Abstract
Penicillium is a diverse genus occurring worldwide and its species play important roles as decomposers of organic materials and cause destructive rots in the food industry where they produce a wide range of mycotoxins. Other species are considered enzyme factories or are common indoor air allergens. Although DNA sequences are essential for robust identification of Penicillium species, there is currently no comprehensive, verified reference database for the genus. To coincide with the move to one fungus one name in the International Code of Nomenclature for algae, fungi and plants, the generic concept of Penicillium was re-defined to accommodate species from other genera, such as Chromocleista, Eladia, Eupenicillium, Torulomyces and Thysanophora, which together comprise a large monophyletic clade. As a result of this, and the many new species described in recent years, it was necessary to update the list of accepted species in Penicillium. The genus currently contains 354 accepted species, including new combinations for Aspergillus crystallinus, A. malodoratus and A. paradoxus, which belong to Penicillium section Paradoxa. To add to the taxonomic value of the list, we also provide information on each accepted species MycoBank number, living ex-type strains and provide GenBank accession numbers to ITS, β-tubulin, calmodulin and RPB2 sequences, thereby supplying a verified set of sequences for each species of the genus. In addition to the nomenclatural list, we recommend a standard working method for species descriptions and identifications to be adopted by laboratories working on this genus.
Key words: Aspergillaceae, Fungal identification, phylogeny, media, nomenclature
Introduction
Penicillium is well known and one of the most common fungi occurring in a diverse range of habitats, from soil to vegetation to air, indoor environments and various food products. It has a worldwide distribution and a large economic impact on human life. Its main function in nature is the decomposition of organic materials, where species cause devastating rots as pre- and postharvest pathogens on food crops (Frisvad & Samson 2004, Pitt & Hocking 2009, Samson et al. 2010), as well as producing a diverse range of mycotoxins (Frisvad et al. 2004). Some species also have positive impacts, with the food industry exploiting some species for the production of speciality cheeses, such as Camembert or Roquefort (Thom 1906, Nelson 1970, Karahadian et al. 1985, Giraud et al. 2010) and fermented sausages (López-Díaz et al. 2001, Ludemann et al. 2010). Their degradative ability has resulted in species being screened for the production of novel enzymes (Raper & Thom 1949, Li et al. 2007, Adsul et al. 2007, Terrasan et al. 2010). Its biggest impact and claim to fame is the production of penicillin, which revolutionised medical approaches to treating bacterial diseases (Fleming 1929, Chain et al. 1940, Abraham et al. 1941, Thom 1945). Many other extrolites have since been discovered that are used for a wide range of applications (Frisvad et al. 2004). Pitt (1979) considered it axiomatic that Penicillium or one of its products has affected every modern human.
It is now more than 200 years since Link (1809) introduced the generic name Penicillium, meaning ‘brush’, and described the three species P. candidum, P. glaucum and the generic type P. expansum. Since then, more than 1000 names were introduced in the genus. Many of these names are not recognisable today because descriptions were incomplete by modern criteria. Some names were published invalidly, or are now considered synonyms of other species. Thom (1930) revised all species described until 1930 and accepted 300 species. In later studies, Raper & Thom (1949) accepted 137 species, Pitt (1979) accepted 150 species, and Ramírez (1982) accepted 252 species (numbers include species described in Eupenicillium). At that time, a morphological species concept was used for Penicillium classification and identification, with DNA sequencing starting to be used during the 1990's. DNA sequencing created the threat that old names previously considered of uncertain application, because their ex-type cultures were no longer morphologically representative, could replace more commonly used but younger names. As such, the List of “Names in Current Use” (NCU) for the family Trichocomaceae (Pitt & Samson 1993) accepted 223 species and disregarded all other names as if not published. This list was updated by Pitt et al. (2000) who accepted 225 species. Species names not accepted on these lists were not to be disregarded permanently, as stated by Pitt et al. (2000), because they were not formally rejected under the nomenclatural code and could still be reintroduced in a revised taxonomy. In fact, this became common practice as many old species were shown to be distinct and were reintroduced (Peterson et al. 2005, Serra et al. 2008, Houbraken et al. 2011a,b, Houbraken et al. 2012a, Visagie et al. 2013).
The abandonment of article 59 in the new International Code of Nomenclature for algae, fungi and plants (ICN) (McNeill et al. 2012) resulted in single name nomenclature for fungi. In anticipation of this change, Houbraken & Samson (2011) redefined the genera in the family Trichocomaceae based on a four gene phylogeny. They segregated the Trichocomaceae into three families, namely the Aspergillaceae (Aspergillus, Hamigera, Leiothecium, Monascus, Penicilliopsis, Penicillium, Phialomyces, Sclerocleista, Warcupiella, Xeromyces), Thermoascaceae (Byssochlamys/Paecilomyces, Thermoascus) and the Trichocomaceae (Rasamsonia, Sagenomella, Talaromyces, Thermomyces, Trichocoma). Penicillium subgenus Biverticillium and Talaromyces were shown to form a monophyletic clade distinct from the other subgenera of Penicillium, with these names recombined as necessary into Talaromyces (Samson et al. 2011). The remaining Penicillium species formed a monophyletic clade together with species classified in Eupenicillium, Eladia, Hemicarpenteles, Torulomyces, Thysanophora and Chromocleista. These generic names were synonymised with Penicillium, while their species were given Penicillium names (Houbraken & Samson 2011). The remaining three Aspergillus species, A. paradoxus (≡ Hemicarpenteles paradoxus), A. malodoratus and A. crystallinus, phylogenetically belonging in Penicillium, are transferred to Penicillium below in the Taxonomy section. To accommodate the morphological variation, the generic diagnosis of Penicillium was amended in Houbraken & Samson (2011). Most importantly, in comparison with the prevailing generic concept (Raper & Thom 1949, Pitt 1979), it now excludes the acerose phialides and usually symmetrically branched conidiophores of species now included in Talaromyces, and was expanded to include the conidiophores with solitary phialides of species in section Torulomyces, and the darkly pigmented stipes that formerly characterised the genus Thysanophora, which show secondary growth by means of the proliferation of an apical penicillus. For infrageneric classification, the genus was divided into two subgenera, Aspergilloides and Penicillium, and 25 sections.
Thom (1954) attempted to explicitly define species concepts used for Penicillium. He was the pioneer of standardised working techniques and emphasised that Penicillium taxonomy demands a consistent, logical approach. He demonstrated these tendencies himself by taking into account infraspecies variation when delineating species. To minimise infraspecies variation, the importance of standardised working techniques were again emphasised by Pitt (1979), Samson & Pitt (1985), Okuda (1994) and Okuda et al. (2000). Although this was relatively effective when dealing with freshly isolated or wild-type strains, comparing strains using only morphology requires experience and nuance, because of the degeneration of characters in old reference material and the large number of species in the genus. New techniques incorporated into taxonomic studies resulted in the physiological species concept (Ciegler & Pitt 1970, Pitt 1973, Frisvad 1981, Frisvad & Filtenborg 1983, Cruickshank & Pitt 1987a,b, El-Banna et al. 1987, Frisvad & Filtenborg 1989, Paterson et al. 1989), phylogenetic species concept, including Genealogical Concordance Phylogenetic Species Recognition (GCPSR) (LoBuglio et al. 1993, Berbee et al. 1995, Boysen et al. 1996, Geiser et al. 1998, Skouboe et al. 1999, O'Donnell et al. 1998, Peterson 2000a, Taylor et al. 2000) and eventually led to the combined approach using morphological, extrolite and genetic data in a polyphasic species concept (Christensen et al. 2000, Frisvad & Samson 2004). In the modern taxonomy, however, sequence data and GCPSR carries more weight than morphology or extrolite data.
Even though Penicillium species are very common and the taxonomic structure of the genus is well defined, species identification is still problematic. Problems include an out-dated accepted species list and a lack of a verified, complete sequence database. The aim of this paper is to address these issues. In honour of Charles Thom and Kenneth B. Raper, the fathers of Penicillium taxonomy, and all the other mycologists who have contributed in the last 200 years towards our current understanding of the genus, we offer this contribution towards a practical, stable, consistent, logical approach to Penicillium taxonomy, identification and nomenclature.
Recommended methods for identification and characterisation of Penicillium
Morphological species recognition
Morphology in the past has been central to the taxonomy of Penicillium and along with multigene phylogenetics and extrolite profiling comprises the polyphasic species concept adopted for Penicillium. Morphology is the physical architecture through which an organism functions in and adapts to its environment, but some aspects may vary or be induced by specific cues in the immediate environment. As a result, strains characterised in one laboratory might look different when grown in another because of subtle differences in nutrients, temperature, lighting or humidity. This sometimes makes comparisons between different studies very difficult. These effects can be minimised using strictly standardised working techniques for medium preparation, inoculation technique and incubation conditions (Samson & Pitt 1985, Okuda 1994, Okuda et al. 2000). We recommend the following standardised methods for laboratories identifying and describing Penicillium species (summarised in Fig. 1).
Fig. 1.
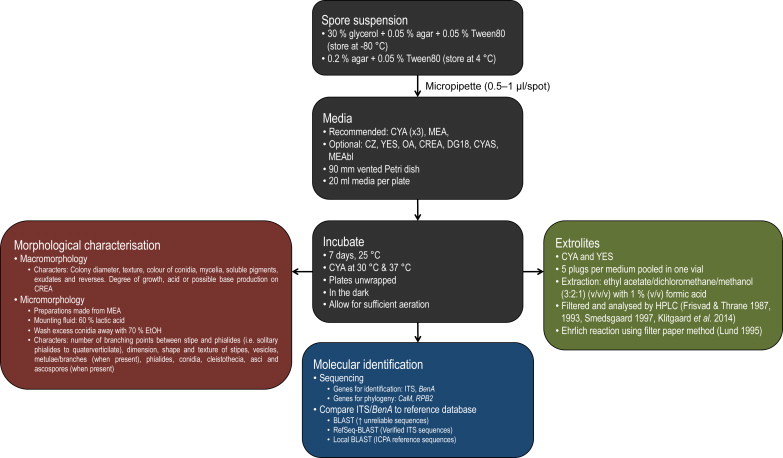
Flow diagram summarising recommended methods for the identification and characterisation of Penicillium. Frisvad & Thrane (1987, 1993), Smedsgaard (1997) and Klitgaard et al. (2014), refer to methods described for detecting extrolites in fungi. Lund (1995) introduced the Ehrlich reaction that tests for production of indole metabolites.
Macromorphology
Colony characters and diameters on specific media are important features for species identification. Czapek Yeast Autolysate agar (CYA) and Malt Extract agar (MEA, Oxoid) is recommended as standard media for Penicillium. Even though malt extract from Oxoid is recommended, many laboratories prefer Difco. It should be noted that two different MEA formulations are widely used in modern taxonomic studies. Blakeslee's MEA was historically widely used, but lately CBS switched to a different formulation (given in Table 1). Both are suitable for characterisation, but studies should state which MEA (Oxoid or Difco) and formulation was used. The following alternative media can be used for observing additional taxonomic characters: Czapek's agar (CZ), Yeast Extract Sucrose agar (YES), Oatmeal agar (OA), Creatine Sucrose agar (CREA), Dichloran 18 % Glycerol agar (DG18), Blakeslee's MEA and CYA with 5 % NaCl (CYAS). CZ was used in the taxonomic treatments of Raper & Thom (1949) and Ramírez (1982) and is chemically well defined. However, it is not widely used for Penicillium studies. YES is the recommended medium for extrolite profiling of species. Sexual reproduction most commonly occurs when strains are grown on OA and thus often provide valuable taxonomic information. OA should be prepared using organic uncooked flakes and not the unsuitable quick cook oats (“3 minute oats”) or prefabricated OA formulations available. Acid production is observed by the colour reaction in CREA (from purple to yellow) and is often useful for distinguishing between closely related species. In some species acid production is followed by base production and is most often observed from the colony reverse. DG18 and CYAS often provide useful information with regards to growth rates on low water activity media. For consistent conidial colours, the addition of zinc-sulphate and copper-sulphate as trace elements (1 g ZnSO4.7H2O and 0.5 g CuSO4.5H2O in 100 ml distilled water) is of utmost importance because these metals vary widely in water sources in different locations and are critical for pigment production. For inorganic chemicals, analytical grade should be used. Experience has shown that the brand of agar used influences colony appearance. As such, it is important to test the agar for consistent character development and note the brand in species descriptions. Even though we do not recommend a brand here as standard, after extensive comparisons CBS opted to use So-BI-Gel agar (Bie & Berntsen, BBB 100030) for medium preparations. Medium formulations are given in Table 1.
Table 1.
Media and stock solutions used for morphological characterisation.
| Czapek stock solution (100 ml) (Pitt 1979) | |
| NaNO3 NaNO3 | 30 g |
| KCl | 5 g |
| MgSO4·7H2OMgSO4·7H2O | 5 g |
| FeSO4·7H2OFeSO4·7H2O | 0.1 g |
| dH2OdH2O | 100 ml |
| *Store at 4–10 °C | |
| Trace elements stock solution (100 ml) | |
| CuSO4·5H2OCuSO4·5H2O | 0.5 g |
| ZnSO4·7H2OZnSO4·7H2O | 0.1 g |
| dH2OdH2O | 100 ml |
| *Store at 4–10 °C | |
| Blakeslee's Malt extract agar (MEAbl,Blakeslee 1915) | |
| Malt extract (Oxoid) | 20 g |
| Peptone (Oxoid) | 1 g |
| Glucose | 20 g |
| Trace elements | 1 ml |
| Agar | 20 g |
| dH2O | 1000 ml |
| *Mix well and autoclave at 121 °C for 15 min. pH 5.3 ± 0.2. | |
| Creatine sucrose agar (CREA,Frisvad 1981) | |
| Sucrose | 30 g |
| Creatine·1H2O | 3 g |
| K3PO4·7H2O | 1.6 g |
| MgSO4·7H2O | 0.5 g |
| KCl | 0.5 g |
| FeSO4·7H2O | 0.01 g |
| Trace elements stock solution | 1 ml |
| Bromocresol purple | 0.05 g |
| Agar | 20 g |
| dH2O | 1000 ml |
| *Mix well and autoclave at 121 °C for 15 min. pH 8.0 ± 0.2. | |
| Czapek's agar (CZ,Raper & Thom 1949) | |
| Czapek concentrate | 10 ml |
| Sucrose | 30 g |
| Trace elements stock solution | 1 ml |
| Agar | 20 g |
| dH2O | 1000 ml |
| *Mix well and autoclave at 121 °C for 15 min. | |
| Czapek Yeast Autolysate agar (CYA,Pitt 1979) | |
| Czapek concentrate | 10 ml |
| Sucrose | 30 g |
| Yeast extract (Difco) | 5 g |
| K2HPO4 | 1 g |
| Trace elements stock solution | 1 ml |
| Agar | 20 g |
| dH2O | 1000 ml |
| *Mix well and autoclave at 121 °C for 15 min. pH 6.2 ± 0.2. | |
| Czapek Yeast Autolysate agar with 5 % NaCl (CYAS) | |
| Czapek concentrate | 10 ml |
| Sucrose | 30 g |
| Yeast extract (Difco) | 5 g |
| K2HPO4K2HPO4 | 1 g |
| Trace elements stock solution | 1 ml |
| NaCl | 50 g |
| Agar | 20 g |
| dH2OdH2O | 1000 ml |
| *Mix well and autoclave at 121 °C for 15 min. pH 6.2 ± 0.2. | |
| Dichloran 18 % Glycerol agar (DG18,Hocking & Pitt 1980) | |
| Dichloran-Glycerol-agar-base (Oxoid) | 31.5 g |
| Glycerol (anhydrous) | 220 g |
| Trace elements stock solution | 1 ml |
| Chloramphenicol | 0.05 g |
| Agar | 20 g |
| dH2O | 1000 ml |
| *Mix well and autoclave at 121 °C for 15 min. After autoclaving, add 0.05 chlortetracycline. pH 5.6 ± 0.2. | |
| Malt Extract agar (MEA,Samson et al. 2010) | |
| Malt extract (Oxoid CM0059) | 50 g |
| Trace elements stock solution | 1 ml |
| dH2O | 1000 ml |
| *Mix well and autoclave at 115 °C for 10 min. pH 5.4 ± 0.2. | |
| Oatmeal agar (OA,Samson et al. 2010) | |
| Oatmeal / flakes | 30 g |
| Trace elements stock solution | 1 ml |
| Agar | 20 g |
| dH2O | 1000 ml |
| *First autoclave flakes (121 °C for 15 min) in 1000 ml dH2O. Squeeze mixture through cheese cloth and use flow through, topping up to 1000 ml with dH2O with 20 g agar. Autoclave at 121 °C for 15 min. pH 6.5 ± 0.2. | |
| Yeast extract sucrose agar (YES,Frisvad 1981) | |
| Yeast extract (Difco) | 20 g |
| Sucrose | 150 g |
| MgSO4·7H2O | 0.5 g |
| Trace elements stock solution | 1 ml |
| Agar | 20 g |
| dH2O | 885 ml |
| *Mix well and autoclave at 121 °C for 15 min. pH 6.5 ± 0.2. | |
Media are prepared in 90 mm Petri-dishes with a volume of 20 ml (Okuda et al. 2000). Glass Petri dishes was traditionally considered best for character observation. However, today it is not feasible using them, with polystyrene Petri dishes recommended. The Petri dishes should preferably be vented which allows for a bit of air exchange. Inoculations are made from spore suspensions in a semi-solid agar solution containing 0.2 % agar and 0.05 % Tween80 (Pitt 1979). In some laboratories, spore suspensions are made in a 30 % glycerol, 0.05 % agar and 0.05 % Tween80 solution. Variation between these two inoculation suspension solutions has not been shown. We recommend using a micropipette for inoculating the spore suspension in three-point fashion (0.5–1 μl per spot). They should not be wrapped with Parafilm, which by restricting air exchange often inhibits growth and sporulation (Okuda et al. 2000). When using walk-in incubators, laboratory rules often require plates to be placed in plastic containers or plastic bags. In this case, the boxes or bags should allow for enough aeration and care should be taken not to have too many plates in one box or in one incubator. For standard bench-top incubators, plates do not need to be incubated in boxes or bags, unless there is a strong air-current in the incubator. All media are incubated at the standard temperature of 25 °C for 7 d, with additional CYA plates at 30 and 37 °C that are useful to distinguish between species. It is crucial that temperatures are carefully checked as small differences have a large impact on colony growth. No effect on colonies has been shown for incubating Petri dishes upside down or vice versa.
After 7 d, colony diameters are measured at the widest part of the colony. Important characters used for describing Penicillium include colony texture, degree of sporulation, the colour of conidia, the abundance, texture and colour of mycelia, the presence and colours of soluble pigments and exudates, colony reverse colours, and degree of growth and acid production (in some species acid production followed by base production) on CREA. The use of a standard colour chart is recommended for descriptions. Although many are available, the most commonly used colour chart in Penicillium is the Methuen Handbook of Color (Kornerup & Wanscher 1967), which although long out of print is available in many libraries. It is worth noting that colour names used by Raper & Thom (1949) were based on Ridgeway (1912) and are also still in use. When colour charts are not available, it is recommended to publish full color photoplates to accompany descriptions of new species. Some species produce specialised structures such as cleistothecia or sclerotia, mostly after longer incubation times, especially on OA. We thus recommend OA plates be incubated for prolonged periods to have a chance of observing these structures. For definitions and explanations of terms used in descriptions of Penicillium colonies, readers are referred to Frisvad & Samson (2004).
Micromorphology
Conidiophore and cleistothecium (when produced) characters of Penicillium are of great taxonomic importance. Conidiophore branching patterns (Fig. 2) were traditionally used in the classification of Penicillium (Thom 1930, Raper & Thom 1949, Pitt 1979). Although these branching patterns do not correspond perfectly with the sections currently accepted for Penicillium, characterising them accurately is still considered important. The conidiophores range from being simple (solitary phialides, Fig. 2A) to very complex patterns with multiple levels of branching resulting in overall symmetrical or asymmetrical patterns. Monoverticillate conidiophores (Fig. 2B) have a terminal whorl of phialides and in some species, the terminal cell of the conidiophore is slightly swollen or vesiculate; such species could be confused with diminutive Aspergillus conidiophores, but they have septa in the stipes unlike species of the latter genus. Divaricate conidiophores, previously also referred to as irregular (Pitt 1979, Fig. 2C), are best described as having a simple to complex branching pattern with numerous subterminal branches formed, but where conidiophore parts are divergent. Biverticillate conidiophores (Fig. 2D, E) have a whorl of three or more metulae between the end of the stipe and the phialides; the metulae may be of unequal or equal length, vary in their degree of divergence, are usually more or less cylindrical but can also be clavate or slightly vesiculate. Terverticillate conidiophores (Fig. 2F) have another level of branching between the stipe and the metulae, often just a continuation of the stipe axis and one side branch, sometimes a true whorl of three or more branches. Quaterverticillate conidiophores (Fig. 2G) are produced by only a few species, and have one extra level of branching beyond the terverticillate pattern. Terverticillate and quaterverticillate conidiophores tend to be conspicuously asymmetrical. In colonies of many species, especially as cultures begin to degenerate, there may be more than one branching pattern or intermediate forms, and it can be challenging to decide which pattern is typical or most developed.
Fig. 2.
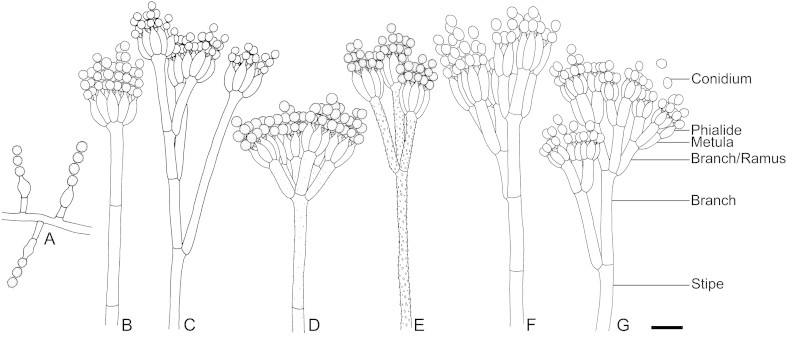
Conidiophore branching patterns observed in Penicillium. A. Conidiophores with solitary phialides. B. Monoverticillate. C. Divaricate. D, E. Biverticillate. F. Terverticillate. G. Quaterverticillate, terms used for describing parts of conidiophores are given. Scale bar = 10 μm.
Other important microscopic characters include the wall texture/ornamentation of stipes and conidia, as well as dimensions, ornamentation and sometimes colours of all elements of the conidiophore (Fig. 1). In our experience, wall textures are very sensitive to minute differences in media composition and aeration. For best observation of conidial ornamentation, differential interference contrast (= Nomarski) is recommended if possible; ornamentation is sometimes most conspicuously visible in air pockets in the preparation.
For microscopic observations, slides are prepared from 7–10 d old MEA colonies. Remove material from the zones where adjacent colonies are closest or growing together, from the part of the colony where the conidial colour is just starting to develop. For observing conidia, material removed from colony centers generally gives more uniform results. Lactic acid (60 %) is the most commonly used mounting fluid, although other solutions such as Shear's solution or lactic acid with cotton blue (Frisvad & Samson 2004, Samson et al. 2010) can also be used. We do not recommend lactophenol because of the corrosiveness and toxicity of phenol. Because of the abundant hydrophobic conidia produced by most species, drops of 70 % ethanol are commonly used to wash away excess conidia and to prevent air being “trapped” when mounted in lactic acid. For photographing conidiophores, we often wash away the spores two or three times. Some species have dense colonies and then it is necessary to tease apart the conidiophores with very fine needles under the dissecting microscope.
DNA barcoding for identification and multilocus sequence typing for phylogenetic species recognition
Sequence markers
During the 1990's, DNA sequencing became one of the most powerful tools for taxonomists, because it created the opportunity for inferring relationships between species without the need for standardising culturing regimes and eliminated problems related to deteriorated cultures. It also created the opportunity for sequence based identifications. DNA barcoding was launched to make species identification of any eukaryotic organism possible for anybody, by using a standardised short DNA sequence and a curated reference database linked to authoritatively identified vouchers (Blaxter 2003, Tautz et al. 2003, Hebert et al. 2003, Blaxter et al. 2005, DeSalle et al. 2005, Ratnasingham & Hebert 2007, Seifert et al. 2007, Min & Hickey 2007a,b, Schoch et al. 2012). Only recently however, was the internal transcribed spacer rDNA area (ITS) accepted as the official barcode for fungi (Schoch et al. 2012). ITS is the most widely sequenced marker for fungi, and universal primers are available (Schoch et al. 2012). In Penicillium, it works well for placing strains into a species complex or one of the 25 sections, and sometimes provides a species identification (Visagie et al. 2014a). Unfortunately, for Penicillium and many other genera of ascomycetes, the ITS is not variable enough for distinguishing all closely related species (Skouboe et al. 1999, Seifert et al. 2007, Schoch et al. 2012). The open source sequence repository GenBank contains a large proportion of incorrectly identified sequences, making identifications of Penicillium using BLAST very tricky for inexperienced workers. This particular problem is addressed in a number of publications (Kõljalg et al. 2005, Santamaria et al. 2012, Kõljalg et al. 2013, Schoch et al. 2014). For Penicillium, the International Commission of Penicillium and Aspergillus (ICPA), in conjunction with the publication of an updated accepted species list presented below, decided to include GenBank accession numbers to reference barcode sequences for each species when available.
Because of the limitations associated with ITS as a species marker in Penicillium, a secondary barcode or identification marker is often needed for identifying isolates to species level. The requirements for a secondary identification marker are clear. It should be easy to amplify, distinguish among closely related species and most importantly, the reference data set should be complete, meaning that there should be representative sequences for all species. It would be an added bonus if this marker is useful for phylogenetic studies, as it will by default become the gene most widely sequenced in future. Based on these criteria, we propose the use of β-tubulin (BenA) as the best option for a secondary identification marker for Penicillium. BenA does, however, have problems associated with it. Although not influencing BLAST identifications, alignments across a diverse genus like Penicillium is difficult and often contains a large proportion ambiguously aligned sites, which can make phylogenies difficult. Also, there is evidence for the amplification of BenA paralogous genes in Aspergillus (Peterson 2008, Hubka & Kolarik 2012) and Talaromyces (Peterson & Jurjević 2013) and it can thus be assumed that the same might be happening in some Penicillium species, although this has not been shown. Other possible secondary marker options include calmodulin (CaM) or the RNA polymerase II second largest subunit (RPB2) genes. Both these genes have similar discriminatory power as BenA. RPB2 has the added advantage of lacking introns in the amplicon, allowing robust and easy alignments when used for phylogenies, but it is sometimes difficult to amplify and the database is incomplete. Similarly, we lack a complete CaM database. Thus, for routine identifications BenA is currently recommended, while for the description of new species, we suggest the use of ITS, BenA, CaM and RPB2 among the markers for multilocus sequence typing and GCPSR. We consider it good practise to include at least ITS and BenA sequences when describing new species, to allow others to more easily recognise the new species.
BenA can successfully be used for accurately identifying Penicillium species. However, as is the case for genes other than BenA, care should be taken in specific groups or situations. Infraspecies variation in BenA occurs in some Penicillium species as is observed in phylogenies published for Penicillium (Frisvad & Samson 2004, Barreto et al. 2011, Peterson et al. 2011, Houbraken et al. 2011b,c, Rivera & Seifert 2011, Rivera et al. 2012, Houbraken et al. 2012a, Visagie et al. 2013, 2014a, 2014b). This variation must be considered for identification purposes and especially when considering whether a strain might represent a new species. This means that in addition to the reference sequences of ex-type cultures sanctioned by ICPA, additional reference sequences are necessary to document sequence variation that differ from the ex-type. ICPA is currently working on populating such a validated database to capture infraspecies variation, but for the time being critical phylogenetic revisions of different sections should be referred to for reliable data. Alternatively, combining ITS, BenA, CaM and RPB2 from a suspected new species with sequences of the same markers from related species will aid in deciding whether a species is new or not, using GCPSR as explained in detail by Taylor et al. (2000). This is in fact common practise in most studies describing and characterising Penicillium species.
β-tubulin as a secondary marker in practise
As discussed above, BenA works well for species identifications in Penicillium. Penicillium chrysogenum and P. allii-sativi is one example where identification based on BenA should be made with care (Houbraken et al. 2012a). This is a consequence of the variation observed among strains of P. chrysogenum, which results in P. allii-sativi forming a clade within the P. chrysogenum clade in BenA species trees. Even though CaM distinguishes P. chrysogenum from P. allii-sativi, it does not distinguish P. chrysogenum and P. rubens. As a result, molecular identifications should be made with great care in section Chrysogena, with the revision by Houbraken et al. (2012a) used as guide. Penicillium kongii was recently introduced as a close relative of P. brevicompactum (Wang & Wang 2013). Even though this species formed a coherent clade distinct from the P. brevicompactum ex-type strain, on examining additional strains previously identified as P. brevicompactum, the P. kongii clade is resolved within the P. brevicompactum clade. This clade thus requires more work to resolve the species from the complex. A similar situation is observed for P. desertorum and the newly described P. glycyrrhizacola (Chen et al. 2013), where more strains and additional genes included in the phylogeny will help to resolve species. Another species that cannot be identified using molecular data is P. camemberti, P. caseifulvum and P. commune, which shares identical BenA and other gene sequences (Giraud et al. 2010). Although these could be considered taxonomic synonyms, their different applications and importance in the cheese industry suggest that it is of more value to keep them as separate species. Morphological identification is necessary in this case, with P. camemberti having white conidia, P. caseifulvum having an orange reverse on YES and P. commune producing green conidia (Frisvad & Samson 2004).
Amplification and identification
Primers used for amplification of the ITS, BenA, CaM and RPB2 genes are included in Table 2 and amplification profiles are given in Table 3. A standard thermal cycle with an annealing temperature of 55 °C is generally used. Sometimes amplification success is low for CaM, especially in sections Canescentia and Ramosa. In this case, dropping the annealing temperature to 52 °C gives good results. The RPB2 amplification is more complicated. We recommend using a touch-up PCR (50–52–55 °C) with primer pair 5Feur and 7CReur for best amplification. When amplification is problematic, the alternative touch-up PCR (48–50–52 °C) profile can be used and/or the alternative primer pair 5F and 7CR.
Table 2.
Primers used for amplification and sequencing.
| Locus | Primer name | Direction | Primer sequence (5′-3′) | Reference |
|---|---|---|---|---|
| Internal Transcribed Spacer (ITS) | ITS1 | Forward | TCC GTA GGT GAA CCT GCG G | White et al. 1990 |
| ITS4 | Reverse | TCC TCC GCT TAT TGA TAT GC | White et al. 1990 | |
| V9G | Forward | TTA CGT CCC TGC CCT TTG TA | de Hoog & Gerrits van den Ende 1998 | |
| LS266 | Reverse | GCA TTC CCA AAC AAC TCG ACT C | Masclaux et al. 1995 | |
| β-tubulin (BenA) | Bt2a | Forward | GGT AAC CAA ATC GGT GCT GCT TTC | Glass & Donaldson 1995 |
| Bt2b | Reverse | ACC CTC AGT GTA GTG ACC CTT GGC | Glass & Donaldson 1995 | |
| Calmodulin (CaM) | CMD5 | Forward | CCG AGT ACA AGG ARG CCT TC | Hong et al. 2006 |
| CMD6 | Reverse | CCG ATR GAG GTC ATR ACG TGG | Hong et al. 2006 | |
| CF1 | Forward | GCC GAC TCT TTG ACY GAR GAR | Peterson et al. 2005 | |
| CF4 | Reverse | TTT YTG CAT CAT RAG YTG GAC | Peterson et al. 2005 | |
| RNA polymerase II second largest subunit (RPB2) | 5F | Forward | GAY GAY MGW GAT CAY TTY GG | Liu et al. 1999 |
| 7CR | Reverse | CCC ATR GCT TGY TTR CCC AT | Liu et al. 1999 | |
| 5Feur | Forward | GAY GAY CGK GAY CAY TTC GG | Houbraken et al. 2012b | |
| 7CReur | Reverse | CCC ATR GCY TGY TTR CCC AT | Houbraken et al. 2012b |
Table 3.
Thermal cycle programs used for amplification.
| Gene | Profile type | Initial denaturing | Cycles | Denaturing | Annealing | Elongation | Final elongation | Rest period |
|---|---|---|---|---|---|---|---|---|
| General ITS, BenA, CaM | standard | 94 °C, 5 min | 35 | 94 °C, 45 s | 55 °C, 45 s | 72 °C, 60 s | 72 °C, 7 min | 10 °C, ∞ |
| General alternative | standard | 94 °C, 5 min | 35 | 94 °C, 45 s | 52 °C, 45 s | 72 °C, 60 s | 72 °C, 7 min | 10 °C, ∞ |
| RPB2 | touch-up | 94 °C, 5 min | 5 | 94 °C, 45 s | 50 °C, 45 s | 72 °C, 60 s | ||
| 5 | 94 °C, 45 s | 52 °C, 45 s | 72 °C, 60 s | |||||
| 30 | 94 °C, 45 s | 55 °C, 45 s | 72 °C, 60 s | 72 °C, 7 min | 10 °C, ∞ | |||
| RPB2 alt. | touch-up | 94 °C, 5 min | 5 | 94 °C, 45 s | 48 °C, 45 s | 72 °C, 60 s | ||
| 5 | 94 °C, 45 s | 50 °C, 45 s | 72 °C, 60 s | |||||
| 30 | 94 °C, 45 s | 52 °C, 45 s | 72 °C, 60 s | 72 °C, 7 min | 10 °C, ∞ |
After sequences are obtained, there is a number of ways to use them to identify the strain. The most widely used method is the BLAST search on NCBI. Using this method, one is able to search all sequences in the database, but as noted above, there are many unidentified and misidentified sequences in the database. For ITS the RefSeq data set, accessible from the NCBI homepage (http://www.ncbi.nlm.nih.gov/refseq/), is now available and can be used to query ITS sequences against a verified ITS database (Schoch et al. 2014). Unfortunately, the RefSeq database does not cover alternative genes. For this, we suggest setting up Local BLAST files and using the verified BenA database provided here.
Extrolite data
Extrolites are produced by the mycelium and sporulating structures of Penicillium species, and exudates, diffusible pigments, and reverse colours are also mixtures of secondary metabolites. Studies of extrolite profiles were very useful for unravelling some morphological species concepts into biologically meaningful segregate species before DNA sequencing provided similar possibilities. As an example, the P. aurantiogriseum complex, critical contaminants of grain, was divided first into species using extrolite profiles by Frisvad & Filtenborg (1983, 1989), delimitations that were subsequently supported by BenA sequencing (Seifert & Louis-Seize 2000).
Identification of unknown strains using extrolites is possible for well-equipped chemical laboratories. The best way of using extrolites as identification aids is to extract and then separate them by HPLC and then partially or fully identify as many of the secondary metabolites as possible, generally using mass spectroscopy based technology (Frisvad et al. 2008). The media used for identification, especially CYA and YES agars, are optimal for production of most major diagnostic extrolites in Penicillium after incubation for 7 d at 25 °C in darkness. Agar plugs are extracted with a mixture of dichloromethane, ethylacetate and methanol. The metabolites extracted can then be analysed using advanced separation and detection techniques, for example ultra high performance liquid chromatography with diode array detection and high resolution mass spectrometric detection (UHPLC-DAD-HRMS) (Kildgaard et al. 2014, Klitgaard et al. 2014). However, simpler HPLC-diode array detection methods can also be used (Frisvad & Thrane 1987, 1993).
Laboratories with less sophisticated equipment can perform valuable confirmatory tests. Although Thin Layer Chromatography (TLC) is no longer considered state of the art for chemical research, it is still a useful technique for detecting coloured or uncoloured extrolites that can be used to confirm the identification of a Penicillium strain. For example, P. brevicompactum consistently produces large amounts of the colourless mycophenolic acid, and this metabolite will make a green colour reaction with ferric chloride (Clutterbuck & Raistrick 1933).
Sometimes only a few extrolites are needed to confirm the identity of a Penicillium isolate. If an isolate produces griseofulvin and roquefortine C, it can only be P. coprophilum, P. griseofulvum or P. sclerotigenum and if the isolate also produces cyclopiazonic acid, it can only be P. griseofulvum (Frisvad & Samson 2004).
Identification based purely on secondary metabolites is not yet possible for all species of Penicillium, but future databases will be better developed and an optimal battery of media for secondary metabolite production may allow this method to be used for identification of all species in the future.
Misidentifications in Penicillium
A few examples demonstrate why Penicillium identification is difficult or how misleading results might be obtained in practise using morphology or sequencing. For example, a fungus producing penicillones A and B and chloctanspirones A, B and terrestrols was identified as P. terrestre (patent strain CCTCC M 204077, strain unavailable) (Liu et al. 2005). The name P. terrestre was used by Raper & Thom (1949), but has since been considered a synonym of P. crustosum. However, additional secondary metabolites produced by the isolate, such as sorbicillin and trichodimerol, are never seen in P. crustosum (Frisvad et al. 2004), suggesting the strain was P. chrysogenum or P. rubens. Liu et al. (2005) did not describe how their isolate was identified. Another strain of P. chrysogenum or P. rubens, SD-118 (HQ652873), was misidentified as a P. commune (Shang et al. 2012, Zhao et al. 2012), based on “100 % sequence identity” with P. commune (FJ499541). The production of chrysogine, sorbicillins and meleagrin indicated that the strain, SD-118, was in fact P. chrysogenum. We propose that the use of the reference sequence databases provided here, will minimise these type of misidentifications.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry fingerprinting (MALDI-TOF MS)
Another detection technique receiving a lot of attention, especially from the medical field, is matrix-assisted laser desorption/ionization time-of-flight mass spectrometry fingerprinting (MALDI-TOF MS). It was successfully applied to the identification of bacteria (Hettick et al. 2006, Siegrist et al. 2007) and yeast species (Amiri-Eliasi & Fenselau 2001, Kolecka et al. 2013). Only a limited number of studies included or focused on Penicillium (Welham et al. 2000, Hettick et al. 2008, Del Chierico et al. 2012, Chalupová et al. 2014) and the closely related Aspergillus (Bille et al. 2011, De Carolis et al. 2011, Iriart et al. 2012, Verwer et al. 2013). Although these studies promisingly report that MALDI-TOF MS distinguish between species, a large degree of variation is observed within a species, even between duplicates of the same strain (Hettick et al. 2008). This means that data from a high number of strains will have to be included in the database to make robust identifications feasible. Difficulties with identifications are also reported in Aspergillus, where not all strains could be identified with 100 % accuracy (Iriart et al. 2012, Verwer et al. 2013). Thus we can conclude that although this technique shows promise, a lot of work remains to make routine identifications feasible.
Taxonomy
Phylogenetic analyses revealed that Hemicarpenteles paradoxus should not be classified in Aspergillus, and together with its closest relatives A. crystallinus and A. malodoratus, should be transferred to Penicillium (Peterson 2000a, 2008, Houbraken & Samson 2011). Houbraken & Samson (2011) included them in Penicillium sect. Paradoxa together with P. atramentosum. The other Hemicarpenteles species were shown to belong in Aspergillus sect. Clavati (H. acanthosporus ≡ A. acanthosporus ≡ Neocarpenteles acanthosporus (Udagawa & Takada) Udagawa & Uchiy.) (Peterson 2000b, Tamura et al. 2000, Varga et al. 2007, Peterson 2008), while H. ornatus and H. thaxteri are classified in Sclerocleista (Pitt et al. 2000, Houbraken & Samson 2011).
Aspergillus paradoxus was described by Fennell & Raper (1955) for strains producing small clavate vesicles suggestive of those observed in aberrant strains of A. clavatus. Later, Raper & Fennell (1965) placed A. paradoxus in the A. ornatus species group, based on the resemblance of its sclerotia to young cleistothecia of A. ornatus and A. citrisporus. Rai et al. (1964, 1967) also isolated A. paradoxus from soil in India, and found that conditions favouring sclerotia production also inhibited the formation of conidial heads and the yellow pigment in the medium. Sarbhoy & Elphick (1968) observed that a strain of A. paradoxus (CBS 793.68 = IMI 117502), isolated from dog dung in the United Kingdom, produced mature unilocular cleistothecia with tough peridial walls, embedding smooth-walled lenticular ascospores. They introduced the name Hemicarpenteles for the sexual morph, with H. paradoxus as type. Houbraken & Samson (2011) noted that the sexual morph most closely resembles Eupenicillium, both producing sclerotoid cleistothecia that ripen from the centre outwards (Sarbhoy & Elphick 1968, Pitt 1979, Stolk & Samson 1983). In our fresh isolates, some strains produce sclerotia abundantly while others have only a few sclerotia. Only two fresh isolates produced ascospores after incubation at 25 °C for three weeks on OA. Aspergillus paradoxus, A. crystallinus and A. malodoratus (Figs 3–6) have a striking similarity with members of section Olsonii and Coronata (Frisvad & Samson 2004, Samson et al. 2004), in particular the long, wide, smooth-walled conidiophores terminating in densely branched penicilli with metulae and phialides. Particularly P. olsonii bears a morphological resemblance with A. paradoxus and A. crystallinus.
Fig. 3.
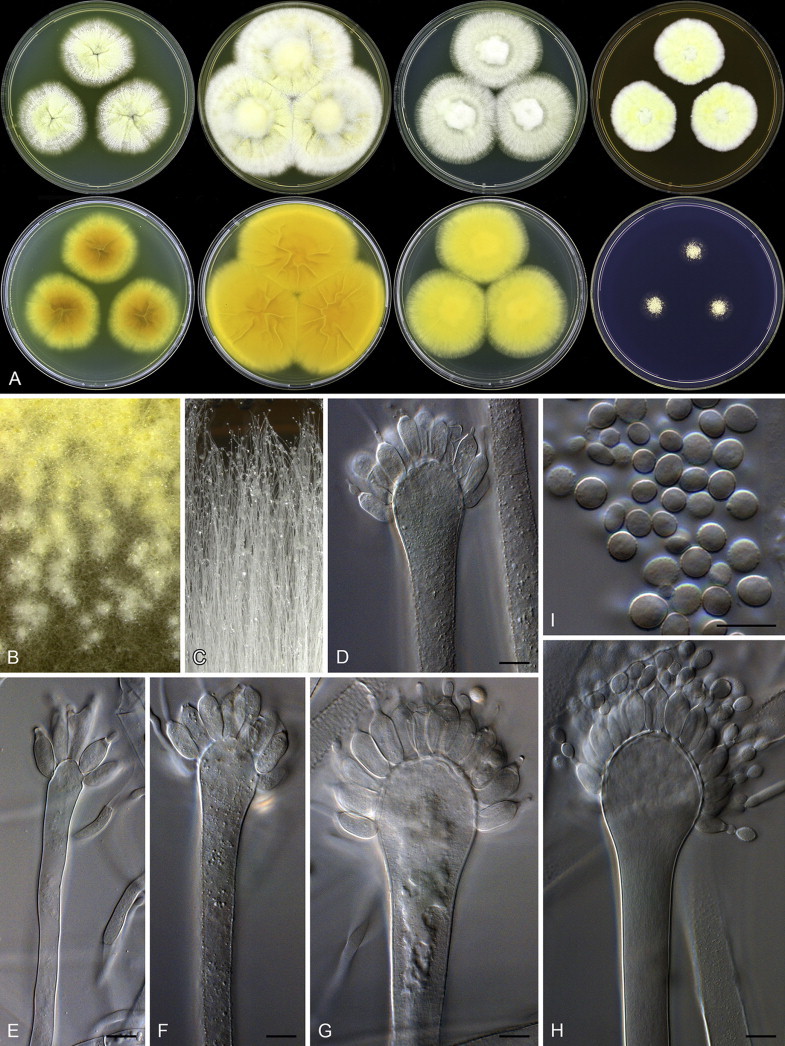
Penicillium paradoxum. A. Colonies: top row left to right, obverse CYA, YES, DG18 and MEA; bottom row left to right, reverse CYA, reverse YES, reverse DG18 and CREA. B. Young sclerotia. C. Phototropic conidiophores after two weaks growth. D–H. Conidiophores. I. Conidia. Scale bars: D–I = 10 μm.
Fig. 4.
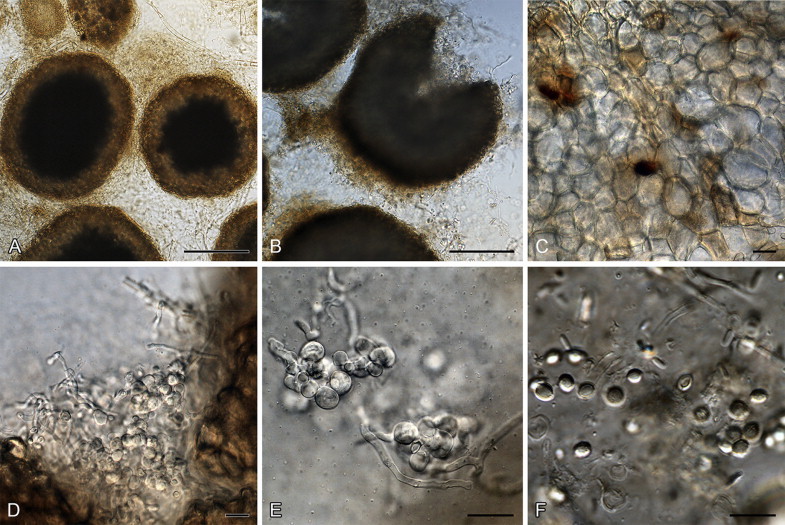
Penicillium paradoxum sexual reproduction. A–C. Cleistothecia. D, E. Asci. F. Ascospores. Scale bars: A, B = 500 μm; C–F = 10 μm.
Fig. 5.
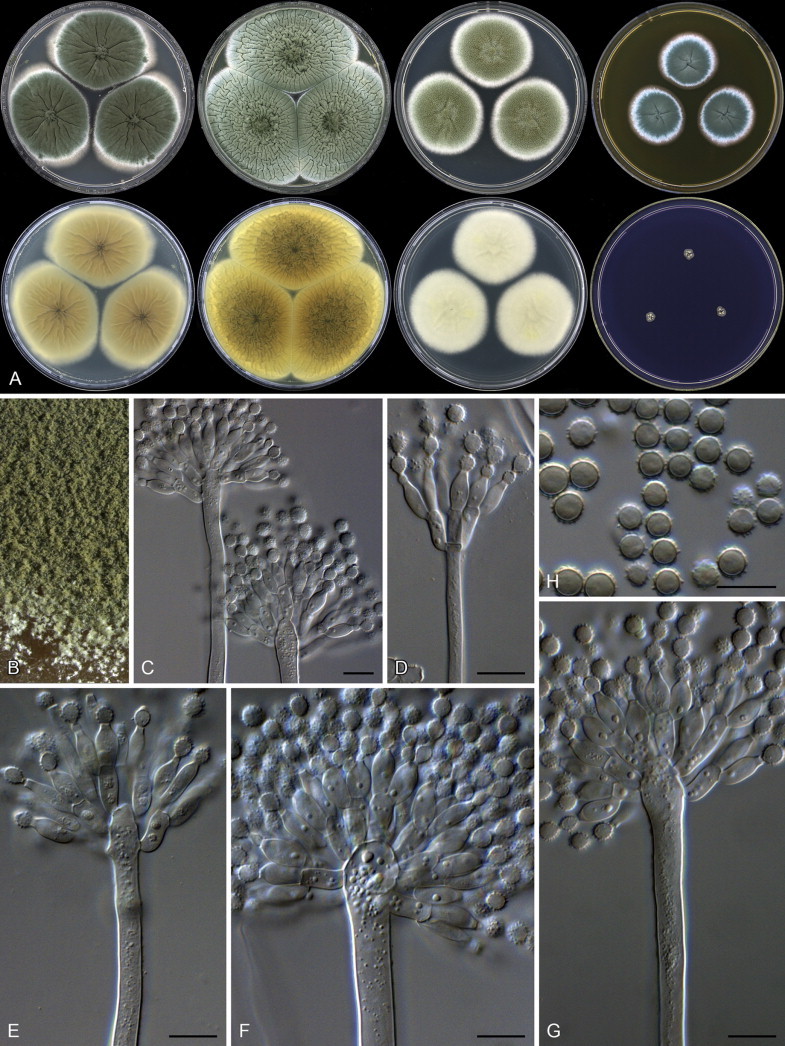
Penicillium crystallinum. A. Colonies: top row left to right, obverse CYA, YES, DG18 and MEA; bottom row left to right, reverse CYA, reverse YES, reverse DG18 and CREA. B. Colony texture on MEA. C–G. Conidiophores. H. Conidia. Scale bars: C–H = 10 μm.
Fig. 6.
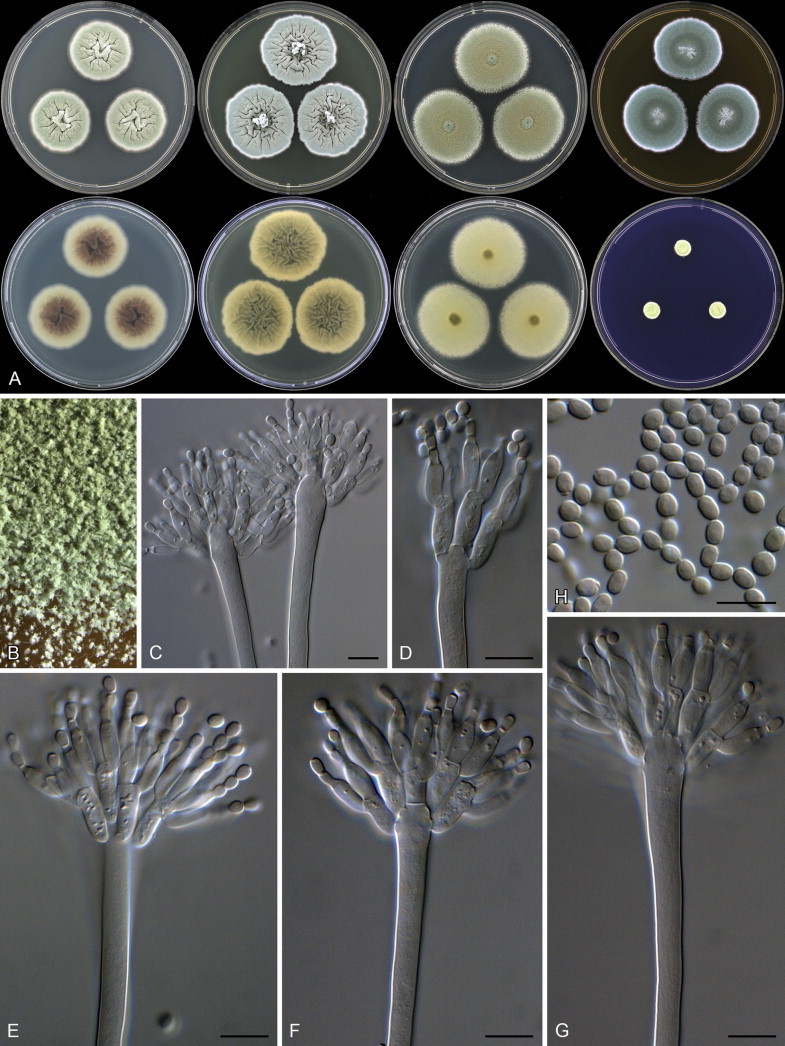
Penicillium malodoratum. A. Colonies: top row left to right, obverse CYA, YES, DG18 and MEA; bottom row left to right, reverse CYA, reverse YES, reverse DG18 and CREA. B. Colony texture on MEA. C–G. Conidiophores. H. Conidia. Scale bars: C–H = 10 μm.
Extrolite data also suggest that A. paradoxus, A. crystallinus and A. malodoratus belong in Penicillium (summarised in Table 4). Pseurotins are found in both Penicillium and Aspergillus species (Frisvad et al. 2004). Sorbicillins were previously only confirmed in P. rubens and P. chrysogenum, and not so far in Aspergillus, although Basaran & Demirbas (2010) reported sorbicillin production in A. parasiticus; we have never observed sorbicillins in any A. parasiticus strain. Penicillium atramentosum, also classified in section Paradoxa, produces andrastin A, atpenins, meleagrin and oxaline, roquefortine C & D and rugulovasine A & B (Frisvad et al. 2004, and reported here). Among Penicillium and Aspergillus species, the atpenins have only been found in P. atramentosum (Omura et al. 1988, Kawada et al. 2009), but the other listed extrolites are common in species of Penicillium sections Paradoxa and Chrysogena. Rugulovasins are an exception and have only been found in P. commune (Frisvad et al. 2004). Aspergillus paradoxus produces brefeldin, an extrolite also reported in P. brefeldianum (Hutchinson et al. 1983), but never in Aspergillus apart from a report for A. clavatus (Wang et al. 2002) that could not be confirmed (Varga et al. 2007). The latter strain was probably actually A. ingratus, first regarded as belonging to Aspergillus section Clavati (Yaguchi et al. 1993) but now considered a synonym of A. paradoxus. Xanthocillins are produced by A. paradoxus strains, and also by P. chrysogenum and P. egyptiacum. Meleagrin is commonly produced in Penicillium sections Paradoxa and Chrysogena, and species of several other sections (Frisvad et al. 2004). Only the closely related extrolite, neoxaline, has been reported in A. japonicus (Hirano et al. 1979). Aspergillus crystallinus produces chrysogine, typical of P. rubens and P. chrysogenum (Frisvad et al. 2004, Houbraken et al. 2012a), and a compound also reported in A. nomius (Varga et al. 2011). Chrysophanic acid, produced in A. crystallinus, was found in Talaromyces islandicus (Howard & Raistrick 1950), and pachybasin was found in P. vulpinum (Frisvad et al. 2004). Aspergillus malodoratus produces andrastin A, meleagrin and oxaline. Andrastin A is produced by several Penicillium species including P. rubens and P. chrysogenum (Houbraken et al. 2011a, 2012a), but has not been found in Aspergillus. On balance, chemotaxonomic evidence points to the placement of A. paradoxus, A. crystallinus and A. malodoratus in Penicillium.
Table 4.
Origin and extrolites of the strains examined in this study.
| Species | Strain | Origin | Extrolites |
|---|---|---|---|
| A. crystallinus | CBS 479.65T = IBT 21947 | Forest soil, Tilaran, Costa Rica | chrysophanic acid, chrysogine, meleagrin, pachybasin |
| A. malodoratus | CBS 490.65T = IBT 21948 | Forest soil, Barranca, Costa Rica | andrastin A, oxaline, meleagrin |
| A. paradoxus | CBS 123898 = IBT 29765 = DTO 76F5 | Dung of dog, the Netherlands | “astyl”, brefeldin A, “SLOF” |
| CBS 123898 = IBT 29768 = DTO 76F6 | Dung of dog, the Netherlands | “astyl”, brefeldin A, pseurotin, “SLOF”, sorbicillins | |
| CBS 527.65T = IBT 19547 | Dung of opossum, New Zealand | “astyl”, brefeldin A, pseurotin, “SLOF”, sorbicillins | |
| CBS 643.95 = IBT 17513 = PF 1116 | Soil, USA, California (ex-type of A. ingratus) | “asyl”, brefeldin A, pseurotin, “SLOF”, sorbicillins | |
| CBS 793.68 = IBT 19572 | Dung of dog, UK | “astyl”, brefeldin A, “PAS”, pseurotin, “SLOF”, sorbicillins | |
| CBS 793.91 = IBT 12297 | Dung of dog, Denmark | brefeldin A, “PAS”, pseurotin, “SLOF”, sorbicillins | |
| IBT 29766 = DTO 76F7 | Dung of dog, the Netherlands | brefeldin A, pseurotin, “SLOF”, sorbicillins | |
| IMI 173765 = IBT 19364 | Jet fuel, New Zealand | “astyl”, brefeldin A, pseurotin, sorbicillins | |
| IMI 320026 = IBT 19365 | Air, UK | brefeldin A, pseurotin, sorbicillins |
From the phenotypic, molecular and extrolite data we can conclude that the three species formerly placed in Aspergillus belong to Penicillium. Consequently we propose the following new combinations:
Penicillium paradoxum (Fennell & Raper) Samson, Houbraken, Visagie & Frisvad, comb. nov. MycoBank MB547045. Figs 3, 4.
Basionym: Aspergillus paradoxus Fennell & Raper, Mycologia 47: 69. 1955. MycoBank MB292853
≡ Hemicarpenteles paradoxus Sarbhoy & Elphick, Trans. Brit. Mycol. Soc. 51: 156. 1968. MycoBank MB265229
= Aspergillus ingratus Yaguchi, Someya & Udagawa, Trans. Mycol. Soc. Japan 34: 305. 1993. MycoBank MB361187
[Non Trichocoma paradoxa Jungh., Praemissa in floram cryptogamicam Javae insulae: 9. 1838. MycoBank MB161024]
Typus: New Zealand, Wellington, dung of opossum, 1948, isolated by J.H. Warcup, Neotype IMI 061446, culture ex-type CBS 527.65 = NRRL 2162 = ATCC 16918 = IMI 061446
Penicillium paradoxum (and P. malodoratum) deviate from other Penicillium species by being phototropic (Raper & Fennell 1965) and occasionally having a well-developed vesicle that makes the conidophore appears like an aspergillum (illustrated by Yaguchi et al. 1993 in the protologue of Aspergillus ingratus). However, stipes were described as small, thin-walled and even septate, and the illustration by Raper & Fennell (1965) indicates a conidial head not unlike P. crystallinum and P. malodoratum. The latter authors also noted that A. crystallinus and A. malodoratus occasionally produced triseriate “sterigmata” and “conidial structures, particularly when small, tend to resemble those of the genus Penicillium”. Most isolates of P. paradoxum are apparently from dung. Penicillium paradoxum, P. malodoratus and P. crystallinum produce strong penetrating odours, similar to those known from coprophilous species in Penicillium series Claviformia, apparently an adaptation to the dung habitat.
Penicillium crystallinum (Kwon-Chung & Fennell) Samson, Houbraken, Visagie & Frisvad, comb. nov. MycoBank MB809315. Fig. 5.
Basionym: Aspergillus crystallinus Kwon-Chung & Fennell, The Genus Aspergillus: 471. 1965. MycoBank MB326624
Typus: Costa Rica, Province of Guanacaste, Tilaran, forest soil, 1962, isolated by K.J. Kwon & D.I. Fennell, Neotype IMI 139270, culture ex-type CBS 479.65 = NRRL 5082 = ATCC 16833 = IMI 139270
Penicillium crystallinum is only known from its ex-type strain (CBS 479.65 = NRRL 5082 = ATCC 16833 = IMI 139270), isolated from forest soil in Costa Rica. The strain shows typical sporulation, but the soft compacted masses of thin-walled cells suggesting sclerotia described by Raper & Fennell (1965) were not observed. The yellow needle crystals of pachybasin observed in the type strain are no longer conspicuously present, but the extrolite profile shows that the strain still produces this compound. The strain also produces the pungent odour noted in P. paradoxum and P. malodoratum.
Penicillium malodoratum (Kwon-Chung & Fennell) Samson, Houbraken, Visagie & Frisvad, comb. nov. MycoBank MB809316. Fig. 6.
Basionym: Aspergillus malodoratus Kwon-Chung & Fennell, The genus Aspergillus: 468. 1965. MycoBank MB326644
Typus: Costa Rica, Province of Puntarenas, Barranca, forest soil, 1962, isolated by K.J. Kwon & D.I. Fennell, Neotype IMI 172289, culture ex-type CBS 490.65 = NRRL 5083 = IMI 172289 = ATCC 16834
Penicillium malodoratum is only known from its ex-type strain (CBS 490.65 = NRRL 5083 = IMI 172289 = ATCC 16834), isolated from forest soil in Costa Rica. Colonies contain numerous long, phototropic conidiophores. Raper & Fennell (1965) observed abundant soft sclerotium-like bodies with thick-walled cells, but we could not detect them. As the name suggests, this fungus produces an unpleasant odour on all media tested. Although A. malodoratus and A. crystallinus were isolated from forest soil in Costa Rica and have similar morphological characters, the taxa can be distinguished by the colony diameter, the length of the conidiophores and the size and shape of the conidia. Conidia of A. crystallinus are globose, 4–7 μm, echinulate, while those of A. malodoratus are subglobose to ellipsoidal, 3–3.5 × 3.5–4 μm and mainly smooth.
Proposed list of accepted species in the genus Penicillium
The following list includes species names that are accepted in the genus Penicillium on 8 August 2014. This list is updated from previous lists published by Pitt & Samson (1993) and Pitt et al. (2000). This revision was considered necessary following changes made to the ICN and the move to single name nomenclature (McNeill et al. 2012), the large number of species described since the 2000 list and new taxonomic information provided by molecular data.
The most dramatic changes in Penicillium are the incorporation of Eupenicillium and several other genera as synonyms (Houbraken & Samson 2011), and the transfer of species of the former Penicillium subgenus Biverticillium species to Talaromyces (Samson et al. 2011). Aspergillus paradoxus, A. crystallinus and A. malodoratus were shown to belong to Penicillium and were transferred above. Several species described as Penicillium belongs to other genera and not to Penicillium (Houbraken & Samson 2011). They are listed below the accepted species list.
Even though the updating of the accepted species list began for nomenclatural purposes, we aim to make this list more functional by including additional information linked to the species names. The list thus includes MycoBank numbers where complete nomenclatural data can be obtained, collection numbers of ex-type strains for future taxonomists requiring authenticated reference material, the species current sectional classification, and GenBank accession numbers for ITS barcodes and, where available, alternative molecular identification markers for BenA, CaM and RPB2.
Despite the considerable amount of time and effort spent on completing this list, there is the possibility of errors or oversights. As such we solicit and gratefully accept any comments on missing names, errors or new data that has become available, especially when publishing a new species. We would also appreciate suggestions for making the list more useful. The active version of the list is currently hosted on the ICPA website (http://www.aspergilluspenicillium.org) where it will be updated as new information comes to light. The website contains a portal for comments, for the convenience of our users. The list published here contains only accepted species; the online version will in future also include data for synonyms. Similar lists are available for Aspergillus (Samson et al. 2014) and Talaromyces (Yilmaz et al. 2014).
Penicillium Link, Mag. Ges. Naturf. Freunde Berlin 3: 16. 1809. MycoBank MB9257.
= Coremium Link, Mag. Ges. Naturf. Freunde Berlin 3: 19. 1809, fide Raper & Thom 1949, Seifert & Samson 1985. [MB7782]. anamorphic synonym.
= Floccaria Grev., Scott. crypt. fl.: pl. 301. 1827, fide Seifert & Samson 1985. [MB8260]. anamorphic synonym.
?= Hormodendrum Bonord., Handbuch allg. Mykol.: 76. 1851 fide de Hoog & Hermanides-Nijhof 1977, but see Hughes 1958. [MB8558]. anamorphic synonym.
?= Walzia Sorokin, Trudy Obshch. Ispyt. Prir. Imp. Kharkov: 47. 1871, fide Constantin 1888. [MB10429]. anamorphic synonym.
= Pritzeliella Henn., Hedwigia Beibl.42: 88. 1903, fide Clements & Shear 1931. [MB9529]. anamorphic synonym.
= Eupenicillium F. Ludw., Lehrbuch der Niederen Kryptogamen: 263. 1892, fide Houbraken & Samson 2011. [MB1933]. teleomorphic synonym.
= Citromyces Wehmer, Ber. Deutsch. Bot. Ges. 11: 333. 1893, fide Thom 1930. [MB7672]. anamorphic synonym.
= Aspergillopsis Sopp, Skr. Vidensk.-Selsk. Christiana Math.-Nat. Kl. 11: 204. 1912, non Aspergillopsis Speg. 1910, fide Pitt 1979. [MB22043]. anamorphic synonym.
= Carpenteles Langeron, Crypt. Fr. Exs.: 344. 1922, fide Stolk & Scott 1967. [MB826]. teleomorphic synonym (= Eupenicillium).
= Torulomyces Delitsch, Systematik der Schimmelpilze: 91. 1943, fide Stolk & Samson 1983 and Houbraken & Samson 2011. [MB10252]. anamorphic synonym.
= Eladia Smith, Trans. Brit. Mycol. Soc. 44: 47. 1961, fide Samson 1981, Houbraken & Samson 2011. [MB8134]. anamorphic synonym.
= Thysanophora Kendrick, Can. J. Bot. 39: 820. 1961, fide Houbraken & Samson 2011. [MB10230]. anamorphic synonym.
= Hemicarpenteles Sarbhoy & Elphick, Trans. Brit. Mycol. Soc. 51: 156. 1968, fide Houbraken & Samson 2011. [MB2279]. teleomorphic synonym.
= Penicillium Link ex Gray sensu Pitt, The Genus Penicillium: 154. 1979 (nom. inval., art 13e). anamorphic synonym.
= Chromocleista Yaguchi & Udagawa, Trans. Mycol. Soc. Japan 34: 101. 1993, fide Houbraken & Samson 2011. [MB25855]. teleomorphic synonym.
Penicillium abidjanum Stolk, Antonie van Leeuwenhoek 34: 49. 1968 ≡ Eupenicillium abidjanum Stolk, Antonie van Leeuwenhoek 34: 49. 1968. [MB335705]. — Herb.: CBS 246.67. Ex-type: CBS 246.67 = ATCC 18385 = FRR 1156 = IMI 136244. Section Lanata-Divaricata. ITS barcode: GU981582. (Alternative markers: BenA = GU981650; RPB2 = JN121469; CaM = KF296383).
Penicillium adametzii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat., 1927: 507. 1927. [MB119777]. — Herb.: IMI 39751. Ex-type: CBS 209.28 = ATCC 10407 = IMI 039751 = MUCL 29106 = NRRL 737. Section Sclerotiora. ITS barcode: JN714929. (Alternative markers: BenA = JN625957; RPB2 = JN121455; CaM = KC773796).
Penicillium adametzioides S. Abe ex G. Sm., Trans. Brit. Mycol. Soc. 46: 335. 1963 ≡ Penicillium adametzioides S. Abe, J. Gen. Appl. Microbiol. 2: 68. 1956 (nom. inval., Art. 36). [MB302372]. — Herb.: CBS 313.59. Ex-type: CBS 313.59 = ATCC 18306 = FAT1302 = IFO 6055 = IMI 068227 = NRRL 3405 = QM 7312. Section Sclerotiora. ITS barcode: JN686433. (Alternative markers: BenA = JN799642; RPB2 = JN406578; CaM = JN686387).
Penicillium albocoremium (Frisvad) Frisvad, Int. Mod. Tax. Meth. Pen. Asp. Clas.: 275. 2000 ≡ Penicillium hirsutum var. albocoremium Frisvad, Mycologia 81: 856. 1990. [MB459817]. — Herb.: IMI 285511. Ex-type: CBS 472.84 = FRR 2931 = IBT 10682 = IBT 21502 = IMI 285511. Section Fasciculata. ITS barcode: AJ004819. (Alternative markers: BenA = AY674326; RPB2 = n.a.; CaM = n.a.).
Penicillium alexiae Visagie Houbraken & Samson, Persoonia 31: 59. 2013. [MB803785]. — Herb.: CBS H-21142. Ex-type: CBS 134558. Section Sclerotiora. ITS barcode: KC790400. (Alternative markers: BenA = KC773778; RPB2 = n.a.; CaM = KC773803).
Penicillium alfredii Visagie, Seifert & Samson, Stud. Mycol. 78: 116. 2014. [MB809180]. — Herb.: CBS H-21800. Ex-type: CBS 138224 = DTO 269A4. Section unclassified. ITS barcode: KJ775684. (Alternative markers: BenA = KJ775177; RPB2 = KJ834520; CaM = KJ775411).
Penicillium allii Vincent & Pitt, Mycologia 81: 300. 1989 ≡ Penicillium hirsutum var. allii (Vincent & Pitt) Frisvad, Mycologia 81: 855. 1989. [MB125498]. — Herb.: MU Vincent 114. Ex-type: CBS 131.89 = IMI 321505 = NRRL 13630 = ATCC 64636 = IMI 321506. Section Fasciculata. ITS barcode: AJ005484. (Alternative markers: BenA = AY674331; RPB2 = n.a.; CaM = n.a.).
Penicillium allii-sativi Frisvad, Houbraken & Samson, Persoonia 29: 89. 2012. [MB801873]. — Herb.: CBS H-21058. Ex-type: CBS 132074 = DTO 149A8 = IBT 26507 = LJC 206. Section Chrysogena. ITS barcode: JX997021. (Alternative markers: BenA = JX996891; RPB2 = JX996627; CaM = JX996232).
Penicillium alutaceum D.B. Scott, Mycopathol. Mycol. Appl. 36: 17. 1968 ≡ Eupenicillium alutaceum D.B. Scott, Mycopathol. Mycol. Appl. 36: 17. 1968. [MB335708]. — Herb.: CBS 317.67. Ex-type: CBS 317.67 = ATCC 18542 = FRR 1158 = IFO 31728 = IMI 136243. Section Exilicaulis. ITS barcode: AF033454. (Alternative markers: BenA = KJ834430; RPB2 = JN121489; CaM = n.a.).
Penicillium amaliae Visagie, Houbraken & K. Jacobs, Persoonia 31: 52. 2013. [MB803784]. — Herb.: CBS H-21141. Ex-type: CBS 134209 = CV 1875 = DTO 183F3 = DAOM 241034. Section Sclerotiora. ITS barcode: JX091443. (Alternative markers: BenA = JX091563; RPB2 = n.a.; CaM = JX141557).
Penicillium anatolicum Stolk, Antonie van Leenwenhoek 34: 46. 1968 ≡ Eupenicillium anatolicum Stolk, Antonie van Leenwenhoek 34: 46. 1968. [MB335710]. — Herb.: CBS 479.66. Ex-type: CBS 479.66 = IBT 30764. Section Citrina. ITS barcode: AF033425. (Alternative markers: BenA = JN606849; RPB2 = JN606593; CaM = JN606571).
Penicillium angulare S.W. Peterson, E.M. Bayer & Wicklow, Mycologia 96: 1289. 2004. [MB487891]. — Herb.: BPI 842268. Ex-type: CBS 130293 = IBT 27051 = NRRL 28157. Section Sclerotiora. ITS barcode: AF125937. (Alternative markers: BenA = KC773779; RPB2 = JN406554; CaM = KC773804).
Penicillium angustiporcatum Takada & Udagawa, Trans. Mycol. Soc. Japan 24: 143. 1983 ≡ Eupenicillium angustiporcatum Takada & Udagawa, Trans. Mycol. Soc. Japan 24: 143. 1983. [MB108322]. — Herb.: NHL 6481. Ex-type: CBS 202.84. Section Gracilenta. ITS barcode: KC411690. (Alternative markers: BenA = KJ834431; RPB2 = JN406617; CaM = n.a.).
Penicillium antarcticum A.D. Hocking & C.F. McRae, Polar Biol. 21: 103. 1999. [MB482749]. — Herb.: DAR 72813. Ex-type: CBS 100492 = FRR 4989. Section Canescentia. ITS barcode: KJ834503. (Alternative markers: BenA = KJ834432; RPB2 = JN406653; CaM = n.a.).
Penicillium araracuaraense Houbraken, et al., Int. J. Syst. Evol. Microbiol. 61: 1469. 2011. [MB518025]. — Herb.: HUA 170334. Ex-type: CBS 113149 = IBT 23247. Section Lanata-Divaricata. ITS barcode: GU981597. (Alternative markers: BenA = GU981642; RPB2 = KF296414; CaM = KF296373).
Penicillium ardesiacum Novobr., Novosti Sist. Nizsh. Rast. 11: 228. 1974. [MB319257]. — Herb.: IMI 174719. Ex-type: CBS 497.73 = ATCC 24719 = FRR 1479 = IFO 30540 = IMI 174719 = VKMF-1749. Section Aspergilloides. ITS barcode: KM189565. (Alternative markers: BenA = KM088805; RPB2 = KM089577; CaM = KM089190).
Penicillium argentinense Houbraken, Frisvad & Samson, Stud. Mycol. 70: 78. 2011. [MB563185]. — Herb.: CBS H-20461. Ex-type: CBS 130371 = IBT 30761. Section Citrina. ITS barcode: JN831361. (Alternative markers: BenA = JN606815; RPB2 = n.a.; CaM = JN606549).
Penicillium arianeae Visagie, Houbraken & Samson, Persoonia 31: 59. 2013. [MB803786]. — Herb.: CBS H-21143. Ex-type: CBS 134559. Section Sclerotiora. ITS barcode: KC773833. (Alternative markers: BenA = KC773784; RPB2 = n.a.; CaM = KC773811).
Penicillium armarii Houbraken, et al. Stud. Mycol. 78: 410. 2014. [MB809955]. — Herb.: CBS H-21870. Ex-type: CBS 138171 = DTO 235-F1. Section Aspergilloides. ITS barcode: KM189758. (Alternative markers: BenA = KM089007; RPB2 = KM089781; CaM = KM089394).
Penicillium astrolabium R. Serra & S.W. Peterson, Mycologia 99: 80. 2007. [MB504766]. — Herb.: BPI 872160. Ex-type: CBS 122427 = NRRL 35611 = MUM 06.161. Section Brevicompacta. ITS barcode: DQ645804. (Alternative markers: BenA = DQ645793; RPB2 = JN406634; CaM = DQ645808).
Penicillium athertonense Houbraken, Stud. Mycol. 78: 412. 2014. [MB809956]. — Herb.: CBS H-21874. Ex-type: CBS 138161 = DTO 030-C2. Section Aspergilloides. ITS barcode: KM189462. (Alternative markers: BenA = KM088690; RPB2 = KM089462; CaM = KM089075).
Penicillium atramentosum Thom, U.S.D.A. Bur. Animal Industr. Bull. 118: 65. 1910. [MB237291]. — Herb.: IMI 39752. Ex-type: CBS 291.48 = ATCC 10104 = FRR 795 = IBT 6616 = IFO 8137 = IMI 039752 = IMI 039752ii = LSHBP 1 = MUCL 29071 = MUCL 29126 = NRRL 795 = QM 7483. Section Paradoxa. ITS barcode: AF033483. (Alternative markers: BenA = AY674402; RPB2 = JN406584; CaM = FJ530964).
Penicillium atrofulvum Houbraken, Frisvad & Samson, Stud. Mycol. 70: 80. 2011. [MB563183]. — Herb.: CBS H-20650. Ex-type: CBS 109.66 = DTO 31B2 = FRR 799 = IBT 30032 = IBT 29667. Section Citrina. ITS barcode: JN617663. (Alternative markers: BenA = JN606677; RPB2 = JN606620; CaM = JN606387).
Penicillium atrosanguineum B.X. Dong, Ceská Mycol. 27: 174. 1973. [MB319260]. — Herb.: CBS H-15524. Ex-type: CBS 380.75 = FRR 1726 = IMI 197488. Section Exilicaulis. ITS barcode: JN617706. (Alternative markers: BenA = KJ834435; RPB2 = JN406557; CaM = n.a.).
Penicillium atrovenetum G. Sm., Trans. Brit. Mycol. Soc. 39: 112. 1956. [MB302377]. — Herb.: IMI 061837. Ex-type: CBS 241.56 = ATCC 13352 = FRR 2571 = IFO 8138 = = IMI 061837 = LSHBSm683 = QM 6963. Section Canescentia. ITS barcode: AF033492. (Alternative markers: BenA = JX140944; RPB2 = JN121467; CaM = KJ867004).
Penicillium aurantiacobrunneum Houbraken, Frisvad & Samson, Stud. Mycol. 70: 80. 2011. [MB563206]. — Herb.: CBS H-20662. Ex-type: CBS 126228 = IBT 18753. Section Citrina. ITS barcode: JN617670. (Alternative markers: BenA = JN606702; RPB2 = n.a.; CaM = JN606522).
Penicillium aurantiogriseum Dierckx, Ann. Soc. Sci. Bruxelles 25: 88. 1901. [MB247956]. — Herb.: IMI 195050. Ex-type: CBS 249.89 = ATCC 48920 = FRR 971 = IBT 14016 = IMI 195050 = MUCL 29090 = NRRL 971. Section Fasciculata. ITS barcode: AF033476. (Alternative markers: BenA = AY674296; RPB2 = JN406573; CaM = n.a.).
Penicillium aurantioviolaceum Biourge, Cellule 33: 282. 1923. [MB257885]. — Herb.: CBS H-21954. Ex-type: CBS 137777 = NRRL 762 = ATCC 14974. Section Aspergilloides. ITS barcode: KM189756. (Alternative markers: BenA = KM089005; RPB2 = KM089779; CaM = KM089392).
Penicillium austroafricanum Houbraken & Visagie, Stud. Mycol. 78: 412. 2014. [MB809957]. — Herb.: CBS H-21864. Ex-type: CBS 137773 = DTO 133-G5. Section Aspergilloides. ITS barcode: KM189610. (Alternative markers: BenA = KM088854; RPB2 = KM089628; CaM = KM089241).
Penicillium bialowiezense K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 450. 1927. [MB258429]. — Herb.: unknown. Ex-type: CBS 227.28 = IBT 23044 = IMI 092237 = LSHBP 71 = NRRL 865. Section Brevicompacta. ITS barcode: EU587315. (Alternative markers: BenA = AY674439; RPB2 = JN406604; CaM = AY484828).
Penicillium biforme Thom, U.S.D.A. Bur. Animal Industr. Bull. 118: 54. 1910. [MB240764]. — Herb.: unnknown. Ex-type: CBS 297.48 = ATCC 10416 = FRR 885 = IFO 7722 = IMI 039820 = LSHB P72 = MUCL 29165 = NRRL 885 = QM 7492. Section Fasciculata. ITS barcode: KC411731. (Alternative markers: BenA = FJ930944; RPB2 = n.a.; CaM = n.a.).
Penicillium bilaiae Chalab., Bot. Mater. Otd. Sporov. Rast. 6: 165. 1950. [MB302379]. — Herb.: IMI 113677. Ex-type: CBS 221.66 = ATCC 22348 = ATCC 48731 = CCRC 31675 = FRR 3391 = IJFM 5025 = IMI 113677 = MUCL 31187 = VKMF-854. Section Sclerotiora. ITS barcode: JN714937. (Alternative markers: BenA = JN625966; RPB2 = JN406610; CaM = JN626009).
Penicillium boreae S.W. Peterson & Sigler, Mycol. Res. 106: 1112. 2002. [MB483980]. — Herb.: BPI 841395. Ex-type: CBS 111717 = NRRL 31002 = UAMH 3896. Section Stolkia. ITS barcode: AF481122. (Alternative markers: BenA = JN617715; RPB2 = n.a.; CaM = AF481138).
Penicillium bovifimosum (Tuthill & Frisvad) Houbraken & Samson, Stud. Mycol. 70: 47. 2011 ≡ Eupenicillium bovifimosum Tuthill & Frisvad, Mycologia 94: 241. 2002. [MB561957]. — Herb.: WY RMF 82071. Ex-type: CBS 102825 = RMF 9598. Section Turbata. ITS barcode: AF263347. (Alternative markers: BenA = KJ834436; RPB2 = JN406649; CaM = FJ530989).
Penicillium brasilianum Bat., Anais Soc. Biol. Pernambuco 15: 162. 1957. [MB302381]. — Herb.: URM IMUR 56. Ex-type: CBS 253.55 = ATCC 12072 = FRR 3466 = QM 6947. Section Lanata-Divaricata. ITS barcode: GU981577. (Alternative markers: BenA = GU981629; RPB2 = KF296420; CaM = AB667857).
Penicillium brefeldianum B.O. Dodge, Mycologia 25: 92. 1933 ≡ Carpenteles brefeldianum (B.O. Dodge) Shear, Mycologia 26: 107. 1934 ≡ Eupenicillium brefeldianum (B.O. Dodge) Stolk & D.B. Scott, Persoonia 4: 400. 1967 ≡ Penicillium dodgei Pitt, Genus Penicillium: 117. 1980. [MB258851]. — Herb.: IMI 216896. Ex-type: CBS 235.81 = NRRL 710 = FRR 710 = IFO 31731 = IMI 216896 = LCP 89.2573 = LCP 89.2578 = MUCL 38762 = QM 1872 = Thom 5296. Section Lanata-Divaricata. ITS barcode: AF033435. (Alternative markers: BenA = GU981623; RPB2 = KF296421; CaM = EU021683).
Penicillium brevicompactum Dierckx, Ann. Soc. Sci. Bruxelles 25: 88. 1901. [MB149773]. — Herb.: IMI 40225. Ex-type: CBS 257.29 = ATCC 10418 = ATCC 9056 = DSM3825 = FRR 862 = IBT 23045 = IMI 040225 = LSHBP 75 = MUCL 28647 = MUCL 28813 = MUCL 28935 = MUCL 30240 = MUCL 30241 = MUCL 30256 = MUCL 30257 = NRRL 2011 = NRRL 862 = NRRL 864 = QM 7496. Section Brevicompacta. ITS barcode: AY484912. (Alternative markers: BenA = AY674437; RPB2 = JN406594; CaM = AY484813).
Penicillium brevistipitatum L. Wang & W.Y. Zhuang, Mycotaxon 93: 234. 2005. [MB356064]. — Herb.: HMAS 130354-1-4. Ex-type: AS 3.6887. Section Penicillium. ITS barcode: DQ221696. (Alternative markers: BenA = DQ221695; RPB2 = JN406528; CaM = n.a.).
Penicillium brocae S.W. Peterson et al., Mycologia 95: 143. 2003. [MB373658]. — Herb.: BPI 841763. Ex-type: CBS 116113 = IBT 26293 = NRRL 31472. Section Sclerotiora. ITS barcode: AF484398. (Alternative markers: BenA = KC773787; RPB2 = JN406639; CaM = KC773814).
Penicillium brunneoconidiatum Visagie, Houbraken & K. Jacobs, Stud. Mycol. 78: 415. 2014. [MB809958]. — Herb.: CBS H-21873. Ex-type: CBS 137732 = DTO 182-E4 = CV 949 = DAOM 241359. Section Aspergilloides. ITS barcode: KM189666. (Alternative markers: BenA = KM088911; RPB2 = KM089685; CaM = KM089298).
Penicillium buchwaldii Frisvad & Samson, FEMS Microbiol. Lett. 339: 86. 2013. [MB800966]. — Herb.: IMI 304286. Ex-type: CBS 117181 = IBT 6005 = IMI 304286. Section Brevicompacta. ITS barcode: JX313164. (Alternative markers: BenA = JX313182; RPB2 = JN406637; CaM = JX313148).
Penicillium burgense Quintan., Av. Aliment. Majora Anim. 30: 176. 1990. [MB130241]. — Herb.: Sáez 1538. Ex-type: CBS 325.89. Section Exilicaulis. ITS barcode: KC411736. (Alternative markers: BenA = KJ834437; RPB2 = JN406572; CaM = n.a.).
Penicillium bussumense Houbraken, Stud. Mycol. 78: 415. 2014. [MB809959]. — Herb.: CBS H-21869. Ex-type: CBS 138160 = DTO 018-B2. Section Aspergilloides. ITS barcode: KM189458. (Alternative markers: BenA = KM088685; RPB2 = KM089457; CaM = KM089070).
Penicillium cainii K.G. Rivera, Malloch & Seifert, Stud. Mycol. 70: 147. 2011. [MB563159]. — Herb.: DAOM 239914. Ex-type: CCFC 239914. Section Sclerotiora. ITS barcode: JN686435. (Alternative markers: BenA = JN686366; RPB2 = n.a.; CaM = JN686389).
Penicillium cairnsense Houbraken, Frisvad & Samson, Stud. Mycol. 70: 83. 2011. [MB563184]. — Herb.: CBS H-20686. Ex-type: CBS 124325 = IBT 29042. Section Citrina. ITS barcode: JN617669. (Alternative markers: BenA = JN606692; RPB2 = n.a.; CaM = JN606512).
Penicillium camemberti Thom, U.S.D.A. Bur. Animal Industr. Bull. 82: 33. 1906. [MB175171]. — Herb.: IMI 27831. Ex-type: CBS 299.48 = ATCC 1105 = ATCC 4845 = FRR 878 = IBT 21508 = IMI 027831 = IMI 092200 = LCP 66.584 = LSHBP 11 = MUCL 29790 = NCTC 582 = NRRL 877 = NRRL 878. Section Fasciculata. ITS barcode: AB479314. (Alternative markers: BenA = FJ930956; RPB2 = JN121484; CaM = n.a.).
Penicillium canariense S.W. Peterson & Sigler, Mycol. Res. 106: 1113. 2002. [MB483981]. — Herb.: BPI 841396. Ex-type: CBS 111720 = NRRL 31003 = IJFM 536 = UAMH 6403. Section Stolkia. ITS barcode: AF481121. (Alternative markers: BenA = JN617714; RPB2 = n.a.; CaM = AF481137).
Penicillium canescens Sopp, Skr. Vidensk.-Selsk. Christiana Math.-Nat. Kl. 11: 181. 1912. [MB153765]. — Herb.: IMI 28260. Ex-type: CBS 300.48 = ATCC 10419 = DSM1215 = FRR 910 = IMI 028260 = MUCL 29169 = NCTC 6607 = NRRL 910 = QM 7550 = VKMF-1148. Section Canescentia. ITS barcode: AF033493. (Alternative markers: BenA = JX140946; RPB2 = JN121485; CaM = KJ867009).
Penicillium canis S.W. Peterson, J. Clin. Microbiol. (in press). [MB807056]. — Herb.: BPI 892763. Ex-type: NRRL 62798. Section Canescentia. ITS barcode: KJ511291. (Alternative markers: BenA = KF900167; RPB2 = KF900196; CaM = KF900177).
Penicillium caperatum Udagawa & Y. Horie, Trans. Mycol. Soc. Japan 14: 371. 1973 ≡ Eupenicillium caperatum Udagawa and Y. Horie, Trans. Mycol. Soc. Japan 14: 371. 1973. [MB319262]. — Herb.: unknown. Ex-type: CBS 443.75 = ATCC 28046 = DSM2209 = NHL 6465. Section Lanata-Divaricata. ITS barcode: KC411761. (Alternative markers: BenA = GU981660; RPB2 = KF296422; CaM = KF296392).
Penicillium capsulatum Raper & Fennell, Mycologia 40: 528. 1948. [MB289079]. — Herb.: IMI 40576. Ex-type: CBS 301.48 = ATCC 10420 = DSM2210 = FRR 2056 = IJFM 5120 = IMI 040576 = NRRL 2056 = QM 4869 = VKMF-445. Section Ramigena. ITS barcode: AF033429. (Alternative markers: BenA = FJ004390; RPB2 = JN406582; CaM = n.a.).
Penicillium carneum (Frisvad) Frisvad, Microbiology, UK, 142: 546. 1996 ≡ Penicillium roqueforti var. carneum Frisvad, Mycologia 81: 858. 1990. [MB415652]. — Herb.: IMI 293204. Ex-type: CBS 112297 = IBT 6884 = IBT 18419 = IMI 293204. Section Roquefortorum. ITS barcode: HQ442338. (Alternative markers: BenA = AY674386; RPB2 = JN406642; CaM = HQ442322).
Penicillium cartierense Houbraken, Stud. Mycol. 78: 415. 2014. [MB809960]. — Herb.: CBS H-21861. Ex-type: CBS 137956 = DTO 092-H9. Section Aspergilloides. ITS barcode: KM189564. (Alternative markers: BenA = KM088804; RPB2 = KM089576; CaM = KM089189).
Penicillium caseifulvum Lund, Filt. & Frisvad, J. Food Mycol. 1: 97. 1998. [MB446013]. — Herb.: C 24999. Ex-type: CBS 101134 = IBT 18282 = IBT 21510. Section Fasciculata. ITS barcode: KJ834504. (Alternative markers: BenA = AY674372; RPB2 = n.a.; CaM = n.a.).
Penicillium catenatum D.B. Scott, Mycopathol. Mycol. Appl. 36: 24. 1968 ≡ Eupenicillium catenatum D.B. Scott, Mycopathol. Mycol. Appl. 36: 24. 1968. [MB335719]. — Herb.: CBS 352.67. Ex-type: CBS 352.67 = ATCC 18543 = CSIR 1097 = IFO 31774 = IMI 136241. Section Exilicaulis. ITS barcode: KC411754. (Alternative markers: BenA = KJ834438; RPB2 = JN121504; CaM = n.a.).
Penicillium cavernicola Frisvad & Samson, Stud. Mycol. 49: 31. 2004. [MB370976]. — Herb.: CBS H-13441. Ex-type: CBS 100540 = IBT 14499. Section Fasciculata. ITS barcode: KJ834505. (Alternative markers: BenA = KJ834439; RPB2 = n.a.; CaM = n.a.).
Penicillium chalybeum Pitt & A.D. Hocking, Mycotaxon 22: 204. 1985. [MB105608]. — Herb.: FRR 2660. Ex-type: CBS 254.87 = FRR 2660. Section Exilicaulis. ITS barcode: KC411713. (Alternative markers: BenA = KJ834440; RPB2 = JN406596; CaM = n.a.).
Penicillium charlesii G. Sm., Trans. Brit. Mycol. Soc. 18: 90. 1933. [MB260433]. — Herb.: unknown. Ex-type: CBS 304.48 = ATCC 8730 = CBS 342.51 = CECT 2277 = FRR 778 = IMI 040232 = LSHBBB127 = LSHBP 146 = NRRL 1887 = NRRL 778 = QM 6338 = QM 6838. Section Charlesia. ITS barcode: AF033400. (Alternative markers: BenA = JX091508; RPB2 = JN121486; CaM = AY741727).
Penicillium chermesinum Biourge, Cellule 33: 284. 1923. [MB260472]. — Herb.: IMI 191730. Ex-type: CBS 231.81 = NRRL 2048 = FRR 2048 = IFO 31745 = IMI 191730. Section Cinnamopurpurea. ITS barcode: AY742693. (Alternative markers: BenA = KJ834441; RPB2 = JN406581; CaM = AY741728).
Penicillium christenseniae Houbraken, Frisvad & Samson, Stud. Mycol. 70: 85. 2011. [MB563187]. — Herb.: CBS H-20656. Ex-type: CBS 126236 = IBT 23355. Section Citrina. ITS barcode: JN617674. (Alternative markers: BenA = JN606680; RPB2 = JN606624; CaM = JN606373).
Penicillium chrysogenum Thom, U.S.D.A. Bur. Animal Industr. Bull. 118: 58. 1910. [MB165757]. — Herb.: IMI 24314. Ex-type: CBS 306.48 = ATCC 10106 = ATHUM2889 = CCRC 30564 = FRR 807 = IBT 5233 = IMI 024314 = IMI 092208 = LSHBAd 3 = LSHBP 19 = MUCL 29079 = MUCL 29145 = NCTC 589 = NRRL 807 = NRRL 810 = QM 7500. Section Chrysogena. ITS barcode: AF033465. (Alternative markers: BenA = AY495981; RPB2 = JN121487; CaM = JX996273).
Penicillium chrzaszczii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 464. 1927. [MB260609]. — Herb.: unknown. Ex-type: CBS 217.28 = FRR 903 = MUCL 29167 = NRRL 1741 = NRRL 903. Section Citrina. ITS barcode: GU944603. (Alternative markers: BenA = JN606758; RPB2 = JN606628; CaM = JN606423).
Penicillium cinerascens Biourge, Cellule 33: 308. 1923. [MB260785]. — Herb.: IMI 92234. Ex-type: NRRL 748 = ATCC 48693 = BIOURGE 90 = FRR 748 = IMI 92234 = QM 7555 = Thom 4733.34. Section Exilicaulis. ITS barcode: AF033455. (Alternative markers: BenA = JX141041; RPB2 = n.a.; CaM = n.a.).
Penicillium cinereoatrum Chalab., Bot. Mater. Otd. Sporov. Rast. 6: 167. 1950. [MB302385]. — Herb.: CBS H-7469. Ex-type: CBS 222.66 = ATCC 22350 = FRR 3390 = IJFM 5024 = IMI 113676 = VKMF-856. Section Exilicaulis. ITS barcode: KC411700. (Alternative markers: BenA = KJ834442; RPB2 = JN406608; CaM = KJ867002).
Penicillium cinnamopurpureum Udagawa, J. Agric. Food Sci., Tokyo 5: 1. 1959. [MB302386]. — Herb.: unknown. Ex-type: CBS 429.65 = CBS 847.68 = NRRL 162 = ATCC 18489 = CSIR 936 = FAT 362 = IAM 7016 = IFO 6032 = NHL 6359 = QM 7888. Section Cinnamopurpurea. ITS barcode: EF626950. (Alternative markers: BenA = EF626948; RPB2 = JN406533; CaM = EF626949).
Penicillium citreonigrum Dierckx, Ann. Soc. Sci. Bruxelles 25: 86. 1901. [MB165197]. — Herb.: IMI 92209i. Ex-type: CBS 258.29 = ATCC 48736 = FRR 761 = IMI 092209 = LSHBP 20 = LSHBP 98 = MUCL 28648 = MUCL 29062 = MUCL 29116 = NRRL 761. Section Exilicaulis. ITS barcode: AF033456. (Alternative markers: BenA = EF198621; RPB2 = JN121474; CaM = EF198628).
Penicillium citrinum Thom, U.S.D.A. Bur. Animal Industr. Bull. 118: 61. 1910. [MB165293]. — Herb.: IMI 92196ii. Ex-type: CBS 139.45 = ATCC 1109 = ATCC 36382 = CECT 2269 = FRR 1841 = IMI 091961 = IMI 092196 = LSHBAd 95 = LSHBP 25 = LSHBP 6 = MUCL 29781 = NRRL 1841 = NRRL 1842. Section Citrina. ITS barcode: AF033422. (Alternative markers: BenA = GU944545; RPB2 = JF417416; CaM = GU944638).
Penicillium clavigerum Demelius, Verh. Zool.-Bot. Ges. Wien 72: 74. 1923. [MB261069]. — Herb.: IMI 39807. Ex-type: CBS 310.48 = ATCC 10427 = CBS 255.94 = FRR 1003 = IMI 039807 = IMI 039807ii = MUCL 15623 = NRRL 1003 = QM 1918. Section Penicillium. ITS barcode: DQ339555. (Alternative markers: BenA = AY674427; RPB2 = n.a.; CaM = n.a.).
Penicillium clavistipitatum Visagie, Houbraken & K. Jacobs, Stud. Mycol. 78: 419. 2014. [MB809961]. — Herb.: CBS H-21882. Ex-type: CBS 138650 = DTO 182-E5 = CV 336 = KAS 4112 = DAOM 241092. Section Aspergilloides. ITS barcode: KM189667. (Alternative markers: BenA = KM088912; RPB2 = KM089686; CaM = KM089299).
Penicillium cluniae Quintan., Av. Aliment. Mejora Anim. 30: 174. 1990. [MB130240]. — Herb.: unknown. Ex-type: CBS 326.89. Section Lanata-Divaricata. ITS barcode: KF296406. (Alternative markers: BenA = KF296471; RPB2 = KF296424; CaM = KF296402).
Penicillium coeruleum Sopp apud Biourge, Cellule 33: 102. 1923. [MB446014]. — Herb.: unknown. Ex-type: CBS 141.45 = NCTC 6595. Section Lanata-Divaricata. ITS barcode: GU981606. (Alternative markers: BenA = GU981655; RPB2 = KF296425; CaM = KF296393).
Penicillium coffeae S.W. Peterson et al., Mycologia 97: 662. 2005. [MB340281]. — Herb.: BPI 863480. Ex-type: CBS 119387 = IBT 27866 = NRRL 35363. Section Charlesia. ITS barcode: AY742702. (Alternative markers: BenA = KJ834443; RPB2 = JN121436; CaM = AY741747).
Penicillium commune Thom, U.S.D.A. Bur. Animal Industr. Bull. 118: 56. 1910. [MB164241]. — Herb.: IMI 39812. Ex-type: CBS 311.48 = ATCC 10428 = ATCC 1111 = CCRC 31554 = DSM2211 = IBT 6200 = IFO 5763 = IMI 039812ii = IMI 039812iii = NRRL 890 = QM 1269 = VKMF-3233. Section Fasciculata. ITS barcode: AY213672. (Alternative markers: BenA = AY674366; RPB2 = n.a.; CaM = n.a.).
Penicillium concentricum Samson, Stolk & Hadlock, Stud. Mycol. 11: 17. 1976. [MB319263]. — Herb.: CBS 477.75. Ex-type: CBS 477.75 = IBT 14571 = IBT 6577. Section Penicillium. ITS barcode: KC411763. (Alternative markers: BenA = AY674413; RPB2 = n.a.; CaM = n.a.).
Penicillium confertum (Frisvad, Filt. & Wicklow) Frisvad, Mycologia 81: 852. 1990 ≡ Penicillium glandicola var. confertum Frisvad, Filt. & Wicklow, Can. J. Bot. 65: 769. 1987. [MB126404]. — Herb.: IMI 296930. Ex-type: CBS 171.87 = IBT 21515 = IBT 3098 = IBT 5672 = IMI 296930 = NRRL 13488 = NRRL A-26904. Section Chrysogena. ITS barcode: JX997081. (Alternative markers: BenA = AY674373; RPB2 = JX996708; CaM = JX996963).
Penicillium contaminatum Houbraken, Stud. Mycol. 78: 419. 2014. [MB809962]. — Herb.: CBS H-21866. Ex-type: CBS 345.52 = DTO 091-A3 = IMI 049057. Section Aspergilloides. ITS barcode: KM189554. (Alternative markers: BenA = KM088793; RPB2 = KM089565; CaM = KM089178).
Penicillium coprobium Frisvad, Mycologia 81: 853. 1990. [MB126405]. — Herb.: IMI 293209. Ex-type: CBS 561.90 = ATCC 58615 = IBT 21516 = IBT 4583 = IBT 6932. Section Penicillium. ITS barcode: DQ339559. (Alternative markers: BenA = AY674425; RPB2 = n.a.; CaM = n.a.).
Penicillium coprophilum (Berk. & M.A. Curtis) Seifert & Samson, Adv. Penicillium Aspergillus Syst.: 145. 1985 ≡ Coremium coprophilum Berk. & M.A. Curtis, J. Linn. Soc., Bot., 10: 363. 1868. [MB114760]. — Herb.: K Cuba Wright 666. Ex-type: CBS 110760 = IBT 5551 = IBT 3064 = NRRL 13627. Section Penicillium. ITS barcode: AF033469. (Alternative markers: BenA = AY674421; RPB2 = JN406645; CaM = n.a.).
Penicillium copticola Houbraken, Frisvad & Samson, Stud. Mycol. 70: 88. 2011. [MB563205]. — Herb.: CBS H-20643. Ex-type: CBS 127355 = IBT 30771. Section Citrina. ITS barcode: JN617685. (Alternative markers: BenA = JN606805; RPB2 = JN606599; CaM = JN606553).
Penicillium coralligerum Nicot & Pionnat, Bull. Soc. Mycol. France 78: 245. 1963 [1962]. [MB335721]. — Herb.: IMI 99159. Ex-type: CBS 123.65 = ATCC 16968 = FRR 3465 = IFO 9578 = IHEM 4511 = IMI 099159 = LCP 58.1674 = NRRL 3465. Section Canescentia. ITS barcode: JN617667. (Alternative markers: BenA = KJ834444; RPB2 = JN406632; CaM = KJ866994).
Penicillium corylophilum Dierckx, Ann. Soc. Sci. Bruxelles 25: 86. 1901. [MB178294]. — Herb.: IMI 39754. Ex-type: CBS 312.48 = TCC9784 = ATHUM2890 = CECT 2270 = FRR 802 = IMI 039754 = MUCL 28671 = MUCL 29073 = MUCL 29131 = NRRL 802 = QM 7510. Section Exilicaulis. ITS barcode: AF033450. (Alternative markers: BenA = JX141042; RPB2 = n.a.; CaM = n.a.).
Penicillium cosmopolitanum Houbraken, Frisvad & Samson, Stud. Mycol. 70: 91. 2011. [MB563188]. — Herb.: CBS H-20665. Ex-type: CBS 126995 = IBT 30681. Section Citrina. ITS barcode: JN617691. (Alternative markers: BenA = JN606733; RPB2 = n.a.; CaM = JN606472).
Penicillium cremeogriseum Chalab., Bot. Mater. Otd. Sporov. Rast. 6: 168. 1950. [MB302390]. — Herb.: CBS 223.60. Ex-type: CBS 223.66 = ATCC 18320 = ATCC 18323 = FRR 1734 = IJFM 5011 = IMI 197492 = NRRL 3389 = VKMF-1034. Section Lanata-Divaricata. ITS barcode: GU981586. (Alternative markers: BenA = GU981624; RPB2 = KF296426; CaM = KF296403).
Penicillium crocicola W. Yamam., Sci. Rep. Hyogo Univ. Agric. 2: 28. 1956. [MB302391]. — Herb.: CBS H-7528. Ex-type: CBS 745.70 = NRRL 6175 = ATCC 18313 = QM 7778. Section Aspergilloides. ITS barcode: KM189581. (Alternative markers: BenA = KJ834445; RPB2 = JN406535; CaM = KM089210).
Penicillium crustosum Thom, The Penicillia: 399. 1930. [MB262401]. — Herb.: IMI 91917. Ex-type: CBS 115503 = ATCC 52044 = FRR 1669 = IBT 5528 = IBT 6175 = IMI 091917 = NCTC 4002. Section Fasciculata. ITS barcode: AF033472. (Alternative markers: BenA = AY674353; RPB2 = n.a.; CaM = DQ911132).
Penicillium cryptum Goch., Mycotaxon 26: 349. 1986 ≡ Eupenicillium cryptum Goch., Mycotaxon 26: 349. 1986. [MB103648]. — Herb.: NY 769. Ex-type: CBS 271.89 = ATCC 60138 = IMI 296794 = NRRL 13460. Section Torulomyces. ITS barcode: KF303647. (Alternative markers: BenA = KF303608; RPB2 = JN121478; CaM = KF303628).
Penicillium crystallinum (Kwon-Chung & Fennell) Samson et al., (published here) ≡ Aspergillus crystallinus Kwon-Chung & Fennell, The Genus Aspergillus: 471 1965. [MB809315]. — Herb.: IMI 139270. Ex-type: CBS 479.65 = NRRL 5082 = ATCC 16833 = IMI 139270. Section Paradoxa. ITS barcode: AF033486. (Alternative markers: BenA = EF669682; RPB2 = EF669669; CaM = FJ530973).
Penicillium cyaneum (Bainier & Sartory) Biourge, Cellule 33: 102. 1923 ≡ Citromyces cyaneus Bainier & Sartory, Bull. Soc. Mycol. France 29: 157. 1913. [MB251712]. — Herb.: IMI 39744. Ex-type: CBS 315.48 = ATCC 10432 = FRR 775 = IFO 5337 = IMI 039744 = NRRL 775 = QM 7516. Section Ramigena. ITS barcode: AF033427. (Alternative markers: BenA = JX091552; RPB2 = JN406575; CaM = n.a.).
Penicillium cyclopium Westling, Ark. Bot. 11: 90. 1911. [MB156739]. — Herb.: unknown. Ex-type: CBS 144.45 = ATCC 8731 = ATHUM2888 = CECT 2264 = DSM1250 = IBT 5130 = IMI 089372 = LSHBP 123 = MUCL 15613 = NRRL 1888 = QM 6839 = VKMF-265. Section Fasciculata. ITS barcode: JN097811. (Alternative markers: BenA = AY674310; RPB2 = JN985388; CaM = n.a.).
Penicillium daejeonium S.H. Yu & H.K. Sang, J. Microbiol. 51: 537. 2013. [MB561572]. — Herb.: KACC 46609. Ex-type: KACC 46609. Section Sclerotiora. ITS barcode: JX436489. (Alternative markers: BenA = JX436493; RPB2 = n.a.; CaM = JX436491).
Penicillium daleae K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 495. 1927. [MB262773]. — Herb.: IMI 89338. Ex-type: CBS 211.28 = ATCC 10435 = DSM 2449 = FRR 2025 = IFO 6087 = IFO 9072 = IMI 034910 = MUCL 29234 = NRRL 2025. Section Lanata-Divaricata. ITS barcode: GU981583. (Alternative markers: BenA = GU981649; RPB2 = KF296427; CaM = KF296385).
Penicillium decaturense S.W. Peterson, E.M. Bayer & Wicklow, Mycologia 96: 1290. 2004. [MB487890]. — Herb.: BPI 842267. Ex-type: CBS 117509 = NRRL 28152 = IBT 27117. Section Citrina. ITS barcode: GU944604. (Alternative markers: BenA = JN606685; RPB2 = JN606621; CaM = JN606413).
Penicillium decumbens Thom, U.S.D.A. Bur. Animal Industr. Bull. 118: 71. 1910. [MB156582]. — Herb.: IMI 190875. Ex-type: CBS 230.81 = FRR 741 = IMI 190875 = MUCL 29107 = NRRL 741. Section Exilicaulis. ITS barcode: AY157490. (Alternative markers: BenA = KJ834446; RPB2 = JN406601; CaM = n.a.).
Penicillium desertorum Frisvad, Houbraken & Samson, Persoonia 29: 90. 2012. [MB801874]. — Herb.: CBS H-21056. Ex-type: CBS 131543 = IBT 16321 = DTO 148I6. Section Chrysogena. ITS barcode: JX997010. (Alternative markers: BenA = JX996818; RPB2 = JX996682; CaM = JX996937).
Penicillium dierckxii Biourge, Cellule 33: 313. 1923. [MB263175]. — Herb.: IMI 92216. Ex-type: CBS 185.81 = IMI 092216 = LSHBP 32 = MUCL 28665 = NRRL 755. Section Ramigena. ITS barcode: EF634444. (Alternative markers: BenA = EF634442; RPB2 = EF634445; CaM = EF634443).
Penicillium digitatum (Pers.: Fr.) Sacc., Fung. Ital.: tab. 894. 1881 ≡ Aspergillus digitatus Pers., Disp. meth. Fung.: 41. 1794 ≡ Monilia digitata Pers., Syn. Meth. Fung.: 693. 1801: Fr., Syst. Mycol. 3: 411. 1832 ≡ Mucor digitata (Pers.) Mérat, Nouvelle flore des environs de Paris 1: 14. 1821. [MB169502]. — Herb.: Lectotype = icon in Saccardo, Fung. Ital.: tab. 894. Jul 1881. Ex-type: CBS 112082 = IBT 13068. Section Digitata. ITS barcode: KJ834506. (Alternative markers: BenA = KJ834447; RPB2 = JN121426; CaM = n.a.).
Penicillium dimorphosporum H.J. Swart, Trans. Brit. Mycol. Soc. 55: 310. 1970. [MB120334]. — Herb.: CBS 456.70. Ex-type: CBS 456.70 = ATCC 22783 = ATCC 52501 = FRR 1120 = IMI 149680. Section Exilicaulis. ITS barcode: AF081804. (Alternative markers: BenA = KJ834448; RPB2 = JN121517; CaM = n.a.).
Penicillium dipodomyicola (Frisvad, Filt. & Wicklow) Frisvad, Int. Mod. Meth. Pen. Asp. Clas.: 275. 2000. [MB459818]. — Herb.: IMI 296935. Ex-type: CBS 173.87 = IBT 21521 = IMI 296935 = ATCC 64187. Section Penicillium. ITS barcode: AY371616. (Alternative markers: BenA = AY674409; RPB2 = n.a.; CaM = n.a.).
Penicillium dipodomyis (Frisvad, Filt. & Wicklow) Banke, Firsvad & S. Rosend, Int. Mod. Meth. Pen. Asp. Clas. 270. 2000. [MB274307]. — Herb.: IMI 296926. Ex-type: IBT 5333 = CBS 110412 = NRRL 13485 = NRRL A-26836 = IMI 296926. Section Chrysogena. ITS barcode: AY371615. (Alternative markers: BenA = AY495991; RPB2 = JF909932; CaM = JX996950).
Penicillium discolor Frisvad & Samson, Antonie van Leeuwenhoek, 72: 120. 1997. [MB442902]. — Herb.: IMI 285513. Ex-type: CBS 474.84 = IBT 21523 = IBT 5738 = IBT 14440 = IMI 285513 = FRR 2933. Section Fasciculata. ITS barcode: AJ004816. (Alternative markers: BenA = AY674348; RPB2 = n.a.; CaM = n.a.).
Penicillium donkii Stolk, Persoonia 7: 333. 1973. [MB319267]. — Herb.: CBS 188.72. Ex-type: CBS 188.72 = NRRL 5562 = ATCC 48439 = CCRC 31694 = FRR 1738 = IFO 31746 = IMI 197489 = MUCL 31188. Section Stolkia. ITS barcode: AF033445. (Alternative markers: BenA = JN617718; RPB2 = n.a.; CaM = AF481136).
Penicillium dravuni Janso, Mycologia 97: 445. 2005. [MB501442]. — Herb.: BPI 844248. Ex-type: F01V25. Section Exilicaulis. ITS barcode: AY494856. (Alternative markers: BenA = n.a.; RPB2 = n.a.; CaM = n.a.).
Penicillium dunedinense Visagie, Seifert & Samson, Stud. Mycol. 78: 121. 2014. [MB809183]. — Herb.: CBS H-21803. Ex-type: CBS 138218 = DTO 244G1. Section Canescentia. ITS barcode: KJ775678. (Alternative markers: BenA = KJ775171; RPB2 = n.a.; CaM = KJ775405).
Penicillium echinulatum Raper & Thom ex Fassat., Acta Univ. Carol., Biol. 1974: 326. 1977. [MB319269]. — Herb.: PRM 778523. Ex-type: CBS 317.48 = IBT 6294 = IMI 040028 = ATCC 10434 = NRRL 1151 = FRR 1151 = IFO 7760 = MUCL 15615 = QM 7519. Section Fasciculata. ITS barcode: AF033473. (Alternative markers: BenA = AY674341; RPB2 = n.a.; CaM = DQ911133).
Penicillium egyptiacum J.F.H. Beyma, Zentralbl. Bakteriol. Parasitenk., Abt. 2 88: 137. 1933 ≡ Eupenicillium egyptiacum (J.F.H. Beyma) Stolk & D.B. Scott, Persoonia 4: 401. 1967. [MB263790]. — Herb.: IMI 040580. Ex-type: CBS 244.32 = ATCC 10441 = CSIR 707 = FRR 2090 = IBT 14684 = IFO 6094 = IFO 8141 = IFO 8847 = IMI 040580 = NRRL 2090 = QM 1875. Section Chrysogena. ITS barcode: AF033467. (Alternative markers: BenA = AY674374; RPB2 = JN406598; CaM = JX996969).
Penicillium ehrlichii Kleb., Ber. Deutsch. Bot. Ges. 48: 374. 1930 ≡ Eupenicillium ehrlichii (Kleb.) Stolk & D.B. Scott, Persoonia 4: 400. 1967 ≡ Penicillium klebahnii Pitt, Genus Penicillium: 122. 1980. [MB319270]. — Herb.: IMI 039737. Ex-type: CBS 324.48 = ATCC 10442 = IMI 039737 = IMI 039737ii = NRRL 708 = QM 1874 = VKMF-273. Section Lanata-Divaricata. ITS barcode: AF033432. (Alternative markers: BenA = KF296464; RPB2 = KF296428; CaM = KF296395).
Penicillium elleniae Houbraken et al., Int. J. Syst. Evol. Microbiol. 61: 1470. 2011. [MB518028]. — Herb.: HUA 170339. Ex-type: CBS 118135 = IBT 23229. Section Lanata-Divaricata. ITS barcode: GU981612. (Alternative markers: BenA = GU981663; RPB2 = KF296429; CaM = KF296389).
Penicillium ellipsoideosporum L. Wang & W.Y. Kong, Mycosystema 19: 463. 2000. [MB467721]. — Herb.: HMAS 71768. Ex-type: CBS 112493 = AS 3.5688. Section Cinnamopurpurea. ITS barcode: JX012224. (Alternative markers: BenA = JQ965104; RPB2 = JN121427; CaM = AY678559).
Penicillium erubescens D.B. Scott, Mycopathol. Mycol. Appl. 36: 14. 1968 ≡ Eupenicillium erubescens D.B. Scott, Mycopathol. Mycol. Appl. 36: 14. 1968. [MB335726]. — Herb.: CBS 318.67. Ex-type: CBS 318.67 = ATCC 18544 = CSIR 1040 = FRR 814 = IFO 31734 = IMI 136204 = NRRL 6223. Section Exilicaulis. ITS barcode: AF033464. (Alternative markers: BenA = HQ646566; RPB2 = JN121490; CaM = EU427281).
Penicillium estinogenum A. Komatsu & S. Abe ex G. Sm., Trans. Brit. Mycol. Soc. 46: 335. 1963 ≡ Penicillium estinogenum A. Komatsu & S. Abe, J. Gen. Appl. Microbiol., Tokyo 2: 132. 1956 (nom. inval., Art. 36). [MB302397]. — Herb.: IMI 68241. Ex-type: CBS 329.59 = ATCC 18310 = CCRC 31557 = FAT1196 = FRR 3428 = IFO 6230 = IMI 068241 = QM 8149 = VKMF-274. Section Gracilenta. ITS barcode: n.a. (Alternative markers: BenA = n.a.; RPB2 = n.a.; CaM = n.a.).
Penicillium euglaucum J.F.H. Beyma, Antonie van Leeuewenhoek 6: 269. 1940 ≡ Eupenicillium euglaucum (J.F.H. Beyma) Stolk & Samson, Stud. Mycol. 23: 90. 1983. [MB289081]. — Herb.: CBS 323.71. Ex-type: CBS 323.71 = IBT 30767. Section Citrina. ITS barcode: JN617699. (Alternative markers: BenA = JN606856; RPB2 = JN606594; CaM = JN606564).
Penicillium expansum Link, Mag. Ges. Naturf. Freunde Berlin 3: 16. 1809. [MB159382]. — Herb.: CBS H-7082. Ex-type: CBS 325.48 = ATCC 7861 = ATHUM2891 = CCRC 30566 = FRR 976 = IBT 3486 = IBT 5101 = IMI 039761 = IMI 039761ii = MUCL 29192 = NRRL 976 = VKMF-275. Section Penicillium. ITS barcode: AY373912. (Alternative markers: BenA = AY674400; RPB2 = JF417427; CaM = n.a.).
Penicillium fagi C. Ramírez & A.T. Martínez, Mycopathologia 63: 57. 1978. [MB283595]. — Herb.: IJFM 3049. Ex-type: CBS 689.77 = CCMF-696 = IJFM 3049 = IMI 253806 = VKMF-2178. Section Exilicaulis. ITS barcode: AF481124. (Alternative markers: BenA = KJ834449; RPB2 = JN406540; CaM = n.a.).
Penicillium fellutanum Biourge, Cellule 33: 262. 1923. [MB264748]. — Herb.: IMI 39734. Ex-type: CBS 229.81 = CBS 326.48 = ATCC 10443 = FRR 746 = IFO 5761 = IMI 039734 = IMI 039734iii = NRRL 746 = QM 7554. Section Charlesia. ITS barcode: AF033399. (Alternative markers: BenA = KJ834450; RPB2 = JN121460; CaM = AY741753).
Penicillium fennelliae Stolk, Antonie van Leeuwenhoek 35: 261. 1969. [MB335728]. — Herb.: CBS 711.68. Ex-type: CBS 711.68 = ATCC 22050 = ATCC 52492 = FRR 521 = IHEM 4389 = IMI 151747 = MUCL 31322. Section Brevicompacta. ITS barcode: JX313169. (Alternative markers: BenA = JX313185; RPB2 = JN406536; CaM = JX313151).
Penicillium flavidostipitatum C. Ramírez & C.C. González, Mycopathologia 88: 3. 1984. [MB106338]. — Herb.: CBS 202.87. Ex-type: CBS 202.87 = IJFM 7824. Section Exilicaulis. ITS barcode: KC411691. (Alternative markers: BenA = KJ834451; RPB2 = n.a.; CaM = n.a.).
Penicillium flavigenum Frisvad & Samson, Mycol. Res. 101: 620. 1997. [MB437441]. — Herb.: CBS 419.89. Ex-type: CBS 419.89 = BT21526 = IBT 3091 = IBT V1035 = IMI 293207. Section Chrysogena. ITS barcode: JX997105. (Alternative markers: BenA = AY495993; RPB2 = JN406551; CaM = JX996281).
Penicillium flavisclerotiatum Visagie, Houbraken & K. Jacobs, Stud. Mycol. 78: 419. 2014. [MB809963]. — Herb.: CBS H-21879. Ex-type: CBS 137750 = DTO 180-I8 = CV 100 = DAOM 241157. Section Aspergilloides. ITS barcode: KM189644. (Alternative markers: BenA = KM088888; RPB2 = KM089662; CaM = KM089275).
Penicillium formosanum H.M. Hsieh, H.J. Su & Tzean, Trans. Mycol. Soc. Republ. China 2: 159. 1987. [MB126488]. — Herb.: PPEH 10001. Ex-type: CBS 211.92 = IBT 19748 = IBT 21527. Section Penicillium. ITS barcode: KC411696. (Alternative markers: BenA = AY674426; RPB2 = JN406615; CaM = n.a.).
Penicillium fractum Udagawa, Trans. Mycol. Soc. Japan 9: 51. 1968 ≡ Eupenicillium fractum Udagawa, Trans. Mycol. Soc. Japan 9: 51. 1968. [MB335729]. — Herb.: CBS H-7086. Ex-type: CBS 124.68 = ATCC 18567 = FRR 3448 = IMI 136701 = NHL 6104 = NRRL 3448. Section Fracta. ITS barcode: KC411674. (Alternative markers: BenA = KJ834452; RPB2 = JN121441; CaM = n.a.).
Penicillium freii Frisvad & Samson, Stud. Mycol. 49: 28. 2004. [MB369274]. — Herb.: IMI 285513. Ex-type: CBS 476.84 = IBT 5137 = IMI 285513. Section Fasciculata. ITS barcode: JN942696. (Alternative markers: BenA = AY674290; RPB2 = JN985430; CaM = n.a.).
Penicillium frequentans Westling, Ark. Bot. 11: 133. 1911. [MB152118]. — Herb.: CBS 105.11. Ex-type: CBS 105.11. Section Aspergilloides. ITS barcode: KM189525. (Alternative markers: BenA = KM088762; RPB2 = KM089534; CaM = KM089147).
Penicillium fuscum (Sopp) Biourge, Cellule 33: 103. 1923 ≡ Citromyces fuscus Sopp, Skr. Vidensk.-Selsk. Christiana Math.-Nat. Kl. 11: 120. 1912 ≡ Eupenicillium pinetorum Stolk, Antonie van Leeuwenhoek 34: 37. 1968. [MB289082]. — Herb.: WIS WSF 15-C. Ex-type: CBS 295.62 = ATCC 14770 = CCRC 31517 = DSM2438 = IFO 7743 = IMI 094209 = MUCL 31196 = NRRL 3008 = WSF15c. Section Aspergilloides. ITS barcode: AF033411. (Alternative markers: BenA = GQ367513; RPB2 = JN121483; CaM = GQ367539).
Penicillium fusisporum L. Wang, PloS ONE 9: e101454-P2. 2014. [MB806119]. — Herb.: HMAS 244961. Ex-type: CBS 137463 = NRRL 62805 = AS 3.15338. Section Aspergilloides. ITS barcode: KF769424. (Alternative markers: BenA = KF769400; RPB2 = n.a.; CaM = KF769413).
Penicillium galliacum C. Ramírez, A.T. Martínez & Berer., Mycopathologia 72: 30. 1980. [MB113021]. — Herb.: CBS H-7464. Ex-type: CBS 167.81 = ATCC 42232 = IJFM 5597. Section Citrina. ITS barcode: JN617690. (Alternative markers: BenA = JN606837; RPB2 = JN606609; CaM = JN606548).
Penicillium georgiense S.W. Peterson & B.W. Horn, Mycologia 101: 79. 2009. [MB509290]. — Herb.: BPI 877332. Ex-type: CBS 132826 = NRRL 35509. Section Charlesii. ITS barcode: EF422852. (Alternative markers: BenA = EF506223; RPB2 = n.a.; CaM = EF506239).
Penicillium glabrum (Wehmer) Westling, Ark. Bot. 11: 131. 1911 ≡ Citromyces glaber Wehmer, Beitr. Einh. Pilze 1: 24. 1893. [MB120545]. — Herb.: IMI 91944. Ex-type: CBS 125543 = IBT 22658 = IMI 91944. Section Aspergilloides. ITS barcode: GU981567. (Alternative markers: BenA = GU981619; RPB2 = JF417447; CaM = GQ367545).
Penicillium gladioli L. McCulloch & Thom, Science 67: 217. 1928 ≡ Eupenicillium crustaceum F. Ludw., Lehrb. Nied. Krypt.: 263. 1892. [MB266048]. — Herb.: IMI 34911. Ex-type: CBS 332.48 = ATCC 10448 = FRR 939 = IBT 14772 = IMI 034911 = IMI 034911ii = LCP 89.202 = MUCL 29174 = NRRL 939 = = QM 1955 = VKMF-2088. Section Penicillium. ITS barcode: AF033480. (Alternative markers: BenA = AY674287; RPB2 = JN406567; CaM = n.a.).
Penicillium glandicola (Oudem.) Seifert & Samson, Adv. Penicillium Aspergillus Syst.: 147. 1985 ≡ Coremiun glandicola Oudem., Ned. Kruidk. Arch. 2: 918. 1903. [MB114761]. — Herb.: Netherlands, Valkenburg, Jul 1901, Rick in herb. Oudemans (L). Ex-type: CBS 498.75 = IBT 21529 = IMI 154241. Section Penicillium. ITS barcode: AB479308. (Alternative markers: BenA = AY674415; RPB2 = n.a.; CaM = n.a.).
Penicillium glaucoroseum Demelius, Verh. Zool.-Bot. Ges. Wien 72: 72. 1923 (1922). [MB158423]. — Herb.: unknown. Ex-type: NRRL 908 (authentic acc. Raper & Thom 1949). Section Lanata-Divaricata. ITS barcode: KF296407. (Alternative markers: BenA = KF296469; RPB2 = KF296430; CaM = KF296400).
Penicillium glycyrrhizacola A.J. Chen, B.D. Sun & W.W. Gao, PloS ONE 8: e78285-P3. 2013. [MB804682]. — Herb.: HMAS 244707. Ex-type: CGMCC3.15271 (G4432). Section Chrysogena. ITS barcode: n.a. (Alternative markers: BenA = KF021538; CaM = n.a.; RPB2 = KF021554).
Penicillium godlewskii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 466. 1927. [MB266206]. — Herb.: unknown. Ex-type: CBS 215.28 = ATCC 10449 = ATCC 48714 = FRR 2111 = IFO 7724 = IMI 040591 = MUCL 29243 = NRRL 2111 = QM 7566 = VKMF-1826. Section Citrina. ITS barcode: JN617692. (Alternative markers: BenA = JN606768; RPB2 = JN606626; CaM = JN606443).
Penicillium goetzii J. Rogers et al., Persoonia 29: 92. 2012. [MB801876]. — Herb.: CBS H-21061. Ex-type: CBS 285.73 = DTO 88G6 = IBT 30199. Section Chrysogena. ITS barcode: JX997091. (Alternative markers: BenA = JX996847; RPB2 = JX996716; CaM = JX996971).
Penicillium gorlenkoanum Baghd., Novosti Sist. Nizsh. Rast. 5: 97. 1968. [MB335731]. — Herb.: CBS H-7490. Ex-type: CBS 408.69 = FRR 511 = IMI 140339 = VKMF-1079. Section Citrina. ITS barcode: GU944581. (Alternative markers: BenA = GU944520; RPB2 = JN606601; CaM = GU944608).
Penicillium gracilentum Udagawa & Y. Horie, Trans. Mycol. Soc. Japan 14: 373. 1973 ≡ Eupenicillium gracilentum Udagawa & Y. Horie, Trans. Mycol. Soc. Japan 14: 373. 1973. [MB319272]. — Herb.: NHL 6452. Ex-type: CBS 599.73 = ATCC 28047 = ATCC 48258 = FRR 1557 = IMI 216900 = NHL 6452. Section Gracilenta. ITS barcode: KC411768. (Alternative markers: BenA = KJ834453; RPB2 = JN121537; CaM = n.a.).
Penicillium grancanariae C. Ramírez, A.T. Martínez & Ferrer, Mycopathologia 66: 79. 1978. [MB319273]. — Herb.: IJFM 3745. Ex-type: CBS 687.77 = IJFM 3745 = IMI 253783. Section Aspergilloides. ITS barcode: KM189529. (Alternative markers: BenA = KM088766; RPB2 = KM089538; CaM = KM089151).
Penicillium grevilleicola Houbraken & Quaedvlieg, Stud. Mycol. 78: 423. 2014. [MB809964]. — Herb.: CBS H-21871. Ex-type: CBS 137775 = DTO 174-E6. Section Aspergilloides. ITS barcode: KM189630. (Alternative markers: BenA = KM088874; RPB2 = KM089648; CaM = KM089261).
Penicillium griseofulvum Dierckx, Ann. Soc. Sci. Bruxelles 25: 88. 1901. [MB120566]. — Herb.: IMI 75832. Ex-type: CBS 185.27 = ATCC 11885 = ATHUM2893 = CECT 2605 = DSM896 = IBT 6740 = IFO 7640 = IFO 7641 = IMI 075832 = IMI 075832ii = LCP 79.3245 = LSHBP 68 = MUCL 28643 = NRRL 2152 = NRRL 2300 = QM 6902 = VKMF-286. Section Penicillium. ITS barcode: AF033468. (Alternative markers: BenA = AY674432; RPB2 = JN121449; CaM = n.a.).
Penicillium griseolum G. Sm., Trans. Br. Mycol. Soc. 40: 485. 1957. [MB302401]. — Herb.: unknown. Ex-type: CBS 277.58 = ATCC 18239 = FRR 2671 = IFO 8175 = IMI 071626 = LSH BB323 = NRRL 2671 = QM 7523. Section unclassified. ITS barcode: EF422848. (Alternative markers: BenA = EF506213; CaM = EF506232; RPB2 = JN121480).
Penicillium griseopurpureum G. Sm., Trans. Brit. Mycol. Soc. 48: 275. 1965. [MB335732]. — Herb.: IMI 96157. Ex-type: CBS 406.65 = ATCC 22353 = FRR 3429 = IFO 9147 = IMI 096157. Section Lanata-Divaricata. ITS barcode: KF296408. (Alternative markers: BenA = KF296467; RPB2 = KF296431; CaM = KF296384).
Penicillium guanacastense K.G. Rivera, Urb & Seifert, Mycotaxon 119: 324. 2011. [MB563044]. — Herb.: DAOM 239912. Ex-type: CCFC 239912. Section Sclerotiora. ITS barcode: JN626098. (Alternative markers: BenA = JN625967; RPB2 = n.a.; CaM = JN626010).
Penicillium guttulosum J.C. Gilman & E.V. Abbott, Iowa St. Coll. J. Sci. 1: 298. 1927. [MB266689]. — Herb.: unknown. Ex-type: ATCC 48734 = NRRL 907 = FRR 907 = Thom 4894.16. Section Exilicaulis. ITS barcode: HQ646592. (Alternative markers: BenA = HQ646576; RPB2 = n.a.; CaM = HQ646587).
Penicillium halotolerans Frisvad, Houbraken & Samson, Persoonia 29: 92. 2012. [MB801875]. — Herb.: CBS H-21060. Ex-type: CBS 131537 = DTO 148H9 = IBT 4315. Section Chrysogena. ITS barcode: JX997005. (Alternative markers: BenA = JX996816; RPB2 = JX996680; CaM = JX996935).
Penicillium hennebertii Houbraken & Samson, Stud. Mycol. 70: 47. 2011 ≡ Thysanophora canadensis Stolk & Hennebert, Persoonia 5: 189. 1968. [MB561964]. — Herb.: CBS H-7854. Ex-type: CBS 334.68 = ATCC 18741 = IMI 137644 = MUCL 21216 = VKMF-2999. Section Thrysanophora. ITS barcode: KJ834507. (Alternative markers: BenA = KJ834454; RPB2 = JN121493; CaM = n.a.).
Penicillium herquei Bainier & Sartory, Bull. Soc. Mycol. France 28: 121. 1912. [MB536431]. — Herb.: IMI 28809. Ex-type: CBS 336.48 = NRRL 1040 = ATCC 10118 = BIOURGE 452 = FRR 1040 = IFO 31747 = IMI 28809 = MUCL 29213 = NCTC 1721 = QM 1926 = Thom 4640.447. Section Sclerotiora. ITS barcode: JN626101. (Alternative markers: BenA = JN625970; RPB2 = JN121494; CaM = JN626013).
Penicillium heteromorphum H.Z. Kong & Z.T. Qi, Mycosystema 1: 107. 1988. [MB135444]. — Herb.: CBS 226.89. Ex-type: CBS 226.89. Section Exilicaulis. ITS barcode: KC411702. (Alternative markers: BenA = KJ834455; RPB2 = JN406605; CaM = n.a.).
Penicillium hetheringtonii Houbraken, Frisvad & Samson, Fungal Divers. 44: 125. 2010. [MB518292]. — Herb.: CBS 122392. Ex-type: CBS 122392 = IBT 29057. Section Citrina. ITS barcode: GU944558. (Alternative markers: BenA = GU944538; RPB2 = JN606606; CaM = GU944642).
Penicillium hirayamae Udagawa, J. Agric. Soc. Tokyo 5: 6. 1959 ≡ Eupenicillium hirayamae D.B. Scott & Stolk, Antonie van Leeuwenhoek 33: 305. 1967. [MB302402]. — Herb.: IMI 78255. Ex-type: CBS 229.60 = ATCC 18312 = IFO 6435 = IMI 078255 = IMI 078255ii = NHL 6046 = NRRL 143 = QM 7885. Section Sclerotiora. ITS barcode: JN626095. (Alternative markers: BenA = JN625955; RPB2 = JN121459; CaM = JN626003).
Penicillium hirsutum Dierckx, Ann. Soc. Sci. Bruxelles 25: 89. 1901. [MB152720]. — Herb.: IMI 40213. Ex-type: CBS 135.41 = ATCC 10429 = FRR 2032 = IBT 21531 = IFO 6092 = IMI 040213 = MUCL 15622 = NRRL 2032. Section Fasciculata. ITS barcode: AY373918. (Alternative markers: BenA = AF003243; RPB2 = JN406629; CaM = n.a.).
Penicillium hispanicum C. Ramírez, A.T. Martínez & Ferrer, Mycopathologia 66: 77. 1978. [MB319274]. — Herb.: IJFM 3223. Ex-type: CBS 691.77 = ATCC 38667 = DSM2416 = IJFM 3223 = IMI 253785 = VKMF-2179. Section Ramigena. ITS barcode: JX841247. (Alternative markers: BenA = KJ834456; RPB2 = JN406539; CaM = n.a.).
Penicillium hoeksii Houbraken, Stud. Mycol. 78: 423. 2014. [MB809965]. — Herb.: CBS H-21860. Ex-type: CBS 137776 = DTO 192-H4. Section Aspergilloides. ITS barcode: KM189707. (Alternative markers: BenA = KM088954; RPB2 = KM089728; CaM = KM089341).
Penicillium hordei Stolk, Antonie van Leeuwenhoek 35: 270. 1969. [MB335734]. — Herb.: CBS 701.68. Ex-type: CBS 701.68 = ATCC 22053 = CECT 2290 = FRR 815 = IBT 17804 = IBT 6980 = IMI 151748 = MUCL 39559. Section Fasciculata. ITS barcode: n.a. (Alternative markers: BenA = AY674347; RPB2 = n.a.; CaM = n.a.).
Penicillium idahoense Paden, Mycopathol. Mycol. Appl. 43: 259. 1971 ≡ Eupenicillium idahoense Paden, Mycopathol. Mycol. appl. 43: 259. 1971. [MB319275]. — Herb.: UVIC JWP 66-32. Ex-type: CBS 341.68 = NRRL 5274 = ATCC 22055 = FRR 881 = IMI 148393. Section Cinnamopurpurea. ITS barcode: KC411747. (Alternative markers: BenA = EF626953; RPB2 = JN121499; CaM = EF626954).
Penicillium incoloratum L.Q. Huang & Z.T. Qi, Acta Mycol. Sin. 13: 264. 1994. [MB363421]. — Herb.: HMAS 65949. Ex-type: CBS 101753 = AS 3.4672. Section Cinnamopurpurea. ITS barcode: KJ834508. (Alternative markers: BenA = KJ834457; RPB2 = JN406651; CaM = KJ866984).
Penicillium indicum D.K. Sandhu & R.S. Sandhu, Can. J. Bot. 41: 1273. 1963. [MB335735]. — Herb.: CBS 7476. Ex-type: CBS 115.63 = NRRL 3387 = ATCC 18324 = FRR 3387 = IFO 31744 = IMI 166620. Section Charlesia. ITS barcode: AY742699. (Alternative markers: BenA = EU427263; RPB2 = JN406640; CaM = AY741744).
Penicillium infra-aurantiacum Visagie, Houbraken & K. Jacobs, Stud. Mycol. 78: 426. 2014. [MB809966]. — Herb.: CBS H-21880. Ex-type: CBS 137747 = DTO 183-C3 = CV 1518 = DAOM 241145. Section Aspergilloides. ITS barcode: KM189684. (Alternative markers: BenA = KM088930; RPB2 = KM089704; CaM = KM089317).
Penicillium infrapurpureum Visagie, Seifert & Samson, Stud. Mycol. 78: 116. 2014. [MB809181]. — Herb.: CBS H-21801. Ex-type: CBS 138219 = DTO 235F6. Section Cinnamopurpurea. ITS barcode: KJ775679. (Alternative markers: BenA = KJ775172; RPB2 = n.a.; CaM = KJ775406).
Penicillium inusitatum D.B. Scott, Mycopathol. Mycol. Appl. 36: 20. 1968 ≡ Eupenicillium inusitatum D.B. Scott, Mycopathol. Mycol. Appl. 36: 20. 1968. [MB335736]. — Herb.: CBS 351.67. Ex-type: CBS 351.67 = ATCC 18622 = CSIR 1096 = FRR 1163 = IMI 136214 = NRRL 5810. Section Fracta. ITS barcode: AF033431. (Alternative markers: BenA = KJ834458; RPB2 = JN121503; CaM = n.a.).
Penicillium isariiforme Stolk & J.A. Mey., Trans. Brit. Mycol. Soc. 40: 187. 1957. [MB302403]. — Herb.: IMI 60371. Ex-type: CBS 247.56 = ATCC 18425 = CCRC 31699 = IFO 6393 = IHEM 4376 = IMI 060371 = LSHBBB308 = MUCL 31191 = MUCL 31323 = NRRL 2638 = QM 1897. Section Ochrosalmonea. ITS barcode: AF454077. (Alternative markers: BenA = KJ834459; RPB2 = JN121470; CaM = n.a.).
Penicillium italicum Wehmer, Hedwigia 33: 211. 1894. [MB162660]. — Herb.: CBS 339.48. Ex-type: CBS 339.48 = ATCC 10454 = DSM2754 = FRR 983 = IBT 23029 = IMI 039760 = MUCL 15608 = NRRL 983 = QM 7572. Section Penicillium. ITS barcode: KJ834509. (Alternative markers: BenA = AY674398; RPB2 = JN121496; CaM = n.a.).
Penicillium jacksonii K.G. Rivera, Houbraken & Seifert, Stud. Mycol. 70: 151. 2011. [MB563160]. — Herb.: DAOM 239937. Ex-type: CCFC 239937. Section Sclerotiora. ITS barcode: JN686437. (Alternative markers: BenA = JN686368; RPB2 = n.a.; CaM = JN686391).
Penicillium jamesonlandense Frisvad & Overy, Int. J. Syst. Evol. Microbiol. 56: 1435. 2006. [MB521421]. — Herb.: DAOM 234087. Ex-type: CBS 102888 = DAOM 234087 = IBT 21984 = IBT 24411. Section Ramosa. ITS barcode: DQ267912. (Alternative markers: BenA = DQ309448; RPB2 = n.a.; CaM = KJ866985).
Penicillium janczewskii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 488. 1927. [MB120703]. — Herb.: IMI 191499. Ex-type: CBS 221.28 = FRR 919 = IMI 191499 = NRRL 919. Section Canescentia. ITS barcode: AY157487. (Alternative markers: BenA = KJ834460; RPB2 = JN406612; CaM = KJ867001).
Penicillium janthinellum Biourge, Cellule 33: 258. 1923. [MB119134]. — Herb.: IMI 40238. Ex-type: CBS 340.48 = ATCC 10455 = IMI 040238 = NRRL 2016 = QM 6865. Section Lanata-Divaricata. ITS barcode: GU981585. (Alternative markers: BenA = GU981625; RPB2 = JN121497; CaM = KF296401).
Penicillium javanicum J.F.H. Beyma, Verh. Kon. Ned. Akad. Wetensch., Afd. Natuurk. 26: 17. 1929 ≡ Carpenteles javanicum (J.F.H. Beyma) Shear, Mycologia 26: 107. 1934 ≡ Eupenicillium javanicum (J.F.H. Beyma) Stolk & D.B. Scott, Persoonia 4: 398. 1967 ≡ Penicillium indonesiae Pitt, Genus Penicillium: 114. 1980. [MB268394]. — Herb.: CBS H-7088. Ex-type: CBS 341.48 = ATCC 9099 = CSIR 831 = FRR 707 = IFO 31735 = IMI 039733 = MUCL 29099 = NRRL 707 = QM 1876. Section Lanata-Divaricata. ITS barcode: GU981613. (Alternative markers: BenA = GU981657; RPB2 = JN121498; CaM = KF296387).
Penicillium jensenii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 494. 1927. [MB120708]. — Herb.: IMI 39768. Ex-type: CBS 327.59 = ATCC 18317 = FRR 909 = IFO 5764 = IMI 039768 = LCP 89.1389 = NRRL 909 = QM 7587. Section Canescentia. ITS barcode: AY443470. (Alternative markers: BenA = JX140954; RPB2 = JN406614; CaM = AY443490).
Penicillium jiangxiense H.Z. Kong & Z.Q. Liang, Mycosystema 22: 4. 2003. [MB489161]. — Herb.: HMAS 82540. Ex-type: AS 3.6521. Section Cinnamopurpurea. ITS barcode: KJ890411. (Alternative markers: BenA = KJ890409; RPB2 = n.a.; CaM = KJ890407).
Penicillium johnkrugii K.G. Rivera, Houbraken & Seifert, Stud. Mycol. 70: 151. 2011. [MB563161]. — Herb.: DAOM 239943. Ex-type: CCFC 239943. Section Sclerotiora. ITS barcode: JN686447. (Alternative markers: BenA = JN686378; RPB2 = n.a.; CaM = JN686401).
Penicillium jugoslavicum C. Ramírez & Munt.-Cvetk., Mycopathologia 88: 65. 1984. [MB124173]. — Herb.: CBS 192.87. Ex-type: CBS 192.87 = IJFM 7785 = IMI 314508. Section Sclerotiora. ITS barcode: KC773836. (Alternative markers: BenA = KC773789; RPB2 = JN406618; CaM = KC773815).
Penicillium kananaskense Seifert, Frisvad & McLean, Can. J. Bot. 72: 20. 1994. [MB362160]. — Herb.: MU-F-39531. Ex-type: CBS 530.93 = ATCC 90282 = DAOM 216105 = IBT 11775 = IMI 356791. Section Aspergilloides. ITS barcode: KM189780. (Alternative markers: BenA = KM089030; RPB2 = KM089804; CaM = KM089417).
Penicillium katangense Stolk, Antonie van Leeuwenhoek 34: 42. 1968 ≡ Eupenicillium katangense Stolk, Antonie van Leenwenhoek 34: 42. 1968. [MB120725]. — Herb.: CBS 247.67. Ex-type: CBS 247.67 = ATCC 18388 = IMI 136206 = NRRL 5182. Section Exilicaulis. ITS barcode: AF033458. (Alternative markers: BenA = n.a.; RPB2 = n.a.; CaM = n.a.).
Penicillium kewense G. Sm., Trans. Brit. Mycol. Soc. 44: 42. 1961. [MB335740]. — Herb.: CBS H-7077. Ex-type: CBS 344.61 = ATCC 18240 = FRR 3441 = IFO 8113 = IMI 086561 = LSHBBB400 = MUCL 2685 = NRRL 3332 = QM 7958. Section Chrysogena. ITS barcode: AF033466. (Alternative markers: BenA = JX996849; RPB2 = JF417428; CaM = JX996973).
Penicillium kiamaense Houbraken & Pitt, Stud. Mycol. 78: 426. 2014. [MB809967]. — Herb.: CBS H-21857. Ex-type: CBS 137947 = FRR 6087 = DTO 056-I6. Section Aspergilloides. ITS barcode: KM189506. (Alternative markers: BenA = KM088743; RPB2 = KM089515; CaM = KM089128).
Penicillium kojigenum G. Sm., Trans. Brit. Mycol. Soc. 44: 43. 1961. [MB335741]. — Herb.: LSHTM BB394. Ex-type: CBS 345.61 = ATCC 18227 = CCRC 31515 = FRR 3442 = IFO 9581 = IMI 086562 = LSHBBB394 = MUCL 2457 = NRRL 3442 = QM 7957. Section Ramosa. ITS barcode: AF033489. (Alternative markers: BenA = KJ834463; RPB2 = JN406564; CaM = KJ867011).
Penicillium kongii L. Wang, Mycologia 105: 1549. 2013. [MB803185]. — Herb.: HMAS 244382. Ex-type: AS 3.15329. Section Brevicompacta. ITS barcode: KC427191. (Alternative markers: BenA = KC427171; RPB2 = n.a.; CaM = KC427151).
Penicillium laeve (K. Ando & Manoch) Houbraken & Samson, Stud. Mycol. 70: 47. 2011 ≡ Torulomyces laevis K. Ando & Manoch, Mycoscience 39: 317. 1998. [MB561960]. — Herb.: TNS-F-238517. Ex-type: CBS 136665 = KY 12727 = NBRC 109724. Section Exilicaulis. ITS barcode: KF667369. (Alternative markers: BenA = KF667365; RPB2 = KF667371; CaM = KF667367).
Penicillium lagena (Delitsch) Stolk & Samson, Stud. Mycol. 23: 100. 1983 ≡ Torulomyces lagena Delitsch, Systematik der Schimmelpilze, Neudamm: 9. 1943. [MB109162]. — Herb.: CBS 185.65. Ex-type: CBS 185.65 = MUCL 8221 = JCM10149 = OAC10034. Section Torulomyces. ITS barcode: KF303665. (Alternative markers: BenA = KF303619; RPB2 = JN121450; CaM = KF303634).
Penicillium lanosocoeruleum Thom, Penicillia: 322. 1930. [MB268949]. — Herb.: unknown. Ex-type: CBS 215.30 = CBS 334.48 = ATCC 10459 = IFO 7761 = IMI 039818 = NRRL 888 = QM 6755 = VKMF-3089. Section Chrysogena. ITS barcode: KC411740. (Alternative markers: BenA = JX996843; RPB2 = JX996723; CaM = JX996967).
Penicillium lanosum Westling, Ark. Bot. 11: 97. 1911. [MB178497]. — Herb.: IMI 40224. Ex-type: CBS 106.11 = ATCC 10458 = FRR 2009 = IFO 5851 = IFO 6099 = IMI 040224 = LSHBP 86 = MUCL 29232 = NRRL 2009 = QM 7591. Section Ramosa. ITS barcode: DQ304540. (Alternative markers: BenA = DQ285627; RPB2 = n.a.; CaM = FJ530974).
Penicillium lapidosum Raper & Fennell, Mycologia 40: 524. 1948 ≡ Eupenicillium lapidosum D.B. Scott & Stolk, Antonie van Leeuwenhoek 33: 298. 1967. [MB289094]. — Herb.: IMI 39743. Ex-type: CBS 343.48 = ATCC 10462 = CCT4477 = IFO 6100 = IMI 039743 = NRRL 718 = QM 1928. Section Exilicaulis. ITS barcode: AF033409. (Alternative markers: BenA = KJ834465; RPB2 = JN121500; CaM = FJ530984).
Penicillium lassenii Paden, Mycopathol. Mycol. Appl. 43: 266. 1971 ≡ Eupenicillium lassenii Paden, Mycopathol. Mycol. Appl. 43: 266. 1971. [MB319281]. — Herb.: UVIC JWP 69-26. Ex-type: CBS 277.70 = NRRL 5272 = ATCC 22054 = FRR 858 = IMI 148395. Section Torulomyces. ITS barcode: KF303648. (Alternative markers: BenA = KF303607; RPB2 = JN121481; CaM = KF303629).
Penicillium lenticrescens Visagie, Seifert & Samson, Stud. Mycol. 78: 123. 2014. [MB809184]. — Herb.: CBS H-21804. Ex-type: CBS 138215 = DTO 129A8. Section Ramosa. ITS barcode: KJ775675. (Alternative markers: BenA = KJ775168; RPB2 = n.a.; CaM = KJ775404).
Penicillium levitum Raper & Fennell, Mycologia 40: 511. 1948 ≡ Carpenteles levitum (Raper & Fennell) C.R. Benj., Mycologia 47: 685. 1955 ≡ Eupenicillium levitum (Raper & Fennell) Stolk & D.B. Scott, Persoonia 4: 402. 1967 ≡ Eupenicillium javanicum var. levitum (Raper & Fennell) Stolk & Samson, Stud. Mycol. 23: 134. 1983. [MB289096]. — Herb.: IMI 039735. Ex-type: CBS 345.48 = ATCC 10464 = IFO 6101 = IFO 8849 = IMI 039735 = NRRL 705 = QM 1877. Section Lanata-Divaricata. ITS barcode: GU981607. (Alternative markers: BenA = GU981654; RPB2 = KF296432; CaM = KF296394).
Penicillium lilacinoechinulatum S. Abe ex G. Sm., Trans. Brit. Mycol. Soc. 46: 335. 1963 ≡ Penicillium lilacinoechinulatum S. Abe, J. Gen. Appl. Microbiol., Tokyo 2: 54. 1956 (nom. inval., Art. 36). [MB120793]. — Herb.: unknown. Ex-type: CBS 454.93 = ATCC 18309 = FAT 84 = FRR 3451 = IFO 6231 = IMI 068211 = QM 7289. Section Sclerotiora. ITS barcode: AY157489. (Alternative markers: BenA = KC773790; RPB2 = n.a.; CaM = KC773816).
Penicillium limosum S. Ueda, Mycoscience 36: 451. 1995 ≡ Eupenicillium limosum S. Ueda, Mycoscience 36: 451. 1995. [MB415136]. — Herb.: CBM NEI-5220. Ex-type: CBS 339.97 = NEI5220. Section Lanata-Divaricata. ITS barcode: GU981568. (Alternative markers: BenA = GU981621; RPB2 = KF296433; CaM = KF296398).
Penicillium lineolatum Udagawa & Y. Horie, Mycotaxon 5: 493. 1977 ≡ Eupenicillium lineolatum Udagawa & Y. Horie, Mycotaxon 5: 493. 1977 ≡ Eupenicillium javanicum var. lineolatum (Udagawa & Y. Horie) Stolk & Samson, Stud. Mycol. 23: 134. 1983. [MB319283]. — Herb.: NHL 2776. Ex-type: CBS 188.77 = NHL 2776. Section Lanata-Divaricata. ITS barcode: GU981579. (Alternative markers: BenA = GU981620; RPB2 = KF296434; CaM = KF296397).
Penicillium lividum Westling, Ark. Bot. 11: 134. 1911. [MB178817]. — Herb.: IMI 39736. Ex-type: CBS 347.48 = ATCC 10102 = CCRC 31286 = DSM1180 = IFO 6102 = IMI 039736 = NRRL 754 = QM 1930 = VKMF-303. Section Aspergilloides. ITS barcode: KM189582. (Alternative markers: BenA = KM088825; RPB2 = KM089598; CaM = KM089211).
Penicillium longicatenatum Visagie, et al., Stud. Mycol. 78: 429. 2014. [MB809968]. — Herb.: CBS H-21875. Ex-type: CBS 137735 = DTO 180-D9 = CV 2847 = DAOM 241119. Section Aspergilloides. ITS barcode: KM189636. (Alternative markers: BenA = KM088880; RPB2 = KM089654; CaM = KM089267).
Penicillium longisporum (W.B. Kend.) Houbraken & Samson, Stud. Mycol. 70: 47. 2011 ≡ Thysanophora longispora W.B. Kend., Can. J. Bot. 39: 826. 1961. [MB561966]. — Herb.: unknown. Ex-type: CBS 354.62 = DAOM 63073 = MUCL 4168. Section Thysanophora. ITS barcode: n.a. (Alternative markers: BenA = KJ834467; RPB2 = n.a.; CaM = n.a.).
Penicillium ludwigii Udagawa, Trans. Mycol. Soc. Japan 10: 2. 1969 ≡ Eupenicillium ludwigii Udagawa, Trans. Mycol. Soc. Japan 10: 2. 1969. [MB335744]. — Herb.: NHL 6118. Ex-type: CBS 417.68 = FRR 559. Section Lanata-Divaricata. ITS barcode: KF296409. (Alternative markers: BenA = KF296468; RPB2 = KF296435; CaM = KF296404).
Penicillium maclennaniae H.Y. Yip, Trans. Brit. Mycol. Soc. 77: 202. 1981. [MB112523]. — Herb.: DAR 35238. Ex-type: CBS 198.81 = DAR35238. Section Exilicaulis. ITS barcode: KC411689. (Alternative markers: BenA = KJ834468; RPB2 = n.a.; CaM = n.a.).
Penicillium macrosclerotiorum L. Wang, X.M. Zhang & W.Y. Zhuang, Mycol. Res. 111: 1244. 2007. [MB492622]. — Herb.: HMAS 133177-1-4. Ex-type: CBS 116871 = AS 3.6581. Section Gracilenta. ITS barcode: KJ834511. (Alternative markers: BenA = KJ834469; RPB2 = JN121432; CaM = DQ911123).
Penicillium madriti G. Sm., Trans. Brit. Mycol. Soc. 44: 44. 1961. [MB335747]. — Herb.: IMI 86563. Ex-type: CBS 347.61 = ATCC 18233 = CCRC 31672 = FRR 3452 = IFO 9148 = IMI 086563 = LSHBBB389 = MUCL 2456 = MUCL 31193 = NRRL 3452 = QM 7959. Section Turbata. ITS barcode: AF033482. (Alternative markers: BenA = KJ834470; RPB2 = JN406561; CaM = EU644076).
Penicillium magnielliptisporum Visagie, Seifert & Samson, Stud. Mycol. 78: 127. 2014. [MB809186]. — Herb.: CBS H-21806. Ex-type: CBS 138225 = DTO 128H8. Section Paradoxa. ITS barcode: KJ775686. (Alternative markers: BenA = KJ775179; RPB2 = n.a.; CaM = KJ775413).
Penicillium malacaense C. Ramírez & A.T. Martínez, Mycopathologia 72: 186. 1980. [MB113025]. — Herb.: IJFM 7093. Ex-type: CBS 160.81 = NRRL 35754 = ATCC 42241 = IJFM 7093 = IMI 253801 = VKMF-2197. Section Cinnamopurpurea. ITS barcode: EU427300. (Alternative markers: BenA = EU427268; RPB2 = JN406626; CaM = KJ866997).
Penicillium malachiteum (Yaguchi & Udagawa) Houbraken & Samson, Stud. Mycol. 70: 47. 2011 ≡ Chromocleista malachitea Yaguchi & Udagawa, Trans. Mycol. Soc. Japan 34: 102. 1993. [MB561971]. — Herb.: CBS 647.95. Ex-type: CBS 647.95 = IBT 17515. Section Sclerotiora. ITS barcode: KC773838. (Alternative markers: BenA = KC773794; RPB2 = n.a.; CaM = KC773820).
Penicillium mallochii K.G. Rivera, Urb & Seifert, Mycotaxon 119: 322. 2012. [MB563043]. — Herb.: DAOM 239917. Ex-type: CCFC 239917. Section Sclerotiora. ITS barcode: JN626104. (Alternative markers: BenA = JN625973; RPB2 = n.a.; CaM = JN626016).
Penicillium malmesburiense Visagie, Houbraken & K. Jacobs, Stud. Mycol. 78: 429. 2014. [MB809969]. — Herb.: CBS H-21872. Ex-type: CBS 137744 = DTO 182-H5 = CV 1180 = DAOM 241144. Section Aspergilloides. ITS barcode: KM189676. (Alternative markers: BenA = KM088921; RPB2 = KM089695; CaM = KM089308).
Penicillium malodoratum (Kwon-Chung & Fennell) Samson et al., (published here) ≡ Aspergillus malodoratus Kwon-Chung & Fennell, Gen. Aspergillus: 468. 1965. [MB809316]. — Herb.: IMI 172289. Ex-type: CBS 490.65 = NRRL 5083 = IMI 172289 = ATCC 16834. Section Paradoxa. ITS barcode: AF033485. (Alternative markers: BenA = EF669681; RPB2 = EF669672; CaM = FJ530972).
Penicillium manginii Duché & R. Heim, Trav. Cryptog.: 450. 1931. [MB270490]. — Herb.: CBS 253.31. Ex-type: CBS 253.31 = NRRL 2134. Section Citrina. ITS barcode: GU944599. (Alternative markers: BenA = JN606651; RPB2 = JN606618; CaM = JN606381).
Penicillium mariae-crucis Quintan., Av. Aliment. Majora Anim. 23: 334. 1982. [MB114171]. — Herb.: CBS 270.83. Ex-type: CBS 271.83 = IMI 256075. Section Lanata-Divaricata. ITS barcode: GU981593. (Alternative markers: BenA = GU981630; RPB2 = KF296439; CaM = KF296374).
Penicillium marinum Frisvad & Samson, Stud. Mycol. 49: 20. 2004. [MB370974]. — Herb.: CBS 109550. Ex-type: CBS 109550 = IBT 14360. Section Penicillium. ITS barcode: KJ834512. (Alternative markers: BenA = AY674392; RPB2 = n.a.; CaM = n.a.).
Penicillium maximae Visagie, Houbraken & Samson, Persoonia 31: 52. 2013. [MB803783]. — Herb.: CBS H-21144. Ex-type: CBS 134565 = NRRL 2060. Section Sclerotiora. ITS barcode: EU427298. (Alternative markers: BenA = KC773795; RPB2 = n.a.; CaM = KC773821).
Penicillium melanoconidium (Frisvad) Frisvad & Samson, Stud. Mycol. 49: 28. 2004 ≡ Penicillium aurantiogriseum var. melanoconidium Frisvad, Mycologia 81: 849. 1989. [MB368219]. — Herb.: IMI 321503. Ex-type: CBS 115506 = IBT 3444 = IMI 321503. Section Fasciculata. ITS barcode: AJ005483. (Alternative markers: BenA = AY674304; RPB2 = n.a.; CaM = n.a.).
Penicillium melinii Thom, Penicillia: 273. 1930. [MB270876]. — Herb.: IMI 40216. Ex-type: CBS 218.30 = ATCC 10469 = FRR 2041 = IFO 7675 = IMI 040216 = MUCL 29235 = NRRL 2041 = QM 7599. Section Exilicaulis. ITS barcode: AF033449. (Alternative markers: BenA = KJ834471; RPB2 = JN406613; CaM = n.a.).
Penicillium meloforme Udagawa & Y. Horie, Trans. Mycol. Soc. Japan 14: 376. 1973 ≡ Eupenicillium meloforme Udagawa & Y. Horie, Trans. Mycol. Soc. Japan 14: 376. 1973 ≡ Eupenicillium javanicum var. meloforme (Udagawa & Y. Horie) Stolk & Samson, Stud. Mycol. 23: 136. 1983. [MB120882]. — Herb.: NHL 6468. Ex-type: CBS 445.74 = ATCC 28049 = IMI 216903 = NHL 6468. Section Lanata-Divaricata. ITS barcode: KC411762. (Alternative markers: BenA = GU981656; RPB2 = KF296440; CaM = KF296396).
Penicillium menonorum S.W. Peterson, IMA Fungus 2: 122. 2011. [MB519297]. — Herb.: BPI 881018. Ex-type: NRRL 50410. Section Exilicaulis. ITS barcode: HQ646591. (Alternative markers: BenA = HQ646573; RPB2 = n.a.; CaM = HQ646584).
Penicillium meridianum D.B. Scott, Mycopathol. Mycol. Appl. 36: 12. 1968 ≡ Eupenicillium meridianum D.B. Scott, Mycopathol. Mycol. Appl. 36: 12. 1968. [MB335750]. — Herb.: CBS 314.67. Ex-type: CBS 314.67 = ATCC 18545 = CSIR 1052 = IMI 136209. Section Exilicaulis. ITS barcode: AF033451. (Alternative markers: BenA = KJ834472; RPB2 = JN406576; CaM = n.a.).
Penicillium mexicanum Visagie, Seifert & Samson, Stud. Mycol. 78: 125. 2014. [MB809185]. — Herb.: CBS H-21805. Ex-type: CBS 138227 = DTO 270F1. Section Paradoxa. ITS barcode: KJ775685. (Alternative markers: BenA = KJ775178; RPB2 = n.a.; CaM = KJ775412).
Penicillium miczynskii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 482. 1927. [MB271171]. — Herb.: IMI 40030. Ex-type: CBS 220.28 = ATCC 10470 = DSM2437 = FRR 1077 = IFO 7730 = IMI 040030 = MUCL 29228 = NRRL 1077 = QM 1957. Section Citrina. ITS barcode: GU944600. (Alternative markers: BenA = JN606706; RPB2 = JN606623; CaM = JN606526).
Penicillium mononematosum (Frisvad, Filt. & Wicklow) Frisvad, Mycologia 81: 857. 1990 ≡ Penicillium glandicola var. mononematosum Frisvad, Filt. & Wicklow, Can. J. Bot. 65: 767. 1987. [MB126406]. — Herb.: IMI 296925. Ex-type: CBS 172.87 = IBT 3072 = IBT 5518 = IBT 21535 = IMI 296925 = NRRL 13482 = NRRL A-26709. Section Chrysogena. ITS barcode: JX997082. (Alternative markers: BenA = AY495997; RPB2 = JX996709; CaM = JX996964).
Penicillium montanense M. Chr. & Backus, Mycologia 54: 574. 1962. [MB335752]. — Herb.: WIS Cryptogamic Herb. No. GW1-6. Ex-type: CBS 310.63 = ATCC 14941 = FRR 3407 = IFO 7740 = IHEM 4375 = IMI 099468 = MUCL 31326 = NRRL 3407. Section Aspergilloides. ITS barcode: KM189551. (Alternative markers: BenA = KM088789; RPB2 = KM089561; CaM = KM089174).
Penicillium multicolor Grig.-Man. & Porad., Arch. des Sciences Biol. Leningrad 19: 120. 1915. [MB119288]. — Herb.: unknown. Ex-type: CBS 501.73 = ATCC 24723 = IMI 174716 = VKM F-1745. Section Charlesia. ITS barcode: KC790402. (Alternative markers: BenA = JN799645; RPB2 = EU427262; CaM = JN799646).
Penicillium nalgiovense Laxa, Zentralbl. Bakteriol. Parasitenk., Abt. 2 86: 160. 1932. [MB114239]. — Herb.: CBS 352.48. Ex-type: CBS 352.48 = ATCC 10472 = IBT 21536 = IMI 039804 = MUCL 31194 = NRRL 911. Section Chrysogena. ITS barcode: AY371617. (Alternative markers: BenA = AY495999; RPB2 = JX996719; CaM = JX996974).
Penicillium namyslowskii K.M. Zalessky, Bull. Int. Aead. Polonc. Sci., Cl. Sci. Math., Sér. B, Sci. Nat. 1927: 479. 1927 ≡ Geosmithia namyslowskii (K.M. Zalessky) Pitt, Can. J. Bot. 57: 2024. 1979. [MB272006]. — Herb.: CBS 353.48. Ex-type: CBS 353.48 = ATCC 11127 = IMI 040033 = MUCL 29226 = NRRL 1070. Section Exilicaulis. ITS barcode: AF033463. (Alternative markers: BenA = JX141067; RPB2 = JF417430; CaM = n.a.).
Penicillium neocrassum R. Serra & S.W. Peterson, Mycologia 99: 81. 2007. [MB504767]. — Herb.: BPI 872161. Ex-type: CBS 122428 = NRRL 35639 = MUM 06.160. Section Brevicompacta. ITS barcode: DQ645805. (Alternative markers: BenA = DQ645794; RPB2 = JN406633; CaM = DQ645809).
Penicillium neoechinulatum (Frisvad, Filt. & Wicklow) Frisvad & Samson, Stud. Mycol. 49: 28. 2004 ≡ Penicillium aurantiogriseum var. neoechinulatum Frisvad, Filt. & Wicklow, Can. J. Bot. 65: 767. 1987. [MB368218]. — Herb.: IMI 296937. Ex-type: CBS 169.87 = CBS 101135 = IBT 3493 = IBT 21537 = IMI 296937 = NRRL 13486. Section Fasciculata. ITS barcode: JN942722. (Alternative markers: BenA = AF003237; RPB2 = JN985406; CaM = n.a.).
Penicillium neomiczynskii A.L.J. Cole et al., Stud. Mycol. 70: 105. 2011. [MB563192]. — Herb.: CBS H-20661. Ex-type: CBS 126231 = IBT 23560. Section Citrina. ITS barcode: JN617671. (Alternative markers: BenA = JN606705; RPB2 = n.a.; CaM = JN606523).
Penicillium nepalense Takada & Udagawa, Trans. Mycol. Soc. Japan 24: 146. 1983 ≡ Eupenicillium nepalense Takada & Udagawa, Trans. Mycol. Soc. Japan 24: 146. 1983. [MB108327]. — Herb.: NHL 6482. Ex-type: CBS 203.84 = NHL 6482. Section Exilicaulis. ITS barcode: KC411692. (Alternative markers: BenA = KJ834474; RPB2 = JN121453; CaM = n.a.).
Penicillium nodulum H.Z. Kong & Z.T. Qi, Mycosystema 1: 108. 1988. [MB135445]. — Herb.: CBS 227.89. Ex-type: CBS 227.89. Section Cinnamopurpurea. ITS barcode: KC411703. (Alternative markers: BenA = KJ834475; RPB2 = JN406603; CaM = KJ867003).
Penicillium nordicum Dragoni & Cantoni ex C. Ramírez, Adv. Penicillium Aspergillus Syst.: 139. 1986 ≡ Penicillium nordicum Dragoni & Cantoni, Ind. Aliment. 155: 283. 1979 (nom. inval., Art. 36). [MB114762]. — Herb.: ATCC 44219. Ex-type: ATCC 44219 = IBT 13307. Section Fasciculata. ITS barcode: KJ834513. (Alternative markers: BenA = KJ834476; RPB2 = n.a.; CaM = n.a.).
Penicillium nothofagi Houbraken, Frisvad & Samson, Stud. Mycol. 70: 105. 2011. [MB563189]. — Herb.: CBS H-20655. Ex-type: CBS 130383 = IBT 23018 = DTO 76C2. Section Citrina. ITS barcode: JN617712. (Alternative markers: BenA = JN606732; RPB2 = n.a.; CaM = JN606507).
Penicillium novae-zeelandiae J.F.H. Beyma, Antonie van Leeuwenhoek 6: 275. 1940. [MB522253]. — Herb.: IMI 40584ii. Ex-type: CBS 137.41 = ATCC 10473 = IFO 31748 = IMI 040584ii = NRRL 2128 = QM 1934 = VKMF-2886. Section Canescentia. ITS barcode: JN617688. (Alternative markers: BenA = KJ834477; RPB2 = JN406628; CaM = KJ866996).
Penicillium ochrochloron Biourge, Cellule 33: 269. 1923. [MB272701]. — Herb.: IMI 39806. Ex-type: CBS 357.48 = ATCC 10540 = IMI 039806 = NRRL 926 = QM 7604. Section Lanata-Divaricata. ITS barcode: GU981604. (Alternative markers: BenA = GU981672; RPB2 = KF296445; CaM = KF296378).
Penicillium ochrosalmoneum Udagawa, J. Agric. Sci. Tokyo Nogyo Daig. 5: 10. 1959 ≡ Eupenicillium ochrosalmoneum D.B. Scott & Stolk, Antonie van Leeuwenhoek 33: 302. 1967. [MB302409]. — Herb.: NHL 6048. Ex-type: CBS 489.66 = ATCC 18338 = CSIR 145 = IMI 116248ii = NRRL 35499. Section Ochrosalmonea. ITS barcode: EF626961. (Alternative markers: BenA = EF506212; RPB2 = JN121524; CaM = EF506237).
Penicillium odoratum M. Chr. & Backus, Mycologia 53: 459. 1961. [MB335755]. — Herb.: WSF 2000. Ex-type: CBS 294.62 = CBS 296.62 = ATCC 14769 = DSM2419 = IFO 7741 = IMI 094208ii = NRRL 3007 = WSF2000. Section Aspergilloides. ITS barcode: KC411730. (Alternative markers: BenA = KJ834478; RPB2 = JN406583; CaM = KM089363).
Penicillium olsonii Bainier & Sartory, Ann. Mycol. 10: 398. 1912. [MB121021]. — Herb.: IMI 192502. Ex-type: CBS 232.60 = IBT 23473 = IMI 192502 = NRRL 13058 = NRRL 13716. Section Brevicompacta. ITS barcode: EU587341. (Alternative markers: BenA = AY674445; RPB2 = JN121464; CaM = DQ658165).
Penicillium onobense C. Ramírez & A.T. Martínez, Mycopathologia 74: 44. 1981. [MB112525]. — Herb.: CBS 174.81. Ex-type: CBS 174.81 = ATCC 42225 = IJFM 3026 = VKMF-2183. Section Lanata-Divaricata. ITS barcode: GU981575. (Alternative markers: BenA = GU981627; RPB2 = KF296447; CaM = KF296371).
Penicillium ornatum Udagawa, Trans. Mycol. Soc. Japan 9: 49. 1968 ≡ Eupenicillium ornatum Udagawa, Trans. Mycol. Soc. Japan 9: 49. 1968. [MB335756]. — Herb.: NHL 6101. Ex-type: CBS 190.68 = ATCC 18608 = IFO 31739 = IMI 137977 = NHL 6101 = NRRL 3471. Section Ramigena. ITS barcode: KC411687. (Alternative markers: BenA = KJ834479; RPB2 = JN121451; CaM = n.a.).
Penicillium osmophilum Stolk & Veenb.-Rijks, Antonie van Leenwenhoek 40: 1. 1974 ≡ Eupenicillium osmophilum Stolk & Veenb.-Rijks, Antonie van Leeuwenhoek 40: 1. 1974. [MB319288]. — Herb.: CBS 462.72. Ex-type: CBS 462.72 = CBS 439.73 = IBT 14678 = NRRL 5922. Section Fasciculata. ITS barcode: EU427295. (Alternative markers: BenA = AY674376; RPB2 = JN121518; CaM = n.a.).
Penicillium ovatum (K. Ando & Nawawi) Houbraken & Samson, Stud. Mycol. 70: 48. 2011 ≡ Torulomyces ovatus K. Ando & Nawawi, Mycoscience 39: 317. 1998. [MB561961]. — Herb.: TNS-F-238518. Ex-type: CBS 136664 = KY 12726. Section Exilicaulis. ITS barcode: KF667370. (Alternative markers: BenA = KF667366; RPB2 = KF667372; CaM = KF667368).
Penicillium oxalicum Currie & Thom, J. Biol. Chem. 22: 289. 1915. [MB121033]. — Herb.: IMI 192332. Ex-type: CBS 219.30 = ATCC 1126 = FRR 787 = IMI 192332 = MUCL 29047 = NRRL 787 = QM 7606. Section Lanata-Divaricata. ITS barcode: AF033438. (Alternative markers: BenA = KF296462; RPB2 = JN121456; CaM = KF296367).
Penicillium palitans Westling, Ark Bot. 11: 83. 1911. [MB203604]. — Herb.: CBS H-7531. Ex-type: CBS 107.11 = ATCC 10477 = IBT 23034 = IMI 040215 = NRRL 2033 = VKMF-3088. Section Fasciculata. ITS barcode: KJ834514. (Alternative markers: BenA = KJ834480; RPB2 = n.a.; CaM = n.a.).
Penicillium palmense C. Ramírez & A.T. Martínez, Mycopathologia 66: 80. 1978. [MB319289]. — Herb.: IJFM 3840. Ex-type: CBS 336.79 = ATCC 38669 = IJFM 3840. Section Aspergilloides. ITS barcode: KJ834515. (Alternative markers: BenA = GQ367508; RPB2 = JN406566; CaM = GQ367534).
Penicillium pancosmium Houbraken, Frisvad & Samson, Stud. Mycol. 70: 108. 2011. [MB563191]. — Herb.: CBS H-20651. Ex-type: CBS 276.75 = DAOM 147467 = IBT 29991. Section Citrina. ITS barcode: JN617660. (Alternative markers: BenA = JN606790; RPB2 = n.a.; CaM = JN606446).
Penicillium paneum Frisvad, Microbiology 142: 546. 1996. [MB415570]. — Herb.: C 25000. Ex-type: CBS 101032 = IBT 21541 = IBT 12407. Section Roquefortorum. ITS barcode: HQ442346. (Alternative markers: BenA = AY674387; RPB2 = n.a.; CaM = HQ442331).
Penicillium paradoxum (Fennell & Raper) Samson et al., (published here) ≡ Aspergillus paradoxus Fennell & Raper, Mycologia 47: 69. 1955 ≡ Hemicarpenteles paradoxus Sarbhoy & Elphick, Trans. Brit. Mycol. Soc. 51: 156. 1968. [MB547045]. — Herb.: IMI 061446. Ex-type: CBS 527.65 = NRRL 2162 = ATCC 16918 = IMI 061446. Section Paradoxa. ITS barcode: EF669707. (Alternative markers: BenA = EF669683; RPB2 = EF669670; CaM = EF669692).
Penicillium paraherquei S. Abe ex G. Sm., Trans. Brit. Mycol. Soc. 46: 335. 1963 ≡ Penicillium paraherquei S. Abe, J. Gen. Appl. Microbiol., Tokyo 2: 131. 1956 (nom. inval., Art. 36). [MB302412]. — Herb.: IMI 68220. Ex-type: CBS 338.59 = ATCC 22354 = ATCC 46903 = FAT964 = FRR 3454 = IFO 6234 = IMI 068220 = NRRL 3454. Section Lanata-Divaricata. ITS barcode: AF178511. (Alternative markers: BenA = KF296465; RPB2 = KF296449; CaM = KF296372).
Penicillium parvulum S.W. Peterson & B.W. Horn, Mycologia 101: 75. 2009. [MB509289]. — Herb.: BPI 877331. Ex-type: CBS 132825 = NRRL 35504. Section Cinnamopurpurea. ITS barcode: EF422845. (Alternative markers: BenA = EF506218; RPB2 = n.a.; CaM = EF506225).
Penicillium parvum Raper & Fennell, Mycologia 40: 508. 1948 ≡ Carpenteles parvum (Raper & Fennell) Udagawa, Trans. Mycol. Soc. Japan 6: 79. 1965 ≡ Eupenicillium parvum (Raper & Fennell) Stolk & D.B. Scott, Persoonia 4: 402. 1967. [MB289101]. — Herb.: CBS H-7105. Ex-type: CBS 359.48 = ATCC 10479 = IFO 7732 = IMI 040587 = NRRL 2095 = QM 1878. Section Exilicaulis. ITS barcode: AF033460. (Alternative markers: BenA = HQ646568; RPB2 = JN406559; CaM = KF900173).
Penicillium pasqualense Houbraken, Frisvad & Samson, Stud. Mycol. 70: 108. 2011. [MB563190]. — Herb.: CBS H-20663. Ex-type: CBS 126330 = IBT 14235. Section Citrina. ITS barcode: JN617676. (Alternative markers: BenA = JN606673; RPB2 = JN606617; CaM = JN606394).
Penicillium patens Pitt & A.D. Hocking, Mycotaxon 22: 205. 1985. [MB105611]. — Herb.: FRR 2661. Ex-type: CBS 260.87 = FRR 2662. Section Aspergilloides. ITS barcode: KC411718. (Alternative markers: BenA = KJ834481; RPB2 = JN406593; CaM = n.a.).
Penicillium paxilli Bainier, Bull. Soc. Mycol. France 23: 95. 1907. [MB203838]. — Herb.: IMI 40226. Ex-type: CBS 360.48 = ATCC 10480 = FRR 2008 = IMI 040226 = NRRL 2008 = QM 725. Section Citrina. ITS barcode: GU944577. (Alternative markers: BenA = JN606844; RPB2 = JN606610; CaM = JN606566).
Penicillium penarojense Houbraken et al., Int. J. Syst. Evol. Microbiol. 61: 1471. 2011. [MB518024]. — Herb.: HUA 170335. Ex-type: CBS 113178 = IBT 23262. Section Lanata-Divaricata. ITS barcode: GU981570. (Alternative markers: BenA = GU981646; RPB2 = KF296450; CaM = KF296381).
Penicillium persicinum L. Wang et al., Antonie van Leeuwenhoek 86: 177. 2004. [MB500259]. — Herb.: HMAS 80638-1-4. Ex-type: CBS 111235 = AS 3.5891 = IBT 24565. Section Chrysogena. ITS barcode: JX997072. (Alternative markers: BenA = AY495982; RPB2 = JN406644; CaM = JX996954).
Penicillium philippinense Udagawa & Y. Horie, J. Jap. Bot. 47: 341. 1972. [MB319291]. — Herb.: CBS H-7099. Ex-type: CBS 623.72 = FRR 1532 = NHL 6130. Section Exilicaulis. ITS barcode: KC411770. (Alternative markers: BenA = KJ834482; RPB2 = JN406543; CaM = n.a.).
Penicillium phoeniceum J.F.H. Beyma, Zentralbl. Bakteriol. Parasitenk., Abt. 2 88: 136. 1933 ≡ Eupenicillium cinnamopurpureum D.B. Scott & Stolk, Antonie van Leeuwenhoek 33: 308. 1967. [MB274284]. — Herb.: IMI 40585. Ex-type: CBS 249.32 = ATCC 10481 = IJFM 5122 = IMI 040585 = NRRL 2070 = QM 7608 = VKMF-321. Section Charlesia. ITS barcode: KC411711. (Alternative markers: BenA = KJ834483; RPB2 = JN406597; CaM = AY741729).
Penicillium pimiteouiense S.W. Peterson, Mycologia 91: 271. 1999. [MB460126]. — Herb.: BPI 806262. Ex-type: CBS 102479 = NRRL 25542. Section Exilicaulis. ITS barcode: AF037431. (Alternative markers: BenA = HQ646569; RPB2 = JN406650; CaM = HQ646580).
Penicillium piscarium Westling, Ark. Bot. 11: 86. 1911. [MB211321]. — Herb.: IMI 40032. Ex-type: CBS 362.48 = ATCC 10482 = FRR 1075 = IFO 8111 = IMI 040032 = NRRL 1075 = VKMF-1823. Section Lanata-Divaricata. ITS barcode: GU981600. (Alternative markers: BenA = GU981668; RPB2 = KF296451; CaM = KF296379).
Penicillium polonicum K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 445. 1927 ≡ Penicillium aurantiogriseum var. polonicum (K.M. Zalessky) Frisvad & Filt., Mycologia 81: 850. 1990. [MB274889]. — Herb.: CBS 222.28. Ex-type: CBS 222.28 = IBT 12821 = IMI 291194 = MUCL 29204 = NRRL 995. Section Fasciculata. ITS barcode: AF033475. (Alternative markers: BenA = AY674305; RPB2 = JN406609; CaM = n.a.).
Penicillium porphyreum Houbraken & Samson, Stud. Mycol. 70: 48. 2011 ≡ Monocillium humicola G.L. Barron, Can. J. Bot. 39: 1575. 1961 ≡ Monocillium humicola var. brunneum M. Chr. & Backus, Mycologia 56: 498. 1964 ≡ Torulomyces brunneus (M. Chr. & Backus) K. Ando, Mycoscience 39: 314. 1998. [MB561959]. — Herb.: CBS 382.64 = WSF 9-c. Ex-type: CBS 382.64 = KY 12723. Section Torulomyces. ITS barcode: KF303666. (Alternative markers: BenA = KF303621; RPB2 = KF303677; CaM = KF303636).
Penicillium psychrosexualis Houbraken & Samson, IMA Fungus 1: 174. 2010. [MB519086]. — Herb.: CBS H-20501. Ex-type: CBS 128137 = IBT 29551. Section Roquefortorum. ITS barcode: HQ442345. (Alternative markers: BenA = HQ442356; RPB2 = n.a.; CaM = HQ442330).
Penicillium pullum S.W. Peterson & Sigler, Mycol. Res. 106: 1115. 2002. [MB483982]. — Herb.: BPI 841398. Ex-type: CBS 331.48 = ATCC 10447 = NRRL 721 = FRR 721 = IFO 6097 = IMI 39747 = QM 1925 = Thom 5179.4. Section Stolkia. ITS barcode: AF033443. (Alternative markers: BenA = JN617719; RPB2 = n.a.; CaM = AF481134).
Penicillium pulvillorum Turfitt, Trans. Brit. Mycol. Soc. 23: 186. 1939. [MB275682]. — Herb.: CBS 280.39. Ex-type: CBS 280.39 = IFO 7763 = NRRL 2026. Section Lanata-Divaricata. ITS barcode: AF178517. (Alternative markers: BenA = GU981670; RPB2 = KF296452; CaM = KF296377).
Penicillium pulvis Houbraken, Visagie, Samson & Seifert, Stud. Mycol. 78: 429. 2014. [MB809970]. — Herb.: CBS H-21878. Ex-type: CBS 138432 = DTO 180-B7. Section Aspergilloides. ITS barcode: KM189632. (Alternative markers: BenA = KM088876; RPB2 = KM089650; CaM = KM089263).
Penicillium purpurescens (Sopp) Raper & Thom, A manual of the Penicillia: 177. 1949 ≡ Citromyces purpurascens Sopp, Skr. Vidensk.-Selsk. Christiana, Math.-Naturvidensk. Kl. 11: 117. 1912. [MB335761]. — Herb.: IMI 39745. Ex-type: CBS 366.48 = NRRL 720 = FRR 720 = ATCC 10485 = IMI 39745. Section Aspergilloides. ITS barcode: KM189561. (Alternative markers: BenA = KM088801; RPB2 = KM089573; CaM = KM089186).
Penicillium quebecense Houbraken, Frisvad & Samson, Stud. Mycol. 70: 111. 2011. [MB563202]. — Herb.: CBS H-20666. Ex-type: CBS 101623 = IBT 29050. Section Citrina. ITS barcode: JN617661. (Alternative markers: BenA = JN606700; RPB2 = JN606622; CaM = JN606509).
Penicillium quercetorum Baghd., Novosti Sist. Nizsh. Rast. 5: 110. 1968. [MB335762]. — Herb.: CBS H-7527. Ex-type: CBS 417.69 = NRRL 3758 = ATCC 48727 = CCRC 31668 = FRR 516 = IFO 31749 = IMI 140342 = MUCL 31203 = VKMF-1074. Section Aspergilloides. ITS barcode: KM189556. (Alternative markers: BenA = KM088795; RPB2 = KM089567; CaM = KM089180).
Penicillium raciborskii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 454. 1927. [MB276002]. — Herb.: IMI 40568. Ex-type: CBS 224.28 = ATCC 10488 = DSM2422 = FRR 2150 = IFO 7676 = IMI 040568 = LSHBP 92 = MUCL 29246 = NRRL 2150. Section Exilicaulis. ITS barcode: AF033447. (Alternative markers: BenA = JX141069; RPB2 = JN406607; CaM = n.a.).
Penicillium radicicola Overy & Frisvad, Syst. Appl. Microbiol. 26: 633. 2003. [MB488233]. — Herb.: C 60161. Ex-type: CBS 112430 = IBT 10696. Section Fasciculata. ITS barcode: KJ834516. (Alternative markers: BenA = AY674357; RPB2 = n.a.; CaM = n.a.).
Penicillium raistrickii G. Sm., Trans. Brit. Mycol. Soc. 18: 90. 1933. [MB276069]. — Herb.: IMI 40221. Ex-type: CBS 261.33 = ATCC 10490 = FRR 1044 = IFO 6104 = IMI 040221 = LSHBB100 = NRRL 1044 = NRRL 2039 = QM 1936 = VKMF-337. Section Ramosa. ITS barcode: AY373927. (Alternative markers: BenA = KJ834485; RPB2 = JN406592; CaM = KJ867006).
Penicillium ramusculum Bat. & H. Maia, Anais Soc. Biol. Pernambuco 13: 27. 1955. [MB302419]. — Herb.: unknown. Ex-type: CBS 251.56 = ATCC 12292 = FRR 3459 = IMI 063546 = IMUR478 = LSHBBB324 = NRRL 3459 = QM 7057. Section Ramigena. ITS barcode: EF433765. (Alternative markers: BenA = EU427269; RPB2 = JN121472; CaM = EU427278).
Penicillium ranomafanaense Houbraken & Hagen, Stud. Mycol. 78: 433. 2014. [MB809971]. — Herb.: CBS H-21862. Ex-type: CBS 137953 = DTO 085-A5. Section Aspergilloides. ITS barcode: KM189541. (Alternative markers: BenA = KM088779; RPB2 = KM089551; CaM = KM089164).
Penicillium raperi G. Sm., Trans. Brit. Mycol. Soc. 40: 486. 1957. [MB302421]. — Herb.: IMI 71625. Ex-type: CBS 281.58 = ATCC 22355 = IFO 8179 = IMI 071625 = LSHBBB338 = NRRL 2674 = QM 7527. Section Lanata-Divaricata. ITS barcode: AF033433. (Alternative markers: BenA = GU981622; RPB2 = KF296453; CaM = KF296399).
Penicillium raphiae Houbraken, Frisvad & Samson, Stud. Mycol. 70: 114. 2011. [MB563203]. — Herb.: CBS H-20660. Ex-type: CBS 126234 = IBT 22407. Section Citrina. ITS barcode: JN617673. (Alternative markers: BenA = JN606657; RPB2 = JN606619; CaM = JN606409).
Penicillium restrictum J.C. Gilman & E.V. Abbott, Iowa St. Coll. J. Sci. 1: 297. 1927. [MB276289]. — Herb.: IMI 40228. Ex-type: CBS 367.48 = ATCC 11257 = FRR 1748 = IMI 040228 = NRRL 1748 = QM 1962. Section Exilicaulis. ITS barcode: AF033457. (Alternative markers: BenA = KJ834486; RPB2 = JN121506; CaM = n.a.).
Penicillium restingae J.P. Andrade et al., Persoonia 32: 293. 2014. [MB807051]. — Herb.: CMR H-12. Ex-type: URM 7075. Section Sclerotiora. ITS barcode: KF803355. (Alternative markers: BenA = KF803349; RPB2 = n.a.; CaM = KF803352).
Penicillium reticulisporum Udagawa, Trans. Mycol. Soc. Japan 9: 52. 1968 ≡ Eupenicillium reticulisporum Udagawa, Trans. Mycol. Soc. Japan 9: 52. 1968. [MB335763]. — Herb.: NHL 6105. Ex-type: CBS 122.68 = ATCC 18566 = IFO 9024 = IMI 136700 = NHL 6105 = NRRL 3447. Section Lanata-Divaricata. ITS barcode: AF033437. (Alternative markers: BenA = GU981665; RPB2 = KF296454; CaM = KF296391).
Penicillium ribium Frisvad & Overy, Int. J. Syst. Evol. Microbiol. 56: 1436. 2006. [MB501061]. — Herb.: DAOM 234091. Ex-type: CBS 127809 = DAOM 234091 = IBT 16537 = IBT 24431. Section Ramosa. ITS barcode: DQ267916. (Alternative markers: BenA = DQ285625; RPB2 = JN406631; CaM = KJ866995).
Penicillium rolfsii Thom, Penicillia: 489. 1930. [MB276674]. — Herb.: IMI 40029. Ex-type: CBS 368.48 = ATCC 10491 = FRR 1078 = IFO 7735 = IMI 040029 = MUCL 29229 = NRRL 1078 = QM 1961. Section Lanata-Divaricata. ITS barcode: JN617705. (Alternative markers: BenA = GU981667; RPB2 = KF296455; CaM = KF296375).
Penicillium roqueforti Thom, U.S.D.A. Bur. Animal Industr. Bull. 82: 35. 1906. [MB213525]. — Herb.: IMI 24313. Ex-type: CBS 221.30 = ATCC 10110 = ATCC 1129 = CECT 2905 = IBT 6754 = IFO 5459 = IMI 024313 = LSHBP 93 = NCTC 588 = NRRL 849 = QM 1937. Section Roquefortorum. ITS barcode: EU427296. (Alternative markers: BenA = AF000303; RPB2 = JN406611; CaM = HQ442332).
Penicillium roseomaculatum Biourge, Cellule 33: 301. 1923. [MB276785]. — Herb.: unknown. Ex-type: CBS 137962 = IMI 189696 = NRRL 728 = FRR 728. Section Aspergilloides. ITS barcode: KM189755. (Alternative markers: BenA = KM089004; RPB2 = KM089778; CaM = KM089391).
Penicillium roseopurpureum Dierckx, Ann. Soc. Sci. Bruxelles 25: 86. 1901. [MB213447]. — Herb.: IMI 40573. Ex-type: CBS 226.29 = ATCC 10492 = ATHUM2895 = FRR 2064 = IMI 040573 = MUCL 28654 = MUCL 29237 = NRRL 2064 = NRRL 2064A. Section Citrina. ITS barcode: GU944605. (Alternative markers: BenA = JN606838; RPB2 = JN606613; CaM = JN606556).
Penicillium roseoviride Stapp & Bortels, Zentralbl. Bakteriol. Parasitenk., Abt. 2 93: 51. 1935. [MB492646]. — Herb.: unknown. Ex-type: CBS 267.35 = ATCC 10412 = IFO 6089 = IMI 039740ii = NRRL 760 = QM 7485. Section Aspergilloides. ITS barcode: KM189549. (Alternative markers: BenA = KM088787; RPB2 = KM089559; CaM = KM089172).
Penicillium rubefaciens Quintan., Mycopathologia 80: 73. 1982. [MB109998]. — Herb.: CBS 145.83. Ex-type: CBS 145.83 = CECT 2752. Section Exilicaulis. ITS barcode: KC411677. (Alternative markers: BenA = KJ834487; RPB2 = JN406627; CaM = n.a.).
Penicillium rubens Biourge, Cellule 33: 265. 1923. [MB276884]. — Herb.: CBS H-20595. Ex-type: CBS 129667 = NRRL 792 = IBT 30129 = ATCC 9783. Section Chrysogena. ITS barcode: JX997057. (Alternative markers: BenA = JF909949; RPB2 = JX996658; CaM = JX996263).
Penicillium rubidurum Udagawa & Y. Horie, Trans. Mycol. Soc. Japan 14: 381. 1973 ≡ Eupenicillium rubidurum Udagawa & Y. Horie, Trans. Mycol. Soc. Japan 14: 381. 1973. [MB319295]. — Herb.: NHL 6460. Ex-type: CBS 609.73 = NRRL 6033 = ATCC 28051 = ATCC 48238 = FRR 1558 = IMI 228551 = NHL 6460. Section Exilicaulis. ITS barcode: AF033462. (Alternative markers: BenA = HQ646574; RPB2 = JN406545; CaM = HQ646585).
Penicillium rudallense Houbraken, Visagie & Pitt, Stud. Mycol. 78: 433. 2014. [MB809972]. — Herb.: CBS H-21867. Ex-type: CBS 138162 = FRR 6085 = DTO 056-I4. Section Aspergilloides. ITS barcode: KM088741. (Alternative markers: BenA = KM089126; RPB2 = KM089513; CaM = KM189504).
Penicillium sacculum E. Dale, Ann. Mycol. 24: 137. 1926 ≡ Eladia saccula (E. Dale) G. Sm., Trans. Brit. Mycol. Soc. 44: 47. 1961. [MB277209]. — Herb.: CBS 231.61. Ex-type: CBS 231.61 = ATCC 18350 = IFO 8114 = IFO 9454 = IMI 051498 = LSHBBB298 = UC4505. Section Eladia. ITS barcode: KC411707. (Alternative markers: BenA = KJ834488; RPB2 = JN121462; CaM = n.a.).
Penicillium sajarovii Quintan., Av. Aliment. Majora Anim. 22: 539. 1981. [MB114172]. — Herb.: CBS 277.83. Ex-type: CBS 277.83 = CECT 2751 = IMI 259992. Section Ramosa. ITS barcode: KC411724. (Alternative markers: BenA = KJ834489; RPB2 = JN406588; CaM = KJ867007).
Penicillium sanguifluum (Sopp) Biourge, Cellule 33: 105. 1923 ≡ Citromyces sanguifluus Sopp, Skr. Vidensk.-Selsk. Christiana Math.-Nat. Kl. 11: 115. 1912. [MB356682]. — Herb.: CBS H-20645. Ex-type: CBS 127032 = IBT 29041. Section Citrina. ITS barcode: JN617681. (Alternative markers: BenA = JN606819; RPB2 = n.a.; CaM = JN606555).
Penicillium saturniforme (L. Wang & W.Y. Zhuang) Houbraken & Samson, Stud. Mycol. 70: 48. 2011 ≡ Eupenicillium saturniforme L. Wang & W.Y. Zhuang Mycopathologia 167: 300. 2009. [MB561958]. — Herb.: AS 3.6886. Ex-type: CBS 122276 = AS 3.6886. Section Aspergilloides. ITS barcode: EU644081. (Alternative markers: BenA = EU644080; RPB2 = JN121439; CaM = EU644062).
Penicillium scabrosum Frisvad, Samson & Stolk, Persoonia 14: 177. 1990. [MB136735]. — Herb.: IMI 285533. Ex-type: CBS 683.89 = FRR 2950 = IBT 3736 = IMI 285533 = DAOM 214786. Section Ramosa. ITS barcode: DQ267906. (Alternative markers: BenA = DQ285610; RPB2 = JN406541; CaM = FJ530987).
Penicillium sclerotigenum W. Yamam., Sci. Rep. Hyogo Univ Agric. 1: 69. 1955. [MB302424]. — Herb.: IMI 68616. Ex-type: CBS 101033 = CBS 343.59 = ATCC 18488 = IBT 14346 = IFO 6167 = IMI 068616 = NRRL 3461 = QM 7779. Section Penicillium. ITS barcode: AF033470. (Alternative markers: BenA = AY674393; RPB2 = JN406652; CaM = n.a.).
Penicillium sclerotiorum J.F.H. Beyma, Zentralbl. Bakteriol. Parasitenk., Abt. 2 96: 418. 1937. [MB277708]. — Herb.: IMI 40569. Ex-type: CBS 287.36 = ATCC 10494 = IFO 6105 = IMI 040569 = NRRL 2074 = QM 1938 = VKMF-353. Section Sclerotiora. ITS barcode: JN626132. (Alternative markers: BenA = JN626001; RPB2 = JN406585; CaM = JN626044).
Penicillium senticosum D.B. Scott, Mycopathol. Mycol. Appl. 36: 5. 1968 ≡ Eupenicillium senticosum D.B. Scott, Mycopathol. Mycol. Appl. 36: 5. 1968. [MB335764]. — Herb.: CBS 316.67. Ex-type: CBS 316.67 = ATCC 18623 = CSIR 1042 = IMI 136211 = IMI 216905. Section Eladia. ITS barcode: KC411733. (Alternative markers: BenA = KJ834490; RPB2 = n.a.; CaM = n.a.).
Penicillium shearii Stolk & D.B. Scott, Persoonia 4: 396. 1967 ≡ Eupenicillium shearii Stolk & D.B. Scott, Persoonia 4: 396. 1967. [MB335765]. — Herb.: CBS 290.48. Ex-type: CBS 290.48 = ATCC 10410 = IFO 6088 = IMI 039739 = IMI 039739iv = NRRL 715 = QM 1870. Section Citrina. ITS barcode: GU944606. (Alternative markers: BenA = JN606840; RPB2 = JN121482; CaM = EU644068).
Penicillium shennangjianum H.Z. Kong & Z.T. Qi, Mycosystema 1: 110. 1988. [MB119509]. — Herb.: CBS 228.89. Ex-type: CBS 228.89. Section Cinnamopurpurea. ITS barcode: KC411705. (Alternative markers: BenA = KJ834491; RPB2 = JN121458; CaM = AY678561).
Penicillium simile Davolos et al., Int. J. Syst. Evol. Microbiol. 62: 457. 2012. [MB509645]. — Herb.: ATCC MYA-4591. Ex-type: CBS 129191 = ATCC MYA-4591. Section Ramosa. ITS barcode: FJ376592. (Alternative markers: BenA = FJ376595; RPB2 = n.a.; CaM = GQ979710).
Penicillium simplicissimum (Oudem.) Thom, Penicillia: 335. 1930 ≡ Spicaria simplicissima Oudem., Ned. Kruidk. Arch. 2: 763. 1902. [MB278201]. — Herb.: CUP Jensen. 1912: No. 5921 (CUP). Ex-type: CBS 372.48 = ATCC 10495 = FRR 902 = IFO 5762 = IMI 039816 = QM 1939. Section Lanata-Divaricata. ITS barcode: GU981588. (Alternative markers: BenA = GU981632; RPB2 = JN121507; CaM = KF296368).
Penicillium sinaicum Udagawa & S. Ueda, Mycotaxon 14: 266. 1982 ≡ Eupenicillium sinaicum Udagawa & S. Ueda, Mycotaxon 14: 266. 1982. [MB110862]. — Herb.: NHL 2894. Ex-type: CBS 279.82 = NHL 2894. Section Chrysogena. ITS barcode: JX997090. (Alternative markers: BenA = JX996846; RPB2 = JN406587; CaM = JX996970).
Penicillium singorense Visagie, Seifert & Samson, Stud. Mycol. 78: 119. 2014. [MB809182]. — Herb.: CBS H-21802. Ex-type: CBS 138214 = DTO 133C6. Section Lanata-Divaricata. ITS barcode: KJ775674. (Alternative markers: BenA = KJ775167; RPB2 = n.a.; CaM = KJ775403).
Penicillium sizovae Baghd., Novosti Sist. Nizsh. Rast. 1968: 103. 1968. [MB335767]. — Herb.: CBS 413.69. Ex-type: CBS 413.69 = FRR 518 = IMI 140344 = VKMF-1073. Section Citrina. ITS barcode: GU944588. (Alternative markers: BenA = GU944535; RPB2 = JN606603; CaM = GU944618).
Penicillium skrjabinii Schmotina & Golovleva, Mikol. Fitopatol. 8: 530. 1974. [MB319296]. — Herb.: IMI 196528. Ex-type: CBS 439.75 = NRRL 13055 = FRR 1945 = IMI 196528 = VKMF-1940. Section Lanata-Divaricata. ITS barcode: GU981576. (Alternative markers: BenA = GU981626; RPB2 = EU427252; CaM = KF296370).
Penicillium smithii Quintan., Av. Aliment. Majora Anim. 23: 340. 1982. [MB114173]. — Herb.: CBS 276.83. Ex-type: CBS 276.83 = CECT 2744 = IMI 259693. Section Exilicaulis. ITS barcode: KC411723. (Alternative markers: BenA = KJ834492; RPB2 = JN406589; CaM = n.a.).
Penicillium solitum Westling, Ark. Bot. 11: 65. 1911. [MB206172]. — Herb.: CBS 424.89. Ex-type: CBS 424.89 = ATCC 9923 = CBS 288.36 = FRR 937 = IBT 3948 = IFO 7765 = IMI 039810 = IMI 092225 = LSHBP 52 = MUCL 28668 = MUCL 29173 = NRRL 937. Section Fasciculata. ITS barcode: AY373932. (Alternative markers: BenA = AY674354; RPB2 = n.a.; CaM = n.a.).
Penicillium soppii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 476. 1927. [MB121424]. — Herb.: IMI 40217. Ex-type: CBS 226.28 = ATCC 10496 = FRR 2023 = IFO 7766 = IMI 040217 = MUCL 29233 = NRRL 2023 = QM 1964 = IBT 18220. Section Ramosa. ITS barcode: AF033488. (Alternative markers: BenA = DQ285616; RPB2 = JN406606; CaM = n.a.).
Penicillium spathulatum Frisvad & Samson, FEMS Microbiol. Lett. 339: 88. 2013. [MB492650]. — Herb.: CBS 117192. Ex-type: CBS 117192 = IBT 22220. Section Brevicompacta. ITS barcode: JX313165. (Alternative markers: BenA = JX313183; RPB2 = JN406636; CaM = JX313149).
Penicillium spinulosum Thom, U.S.D.A. Bur. Animal Industr. Bull. 118: 76. 1910. [MB215401]. — Herb.: IMI 24316i. Ex-type: CBS 374.48 = ATCC 10498 = FRR 1750 = IMI 024316 = LSHBAd 29 = MUCL 13910 = MUCL 13911 = NCTC 591 = NRRL 1750 = QM 7654. Section Aspergilloides. ITS barcode: AF033410. (Alternative markers: BenA = KJ834493; RPB2 = JN406558; CaM = GQ367524).
Penicillium steckii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 469. 1927. [MB278769]. — Herb.: IMI 40583. Ex-type: CBS 260.55 = ATCC 10499 = CECT 2268 = DSM1252 = IMI 040583 = NRRL 2140 = QM 6413. Section Citrina. ITS barcode: GU944597. (Alternative markers: BenA = GU944522; RPB2 = JN606602; CaM = GU944611).
Penicillium sterculiniicola Houbraken, Stud. Mycol. 78: 436. 2014. [MB809973]. — Herb.: CBS H-21877. Ex-type: CBS 122426 = DTO 031-A4. Section Aspergilloides. ITS barcode: KM189464. (Alternative markers: BenA = KM088693; RPB2 = KM089465; CaM = KM089078).
Penicillium stolkiae D.B. Scott, Mycopathol. Mycol. Appl. 36: 8. 1968 ≡ Eupenicillium stolkiae D.B. Scott, Mycopathol. Mycol. Appl. 36: 8. 1968. [MB335768]. — Herb.: CBS 315.67. Ex-type: CBS 315.67 = ATCC 18546 = CSIR 1041 = FRR 534 = IMI 136210 = NRRL 5816. Section Stolkia. ITS barcode: AF033444. (Alternative markers: BenA = JN617717; RPB2 = JN121488; CaM = AF481135).
Penicillium striatisporum Stolk, Antonie van Leeuwenhoek 35: 268. 1969. [MB335769]. — Herb.: CBS 705.68. Ex-type: CBS 705.68 = ATCC 22052 = CCRC 31679 = FRR 827 = IMI 151749 = MUCL 31202. Section Exilicaulis. ITS barcode: AF038938. (Alternative markers: BenA = JX141156; RPB2 = JN406538; CaM = n.a.).
Penicillium subarcticum S.W. Peterson & Sigler, Mycol. Res. 106: 1116. 2002. [MB483983]. — Herb.: BPI 841397. Ex-type: CBS 111719 = NRRL 31108 = UAMH 3897. Section Stolkia. ITS barcode: AF481120. (Alternative markers: BenA = JN617716; RPB2 = n.a.; CaM = AF481141).
Penicillium sublateritium Biourge, Cellule 33: 315. 1923. [MB279265]. — Herb.: IMI 40594. Ex-type: CBS 267.29 = ATCC 10502 = FRR 2071 = IFO 6107 = IMI 040594 = LSHBP 55 = MUCL 28655 = NRRL 2071 = QM 7652 = NBRC 6107. Section Ramigena. ITS barcode: EU427288. (Alternative markers: BenA = JX091557; RPB2 = JN406590; CaM = AB566102).
Penicillium sublectaticum Houbraken, et al., Stud. Mycol. 78: 436. 2014. [MB809974]. — Herb.: CBS H-21955. Ex-type: CBS 138217 = DTO 244-G2. Section Aspergilloides. ITS barcode: KM189761. (Alternative markers: BenA = KM089010; RPB2 = KM089784; CaM = KM089397).
Penicillium subrubescens Houbraken et al., Antonie van Leeuwenhoek 103: 1354. 2013. [MB801306]. — Herb.: CBS H-21029. Ex-type: CBS 132785 = DTO 188D6 = FBCC 1632 = IBT 31985. Section Lanata-Divaricata. ITS barcode: KC346350. (Alternative markers: BenA = KC346327; RPB2 = KC346306; CaM = KC346330).
Penicillium subspinulosum Houbraken, Stud. Mycol. 78: 436. 2014. [MB809975]. — Herb.: CBS H-21856. Ex-type: CBS 137946 = DTO 041-F2. Section Aspergilloides. ITS barcode: KM189483. (Alternative markers: BenA = KM088719; RPB2 = KM089491; CaM = KM089104).
Penicillium sucrivorum Visagie & K. Jacobs, Mycologia 106: 546. 2014. [MB804723]. — Herb.: CBS H-21331. Ex-type: CBS 135116 = DAOM 241042 = DTO 183E5. Section Citrina. ITS barcode: JX140872. (Alternative markers: BenA = JX141015; RPB2 = n.a.; CaM = JX141506).
Penicillium sumatraense Szilvinyi [as ‘sumatrense’], Archiv. Hydrobiol. 14, Suppl. 6: 535. 1936. [MB319297]. — Herb.: unknown. Ex-type: CBS 281.36 = NRRL 779 = FRR 779. Section Citrina. ITS barcode: GU944578. (Alternative markers: BenA = JN606639; RPB2 = n.a.; CaM = JN606368).
Penicillium svalbardense Frisvad, Sonjak & Gundae-Cim, Antonie van Leeuwenhoek 92: 48. 2007. [MB529943]. — Herb.: EX-F 1307. Ex-type: CBS 122416 = IBT 23856. Section Lanata-Divaricata. ITS barcode: GU981603. (Alternative markers: BenA = KC346325; RPB2 = KF296457; CaM = KC346338).
Penicillium swiecickii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 474. 1927. [MB279618]. — Herb.: unknown. Ex-type: CBS 119391 = FRR 918 = IBT 27865 = IMI 191500 = NRRL 918. Section Ramosa. ITS barcode: AF033490. (Alternative markers: BenA = KJ834494; RPB2 = n.a.; CaM = KJ866993).
Penicillium tardochrysogenum Frisvad, Houbraken & Samson, Persoonia 29: 93. 2012. [MB801877]. — Herb.: CBS H-21057. Ex-type: CBS 132200 = IBT 30075 = DTO 149B9. Section Chrysogena. ITS barcode: JX997027. (Alternative markers: BenA = JX996898; RPB2 = JX996634; CaM = JX996239).
Penicillium taxi R. Schneid., Zentralbl. Bakteriol. Parasitenk., Abt. 2 110: 43. 1956 ≡ Thysanophora taxi (R. Schneid.) Stolk & Hennebert, Persoonia 5: 193. 1968. [MB282799]. — Herb.: unknown. Ex-type: CBS 206.57 = ATCC 18484 = BBA 7480 = MUCL 11402 = QM 8153. Section Thysanophora. ITS barcode: KJ834517. (Alternative markers: BenA = KJ834495; RPB2 = JN121454; CaM = n.a.).
Penicillium terrenum D.B. Scott, Mycopathol. Mycol. Appl. 36: 1. 1968 ≡ Eupenicillium terrenum D.B. Scott, Mycopathol. Mycol. Appl. 36: 1. 1968. [MB335771]. — Herb.: CBS 313.67. Ex-type: CBS 313.67 = ATCC 18547 = CSIR 1022 = IMI 136208. Section Exilicaulis. ITS barcode: AM992111. (Alternative markers: BenA = KJ834496; RPB2 = JN406577; CaM = n.a.).
Penicillium terrigenum Seifert et al., Stud. Mycol. 70: 125. 2011. [MB563204]. — Herb.: CBS H-20667. Ex-type: CBS 127354 = IBT 30769. Section Citrina. ITS barcode: JN617684. (Alternative markers: BenA = JN606810; RPB2 = JN606600; CaM = JN606583).
Penicillium thiersii S.W. Peterson, E.M. Bayer & Wicklow, Mycologia 96: 1283. 2004. [MB487738]. — Herb.: BPI 842269. Ex-type: CBS 117503 = IBT 27050 = NRRL 28162. Section Aspergilloides. ITS barcode: AF125936. (Alternative markers: BenA = KJ834497; RPB2 = JN121434; CaM = AY741726).
Penicillium thomii Maire, Bull. Soc. Hist. Nat. Afrique N. 8: 189. 1917 ≡ Citromyces thomii (Maire) Sacc., Syll. Fung. 25: 683. 1931 ≡ Penicillium lividum var. thomii (Maire) Stolk & Samson, Adv. Penicillium Aspergillus Syst.: 170. 1986. [MB202819]. — Herb.: IMI 189694. Ex-type: CBS 225.81 = IMI 189694 = NRRL 2077. Section Aspergilloides. ITS barcode: KM189560. (Alternative markers: BenA = KM088799; RPB2 = KM089571; CaM = KM089184).
Penicillium thymicola Frisvad & Samson, Stud. Mycol. 49: 29. 2004. [MB370969]. — Herb.: CBS 111225. Ex-type: CBS 111225 = IBT 5891. Section Fasciculata. ITS barcode: KJ834518. (Alternative markers: BenA = AY674321; RPB2 = n.a.; CaM = FJ530990).
Penicillium tricolor Frisvad et al., Can. J. Bot. 72: 937. 1994. [MB541710]. — Herb.: DAOM 216240. Ex-type: CBS 635.93 = IBT 12493 = DAOM 216240. Section Fasciculata. ITS barcode: JN942704. (Alternative markers: BenA = AY674313; RPB2 = JN985422; CaM = n.a.).
Penicillium tropicoides Houbraken, Frisvad & Samson, Fungal Divers. 44: 127. 2010. [MB518293]. — Herb.: CBS 122410. Ex-type: CBS 122410 = IBT 29043. Section Citrina. ITS barcode: GU944584. (Alternative markers: BenA = GU944531; RPB2 = JN606608; CaM = GU944624).
Penicillium tropicum Houbraken, Frisvad & Samson, Fungal Divers. 44: 129. 2010 ≡ Eupenicillium tropicum Tuthill & Frisvad, Mycol. Progress 3: 14. 2004. [MB518294]. — Herb.: SC42-1. Ex-type: CBS 112584 = IBT 24580. Section Citrina. ITS barcode: GU944582. (Alternative markers: BenA = GU944532; RPB2 = JN606607; CaM = GU944625).
Penicillium trzebinskii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 498. 1927. [MB280795]. — Herb.: unknown. Ex-type: CBS 382.48 = ATCC 10507 = FRR 731 = IFO 6110 = IMI 039749 = MUCL 29102 = NRRL 731 = QM 7678. Section Aspergilloides. ITS barcode: KM189784. (Alternative markers: BenA = KM089034; RPB2 = KM089808; CaM = KM089421).
Penicillium tsitsikammaense Houbraken, Stud. Mycol. 78: 440. 2014. [MB809976]. — Herb.: CBS H-21881. Ex-type: CBS 328.71 = DTO 006-I3 = CSIR 1092. Section Aspergilloides. ITS barcode: KM189451. (Alternative markers: BenA = KM088675; RPB2 = KM089447; CaM = KM089060).
Penicillium tularense Paden, Mycopathol. Mycol. Appl. 43: 264. 1971 ≡ Eupenicillium tularense Paden, Mycopathol. Mycol. Appl. 43: 264. 1971. [MB319298]. — Herb.: UVIC JWP 68-31. Ex-type: CBS 430.69 = ATCC 22056 = FRR 899 = IFO 31740 = IMI 148394 = NRRL 5273 = AS 3.14006. Section Brevicompacta. ITS barcode: AF033487. (Alternative markers: BenA = KC427175; RPB2 = JN121516; CaM = JX313135).
Penicillium tulipae Overy & Frisvad, Syst. Appl. Microbiol. 26: 634. 2003. [MB488954]. — Herb.: C 60162. Ex-type: CBS 109555 = CBS 187.88 = IBT 3458. Section Fasciculata. ITS barcode: KJ834519. (Alternative markers: BenA = AY674344; RPB2 = n.a.; CaM = n.a.).
Penicillium turbatum Westling, Ark. Bot. 11: 128. 1911. [MB202895]. — Herb.: IMI 39738. Ex-type: CBS 383.48 = CBS 237.60 = ATCC 9782 = DSM2426 = FRR 757 = IFO 7767 = IMI 039738 = MUCL 29115 = NRRL 757 = NRRL 758 = QM 1941. Section Turbata. ITS barcode: AF034454. (Alternative markers: BenA = KJ834499; RPB2 = JN406556; CaM = n.a.).
Penicillium turcosoconidiatum Visagie, Houbraken & K. Jacobs, Stud. Mycol. 78: 440. 2014. [MB809977]. — Herb.: CBS H-21876. Ex-type: CBS 138557 = DTO 181-A3 = CV 110 = DAOM 241130. Section Aspergilloides. ITS barcode: KM189645. (Alternative markers: BenA = KM088889; RPB2 = KM089663; CaM = KM089276).
Penicillium ubiquetum Houbraken, Frisvad & Samson, Stud. Mycol. 70: 127. 2011. [MB563201]. — Herb.: CBS H-20659. Ex-type: CBS 126437 = IBT 22226. Section Citrina. ITS barcode: JN617680. (Alternative markers: BenA = JN606800; RPB2 = n.a.; CaM = JN606460).
Penicillium ulaiense H.M. Hsieh, H.J. Su & Tzean, Trans. Mycol. Soc. Rep. China 2: 161. 1987. [MB126489]. — Herb.: PPEH 29001.87. Ex-type: CBS 210.92 = CBS 261.94 = CCRC 32655 = IBT 18387 = IBT 23037. Section Penicillium. ITS barcode: KC411695. (Alternative markers: BenA = AY674408; RPB2 = n.a.; CaM = n.a.).
Penicillium vagum Houbraken, et al., Stud. Mycol. 78: 443. 2014. [MB809978]. — Herb.: CBS H-21926. Ex-type: CBS 137728 = DTO 180-G3 = CV 25 = DAOM 241357. Section Aspergilloides. ITS barcode: KM189642. (Alternative markers: BenA = KM088886; RPB2 = KM089660; CaM = KM089273).
Penicillium valentinum C. Ramírez and A.T. Martínez, Mycopathologia 72: 183. 1980. [MB113027]. — Herb.: unknown. Ex-type: CBS 172.81 = ATCC 42227 = IJFM 5071. Section Aspergilloides. ITS barcode: KM189550. (Alternative markers: BenA = KM088788; RPB2 = KM089560; CaM = KM089173).
Penicillium vancouverense Houbraken, Frisvad & Samson, Stud. Mycol. 70: 131. 2011. [MB563207]. — Herb.: CBS H-20646. Ex-type: CBS 126323 = IBT 20700. Section Citrina. ITS barcode: JN617675. (Alternative markers: BenA = JN606663; RPB2 = n.a.; CaM = JN606399).
Penicillium vanderhammenii Houbraken et al., Int. J. Syst. Evol. Microbiol. 61: 1473. 2011. [MB518027]. — Herb.: HUA 170337. Ex-type: CBS 126216 = IBT 23203. Section Lanata-Divaricata. ITS barcode: GU981574. (Alternative markers: BenA = GU981647; RPB2 = KF296458; CaM = KF296382).
Penicillium vanluykii Frisvad, Houbraken & Samson, Persoonia 29: 97. 2012. [MB801878]. — Herb.: CBS H-21059. Ex-type: CBS 131539 = DTO 148I2 = IBT 14505. Section Chrysogena. ITS barcode: JX997007. (Alternative markers: BenA = JX996879; RPB2 = JX996615; CaM = JX996220).
Penicillium vanoranjei Visagie, Houbraken & Samson, Persoonia 31: 46. 2013. [MB803782]. — Herb.: CBS H-21145. Ex-type: CBS134406. Section Sclerotiora. ITS barcode: KC695696. (Alternative markers: BenA = KC695686; RPB2 = n.a.; CaM = KC695691).
Penicillium vasconiae C. Ramírez & A.T. Martínez, Mycopathologia 72: 189. 1980. [MB113028]. — Herb.: CBS 339.79. Ex-type: CBS 339.79 = ATCC 42224 = IJFM 3008. Section Lanata-Divaricata. ITS barcode: GU981599. (Alternative markers: BenA = GU981653; RPB2 = KF296459; CaM = KF296386).
Penicillium velutinum J.F.H. Beyma, Zentralbl. Bakteriol. Parasitenk., Abt. 2 91: 353. 1935. [MB283175]. — Herb.: IMI 40571. Ex-type: CBS 250.32 = ATCC 10510 = CECT 2318 = IJFM 5108 = IMI 040571 = NRRL 2069 = QM 7686 = VKMF-379. Section Exilicaulis. ITS barcode: AF033448. (Alternative markers: BenA = JX141170; RPB2 = n.a.; CaM = n.a.).
Penicillium venetum (Frisvad) Frisvad, Adv. Penicillium Aspergillus Syst.: 275. 2000 ≡ Penicillium hirsutum var. venetum Frisvad, Mycologia 81: 856. 1990. [MB459816]. — Herb.: IMI 321520. Ex-type: IBT 10661 = IMI 321520. Section Fasciculata. ITS barcode: AJ005485. (Alternative markers: BenA = AY674335; RPB2 = n.a.; CaM = n.a.).
Penicillium verhagenii Houbraken, Stud. Mycol. 78: 443. 2014. [MB809979]. — Herb.: CBS H-21865. Ex-type: CBS 137959 = DTO 193-A1. Section Aspergilloides. ITS barcode: KM189708. (Alternative markers: BenA = KM088955; RPB2 = KM089729; CaM = KM089342).
Penicillium verrucosum Dierckx, Ann. Soc. Sci. Bruxelles 25: 88. 1901. [MB212252]. — Herb.: IMI 200310. Ex-type: CBS 603.74 = ATCC 48957 = ATHUM2897 = CECT 2906 = FRR 965 = IBT 12809 = IBT 4733 = IMI 200310 = IMI 200310ii = MUCL 28674 = MUCL 29089 = MUCL 29186 = NRRL 965. Section Fasciculata. ITS barcode: AY373938. (Alternative markers: BenA = AY674323; RPB2 = JN121539; CaM = DQ911138).
Penicillium vinaceum J.C. Gilman & E.V. Abbott, Iowa St. Coll. J. Sci. 1: 299. 1927. [MB281754]. — Herb.: IMI 29189. Ex-type: CBS 389.48 = ATCC 10514 = FRR 739 = IMI 029189 = NRRL 739 = QM 6746. Section Exilicaulis. ITS barcode: AF033461. (Alternative markers: BenA = HQ646575; RPB2 = JN406555; CaM = HQ646586).
Penicillium virgatum Nirenberg & Kwasna, Mycol. Res. 109: 977. 2005. [MB341488]. — Herb.: BBA 65745. Ex-type: CBS 114838 = BBA 65745. Section Ramosa. ITS barcode: AJ748692. (Alternative markers: BenA = KJ834500; RPB2 = JN406641; CaM = KJ866992).
Penicillium viridicatum Westling, Ark. Bot. 11: 88. 1911 ≡ Penicillium aurantiogriseum var. viridicatum (Westling) Frisvad & Filt., Mycologia 81: 850. 1990. [MB163349]. — Herb.: IMI 39758ii. Ex-type: CBS 390.48 = ATCC 10515 = IBT 23041 = IFO 7736 = IMI 039758 = IMI 039758ii = NRRL 963 = QM 7683. Section Fasciculata. ITS barcode: AY373939. (Alternative markers: BenA = AY674295; RPB2 = JN121511; CaM = n.a.).
Penicillium viticola Nonaka & Masuma, Mycoscience 52: 339. 2011. [MB516048]. — Herb.: TNS-F38702. Ex-type: JCM 17636 = FKI-4410. Section Sclerotiora. ITS barcode: AB606414. (Alternative markers: BenA = AB540174; RPB2 = n.a.; CaM = n.a.).
Penicillium vulpinum (Cooke & Massee) Seifert & Samson, Adv. Penicillium Aspergillus Syst.: 144. 1985 ≡ Coremium vulpinum Cooke & Massee, Grevillea 16: 81. 1888. [MB114763]. — Herb.: “on dung”, s. coll., in herb. Cooke (K). Ex-type: CBS 126.23 = ATCC 10426 = IMI 040237 = NRRL 2031 = VKMF-257. Section Penicillium. ITS barcode: AF506012. (Alternative markers: BenA = KJ834501; RPB2 = n.a.; CaM = n.a.).
Penicillium waksmanii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat.: 468. 1927. [MB121677]. — Herb.: IMI 39746i. Ex-type: CBS 230.28 = ATCC 10516 = FRR 777 = IFO 7737 = IMI 039746 = IMI 039746i = MUCL 29120 = NRRL 777 = QM 7681. Section Citrina. ITS barcode: GU944602. (Alternative markers: BenA = JN606779; RPB2 = JN606627; CaM = JN606431).
Penicillium wellingtonense A.L.J. Cole et al., Stud. Mycol. 70: 133. 2011. [MB563208]. — Herb.: CBS H-20657. Ex-type: CBS 130375 = IBT 23557 = DTO 76C6. Section Citrina. ITS barcode: JN617713. (Alternative markers: BenA = JN606670; RPB2 = JN606616; CaM = JN606395).
Penicillium westlingii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 473. 1927. [MB282076]. — Herb.: IMI 92272. Ex-type: CBS 231.28 = IMI 092272. Section Citrina. ITS barcode: GU944601. (Alternative markers: BenA = JN606718; RPB2 = JN606625; CaM = JN606500).
Penicillium wotroi Houbraken et al., Int. J. Syst. Evol. Microbiol. 61: 1474. 2011. [MB518026]. — Herb.: HUA 170336. Ex-type: CBS 118171 = IBT 23253. Section Lanata-Divaricata. ITS barcode: GU981591. (Alternative markers: BenA = GU981637; RPB2 = KF296460; CaM = KF296369).
Penicillium yarmokense Baghd., Novosti Sist. Nizsh. Rast. 5: 99. 1968. [MB335774]. — Herb.: CBS H-7536. Ex-type: CBS 410.69 = FRR 520 = IMI 140346 = VKMF-1076. Section Canescentia. ITS barcode: KC411757. (Alternative markers: BenA = KJ834502; RPB2 = JN406553; CaM = KJ867013).
Penicillium yezoense Hanzawa ex Houbraken, Stud. Mycol. 78: 443. 2014. [MB809980]. — Herb.: CBS H-21863. Ex-type: CBS 350.59 = ATCC 18333 = FRR 3395 = IFO 5362 = IMI 068615. Section Aspergilloides. ITS barcode: KM189553. (Alternative markers: BenA = KM088792; RPB2 = KM089564; CaM = KM089177).
Penicillium zonatum Hodges & J.J. Perry, Mycologia 65: 697. 1973 ≡ Eupenicillium zonatum Hodges & J.J. Perry, Mycologia 65: 697. 1973. [MB319303]. — Herb.: BPI FSL 525. Ex-type: CBS 992.72 = ATCC 24353. Section Lanata-Divaricata. ITS barcode: GU981581. (Alternative markers: BenA = GU981651; RPB2 = KF296461; CaM = KF296380).
Penicillium zhuangii L. Wang, PloS ONE 9: e101454-P4. 2014. [MB805945]. — Herb.: HMAS 244922. Ex-type: CBS 137464 = NRRL 62806 = AS 3.15341. Section Aspergilloides. ITS barcode: KF769435. (Alternative markers: BenA = KF769411; RPB2 = n.a.; CaM = KF769422).
Doubtful species
Penicillium asymmetricum (Subram. & Sudha) Houbraken & Samson, Stud. Mycol. 70: 47. 2011 ≡ Thysanophora asymmetrica Subram. & Sudha, Kavaka 12: 88. 1985. [MB561963]. — Herb.: unknown. Ex-type: No culture available. Note: No material is available for this species. As such, we cannot confirm the taxonomic position of the species and consider it as doubtful.
Penicillium coniferophilum Houbraken & Samson, Stud. Mycol. 70: 47. 2011 ≡ Thysanophora striatispora Barron & Cooke, Mycopathol. Mycol. Appl. 40: 353. 1970. [MB561968]. — Herb.: unknown. Ex-type: No culture available. Note: No material is available for this species. As such, we cannot confirm the taxonomic position of the species and consider it as doubtful.
Penicillium glaucoalbidum (Desm.) Houbraken & Samson, Stud. Mycol. 70: 47. 2011 ≡ Sclerotium glaucoalbidum Desm., Ann. Sci. Nat., Bot. 16: 329. 1851 ≡ Thysanophora glaucoalbida (Desm.) M. Morelet, Ann. Soc. Sci. Nat. Archéol. Toulon Var 20: 104. 1968. [MB561965]. — Herb.: unknown. Ex-type: No culture available. Note: No material is available for this species. As such, we cannot confirm the taxonomic position of the species and consider it as doubtful.
Penicillium melanostipe Houbraken & Samson, Stud. Mycol. 70: 47. 2011 ≡ Thysanophora verrucosa Mercado, Gené & Guarro, Mycotaxon 67: 419. 1998. [MB561970]. — Herb.: HAC (M) 9165. Ex-type: No culture available. Note: No material is available for this species. As such, we cannot confirm the taxonomic position of the species and consider it as doubtful.
Penicillium parviverrucosum (K. Ando & Pitt) Houbraken & Samson, Stud. Mycol. 70: 48. 2011 ≡ Torulomyces parviverrucosus K. Ando & Pitt, Mycoscience 39: 317. 1998. — Herb.: TNS-F-238516. Ex-type: KY 12720. Note: Material for this species is not available for study. As such, we cannot confirm the taxonomic position of the species and consider it as doubtful.
Penicillium taiwanense (Matsush.) Houbraken & Samson, Stud. Mycol. 70: 48. 2011 ≡ Phialomyces taiwanensis Matsush., Matsushima Mycol. Mem. 4: 12. 1985 ≡ Thysanophora taiwanensis (Matsush.) Mercado, Gené & Guarro, Mycotaxon 67: 421. 1998. [MB561969]. — Herb.: unknown. Ex-type: No culture available. Section Thysanophora. Note: No material is available for this species. As such, we cannot confirm the taxonomic position of the species and consider it as doubtful.
Excluded species
Penicillium arenicola Chalabuda, Bot. Mater. Otd. Sporov. Rast. 6: 162. 1950. [MB302375]. — Herb.: IMI 117658. Ex-type: CBS 220.66 = ATCC 18321 = ATCC 18330 = DSM 2435 = FRR 3392 = IMI 117658 = NRRL 3392 = VKM F-1035. ITS barcode: GU092964. (Alternative markers: BenA = n.a.; CaM = n.a.; RPB2 = JN121457). Note: Molecular data suggest that this species is related to Phialomyces (Houbraken & Samson 2011).
Penicillium canadense G. Sm., Trans. Br. Mycol. Soc. 39: 113. 1956 = Penicillium arenicola Chalab., Bot. Mater. Otd. Sporov. Rast. 6: 162. 1950. [MB302384]. — Herb.: unknown. Ex-type: CBS 245.56 = ATCC 18424 = FRR 2553 = IMI 061834 = LSHB BB300 = NRRL 2553 = QM 6970 = WB 4155. ITS barcode: GU092963. (Alternative markers: BenA = GU092758; CaM = n.a.; RPB2 = n.a). Note: Molecular data suggest that this species is a synonym of P. arenicola and is related to Phialomyces (Houbraken & Samson 2011).
Penicillium kabunicum Baghdadi, Novosti Sist. Nizs. Rast.: 98. 1968. [MB335738]. — Herb.: CBS 575.90. Ex-type: CBS 575.90 = CBS 409.69 = FRR 513 = IMI 140341 = VKM F-1072. ITS barcode: n.a.. (Alternative markers: BenA = n.a.; CaM = n.a.; RPB2 = n.a.). Note: Molecular data suggest that this species does not belong to Penicillium (Houbraken, unpubl. data).
Penicillium moldavicum Milko & Beliakova, Novosti Sist. Nizs. Rast. 1967: 255. 1967. [MB335751]. — Herb.: unknown. Ex-type: CBS 574.90 = ATCC 18355 = CBS 627.67 = FRR 665 = IMI 129966 = VKM F-922. ITS barcode: n.a.. (Alternative markers: BenA = n.a.; CaM = n.a.; RPB2 = n.a.). Note: Molecular data suggest that this species does not belong to Penicillium (Houbraken, unpubl. data).
Penicillium nodositatum Valla, Pl. Soil 114: 146. 1989. [MB126535]. — Herb.: CBS 330.90. Ex-type: CBS 333.90. ITS barcode: KC790403. (Alternative markers: BenA = KC790399; CaM = n.a.; RPB2 = n.a.). Note: Molecular data suggest that this species is closely related to P. kabunicum and does not belong in Penicillium (Visagie et al. 2013).
Penicillium resedanum McLennan & Ducker, Aust. J. Bot. 2: 360. 1954. [MB302422]. — Herb.: IMI 062877. Ex-type: CBS 181.71 = ATCC 22356 = FRR 578 = IMI 062877 = NRRL 578. ITS barcode: AF033398. (Alternative markers: BenA = n.a.; CaM = n.a.; RPB2 = n.a.). Note: Pitt (1979) noted that this species form acerose phialides with weak growth on G25N, suggesting a relationship with Talaromyces, which is confirmed by ITS data (Houbraken & Samson 2011).
Acknowledgements
This research was supported by grants from the Alfred P. Sloan Foundation Program on the Microbiology of the Built Environment.
Footnotes
Peer review under responsibility of CBS-KNAW Fungal Biodiversity Centre.
Contributor Information
J. Houbraken, Email: j.houbraken@cbs.knaw.nl.
J.C. Frisvad, Email: jcf@bio.dtu.dk.
References
- Abraham E., Chain E., Fletcher C. Further observations on penicillin. The Lancet. 1941;238:177–188. [Google Scholar]
- Adsul M.G., Bastawde K.B., Varma A.J. Strain improvement of Penicillium janthinellum NCIM 1171 for increased cellulase production. Bioresource Technology. 2007;98:1467–1473. doi: 10.1016/j.biortech.2006.02.036. [DOI] [PubMed] [Google Scholar]
- Amiri-Eliasi B., Fenselau C. Characterization of protein biomarkers desorbed by MALDI from whole fungal cells. Analytical Chemistry. 2001;73:5228–5231. doi: 10.1021/ac010651t. [DOI] [PubMed] [Google Scholar]
- Barreto M.C., Houbraken J., Samson R.A. Taxonomic studies of the Penicillium glabrum complex and the description of a new species P. subericola. Fungal Diversity. 2011;49:23–33. [Google Scholar]
- Basaran P., Demirbas R.M. Spectroscopic detection of pharmaceutical compounds from an aflatoxigenic strain of Aspergillus parasiticus. Microbiological Research. 2010;165:516–522. doi: 10.1016/j.micres.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 137.Berbee M., Yoshimura A., Sugiyama J. Is Penicillium Monophyletic? An evaluation of phylogeny in the family Trichocomaceae from 18S, 5.8S and ITS ribosomal DNA sequence data. Mycologia. 1995;87:210–222. [Google Scholar]
- Bille E., Dauphin B., Leto J. MALDI-TOF MS Andromas strategy for the routine identification of bacteria, mycobacteria, yeasts, Aspergillus spp. and positive blood cultures. Clinical Microbiology and Infection. 2011;18:1117–1125. doi: 10.1111/j.1469-0691.2011.03688.x. [DOI] [PubMed] [Google Scholar]
- Blakeslee A. Lindner's roll tube method of separation cultures. Phytopathology. 1915;5:68–69. [Google Scholar]
- Blaxter M. Counting angels with DNA. Nature. 2003;421:1–3. doi: 10.1038/421122a. [DOI] [PubMed] [Google Scholar]
- Blaxter M., Mann J., Chapman T. Defining operational taxonomic units with DNA barcodes data. Philosophical Transactions of the Royal Society B. 2005;360:1935–1943. doi: 10.1098/rstb.2005.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boysen M., Skouboe P., Frisvad J.C., Rossen L. Reclassification of the Penicillium roqueforti group into three species on the basis of molecular genetic and biochemical profiles. Microbiology. 1996;142:541–549. doi: 10.1099/13500872-142-3-541. [DOI] [PubMed] [Google Scholar]
- Chain E., Florey H., Gardner A. Penicillin as a chemotherapeutic agent. The Lancet. 1940;236:226–228. [Google Scholar]
- Chalupová J., Raus M., Sedlářová M. Identification of fungal microorganisms by MALDI-TOF mass spectrometry. Biotechnology Advances. 2014;32:230–241. doi: 10.1016/j.biotechadv.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Chen A.J., Tang D., Zhou Y.Q. Identification of Ochratoxin A producing fungi associated with fresh and dry liquorice. PLoS One. 2013;8:e78285. doi: 10.1371/journal.pone.0078285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M., Frisvad J.C., Tuthill D.E. Penicillium species diversity in soil and some taxonomic and ecological notes. In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 309–321. [Google Scholar]
- Ciegler A., Pitt J.I. Survey of the genus Penicillium for tremorgenic toxin production. Mycopathologia et Mycologia applicata. 1970;42:119–124. doi: 10.1007/BF02051832. [DOI] [PubMed] [Google Scholar]
- Clutterbuck P.W., Raistrick H. Studies in the biochemistry of microorganisms XXXI. The molecular constitution of the metabolic products of Penicillium brevicompactum Dierckx and related species. II. Mycophenolic acid. Biochemical Journal. 1933;27:654–667. doi: 10.1042/bj0270654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank R.H., Pitt J.I. Identification of species in Penicillium subgenus Penicillium by enzyme electrophoresis. Mycologia. 1987:614–620. [Google Scholar]
- Cruickshank R.H., Pitt J.I. The zymogram technique – isoenzyme patterns as an aid in Penicillium classification. Microbiological Sciences. 1987;4:14–17. [PubMed] [Google Scholar]
- De Carolis E., Posteraro B., Lass-Flörl C. Species identification of Aspergillus, Fusarium and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clinical Microbiology and Infection. 2011;18:475–484. doi: 10.1111/j.1469-0691.2011.03599.x. [DOI] [PubMed] [Google Scholar]
- Del Chierico F., Masotti A., Onori M. MALDI-TOF MS proteomic phenotyping of filamentous and other fungi from clinical origin. Journal of Proteomics. 2012;75:3314–3330. doi: 10.1016/j.jprot.2012.03.048. [DOI] [PubMed] [Google Scholar]
- DeSalle R., Egan M.G., Siddal M. The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philosophical Transactions of the Royal Society B. 2005;360:1905–1916. doi: 10.1098/rstb.2005.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Banna A.A., Pitt J.I., Leistner L. Production of mycotoxins by Penicillium species. Systematic and Applied Microbiology. 1987;10:42–46. [Google Scholar]
- Fennell D.I., Raper K.B. New species and varieties of Aspergillus. Mycologia. 1955;47:68–89. [Google Scholar]
- Fleming A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzae. British Journal of Experimental Pathology. 1929;10:226–236. [PubMed] [Google Scholar]
- Frisvad J.C. Physiological criteria and mycotoxin production as aids in identification of common asymmetric Penicillia. Applied and Environmental Microbiology. 1981;41:568–579. doi: 10.1128/aem.41.3.568-579.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Andersen B., Thrane U. The use of secondary metabolite profiling in fungal taxonomy. Mycological Research. 2008;112:231–240. doi: 10.1016/j.mycres.2007.08.018. [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Filtenborg O. Classification of terverticillate Penicillia based on profiles of mycotoxins and other secondary metabolites. Applied and Environmental Microbiology. 1983;46:1301–1310. doi: 10.1128/aem.46.6.1301-1310.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Filtenborg O. Terverticillate Penicillia: chemotaxonomy and mycotoxin production. Mycologia. 1989;81:837–861. [Google Scholar]
- Frisvad J.C., Samson R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology. 2004;49:1–174. [Google Scholar]
- Frisvad J.C., Smedsgaard J., Larsen T.O. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Studies in Mycology. 2004;49:201–241. [Google Scholar]
- Frisvad J.C., Thrane U. Standardized High Performance Liquid Chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone indices and UV VIS spectra (diode array detection) Journal of Chromatography. 1987;404:195–214. doi: 10.1016/s0021-9673(01)86850-3. [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Thrane U. Liquid column chromatography of mycotoxins. In: Betina V., editor. Chromatography of mycotoxins: techniques and applications. Vol. 54. Elsevier; Amsterdam: 1993. pp. 253–372. (Journal of Chromatography Library). [Google Scholar]
- Geiser D.M., Pitt J.I., Taylor J.W. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud F., Giraud T., Aguileta G. Microsatellite loci to recognize species for the cheese starter and contaminating strains associated with cheese manufacturing. International Journal of Food Microbiology. 2010;137:204–213. doi: 10.1016/j.ijfoodmicro.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Glass N.L., Donaldson G.C. Development of premier sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N., Ratnasingham S., deWaard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society London B (Suppl.) 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettick J.M., Kashon M.L., Slaven J.E. Discrimination of intact mycobacteria at the strain level: A combined MALDI-TOF MS and biostatistical analysis. Proteomics. 2006;6:6416–6425. doi: 10.1002/pmic.200600335. [DOI] [PubMed] [Google Scholar]
- Hettick J.M., Green B.J., Buskirk A.D. Discrimination of Penicillium isolates by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry fingerprinting. Rapid Communications in Mass Spectrometry. 2008;22:2555–2560. doi: 10.1002/rcm.3649. [DOI] [PubMed] [Google Scholar]
- Hirano A., Iwai Y., Masuma R. Neoxaline, a new alkaloid produced by Aspergillus japonicus. Production, isolation and properties. The Journal of Antibiotics. 1979;32:781–785. doi: 10.7164/antibiotics.32.781. [DOI] [PubMed] [Google Scholar]
- Hocking A.D., Pitt J.I. Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Applied and Environmental Microbiology. 1980;39:488–492. doi: 10.1128/aem.39.3.488-492.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoog G.S. de, Gerrits van den Ende A.H. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Hong S.-B., Cho H.-S., Shin H.-D. Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology. 2006;56:477–486. doi: 10.1099/ijs.0.63980-0. [DOI] [PubMed] [Google Scholar]
- Houbraken J., Frisvad J.C., Samson R.A. Fleming's penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus. 2011;2:87–95. doi: 10.5598/imafungus.2011.02.01.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Frisvad J.C., Samson R.A. Taxonomy of Penicillium section Citrina. Studies in Mycology. 2011;70:53–138. doi: 10.3114/sim.2011.70.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Frisvad J.C., Seifert K.A. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia. 2012;29:78–100. doi: 10.3767/003158512X660571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., López-Quintero C.A., Frisvad J.C. Penicillium araracuarense sp. nov., Penicillium elleniae sp. nov., Penicillium penarojense sp. nov., Penicillium vanderhammenii sp. nov. and Penicillium wotroi sp. nov., isolated from leaf litter. International Journal of Systematic and Evolutionary Microbiology. 2011;61:1462–1475. doi: 10.1099/ijs.0.025098-0. [DOI] [PubMed] [Google Scholar]
- Houbraken J., Samson R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Spierenburg H., Frisvad J.C. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie van Leeuwenhoek. 2012;101:403–421. doi: 10.1007/s10482-011-9647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B.H., Raistrick H. Studies in the biochemistry of micro-organisms. 81. The colouring matters of Penicillium islandicum Sopp. Part 2. Chrysophanic acid, 4:5-dihydroxy-2-methylanthraquinone. Biochemical Journal. 1950;46:49–53. doi: 10.1042/bj0460049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V., Kolarik M. β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: primer specificity testing and taxonomic consequences. Persoonia. 2012;29:1–10. doi: 10.3767/003158512X658123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson C.R., Li S.-W., McInnes A.G. Comparative biochemistry of fatty acid and macrolide antibiotic (brefeldin-A) formation in Penicillium brefeldianum. Tetrahedron. 1983;39:3507–3513. [Google Scholar]
- Iriart X., Lavergne R.A., Fillaux J. Routine identification of medical fungi by the new Vitek MS Matrix-Assisted Laser Desorption Ionization-Time of Flight System with a new time-effective strategy. Journal of Clinical Microbiology. 2012;50:2107–2110. doi: 10.1128/JCM.06713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahadian C., Josephson D.B., Lindsay R.C. Volatile compounds from Penicillium sp. contributing musty-earthy notes to Brie and Camembert cheese flavors. Journal of Agricultural and Food Chemistry. 1985;33:339–343. [Google Scholar]
- Kawada M., Momose I., Someno T. New atpenins, NBRI23477 A and B, inhibit the growth of human prostrate cancer cells. Journal of Antibiotics. 2009;62:243–246. doi: 10.1038/ja.2009.20. [DOI] [PubMed] [Google Scholar]
- Kildgaard S., Mansson M., Dosen I. Accurate dereplication of bioactive secondary metabolites from marine-derived fungi by UHPLC-DAD-QTOFMS and MS/HRMS library. Marine Drugs. 2014;12:3681–3705. doi: 10.3390/md12063681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitgaard A., Iversen A., Andersen M.R. Aggressive dereplication using UHPLC-DAD-QTOF – screening extracts for up to 3000 fungal secondary metabolites. Analytical and Bioanalytical Chemistry. 2014;406:1933–1943. doi: 10.1007/s00216-013-7582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolecka A., Khayhan K., Groenewald M. Identification of medically relevant species of arthroconidial yeasts by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. Journal of Clinical Microbiology. 2013;51:2491–2500. doi: 10.1128/JCM.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A., Wanscher J.H. Methuen & Co Ltd; Copenhagen: 1967. Methuen handbook of colour. [Google Scholar]
- Kõljalg U., Larsson K.-H., Abarenkov K. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytologist. 2005;166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- Kõljalg U., Nilsson R.H., Abarenkov K. Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology. 2013;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Li Y., Cui F., Liu Z. Improvement of xylanase production by Penicillium oxalicum ZH-30 using response surface methodology. Enzyme and Microbial Technology. 2007;40:1381–1388. [Google Scholar]
- Link H.F. Observationes in Ordines plantarum naturales. Dissertatio 1ma. Magazin der Gesellschaft Naturforschenden Freunde Berlin. 1809;3:3–42. [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Liu W., Gu Q., Zhu W. Penicillones A and B, two novel polyketides with tricyclo [5.3.1.03,8] undecane skeleton, from amarine-derived fungus Penicillium terrestre. Tehtrahedron Letters. 2005;46:4993–4996. [Google Scholar]
- López-Díaz T.-M., Santos J.-A., García-López M.-L. Surface mycoflora of a Spanish fermented meat sausage and toxigenicity of Penicillium isolates. International Journal of Food Microbiology. 2001;68:69–74. doi: 10.1016/s0168-1605(01)00472-x. [DOI] [PubMed] [Google Scholar]
- LoBuglio K.F., Pitt J.I., Taylor J.W. Phylogenetic analysis of two ribosomal DNA regions indicates multiple independent losses of a sexual Talaromyces state among asexual Penicillium species in subgenus Biverticillium. Mycologica. 1993;85:592–604. [Google Scholar]
- Ludemann V., Greco M., Rodríguez M.P. Conidial production by Penicillium nalgiovense for use as starter cultures in dry fermented sausages by solid state fermentation. LWT – Food Science and Technology. 2010;43:315–318. [Google Scholar]
- Lund F. Differentiating Penicillium species by detection of indole metabolites using a filter paper method. Letters in Applied Microbiology. 1995;20:228–231. [Google Scholar]
- Masclaux F., Guého E., Hoog G.S. de. Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. Medical Mycology. 1995;33:327–338. doi: 10.1080/02681219580000651. [DOI] [PubMed] [Google Scholar]
- McNeill J., Barrie F.F., Buck W.R., editors. International code of nomenclature for algae, fungi, and plants (Melbourne Code) Koeltz Scientific Books; Königstein: 2012. [Regnum vegetabile no. 154.] [Google Scholar]
- Min X.J., Hickey D.A. Assessing the effect of varying sequence length on DNA barcoding of fungi. Molecular Notes. 2007;7:365–373. doi: 10.1111/j.1471-8286.2007.01698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min X.J., Hickey D.A. DNA barcodes provide a quick preview of mitochondrial genome composition. PLoS One. 2007;3:e325. doi: 10.1371/journal.pone.0000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.H. Production of blue cheese flavor via submerged fermentation by Penicillium roqueforti. Journal of Agricultural and Food Chemistry. 1970;18:567–569. [Google Scholar]
- O'Donnell K., Cigelnik E., Nirenberg H.I. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 1998;90:465. [Google Scholar]
- Okuda T. Variation in colony characteristics of Penicillium strains resulting from minor variations in culture conditions. Mycologia. 1994;86:259–262. [Google Scholar]
- Okuda T., Klich M.A., Seifert K.A. Media and incubation effect on morphological characteristics of Penicillium and Aspergillus. In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 83–99. [Google Scholar]
- Omura S., Tomoda H., Kimura K. Atpenins, new antifungal antibiotics produced by Penicillium sp. — production, isolation, physicochemical and biological properties. Journal of Antibiotics. 1988;41:1769–1774. doi: 10.7164/antibiotics.41.1769. [DOI] [PubMed] [Google Scholar]
- Paterson R.R.M., Bridge P.D., Crosswaite M.J. A reappraisal of the terverticillate Penicillia using biochemical, physiological and morphological features iii. An evaluation of pectinase and amylase isoenzymes for species characterization. Journal of General Microbiology Journal of General Microbiology. 1989;135:2979–2991. doi: 10.1099/00221287-135-11-2979. [DOI] [PubMed] [Google Scholar]
- Peterson S.W. Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences. In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 163–178. [Google Scholar]
- Peterson S.W. Phylogenetic relationships in Aspergillus based on rDNA sequence analysis. In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 323–355. [Google Scholar]
- Peterson S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.3852/mycologia.100.2.205. [DOI] [PubMed] [Google Scholar]
- Peterson S.W., Jurjević Z. Talaromyces columbinus sp. nov., and genealogical concordance analysis in Talaromyces Clade 2a. PLoS One. 2013;8:e78084. doi: 10.1371/journal.pone.0078084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.W., Orchard S.S., Menon S. Penicillium menonorum, a new species related to P. pimiteouiense. IMA Fungus. 2011;2:121–125. doi: 10.5598/imafungus.2011.02.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.W., Vega F., Posada F. Penicillium coffeae, a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia. 2005;97:659–666. doi: 10.3852/mycologia.97.3.659. [DOI] [PubMed] [Google Scholar]
- Pitt J.I. An appraisal of identification methods for Penicillium species Novel taxonomic criteria based on temperature and water relations. Mycologia. 1973;65:1135–1157. [PubMed] [Google Scholar]
- Pitt J.I. Academic Press Inc; London: 1979. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. [Google Scholar]
- Pitt J.I., Hocking A.D. Springer; New York: 2009. Fungi and Food Spoilage. [Google Scholar]
- Pitt J.I., Samson R.A. Names in current use in the family Trichocomaceae. In: Greuter W., editor. Names in current use in the family Trichocomaceae, Cladoniaceae, Pinaceae, and Lemnaceae. Koeltz Scientific Books; Konigstein: 1993. pp. 13–57. [Google Scholar]
- Pitt J.I., Samson R.A., Frisvad J.C. List of accepted species and their synonyms in the family Trichocomaceae. In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 9–79. [Google Scholar]
- Rai J.N., Tewari J.P., Mukerji K.G. A new Aspergillus from Indian soils: A. striatus spec. nov. Canadian Journal of Botany. 1964;42:1521–1524. [Google Scholar]
- Rai J.N., Tewari J.P., Sinha A.K. Effect of environmental conditions on sclerotia and cleistothecia production in Aspergillus. Mycopathologia et mycologia applicata. 1967;31:209–224. doi: 10.1007/BF02053418. [DOI] [PubMed] [Google Scholar]
- Ramírez C. Elsevier Biomedical Press; Amsterdam: 1982. Manual and atlas of the Penicillia. [Google Scholar]
- Raper K.B., Fennell D.I. The Williams & Wilkins Company; Baltimore: 1965. The genus Aspergillus. [Google Scholar]
- Raper K.B., Thom C. The Williams & Wilkins Company; Baltimore: 1949. A manual of the penicillia. [Google Scholar]
- Ratnasingham S., Hebert P.D.N. Bold: The Barcode of Life Data System. Molecular Ecology Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgeway R. 1912. Color standards and color nomenclature. Washington DC. [Google Scholar]
- Rivera K.G., Díaz J., Chavarría-Díaz F. Penicillium mallochii and P. guanacastense, two new species isolated from Costa Rican caterpillars. Mycotaxon. 2012;119:315–328. [Google Scholar]
- Rivera K.G., Seifert K.A. A taxonomic and phylogenetic revision of the Penicillium sclerotiorum complex. Studies in Mycology. 2011;70:139–158. doi: 10.3114/sim.2011.70.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Houbraken J., Thrane U. CBS KNAW Biodiversity Center; Utrecht: 2010. Food and indoor fungi. [Google Scholar]
- Samson R.A., Pitt J.I. General recommendations. In: Samson R.A., Pitt J.I., editors. Advances in Penicillium and Aspergillus systematics. Plenum Press; London: 1985. pp. 455–460. [Google Scholar]
- Samson R.A., Seifert K.A., Kuijpers A. Phylogenetic analysis of Penicillium subgenus Penicillium using partial B-tubulin sequences. Studies in Mycology. 2004;49:175–200. [Google Scholar]
- Samson R.A., Visagie C.M., Houbraken J. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Yilmaz N., Houbraken J. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Studies in Mycology. 2011;70:159–183. doi: 10.3114/sim.2011.70.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria M., Fosso B., Consiglio A. Reference databases for taxonomic assignment in metagenomics. Briefings in Bioinformatics. 2012;13:682–695. doi: 10.1093/bib/bbs036. [DOI] [PubMed] [Google Scholar]
- Sarbhoy A.K., Elphick J.J. Hemicarpenteles paradoxus gen. & sp.nov.: the perfect state of Aspergillus paradoxus. Transactions of the British Mycological Society. 1968;51:155–157. [Google Scholar]
- Schoch C.L., Robbertse B., Robert V. Finding needles in haystacks: linking scientific names, reference specimens and molecular data for fungi. Database. 2014;2014 doi: 10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C.L., Seifert K.A., Huhndorf S. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K.A., Louis-Seize G. Phylogeny and species concepts in the Penicillium aurantiogriseum complex as inferred from partial β-tubulin gene DNA sequences. In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 189–198. [Google Scholar]
- Seifert K.A., Samson R.A., deWaard J.R. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3901–3906. doi: 10.1073/pnas.0611691104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra R., Peterson S.W., CTCOR Multilocus sequence identification of Penicillium species in cork bark during plank preparation for the manufacture of stoppers. Research in Microbiology. 2008;159:178–186. doi: 10.1016/j.resmic.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Shang Z., Li X., Meng L. Chemical profile of the secondary metabolites produced by a deep-sea sediment derived fungus Penicillium commune. Chinese Journal of Oceanology and Limnology. 2012;30:305–314. [Google Scholar]
- Siegrist T.J., Anderson P.D., Huen W.H. Discrimination and characterization of environmental strains of Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) Journal of Microbiological Methods. 2007;68:554–562. doi: 10.1016/j.mimet.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Skouboe P., Frisvad J.C., Taylor J.W. Phylogenetic analysis of nucleotide sequences from the ITS region of terverticillate Penicillium species. Mycological Research. 1999;103:873–881. [Google Scholar]
- Smedsgaard J. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. Journal of Chromatography A. 1997;760:264–270. doi: 10.1016/s0021-9673(96)00803-5. [DOI] [PubMed] [Google Scholar]
- Stolk A.C., Samson R.A. The Ascomycete genus Eupenicillium and related Penicillium anamorphs. Studies in Mycology. 1983;23:1–149. [Google Scholar]
- Tamura M., Kawahara K., Sugiyama J. Molecular phylogeny of Aspergillus and associated teleomorphs in the Trichocomaceae (Eurotiales) In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 357–372. [Google Scholar]
- Tautz D., Arctander P., Minelli A. A plea for DNA taxonomy. Trends in Ecology and Evolution. 2003;18:70–74. [Google Scholar]
- Taylor J.W., Jacobson D.J., Kroken S. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- Terrasan C.R.F., Temer B., Duarte M.C.T. Production of xylanolytic enzymes by Penicillium janczewskii. Bioresource Technology. 2010;101:4139–4143. doi: 10.1016/j.biortech.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Thom C. Fungi in cheese ripening: Camembert and Roquefort. U.S. Department of Agriculture, Bureau of Animal Industry – Bulletin. 1906;82:1–39. [Google Scholar]
- Thom C. The Williams & Wilkins Company; Baltimore: 1930. The Penicillia. [Google Scholar]
- Thom C. Mycology present penicillin. Mycologia. 1945;37:460–475. [Google Scholar]
- Thom C. The evolution of species concepts in Aspergillus and Penicillium. Annals of the New York Academy of Sciences. 1954;60:24–34. doi: 10.1111/j.1749-6632.1954.tb39995.x. [DOI] [PubMed] [Google Scholar]
- Varga J., Due M., Frisvad J.C. Taxonomic revision of Aspergillus section Clavati based on molecular, morphological and physiological data. Studies in Mycology. 2007;59:89–106. doi: 10.3114/sim.2007.59.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Frisvad J.C., Samson R.A. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Studies in Mycology. 2011;69:57–80. doi: 10.3114/sim.2011.69.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwer P.E.B., Leeuwen W.B., Girard V. Discrimination of Aspergillus lentulus from Aspergillus fumigatus by Raman spectroscopy and MALDI-TOF MS. European Journal of Clinical Microbiology & Infectious Diseases. 2013;33:245–251. doi: 10.1007/s10096-013-1951-4. [DOI] [PubMed] [Google Scholar]
- Visagie C.M., Hirooka Y., Tanney J.B. Aspergillus, Penicillium and Talaromyces isolated from in house dust samples collected around the world. Studies in Mycology. 2014;78:63–139. doi: 10.1016/j.simyco.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.M., Houbraken J., Rodriques C. Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia. 2013;31:42–62. doi: 10.3767/003158513X667410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.M., Seifert K.A., Houbraken J. Diversity of Penicillium section Citrina within the fynbos biome of South Africa, including a new species from a Protea repens infructescence. Mycologia. 2014;106:537–552. doi: 10.3852/13-256. [DOI] [PubMed] [Google Scholar]
- Wang B., Wang L. Penicillium kongii, a new terverticillate species isolated from plant leaves in China. Mycologia. 2013;105:1547–1554. doi: 10.3852/13-022. [DOI] [PubMed] [Google Scholar]
- Wang J., Huang Y., Fang M. Brefeldin A, a cytotoxin produced by Paecilomyces sp. and Aspergillus clavatus isolated from Taxus mairei and Torreya grandis. FEMS Immunology & Medical Microbiology. 2002;34:51–57. doi: 10.1111/j.1574-695X.2002.tb00602.x. [DOI] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Shinsky T.J., White T.J., editors. PCR protocols: a guide to methods and applications. Academic Press Inc; New York: 1990. pp. 315–322. [Google Scholar]
- Welham K.J., Domin M.A., Johnson K. Characterization of fungal spores by laser desorption/ionization time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2000;14:307–310. doi: 10.1002/(SICI)1097-0231(20000315)14:5<307::AID-RCM823>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Yaguchi T., Someya A., Miyadoh S. Aspergillus ingratus, a new species in Aspergillus section Clavati. Transactions of the Mycological Society of Japan. 1993;34:305–310. [Google Scholar]
- Yilmaz N., Visagie C.M., Houbraken J. Polyphasic taxonomy of the genus Talaromyces. Studies in Mycology. 2014;78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chen H., Shang Z. SD-118 – Xanthocillin X (1). A novel marine agent extracted from Penicillium commune, induces autophagy through the inhibition of the MEK/ERK pathway. Marine Drugs. 2012;10:1345–1359. doi: 10.3390/md10061345. [DOI] [PMC free article] [PubMed] [Google Scholar]


