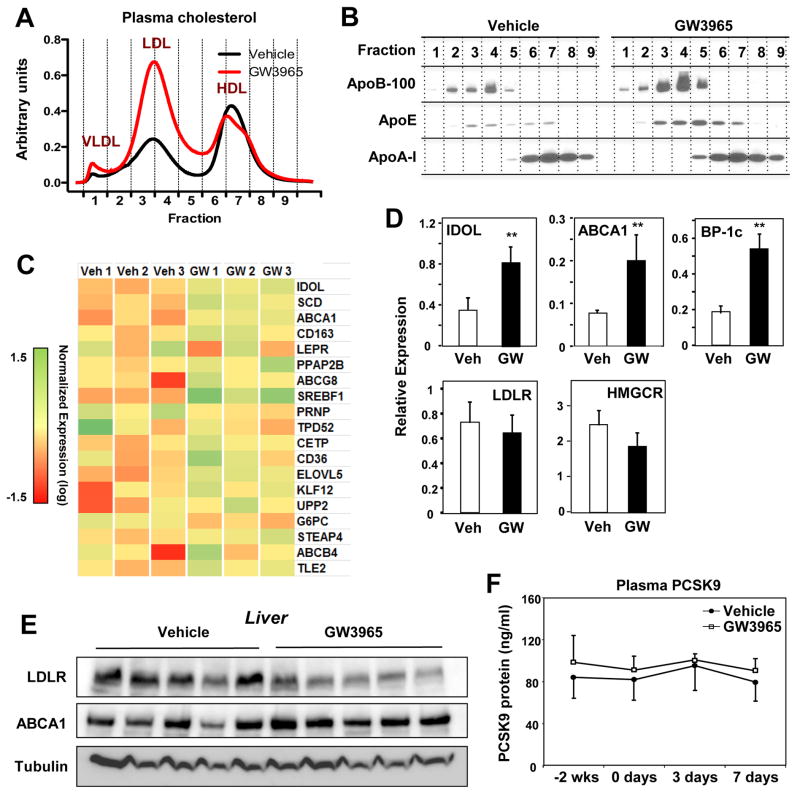
To address whether LXR activation might have different consequences for hepatic LDLR protein levels in mice and primates, we treated high-fat diet–fed cynomolgus monkeys with a synthetic LXR agonist. GW3965 treatment for seven days led to marked increases in total cholesterol and LDL cholesterol levels with no changes in HDL or VLDL cholesterol levels (Figure 3A and Figure S3A). Plasma apolipoprotein B-100 levels were increased (Figure 3B). Hepatic cholesterol levels were not different between groups (Figure S3B). Plasma triglyceride levels were also not different (Fig. S3C), although there was a trend towards increased hepatic triglyceride content with GW3965 treatment (Fig. S3B).
Figure 3. LXR activation in nonhuman primates reduces hepatic LDLR protein and raises plasma LDL levels.
(A) Plasma lipoprotein cholesterol distribution of cynomolgus monkeys treated with vehicle or GW3965 (10 mg/kg for 7 days) (n = 5 pooled samples/group). (B) Plasma apolipoprotein (apoB-100, apoE, and apoA-I) distributions for animals shown in A. (C) Transcriptional profiling of mRNA from the livers of cynomolgus monkeys treated for 2 days with vehicle or GW3965. Tissues were obtained 6 h after the last dose and transcript levels were analyzed with Affymetrix arrays. (D) Hepatic gene expression from the animals in D as determined by real-time PCR; BP-1C = SREBP-1c (n=5/group; ** p < 0.01). (E) Protein lysates of the liver from cynomolgus monkeys treated with vehicle or GW3965 (10 mg/kg for 7 days). Tissues were obtained 24 h after the final dose; immunoblots were performed for LDLR, ABCA1, and tubulin. (F) ELISA of plasma levels of PCSK9 in cynomolgus monkeys treated with vehicle or GW3965 (10 mg/kg for 7 days) (n = 5/group). Error bars represent SEM
Genome-wide transcriptional profiling of hepatic gene expression confirmed the induction of multiple LXR target genes in response to GW3965 treatment (Figure 3C). In contrast to the findings in mouse liver in vivo (Figure 1), LXR activation substantially induced IDOL mRNA expression in monkey liver in vivo (Figure 3D). mRNAs encoding the established LXR targets ABCA1 and SREBP-1c were also induced by LXR agonist in parallel. Consistent with increased IDOL expression and higher plasma LDL levels, hepatic LDLR protein levels were reduced in GW3965-treated monkeys (Figure 3E). There was no change in LDLR or HMCGR transcript levels in response to LXR activation, strongly suggesting that the observed changes stemmed from IDOL-mediated post-transcriptional effects (Figure 3D and data not shown). We also found no effect of GW3965 treatment on plasma PCSK9 protein levels in cynomolgus monkeys (Fig. 3F).
IDOL contributes to the effect of LXR agonists on plasma LDL cholesterol
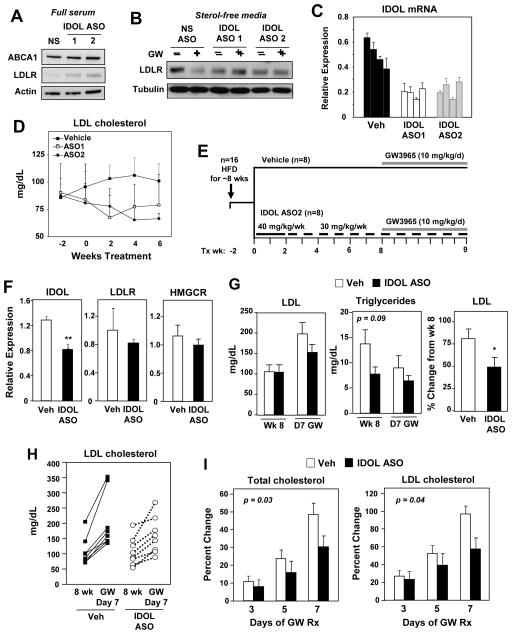
To definitively address the involvement of IDOL in the effects of LXR agonists on plasma lipid levels in cynomolgus monkeys, we generated and characterized high-affinity, generation 2.5 antisense oligonucleotides (ASOs). We identified two ASOs that significantly reduced IDOL mRNA levels in human Huh-7 cells, human Hep3B cells, and primary monkey hepatocytes (Figure S4A). Furthermore, when HepG2 cells were cultured in full serum-containing (sterol-rich) media (conditions under which the LXR-IDOL pathway is active) IDOL ASO treatment raised LDLR protein levels (Figure 4A). When HepG2 cells were cultured in sterol-deficient media (conditions under which the endogenous LXR pathway is inactive and LDLR is maximally expressed) IDOL ASO-treatment blocked the ability of synthetic LXR agonist to stimulate LDLR degradation (Figure 4B). Note, IDOL ASOs would not be expected to raise basal LDLR expression in sterol-deficient media because IDOL is not expressed in the absence of LXR agonist.
Figure 4. IDOL knockdown modulates hepatic LDLR expression in nonhuman primates.
(A,B) Immunoblot analysis of HepG2 cells transfected with control or IDOL ASOs. (A) Cells were cultured in full serum. (B) Cells were cultured in sterol-deficient media containing 5 μM simvastatin plus 100 μM mevalonic and treated for 24 h with GW3965 (1 μM). Data are representative of three independent experiments. (C) Activity of IDOL ASOs in vivo. Cynomolgus monkeys were treated for 8 weeks with vehicle or IDOL ASOs, and hepatic IDOL expression levels were quantified by real-time PCR. Each bar represents an individual animal. (D) Plasma LDL cholesterol was measured at the designated time points for animals shown in C. (E) Schematic showing the experimental design for evaluating the effects of IDOL ASO treatment on the response to LXR activation in monkeys. (F) Liver gene expression determined by real-time PCR (n = 8/group). *** p < 0.001. (G) Average plasma LDL cholesterol level (Left) and average triglyceride level (Middle) before and after GW3965 treatment. (Right) Percent change in LDL cholesterol levels (average for group) after GW3965 treatment. (H) Plasma LDL levels of each nonhuman primate at week 8 and day 7 of GW3965 treatment. (I) Percent change in total and LDL cholesterol levels over the course of the entire GW3965 treatment period. Two-way ANOVA was used to assess statistical significance. Error bars represent SEM.
In vivo studies in a cohort of cynomolgus monkeys (n = 4/group) confirmed the ability of IDOL ASOs to reduce hepatic IDOL mRNA expression (Figure S4B, 4C). There was also a trend towards lower total plasma cholesterol and plasma LDL cholesterol levels in the animals receiving IDOL ASOs (Figure 4D and S4C). HDL cholesterol levels were not different between groups (Figure S4C).
To test the ability of IDOL inhibition to affect LXR-dependent changes in plasma cholesterol levels, we conducted a study on a larger cohort of high-fat diet–fed monkeys (study design is shown in Figure 4E). The animals were treated for nine weeks with vehicle or the IDOL ASO (n = 8/group). During the ninth week, all of the animals were administered GW3965 to activate the LXR–IDOL pathway. No overt hepatotoxicity was observed in response to IDOL ASO administration as assessed by plasma AST and ALT levels (Figure S4D). The IDOL ASO led to a partial (~45–50%) reduction in IDOL mRNA expression in the liver (Figure 4F). This partial IDOL knockdown was associated with a blunted effect of the LXR agonist on plasma LDL cholesterol levels on day 7 of treatment (Figure 4G–I). There was substantial variability in the individual plasma LDL cholesterol values, prompting us to evaluate the effect of LXR agonist treatment for each individual animal (Figure 4H). Although the difference in average plasma LDL cholesterol levels between groups did not reach statistical significance, the average percent change in LDL cholesterol for each individual in response to LXR agonist was significantly different (p < 0.05) (Figure 4G). Moreover, the change in LDL cholesterol and total cholesterol in response to GW3965 over the entire course of LXR agonist treatment (Day 3, 5, 7) was also significantly different between vehicle and IDOL ASO-treated animals (Figure 4I). Plasma and hepatic triglyceride levels, and hepatic cholesterol levels were not significantly different between groups (Figure 4G and S4E). HPLC analysis of plasma cholesterol distributions at the beginning and at the end of the study (pre-treatment baseline versus Day 7 GW treatment) revealed a smaller change from baseline in the IDOL-ASO group in both the LDL and VLDL fractions. There was no change in hepatic mRNA expression of LDLR or HMGCR suggesting that IDOL inhibition can modulate plasma LDL levels independent of those parameters (Figure 4F).
Discussion
We have shown here that regulation of IDOL expression by the LXR signaling pathway has distinct consequences for plasma lipid levels in mice and cynomolgus monkeys. Ligand activation of LXR more strongly induces hepatic IDOL expression in primates compared to mice. As a consequence, LXR agonists raise plasma LDL levels in primates but not mice. These results have implications for our understanding of the role of LXR nuclear receptors in orchestrating lipid homeostasis as well as the ongoing effort to target LXR or its downstream effectors in human disease.
Murine models can be valuable tools for the development of new drug strategies. However, differences in hepatic lipid metabolism between mice and primates complicate the use of mice as models for human therapeutics. In the case of the LXR–IDOL pathway, the study of mice was not predictive of the in vivo relevance of this pathway in primates. The fact that mice carry most of their cholesterol on HDL makes it difficult to assess LDL metabolism in this model without dietary or genetic manipulations. In the future, it will be interesting to examine the potential effect of increased peripheral LDLR expression in a more humanized mouse model such as CETP transgenic mice (Agellon et al., 1991). Furthermore, although our data indicate that the LXR-IDOL pathway is not a strong determinant of LDLR pathway activity in mouse liver, IDOL clearly affects LDLR protein levels in a number of peripheral tissues. The question of whether altered IDOL target expression in peripheral tissues can influence cellular or systemic cholesterol metabolism will be an interesting one for future investigation.
Genome-wide association studies have linked polymorphisms at the IDOL locus to plasma LDL cholesterol levels in humans (Chasman et al., 2012; Teslovich et al., 2010; Weissglas-Volkov et al., 2011). Most recently, individuals with a complete loss-of-function mutation in the IDOL gene were identified and shown to have reduced levels of LDL cholesterol, suggestive of increased LDLR-dependent LDL clearance (Sorrentino et al., 2013a). Our results help to explain why genome-wide association studies have linked variation in the IDOL locus with changes in human plasma lipid levels, despite the low activity of the LXR-IDOL pathway in mouse liver.
Synthetic LXR agonists are powerful regulators of reverse cholesterol transport that inhibit the development of atherosclerosis in mice (Naik et al., 2006; Tangirala et al., 2002). The development of LXR agonists as therapeutics, however, has been hampered by the induction of hepatic lipogenesis, an undesirable side effect of LXR agonists. Our results identify elevation of plasma LDL cholesterol levels as an additional unexpected complication of systemic LXR agonism in primates. Importantly, this effect was not predicted from preclinical studies in rodents due to the species-specific differences in hepatic activity of the LXR-IDOL pathway.
Three pathways are known to exert a strong influence on LDLR expression: SREBP, PCSK9, and IDOL (Abifadel et al., 2003; Brown and Goldstein, 1986; Cohen et al., 2005; Zelcer et al., 2009). Statins, which act indirectly through SREBP, and antibodies directed against PCSK9, which are in late-stage clinical development, increase LDLR (Stein and Raal, 2014; Young and Fong, 2012). As an independent mechanism for regulation of the LDLR, the LXR–IDOL pathway could represent a potential target for the development of a new class of lipid-lowering drugs. Interestingly, statins have additive effects with IDOL inhibition on LDLR expression in cells (Scotti et al., 2011).
Our results establish that changing the activity of the LXR– IDOL pathway in nonhuman primates changes LDLR protein expression and plasma LDL levels. An unresolved issue at this stage is whether IDOL inhibition would be predicted to lower plasma LDL cholesterol in humans in the absence of LXR activation. Although there was a trend for reduction in cholesterol levels in response to IDOL ASOs (Figure 4D), the high variability in plasma lipids in our cohorts, as well as the modest degree of knockdown of IDOL expression by our ASOs (Figure 4F), prevents us from being able to answer the question at this time. Given the variability in plasma LDL levels in our ASO study, it would have been interesting to assess LDL protein levels in individual animals before and after ASO treatment. Unfortunately, pre-treatment liver biopsies were not obtained under the current experimental protocol. Nevertheless, these observations support further investigation of the LXR– IDOL pathway as a potential target for the modulation of LDL cholesterol levels. Our results further suggest that IDOL inhibition may mitigate undesirable effects of synthetic LXR agonists on plasma LDL cholesterol levels in humans.
Experimental Procedures
Cell Culture and LXR agonists
The LXR agonists GW3965 and T0901317 were synthesized as described (Collins et al., 2002; Collins, 2002; Li, 2000). Primary peritoneal macrophages were collected four days after thioglycollate injection (Hong et al., 2011). Primary murine hepatocytes were collected as described (Rong et al., 2013). Human and nonhuman primate primary hepatocytes were purchased from Corning and Life Technologies. Cells were placed in 0.5% serum or 10% LPDS supplemented with 5 μM simvastatin plus 100 μM mevalonic acid for 5 h and then treated overnight with GW3965. ASOs were transfected using Lipofectamine or FUGENE 9. siOligos targeting human LXRα or control were purchased from ThermoScientific and transfected using Dharmafect 4.
RNA Analysis
Total RNA was isolated from tissues using TRIzol (Invitrogen) reverse transcribed with random hexamers with the Taqman Reverse Transcription Reagents Kit (Applied Biosystems). Real-time quantitative PCR assays were performed using an Applied Biosystems 7900HT sequence detector. For human and mouse experiments, Sybergreen (Diagenode) was used. Gene-specific Applied Biosystems Taqman assays and iTaq were used to quantify nonhuman primate gene expression. Results show averages of duplicate experiments normalized to 36B4 or Gapdh. The primer sequences and Taqman assay IDs are available upon request. The microarray profiling studies were performed with Affymetrix Human U133 Plus 2.0 Array chips by the UCLA Clinical Microarray Core (GEO accession # GSE61381).
Antibodies and Immunoblots
Total cell lysates were prepared in RIPA buffer supplemented with protease inhibitors (Roche Molecular Biochemicals) and PMSF. Samples (10–40 μg) were separated on NuPAGE Bis-Tris gels (Invitrogen) and transferred to nitrocellulose. Membranes were probed with the following antibodies: LDLR (1:1000; Cayman Chemical or AbCam), ABCA1 (1:1000; Novus), actin (1:10,000; Sigma), and tubulin (1:3000 Calbiochem). Horseradish peroxidase (HRP)–conjugated secondary antibodies (Zymed) were used, and antibody binding was visualized by chemiluminescence (ECL Plus GE Healthcare). Blots were quantified by densitometry with ImageJ software (version 1.42q; National Institutes of Health).
Murine Studies
Mice were housed under pathogen-free conditions in a temperature-controlled room with a 12-h light/dark cycle. Idol−/− mice were generated through injection of previously described SV129 Idol-targeted ES cells (Scotti et al., 2011) into C57BL/6 blastocysts. Mice and were studied on a mixed SV129/C57BL/6 background as well as after backcrossing seven generations to C57BL/6 mice. Mice were fed a standard chow diet or a western diet (Research Diets #D12079B). For ligand treatment studies, mice were gavaged with either vehicle or GW3965 or T0901317 (40 mg/kg) daily for 3 days. Tissues were harvested 6 h after the last gavage. All mouse experiments were approved by the UCLA Animal Care and Research Advisory Committee.
Antisense Oligonucleotides
Antisense oligonucleotides (ASO) were designed to target cynomolgus monkey IDOL mRNA (ASO1, ASO 549069, 5′–CTCCATGACCATACAA–3′ and ASO2, ASO 549127, 5′–AAGTTTAAGTAACCCA–3′). ASOs consisted of a chimeric 16-mer phosphorothioate backbone containing 2-O-methoxyethyl groups at positions 1, 2, and 16 and 2′4′-constrained ethyl BNAs at positions 3, 14, and 15 targeting cynomolgus monkey IDOL sequences. The ASOs were synthesized and purified on an automated DNA synthesizer using phosphoramidite chemistry (Murray et al., 2012).
Nonhuman Primate Studies
All experiments were approved by the WFUHS Institutional Animal Care and Use Committee (IACUC). Male cynomolgus were pair-housed when possible in climate-controlled conditions with 12-h light/dark cycles. Monkeys were provided water ad libitum and fed twice a day with high-fat, moderate cholesterol semisynthetic diet. For the 7-day LXR agonist treatment study, 10 monkeys were fasted overnight, anesthetized with 5 mg/kg ketamine and 0.0075 mg/kg dexmedetomidine and gavaged with either vehicle (1% hydroxypropyl methylcellulose) or 10 mg/kg. The anesthesia was then reversed with atipamezole (0.075 mg/kg). The treatment regimen was conducted daily for 7 days. On day 7, fasted monkeys were anesthetized with 25 mg/kg ketamine and isoflurane. The sedated monkeys were euthanized by exsanguination followed by whole-body perfusion with saline and tissues were isolated. For the 2-day LXR agonist treatment study, 10 monkeys were fasted, anesthetized, and gavaged with either vehicle or 10 mg/kg GW3965. Exactly 6 h after dosing on day 2, fasted monkeys were euthanized and tissues collected.
For in vivo validation of the IDOL ASOs, monkeys were fed the high-fat diet for 9 weeks and subcutaneously injected once weekly for 9 weeks with vehicle, 20 mg/kg ASO1, or 20 mg/kg ASO2 (n = 4/group). Two days after the 9th treatment, fasted monkeys were euthanized and tissues were collected. For the combined LXR agonist and ASO treatment study, 16 monkeys were fed high-fat diet for 4 weeks and subcutaneously injected once during study week 1 with vehicle or 40 mg/kg ASO2 and once during study week 2–9 with vehicle or 30 mg/kg ASO2. Starting on study week 8, monkeys were fasted overnight, anesthetized as above and gavaged with 10 mg/kg GW3965. The treatment regimen was conducted once daily for 8 days. On the 8th day, fasted monkeys were euthanized exactly 6 h after dosing and tissues were collected.
Lipid and Lipoprotein Analysis
Lipoprotein cholesterol distributions were determined by on-line, high-performance gel-filtration chromatography with Infinity Cholesterol reagent (Thermo) at the Wake Forest University Lipid Core. Plasma total cholesterol and triglyceride were measured using the Cholesterol Reagent Set (Pointe Scientific) and Triglyceride Reagent and Free Glycerol Reagent (Sigma). Serum levels of PCSK9 were measured using the Human PCSK9 Quantikine ELISA kit from R&D Systems. For apolipoprotein analysis, an equal volume of plasma from each monkey was pooled and separated on a Superose 6 10/300 GL column (GE Healthcare). Fractions were resolved on SDS-PAGE gels, the proteins were transferred to a nitrocellulose membrane, and apolipoproteins were detected with goat anti-monkey affinity-purified antibodies.
Statistical Analysis
Statistics were performed using Student’s t test (2 groups) or ANOVA (>2 groups), with post-hoc tests to compare to the control group. Data are presented as means ± SEM or means ± SD as indicated and considered statistically significant at p < 0.05.
Supplementary Material
Acknowledgments
We are grateful for the technical contributions of Kathryn L. Kelley and Janet K. Sawyer at Wake Forest University School of Medicine, Jon Salazar at UCLA and Matthew Hallowell from Tulane University. This work was supported by NIH grants HL088528, HL111932 (R.E.T), HL066088 (P.T.) and HL090553 (S.G.Y. and P.T.) and ADA grant 1-14-JF-33 (C.H.). P.T. in an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
C.H. designed and performed experiments, interpreted results and wrote the manuscript. S.M.M., A.L.M., and M.G designed and performed experiments and interpreted results. J.D.L., L.C., E.S., R.B., J.K., and R.K.T. performed experiments. B.T.C. and M.E.J. synthesized reagents. L.F., R.L., and S.G.Y. designed experiments and interpreted results. R.E.T. and P.T. designed experiments, interpreted results and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- Agellon LB, Walsh A, Hayek T, Moulin P, Jiang XC, Shelanski SA, Breslow JL, Tall AR. Reduced high density lipoprotein cholesterol in human cholesteryl ester transfer protein transgenic mice. J Biol Chem. 1991;266:10796–10801. [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Calkin AC, Goult BT, Zhang L, Fairall L, Hong C, Schwabe JW, Tontonoz P. FERM-dependent E3 ligase recognition is a conserved mechanism for targeted degradation of lipoprotein receptors. Proc Natl Acad Sci U S A. 2011;108:20107–20112. doi: 10.1073/pnas.1111589108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman DI, Giulianini F, MacFadyen J, Barratt BJ, Nyberg F, Ridker PM. Genetic determinants of statin-induced low-density lipoprotein cholesterol reduction: the Justification for the Use of Statins in Prevention: an Intervention Trial Evaluating Rosuvastatin (JUPITER) trial. Circ Cardiovasc Genet. 2012;5:257–264. doi: 10.1161/CIRCGENETICS.111.961144. [DOI] [PubMed] [Google Scholar]
- Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- Collins JL, Fivush AM, Watson MA, Galardi CM, Lewis MC, Moore LB, Parks DJ, Wilson JG, Tippin TK, Binz JG, et al. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J Med Chem. 2002;45:1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- Collins JL, Fivush AM, Maloney PR, Stewart EL, Willson TM. Preparation of substituted phenylacetamides and benzamides as agonists for Liver X receptors (LXR) 2002. [Google Scholar]
- Grefhorst A, Elzinga BM, Voshol PJ, Plosch T, Kok T, Bloks VW, van der Sluijs FH, Havekes LM, Romijn JA, Verkade HJ, et al. Stimulation of lipogenesis by pharmacological activation of the liver X receptor leads to production of large, triglyceride-rich very low density lipoprotein particles. J Biol Chem. 2002;277:34182–34190. doi: 10.1074/jbc.M204887200. [DOI] [PubMed] [Google Scholar]
- Honzumi S, Shima A, Hiroshima A, Koieyama T, Ubukata N, Terasaka N. LXRalpha regulates human CETP expression in vitro and in transgenic mice. Atherosclerosis. 2010;212:139–145. doi: 10.1016/j.atherosclerosis.2010.04.025. [DOI] [PubMed] [Google Scholar]
- Laffitte BA, Joseph SB, Walczak R, Pei L, Wilpitz DC, Collins JL, Tontonoz P. Autoregulation of the human liver X receptor alpha promoter. Mol Cell Biol. 2001;21:7558–7568. doi: 10.1128/MCB.21.22.7558-7568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Medina JC, Hasegawa H, Cutler ST, Liu J, Zhu L, Shan B, Lustig K. Preparation of bis(trifluoromethyl)hydroxymethylbenzenesulfonamides, -ureas, and -carbamates as liver X receptor modulators. 2000. [Google Scholar]
- Luo Y, Tall AR. Sterol upregulation of human CETP expression in vitro and in transgenic mice by an LXR element. J Clin Invest. 2000;105:513–520. doi: 10.1172/JCI8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S, Ittig D, Koller E, Berdeja A, Chappell A, Prakash TP, Norrbom M, Swayze EE, Leumann CJ, Seth PP. TricycloDNA-modified oligo-2′-deoxyribonucleotides reduce scavenger receptor B1 mRNA in hepatic and extra-hepatic tissues--a comparative study of oligonucleotide length, design and chemistry. Nucleic Acids Res. 2012;40:6135–6143. doi: 10.1093/nar/gks273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik SU, Wang X, Da Silva JS, Jaye M, Macphee CH, Reilly MP, Billheimer JT, Rothblat GH, Rader DJ. Pharmacological activation of liver X receptors promotes reverse cholesterol transport in vivo. Circulation. 2006;113:90–97. doi: 10.1161/CIRCULATIONAHA.105.560177. [DOI] [PubMed] [Google Scholar]
- Peet DJ, Janowski BA, Mangelsdorf DJ. The LXRs: a new class of oxysterol receptors. Curr Opin Genet Dev. 1998;8:571–575. doi: 10.1016/s0959-437x(98)80013-0. [DOI] [PubMed] [Google Scholar]
- Repa JJ, Turley SD, Lobaccaro JA, Medina J, Li L, Lustig K, Shan B, Heyman RA, Dietschy JM, Mangelsdorf DJ. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- Scotti E, Calamai M, Goulbourne CN, Zhang L, Hong C, Lin RR, Choi J, Pilch PF, Fong LG, Zou P, et al. IDOL stimulates clathrin-independent endocytosis and multivesicular body-mediated lysosomal degradation of the low-density lipoprotein receptor. Mol Cell Biol. 2013;33:1503–1514. doi: 10.1128/MCB.01716-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti E, Hong C, Yoshinaga Y, Tu Y, Hu Y, Zelcer N, Boyadjian R, de Jong PJ, Young SG, Fong LG, et al. Targeted disruption of the idol gene alters cellular regulation of the low-density lipoprotein receptor by sterols and liver x receptor agonists. Mol Cell Biol. 2011;31:1885–1893. doi: 10.1128/MCB.01469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino V, Fouchier SW, Motazacker MM, Nelson JK, Defesche JC, Dallinga-Thie GM, Kastelein JJ, Kees Hovingh G, Zelcer N. Identification of a loss-of-function inducible degrader of the low-density lipoprotein receptor variant in individuals with low circulating low-density lipoprotein. Eur Heart J. 2013a;34:1292–1297. doi: 10.1093/eurheartj/ehs472. [DOI] [PubMed] [Google Scholar]
- Sorrentino V, Nelson JK, Maspero E, Marques AR, Scheer L, Polo S, Zelcer N. The LXR-IDOL axis defines a clathrin, caveolae, and dynamin-independent endocytic route for LDLR internalization and lysosomal degradation. J Lipid Res. 2013b;54:2174–84. doi: 10.1194/jlr.M037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein EA, Raal F. Reduction of Low-Density Lipoprotein Cholesterol by Monoclonal Antibody Inhibition of PCSK9. Annu Rev Med. 2014;65:417–431. doi: 10.1146/annurev-med-022613-090402. [DOI] [PubMed] [Google Scholar]
- Tangirala RK, Bischoff ED, Joseph SB, Wagner BL, Walczak R, Laffitte BA, Daige CL, Thomas D, Heyman RA, Mangelsdorf DJ, et al. Identification of macrophage liver X receptors as inhibitors of atherosclerosis. Proc Natl Acad Sci U S A. 2002;99:11896–11901. doi: 10.1073/pnas.182199799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissglas-Volkov D, Calkin AC, Tusie-Luna T, Sinsheimer JS, Zelcer N, Riba L, Tino AM, Ordonez-Sanchez ML, Cruz-Bautista I, Aguilar-Salinas CA, et al. The N342S MYLIP polymorphism is associated with high total cholesterol and increased LDL receptor degradation in humans. J Clin Invest. 2011;121:3062–3071. doi: 10.1172/JCI45504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SG, Fong LG. Lowering plasma cholesterol by raising LDL receptors--revisited. N Engl J Med. 2012;366:1154–1155. doi: 10.1056/NEJMe1202168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Fairall L, Goult BT, Calkin AC, Hong C, Millard CJ, Tontonoz P, Schwabe JW. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 2011;25:1262–1274. doi: 10.1101/gad.2056211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.