Abstract
Indoleamine 2,3-dioxygenases (IDOs) are tryptophan-catabolizing enzymes with immunomodulatory functions. However, the biological role of IDO2 and its relationship with IDO1 are unknown. To assess the relationship between IDO2 and IDO1, we investigated the effects of co-expression of human (h) IDO2 on hIDO1 activity. Cells co-expressing hIDO1 and hIDO2 showed reduced tryptophan metabolic activity compared with those expressing hIDO1 only. In a proteomic analysis, hIDO1-expressing cells exhibited enhanced expression of proteins related to the cell cycle and amino acid metabolism, and decreased expression of proteins related to cell survival. However, cells co-expressing hIDO1 and hIDO2 showed enhanced expression of negative regulators of cell apoptosis compared with those expressing hIDO1 only. Co-expression of hIDO1 and hIDO2 rescued the cell death induced by tryptophan-depletion through hIDO1 activity. Cells expressing only hIDO2 exhibited no marked differences in proteome profiles or cell growth compared with mock-transfectants. Cellular tryptophan metabolic activity and cell death were restored by co-expressing the hIDO2 mutant substituting the histidine 360 residue for alanine. These results demonstrate that hIDO2 plays a novel role as a negative regulator of hIDO1 by competing for heme-binding with hIDO1, and provide information useful for development of therapeutic strategies to control cancer and immunological disorders that target IDO molecules.
Introduction
Indoleamine 2,3-dioxygenase 1 (IDO1) and its paralog IDO21,2 are involved in tryptophan catabolism. IDO molecules have immunosuppressive functions, as IDO1 is required for maternal tolerance to fetal tissue and L, D-1-methyl tryptophan (1MT)-triggered rejection of the fetal tissue.3 IDO1 has been implicated in various diseases, including cancers, chronic infection and autoimmunity.4 Genetic ablation of Ido1 leads to upregulation of Ido2 in the epididymis, suggesting a possible functional overlap between these two molecules.5 In this study, we evaluated the function interaction between IDO1 and IDO2, focusing on the impact of IDO2 co-expression on IDO1 catalytic activity.
IDO1 is the major rate-limiting enzyme that catalyzes conversion of tryptophan to kynurenine, an initial step in tryptophan degradation by the kynurenine pathway.6,7 Human (h) IDO1 shares 58% sequence homology with mouse IDO1.8 IDO1 is widely expressed in various tissues, including lung, small intestine, placenta, spleen, central nervous system and epididymis,5,9,10 and its expression is increased markedly in various cancers. IDO1 plays a role in immune tolerance,11,12 promoting tumorigenesis.13, 14, 15 IDO1-mediated tryptophan depletion is considered to be one of the immune suppressive mechanisms and a target pathway for development of anticancer drugs.
IDO2 was identified by in silico mapping of the National Center for Biotechnology Information human genome sequence using the IDO1 sequence as a probe.2 It was mapped to chromosome 8p12, immediately downstream of the IDO1 gene. The hIDO2 protein shows 43% amino acid homology with hIDO1.16 IDO2 is detectable in placenta, brain, liver, kidney and the epididymis of mice5 and some human gastric, colon and pancreatic cancer cell tumors.17,18 However, the functional activity of IDO2 is obscure. IDO2 is considered to be biologically inactive or active only under specific (albeit uncharacterized) conditions.19 Kynurenine is not detectable in HEK293 cells transfected with hIDO2,20,21 but is present at low levels in a cell line transfected with an inducible construct.2 IDO1 and IDO2 seem to be regulated differently because the levo (L)-1MT isomer eliminates IDO1 activity,2,18,19 whereas the dextro (D)-1MT stereoisomer exclusively blocks IDO2 activity.2 Because IDO2 has a very-low level of catalytic activity, it may have a distinct biological role. IDO2, but not IDO1, is a critical mediator of arthritis development and autoantibody production,22 suggesting separate in vivo functions for IDO1 and IDO2. However, the IDO2 blocker D-1MT has been evaluated in clinical trials as an anticancer treatment, suggesting that IDO2 may play a cooperative role with IDO1 in immune regulation. Both IDO1 and IDO2 are expressed in normal mouse epididymis, but IDO2 is highly upregulated in the epididymis of IDO1-deficient mice.5 High IDO2 expression is insufficient to compensate for kynurenine production. Upregulation/alteration of IDO2 splice variants was detected in peritoneal macrophages of IDO1-deficient mice, but no such changes in IDO1 expression or activity were observed in IDO2-deficient mice.23 Therefore, we presume that IDO2 plays a particular biological role and that interplay and/or interactive-regulation between IDO1 and IDO2 is possible.
In this study, we investigated the influence of hIDO2 co-expression on hIDO1 catalytic activity to assess the interplay between these two molecules, using a hIDO1 and hIDO2-overexpressing cell line. The results reveal a novel regulatory effect of hIDO2 on hIDO1 catalytic activity and the possible underlying mechanism.
Materials and methods
Plasmid DNA
hIDO1 and hIDO2 (named hIDO1-2) recombinant DNA, the hIDO1-coding region tagged with Flag, or hIDO2 tagged with hemagglutinin (HA) were subcloned into bicistronic vectors including the internal ribosomal entry site or the porcine teschovirus-1 (P2A) peptide to construct hIDO1 and hIDO2.24 The enhanced green fluorescent protein (eGFP) or mCherry was included as a reporter gene. The hIDO2 (H; H360A) mutant, with alanine replacing histidine at position 360, was constructed by polymerase chain reaction-based site-directed mutagenesis using the forward primer 5′–CTGCGGAGCTATGCCATCACCATG–3′ and the reverse primer, 5′–CATGGTGATGGCATAGCTCCGAG–3′.
Cell culture and construction of hIDO-expressing HEK293 stable cell lines
Human embryonic kidney (HEK293) cells were maintained in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY, USA) supplemented with 10% bovine calf serum, penicillin, streptomycin and gentamycin. HEK293 cells were transfected with 2 μg of the hIDO1, hIDO2, hIDO1-2 or hIDO1-2 (H) hIDO DNA constructs using the calcium phosphate or polyethylenimine (PEI) (Sigma-Aldrich, St Louis, MO, USA) transfection method to construct hIDO1-, hIDO2- and hIDO1-hIDO2-expressing HEK293 stable cell lines. Transfected HEK293 cells were selected with 20 μgml−1 blasticidin (Invitrogen, Carlsbad, CA, USA) including 50 μM L-tryptophan (L-Trp) (Sigma-Aldrich) for 2–4 weeks, and single-cell originated hIDO-expressing stable cell lines were isolated by flow cytometry analysis and western blotting.
Kynurenine assay
The catalytic activity of the hIDOs was measured by means of a colorimetric kynurenine assay.25 Briefly, stable hIDO1-expressing HEK293 cells were transfected with hIDO2 wild-type and hIDO2 mutant DNAs using PEI. The next day, the medium was exchanged for fresh medium containing 100 μM L-Trp, and the culture supernatants were harvested after additional 24 h incubation. The hIDO-expressing HEK293 stable cells were seeded onto a 96-well plate at 2 × 105 cells per well in the absence or presence of 100 μM L-Trp, and the culture supernatants were harvested after 24 h incubation. A 160 μl aliquot of culture supernatant was transferred to a 96-well round-bottom plate (SPL Life Sciences, Pocheon, Korea); 10 μl of 30% trichloroacetic acid (Sigma-Aldrich) were added to each well and plates were incubated for 30 min at 50 °C to hydrolyze N-formylkynurenine to kynurenine. The samples were then centrifuged at 3000 g for 10 min. Then, 100 μl aliquots of the supernatant were harvested and mixed with 100 μl of freshly prepared Ehrlich's reagent (2% 4-demethylaminobenzaldehyde in glacial acetic acid; Tokyo Chemical Industry, Tokyo, Japan). Absorbance at 492 nm was measured using a microplate reader (BioTek, Winooski, VT, USA) with the Gen5 software (BioTek) after 10 min incubation.
Proteomic analysis
The hIDO1-, hIDO2- and hIDO1-2-expressing HEK293 stable cell lines were subjected to proteomic analysis. Proteins were quantitated and identified using isobaric tags for relative and absolute quantitation 26 and mass spectrometry. A total of 2557 proteins were analyzed using the International Protein Index Human Database with the proteomic tools. Normalized intensities for those proteins were determined using a hierarchical clustering algorithm. Analysis of variance was performed to identify differentially expressed proteins with a threshold false discovery rate. Significantly differentially expressed proteins were classified based on their biological functions. Enrichment of each biological category was examined statistically using Fisher's exact test.
Cell proliferation assay
The stable hIDO-expressing HEK293 cell lines were seeded in 96-well plates at a density of 4 × 104 per well. Cell proliferation rates were then measured with the colorimetric WST-1 assay (ITSBio, Seoul, Korea) over 3 days in the absence or presence of 25 μM L-Trp. Absorbance at 450 nm was measured using a microplate reader with the Gen5 software.
Flow cytometry analysis
The stable hIDO1-expressing HEK293 cell lines transfected with hIDO2 and hIDO2 (H) mutant DNAs were fixed in 4% paraformaldehyde for 10 min at 48 h posttransfection. Fixed cells were washed with Triton buffer (0.5% Triton X-100, 0.1% BSA in PBS) and stained with anti-HA epitope (Applied Biological Materials, Richmond, BC, Canada) primary antibodies, and an allophycocyanin-conjugated rat anti-mouse secondary IgG antibody (BD Biosciences, Franklin Lakes, NJ, USA). The stained cells were washed and analyzed by FACSCalibur (BD Bioscience) running the FlowJo software (Tree Star, Ashland, OR, USA).
Western blotting assay
The stable hIDO-expressing HEK293 cell lines were harvested and lysed with RIPA buffer (1 ml/1 × 107 cells; 150 mM NaCl, 50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.1% sodium deoxycholate, 1 mM EDTA and protease inhibitors). Cell lysates were loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and stained with mouse Flag epitope (Sigma-Aldrich) and anti-HA epitope (Applied Biological Materials) primary antibodies and a horseradish peroxidase-conjugated goat anti-mouse secondary IgG antibody (KOMA Biotechnology, Seoul, Korea). A monoclonal anti-β-actin (Sigma-Aldrich) antibody was used as the loading control. An enhanced chemiluminescence reagent (Intron, Seoul, Korea) was used to visualize the protein signals.
Statistical procedures
The statistical analysis was performed using the two-tailed t-test with the GraphPad Prism ver. 5 software (GraphPad Software, San Diego, CA, USA). A value of P<0.05 was taken to indicate statistical significance.
Results
Overexpression of hIDO2 leads to decreased hIDO1 activity at the single-cell level
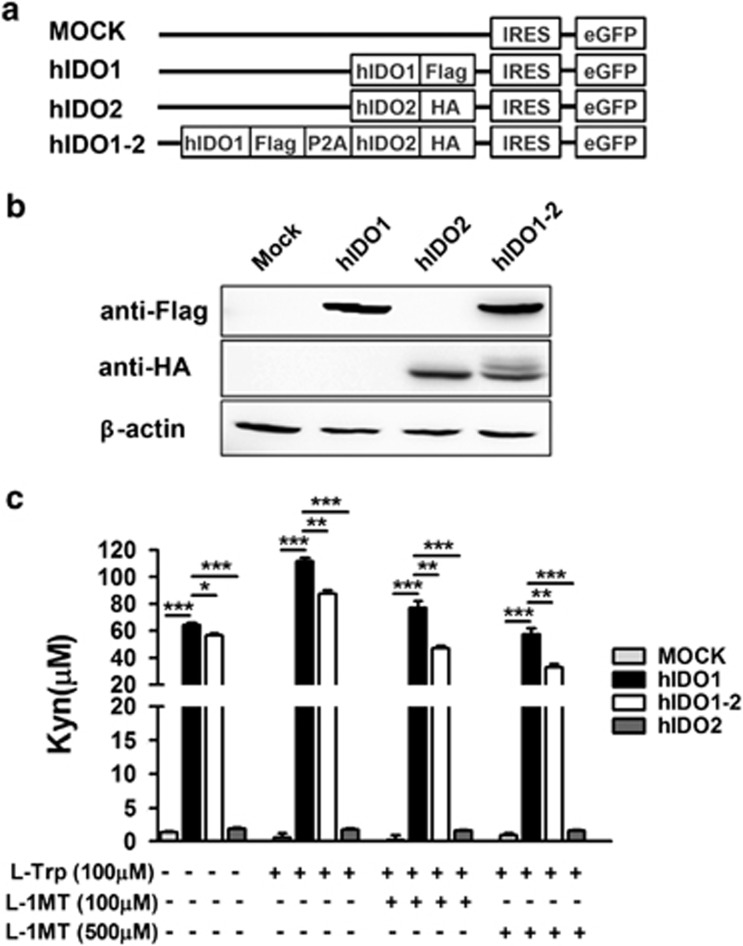
We measured kynurenine production by hIDO1 and hIDO2 (hIDO1-2)-overexpressing HEK293 cell lines in comparison with those produced by HEK293 cells expressing either hIDO1 or hIDO2 to investigate the influence of hIDO2 on hIDO1 functional activity. Transfection with a plasmid in which recombinant DNAs encoding Flag-tagged hIDO1 and HA-tagged hIDO2 were linked with the 2A peptide was used to establish hIDO1 and hIDO2-co-expressing HEK293 cells (Figure 1a). HEK293 cells transfected to co-express Flag-tagged hIDO1 and HA-tagged hIDO2, and control cells transfected to express either Flag-tagged hIDO1 or HA-tagged hIDO2, were cultured with 20 μgml−1 blasticidin for 2–4 weeks. More than five single-cell originating cell lines expressing hIDO1 and hIDO2 at similar levels were selected from each group of transfectants, and their expression levels were confirmed by Western blotting using anti-Flag and HA antibodies (Figure 1b).
Figure 1.
Inhibition of human indoleamine 2,3-dioxygenase 1 (hIDO1) enzymatic activity by co-expressing hIDO2 at the single-cell level. (a) Diagram of DNA constructs for expression of hIDO1, hIDO2, or both hIDO1 and hIDO2 (hIDO1-2) tagged with Flag, hemagglutinin epitopes, and fused with the eGFP gene. A self-cleaving 2A peptide (26 amino acids) between hIDO1-Flag and hIDO2-hemagglutinin was used for efficient expression of multiple genes. (b) Western blotting was performed using extracts of HEK293 cells expressing hIDO1-Flag, hIDO2-HA, or both proteins. HEK293 cells were transfected with plasmid DNA as shown in a, and subsequently selected with 2 μgml−1 blasticidin for 2–4 weeks to generate stable cell lines. hIDO1 or hIDO2 protein levels were determined by staining with anti-Flag and -hemagglutinin monoclonal antibodies, respectively, using the LAS 4000 Mini. β-actin was used as the internal control. (c) Kynurenine production was measured with a quantitative colorimetric assay using Ehrlich's reagent. hIDO-expressing stable cell lines were incubated for 24 h with complete medium including 100 μM L-tryptophan (L-Trp) in the absence or presence of 100 or 500 μM of the IDO inhibitor L-1MT. *P<0.05, **P<0.01, ***P<0.001. Data shown (b and c) represent at least three independent experiments.
The stable cell lines were subjected to kynurenine assay after an overnight incubation. Cells expressing hIDO1 only had high tryptophan catalytic activity and produced 64.54±2.19 μM kynurenine, whereas those expressing only hIDO2 showed significantly lower tryptophan metabolic activity, as evidenced by their production of ⩽2 μM kynurenine. These results agree with those of previous studies.20,21 Interestingly, kynurenine production by HEK293 cells co-expressing hIDO1 and hIDO2 was significantly lower (mean, 56.40±2.75 μM) than that by cells expressing only hIDO1, despite the similar hIDO1 expression of these two cell lines. This difference in L-Trp catalytic activity between the two cell lines was confirmed upon addition of 100 μM L-Trp to the culture medium. L-Trp supplementation increased kynurenine production by both co-expressing cells and those expressing only hIDO1 (87.57±4.01 and 111.67±4.43 μM, respectively) (Figure 1c). This difference remained when the IDO1 inhibitor L-1MT was added to the medium; however, this treatment decreased kynurenine production by both cell types in a dose-dependent manner. Therefore, co-expression of hIDO2 reduced the catalytic activity of hIDO1.
Differential protein expression profiles of the hIDO1-, hIDO2- and hIDO1-2-expressing stable cell lines and implications for cell survival
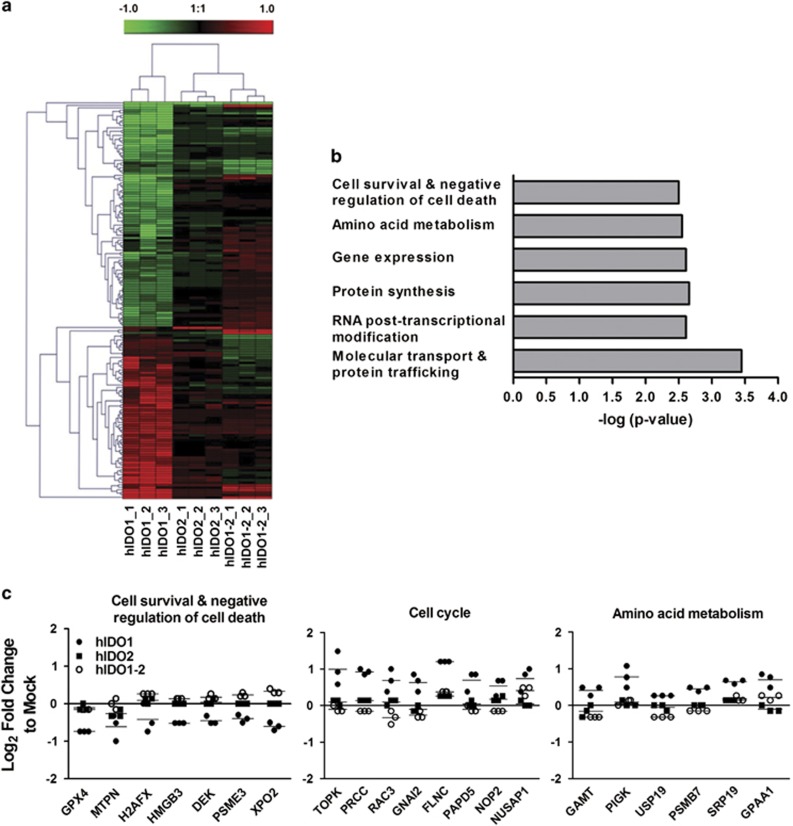
We compared the protein profiles of cells expressing either hIDO1 or hIDO2 with those co-expressing hIDO1 and hIDO2 (hIDO1-2) to evaluate the effect of hIDO2 co-expression on the catalytic activity of hIDO1 (Figure 2). Among the 2557 proteins analyzed, 159 were identified by analysis of variance as differentially expressed among the groups with fold-changes >1.5. A heat map analysis showed differential protein expression among the stable hIDO1-, hIDO2- and hIDO1-2-expressing HEK293 cell lines (Figure 2a). The top six biological classifications were identified by Fisher's exact test (Figure 2b). Unexpectedly, hIDO2 overexpression did not result in marked changes in protein profile compared with mock-transfected control cells. However, hIDO1 overexpression enhanced the expression of proteins related to the cell cycle and amino acid metabolism compared with mock-transfected cells, suggesting enhanced metabolic activity in hIDO1-expressing cells. Additionally, the expression of cell survival/negative regulation of cell death-related proteins also showed considerable changes. Among the 159 proteins, 69 were expressed at higher levels by cells co-expressing hIDO1 and hIDO2 compared with cells expressing only hIDO1 (Table 1); 46 proteins were expressed at lower levels (Table 2). Seven of the 67 proteins whose expression was enhanced in cells co-expressing hIDO1 and hIDO2 (GPX4,27 MTPN,28 H2AFX,29 HMGB3,30 DEK,31 PSME332 and XPO233) were related to cell morphogenesis, proliferation or negative regulation of cell death/apoptosis (Figure 2c). The expression levels of these proteins were not enhanced markedly in cells expressing only IDO2. IDO1 levels were slightly (1.5 × ) higher in the co-expressing cells than in those expressing hIDO1 only. Therefore, the proteins whose expression was enhanced in co-expressing cells would likely be those suppressed by hIDO1 over-expression, and suppression of which was reversed in the presence of hIDO2, regardless of its function.
Figure 2.
Evaluation of the protein profiles of human indoleamine 2,3-dioxygenase 1 (hIDO1)-, hIDO2-, or hIDO1-2-overexpressing HEK293 stable cells using a proteomic approach. (a) Hierarchical clustering analysis of 159 differentially expressed proteins identified by analysis of variance at a false discovery rate <0.05. Scale bar at the top indicates fold change compared to mock control on a log2 scale. (b) Functional classifications of the 159 proteins. Significant biological classes were identified by Fisher's exact test. The top six significant categories with P-values are shown. (c) Fold changes in expression of proteins related to cell survival/negative regulation of cell death, amino acid metabolism, and cell cycle compared with the mock control are plotted on a log2 scale.
Table 1. The 69 proteins detected at higher levels (>1.5-fold) in cells co-expressing human indoleamine 2,3-dioxygenase 1 (hIDO1) and hIDO2 than in cells expressing only hIDO1.
| Fold changes | |||
|---|---|---|---|
| Accession no. | Symbol | Protein names | hIDO1-2 vs hIDO1 |
| Q71U36 | TUBA1A | Tubulin alpha-1A chain | 8.316 |
| P84077 | ARF1 | ADP-ribosylation factor 1 | 3.249 |
| Q96P16 | RPRD1A | Isoform 1 of regulation of nuclear pre-mRNA domain-containing protein 1A | 2.644 |
| Q15758 | ATB(0) | Neutral amino acid transporter B(0) | 2.598 |
| P08107 | HSP71 | Heat shock 70-kDa protein 1A/1B | 2.321 |
| P49207 | RPL34 | 60S ribosomal protein L34 | 2.224 |
| Q13561 | DCTN2 | Isoform 2 of dynactin subunit 2 | 2.014 |
| P30046 | DOPD | D-dopachrome decarboxylase | 2.008 |
| O60333 | KIF1B | Isoform 3 of kinesin-like protein KIF1B | 1.988 |
| O75503 | CLN5 | ceroid-lipofuscinosis neuronal protein 5 | 1.919 |
| P61970 | NUTF2 | Nuclear transport factor 2 | 1.908 |
| Q14202 | ZMYM3 | Isoform 1 of zinc finger MYM-type protein 3 | 1.904 |
| P00918 | CA2 | Carbonic anhydrase 2 | 1.904 |
| P05387 | RPLP2 | 60S acidic ribosomal protein P2 | 1.904 |
| P49643 | PRIM2 | Isoform 1 of DNA primase large subunit | 1.888 |
| O75832 | PSD10 | 26S proteasome non-ATPase regulatory subunit 10 | 1.866 |
| P55060 | XPO2 | Isoform 1 of Exportin-2 | 1.853 |
| P62495 | ERF1 | Eukaryotic peptide chain release factor subunit 1 | 1.833 |
| Q9BWJ5 | SF3B5 | Splicing factor 3B subunit 5 | 1.800 |
| P39687 | ANP32A | Acidic leucine-rich nuclear phosphoprotein 32 family member A | 1.798 |
| P41567 | eIF1 | Eukaryotic translation initiation factor 1 | 1.794 |
| P15954 | COX7C | Cytochrome c oxidase subunit 7C, mitochondrial | 1.788 |
| O01805 | ACBP1 | Isoform 1 of Acyl-CoA-binding protein | 1.788 |
| A5D904 | RPS9 | 40S ribosomal protein S9 | 1.771 |
| A6NKZ8 | YI016 | Putative tubulin beta chain-like protein ENSP00000290377 | 1.771 |
| Q96AG4 | LRRC59 | Leucine-rich repeat-containing protein 59 | 1.761 |
| O60762 | DPM1 | Dolichol-phosphate mannosyltransferase | 1.752 |
| O95248 | MTMR5 | Isoform 1 of myotubularin-related protein 5 | 1.726 |
| Q9Y224 | C14orf166 | UPF0568 protein C14orf166 | 1.726 |
| P62851 | RPS25 | 40S ribosomal protein S25 | 1.720 |
| Q9NTK5 | OLA1 | Isoform 1 of Obg-like ATPase 1 | 1.714 |
| H0Y9X3 | PDCD6 | Programmed cell death protein 6 | 1.699 |
| Q9BTT0 | ANP32E | Acidic leucine-rich nuclear phosphoprotein 32 family member E | 1.699 |
| Q9UBX3 | DIC | Isoform 1 of mitochondrial dicarboxylate carrier | 1.667 |
| Q6PD74 | AAGAB | Alpha- and gamma-adaptin-binding protein p34 | 1.667 |
| Q7Z524 | HUMEEP | HUMEEP | 1.667 |
| Q9UL15 | BAG5 | Isoform 1 of BAG family molecular chaperone regulator 5 | 1.654 |
| P61289 | PSME3 | Isoform 1 of Proteasome activator complex subunit 3 | 1.640 |
| Q15008 | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 1.640 |
| Q7L5N1 | CSN6 | COP9 signalosome complex subunit 6 | 1.603 |
| P16104 | H2AFX | Histone H2A.x | 1.602 |
| O15145 | ARPC3 | Actin-related protein 2/3 complex subunit 3 | 1.594 |
| Q99471 | PFDN5 | Prefoldin subunit 5 | 1.594 |
| O43808 | PMP34 | Peroxisomal membrane protein PMP34 | 1.587 |
| Q96IL1 | DIAPH1 | Isoform 1 of protein diaphanous homolog 1 | 1.583 |
| Q00688 | FKBP3 | Peptidyl-prolyl cis-trans isomerase FKBP3 | 1.582 |
| Q86XI2 | CNDG2 | Isoform 2 of condensin-2 complex subunit G2 | 1.578 |
| P99999 | CYC | Cytochrome c | 1.571 |
| P62805 | HIST4H4 | Histone H4 | 1.571 |
| P62913 | RPL11 | Isoform 1 of 60S ribosomal protein L11 | 1.571 |
| O15347 | HMGB3 | High mobility group protein B3 | 1.571 |
| A5D7K0 | BLVRA | Biliverdin reductase A | 1.565 |
| Q12125 | GET4 | Isoform 2 of Golgi to ER traffic protein 4 homolog | 1.552 |
| P35659 | DEK | Protein DEK | 1.547 |
| Q96CD2 | PPCDC | Isoform 1 of Phosphopantothenoylcysteine decarboxylase | 1.547 |
| P31949 | S100-A11 | Protein S100-A11 | 1.541 |
| Q96IX5 | USMG5 | Up-regulated during skeletal muscle growth protein 5 | 1.539 |
| P62330 | ARF6 | ADP-ribosylation factor 6 | 1.529 |
| P58546 | MTPN | Myotrophin | 1.523 |
| Q53H12 | AGK | Isoform 1 of acylglycerol kinase, mitochondrial | 1.523 |
| Q13084 | MRPL28 | 39S ribosomal protein L28, mitochondrial | 1.511 |
| Q9C005 | DPY30 | Protein dpy-30 homolog | 1.508 |
| P62277 | RPS13 | 40S ribosomal protein S13 | 1.504 |
| Q99986 | VRK1 | Serine/threonine-protein kinase VRK1 | 1.504 |
| B4DJ38 | B4DJ38 | cDNA FLJ56092, highly similar to pentatricopeptide repeat protein 1 | 1.504 |
| Q9Y5S9 | RBM8A | Isoform 1 of RNA-binding protein 8A | 1.503 |
| Q92973 | TNPO1 | Isoform 1 of transportin-1 | 1.503 |
| Q5BJH1 | PSAP | Isoform Sap-mu-0 of proactivator polypeptide | 1.503 |
| P36969 | GPX4 | phospholipid hydroperoxide glutathione peroxidase | 1.500 |
Table 2. Forty-six proteins detected at lower levels (>1.5-fold) in cells co-expressing human indoleamine 2,3-dioxygenase 1 (hIDO1) and hIDO2 than in cells expressing hIDO1 only.
| Fold changes | |||
|---|---|---|---|
| Accession no. | Symbol | Protein names | hIDO1-2 vs hIDO1 |
| Q15427 | SF3B4 | Splicing factor 3B subunit 4 | 2.61 |
| P21333 | FLNA | Isoform 2 of Filamin-A | 2.538 |
| P05204 | HMG-17 | Non-histone chromosomal protein HMG-17 | 2.263 |
| Q96KB5 | TOPK | Lymphokine-activated killer T-cell-originated protein kinase | 2.144 |
| Q92733 | PRCC | Proline-rich protein PRCC | 2.109 |
| P60763 | RAC3 | Ras-related C3 botulinum toxin substrate 3 | 2.021 |
| Q9H2H9 | SLC38A1 | Sodium-coupled neutral amino acid transporter 1 | 2.003 |
| Q8ND24 | RNF214 | RING finger protein 214 | 1.945 |
| P67936 | TPM4 | Isoform 1 of tropomyosin alpha-4 chain | 1.921 |
| P04897 | GNAI2 | Isoform 2 of Guanine nucleotide-binding protein G(i) subunit alpha-2 | 1.866 |
| P49069 | CAMLG | Calcium signal-modulating cyclophilin ligand | 1.775 |
| Q14315 | FLNC | Isoform 1 of Filamin-C | 1.769 |
| Q9H7N4 | SFR19 | Splicing factor, arginine/serine-rich 19 | 1.753 |
| Q8NDF8 | PAPD5 | PAP-associated domain-containing protein 5 isoform b | 1.732 |
| P05114 | HMG-14 | Non-histone chromosomal protein HMG-14 | 1.727 |
| O43157 | PLXNB1 | Isoform 2 of Plexin-B1 | 1.685 |
| Q14353 | GAMT | Guanidinoacetate N-methyltransferase | 1.662 |
| O43542 | XRCC3 | DNA repair protein XRCC3 | 1.655 |
| A4D105 | RPA3 | Replication protein A 14 kDa subunit | 1.65 |
| Q8NEY8 | PPHLN1 | Isoform 1 of Periphilin-1 | 1.65 |
| Q9Y679 | AUP1 | Isoform short of ancient ubiquitous protein 1 | 1.644 |
| P37802 | TAGLN2 | Transgelin-2 | 1.625 |
| Q9BZX2 | UCK2 | Isoform 1 of Uridine-cytidine kinase 2 | 1.625 |
| P46087 | NOP2 | Putative ribosomal RNA methyltransferase 1 | 1.615 |
| Q6VN20 | RANBP10 | Ran-binding protein 10 | 1.615 |
| Q9Y2R4 | DDX52 | Probable ATP-dependent RNA helicase DDX52 | 1.614 |
| Q9H061 | TMEM126A | Transmembrane protein 126A | 1.61 |
| Q9NZT2 | OGFR | Isoform 1 of opioid growth factor receptor | 1.609 |
| Q9BYB4 | GNB1L | Isoform 1 of guanine nucleotide-binding protein subunit beta-like protein 1 | 1.609 |
| Q92643 | PIGK | GPI-anchor transamidase | 1.604 |
| Q9UPT8 | ZC3H4 | Zinc finger CCCH domain-containing protein 4 | 1.6 |
| Q9UBV2 | SE1L1 | Isoform 1 of protein sel-1 homolog 1 | 1.594 |
| Q9UPN9 | TRIM33 | Isoform alpha of E3 ubiquitin-protein ligase TRIM33 | 1.589 |
| Q9H6R0 | DHX33 | Highly similar to putative ATP-dependent RNA helicase DHX33 | 1.583 |
| O95453 | PARN | Poly(A)-specific ribonuclease PARN | 1.572 |
| P11021 | GRP78 | 78 kDa glucose-regulated protein | 1.571 |
| Q9NXE4 | SMPD4 | Isoform 4 of sphingomyelin phosphodiesterase 4 | 1.542 |
| Q9NRL3 | STRN4 | Striatin-4 | 1.534 |
| Q9NYV4 | CDK12 | Isoform 1 of cyclin-dependent kinase 12 | 1.53 |
| Q13610 | PWP1 | Periodic tryptophan protein 1 homolog | 1.521 |
| Q99436 | PSMB7 | Proteasome subunit beta type-7 | 1.518 |
| P24390 | KDELR1 | ER lumen protein retaining receptor 1 | 1.515 |
| Q6P9B9 | INTS5 | Integrator complex subunit 5 | 1.515 |
| P49821 | NDUFV1 | Isoform 2 of Nadh dehydrogenase (ubiquinone) flavoprotein 3 | 1.513 |
| Q9ULV3 | CIZ1 | Isoform 2 of cip1-interacting zinc finger protein | 1.5 |
| O94966 | USP19 | Isoform 1 of ubiquitin carboxyl-terminal hydrolase 19 | 1.5 |
Co-expression of hIDO2 alleviate cell growth arrest and death
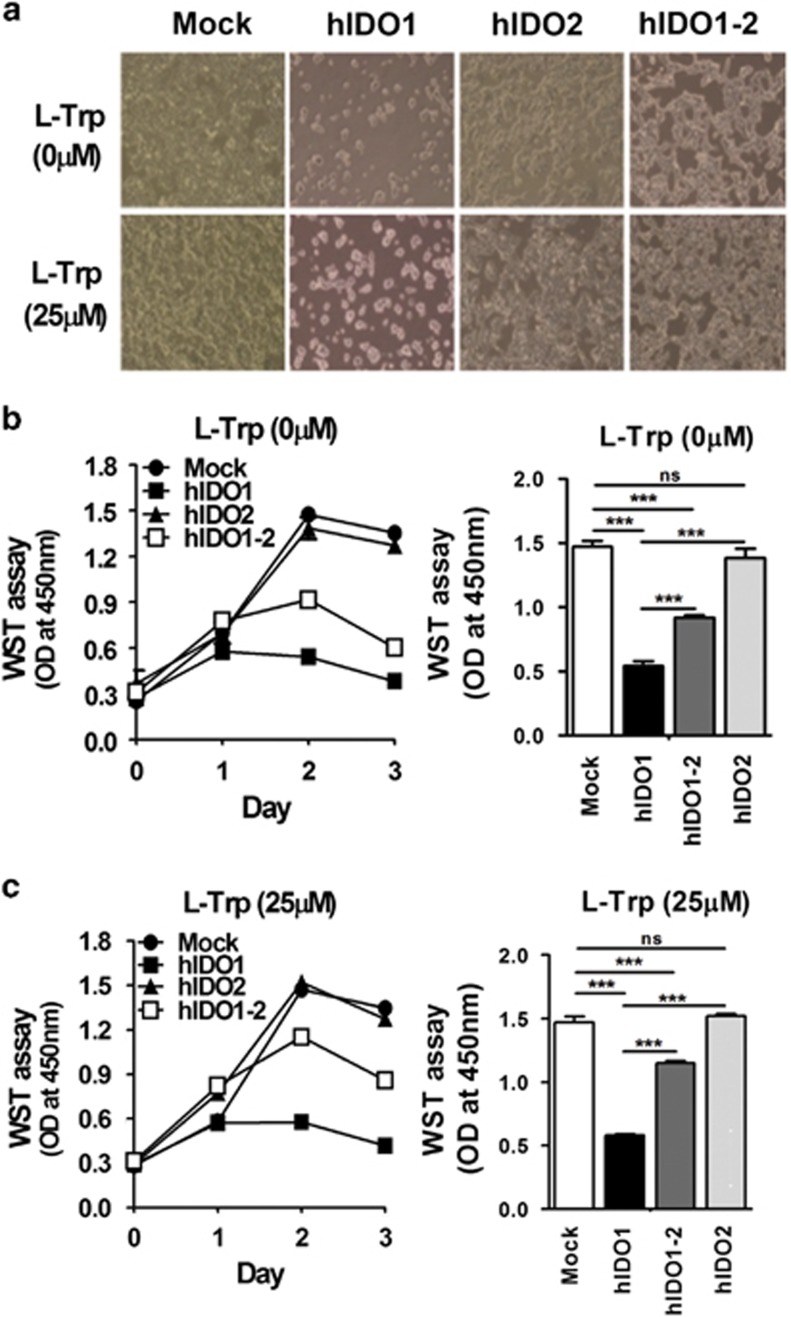
The proteomic analysis indicated that the expression levels of negative regulators of cell death were enhanced in cells co-expressing hIDO1 and hIDO2. Therefore, we postulated that high catalytic activity of hIDO1 would increase susceptibility to cell death (likely because of rapid depletion of tryptophan in the culture medium). Moreover, the reduced hIDO1 activity because of hIDO2 co-expression would rescue the cells from death. To test this, we compared the growth and death of cells co-expressing hIDO1 and hIDO2 and cells expressing hIDO1 only. Cell proliferation and morphological changes were examined under a microscope (Figure 3a); proliferation was also assessed using the quantitative colorimetric WST assay (Figure 3b). Microscopic observations and the WST assay showed that the proliferation and growth of cells over-expressing only hIDO1 were limited, whereas cells over-expressing hIDO2 and mock-transfected cells continued to proliferate. Interestingly, cells that over-expressed hIDO1 together with hIDO2 showed two-fold greater proliferation and enhanced survival compared to cells overexpressing only hIDO1. Supplementation of the culture medium with L-Trp increased the difference in survival between the two cell lines (Figure 3c). These results indicate that hIDO1 overexpression rapidly induced cell death, which was alleviated by hIDO2 co-expression because of a reduction in the catalytic activity of hIDO1.
Figure 3.
Alleviation of cell growth arrest and death by co-expression of hIDO2. (a) Morphological changes in hIDO1-, hIDO2- or hIDO1-2-expressing HEK293 stable cells in the absence or presence of 25 μM L-Trp were detected on day 3 during the cell proliferation assay ( × 100 magnification). Cell proliferation was measured by WST colorimetric assay during the 3-day culture period in the (b) absence or (c) presence of 25 μM L-Trp. Absorbance at 450 nm was measured using a microplate reader. Cell proliferation values on day 2 were subjected to statistical analysis. All experiments were performed independently three times. ns, not significant, ***P<0.001.
hIDO1 functional activity is modulated via the hIDO2-heme-binding site
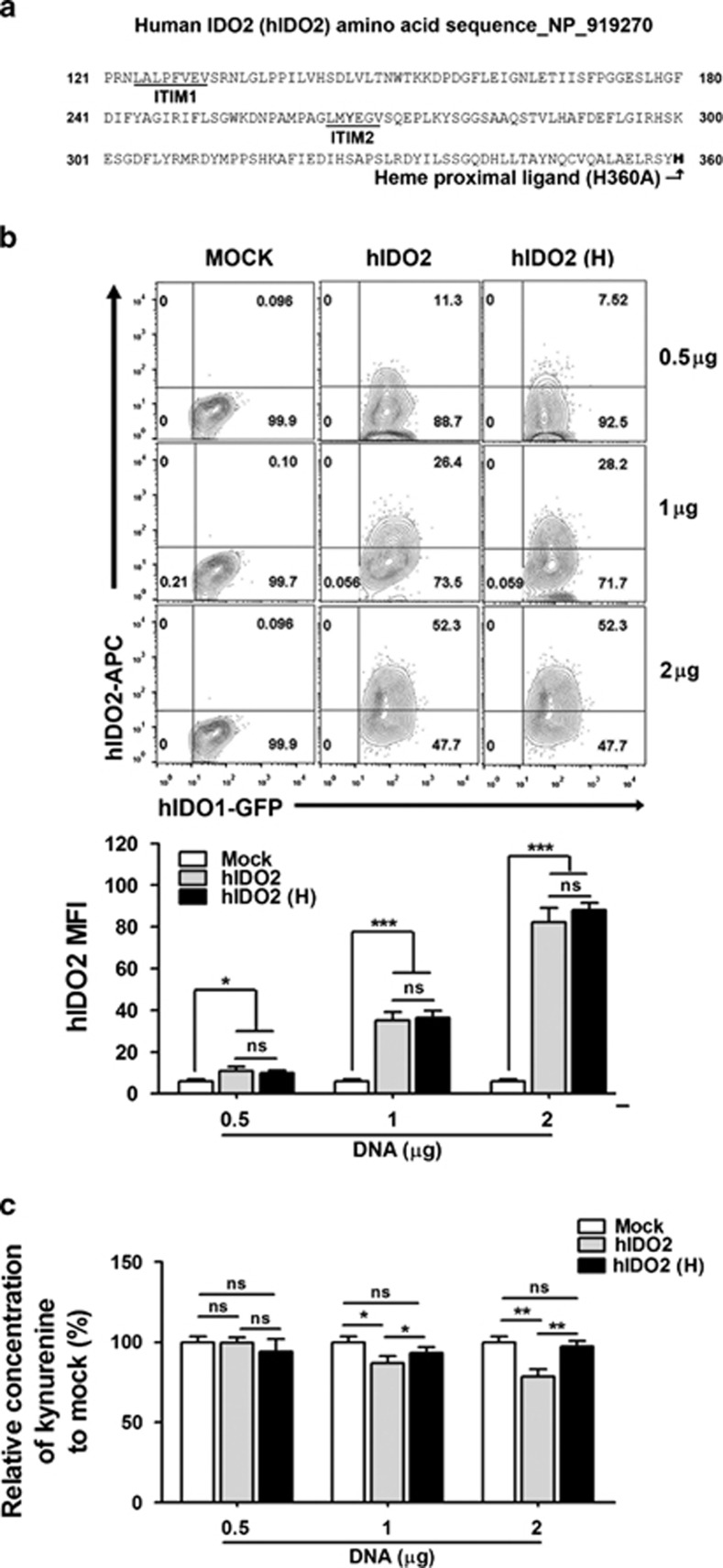
Based on our results, we hypothesized that the negative regulation of hIDO1 activity by hIDO2 co-expression may be because of competition rather than any specific hIDO2 functional activity. hIDO2 has two immunoreceptor tyrosine-based inhibition (ITIM) motifs, which play roles in immune cell signal transduction;34,35 and a heme-binding site, which is essential for hIDO1 catalytic activity.34,36,37 These two sites correspond to regions in the IDO2 sequence, in which mutations or alternative splicing occur.2 Therefore, we postulated that hIDO2 suppresses hIDO1 catalytic activity through heme-binding site-based competition with hIDO1. To test this, we generated a hIDO2 mutant, in which the histidine residue at position 360 was substituted for alanine (H360A) to abrogate hIDO2 heme-binding activity. Then, we examined the effects of co-expression of this hIDO2 mutant with hIDO1 (Figure 4a). A DNA construct harboring the HA-tagged hIDO2 mutant was transfected into stable hIDO1-expressing cell lines at various doses, and kynurenine production by cells expressing H360A was compared with that of control cells transfected with wild-type hIDO2 and mock-transfected control cells. Flow cytometry analysis confirmed dose-dependent expression of the wide type or mutant form of hIDO2 after transfection; however, the levels were consistent among the hIDO2 forms transfected with an identical dose (Figure 4b). The hIDO2 did not exert suppressive effects when H360A (hIDO2 heme-binding mutant) was co-expressed with hIDO1, as kynurenine production increased significantly compared with that in hIDO2-transfected control cells in a DNA-dose-dependent manner (Figure 4c). These results suggest that negative regulation by hIDO2 is based on competition with hIDO1 for heme binding.
Figure 4.
Human indoleamine 2,3-dioxygenase 2 (hIDO2) modulates hIDO1 functional activity through the heme-binding histidine residue 360 in a dose-dependent manner. (a) The hIDO2 amino acid sequence obtained from the National Center for Biotechnology Information (GenBank accession number: NP_919270) annotated with the putative immunoreceptor tyrosine-based inhibition ITIM1 and ITIM2 motifs, and heme proximal ligand. The hIDO2 (H) mutant was constructed by substituting histidine 360 for alanine (H360A). Mutated residues are shown in bold. (b) Flow cytometric analysis of protein expression after transient transfection with hIDO2 or hIDO2 (H) constructs into the green florescent protein-expressing hIDO1 stable cell line. Plasmid DNAs (0.5, 1, and 2 μg) were subjected to transfection using polyethyleneimine. hIDO2 and hIDO2 (H) protein levels were compared after gating green florescent protein-positive hIDO1-expressing cells stained with an anti-hemagglutinin primary antibody and allophycocyanin-conjugated secondary antibodies 48 h after transfection. The mean hIDO2 fluorescence intensity values of all transfected cells were plotted. (c) Kynurenine production was measured using Ehrlich's reagent and calculated compared to the mock-transfectant at 48 h posttransfection. Empty vector was used as the mock control. ns, not significant, *P<0.05, **P<0.01, ***P<0.001.
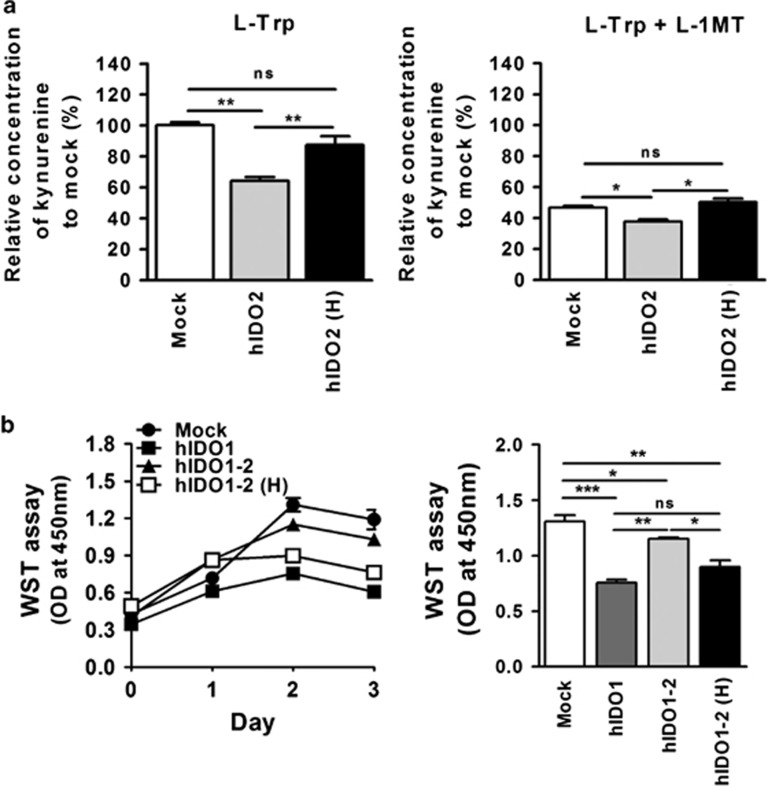
Effect of the IDO inhibitor L-1MT on hIDO1 functional activity upon co-expression with hIDO2 or hIDO2 (H) genes
Next, we generated stable cell lines from hIDO1-expressing cells that stably co-expressed the hIDO2 wild-type or the H360A mutant. These cells were subjected to kynurenine assay following growth in medium supplemented with L-Trp. Cell lines expressing hIDO2 at similar levels were selected after flow cytometric analysis of levels of HA-tagged, wild-type and mutant hIDO2. Co-expression of the H360A mutant did not suppress hIDO1 catalytic activity to the same degree as did co-expression of wild-type IDO2 (Figure 5a). Although L-1MT co-treatment significantly reduced hIDO1 activity, such trends were still maintained (Figure 5a). Therefore, the suppressive effects of hIDO2 were focused on hIDO1 catalytic activity.
Figure 5.
Effects of the indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor L-1MT on human (h) IDO1 activity in cells co-expressing hIDO2 (H) mutants. (a) Kynurenine production was measured in culture supernatants of stable hIDO1-expressing cells transfected with 2 μg hIDO2 or hIDO2 (H) mutant DNAs. Transfected cells were incubated for 24 h with complete medium including 100 μM L-Trp in the absence or presence of 100 μM L-1MT. (b) Proliferation of stable hIDO1-, hIDO1-2- and hIDO1-2 (H)-expressing HEK293 cells was measured by WST assay during the 3-day culture period. Cell proliferation values on day 2 were subjected to statistical analysis. Absorbance at 450 nm was measured using a microplate reader with the Gen5 software. ns, not significant, *P<0.05, **P<0.01, ***P<0.001.
Next, we investigated whether the H360A mutant could restore the reduction in growth rate caused by hIDO1-mediated tryptophan depletion. WST proliferation assays demonstrated that co-expression of the H360A mutant did not negatively affect hIDO1-mediated cell death compared with co-expression of wild-type hIDO2 during the 3-day culture period. This resulted in early arrest of the growth of cells co-expressing the H360A mutant and hIDO1, as well as cells expressing hIDO1 only (Figure 5b). These results confirm that hIDO2 negatively regulates hIDO1 catalytic activity via its heme-binding domain.
Discussion
Our findings demonstrated that hIDO2 functioned as a negative regulator of hIDO1 catalytic activity, an effect mediated by its heme-binding activity.
In contrast to the established biological role of IDO1 in tryptophan catabolism and as an immune modulator implicated in tumors and inflammatory diseases, the biological relevance of IDO2 is unknown. Here, we demonstrated that hIDO2 suppressed the catalytic activity of hIDO1. Kynurenine assays revealed that IDO2 had negligible catalytic activity, in agreement with previous reports that IDO2 is inactive under physiological conditions and that its catalytic activity corresponds to only 3–5% of that of IDO1.16,38 Although IDO2 is a candidate target of D-1MT, which has been evaluated in clinical trials for treatment of cancers,39 the physical evidence supporting this notion is reliant on inhibition of the already very-low catalytic activity of IDO2. We did not detect inhibition by D-1MT of hIDO1 catalytic activity (data not shown), consistent with a previous report that IDO1 is not a D-1MT target.19 We could not determine whether hIDO2 catalytic activity is the D-1MT target because hIDO2 catalytic activity was negligible. These results suggest that IDO2 does not play a role as an active catalytic enzyme, and suggest that the anticancer effect of D-1MT may be dependent on negative regulation of IDO1 catalytic activity by IDO2, as IDO2 is a target of D-1MT.
Co-expression of IDO1 and IDO2 has been detected in tumor cells, dendritic cells and the epididymis.5,9 The IDO2 protein is highly upregulated in the epididymis of IDO1-deficient mice, suggesting that it contributes to immune modulation or maintenance of IDO1 tryptophan metabolism5. IDO2 upregulation in IDO1-deficient mice suggests a functional interaction between these two molecules. Here, we provide evidence that IDO2 co-expression suppresses IDO1 catalytic activity, and that this effect is dependent on the presence of an intact heme-binding site. This finding indicates that the IDO2 heme-binding site is the active site, suggesting the presence of competition for heme-binding upon co-expression of IDO1 and IDO2. Therefore, although IDO2 is inactive during tryptophan metabolism, a mutation in the heme-binding site or a change in its expression level could regulate IDO1 catalytic activity, even if the IDO1 expression level is unaffected.
In addition to their catalytic activity, IDO1 and IDO2 have signaling activities because of their two putative ITIM-motifs.34,35 The ITIM-motif is responsible for transforming growth factor (TGF-β)-mediated signaling, and the TGF-β-IDO axis activates the noncanonical nuclear factor-κB pathway through SHP.34 Signaling activity regulates IDO expression. In this study, mutation of the active ITIM-motif at position 26634 resulted in maintenance of the heme-binding-mediated negative regulation by hIDO2 of hIDO1 catalytic activity (data not shown). Therefore, the hIDO2-ITIM-motif may be irrelevant to the catalytic function and suppression of hIDO1 catalytic activity by hIDO2.
Our data demonstrate that hIDO2 plays a role as a negative modulator of hIDO1 activity, rather than as a catalytic enzyme. This is further supported by the independent and separately grouped protein profiles of hIDO2- and hIDO1-expressing cells. Although we did not perform a functional assessment of these molecules in immunological tolerance in this study, IDO1-dependent generation of T-regulatory cells has been reported to be defective in IDO2-deficient mice,23 supporting our suggestion of a functional interaction between IDO1 and IDO2. However, the fact that IDO2 deficiency reduces inflammatory cytokine production after immune stimulation suggests a non-redundant function.23 In addition, IDO1- and IDO2-deficient mice showed different susceptibilities to antibody-mediated autoimmunity.22 Therefore, IDO2 might play a separate role. The proteomes of cells expressing only hIDO2 showed few changes compared with those of mock-transfectants, suggesting that hIDO2 rarely functions under nonstimulatory normal conditions (Figure 2a). Therefore, it is possible that hIDO2 expression and function are induced by environmental changes such as inflammation or increased needs for tryptophan catabolism. One possible scenario is that hIDO2, when over-expressed in the presence of hIDO1, reduces hIDO1 catalytic activity through competition for heme-binding, but may play independent roles related to immune modulation through its ITIM-motif under activating conditions.
In summary, our results suggest a novel role for hIDO2 as a negative regulator of hIDO1 catalytic activity, an effect associated with the heme-binding site. These results provide information useful for both understanding the role of IDO2 and the development of IDO1-targeted therapeutic strategies for cancers and immunological diseases.
Acknowledgments
This study was supported by a grant from the Korea Healthcare Technology R &D project, Ministry of Health, Welfare and Family Affairs of Korea (HT 12C0223).
The authors declare no conflict of interest.
References
- Murray MF. The human indoleamine 2,3-dioxygenase gene and related human genes. Curr Drug Metab. 2007;8:197–200. doi: 10.2174/138920007780362509. [DOI] [PubMed] [Google Scholar]
- Metz R, Duhadaway JB, Kamasani U, Laury-Kleintop L, Muller AJ, Prendergast GC. Novel tryptophan catabolic enzyme IDO2 is the preferred biochemical target of the antitumor indoleamine 2,3-dioxygenase inhibitory compound D-1-methyl-tryptophan. Cancer Res. 2007;67:7082–7087. doi: 10.1158/0008-5472.CAN-07-1872. [DOI] [PubMed] [Google Scholar]
- Munn DH, Sharma MD, Lee JR, Jhaver KG, Johnson TS, Keskin DB, et al. Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science. 2002;297:1867–1870. doi: 10.1126/science.1073514. [DOI] [PubMed] [Google Scholar]
- Prendergast GC. Immune escape as a fundamental trait of cancer: focus on IDO. Oncogene. 2008;27:3889–3900. doi: 10.1038/onc.2008.35. [DOI] [PubMed] [Google Scholar]
- Fukunaga M, Yamamoto Y, Kawasoe M, Arioka Y, Murakami Y, Hoshi M, et al. Studies on tissue and cellular distribution of indoleamine 2,3-dioxygenase 2: the absence of IDO1 upregulates IDO2 expression in the epididymis. J Histochem Cytochem. 2012;60:854–860. doi: 10.1369/0022155412458926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Nomiyama S, Hirata F, Hayaishi O. Indoleamine 2,3-dioxygenase. Purification and some properties. J Biol Chem. 1978;253:4700–4706. [PubMed] [Google Scholar]
- Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- Tone S, Takikawa O, Habara-Ohkubo A, Kadoya A, Yoshida R, Kido R. Primary structure of human indoleamine 2,3-dioxygenase deduced from the nucleotide sequence of its cDNA. Nucleic Acids Res. 1990;18:367. doi: 10.1093/nar/18.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhu BT. Indoleamine 2,3-dioxygenase tissue distribution and cellular localization in mice: implications for its biological functions. J Histochem Cytochem. 2010;58:17–28. doi: 10.1369/jhc.2009.953604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AM, Driussi C, Turner V, Takikawa O, Hunt NH. Tissue distribution of indoleamine 2,3-dioxygenase in normal and malaria-infected tissue. Redox Rep. 2000;5:112–115. doi: 10.1179/135100000101535384. [DOI] [PubMed] [Google Scholar]
- Munn DH. Indoleamine 2,3-dioxygenase, tumor-induced tolerance and counter-regulation. Curr Opin Immunol. 2006;18:220–225. doi: 10.1016/j.coi.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Moretti S, Menicali E, Voce P, Morelli S, Cantarelli S, Sponziello M, et al. Indoleamine 2,3-dioxygenase 1 (IDO1) is up-regulated in thyroid carcinoma and drives the development of an immunosuppressant tumor microenvironment. J Clin Endocrinol Metab. 2014;99:E832–E840. doi: 10.1210/jc.2013-3351. [DOI] [PubMed] [Google Scholar]
- Friberg M, Jennings R, Alsarraj M, Dessureault S, Cantor A, Extermann M, et al. Indoleamine 2,3-dioxygenase contributes to tumor cell evasion of T cell-mediated rejection. Int J Cancer. 2002;101:151–155. doi: 10.1002/ijc.10645. [DOI] [PubMed] [Google Scholar]
- Thaker AI, Rao MS, Bishnupuri KS, Kerr TA, Foster L, Marinshaw JM, et al. IDO1 metabolites activate beta-catenin signaling to promote cancer cell proliferation and colon tumorigenesis in mice. Gastroenterology. 2013;145 (416-425:e411–e414. doi: 10.1053/j.gastro.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ino K, Yoshida N, Kajiyama H, Shibata K, Yamamoto E, Kidokoro K, et al. Indoleamine 2,3-dioxygenase is a novel prognostic indicator for endometrial cancer. Br J Cancer. 2006;95:1555–1561. doi: 10.1038/sj.bjc.6603477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HJ, Sanchez-Perez A, Weiser S, Austin CJ, Astelbauer F, Miu J, et al. Characterization of an indoleamine 2,3-dioxygenase-like protein found in humans and mice. Gene. 2007;396:203–213. doi: 10.1016/j.gene.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Witkiewicz AK, Costantino CL, Metz R, Muller AJ, Prendergast GC, Yeo CJ, et al. Genotyping and expression analysis of IDO2 in human pancreatic cancer: a novel, active target. J Am Coll Surg. 2009;208:781–787. doi: 10.1016/j.jamcollsurg.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lob S, Konigsrainer A, Zieker D, Brücher BL, Rammensee HG, Opelz G, et al. IDO1 and IDO2 are expressed in human tumors: levo- but not dextro-1-methyl tryptophan inhibits tryptophan catabolism. Cancer Immunol Immunother. 2009;58:153–157. doi: 10.1007/s00262-008-0513-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lob S, Konigsrainer A, Schafer R, Rammensee HG, Opelz G, Terness P. Levo- but not dextro-1-methyl tryptophan abrogates the IDO activity of human dendritic cells. Blood. 2008;111:2152–2154. doi: 10.1182/blood-2007-10-116111. [DOI] [PubMed] [Google Scholar]
- Liu X, Shin N, Koblish HK, Yang G, Wang Q, Wang K, et al. Selective inhibition of IDO1 effectively regulates mediators of antitumor immunity. Blood. 2010;115:3520–3530. doi: 10.1182/blood-2009-09-246124. [DOI] [PubMed] [Google Scholar]
- Qian F, Villella J, Wallace PK, Mhawech-Fauceglia P, Tario JD, Jr, Andrews C, et al. Efficacy of levo-1-methyl tryptophan and dextro-1-methyl tryptophan in reversing indoleamine-2,3-dioxygenase-mediated arrest of T-cell proliferation in human epithelial ovarian cancer. Cancer Res. 2009;69:5498–5504. doi: 10.1158/0008-5472.CAN-08-2106. [DOI] [PubMed] [Google Scholar]
- Merlo LM, Pigott E, DuHadaway JB, Grabler S, Metz R, Prendergast GC, et al. IDO2 is a critical mediator of autoantibody production and inflammatory pathogenesis in a mouse model of autoimmune arthritis. J Immunol. 2014;192:2082–2090. doi: 10.4049/jimmunol.1303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz R, Smith C, DuHadaway JB, Chandler P, Baban B, Merlo LM, et al. IDO2 is critical for IDO1-mediated T-cell regulation and exerts a non-redundant function in inflammation. Int Immunol. 2014;26:357–367. doi: 10.1093/intimm/dxt073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samikkannu T, Saiyed ZM, Rao KV, Babu DK, Rodriguez JW, Papuashvili MN, et al. Differential regulation of indoleamine-2,3-dioxygenase (IDO) by HIV type 1 clade B and C Tat protein. AIDS Res Hum Retroviruses. 2009;25:329–335. doi: 10.1089/aid.2008.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Arai M, Imai H, Koumura T, Yoshida M, Emoto K, Umeda M, et al. Mitochondrial phospholipid hydroperoxide glutathione peroxidase plays a major role in preventing oxidative injury to cells. J Biol Chem. 1999;274:4924–4933. doi: 10.1074/jbc.274.8.4924. [DOI] [PubMed] [Google Scholar]
- Anderson KM, Berrebi-Bertrand I, Kirkpatrick RB, McQueney MS, Underwood DC, Rouanet S, et al. cDNA sequence and characterization of the gene that encodes human myotrophin/V-1 protein, a mediator of cardiac hypertrophy. J Mol Cell Cardiol. 1999;31:705–719. doi: 10.1006/jmcc.1998.0903. [DOI] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Nemeth MJ, Cline AP, Anderson SM, Garrett-Beal LJ, Bodine DM. Hmgb3 deficiency deregulates proliferation and differentiation of common lymphoid and myeloid progenitors. Blood. 2005;105:627–634. doi: 10.1182/blood-2004-07-2551. [DOI] [PubMed] [Google Scholar]
- Broxmeyer HE, Kappes F, Mor-Vaknin N, Legendre M, Kinzfogl J, Cooper S, et al. DEK regulates hematopoietic stem engraftment and progenitor cell proliferation. Stem Cells Dev. 2012;21:1449–1454. doi: 10.1089/scd.2011.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang R. Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBO J. 2008;27:852–864. doi: 10.1038/emboj.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann U, Brinkmann E, Gallo M, Pastan I. Cloning and characterization of a cellular apoptosis susceptibility gene, the human homologue to the yeast chromosome segregation gene CSE1. Proc Natl Acad Sci USA. 1995;92:10427–10431. doi: 10.1073/pnas.92.22.10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotta MT, Orabona C, Volpi C, Vacca C, Belladonna ML, Bianchi R, et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat Immunol. 2011;12:870–878. doi: 10.1038/ni.2077. [DOI] [PubMed] [Google Scholar]
- Orabona C, Pallotta MT, Grohmann U. Different partners, opposite outcomes: a new perspective of the immunobiology of indoleamine 2,3-dioxygenase. Mol Med. 2012;18:834–842. doi: 10.2119/molmed.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohn TK, Takikawa O, Truscott RJ, Walker MJ. Asp274 and his346 are essential for heme binding and catalytic function of human indoleamine 2,3-dioxygenase. J Biol Chem. 2003;278:29525–29531. doi: 10.1074/jbc.M301700200. [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Oda S, Otsuki T, Hino T, Yoshida T, Shiro Y. Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase. Proc Natl Acad Sci USA. 2006;103:2611–2616. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takikawa O, Kuroiwa T, Yamazaki F, Kido R. Mechanism of interferon-gamma action. Characterization of indoleamine 2,3-dioxygenase in cultured human cells induced by interferon-gamma and evaluation of the enzyme-mediated tryptophan degradation in its anticellular activity. J Biol Chem. 1988;263:2041–2048. [PubMed] [Google Scholar]
- Hou DY, Muller AJ, Sharma MD, DuHadaway J, Banerjee T, Johnson M, et al. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res. 2007;67:792–801. doi: 10.1158/0008-5472.CAN-06-2925. [DOI] [PubMed] [Google Scholar]