Abstract
B-RafV600E mutant is found in 40–70% of papillary thyroid carcinoma (PTC) and has an important role in the pathogenesis of PTC. The sodium iodide symporter (NIS) is an integral plasma membrane glycoprotein that mediates active iodide transport into the thyroid follicular cells, and B-RafV600E has been known to be associated with the loss of NIS expression. In this study, we found that B-RafV600E inhibited NIS expression by the upregulation of its promoter methylation, and that specific regions of CpG islands of NIS promoter in B-RafV600E harboring PTC were highly methylated compared with surrounding normal tissue. Although DNA methyltransferase 3a and 3b (DNMT3a,3b) were not increased by B-RafV600E, DNMT1 expression was markedly upregulated in PTC and B-RafV600E expressing thyrocytes. Furthermore, DNMT1 expression was upregulated by B-RafV600E induced NF-κB activation. These results led us to conclude that NIS promoter methylation, which was induced by B-RafV600E, is one of the possible mechanisms involved in NIS downregulation in PTC.
Introduction
Thyroid cancer, a common endocrine malignancy, has rapidly increased worldwide in recent decades.1 Papillary thyroid carcinoma (PTC) is the most prevalent cancer derived from follicular cells.2 Although 10-year survival rate of PTC following thyroidectomy and radioiodine ablation exceeds 90%, 5-year survival rate decreases to ⩽50% when repeated recurrence and/or distant metastases occur owing to the failure of radioactive iodine therapy.3,4 Decreased expression of thyroid iodide-metabolizing genes such as thyroperoxidase, thyroid stimulating hormone (TSH) receptor, sodium iodide symporter (NIS), thyroid transcription factor 1 through dedifferentiation process has been suggested as the possible mechanism of iodine radiotherapy resistance.5, 6, 7, 8, 9
B-RafV600E, which causes constitutive Erk1/2 activation, is the most prevalent genomic alteration in PTC. Along with Ras oncogene mutations and RET gene rearrangements, B-RafV600E can activate MAP kinase pathway that has an essential role in mediating cellular differentiation, proliferation, senescence and survival. B-RafV600E is correlated with aggressive clinicopathologic characteristics10,11 as well as radioiodine treatment failure in PTC.12,13 Indeed, the prevalence of B-RafV600E mutation in recurrent radioiodine-refractory PTC is higher than that in primary PTC,14 and B-RafV600E has been shown to be associated with the loss of thyroid iodide-metabolizing genes.15, 16, 17 Furthermore, the suppression of B-RafV600E was found to restore the expression of these genes in thyroid cells in vitro18 and transgenic mouse model.19 Therefore, the understanding of molecular mechanism of how B-RafV600E regulates thyroid iodide-metabolizing gene expression is important for improving the overall survival of radioiodine-refractory PTC by overcoming radioiodine therapy resistance.
NIS is one of the thyroid iodide-metabolizing genes regulated by B-RafV600E. Although several regulation mechanisms of NIS expression in normal thyroid have been evaluated, the regulation by B-RafV600E during carcinogenesis has not yet been fully studied. As CpG islands methylation at promoter regions is a major epigenetic alteration to silence several genes in various cancers20 and the presence of B-RafV600E has been strongly associated with the ‘CpG island methylator phenotype' in colorectal cancer,21 we thought that NIS expression can be regulated epigenetically by B-RafV600E in PTC.
In this study, we observed that the expression of NIS was negatively correlated with B-RafV600E expression, whereas CpG island methylation of NIS promoter regions was positively correlated with B-RafV600E in human PTC tissues. By using B-RafV600E expression model system in normal thyroid epithelium, we therefore attempted to elucidate molecular mechanism of B-RafV600E induced CpG island methylation of NIS promoter.
Materials and Methods
Tumor samples from patients
Fresh tissues from PTC and surrounding normal thyroid were obtained from patients at the Ajou University Hospital after surgical resection with informed consent. Fresh tumor and normal tissues were separately sampled in the representative areas by an experienced pathologist and snap frozen in liquid nitrogen immediately after resection, according to the specimen regulation of the Ajou University Hospital. Patients who had a past history of chemotherapy or radiation therapy before the surgery were excluded from the study.
Cloning of B-RafV600E and lentivirus preparation
B-RafV600E mutant was cloned from PTC tissue in our laboratory. After insertion of cDNA into TOPO cloning vector (Invitrogen, Carlsbad, CA, USA) and subsequently sequenced. cDNAs were inserted to pCDH-CMV-MCS-EF1-Puro lentivirus vector (System Biosciences, Mountain View, CA, USA). To generate lentiviral particles, HEK293TN cells were transfected with plasmid DNA (pGag-pol, pVSV-G and pCDH-B-RafV600E).
Thyrocytes isolation and culture
Normal thyrocytes were isolated from human tissues. Briefly, thyroid tissue was cut into small pieces and then enzymatic digestion was carried out by adding 0.2% collagenase I (Worthington Biochemical, Lakewood, NJ, USA) in HEPES buffer (30 mM HEPES, 130 mM NaCl, 4 mM glucose), with gentle shaking for 6 h at 32 °C. The cells were centrifuged at 1500 r.p.m. for 5 min and washed with phosphate-buffered saline (PBS) three times. The cell pellet was resuspended in completed media (F12K; Gibco BRL, Bethesda, MD, USA) (TSH 10 mU ml−1, insulin; 0.01 mU ml−1, hydrocortisone; 10 nM, transferrin; 0.005 mg ml−1, somatostatin; 10 ng ml−1, glycyl-L-histidyl-L-lysine acetate; 10 ng ml−1, Sigma, St Louis, MO, USA) and distributed into 10-cm tissue culture dishes. We performed immunocytochemistry to detect two kinds of thyrocyte specific proteins including thyroglobulin and thyroid transcription factor 1 to confirm the identity of thyrocytes (data not shown).
Real-time PCR
Total cellular RNAs were isolated from thyrocytes. First strand cDNA was synthesized by reverse transcription reaction using oligo-dT primers from 1 μg of total cellular RNA in 10 μl reaction volume. The primers used for real-time PCR were as follows: 5′-acactgcatgcgaccctctcct-3′ and 5′-tgctgagggtgccactgtaa-3′, NIS; 5′-gaggaagctaaggactagttc-3′ and 5′-actccacaatttgatcactaaatc-3′, DNMT (DNA methyltransferase)-1; 5′-gccgaattgtgtcttggtggatgaca-3′ and 5′-cctggtggaatgcactgcagaagga-3′, DNMT3a; 5′-tacacagacgtgtccaacatgggc-3′ and 5′-ggatgccttcaggaatcacacctc-3′, DNMT3b; 5′-gcctccagaccattaacctcagt-3′ and 5′-gctccatgatcacctggggcat-3′, SP1; 5′-tttgcttggctctttctacacctc-3′ and 5′-gaatgtctgttttaagctccctgg-3′, PAX8; 5′-ccctggcacccagcac-3′, 5′-gccgatccacacggagtac-3′, β-actin, respectively.
Immunohistochemistry and immunocytochemistry
Immunohistochemical stainings were performed with primary antibodies on 4-μm-thickness representative tissue sections of formalin-fixed, paraffin-embedded tissue section in the Benchmark XT automated immunohistochemistry stainer (Ventana Medical Systems Inc., Tucson, AZ, USA). The primary antibodies used were as follows: B-RafV600E, 1:50 (CloneVE1; Spring Bioscience, Pleasanton, CA, USA); DNMT1 (Abcam, Cambridge, MA, USA), DNMT3a (Abcam), DNMT3b (Abcam) and NIS (Abcam). Detection was done using the Ventana Optiview DAB Kit (Ventana Medical Systems).
Thyrocytes were cultured on a cover glass, fixed with 4% paraformaldehyde in PBS at 4 °C for 20 min, and then washed three times with PBS containing 0.1% Tween-20. Cells were incubated with 1.5% horse serum in PBST for 1 h at room temperature, and anti-NF-κB p65 (Cell Signaling, Danvers, MA, USA) was then applied overnight at 4 °C. The secondary antibody (1:500) was applied for 1 h, and the cells were observed under a fluorescence microscope (Axio Imager M1, Carl Zeiss, Oberkochen, Germany).
Western blotting
The cell lysates were resolved on 8–13% SDS-polyacrylamide gel electrophoresis. The gel-separated proteins were transferred to nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany), and then incubated with antibodies raised against total Erk1/2 (Cell Signaling), p-Erk1/2 (Cell Signaling), actin (Santa Cruz, Santa Cruz, CA, USA), B-Raf (Cell Signaling), tubulin (Santa Cruz), DNMT1 (Abcam), DNMT3a (Abcam), DNMT3b (Abcam), IκBα (Santa Cruz) and NIS (Abcam) for 1 h at room temperature or overnight at 4 °C.
DNMT activity assay
Nucleus was isolated from control or B-RafV600E lentivirus infected thyrocytes, surrounding normal tissue and PTC, respectively. The enzyme activity was then assayed by using DNA methyltransferase activity assay kit (Epigentek, Farmingdale, NY, USA) according to the manufacturer's instruction. Absorbance was read at 450 nm on a spectrophotometer.
Measurement of iodide uptake
Iodide uptake was measured using Premo Halide Sensor (Invitrogen) as specified by the manufacturer. Briefly, thyrocytes were coinfected with baculovirus encoding halide-sensitive YFP and control or B-RafV600E lentivirus, and then mixed with an equal volume of 2 × Premo halide stimulus buffer containing 150 mM NaI, and the fluorescence intensities were monitored every 2 s for 60 s at an excitation wavelength of 480 nm and an emission wavelength of 560 nm to record YFP quenching, evoked by inward currents of iodide ion.
Sequencing analysis of bisulfate-treated DNA and methylation-specific PCR
NIS promoter region in genomic DNA from surrounding normal and PTC regions was sequenced after bisulfite treatment. Genomic DNA (1 μg) from surrounding normal region and PTC was treated with bisulfite, and then PCR was carried out with 5′-gatagtttatagatttttaatttagggagt-3′ and 5′-aaaacataaaaacaaaaacccc-3′ primers. Sequence was analyzed with specific nucleotide peak at CpG islands region. Same bisulfite-treated genomic DNA was used for methylation-specific PCR analysis. The primers used were 5′-gagttgttttcgtaagttttaaggc-3′ and 5′-tatccccgctatctatctctacgt-3′ for methylation PCR and 5′-gttgtttttgtaagttttaaggtga-3′ and 5′-tatccccactatctatctctacatc-3′ for unmethylation PCR.
Statistical analysis
Numerical data are presented as mean±s.d. of independent determinations. Statistical analysis of differences was performed by Student's t-test, and a P-value <0.05 was considered as significant.
Results
NIS expression in PTC
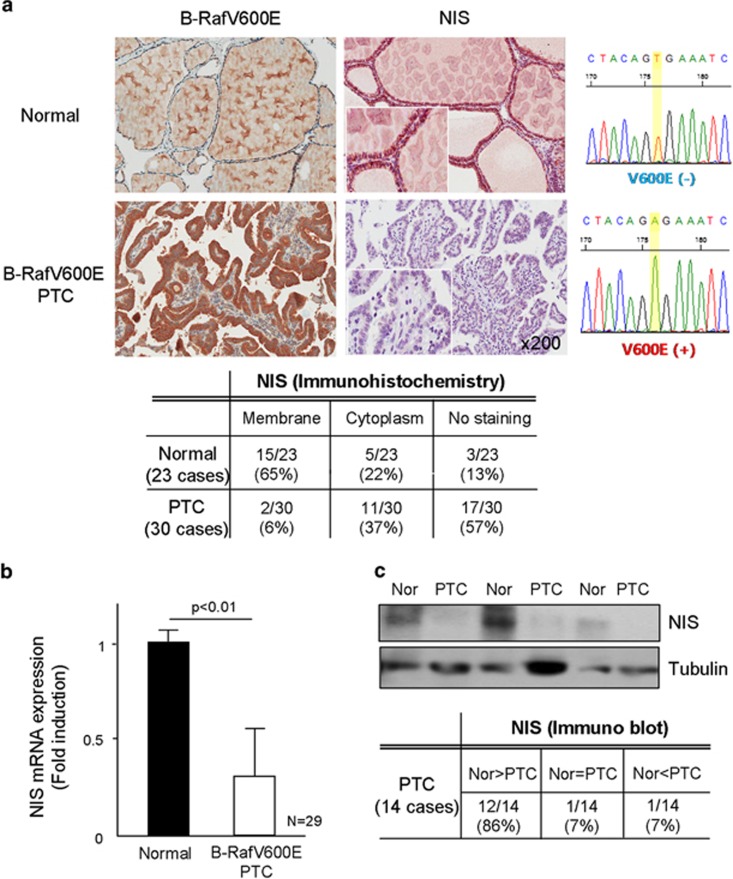
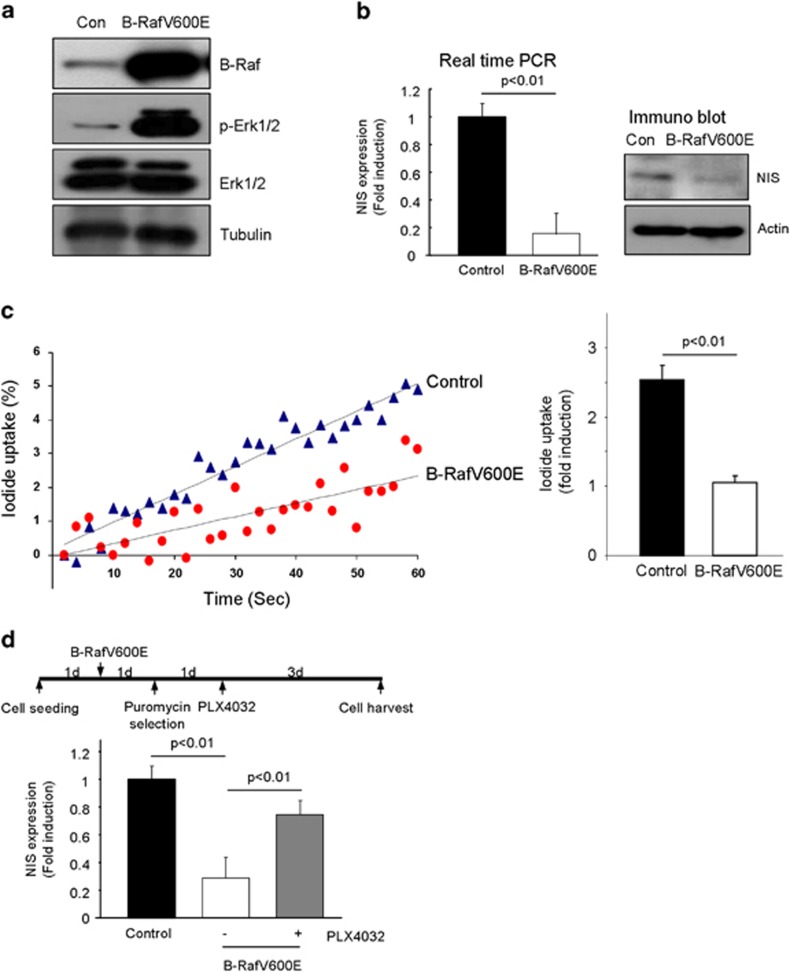
For analysis of NIS expression, we employed three kinds of molecular biological methods, such as immunohistochemistry, real-time PCR and western blotting. NIS was found staining in the plasma membrane region of normal thyroid follicle (15/23, 65%); however, its membrane staining was markedly downregulated in B-RafV600E PTC (2/30, 6%), whereas 37% of PTC tissues showed diffuse cytoplasmic staining, revealed by immunohistochemical analysis (Figure 1a). We further analyzed NIS expression by western blot and real-time PCR, and Figures 1b and c they showed markedly downregulated NIS expression in B-RafV600E harboring PTC. When B-RafV600E was expressed in normal primary thyrocytes isolated from surrounding tissues by lentivirus transduction, marked upregulation of p-Erk1/2 (Figure 2a) and downregulation of NIS expression was observed (Figure 2b). We further confirmed the NIS expression by the uptake of iodide in thyrocytes. Figure 2c clearly shows that B-RafV600E expressing thyrocytes did not efficiently take up iodide. Furthermore, B-Raf inhibitor (PLX4032) restored NIS expression in B-RafV600E expressing thyrocytes (Figure 2d). These data clearly suggested that B-RafV600E inhibited NIS expression in PTC and isolated primary thyrocytes.
Figure 1.
Sodium iodide symporter (NIS) expression in papillary thyroid carcinoma (PTC). (a) Immunohistochemical analysis of NIS. B-RafV600E mutant and NIS protein were analyzed by immunohistochemical analysis (left figure), and its staining pattern is summarized in a table (lower table). Twenty-three surrounding normal region and 30 cases of PTC were analyzed NIS and B-RafV600E, respectively. B-Raf was sequenced (right figure). NIS expression was analyzed by real-time PCR (b) and western blotting (c). Twenty-nine cases of surrounding normal region and PTC were analyzed for NIS expression by real-time PCR and 14 cases of fresh tissues were analyzed for NIS protein expression by western blotting.
Figure 2.
B-RafV600E inhibited sodium iodide symporter (NIS) expression in primary isolated thyrocytes. (a) B-RafV600E induced Erk1/2 phosphorylation in thyrocytes. (b) B-RafV600E inhibited NIS expression. Thyrocytes were infected with B-RafV600E lentivirus and then selected with 3.5 μg ml−1 puromycin for 1 week. NIS expression was analyzed by real-time PCR (left panel) and western blotting (right panel). (c) B-RafV600E inhibited iodide uptake in thyrocytes. Control and B-RafV600E expressing thyrocytes were analyzed for iodide uptake by using Premo Halide Sensor (Invitrogen) for 1 min, and the results are presented as dot graph (left panel). Three independent experiments are presented as a bar graph (right panel). (d) PLX4032 inhibited B-RafV600E induced NIS downregulation. Schematic drawing of an experiment (upper panel). Thyrocytes were infected with control or B-RafV600E lentivirus for 24 h and selected with 3.5 μg ml−1 puromycin. PLX4032 (20 μM) was applied and the cells were harvested 3 days later.
NIS promoter region in PTC was methylated
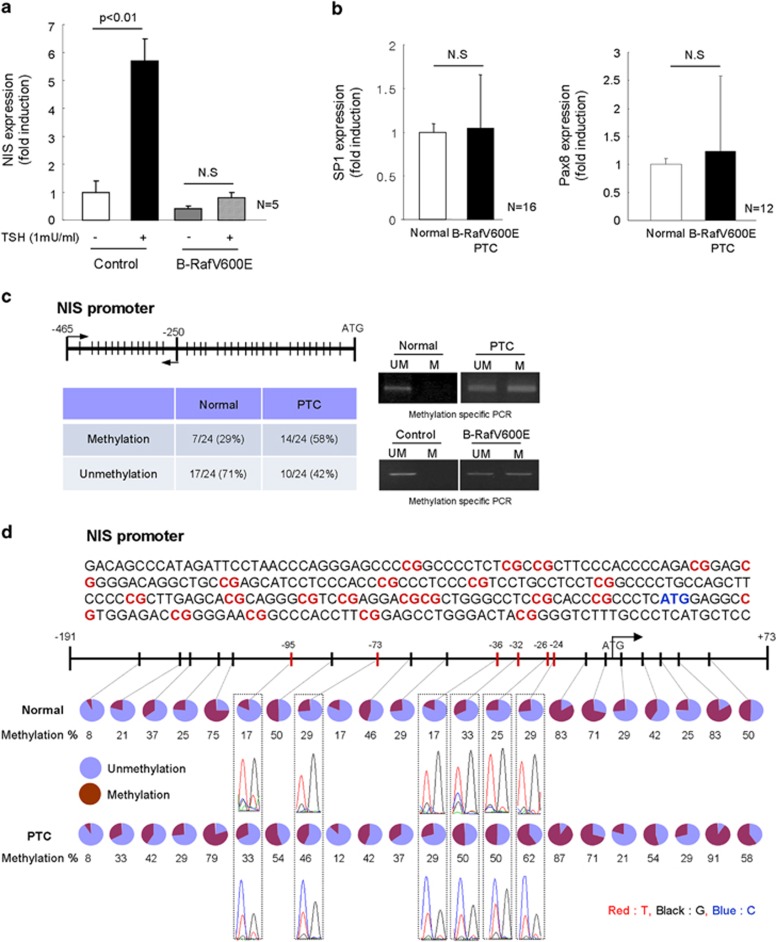
NIS expression is regulated by several kinds of transcription factors including SP1, PAX8, cAMP-responsive element binding protein and epigenetic regulation such as its promoter CpG islands methylation. Therefore, we first analyzed NIS expression after TSH treatment. It is well known that TSH can increase NIS expression via cAMP-PKA signal pathway. Figure 3a shows that the NIS expression is upregulated by TSH treatment; however, its expression was not significantly induced in B-RafV600E expressing thyrocytes. Furthermore, SP1 and PAX8 expressions were not induced in B-RafV600E harboring PTC (Figure 3b). These data suggest that B-RafV600E did not alter SP1 or PAX8-dependent pathway, but it might rather be involved in other inhibition pathway. Consequently, therefore, we considered epigenetic alteration such as promoter methylation by B-RafV600E. Thus, we analyzed promoter methylation by methylation-specific PCR and found more frequent promoter methylation in B-RafV600E expressing thyrocytes and PTC tissues (normal 29% vs cancer 58% Figure 3c). To further confirm NIS promoter methylation, we analyzed 24 cases of surrounding normal and B-RafV600E harboring PTC regions, including exon 1 (−191 to +73), by sequencing after bisulfite modification, and found that 6 CpG islands (−24 CpG; 29% vs 62%, −26 CpG; 25% vs 50%, −32 CpG; 33% vs 50%, −36 CpG; 17% vs 29%, −73 CpG; 29% vs 46%, −95 CpG; 17% vs 33%, normal and caner, respectively) of tumor region were more highly methylated than normal regions (⩾1.5 folds upregulation) (Figure 3d): Specially, −24 and −26 CpG sites in B-RafV600E PTC were more highly methylated (twofold upregulation). These data suggested that B-RafV600E inhibited NIS expression by induction of NIS promoter CpG methylation.
Figure 3.
Sodium iodide symporter (NIS) promoter region was highly methylated in papillary thyroid carcinoma (PTC). (a) Thyroid stimulating hormone (TSH) did not induce NIS expression in B-RafV600E expressing thyrocytes. Thyrocytes were infected with control or B-RafV600E lentivirus for 1 week without TSH in the media. Then, they were treated with TSH (1 mU ml−1) for 24 h and analyzed for NIS expression by real-time PCR. Bar graph indicates five independent experiments. (b) SP1 and PAX8 expressions in surrounding normal region and PTC were analyzed by real-time PCR. (c) Methylation-specific PCR analysis of NIS promoter region. Twenty-four cases of normal and PTC regions were analyzed for CpG islands methylation of NIS promoter. Methylation and unmethylation PCR primer were used for the regions indicated (arrow). Lines indicate CpG islands in NIS promoter. Normal and PTC regions methylation PCR is shown in the right upper panel, and B-RafV600E induced methylation in isolated thyrocytes is shown in the right lower panel, respectively. (d) Bisulfite-treated DNA sequencing analysis of NIS promoter region. Twenty-four cases of surrounding normal and PTC region were analyzed for NIS promoter region CpG islands methylation by bisulfite sequencing. Twenty-two CpG islands in NIS promoter and exon1 region (−191 to +73) were analyzed and methylation status is shown as a percentage. Blue circle indicates unmethylation status and red circle for methylation.
DNMTs expression in B-RafV600E PTC
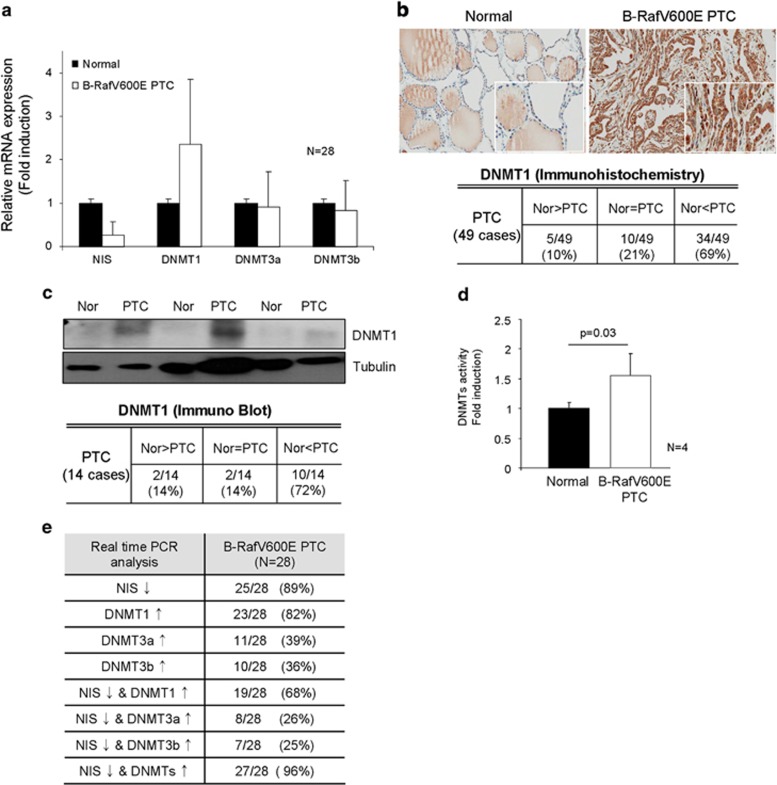
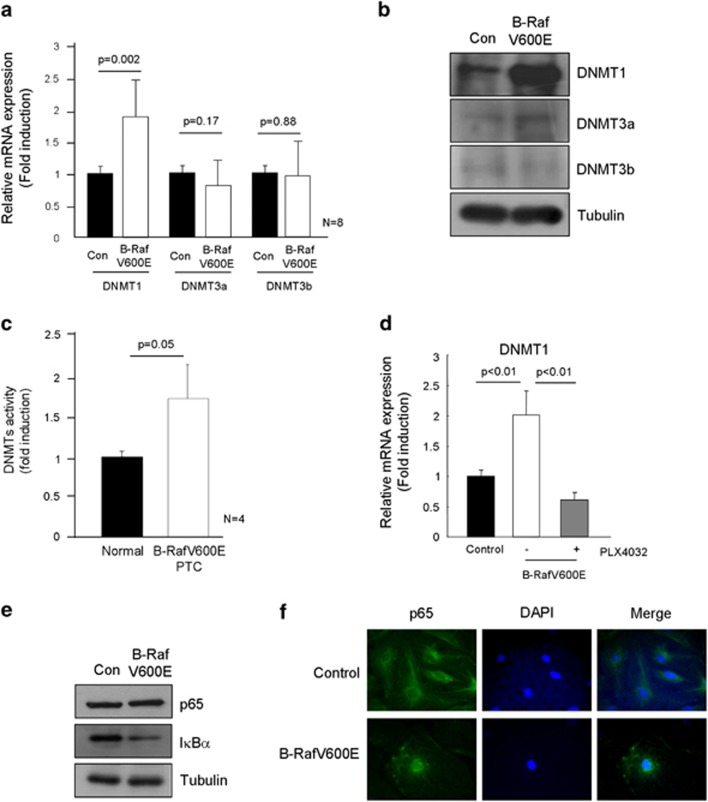
Promoter methylation in CpG islands is induced and maintained by DNMT1, 3a and 3b. However, as shown in Figure 4a, only DNMT1 expression but not DNMT3a and 3b, was markedly upregulated in PTC compared with normal region, revealed by real-time PCR analysis. Immunohistochemical analysis showed dense cytoplasmic and nucleus immunostaining of DNMT1 in B-RafV600E PTC (Figure 4b, upper panel), and 69% (34/49 cases) of B-RafV600E PTC with higher DNMT1 expression than surrounding normal thyroid follicle (Figure 4b, lower table). However, DNMT3a and 3b immunostaining intensity was not different between normal region and PTC (Supplementary Figure 1). We further analyzed DNMT1 protein expression in B-RafV600E PTC tissues by western blotting and found the induction of DNMT1 (Figure 4c), and the enzyme activity was increased in PTC compared with normal regions (Figure 4d). Furthermore, real-time PCR showed that DNMT1 and NIS expressions were highly inversely correlated in B-RafV600E PTC (Figure 4e,Table 1).
Figure 4.
DNA methyltransferase 1 (DNMT1) expression in B-RafV600E harboring papillary thyroid carcinoma (PTC). (a) DNMT1, but not DNMT3a,3b, was highly expressed in B-RafV600E harboring PTC. Twenty-eight cases of normal and PTC were analyzed for DNMTs expression by real-time PCR. (b) Immunohistochemical analysis of DNMT1. Forty-nine cases of normal and PTC were analyzed for DNMT1 expression by immunohistochemistry. Dense nucleal and cytoplasmic staining pattern was observed in B-RafV600E harboring PTC. Comparison of DNMT1 expression between PTCs and normal regions of the same slide is presented as a table. Nor and PTC indicate normal and cancer, respectively. (c) Western blotting of DNMT1. Fourteen cases of B-RafV600E harboring PTC tissue were further analyzed by western blotting. DNMT1 expression is presented as a table. (d) B-RafV600E increased DNMTs activity. DNMTs activity was measured in normal surrounding tissue and B-RafV600E harboring PTC. (e) Comparison between sodium iodide symporter (NIS) and DNMTs expression.
Table 1. NIS and DNMT expression in PTC.
| NIS | DNMT1 | DNMT3a | DNMT3b | |||||
|---|---|---|---|---|---|---|---|---|
| No. 1 | ↓↓ | 0.07 | ↑↑ | 2.25 | ↓ | 0.35 | ↓ | 0.6 |
| No. 2 | ↓↓ | 0.01 | ↑↑ | 3.25 | ↑ | 1.67 | ↑↑ | 2.03 |
| No. 3 | ↓↓ | 0.01 | ↑↑ | 2.82 | ↓↓ | 0.09 | ↑ | 1.12 |
| No. 4 | ↓↓ | 0.01 | ↑↑ | 2.67 | ↓ | 0.66 | ↓ | 0.32 |
| No. 5 | ↓↓ | 0.06 | ↑↑ | 2.28 | ↑ | 1.42 | N.D | |
| No. 6 | ↓↓ | 0.15 | ↑↑ | 2.22 | ↑ | 1.07 | ↓ | 0.85 |
| No. 7 | ↓↓ | 0.21 | ↓ | 0.92 | ↑ | 1.93 | ↑↑ | 2.9 |
| No. 8 | ↓↓ | 0.01 | ↓ | 0.79 | ↓ | 0.4 | ↓↓ | 0.13 |
| No. 9 | ↓↓ | 0.05 | ↑ | 1.38 | ↑ | 1.17 | ↓↓ | 0.2 |
| No. 10 | ↓↓ | 0.29 | ↑↑ | 2.54 | ↓↓ | 0.21 | 1.08 | |
| No. 11 | ↓ | 0.48 | ↑↑ | 10.05 | ↓ | 0.68 | ↓↓ | 0.12 |
| No. 12 | ↓↓ | 0.11 | ↑↑ | 2.9 | ↑↑ | 3.55 | ↑ | 1.42 |
| No. 13 | ↓ | 0.34 | ↑ | 1.24 | ↓ | 0.62 | ↓↓ | 0.28 |
| No. 14 | ↑ | 1.32 | ↓ | 0.46 | ↑ | 1.46 | ↑ | 1.43 |
| No. 15 | ↓↓ | 0.04 | ↑↑ | 2.82 | ↓ | 0.44 | ↓↓ | 0.16 |
| No. 16 | ↓↓ | 0.14 | ↑↑ | 8.63 | ↑↑ | 5.09 | ↑ | 1.67 |
| No. 17 | ↓↓ | 0.12 | ↑↑ | 3.38 | 1.09 | ↓ | 0.79 | |
| No. 18 | ↓↓ | 0.15 | ↑ | 1.48 | ↑ | 1.86 | ↑↑ | 3.78 |
| No. 19 | ↓↓ | 0.01 | ↑↑ | 5.73 | ↓ | 0.76 | 0.99 | |
| No. 20 | ↑↑ | 2.98 | 1.08 | ↓↓ | 0.14 | ↓↓ | 0.26 | |
| No. 21 | ↓↓ | 0.01 | ↓ | 0.72 | ↓↓ | 0.12 | ↑ | 1.58 |
| No. 22 | ↑ | 1.54 | ↑ | 1.54 | ↑ | 1.39 | ↓ | 0.47 |
| No. 23 | ↓ | 0.40 | ↓ | 0.76 | ↓ | 0.76 | 1.04 | |
| No. 24 | ↓↓ | 0.04 | ↑↑ | 4.92 | ↓ | 0.65 | ↓ | 0.36 |
| No. 25 | ↓↓ | 0.05 | ↑ | 1.23 | ↓↓ | 0.03 | ↓↓ | 0.14 |
| No. 26 | ↓ | 0.59 | ↑↑ | 4.06 | ↓ | 0.33 | 0.93 | |
| No. 27 | ↓↓ | 0.01 | ↓ | 0.82 | ↓ | 0.88 | 0.93 | |
| No. 28 | ↓↓ | 0.02 | ↓ | 0.74 | ↓↓ | 0.01 | ↓↓ | 0.05 |
Abbreviations: DNMT, DNA methyltransferase; NIS, sodium iodide symporter; PTC, papillary thyroid carcinoma.
NIS and DNMT expression levels were analyzed in 28 cases of PTC and surrounding normal tissue.
B-RafV600E increased DNMT1 expression in isolated thyrocytes
Next, we evaluated the DNMTs expression in B-RafV600E expressing isolated primary thyrocytes. Figure 5a shows that B-RafV600E increased DNMT1 expression in the primary thyrocytes, but not DNMT3a and 3b. Furthermore, its protein expression and activity were also increased in B-RafV600E expressing cells (Figures 5b and c); Even though marked induction of DNMT1 was observed, only a slight increase of DNMT3a and very low amount of DNMT3b were detected in B-RafV600E expressing cells. Furthermore, B-Raf inhibitor completely inhibited DNMT1 expression (Figure 5d). Next, we explored possible mechanism of B-RafV600E regulated DNMT1 expression. As DNMT1 expression is regulated by several transcription factors including SP1 and NF-κB, and we already found no alteration of SP1 expression in B-RafV600E harboring PTC (Figure 3b) and isolated thyrocytes (Supplementary Figure 2), we therefore analyzed NF-κB pathway. It is well known that in the signaling pathway B-RafV600E directly regulated NF-κB transcriptional activity via Iκ-B degradation.2,22 Figures 5e and f clearly show that NF-κB activation occurs in B-RafV600E expressing thyrocytes.
Figure 5.
DNA methyltransferase 1 (DNMT1) expression in B-RafV600E expressing thyrocytes. B-RafV600E induced DNMT1 expression, but not DNMT3a,3b. Thyrocytes were infected with control or B-RafV600E lentivirus for 1 week and then analyzed for DNMTs expression by real-time PCR (a), and western blot (b). (c) DNMTs activity in B-RafV600E harboring PTC. DNMTs activity was measured in normal surrounding regions and B-RafV600E harboring PTC, respectively. (d) PLX4032 inhibited B-RafV600E-induced DNMT1 upregulation. Experimental scheme was same as in Figure 2d. Thyrocytes were infected with control or B-RafV600E lentivirus for 24 h and selected with 3.5 μg ml−1 puromycin. PLX4032 (20 μM) was applied and the cells were harvested 3 days later. B-RafV600E induced NF-κB activation. IκBα degradation was accelerated in B-RafV600E expressing thyrocytes (e) and p65 nucleus localization was more frequently observed in B-RafV600E expressing thyrocytes (f).
Discussion
NIS, integral plasma membrane glycoprotein, is expressed at the highest level in the thyroid and lactating breast.23 NIS mRNA expression in thyroid follicular cells is regulated by the binding of transcription factors such as PAX8 and cAMP-responsive element binding protein to the NIS upstream enhancer in response to TSH stimulation.24 Iodide accumulation mediated by the above processes is reduced in thyroid carcinoma, particularly in dedifferentiated carcinoma, and the expression of iodide-metabolizing genes including NIS is also reduced in thyroid cancer. As reduced ability of iodide accumulation owing to the downrelugation of NIS is associated with radioiodine therapy resistance, which leads to treatment failure, an understanding of the involved molecular mechanism is important to overcome radioiodine therapy resistance. Furthermore, evaluation of iodine uptake by remaining or recurrent cancer cells is one of the most important tools during the follow-up and for the treatment of PTC.
There have been a few studies to suggest the molecular mechanism of how B-RafV600E represses NIS expression. Mitsutake et al.25 showed that RET/PTC induced MAPK activation due to B-Raf and then inhibited NIS expression, and this was further confirmed by siRNA targeting B-Raf, which blocked the NIS downregulation. However, Riesco-Eizaguirre et al.26 reported that MAPK inhibitor, U0126, showed limited recovery of NIS. In the present study also, B-RafV600E induced NIS repression was restored by PLX4032, a B-RafV600E kinase specific inhibitor, but not by U0126 MAPK inhibitor (Supplementary Figure 3). Therefore, B-RafV600E induced repression of NIS appears to be not mediated by MAPK signal pathway. Recently, Riesco-Eizaguirre et al.27 showed a possibility that B-RafV600E induced repression of NIS might be dependent on increased TGFβ1 secretion and its paracrine signaling, by showing that NIS was restored by inhibiting this signaling pathway with the overexpression of inhibitory SMAD7. Therefore, more studies are obviously needed to clarify how B-RafV600E signaling pathway regulates NIS expression.
In the present study, we observed limited upregulation of NIS by TSH stimulation in B-RafV600E expressing primary thyrocytes. Furthermore, the level of known transcription factors for NIS transcription such as SP1 and PAX8 was not different between normal thyroid and B-RafV600E harboring PTC. As CpG-rich regions exist in the NIS promoter as well as downstream from the transcription start site, epigenetic mechanisms through methylation of CpG could be responsible for alterations of NIS expression in thyroid cancer. Indeed, aberrant hypermethylation of NIS promoter region have been found in several human thyroid cancer cell lines along with the restoration of NIS expression after demethylating agent treatment.28, 29, 30, 31 However, there has been no report on the methylation status of NIS promoter exclusively in B-RafV600E harboring PTC. We observed in the present study that in 24 cases of B-RafV600E harboring PTC samples, six specific CpG sites in the NIS promoter region (−191 to +73) were more highly methylated than surrounding normal tissue. Furthermore, B-RafV600E expressing primary thyrocytes showed increased methylation status in the promoter of NIS. These data convinced us that B-RafV600E signaling pathway is the primary cause of increased methylation of NIS promoter, thus leading us to investigate how B-RafV600E can epigenetically regulate transcription of NIS.
DNMT1 and DNMT3a,b are known as major mediators of maintenance methylation and de novo methylation of DNA in embryogenesis and carcinogenesis.20 Therefore, we measured mRNA levels of DNMT1 and DNMT3a,b in B-RafV600E harboring PTC along with adjacent normal tissues, and found that only DNMT1 expression was upregulated. As DNMT3a,b levels were similar between normal thyroid tissue and B-RafV600E harboring PTC, increased DNMTs activity can be interpreted as a result from increased DNMT1 in B-RafV600E harboring PTC. As DNMT3a,b, but not DNMT1, are known to catalyze de novo methylation of DNA, the upregulation of DNMT1 in B-RafV600E harboring PTC cannot easily explain the increased methylation of NIS promoter region. Nonetheless, DNMT1 has been reported to function together with DNMT3 synergistically, and is important in maintaining the methylation status of promoter regions for repression of important tumor suppressor genes in various cancers.32, 33, 34 Although it was highly likely that NIS expression was epigenetically regulated, we measured the expressions of NIS and DNMTs. As expected, DNMTs expression was negatively correlated with NIS expression in B-RafV600E harboring PTC samples (96%). Similarly, B-RafV600E expressing primary thyrocytes showed induction of DNMT1 mRNA and protein.
B-RafV600E has been shown to be able to activate NF-κB.22 Furthermore, Liu et al.35 showed that bortezomib, a proteasome inhibitor that prevents proteasomal degradation of IκB, inhibited NF-κB activation and decreased DNMT1 expression through the abrogation of SP1/NF-κB complex and disruption of binding to the DNMT1 promoter. This study further indicates that B-RafV600E induced NF-κB activation can increase SP1 transcription activity on the promoter of DNMT1. Indeed, it is highly possible that NF-κB pathway was activated, because we observed degradation of IκB and nuclear translocation of NF-κB in the B-RafV600E expressed primary thyrocytes (Figure 5e).
In summary, the repression of NIS in B-RafV600E harboring PTC is due to epigenetic suppression of transcription by increased DNMT1 expression. B-RafV600E induced NIS suppression was dependent on B-RafV600E kinase activity, but not dependent on MAPK signaling. Instead, NF-κB pathway activation from B-RafV600E signaling can be the main cause of NIS suppression through the induction of DNMT1 in B-RafV600E harboring PTC.
Acknowledgments
We appreciate Professor Woon Ki Paik for his careful reading of this manuscript. This work was supported by a grant (2012R1A1A2007267, NRF-2012R1A5A2048183) of National Research Foundation of Korea to TJP and a grant (2012R1A1A2004721) to J-HK.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13:184–199. doi: 10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, Noguchi S, Yamashita H, Murakami N, Watanabe S, Uchino S, et al. Changing trends and prognoses for patients with papillary thyroid cancer. Arch Surg. 1998;133:1058–1065. doi: 10.1001/archsurg.133.10.1058. [DOI] [PubMed] [Google Scholar]
- Ezaki H, Ebihara S, Fujimoto Y, Iida F, Ito K, Kuma K, et al. Analysis of thyroid carcinoma based on material registered in Japan during 1977-1986 with special reference to predominance of papillary type. Cancer. 1992;70:808–814. doi: 10.1002/1097-0142(19920815)70:4<808::aid-cncr2820700415>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Ros P, Rossi DL, Acebron A, Santisteban P. Thyroid-specific gene expression in the multi-step process of thyroid carcinogenesis. Biochimie. 1999;81:389–396. doi: 10.1016/s0300-9084(99)80086-8. [DOI] [PubMed] [Google Scholar]
- Fabbro D, Di Loreto C, Beltrami CA, Belfiore A, Di Lauro R, Damante G. Expression of thyroid-specific transcription factors TTF-1 and PAX-8 in human thyroid neoplasms. Cancer Res. 1994;54:4744–4749. [PubMed] [Google Scholar]
- Brabant G, Maenhaut C, Kohrle J, Scheumann G, Dralle H, Hoang-Vu C, et al. Human thyrotropin receptor gene: expression in thyroid tumors and correlation to markers of thyroid differentiation and dedifferentiation. Mol Cell Endocrinol. 1991;82:R7–R12. doi: 10.1016/0303-7207(91)90018-n. [DOI] [PubMed] [Google Scholar]
- Ohta K, Endo T, Onaya T. The mRNA levels of thyrotropin receptor, thyroglobulin and thyroid peroxidase in neoplastic human thyroid tissues. Biochem Biophys Res Commun. 1991;174:1148–1153. doi: 10.1016/0006-291x(91)91540-s. [DOI] [PubMed] [Google Scholar]
- Arturi F, Russo D, Schlumberger M, du Villard JA, Caillou B, Vigneri P, et al. Iodide symporter gene expression in human thyroid tumors. J Clin Endocrinol Metab. 1998;83:2493–2496. doi: 10.1210/jcem.83.7.4974. [DOI] [PubMed] [Google Scholar]
- Lin KL, Wang OC, Zhang XH, Dai XX, Hu XQ, Qu JM. The BRAF mutation is predictive of aggressive clinicopathological characteristics in papillary thyroid microcarcinoma. Ann Surg Oncol. 2010;17:3294–3300. doi: 10.1245/s10434-010-1129-6. [DOI] [PubMed] [Google Scholar]
- Lee X, Gao M, Ji Y, Yu Y, Feng Y, Li Y, et al. Analysis of differential BRAF(V600E) mutational status in high aggressive papillary thyroid microcarcinoma. Ann Surg Oncol. 2009;16:240–245. doi: 10.1245/s10434-008-0233-3. [DOI] [PubMed] [Google Scholar]
- Xing M, Westra WH, Tufano RP, Cohen Y, Rosenbaum E, Rhoden KJ, et al. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. J Clin Endocrinol Metab. 2005;90:6373–6379. doi: 10.1210/jc.2005-0987. [DOI] [PubMed] [Google Scholar]
- Mian C, Barollo S, Pennelli G, Pavan N, Rugge M, Pelizzo MR, et al. Molecular characteristics in papillary thyroid cancers (PTCs) with no 131I uptake. Clin Endocrinol (Oxf) 2008;68:108–116. doi: 10.1111/j.1365-2265.2007.03008.x. [DOI] [PubMed] [Google Scholar]
- Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer. 2005;12:245–262. doi: 10.1677/erc.1.0978. [DOI] [PubMed] [Google Scholar]
- Durante C, Puxeddu E, Ferretti E, Morisi R, Moretti S, Bruno R, et al. BRAF mutations in papillary thyroid carcinomas inhibit genes involved in iodine metabolism. J Clin Endocrinol Metab. 2007;92:2840–2843. doi: 10.1210/jc.2006-2707. [DOI] [PubMed] [Google Scholar]
- Romei C, Ciampi R, Faviana P, Agate L, Molinaro E, Bottici V, et al. BRAFV600E mutation, but not RET/PTC rearrangements, is correlated with a lower expression of both thyroperoxidase and sodium iodide symporter genes in papillary thyroid cancer. Endocr Relat Cancer. 2008;15:511–520. doi: 10.1677/ERC-07-0130. [DOI] [PubMed] [Google Scholar]
- Espadinha C, Santos JR, Sobrinho LG, Bugalho MJ. Expression of iodine metabolism genes in human thyroid tissues: evidence for age and BRAFV600E mutation dependency. Clin Endocrinol (Oxf) 2009;70:629–635. doi: 10.1111/j.1365-2265.2008.03376.x. [DOI] [PubMed] [Google Scholar]
- Liu D, Hu S, Hou P, Jiang D, Condouris S, Xing M. Suppression of BRAF/MEK/MAP kinase pathway restores expression of iodide-metabolizing genes in thyroid cells expressing the V600E BRAF mutant. Clin Cancer Res. 2007;13:1341–1349. doi: 10.1158/1078-0432.CCR-06-1753. [DOI] [PubMed] [Google Scholar]
- Chakravarty D, Santos E, Ryder M, Knauf JA, Liao XH, West BL, et al. Small-molecule MAPK inhibitors restore radioiodine incorporation in mouse thyroid cancers with conditional BRAF activation. J Clin Invest. 2011;121:4700–4711. doi: 10.1172/JCI46382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Palona I, Namba H, Mitsutake N, Starenki D, Podtcheko A, Sedliarou I, et al. BRAFV600E promotes invasiveness of thyroid cancer cells through nuclear factor kappaB activation. Endocrinology. 2006;147:5699–5707. doi: 10.1210/en.2006-0400. [DOI] [PubMed] [Google Scholar]
- Dohan O, De la Vieja A, Paroder V, Riedel C, Artani M, Reed M, et al. The sodium/iodide symporter (NIS): characterization, regulation, and medical significance. Endocr Rev. 2003;24:48–77. doi: 10.1210/er.2001-0029. [DOI] [PubMed] [Google Scholar]
- Kogai T, Brent GA. The sodium iodide symporter (NIS): regulation and approaches to targeting for cancer therapeutics. Pharmacol Ther. 2012;135:355–370. doi: 10.1016/j.pharmthera.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsutake N, Miyagishi M, Mitsutake S, Akeno N, Mesa C, Jr, Knauf JA, et al. BRAF mediates RET/PTC-induced mitogen-activated protein kinase activation in thyroid cells: functional support for requirement of the RET/PTC-RAS-BRAF pathway in papillary thyroid carcinogenesis. Endocrinology. 2006;147:1014–1019. doi: 10.1210/en.2005-0280. [DOI] [PubMed] [Google Scholar]
- Riesco-Eizaguirre G, Gutierrez-Martinez P, Garcia-Cabezas MA, Nistal M, Santisteban P. The oncogene BRAF V600E is associated with a high risk of recurrence and less differentiated papillary thyroid carcinoma due to the impairment of Na+/I- targeting to the membrane. Endocr Relat Cancer. 2006;13:257–269. doi: 10.1677/erc.1.01119. [DOI] [PubMed] [Google Scholar]
- Riesco-Eizaguirre G, Rodriguez I, De la Vieja A, Costamagna E, Carrasco N, Nistal M, et al. The BRAFV600E oncogene induces transforming growth factor beta secretion leading to sodium iodide symporter repression and increased malignancy in thyroid cancer. Cancer Res. 2009;69:8317–8325. doi: 10.1158/0008-5472.CAN-09-1248. [DOI] [PubMed] [Google Scholar]
- Galrao AL, Sodre AK, Camargo RY, Friguglietti CU, Kulcsar MA, Lima EU, et al. Methylation levels of sodium-iodide symporter (NIS) promoter in benign and malignant thyroid tumors with reduced NIS expression. Endocrine. 2013;43:225–229. doi: 10.1007/s12020-012-9779-8. [DOI] [PubMed] [Google Scholar]
- Venkataraman GM, Yatin M, Marcinek R, Ain KB. Restoration of iodide uptake in dedifferentiated thyroid carcinoma: relationship to human Na+/I-symporter gene methylation status. J Clin Endocrinol Metab. 1999;84:2449–2457. doi: 10.1210/jcem.84.7.5815. [DOI] [PubMed] [Google Scholar]
- Provenzano MJ, Fitzgerald MP, Krager K, Domann FE. Increased iodine uptake in thyroid carcinoma after treatment with sodium butyrate and decitabine (5-Aza-dC) Otolaryngol Head Neck Surg. 2007;137:722–728. doi: 10.1016/j.otohns.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Li W, Venkataraman GM, Ain KB. Protein synthesis inhibitors, in synergy with 5-azacytidine, restore sodium/iodide symporter gene expression in human thyroid adenoma cell line, KAK-1, suggesting trans-active transcriptional repressor. J Clin Endocrinol Metab. 2007;92:1080–1087. doi: 10.1210/jc.2006-2106. [DOI] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kitazawa S, et al. DNA methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinogenesis. 2006;27:1160–1168. doi: 10.1093/carcin/bgi361. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Sunaga N, Shames DS, Toyooka S, Gazdar AF, Minna JD. RNA interference-mediated knockdown of DNA methyltransferase 1 leads to promoter demethylation and gene re-expression in human lung and breast cancer cells. Cancer Res. 2004;64:3137–3143. doi: 10.1158/0008-5472.can-03-3046. [DOI] [PubMed] [Google Scholar]
- Liu S, Liu Z, Xie Z, Pang J, Yu J, Lehmann E, et al. Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-kappaB-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood. 2008;111:2364–2373. doi: 10.1182/blood-2007-08-110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.