Dear Editor,
What does the evolutionary origin of a plant protein tell about its subcellular localization? Naively thinking, one would assume that plant proteins that were originally encoded in the endosymbiont genome are targeted to the chloroplast. However, published data seem to support only a loose link between evolutionary origin and subcellular localization. About half of the Arabidopsis proteins with a detectable cyanobacterial ortholog are targeted to subcellular compartments other than the chloroplast (Martin et al., 2002). Here we show that the naive view is valid when considering the full phylogenetic profile of plant genes with cyanobacterial orthologs. Genes that are present also in non-photosynthesizing lineages presumably trace back to a primordial eukaryote. They have been inherited largely vertically and show no evidence for a preferential chloroplast targeting. In contrast, genes that are among eukaryotes confined to lineages that have undergone primary or secondary endosymbiosis are likely to be of true cyanobacterial origin. They are indeed mostly targeted to chloroplasts in plants.
Unraveling the composition of the chloroplast proteome is crucial for understanding the function and integration of this organelle into the metabolic network of photosynthesizing organisms. Besides photosynthesis, chloroplasts play essential roles in the biosynthesis of amino acids and vitamins, lipids and isoprenoids, the storage of fixed carbon, and other processes. Thus, there is a strong need to tightly coordinate chloroplast activities with the overall metabolism of the cell, and accordingly different retrograde chloroplast-to-nucleus signaling pathways have evolved (Jarvis and Lopez-Juez, 2013). During evolution—after the initial uptake of a cyanobacterium by a heterotrophic host—the cyanobacterium evolved into the contemporary plastid and most of the originally cyanobacterial genes were transferred into the nuclear genome of the host (Martin et al., 2002). Current estimates suggest that 4300–4500 Arabidopsis proteins were acquired from the ancestral plastid. This genetic reorganization created the necessity to establish an effective ‘back-transport’ of the encoded proteins to their original location using an N-terminal signal sequence called cTP (chloroplast transit peptide) (Jarvis and Lopez-Juez, 2013).
Technological advances in high-throughput genome sequencing and proteomics have boosted the analysis of the chloroplast proteome. To date, the curated reference plastid proteomes for maize and Arabidopsis (http://ppdb.tc.cornell.edu) comprise 1564 and 1559 proteins, respectively. These numbers are contrasted by those obtained from bioinformatics analysis of the sequenced genomes. About twice as many proteins in these species carry a cTP. This already suggests that a considerable number of chloroplast proteins still remain to be discovered. Unfortunately, the presence of a cTP provides only ambiguous evidence to infer chloroplast localization. For example, of 1325 experimentally identified chloroplast proteins in Arabidopsis, 14% lack an identifiable cTP at their N-terminus (Zybailov et al., 2008). Extrapolations from systematic studies revealed that the fraction of chloroplast proteins that lack a cTP could be about 11% of the total chloroplast proteome (Armbruster et al., 2009). In turn, targeting prediction algorithms, such as TargetP (www.cbs.dtu/dk/services/TargetP), do not consider N-terminal protein acylation, which can overwrite chloroplast targeting signals resulting in deviating subcellular localizations (Stael et al., 2011). Thus, alternative approaches need to be sought to complement existing information in the prediction of chloroplast targeting. Directed experimental approaches focusing on signaling components had only limited success (Bayer et al., 2011, 2012), possibly due to the low abundance of such proteins in the chloroplast.
Considering the evolutionary origin of plant proteins has also been proposed to help predicting their subcellular localization. An orthogenomics approach using 17 species identified 56 Arabidopsis proteins of endosymbiotic origin of which 54 were targeted to chloroplasts (Ishikawa et al., 2009). However, the small number of analyzed proteins makes this finding hard to generalize. Moreover, it is contrasted by a previous notion based on a much larger data set that only about half of the Arabidopsis proteins with putative cyanobacterial origins are targeted to the chloroplast (Martin et al., 2002). Thus, it remains still unclear to what extent the endosymbiotic origin of a plant protein provides information about its localization.
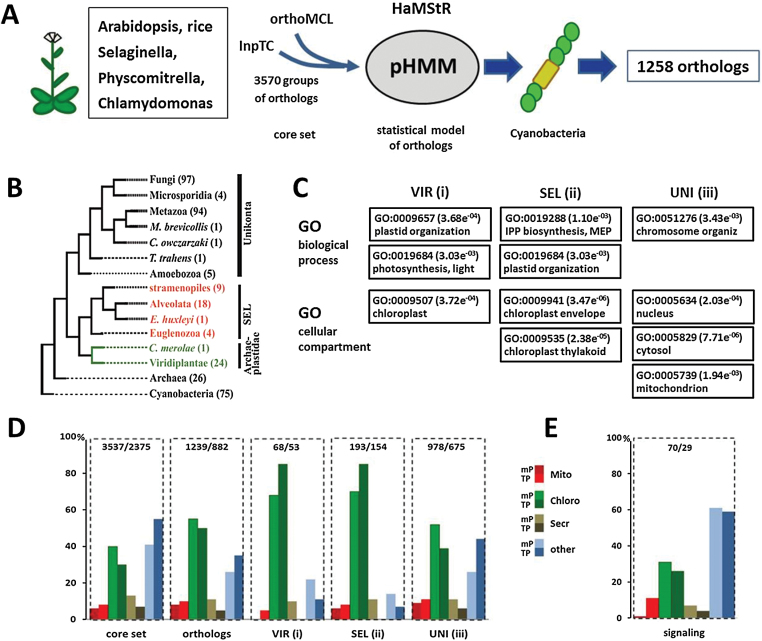
Here, we addressed this question taking advantage of the massively increased number of complete genome sequences across the tree of life. We identified higher plant proteins of cyanobacterial origin using the approach illustrated in the flow scheme in Figure 1A (see also Supplementary Data online). First, we extracted a non-redundant core set of 3570 plant orthologs present in the genomes of Arabidopsis thaliana (At), Oryza sativa (Os), Selaginella moellendorfii (Sm), Physcomitrella patens (Pp), and Chlamydomonas reinhardtii (Cr). This core set was subsequently used as input for a HaMStR ortholog search (http://sourceforge.net/projects/hamstr) (Ebersberger et al., 2009) in 260 eukaryotes, 26 archaea, and 75 cyanobacteria (Supplemental Table 1). For 1750 proteins, we could trace ortholog candidates in at least one cyanobacterial species. However, orthology prediction over these evolutionary distances is hard, posing the risk of including false positives (Ebersberger et al., 2014). To reduce the risk that spurious orthology assignments confound our data, we considered only those 1258 Arabidopsis proteins with a detectable ortholog in at least 10% of the Cyanobacteria for further analysis (Supplemental Table 2).
Figure 1.
Analysis Work Flow.
(A) A plant core set comprising 3570 proteins was extracted from Arabidopsis, rice, Selaginella, Physcomitrella, and Chlamydomonas to build a profile hidden Markov model (pHMM) for searching orthologs in 75 cyanobacterial genomes as described in the Supplementary Methods online.
(B) Phylogenetic profiles of the identified orthologs were established across 260 eukaryotes, 26 archaea, and 75 cyanobacteria as described online. Species of the green lineage are depicted in green and species that have undergone secondary endosymbiosis are depicted in red. Genes with orthologs in at least eight cyanobacterial species (1258 orthologs) were then further divided into three categories according to the presence of orthologs only in (1) Viridiplantae (VIR), (2) in Viridiplantae and the Secondary Endosymbiosis Lineage (SEL), and (3) also in unikonts (UNI).
(C) Distribution patterns for gene ontology terms linked to biological processes and cellular compartments in the three categories. IPP, isopentenyl diphosphate; MEP, Methylerythritol Phosphate pathway of isoprenoid biosynthesis.
(D) Fraction of proteins assigned to four different subcellular localizations: Mito, mitochondrion; Chloro, chloroplast; Secr, secretory pathway; other, any other location. mP and TP denote predictions based on Plant-mPLoc and TargetP, respectively. Only proteins with a TargetP confidence score of 3 or better were considered. The total number of proteins with predicted localization is indicated on top of each histogram (first: Plant-mPLoc, second: TargetP).
(E) Targeting prediction for the signaling proteins with cyanobacterial orthologs (Supplemental Table 3) as described for (D).
In the next step, we sought to identify what fraction of the genes that are shared between plants and cyanobacteria have been acquired via endosymbiotic gene transfer, and which represent genes with an evolutionary ancestry predating the emergence of phototrophic eukaryotes. To this end, we distinguished three categories (Figure 1B): (1) 68 genes restricted to the green lineage (Viridiplantae, VIR) which were acquired presumably via the primary endosymbiotic event; (2) 198 genes present in the green lineage and in species that have undergone secondary endosymbiosis (Secondary Endosymbiosis Lineage, SEL); and (3) 992 genes that are found throughout the eukaryotic tree including the unikonts (UNI). Most likely, these genes are evolutionary ancient and followed a vertical line of inheritance.
We next focused on the distribution of gene ontology (GO) terms describing protein function and localization (Figure 1C). GO terms associated with chloroplast-specific processes like photosynthesis and isoprenoid biosynthesis via the MEP pathway, as well as plastid organization, are significantly over-represented in categories VIR and SEL. Not unexpectedly, most proteins participating in photosynthetic core processes, chloroplast metabolism, and maintenance are found in SEL. Additionally, this category harbors many proteins with a regulatory function like the ABC1 kinases, GTP-binding proteins, or proteases like FtsH or Clp subunits (Supplemental Table 2). In contrast, proteins acting in the nucleus, the cytosol, and mitochondria are enriched in category UNI. Among these are proteins involved in chromosome organization and house-keeping like metabolic enzymes or protein synthesis (Supplemental Table 2). So far, we have shown that proteins with a chloroplast-related function are preferentially found in categories VIR and SEL. However, this does not yet provide information about what fraction of proteins in these two categories are localized in chloroplasts. To address this question, we predicted the subcellular localization of the 3570 proteins in our core set using two complementary tools: Plant-mPLoc (www.csbio.sjtu.edu.cn/bioinf/plant-multi) and TargetP (www.cbs.dtu/dk/services/TargetP). Plant-mPLoc integrates GO annotation, functional domain content, and sequence similarity scoring to assign proteins to 12 different subcellular localizations. TargetP uses neuronal networks to identify putative targeting signals within the query protein. Forty percent (TargetP: 30%) of the core set is predicted to be targeted to chloroplasts (Figure 1D). When we now focus on the subset of 1258 proteins with identifiable orthologs in cyanobacteria, this number does—not unexpectedly—increase to 55% (50%). This closely resembles previous findings (Martin et al., 2002) and, when taken at face value, this could suggest that about half of originally cyanobacterial proteins have been neo-localized to compartments other than the chloroplast. However, the scenario completely changes when we take our phylogenetic profiles into account: the majority of proteins in categories VIR and SEL have a predicted chloroplast localization (Plant-mPLoc: 70%; TargetP: 85%), while this applies to only 51% (TargetP: 39%) of the proteins in category UNI. In summary, our results show that plant proteins of true cyanobacterial origin have indeed a high probability for functioning in chloroplasts, even when their role is not in photosynthesis. Thus, evolutionary descent is indeed a fairly good predictor of chloroplast targeting.
To investigate the evolutionary origins of intracellular communication, we concentrated on the 70 signaling factors (protein kinases and -phosphatases, GTP- and Ca2+-binding proteins) in the set of 1258 plant proteins with cyanobacterial orthologs (Supplemental Table 3). At first sight, one would expect a prevalence of bacterial-type signaling factors involved in chloroplast communication. However, we find only nine of these genes in our data set. They represent key factors of chloroplast signaling: STN8, two ABC1 kinases, three GTP-binding proteins, the ABA biosynthesis protein ABA4, and the two retrograde signaling proteins GUN4 and GUN5. The other 61 proteins include a considerable number of typical eukaryote-type serine/threonine kinases such as mitogen-activated protein (MAP) kinases or a Ca2+-dependent protein kinase (CDPK). Based on targeting prediction and database searches (Supplemental Table 3), only 31% (TargetP: 26%) of these proteins are targeted to the chloroplast and the majority (61%; TargetP 59%) is targeted to other compartments (cytosol, membranes, nucleus, Figure 1E). Experimental localization of 10 candidates confirmed that indeed two-thirds are not targeted to the chloroplast (not shown). These findings nicely integrate with our results from the phylogenetic profiling. The nine proteins involved in chloroplast signaling belong to the category SEL and are likely of true cyanobacterial origin. The remaining 61 proteins belong to category UNI with orthologs present in all eukaryotes indicating that they represent evolutionary very ancient proteins. A connection to chloroplast signaling is therefore not necessarily given.
In summary, our analysis demonstrates that the presence of a cyanobacterial ortholog for a plant protein alone does not reliably indicate its chloroplast localization. Only a phylogenetic profiling, distinguishing horizontally acquired genes from evolutionary ancient and vertically inherited genes, can form the basis for informed predictions about the localization and potentially also the function of the corresponding proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work has been supported by the EU Marie Curie project CALIPSO (GA 2013–607607) and the Austrian Research Foundation (FWF) project P-25359-B21 to M.T. I.E. acknowledges financial support by the Biodiversity and Climate Research Center Frankfurt (BIK-F).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Wolfgang Löffelhardt (University of Vienna) for critical reading and comments on the manuscript. No conflict of interest declared.
REFERENCES
- Armbruster U., Hertle A., Makarenko E., Zuhlke J., Pribil M., Dietzmann A., Schliebner I., Aseeva E., Fenino E., Scharfenberg M., et al. (2009). Chloroplast proteins without cleavable transit peptides: rare exceptions or a major constituent of the chloroplast proteome? Mol. Plant. 2, 1325–1335. [DOI] [PubMed] [Google Scholar]
- Bayer R.G., Stael S., Csaszar E., Teige M. (2011). Mining the soluble chloroplast proteome by affinity chromatography. Proteomics. 11, 1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer R.G., Stael S., Rocha A.G., Mair A., Vothknecht U.C., Teige M. (2012). Chloroplast-localized protein kinases: a step forward towards a complete inventory. J. Exp. Bot. 63, 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger I., Simm S., Leisegang M.S., Schmitzberger P., Mirus O., von Haeseler A., Bohnsack M.T., Schleiff E. (2014). The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res. 42, 1509–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger I., Strauss S., von Haeseler A. (2009). HaMStR: profile hidden Markov model based search for orthologs in ESTs. BMC Evol. Biol. 9, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Fujiwara M., Sonoike K., Sato N. (2009). Orthogenomics of photosynthetic organisms: bioinformatic and experimental analysis of chloroplast proteins of endosymbiont origin in Arabidopsis and their counterparts in Synechocystis. Plant Cell Physiol. 50, 773–788. [DOI] [PubMed] [Google Scholar]
- Jarvis P., Lopez-Juez E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. [DOI] [PubMed] [Google Scholar]
- Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., Leister D., Stoebe B., Hasegawa M., Penny D. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. U S A. 99, 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stael S., Bayer R.G., Mehlmer N., Teige M. (2011). Protein N-acylation overrides differing targeting signals. FEBS Lett. 585, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PloS One. 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.