Abstract
Testosterone is an important hormone that has been shown to have sex-specific links to fitness in numerous species. Although testosterone concentrations vary substantially between individuals in a population, little is known about its heritable genetic basis or between-sex genetic correlations that determine its evolutionary potential. We found circulating neonatal testosterone levels to be both heritable (0.160 ± 0.064 s.e.) and correlated between the sexes (0.942 ± 0.648 s.e.) in wild red deer calves (Cervus elaphus). This may have important evolutionary implications if, as in adults, the sexes have divergent optima for circulating testosterone levels.
Keywords: androgens, interindividual variation, quantitative genetics, reproductive endocrinology, selection, steroid hormones
1. Introduction
Hormone concentrations vary between individuals in a population [1]; however, the causes of this variation remain poorly understood despite many hormones being important in mediating fitness-related traits [2–4]. Circulating testosterone concentrations are often associated with male fitness (see reviews in [4,5]), although variation in levels, and subsequent fitness consequences, are now also recognized as important in females [3]. Although most studies of interindividual testosterone variation have focused on adults, early-life concentrations may also be important for juvenile fitness [6,7].
If selection on testosterone concentration is to generate an evolutionary response (a genetic change in the population mean), then it must have a heritable genetic basis. Evidence suggests that testosterone levels are heritable in humans (average h2 = 0.45, range = 0–0.91, electronic supplementary material, table S1) and captive populations of several taxa (electronic supplementary material, table S1), but levels of genetic variance for testosterone concentration have rarely been investigated in the wild, where heritability estimates might be lower because individuals are subjected to greater environmental variation [8]. Furthermore, most studies focus on adult concentrations despite substantial age-related variation in heritability estimates in humans (electronic supplementary material, table S1). Such studies also tend to estimate heritability using traditional methods that rely on paired-relative relationships such as parent–offspring; however, these risk inflating heritability estimates due to shared environmental/maternal effects [9]. ‘Animal models’ allow the use of relatedness information across complex pedigrees, which both reduces the potential for bias and allows more efficient use of the data typical of natural populations (see [9,10] and references therein).
Despite growing interest in interindividual variation in female testosterone levels [3], sex differences in genetic variation for this trait remain understudied, although females might be expected to have lower genetic variance [11] because their circulating testosterone levels also tend to be lower [3,7]. The genetic correlation between the levels of testosterone in the sexes is also poorly understood, with only one direct estimate in humans, where a positive cross-sex genetic correlation was recorded [12]. A positive correlation has also been inferred in adult mammals [13,14], birds [3,15] and fish [16], but not directly estimated. In adults, selection would be expected to favour higher testosterone concentrations in males, and lower concentrations in females [13,15]. Where these sexually antagonistic selection pressures exist, a shared genetic architecture (i.e. a positive genetic correlation [17]) would constrain either sex from reaching their optimum. Testosterone levels may also be important to sex-specific trait development in early life (see [18] and references therein). For example, neonatal male rats with suppressed testosterone develop feminized adult behaviour, whereas neonatal females exposed to high levels of testosterone are less receptive to males as adults [19]. This again suggests sexually antagonistic selection pressures that could constrain trait evolution in the presence of a positive genetic correlation.
In a previous study of red deer calves (Cervus elaphus), neonatal testosterone levels showed substantial interindividual variation and weak sex-specific associations with survival [7]. Here, we extend this to investigate (i) the heritability and (ii) the between-sex genetic correlation of circulating neonatal testosterone concentrations in these red deer calves.
2. Material and methods
Our study population of red deer on the Isle of Rum, Scotland has been monitored continuously at the individual level since 1972 [20], allowing a complex pedigree to be constructed based on behavioural and genetic data [21]. Plasma testosterone concentration was measured in 854 0- to 14-day-old calves born 1996–2012 (mean age: 1.90 ± 0.07 s.e. days). Maternity was known for all calves, and paternity had been assigned to 786 calves. In total, the informative pedigree for these calves contained 1293 maternal links and 1137 paternal links, had a modal depth of five generations, and a maximum depth of nine generations. Blood was collected into lithium heparin tubes, centrifuged at 3000 rpm for 10 min and plasma removed and frozen within 24 h of capture. All work was carried out under a UK Home Office licence.
(a). Testosterone extraction and immunoassay
Hormones were extracted and testosterone concentrations measured in duplicate (in pg ml−1) using a commercially available testosterone competitive-binding enzyme immunoassay (EIA) kit (582701, Cayman Chemicals Ltd., USA) (see [7] for further details). Serial dilutions of pooled samples showed high parallelism with the standard curve (p < 0.001), and no samples fell below the limit of detection (5.78 pg ml−1). The intra- and interassay coefficients of variance were 5.13% and 11.66%, respectively.
(b). Statistical analyses
Testosterone concentration was log-transformed (to normalize residuals) and analysed using an animal model in ASReml-R [22]. The following fixed effects were included because they had previously been found to explain substantial interindividual variation [7]: calf age at capture ((i) before or after 24 h old, and (ii) estimated age of calf, in hours, nested within (i)); calf sex; sex of the mother's previous calf (whether or not the maternal sibling born prior to the focal calf was male); sample collection time (decimal hours from 1 (01.00) to 24 (00. 00)); and date of assay.
After accounting for fixed effects, variance in testosterone concentration (VP) was partitioned into three components: additive genetic variance (VA), calf birth year (VYOB) and the remaining unexplained residual variance (VR). Variance due to maternal effects can be distinguished from VA for other traits in this population [9]; however, in this study, maternal identity was tested but excluded because effects were negligible (p = 1). Random effects were tested for significance using likelihood ratio tests of models with and without each effect, assuming a chi-squared distribution with 1 d.f. Heritability (h2) was calculated as VA/VP.
A bivariate model was then fitted to estimate components of (co)variance in male (n = 420) and female (n = 434) testosterone concentrations using the same fixed effects as in the univariate animal model. Because there was greater phenotypic variance in male than female levels [7], a second bivariate model was fitted with the sex-specific (log-transformed) testosterone concentrations standardized to have a variance of 1 by dividing by the standard deviation after correcting for calf age, sex of previous calf, collection time and assay date. Both models were also fitted to estimate sex-specific VA, VYOB and VR, along with associated between-sex correlations for VA and VYOB (note that a cross-sex residual covariance does not exist). To examine whether male and female testosterone concentrations had the same underlying genetic structure, likelihood ratio tests (assuming chi-squared distribution with 2 d.f.) were used to compare a model where VA estimates were allowed to vary between the sexes, with one where the two VA estimates were constrained to be equal and the correlation between them to be 1.
3. Results
Both VA and VYOB explained significant variation in circulating testosterone levels (16% and 4.3%, respectively; table 1). Before standardizing for sex-specific phenotypic differences, VA appeared lower in males than females (table 2a), although the genetic correlation was close to 1, and constraining the model to have equal VA and a between-sex genetic correlation of 1 did not generate a worse model (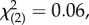 p = 0.971). After standardizing for the sex-specific variance, the genetic correlation remained close to 1 (table 2b) and VA still did not differ significantly between the sexes (
p = 0.971). After standardizing for the sex-specific variance, the genetic correlation remained close to 1 (table 2b) and VA still did not differ significantly between the sexes (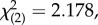 p = 0.336).
p = 0.336).
Table 1.
Components of variance in circulating neonatal testosterone concentrations from univariate models (n = 854 calves, 17 years). See electronic supplementary material, table S2 for details of fixed effects.
| component (s.e.) | p-value | proportion of variance (s.e.) | |
|---|---|---|---|
| VA | 0.049 (0.020) | 0.003 | 0.160 (0.064) |
| VYOB | 0.013 (0.008) | 0.042 | 0.043 (0.024) |
| VR | 0.243 (0.021) | 0.797 (0.076) |
Table 2.
Estimated variance (diagonal), covariance (below diagonal) and correlation (above diagonal) components between male (n = 420) and female (n = 434) testosterone concentrations from bivariate animal models using (a) unstandardized (i.e. with sex-specific phenotypic variances) or (b) sex-standardized data. Standard errors are in parentheses.
| (a) unstandardized |
(b) standardized |
||||
|---|---|---|---|---|---|
| VA | ♂ | ♀ | VA | ♂ | ♀ |
| ♂ | 0.060 (0.044) | 0.628 (0.399) | ♂ | 0.094 (0.115) | 0.942 (0.648) |
| ♀ | 0.044 (0.027) | 0.082 (0.033) | ♀ | 0.157 (0.083) | 0.295 (0.121) |
| VYOB | ♂ | ♀ | VYOB | ♂ | ♀ |
| ♂ | 0.019 (0.014) | 0.986 (0.407) | ♂ | 0.064 (0.045) | 0.998 (0.395) |
| ♀ | 0.015 (0.009) | 0.012 (0.009) | ♀ | 0.050 (0.029) | 0.039 (0.030) |
| VR | ♂ | ♀ | VR | ♂ | ♀ |
| ♂ | 0.266 (0.041) | n.a. | ♂ | 0.871 (0.119) | n.a. |
| ♀ | n.a. | 0.181 (0.029) | ♀ | n.a. | 0.688 (0.109) |
4. Discussion
We present one of the first estimates of testosterone heritability (0.160 ± 0.064 s.e.) in a wild population, and the first (to the best of our knowledge) in young wild animals. This estimate is lower than has been reported for other species (electronic supplementary material, table S1), which may reflect inherent species differences, but could also be due to other factors. While most studies involve adult testosterone, we focused on neonatal levels, which have shown lower h2 estimates in the few other studies to estimate them (e.g. humans; see the electronic supplementary material, table S1, for citations). Different methods of calculating h2 may also give different estimates, with the animal model able to account for shared maternal/environmental effects that may inflate cross-relative studies [10]. Estimates of h2 may be further reduced in wild populations because of exposure to more environmental variability than captive populations [23]. In addition to the effects of VA, there was also a small but significant calf cohort effect (table 1) indicating the importance of environmental effects during fetal development/early life in determining individual testosterone levels. It was surprising that maternal identity did not also account for any variance, as strong maternal effects have been associated with other birth traits within this population [9].
We also provide the first direct estimate of the magnitude of the cross-sex genetic correlation in this trait outside of humans (0.942 ± 0.648 s.e.). Additive genetic variance appeared to be somewhat higher in females than in males, which might allow independent evolution of neonatal female testosterone concentrations if a response to selection were based on genes that did not influence male concentrations. Overall, however, our results concur with previous estimates [12] and inferences from adults of other taxa [3,13–16], suggesting that testosterone concentrations are determined by the same genes in both sexes.
The significantly non-zero heritability estimate means that there is scope for circulating neonatal testosterone concentrations to evolve under appropriate selection pressures; however, the high cross-sex genetic correlation means that sexes could not evolve independently. While neonatal testosterone has previously been linked to early-life survival in the population [7], given that it was only significant for a subset of animals (firstborn males), selection on this trait may be relatively weak. The absence of sexually antagonistic selection via juvenile survival on neonatal levels in this population [7] could be indicative of resolved sexual conflict [24]. There does, however, remain evidence of differences in sexual optima amongst neonates (e.g. rats [20]), and for sexually antagonistic selection in adults [13,15] within other systems. We also do not know how neonatal testosterone levels in any taxa relate to their hormone levels later in life, which may be under stronger selection. While little is known about the magnitude of between-sex genetic correlations in hormone traits overall, the correlation found in this study is unexpectedly strong given that physiological traits typically have the lowest cross-sex genetic correlation for any trait type [25]. Thus, to date, the strength of selection and the degree of sexual antagonism for this trait remain unresolved, and predicting its likely evolutionary dynamics requires further research.
In summary, this study is among the first to use an animal model to estimate hormone heritability (see also [13]), and the first, to the best of our knowledge, in wild mammals. We show circulating neonatal testosterone levels to be both heritable and to have similar genetic architecture in both sexes. There is, therefore, scope for this trait to evolve should the selection pressures exist; however, the sexes cannot evolve independently. This may have important implications if the sexes have divergent optima that could constrain the evolution of this trait.
Supplementary Material
Acknowledgements
We thank Tim Clutton-Brock, field assistants and volunteers, Forbes Howie (Queen's Medical Research Institute, University of Edinburgh) for laboratory support, and Scottish Natural Heritage for permission to work on the Isle of Rum.
Data accessibility
Calf data deposited in the Dryad repository: http://doi.org/10.5061/dryad.qb190. Pedigree data deposited in the Dryad repository: http://doi:10.5061/dryad.7jn8t.
Funding statement
This research was supported by a Natural Environmental Research Council PhD studentship to A.T.P., and an Australian Research Council Future Fellowship to L.E.B.K.
References
- 1.Kempenaers B, Peters A, Foerster K. 2008. Sources of individual variation in plasma testosterone levels. Phil. Trans. R. Soc. B 363, 2722–2723. ( 10.1098/rstb.2007.0001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owen-Ashley N, Hasselquist D, Wingfield J. 2004. Androgens and immunocompetence handicap hypothesis: unravelling direct and indirect pathways of immunosuppression in song sparrows. Am. Nat. 164, 490–505. ( 10.1086/423714) [DOI] [PubMed] [Google Scholar]
- 3.Ketterson E, Nolan V, Jr, Sandell M. 2005. Testosterone in females: mediator of adaptive traits, constraint on sexual dimprohism, or both? Am. Nat. 166, s85–s95. ( 10.1086/444602) [DOI] [PubMed] [Google Scholar]
- 4.Hau M. 2007. Regulation of male traits by testosterone: implications for the evolution of vertebrate life histories. Bioessays 29, 133–144. ( 10.1002/bies.20524) [DOI] [PubMed] [Google Scholar]
- 5.Wingfield J, Lynn S, Soma K. 2001. Avoiding the ‘costs’ of testosterone: ecological bases of hormone–behavior interactions. Brain Behav. 57, 239–251. ( 10.1159/000047243) [DOI] [PubMed] [Google Scholar]
- 6.Cox R, Skelly S, John-Alcer H. 2005. Testosterone inhibits growth in juvenile male eastern fence lizards (Sceloporus undulatus): implications for energy allocation and sexual size dimorphism. Physiol. Biochem. Zool. 78, 531–545. ( 10.1086/430226) [DOI] [PubMed] [Google Scholar]
- 7.Pavitt A, Walling C, Pemberton J, Kruuk L. 2014. Causes and consequences of variation in early life testosterone in a wild population of red deer. Funct. Ecol. 28, 1224–1234. ( 10.1111/1365-2435.12260) [DOI] [Google Scholar]
- 8.Calisi R, Bentley G. 2009. Lab and field experiments: are they the same animal? Horm. Behav. 56, 1–10. ( 10.1016/j.yhbeh.2009.02.010) [DOI] [PubMed] [Google Scholar]
- 9.Kruuk L, Hadfield J. 2007. How to separate genetic and environmental causes of similarity between relatives. J. Evol. Biol. 20, 1890–1903. ( 10.1111/j.1420-9101.2007.01377.x) [DOI] [PubMed] [Google Scholar]
- 10.Kruuk L. 2004. Estimating genetic parameters in natural animal populations using the ‘animal model’. Phil. Trans. R. Soc. B 359, 873–890. ( 10.1098/rstb.2003.1437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyman M, Rowe L. 2014. Male bias in distribution of additive genetic, residual and phenotypic variances of shared traits. Am. Nat. 184, 326–337. ( 10.1086/677310) [DOI] [PubMed] [Google Scholar]
- 12.Hoekstra R, Bartels M, Boomsma D. 2006. Heritability of testosterone levels in 12-year-old twins and its relation to pubertal development. Twin Res. Hum. Genet. 9, 558–565. ( 10.1375/twin.9.4.558) [DOI] [PubMed] [Google Scholar]
- 13.Schroderus E, Jokinen I, Koivula M, Mappes T, Mills S, Oksanen T, Poikonen T. 2010. Intra- and intersexual trade-offs between testosterone and immune system: implications for sexual and sexually antagonistic selection. Am. Nat. 176, E90–E97. ( 10.1086/656264) [DOI] [PubMed] [Google Scholar]
- 14.Mills S, Koskela E, Mappes T. 2012. Intralocus sexual conflict for fitness: sexually antagonistic alleles for testosterone. Proc. R. Soc. B 279, 1889–1895. ( 10.1098/rspb.2011.2340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moller A, Garamszegi L, Gil D, Hurtrez-Bousses S, Eens M. 2005. Correlated evolution of male and female testosterone profiles in birds and its consequences. Behav. Ecol. Sociobiol. 58, 534–544. ( 10.1007/s00265-005-0962-2) [DOI] [Google Scholar]
- 16.Mank J. 2007. The evolution of sexually selected traits and antagonistic androgen expression in Actinopterygiian fishes. Am. Nat. 169, 142–149. ( 10.1086/510103) [DOI] [PubMed] [Google Scholar]
- 17.Lande R. 1987. Genetic correlations between the sexes in the evolution of sexual dimorphism and mating preferences. In Sexual selectional testing the alternatives (eds Bradbury JW, Andersson MB.), pp. 83–94. Chichester, UK: John Wiley & Sons Ltd. [Google Scholar]
- 18.Lummaa V, Pettay J, Russell A. 2007. Male twins reduce fitness of female co-twins in humans. Proc. Natl Acad. Sci. USA 104, 10 915–10 920. ( 10.1073/pnas.0605875104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaff D, Zigmond R. 1971. Neonatal androgen effects on sexual and non-sexual behavior of adult rats tested under various hormone regimes. Neuroendocrinology 7, 129–145. ( 10.1159/000121961) [DOI] [PubMed] [Google Scholar]
- 20.Clutton-Brock T, Guinness F, Albon S. 1982. Red deer: behavior and ecology of two sexes. Chicago, IL: The University of Chicago Press. [Google Scholar]
- 21.Walling C, Pemberton J, Hadfield J, Kruuk L. 2010. Comparing parentage inference software: reanalysis of a red deer pedigree. Mol. Ecol. 19, 1914–1928. ( 10.1111/j.1365-294X.2010.04604.x) [DOI] [PubMed] [Google Scholar]
- 22.Butler D. 2009. asreml: asreml() fits the linear mixed model. R package v. 3.0 ed20093.
- 23.Charmantier A, Garant D. 2005. Environmental quality and evolutionary potential: lessors from wild populations. Proc. R. Soc. B 272, 1415–1425. ( 10.1098/rspb.2005.3117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox R, Calsbeek R. 2009. Sexually antagonistic selection, sexual dimorphism, and the resolution of intralocus sexual conflict. Am. Nat. 173, 176–187. ( 10.1086/595841) [DOI] [PubMed] [Google Scholar]
- 25.Poissant J, Wilson A, Coltman D. 2009. Sex-specific genetic variance and the evolution of sexual dimorphism: a systematic review of cross-sex genetic correlations. Evolution 64, 97–107. ( 10.1111/j.1558-5646.2009.00793.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Calf data deposited in the Dryad repository: http://doi.org/10.5061/dryad.qb190. Pedigree data deposited in the Dryad repository: http://doi:10.5061/dryad.7jn8t.


