Abstract
Mitochondrial morphology is dynamically regulated by forming small, fragmented units or interconnected networks, and this is a pivotal process that is used to maintain mitochondrial homeostasis. Although dysregulation of mitochondrial dynamics is related to the pathogenesis of several human diseases, its molecular mechanism is not fully elucidated. In this study, we demonstrate the potential role of miR-27 in the regulation of mitochondrial dynamics. Mitochondrial fission factor (MFF) mRNA is a direct target of miR-27, whose ectopic expression decreases MFF expression through binding to its 3′-untranslated region. Expression of miR-27 results in the elongation of mitochondria as well as an increased mitochondrial membrane potential and mitochondrial ATP level. Our results suggest that miR-27 is a novel regulator affecting morphological mitochondrial changes by targeting MFF.
Introduction
MicroRNAs (miRNAs) are a type of small noncoding RNA that regulates numerous cellular activities by suppressing gene expression.1, 2, 3 These RNAs are involved in various cellular processes, including cellular proliferation, differentiation, death and development via imperfect base pairing with target mRNAs.4, 5, 6 Aberrant expression of miRNAs has been associated with many pathological conditions, such as malignancies and metabolic disorders.7, 8, 9
Mitochondria continuously change their morphology by fusing or dividing in response to the different physiological needs of the cells,10 and several studies have shown that the tight regulation of mitochondrial morphology is critical for the maintenance of mitochondrial structures and functions affecting cell fate.11, 12, 13 Mitochondrial dynamics is governed by several core proteins, including mitofusin 1 (MFN1), mitofusin 2 (MFN2), dynamin-related protein 1 (DRP1), mitochondrial fission factor (MFF), mitochondrial fission 1, mitochondrial dynamics 51 and optic atrophy protein 1 (OPA1).14, 15, 16, 17, 18 Although disruption of the dynamic mitochondrial balance is known to be related to several physiological and pathological conditions such as aging, apoptosis, cancer, neurodegenerative diseases and diabetes, the regulatory mechanisms involved in mitochondrial dynamics remain largely unknown.19, 20, 21 Recently, several studies have indicated the involvement of miRNAs in the regulation of mitochondrial dynamics. For example, miR-499 and miR-30 regulate the mitochondrial fission machinery by directly targeting DRP1;22, 23 miR-484 and miR-761 are responsible for regulating mitochondrial fission 1 and MFF, respectively;24, 25 miR-140 and miR-19b negatively regulate mitochondrial fusion by downregulating MFN1;26, 27 and miR-106b is responsible for mitochondrial dysfunction by targeting MFN2.28
Results from this study reveal that miR-27 functions as a novel factor regulating mitochondrial dynamics by suppressing MFF expression. We show that miR-27 suppresses the association of MFF mRNA with polysomes via its 3′-untranslated region (UTR). Ectopic expression of the miR-27 precursor resulted in mitochondrial fusion, thereby increasing the mitochondrial membrane potential as well as the mitochondrial ATP level. Taken together, our data provide experimental evidence, suggesting that miR-27 is involved in negatively regulating mitochondrial fission by directly targeting MFF.
Materials and methods
Cell culture, transfection, plasmids and miRNAs
Human CHANG liver cells stably overexpressing mitochondria-targeted yellow fluorescent protein (mtYFP) were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and antibiotics. Enhanced green fluorescent protein (EGFP) reporters were cloned by inserting 3'-UTR fragments from the MFF mRNA into pEGFP-C1 (BD Bioscience, San Jose, CA, USA). A mutant reporter lacking the binding sites for the miR-27 seed region was generated by site-directed mutagenesis using the KOD-Plus-Mutagenesis Kit (Toyobo, Osaka, Japan). The plasmids, miRNAs [control miRNA (Ctrl)], as well as the precursor and an inhibitor of miR-27 (Bioneer, Daejeon, Korea) were transiently transfected using Lipofectamine 2000 (Invitrogen).
Western blot analysis
Whole-cell lysates were prepared using RIPA buffer (10 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA and 0.1% sodium dodecyl sulfate) containing 1 × protease inhibitor cocktail (Roche, Basel, Switzerland), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore, Darmstadt, Germany). The membranes were incubated with primary antibodies against MFF (Abcam, Cambridge, MA, USA), GFP (Santa Cruz Biotech, Santa Cruz, CA, USA) or β-actin (Abcam) and then further incubated with the appropriate secondary antibodies conjugated to horseradish peroxidase (Santa Cruz Biotech). The signals were detected using enhanced luminescence (Bio-Rad, Hercules, CA, USA).
RNA analysis
Total RNA was prepared from whole-cell lysates using Trizol (Invitrogen). After reverse transcription (RT) using random hexamers and reverse transcriptase (Toyobo), the mRNA abundance was assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis using the SYBR green PCR master mix (Kapa Biosystems, Wilmington, MA, USA) and gene-specific primer sets (Table 1). RT-qPCR analysis was performed using the StepOne Plus system (Life Technologies, Waltham, MA, USA).
Table 1. Primer list used in this study.
| Primer name | Sequences |
|---|---|
| Human MFF-3U-F | 5′-AAAAAGATCTTAA CACGTCTGAGCA-3′ |
| Human MFF-3U-R | 5′-AAAAGGTACCTCTGGCACCAGA-3′ |
| Human MFF-3UM-F | 5′-AAGTGTGACACAAAGAAAAACAATTATT-3′ |
| Human MFF-3UM-R | 5′-TCTTTGTGTCACATTTTCTGAATCAAT-3′ |
| Human MFF-F | 5′-CACCACCTCGTGTACTTACGC-3′ |
| Human MFF-R | 5′-GTCTGCCAACTGCTCGGATTT-3′ |
| Human GAPDH-F | 5′-TGCACCACCAACTGCTTAGC-3′ |
| Human GAPDH-R | 5′-GGCATGGACTGTGGTCATGAG-3′ |
Abbreviations: F, forward; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MFF, mitochondrial fission factor; R, reverse.
Fluorescence microscopy
To visualize the changes in mitochondrial morphology, YFP signals from CHANG liver cells stably expressing mtYFP or cells incubated with 100 nM MitoTracker Red CMXRos (Invitrogen) for 30 min at 37 °C were observed under the fluorescence microscope. Images were acquired using an Axiovertcam mRM camera attached to an Axiovert 200M microscope (Carl Zeiss, Oberkochen, Germany).
Measurement of the mitochondrial membrane potential and ATP level
The mitochondrial membrane potential was measured using the JC1 Mitochondrial Membrane Potential Assay Kit (Abcam). Cells were incubated with tetraethyl benzimidazoly carbocyanine iodide (JC-1) staining solution (Abcam) for 10 min at 37 °C in the dark, and the fluorescence was measured at 535 nm (excitation)/590 nm (emission) using a Victor3 fluorescent plate reader (Perkin-Elmer, Waltham, MA, USA).
The cellular mitochondrial ATP level was measured using the Mitochondrial ToxGlo assay (Promega, Madison, WI, USA) according to the manufacturer's procedure. Briefly, CHANG liver cells were harvested and suspended by pipetting until they were evenly dispersed. The resuspended cells were then incubated with galactose-containing media at 37 °C for 90 min and then further incubated with ATP detection reagent. Luminescence was measured using a Victor3 plate reader (Perkin-Elmer).
Polysome analysis
Forty-eight hours after transfection of the precursor of miR-27 or the inhibitor of miR-27 with the control miRNA, CHANG liver cells were preincubated with cycloheximide (100 μg ml−1, 15 min) and then lysed with polysome extraction buffer containing 20 mM Tris-HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.5% (v v−1) Nonidet P-40, 1 × protease inhibitor cocktail and RNase inhibitor, followed by centrifugation at 10 000 g for 10 min. The lysates were further fractionated by ultracentrifugation through linear sucrose gradients as described in the previous studies.29, 30 RNAs from each fraction were isolated, and cDNA was synthesized as described above. The relative levels of MFF and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNAs were analyzed by RT-qPCR using specific primer sets.
Results
Identification of miRNAs targeting MFF
Although MFF is one of the critical factors regulating mitochondrial morphological changes, the mechanisms involved in the regulation of MFF expression are not fully understood.16, 17, 25, 31 To identify miRNAs affecting the morphological changes of mitochondria via MFF, we analyzed the MFF mRNA using TargetScan 4.2 and microrna.org. Human MFF mRNA (NM_020194) is composed of a 266-bp 5′-UTR, the MFF coding sequence, and an ~700-bp 3′-UTR, which harbors miRNA binding sites, as shown in Figure 1a. In silico analysis revealed that several miRNAs can potentially interact with the MFF mRNA 3′-UTR. Among these miRNAs, we examined the effects of miR-141, miR-27b and miR-200c on MFF expression using western blot analysis (Figure 1b). miR-27b and miR-200c resulted in a significant downregulation of MFF expression, whereas miR-141 did not alter the MFF level in CHANG liver cells, despite the positive prediction of binding between miR-141 and the MFF mRNA. Because miR-200c did not show reproducible effects on MFF expression in CHANG liver cells, we decided to further examine the relationship between miR-27b and MFF expression. Although miR-27a and miR-27b are located on different chromosomal loci, their mature forms present the same sequence, except for two nucleotides at their 3′ ends, which may allow them to share common target mRNAs. In silico analysis via TargetScan 4.2 and microrna.org showed that both miR-27a and miR-27b could undergo seed region base pairing with the MFF mRNA (positions 1656–1661) (Figure 1c). miR-27a and miR-27b expression decreased MFF expression, as shown in Figure 1d, which indicated that miR-27 (miR-27a/b) has the potential to regulate MFF expression.
Figure 1.
miR-27 targets the mitochondrial fission factor (MFF). (a) Schematic of the MFF mRNA showing the miR-27 binding sites in its 3′-untranslated region (UTR). 5′-UTR (1–266 bp), coding region (267–1295 bp) and 3′-UTR (1296–2014 bp). miRNA binding sites are located in the MFF mRNA 3′-UTR (b and d) CHANG liver cells were transfected with miR-141, miR-27a, miR-27b, miR-200c or control miRNA, and 48 h after transfection, MFF expression was analyzed by western blotting using an anti-MFF antibody. The β-actin level is shown as a loading control. The results are representative of three independent experiments, and the number represents the mean±s.e.m. of three independent experiments. (c) Prediction of miR-27a and miR-27b binding to MFF mRNA using Targetscan.
MFF translational repression by miR-27
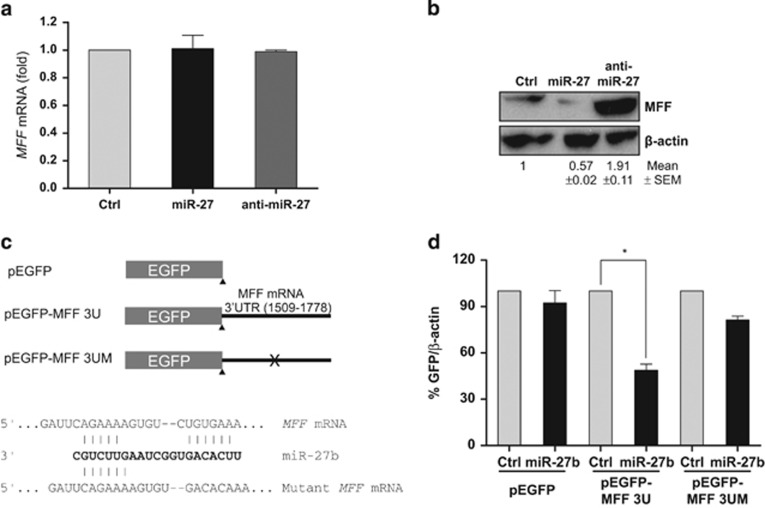
miRNAs inhibit protein synthesis by either suppressing translation or by destabilizing the target mRNAs.3, 32, 33 To understand the regulation of MFF expression by miR-27, we assessed the levels of the MFF mRNA and protein by RT-qPCR and western blotting, respectively, after expressing the miR-27 precursor or inhibitor (anti-miR-27) in CHANG liver cells. Neither the miR-27 precursor nor the inhibitor altered the MFF mRNA level (Figure 2a). However, the miR-27 precursor downregulated the MFF protein level, whereas anti-miR-27 increased the protein level (Figure 2b). To further analyze the regulation of MFF expression by miR-27, we generated an EGFP reporter (pEGFP-MFF 3U) harboring the MFF mRNA miR-27 binding sites (1509–1778), and we also generated a mutant construct (pEGFP-MFF 3UM) lacking the ‘seed' region for miR-27 binding using site-directed mutagenesis, as shown in Figure 2c. The reporter constructs were sequentially transfected into CHANG liver cells, and miR-27 expression and EGFP levels were analyzed by western blotting using an anti-EGFP antibody and statistical analysis. As observed in Figure 2d, miR-27 expression decreased the expression of EGFP harboring miR-27 binding sites, but it did not affect the expression of mutant EGFP. These findings indicate that miR-27 interacts with the MFF 3′-UTR, thereby repressing MFF expression.
Figure 2.
miR-27 regulates the mitochondrial fission factor (MFF) expression. After transfection of pre-miR-27, anti-miR-27 and control miRNA, the MFF mRNA and protein levels were analyzed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (a) and western blotting (b), respectively. (a) The relative expression of MFF mRNA was analyzed using GAPDH mRNA for normalization. The data represent the mean±s.e.m. from three independent experiments. (b) The MFF protein level was analyzed by western blotting using an anti-MFF antibody, and the β-actin level is shown as a loading control. Images are representative of three independent experiments, and the numbers represent the mean±s.e.m. from three independent experiments. (c) Schematic representation of the reporter plasmids pEGFP (control), pEGFP-MFF 3U and pEGFP-MFF 3UM, the last of which bears five mutated nucleotides in the MFF mRNA that correspond to the miR-27 seed region. (d) CHANG liver cells were co-transfected with the plasmids presented in panel c and with the indicated miRNAs. Forty-eight hours after transfection, the EGFP expression levels were assessed by western blotting. The data represent the mean±s.e.m. of three independent experiments. *P<0.05. EGFP, enhanced green fluorescent protein.
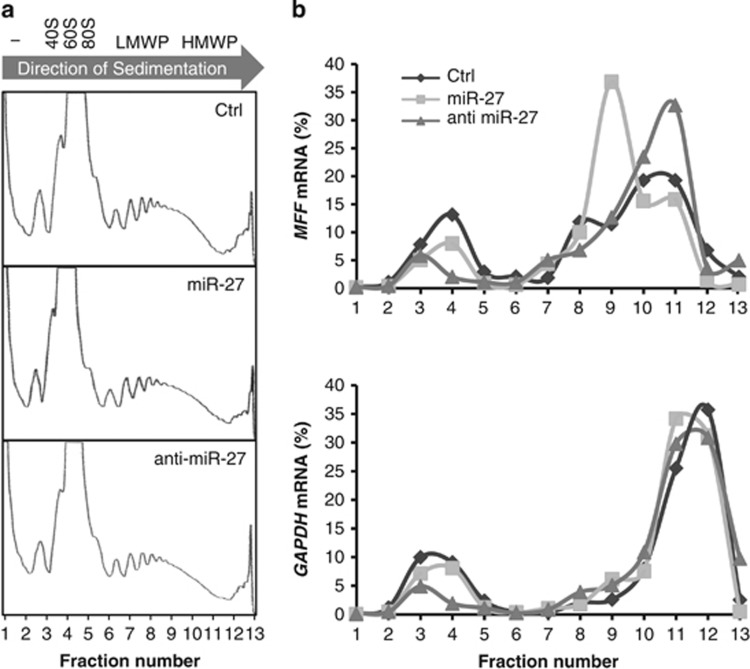
Because miR-27 negatively regulated MFF expression without significantly changing the MFF mRNA levels, we investigated whether miR-27 was involved in MFF mRNA translational repression. The relative association of the MFF mRNA with polyribosomes (polysomes) was analyzed using polysome fractionation on sucrose gradients, as described in the previous studies.29, 30, 34, 35, 36 The levels of MFF mRNA in each fraction, that is, untranslated (fractions 1 and 2), ribosome subunits and monoribosomes (fractions 3–5), low-molecular-weight polysomes (fractions 6–8) and high-molecular-weight polysomes (fractions 9–13), were then measured by RT-qPCR. The polysome profiles were not affected by transfection of the miR-27 precursor or inhibitor (Figure 3a). Compared with the distribution of the MFF mRNA in control cells (peaking at fractions 7 and 8), the expression of the miR-27 precursor resulted in a shift of the MFF mRNA distribution to the lower portions of the gradient, with much of the MFF mRNA peaking at fraction 7 (Figure 3b, top). Conversely, expression of the miR-27 inhibitor increased the relative abundance of the MFF mRNA in the highly translated fractions (fractions 8 and 9). In contrast, the distribution of GAPDH mRNA was not affected by miR-27 expression (Figure 3b, bottom). These data revealed that miR-27 altered the MFF mRNA translational status. Taken together, these observations imply that miR-27 represses MFF mRNA translation by interacting with its 3′-UTR.
Figure 3.
miR-27 inhibits the mitochondrial fission factor (MFF) mRNA translation. (a) Polysome profiles. Cell lysates, prepared from CHANG liver cells transfected with pre-miR-27, anti-miR-27 or control miRNA, were fractionated through sucrose gradients. The arrow indicates the direction of sedimentation; − indicates fractions with no ribosomal components; 40S and 60S indicate small and large ribosomal subunits, respectively; 80S indicates monosomes; and LMWP and HMWP indicate low- and high-molecular-weight polysomes, respectively. (b) The relative distributions of the MFF (top) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNAs (bottom) were studied by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis of each of the 13 fraction. The data are representative of two independent experiments.
Reduction of mitochondrial fission by miR-27
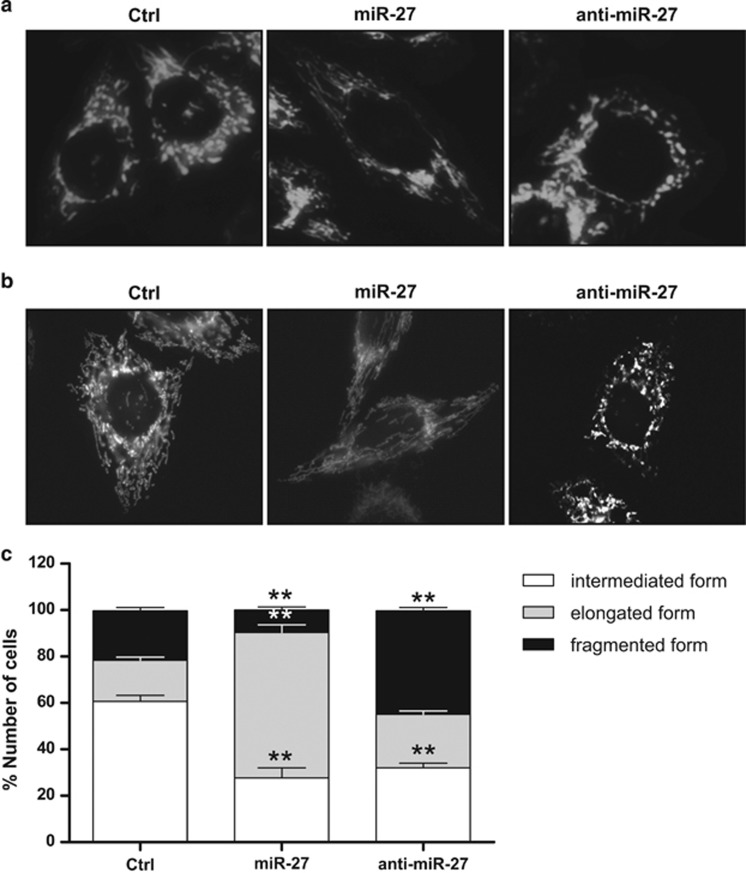
MFF mediates mitochondrial fission by recruiting DRP1, and MFF downregulation is responsible for mitochondrial elongation.16, 17, 33 Because miR-27 regulates MFF expression, we further tested whether miR-27 affected changes in mitochondria morphology. The miR-27 precursor or inhibitor with control miRNA were transfected into CHANG liver cells stably expressing mtYFP.37 After 48 h of transfection, mitochondrial morphology was observed by observing the YFP signals under a fluorescence microscope. miR-27 precursor expression increased the number of cells with elongated mtYFP fluorescent structures, whereas the miR-27 inhibitor decreased the number of cells with these structures (Figure 4a). Additionally, miR-27 expression resulted in mitochondrial elongation, as visualized using Mitotracker, whereas miR-27 inhibition was responsible for mitochondrial fragmentation (Figure 4b). Owing to these observations, we evaluated the effects of miR-27 on mitochondria morphology. The expression of the miR-27 precursor reduced the portion of cells showing fragmented mitochondria and enhanced mitochondrial elongation. However, inhibition of miR-27 using antagomiR resulted in significant enhancement of mitochondrial fission (Figure 4c). Taken together, these results indicate that miR-27 expression results in the reduction of mitochondrial fission by targeting MFF.
Figure 4.
miR-27 inhibits mitochondrial fission. CHANG liver cells expressing mitochondria-targeted YFP (mtYFP) were transiently transfected with pre-miR-27, anti-miR-27 or control miRNA. Forty-eight hours after transfection, the mtYFP (a) or Mitotracker (b) signals were observed under a fluorescence microscope. The data are representative of three independent experiments. (c) Scoring of mitochondrial morphology for the indicated cells. Forty-eight hours after transfection, each cell was placed into one of three morphological categories, and the percentages of cells with the indicated mitochondrial morphologies (intermediate, elongated or fragmented forms) from 100 cells were calculated from three independent experiments. The data represent the mean±s.e.m. from three independent experiments. **P<0.01.
Enhancement of the mitochondrial potential by miR-27
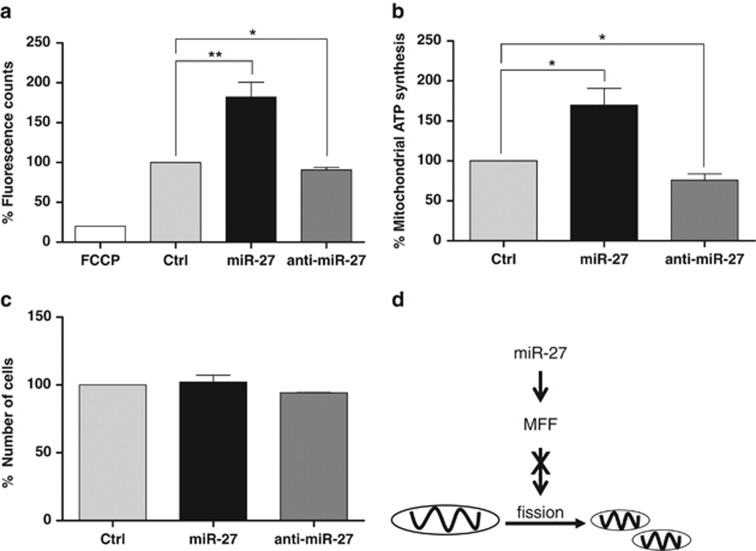
The dynamic regulation of the mitochondrial network is important for several mitochondrial functions such as mitochondrial Ca2+ buffering, the activity of the electron transfer chain, mitochondrial metabolism and the maintenance of mitochondrial DNA.19, 38, 39, 40 To assess whether the changes in mitochondrial morphology induced by miR-27 expression affected the mitochondrial membrane potential, CHANG liver cells were transfected with miR-27 precursor or inhibitor with control miRNA, and the mitochondrial potentials were determined by JC-1 dye staining. As shown in Figure 5a, ectopic miR-27 expression resulted in an increase in the mitochondrial membrane potential, whereas miR-27 inhibition decreased the mitochondrial membrane potential.
Figure 5.
miR-27 increases mitochondrial activity. Mitochondrial membrane potentials (a) and mitochondrial ATP levels (b) after miR-27 regulation. (a) CHANG cells were transiently transfected with pre-miR-27, anti-miR-27 or control miRNA. Forty-eight hours after transfection, the cells were incubated with tetraethyl benzimidazoly carbocyanine iodide (JC-1) dye, and the relative mitochondrial membrane potentials were determined by measuring the fluorescence. (b) After transfection of pre-miR-27, anti-miR-27 or control miRNA, the cells were further incubated with galactose-containing medium, and the mitochondrial ATP levels were analyzed by measuring the luminescence after adding an ATP Detection Reagent. (c) The cell number was determined 48 h after transfection. The data represent the mean±s.e.m. from three independent experiments. *P<0.05 and **P<0.01 (d) Schematic diagram of the role of miR-27 in the regulation of mitochondrial dynamics in CHANG liver cells.
To disprove that the increase in mitochondrial activity induced by miR-27 was because of a change in cell number, mitochondrial ATP synthesis after miR-27 expression was analyzed using the ToxGlo assay. The mitochondrial ATP level was increased by ectopic miR-27 expression (P=0.028) and was decreased by inhibitor expression (P=0.021) (Figure 5b). However, there were no significant effects on cell number (Figure 5c). These results indicate that the effects of miR-27 on mitochondrial activity are independent of cell viability. Taken together, these results suggest that miR-27 enhances mitochondrial function by downregulating MFF expression.
Discussion
The dynamic control of mitochondrial morphology is a pivotal process for maintaining cellular homeostasis, and its dysregulation is associated with several human diseases such as cancer, diabetes and neurodegenerative diseases.11, 12, 13 Although several efforts have been made to identify novel factors that regulate mitochondrial dynamics, including miRNAs, and to elucidate the regulatory mechanisms governing mitochondrial dynamics, these factors and mechanisms remain largely unknown.19, 20, 21, 22, 23, 24, 25, 26, 27, 28
The present study demonstrates that miR-27 negatively regulates mitochondrial fission by inhibiting MFF mRNA translation. Ectopic miR-27 expression resulted in increases in mitochondrial fusion and mitochondrial activity, whereas miR-27 inhibition enhanced mitochondrial fission and reduced the mitochondrial membrane potential. We performed an in silico analysis using TargetScan 4.2 and microrna.org to examine whether miR-27 affected the expression of other factors that regulate mitochondrial morphology, and we confirmed that miR-27 could not regulate DRP1, mitochondrial fission 1 or MFN1/2. To our knowledge, this is the first study demonstrating the direct regulation of the mitochondrial fission machinery and mitochondrial activity by miR-27 via MFF regulation.
MFF is one of the critical regulators promoting mitochondrial fission by recruiting DRP1 to the mitochondria, and fine-tuning of its expression is responsible for mitochondrial dynamics.16, 17, 31 However, the detailed mechanisms involved in MFF expression are not fully elucidated. It was recently shown that MFF downregulation by miR-761 promotes mitochondrial fusion in rat cardiomyocytes and protects those cells from hydrogen peroxide-induced apoptosis.25 In addition, in this study, we demonstrated a novel function of miR-27 as a MFF mRNA translational suppressor that enhances mitochondrial fusion. Further studies may enable us to identify novel regulators and to understand the molecular axis affecting MFF expression at the transcriptional, posttranscriptional and posttranslational levels (Figure 5d).
In contrast to our findings, miR-27 was recently reported to impair adipocyte differentiation and mitochondrial function by targeting prohibitins in human adipose-derived stem cells.41 Prohibitins have a role as protein scaffolds in the mitochondria and are involved in diverse cellular processes, including the processing of OPA1 by m-AAA protease, thereby enhancing mitochondrial fusion.42, 43 Previous studies have shown that prohibitin levels increase and that miR-27 levels decrease during adipogenesis.36, 41, 44, 45, 46 The inverse correlation between prohibitins and miR-27 is responsible for the efficient mitochondrial fusion during adipogenesis in response to the cellular energy demand in adipocytes.
The dynamin-like GTPase OPA1, a gene product of human dominant optic atrophy, is involved in mitochondrial fusion and remodeling, and downregulation of OPA1 results in mitochondrial fragmentation.43, 47, 48 However, a recent study by Otera et al.16 demonstrated that depletion of MFF suppresses OPA1 knockdown-induced mitochondrial fragmentation, indicating that MFF limits OPA1 silencing-induced mitochondrial fission. Another study showed that OPA1 normally counteracts the proapoptotic action of mitochondrial fission 1, which promotes mitochondrial fission.48 We did not examine the expression levels of prohibitin or OPA1 in our system; therefore, it is difficult to conclude what would be the predominant mechanism governing mitochondrial dynamics with this limited information. Additionally, in silico analysis indicated that miR-27 potentially targets OPA1; however, we did not see significant downregulation of OPA1 after overexpression of miR-27 in our system. The discrepancy between the results of a previous study and our findings showing the effect of miR-27 on mitochondrial morphology may result from the use of various assays and systems to measure the relative levels of MFF, prohibitin and OPA1. Additional studies must be designed to further explore and understand the role of miR-27 in the regulation of mitochondrial dynamics in various systems.
miRNA expression alterations have been reported to be involved in several physiological and pathological processes such as aging, tumorigenesis, metabolism and inflammation,8, 49, 50, 51 and several studies have demonstrated differential expression of miR-27 in various diseases. miR-27 is upregulated in breast cancer, oral cancer, glioma and peripheral arterial disease41, 52, 53, 54, 55 and is downregulated in prostate cancer and head and neck squamous cancer.56, 57 Several reports have shown that mitochondrial fusion can inhibit cell death, whereas mitochondrial fission is involved in the promotion of apoptosis.58, 59, 60, 61 From this interpretation, it would be assumed that the increased expression of miR-27 in various cancers may enhance mitochondrial fusion via targeting MFF, thereby promoting mitochondrial dysfunction and tumor progression. Further studies are necessary to explore the relationship between miR-27 and MFF in health and disease.
Acknowledgments
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korea government (MEST) (2012M3A9D1054517, 2012R1A5A2047939 for EKL and 2009-0093826 for WK).
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight. Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Ke XS, Liu CM, Liu DP, Liang CC. MicroRNAs: key participants in gene regulatory networks. Curr Opin Chem Biol. 2003;7:516–523. doi: 10.1016/s1367-5931(03)00075-9. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello S, Scorrano L. Mitochondrial shape changes: orchestrating cell pathophysiology. EMBO Rep. 2010;11:678–684. doi: 10.1038/embor.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Liesa M, Palacin M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chan DC. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc Natl Acad Sci USA. 2007;104:18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokata S, Youle RJ, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Frank S, Gaume B, Herrler M, Youle RJ, Fuller MT. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Grandemange S, Herzig S, Martinou JC. Mitochondrial dynamics and cancer. Semin Cancer Biol. 2009;19:50–56. doi: 10.1016/j.semcancer.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, et al. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011;17:71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010;6:e1000795. doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Jiao JQ, Wang JX, Liu JP, Li Q, et al. miR-484 regulates mitochondrial network through targeting Fis1. Nat Commun. 2012;3:781. doi: 10.1038/ncomms1770. [DOI] [PubMed] [Google Scholar]
- Long B, Wang K, Li N, Murtaza I, Xiao JY, Fan YY, et al. miR-761 regulates the mitochondrial network by targeting mitochondrial fission factor. Free Radic Biol Med. 2013;65:371–379. doi: 10.1016/j.freeradbiomed.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li Y, Jiao J, Wang J, Li Y, Qin D, et al. Mitofusin 1 is negatively regulated by microRNA 140 in cardiomyocyte apoptosis. Mol Cell Biol. 2014;34:1788–1799. doi: 10.1128/MCB.00774-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang FS, Wu ZY, Lin JL, Lan WB, Lin JH, et al. MicroRNA-19b targets Mfn1 to inhibit Mfn1-induced apoptosis in osteosarcoma cells. Neoplasma. 2014;61:265–273. doi: 10.4149/neo_2014_034. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang L, Gao YF, Fam ZM, Cai XY, Liu MY, et al. MicroRNA-106b induces mitochondrial dysfunction and insulin resistance in C2C12 myotubes by targeting mitofusin-2. Mol Cell Endocrinol. 2013;381:230–24. doi: 10.1016/j.mce.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, et al. Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol Cell. 2006;22:117–128. doi: 10.1016/j.molcel.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Lee EK, Kim HH, Kuwano Y, Abdelmohsen K, Srikantan S, Subarab SS, et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol. 2010;17:732–739. doi: 10.1038/nsmb.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Srikantan S, et al. p16(INK4a) translation suppressed by miR-24. PLoS ONE. 2008;3:e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci USA. 2008;105:20297–20302. doi: 10.1073/pnas.0809376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2011;31:626–638. doi: 10.1128/MCB.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YS, Yoon DS, Lim IK, Yoon SH, Chung HY, Rojo M, et al. Formation of elongated giant mitochondria in DFO-induced cellular senescence: involvement of enhanced fusion process through modulation of Fis1. J Cell Physiol. 2006;209:468–480. doi: 10.1002/jcp.20753. [DOI] [PubMed] [Google Scholar]
- Szabadkai G, Simoni AM, Chami M, Weickowski MR, Youle RJ, Rizzuto R, et al. Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell. 2004;16:59–68. doi: 10.1016/j.molcel.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Ono T, Isobe K, Nakada K, Hayashi JI. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat Genet. 2001;28:272–275. doi: 10.1038/90116. [DOI] [PubMed] [Google Scholar]
- Pich S, Bach D, Briones P, Liesa M, Camps M, Testar X, et al. The Charcot–Marie–Tooth type 2A gene product, Mfn2, up-regulates fuel oxidation through expression of OXPHOS system. Hum Mol Genet. 2005;14:1405–1415. doi: 10.1093/hmg/ddi149. [DOI] [PubMed] [Google Scholar]
- Kang T, Lu W, Xu W, Anderson L, Bacanamwo M, Thompson W, et al. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. J Biol Chem. 2012;288:34394–34402. doi: 10.1074/jbc.M113.514372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Merkwirth C, Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J Cell Sci. 2009;122:3823–3830. doi: 10.1242/jcs.037655. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Lin Y, Kang Y, Huang B, Xu W, Garcia-Barrio M, et al. Mitochondrial dysfunction and adipogenic reduction by prohibitin silencing in 3T3-L1 cells. PLoS ONE. 2012;7:e34315. doi: 10.1371/journal.pone.0034315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Kim AY, Lee HW, Son YH, Lee GY, Lee JW, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun. 2010;392:323–328. doi: 10.1016/j.bbrc.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, Ailhaud G, et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun. 2009;390:247–251. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- Wong ED, Wagner JA, Scott SV, Okreglak V, Holewinske TJ, Cassidy-Stone A, et al. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J Cell Biol. 2003;160:303–311. doi: 10.1083/jcb.200209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Yoon Y. Transient contraction of mitochondria induces depolarization through the inner membrane dynamin OPA1 protein. J Biol Chem. 2014;289:11862–11872. doi: 10.1074/jbc.M113.533299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nature reviews. Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li H, Ham L, Zhang K, Wang G, Wang Y, et al. Expression and function of miR-27b in human glioma. Oncol Rep. 2011;26:1617–1621. doi: 10.3892/or.2011.1458. [DOI] [PubMed] [Google Scholar]
- Jin L, Wessely O, Marcusson EG, Ivan C, Calin GA, Alahari SK, et al. Prooncogenic factors miR-23b and miR-27b are regulated by Her2/Neu, EGF, and TNF-alpha in breast cancer. Cancer Res. 2013;73:2884–2896. doi: 10.1158/0008-5472.CAN-12-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Cao H, Zhuang J, Wan J, Guan M, Yu B, et al. Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin Chim Acta. 2011;412:66–70. doi: 10.1016/j.cca.2010.09.029. [DOI] [PubMed] [Google Scholar]
- Lo WY, Wang HJ, Chiu CW, Chen SF. miR-27b-regulated TCTP as a novel plasma biomarker for oral cancer: from quantitative proteomics to post-transcriptional study. J Proteom. 2012;77:154–166. doi: 10.1016/j.jprot.2012.07.039. [DOI] [PubMed] [Google Scholar]
- Susuki D, Kimura S, Naganuma S, Tsuchiyama K, Tanaka T, Kitamura N, et al. Regulation of microRNA expression by hepatocyte growth factor in human head and neck squamous cell carcinoma. Cancer Sci. 2011;102:2164–2171. doi: 10.1111/j.1349-7006.2011.02096.x. [DOI] [PubMed] [Google Scholar]
- Sun T, Yang M, Che S, Balk S, Pomerantz M, Hsieh CL, et al. The altered expression of MiR-221/-222 and MiR-23b/-27b is associated with the development of human castration resistant prostate cancer. Prostate. 2012;72:1093–1103. doi: 10.1002/pros.22456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lazaro M, Bonekamp NA, Galindo MF, Jordan J, Schrader M. 6-Hydroxydopamine (6-OHDA) induces Drp1-dependent mitochondrial fragmentation in SH-SY5Y cells. Free Radic Biol Med. 2008;44:1960–1969. doi: 10.1016/j.freeradbiomed.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Frank S, Gaume B, Herrler M, Youle RJ, Fuller MT, et al. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]