Abstract
Bacteria in natural and artificial environments often reside in self-organized, integrated communities known as biofilms. Biofilms are highly structured entities consisting of bacterial cells embedded in a matrix of self-produced extracellular polymeric substances (EPS). The EPS matrix acts like a biological ‘glue’ enabling microbes to adhere to and colonize a wide range of surfaces. Once integrated into biofilms, bacterial cells can withstand various forms of stress such as antibiotics, hydrodynamic shear and other environmental challenges. Because of this, biofilms of pathogenic bacteria can be a significant health hazard often leading to recurrent infections. Biofilms can also lead to clogging and material degradation; on the other hand they are an integral part of various environmental processes such as carbon sequestration and nitrogen cycles. There are several determinants of biofilm morphology and dynamics, including the genotypic and phenotypic states of constituent cells and various environmental conditions. Here, we present an overview of the role of relevant physical processes in biofilm formation, including propulsion mechanisms, hydrodynamic effects, and transport of quorum sensing signals. We also provide a survey of microfluidic techniques utilized to unravel the associated physical mechanisms. Further, we discuss the future research areas for exploring new ways to extend the scope of the microfluidic approach in biofilm studies.
I. INTRODUCTION
In the environment, bacteria are often found in close-knit communities encased in an extracellular matrix and attached to a surface, forming what are known as microbial biofilms [1]. This aggregation and the consequent self-secretion of polymeric substances are associated with several physiological and phenotypic features, such as higher resistance to external stresses [2], higher accessibility of nutrients [3], and altered gene transcription [4]. Due to these effects, biofilms introduce important challenges in clinical and industrial settings, including persistent infection of human tissues, increased tolerance to bactericides, and biofouling and clogging problems in flow systems and pipelines [5]. On the other hand, biofilms of particular bacterial species bear important advantages in certain applications such as biomineralization [6], wastewater treatment [7], and bioremediation of oil and gasoline spills [8]. Thus, it is critically important to elucidate and characterize the physical mechanisms involved in the formation, development, and dispersion of bacterial biofilms.
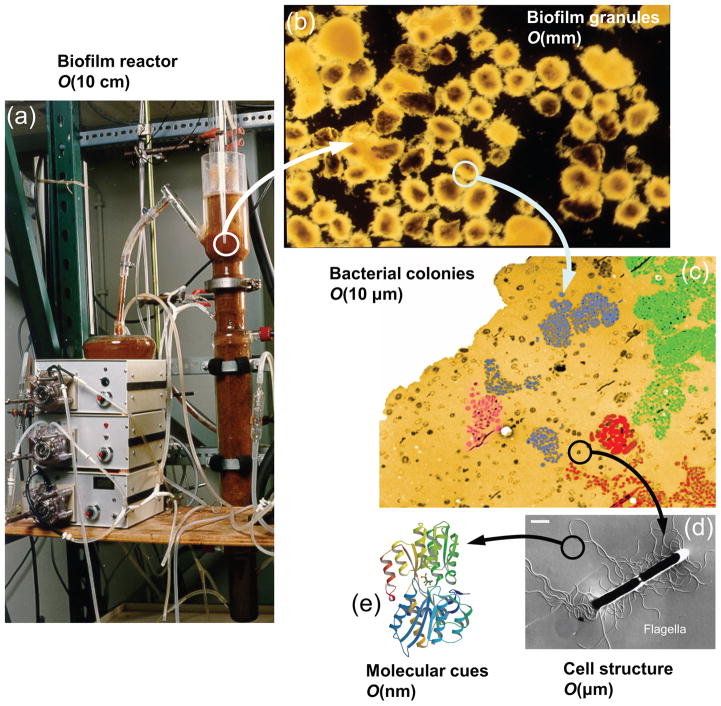
Bacteria in the planktonic state have been the focus of numerous studies, but the biofilm state represents a daunting challenge for researchers. This is primarily due to the fact that biofilms are comprised of complex, heterogenous structures with a wide-ranging versatility in morphology, mechanical properties, and chemical composition. Further, there are multiple scales involved in the framework of biofilms [9] as depicted in Fig. 1. On the microscale, clusters of microbes, consisting of several hundred to several thousand cells adhered together, form microcolonies which are the building blocks of biofilm structure. At the nanoscale, enzymes and molecular cues are used for communication and coordination among microbes. At the mesoscale, microcolonies may aggregate, and in subsequent developmental stages, evolve into more complex configurations such as granules, ripples, or streamers. In practical applications, we generally deal with the implications of biofilms at the macroscale, which corresponds to the size of the chemical reactors (e.g. fluidized beds and biofilm airlift reactors) or transmission pipelines. On the other hand, biofilm processes which are physical, chemical, and biological in nature, occur over a broad range of time scales [10] as demonstrated schematically in Fig. 2. While the characteristic time for the convective transport is on the order of milliseconds, some other crucial developments, such as cell growth and biomass detachment, take place over the course of days. The sophisticated diversity of biofilm processes, spanning a broad spectrum of temporal and spatial scales, increases the complications of biofilm modeling and constrains the scope and validity of the models suggested so far.
FIG. 1.
Different length scales present in the framework of biofilms. At macroscale, biofilms appear in chemical reactors and industrial equipment. The biomass, on the mesoscale, is comprised of smaller aggregates of bacteria adhered together via production of EPS. The constituent elements of these microcolonies, at microscale, are bacterial cells attached to the substratum or to other cells. The communication among these cells is carried out via secretion of signaling molecular cues which are at nanoscale. Images (a),(b),(c) are courtesy of Cristian Picioreanu. Images (d) and (e) are adapted and reproduced with permission from ref. [11, 12].
FIG. 2.
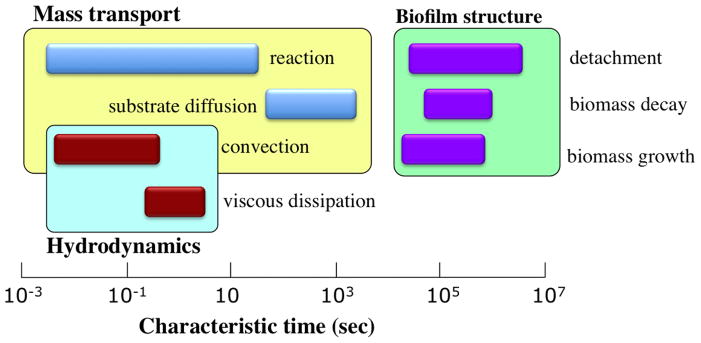
Characteristic time scales for processes occurring in the framework of biofilms. The momentum transfer processes are associated with the shortest time scales. The substrate mass transport phenomena are slower and occur on the order of minutes. The processes that alter the morphology and volume of the biomass take place on the most prolonged time scales. Image adapted and reproduced with permission from ref. [10].
To understand biofilm formation and maturation, it is necessary to study aggregation of bacteria at ecologically relevant spatiotemporal scales and in the presence of both flow and interactions with extracellular polymeric substances. Microfluidic and/or lab-on-a-chip (LOC) techniques have emerged as a promising approach in biofilm research due to their remarkable advantages including high throughput screening capabilities, low sample volume requirements, compatibility with the length and time scale of processes involved in biofilm formation, and the ability to integrate several laboratory functions on a miniaturized chip [13]. For example, one could build multiple bioreactors on a single biomicrofluidic platform and simultaneously conduct real-time monitoring and/or incorporation of sensors. The capacity to implement multiple laboratory experiments in one device and the ability to collect significant pertinent data for accurate statistical analysis renders microfluidics desirable for exploring various aspects of microbial biofilms [14–18].
In the current review article, we focus on the interplay of various physical processes affecting the formation and development of biofilms. In addition, we present a survey of various LOC techniques utilized to explore and analyze these physical mechanisms. The swimming behavior of motile bacteria is heavily modified in the proximity of rigid surfaces and this phenotypical change substantially influences the adhesion rate and the associated attachment process. In Sec. II, we outline different near-wall swimming strategies and the role of cell appendages in formation of biofilms. Sec. III is devoted to elucidating the rheological properties of mature biofilms behaving as viscoelastic materials and to reviewing various techniques which have been employed to measure the mechanical characteristics of bacterial aggregates. In Sec. IV, we discuss hydrodynamic and environmental conditions favoring the formation of biofilm streamers, i.e. filamentous biomass structures attached locally to the substratum and oscillating freely with fluid flow. In Sec. V, the mechanisms of chemical signaling among sessile cells inside a biofilm have been discussed and the effects of hydrodynamics and confinement on the characteristics of quorum sensing have been delineated. In Sec. VI, we present a survey of microscale fabrication techniques utilized to make engineered topographies that inhibit the formation of biofilms. Concluding remarks and suggestion for future research directions have been presented in Sec. VII.
II. NEAR-SURFACE MOTION OF BACTERIA
In the initial stages of biofilm formation, motile bacteria swim around and adhere to a variety of surfaces, such as host cells and tissues [19–21], biomedical devices [22–24], and industrial equipments [25–27]. Understanding the swimming strategy of the microorganisms has been shown to be essential for identifying the adhesion rate and elucidating the subsequent colonization process. In particular, scrutinizing the near-surface motility of different types of bacteria is of great importance, not only to enhance our physical understanding of the impact of surfaces on the motility, but also to devise more effective scenarios in order to influence and control the phenomena concerning the formation of biofilms. In this regard, numerous experimental, analytical, and numerical investigations have been conducted to shed light on the physical interactions between bacteria and nearby surfaces in different flow regimes.
The motion of bacteria in fluid environments, such as water, mucus, and blood, is chiefly dominated by viscous forces. Correspondingly, the typical values of Reynolds number, representing the relative magnitude of inertial and viscous forces, can be as small as 10−5 [28, 29]. In the limit of low Reynolds number, a swimming bacterium as a whole is both force-free and torque-free [30]. Further, the associated governing equations for Newtonian fluids are linear and thus, the effect of various forces can be incorporated via superposition. These important ramifications facilitate the theoretical study of swimming dynamics through analytical and numerical methods.
A. Flagella-mediated Motility Near Walls
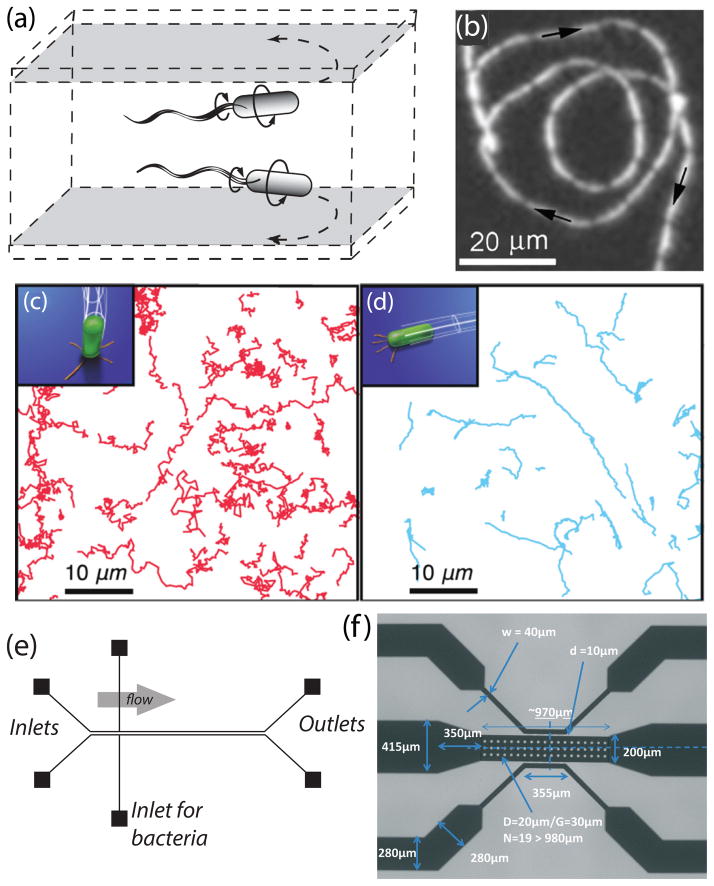
The motility of peritrichously flagellated bacteria such as Escherichia coli has been extensively scrutinized over the past decades. It is known that these bacterial cells propel forward due to the thrust force generated by the rotary motion of their flagellar bundle [31]. Their swimming strategy, termed as run-and-tumble, consists of prolonged straight trajectories interrupted by rapid changes of orientation [32–34]. The trajectory of bacteria, however, is significantly modified in the vicinity of a solid boundary where the bacteria start to swim in circular trajectories [35, 36]. Since a large body of information exists for the specific case of E. coli, we first focus on the near-surface motility of this multiply-flagellated bacterium. During a run, the flagella of E. coli cells form a helical bundle rotating counter-clockwise at a rate of ~ 100 Hz, while the cell body rotates clockwise at ~ 25 Hz [37]. In the absence of external flows, the forward propulsion stems from the anisotropy of viscous drag force in axial and transverse directions of the flagellar bundle [38]. The resulting thrust drives the cell in a straight path in the bulk fluid until tumbling occurs via the reversal of flagellar rotation [39]. The presence of a nearby wall, however, modifies the physics of swimming associated with hydrodynamic interactions and leads to a curvilinear motion [35, 40]. This “wall effect” can be explained by considering a torque arising from the asymmetrical spatial distribution of drag forces across the flagellated filament and the cell body [41]. The portions of a helical flagellum closest to the wall feel larger drag forces compared to the parts further away, leading to a net local force on the filament. To satisfy the force-free condition, the viscous flow exerts an equal and opposite force on the counter-rotating cell body, thereby rendering a torque in clockwise direction when viewed from above the surface. This mechanism explains the right-hand side swimming of E. coli cells in the proximity of a bottom surface as observed in experimental studies [42] (see Fig. 3(a)). Swimming in circular trajectories enhances the surface coverage and the rate of adhesion as the bacteria wander longer durations in the vicinity of the surface [43]. The radius of curvature of the circular paths depends on the ratio of axial swimming speed to the rate of rotation about the normal direction to the surface [41]. These parameters are functions of the geometry and the physiology of the organisms, the physical properties of the surrounding fluid, and distance to the nearby wall [44, 45]. For example, it has been predicted that by growing the cell’s aspect ratio the radius of curvature will increase [45]. Also, experimental observations have indicated that E. coli cells sense the wall effect when they are within one body length (~ 10 μm) away from the surface [42, 46, 47]. The circular motion of bacteria continues for a while until disrupted by tumbling, cell-cell interactions, or Brownian motion [48].
FIG. 3.
(a) Schematic depiction of right-hand side swimming of E. coli cells in proximity of a rigid surface. (b) A typical swimming trajectory of a Caulobacter crescentus swarmer cell near a wall. The cell follows a circular trajectory while swimming backward. (c,d) Schematics and trajectories of TFP-mediated (c) upright walking and (d) lengthwise crawling observed in strains of P. aeruginosa. (e) Schematic of the microfluidic chamber design used by De La Fuente et al. [87] to estimate the adhesion forces of type I and type IV pili. (f) Scanning electron microscopy image of the microfluidic device used by Wright et al. [88] to study the effect of acetic acid on the motility of LM. Images adapted and reproduced with permission from ref. 42, 86, 78, 87 and 88, respectively.
The motility of monotrichous bacteria differs from E. coli. Monotrichous bacteria have been reported to follow a swimming pattern termed as run-and-reverse, i.e. in the bulk fluid they change the orientation of their straight swimming path by alternating the direction of the rotary movement of their single flagellum [49–51]. For example, cells of Pseudomonas aeruginosa propel forward when their protein motor rotates the flagellum counter-clockwise and move backward when the filament rotates clockwise [51]. Each occurrence of swimming direction reversal is accompanied by a slight change in the trajectory of the cells, mainly due to the Brownian motion [52] and the flicking of the flagellum upon resuming the run [53, 54]. Using this swimming strategy, the singly-flagellated bacteria can follow the favorable chemoattractants in the bulk fluid [55]. This motility behavior is modified in the proximity of rigid surfaces where the bacteria perform circular motion when swimming backward. This “run-and-arc” swimming pattern has been observed for a wide variety of monotrichous bacteria, such as Vibrio alginolyticus [52, 55, 56], Caulobacter crescents [57, 58], and P. aeruginosa [50] (see Fig. 3(b)), and has been suggested to require less energy compared to other swimming strategies in case of large-sized bacteria [34]. Nonetheless, these bacteria pursue straight paths when swimming in the forward direction close to a wall [55, 56]. The rationale given for the asymmetry between forward and backward trajectories near a solid surface is based on the stability of the pitching angle, i.e. the orientation of the cell with respect to the wall [52]. Numerical simulations of a single bacterium performed using the boundary element method have revealed that in forward swimming the pitching motion (rotation around an axis parallel to the surface and normal to the cell) is stable, thereby the cell maintains an almost fixed orientation aligned to the wall. Consequently, a straight path is preferred as the yaw rate (angular velocity about an axis normal to the surface) is negligible when the pitch angle is small. On the other hand, when the cell is swimming backward, the pitching motion becomes unstable and the trajectory of the cell will depend on inclination of its flagellum with respect to the wall. If the cell is initially oriented away from the wall, the pitch angle increases over time and the cell departs the region in the proximity of the surface, moving along a straight line. Otherwise, if the cell swims toward the wall, it approaches the surface quickly with an average speed larger than that achieved in the bulk fluid. Experimental studies yield up to 50% boost in speed of bacteria when moving in the backward direction adjacent to the rigid boundaries [59, 60]. This is postulated to be the result of higher torque exerted on flagellum when the filament precedes the cell body in swimming towards the wall [56]. This approaching motion induces a rotary movement in the yaw direction, thereby leading to a curvilinear trajectory of the cell parallel to the surface [52]. The aforementioned dynamical disparity results in a longer residence time of the bacteria near a wall which, in turn, increases the probability of adhesion to the surface and formation of biofilm [52, 56].
B. Role of Flagella in Formation of Biofilms
Flagellar motility, in general, plays an important role in the evolution of biofilms by empowering the cells to access abiotic surfaces and to penetrate mucosal layers, providing the force required to overcome the near-surface repulsive effects, facilitating biofilm growth via recruitment of planktonic cells, and promoting the dispersal of biofilms. The presence of flagella enhances the probability of cell-surface encounter and reversible attachment prior to the formation of EPS matrix. In addition flagella foster the adhesion process in some bacterial species as they tether to various surfaces via the adhesin binding [61].
Despite the overall significance of flagella, their impact on the formation and morphology of bacterial colonies is widely versatile and depends on several factors, including culture conditions, type of surface material, biofilm age, and the phenotype of bacterial species [62]. For example, while flagellar-deficient mutants of Agrobacterium tumefaciens are significantly incompetent in forming biofilms in static cultures, under flow conditions the size of their microcolonies is markedly enhanced relative to the wild-type strains [63]. The expression of genes associated with flagellar filaments is repressed during biofilm maturation phase [64] as endurance of motility might disrupt the microcolony structure. This transcription change is accompanied by a transition in attachment location from the cell pole to the cell side in strains of P. aeruginosa [65]. The inhibition of motility and transition to a nonmotile state is controlled through the intracellular level of signaling molecule c-di-GMP [66]. Also, at the functional level, the secretion of the protein EpsE leads to the rapid loss of motility during early stages of biofilm formation via a clutch-like mechanism [64]. The conventional notion of sessility of bacterial cells encased within a biofilm matrix has been recently challenged by the discovery of “stealth swimmers” [67, 68], i.e. a subpopulation of microorganisms inside the biofilm which maintain the motility phenotype. Although they constitute 0.1–1% of the cells, they impact the nutrient diffusion by tunneling into the biofilm structure.
The interplay of surface properties and flagellar motility greatly affects the size and morphology of corresponding biofilms. For example, hydrophobic surfaces are associated with uniform adhesion of bacterial cells and tightly packed biofilms, as opposed to hydrophilic surfaces which yield lower attachment rate and unstructured microcolonies [61]. In the absence of flagella, the mutants of Vibrio cholerae form rugose colonies instead of smooth-edged colonies of the wild-type strain [69]. In some species of bacteria, such as Aeromonas spp. [63], the flagella function primarily as adhesins and attach to a variety of abiotic and biological surfaces, while in some other cell types, the flagella might even play a negative role in the adhesion process, e.g. the attachment of opportunistic pathogen Bacillus cereus to glass surfaces [62].
C. Pili-mediated Motility Near Walls
Besides flagella, other appendages of bacteria also play a crucial role in the near-surface motility and biofilm formation. In particular, in some pathogens such as P. aeruginosa type IV pili (TFP) mediate bacterial motion through twitching motility [70]. These protein filaments, which are polymerized around the cell poles, undergo a cycle of adhesion and retraction, thereby generating quite substantial forces to pull the bacterium along the surface [71]. This force is on the order of 100 pN for a single pilus [72] and can reach up to 1 nN if the pili retract in a bundle [73]. The retraction occurs while the distal tip of TFP adhered to the surface via excretion of polar holdfast adhesive polysaccharides [74]. Exploiting multiple pili and through a tug-of-war mechanism, bacteria can cover distances that are larger than the extension length of individual pili [75]. Interestingly, the force generated by these pili can influence the dynamics of pilus retraction by acting as a switch in some cases leading to a transition from retraction to elongation [76]. This peculiar behavior has recently been shown to possibly arise due to the deformation of the TFP itself [77].
In the vicinity of surfaces, TFP mediates two important motility modes: upright walking in which the cell moves normal to the surface plane by deploying extended pili (see Fig. 3(c)), and lengthwise crawling in which the cell moves parallel to the surface plane by retracting pili adhered to the wall [78, 79] (see Fig. 3(d)). The two mechanisms offer their own advantages, and bacteria reversibly switch between them [78]. Walking mode, associated with higher speeds, jagged trajectories, and low directional persistence, is more suited for quick exploration of the environment [79]. On the other hand, in crawling mode, cells move more slowly and more persistently in terms of the inclination of their swimming trajectories [79]. Using the crawling mechanism, bacteria can traverse a specific direction in an efficient fashion [79]. The crawling strategy is interrupted infrequently by another motility mode termed as slingshot motion [80]. In this mode, while most of the pili are under tension, a single pilus which is directed sideways to the cell’s main axis is released and consequently the cell abruptly reorients and moves to a new location [80]. The slingshot motility occurs within a short duration compared to crawling mode and is associated with a 20-fold increase in instantaneous velocity. Accordingly, slingshot and crawling modes contribute almost equally in displacing piliated bacteria [80].
The role of pili fibers in establishing initial interactions with and adherence to abiotic surfaces, such as polyvinyl chloride (PVC), is critical. Pili also have a significant contribution in attachment to eukaryotic cell surfaces and therefore, pathogenesis [81]. In particular, in E. coli [82] and Xyllela fastidious [83] strains, type I pili form stable cell-to-surface attachment using the mannose-specific adhesin FimH which causes pathogenesis as well. Contrary to type I pili which are essential for biofilm formation, TFP and the associated twitching motility are of lesser importance and only affect the morphology of biofilms. For example, twitching defective strains of P. aeruginosa produce smooth, rounded microcolonies, while the biofilms resulting from the wild-type strain have spreading morphology with highly irregular edges [81]. Experiments conducted in flow cells have revealed that the TFP are the major adhesins for initial attachment of P. aeruginosa cells to glass surfaces [84]. It has been shown that hyperpiliated twitching deficient strains of P. aeruginosa form localized multilayer microcolonies which are more dense than normal [84]. These observations suggest the idea that the twitching motility significantly promotes cell migration along the surface and actively contributes in maturation of biofilms. In general, the presence of pili causes more cells to accumulate and form larger aggregates of bacteria [83] either via twitching function or by acting as retractable adhesins for surface anchoring. For instance, curved Caulobacter crescentus cells do not twitch, however, they employ their curvature to orient their polar pili towards the nearby surface in order to facilitate their adhesion to the substratum under flow conditions [85].
D. Microfluidic Assays
Microfluidic devices enable researchers to study the complex interplay of motile bacteria with nearby substrata via providing a miniaturized test bed which mimics the physical and biological conditions pertinent to the microorganism of interest. In this regard, microfluidic flow cells have emerged in recent years as a promising tool to investigate the role of various types of cell appendages in motility and adhesion of bacterial cells to adjacent surfaces. For example, De La Fuente et al. [87] employed a dual-channel microfluidic device (see Fig. 3(e)) to assess the effect of type I and type IV pili on attachment of Xylella fastidiosa cells to a glass substratum under a relatively wide range of flow conditions. To estimate the associated adhesion force, they measured the flow rate required to detach the anchored wild type and pili-deficient cells and calculated the corresponding drag force. Their observations indicate that the attachment process is predominantly dependent on type I pili and the presence of TFP limits the surface colonization due to introduction of twitching motility modes. The same microfluidic setup has been utilized to elucidate the autoaggregation phenomenon of X. fastidiosa cells [89], probe the impact of calcium on surface attachment by pathogenic bacteria [90], and compare the contribution of TFP and polar flagella in biofilm formation by Acidovorax citrulli cells [91]. In the aforementioned studies, the microfluidic chambers, to some extent, mimic the environmental conditions of water conducting conduits (xylem vessels) of plant hosts and provide the botanists invaluable insights about the physical behavior of virulent bacteria. Most recently, Wright et al. [88] designed a nanoporous microfluidic device to analyze the effect of acetic acid and temperature on the swimming trajectory and speed of food-borne Listeria monocytogenes (LM) cells. Their device is composed of one central and two feeder microchannels connected together via nanopores to allow diffusion of acetic acid into the medium of bacteria (see Fig. 3(f)). In this study, it is determined that a relatively high concentration of acetic acid is required to impede the flagellar motility and subsequently, the biofilm-forming capability of LM cells.
III. RHEOLOGY OF BIOFILMS
The microorganisms constituting a biofilm are bound together by a protective matrix which is comprised of extracellular polymeric substances (EPS) including polysaccharides, alginate, nucleic acids, and lipids [92]. The constituent components of EPS altogether compose up to 50–90% of the total organic matter in biofilms [93] and create a highly hydrated viscoelastic gel [94] that provides increased flexibility to cope with environmental stresses [95, 96]. In particular, due to inherent characteristics of viscoelasticity, biofilms exhibit enhanced mechanical stability against fragmentation by shear flows [97]. Despite the abundance of studies regarding the biochemistry, genetics, and biology of biofilms [1], few investigations have been conducted to examine the correlation of the rheological characteristics of the biofilms with the morphology of microcolonies and the ambient conditions.
A. Conventional Rheometry
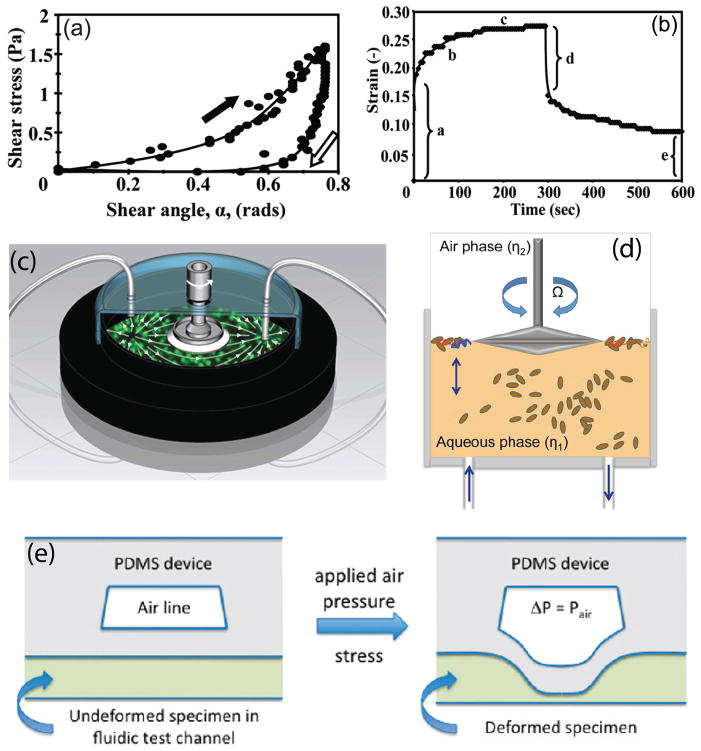
The structure and mechanical behavior of various types of biofilms have been intensely studied over the past few decades using the conventional rheometry techniques, such as stress-strain and creep tests. In a stress-strain test, the biofilm sample undergoes shear stress loading for a relatively short period of time followed by an unloading process. The resulting stress-strain curves typically exhibit a hysteresis loop, resembling a ‘J’ shape, which is a characteristic of viscoelastic materials [98] (see Fig. 4(a)). The existence of hysteresis indicates the occurrence of viscous flow and dissipation of mechanical energy through irreversible residual deformation. Also, based on the outcomes of this test, the shear modulus is observed to rise with increasing shear stress and thus the biofilms show elevated stiffness in high shear regime [99]. In creep tests, the biofilm positioned inside a flow cell is exposed to fluid shear stress for an extended period of time and then the stress is removed in order to observe the recovery behavior. The associated creep curves typically display five characteristic regions (see Fig. 4(b)): (1) an immediate elastic deformation, (2) a transient, nonlinear viscoelastic response, (3) a steady-state viscous flow with constant viscosity, (4) an instantaneous partial elastic recoil upon removal of shear stress, and (5) a residual deformation due to fluid-like behavior [95, 98, 100]. When shear stresses larger than a certain threshold, referred to as the yield point, are applied, the biofilm flows entirely analogously to a liquid [101] and the biofilm stiffness drops by several orders of magnitude [102]. Contrary to our intuition based on ordinary materials, this yielding event is recoverable and the biofilms regain their viscoelastic behavior and original stiffness only a few minutes after stress removal [102]. It is hypothesized that the restoring behavior of the biofilms occurs due to the transient nature of the interconnections between the cells and biopolymers which facilitates the reestablishment of those bonds after they are ruptured [102]. This process protects the integrity of the biofilm fragments in high shear flows and promotes the possibility of dispersal of intact biofilms.
FIG. 4.
(a) Typical stress-strain curve of a biofilm created by aggregation of Staphylococcus aureus cells. The curve exhibits a characteristic J shape due to higher stiffness of the biofilm at elevated values of shear stress. The arrows indicate the hysteresis loop and the irreversible deformation of the biofilm. (b) Creep curve of a biofilm of S. aureus. The characteristic regions are highlighted in the diagram. (c) Base of a parallel plate rheometer after draining media. The microcolonies are evident in the image. (d) Schematic overview of a subphase rheometer modified to conduct real-time measurements of the viscoelastic properties of different types of the interfacial biofilms (pellicles). (e) Schematics portraying the operating mechanism of the microfluidic rheometer with a flexible membrane. By applying air pressure, the PDMS layer and subsequently, the biofilm specimen is deformed. This deformation is detected using confocal microscopy. Images adapted and reproduced with permission from ref. [98, 101, 103, 107].
In addition to the aforementioned tests, small-amplitude oscillatory shear test is also used to characterize the dynamical behavior of the biofilms in response to periodic loading. By imposing a defined deformation which is sinusoidally varying in time, an out-of-phase stress response can be measured. Using the corresponding phase shift, one can quantify the viscoelastic properties of the sample, i.e. the elastic storage modulus G′ (ω) and the viscous loss modulus G″ (ω) as functions of the angular frequency ω. Several rheological investigations [101–103] conducted on a wide variety of bacterial biofilms have indicated that in low and intermediate frequencies, the elastic modulus surpasses the loss modulus and thus the biofilm exhibits solid-like elastic behavior. However, by increasing the frequency, the viscous modulus dominates [102] and the stress relaxation occurs, leading to the flow of the biofilm material. The reciprocal of the crossover frequency describes the average relaxation time which characterizes the lifetime of the transient polymeric interconnections within the biofilm matrix [104, 105]. The spectrum of relaxation time as qualified by Shaw et al. [104] lies in the range of 350–2600 s for a wide sample of natural biofilms. Over longer time scales, the stored energy associated with reversible elastic deformation is entirely dissipated by irreversible viscous flow of the biomass [105]. This relaxation mechanism provides the cells within a biofilm with sufficient time to react phenotypically in response to external shear stresses and adapt themselves to new environmental conditions via, for example, increasing the secretion of EPS [98, 104]. Experimental observations have delineated stronger adhesion and lower detachment rates in the case of biofilms created by cells that are grown under high shear conditions [99]. Furthermore, in the event of prolonged exposure to mechanical stress, viscoelasticity impedes the rupture process and reduces the probability of structural failure.
B. In-situ Rheometry
Utilizing conventional techniques for characterizing the rheological properties of biofilms has several disadvantages. First and foremost, scraping fragments of a biofilm in order to test them in a rheometer will disrupt the structural integrity of the biofilm and adversely affect the measurement procedure. In addition, due to strong dependence of the mechanical characteristics of the biofilms on the cultivation conditions [106], the rheological response of the scraped fragments are significantly different than those biofilms grown in natural environments. As a result, a large variability is seen in the values of elastic modulus reported in the literature [95, 107–109]. To tackle these practical challenges, in-situ rheological techniques have emerged in recent years. In this approach, the biofilm is cultured inside a rheometer fixture, thereby allowing real-time measurement of its structural specifications during the development of the matrix. By transforming a rheometer into a bioreactor, one can instantaneously evaluate the mechanical behavior of the bacterial biofilms in response to various types of physical stimuli, such as shear flow, osmotic pressure, and chemical treatment. For example, using a continuous-flow bioreactor incorporated into a parallel plate rheometer, Pavlovsky et al. [101] estimated the elastic and viscous moduli of Staphylococcus epidermidis biofilms over a relatively broad range of angular frequencies (see Fig. 4(c)). The profound difference between the associated outcomes and the results previously reported for the same strains of the bacterium has been attributed to in situ measurement and the higher degree of hydration resulting from continuous influx of an aqueous solution of nutrients. In another study, Rühs et al. [103] utilized interfacial rheology in conjunction with in-situ cultivation to quantify the viscoelastic properties of the pellicles, i.e. biofilms grown at an air-water interface. Using a modified subphase rheometer, they specified the effect of nutrient concentration, temperature, surfactants, and pH of the growth medium on the rigidity of the biofilms formed by B. subtilis, P. fluorescent, and E. coli (see Fig. 4(d)).
C. Microrheology
Another issue regarding the rheology of the biofilms concerns the nonuniform and heterogenous configuration of these microscopic bacterial communities. The aforementioned experimental techniques are unable to shed light on the internal structure of the microcolonies and the bonds between cell clusters. The methodologies developed in the context of microrheology are generally helpful in overcoming this challenge. For example, Rogers et al. [110] employed particle tracking microrheology (PTM) to measure the temporal and spatial variation of stiffness of the biofilms during growth and starvation periods. In this method, the shear compliance of the biofilms can be calculated using the mean square displacement of individual bacteria which are tracked by analyzing a series of images captured by an inverted microscope. Although this technique offers valuable information in terms of viscoelastic properties as sensed by an individual cell moving inside a biofilm, its application to motile cells is not as robust as nonmotile cells due to confusion between thermal and active motion of the bacteria. In a more recent study, Galy et al. [111] developed a novel technique based on magnetic tweezers and micron-sized magnetic particles to reveal the three-dimensional spatial mapping of the elastic compliance of E. coli biofilms. Using this magnetic setup allows us to fine-tune the applied force at any position within the biofilm structure and to derive the associated creep curves by measuring the deflection of inserted probe particles via processing of images captured by a confocal microscope. The outcomes of this experiment indicates over two orders of magnitude variation in the magnitude of shear compliance depending on the measurement site inside the biofilm. Despite the power of this approach for characterizing the local mechanics of the biofilms, it might disrupt the structural integrity of the microcolonies due to insertion of a large number of magnetic particles into the living texture of the bacterial community. Further studies are needed to specify the viability conditions required for microrheology measurements.
In recent investigations, advanced methodologies are emerging to probe the mechanical properties of the biofilm microstructure and the rheological characteristics of the matrix and EPS. The heterogeneous and fragile nature of the biofilms necessitates developing innovative rheometry techniques in order to quantify their behavior under applied stress with little error margin. For example, microbead force spectroscopy has been employed to quantify the variation of the adhesive force between a biofilm patch grown on the tip of an atomic force microscope (AFM) and a substratum as the biofilm evolves into a matured state [112]. Nonetheless, techniques invented to characterize the rheological properties of the soft materials [113] such as two-point microrheology [114] and nonlinear microrheology [115] still await applications in the mechanical studies of biofilms.
D. Microfluidic Rheometry
In the past decade, microfluidics has been an integral part of rheometry techniques [116] and various microdevices have been developed to probe the mechanical properties of human blood plasma [117], circulating cells [118], Newtonian and non-Newtonian liquids [119], power-law fluids [120], etc. However, few microfluidic rheometers are specialized to measure the rheological characteristics of bacterial biofilms. The microfluidic techniques are advantageous in rheological applications as they allow for in situ, microscale characterization of the mechanical properties of the biofilms. In this regard, Hohne et al. [107] developed a new microfluidic approach in which, the mechanical strain of a biofilm specimen, positioned inside a microchannel and subjected to a fixed air pressure, is determined by measuring the deflection of an adjacent membrane using a confocal laser scanning microscope (CLSM) [107] (see Fig. 4(e)). Using this method, they could estimate the Young modulus and relaxation time of biofilms developed by strains of Staphylococcus epidermidis and Klebsiella pneumoniae inside a microchannel. Moreover, they observed hardening behavior, i.e. elevated rigidity, when the biofilms were subjected to large deformations. In a more recent study, Mosier et al. [18] developed a microfluidic flowcell device integrated with a combined AFM/CLSM to conduct in situ optical and mechanical characterization of bacterial biofilms. Their device is composed of multiple inlets/outlets and a central growth chamber mounted on a glass microscope slide. Using force spectroscopy, i.e. nanoindentation by an AFM, they measured the elastic modulus of a biofilm sample at various spots in static condition. Further investigations are needed to explore the full potential of microfluidic devices in rheological applications pertinent to the mechanical behavior of biofilms under complex dynamic conditions.
IV. BIOFILM STREAMERS
Viscoelastic biofilms can exhibit a range of interesting responses to an external applied force. A common source of external force on biofilms is hydrodynamic flow. When there is a sustained hydrodynamic flow over a biofilm, filamentous structures called streamers can form. These streamers are generally tethered at one end to a surface while the rest of the structure floats freely in the bulk fluid.
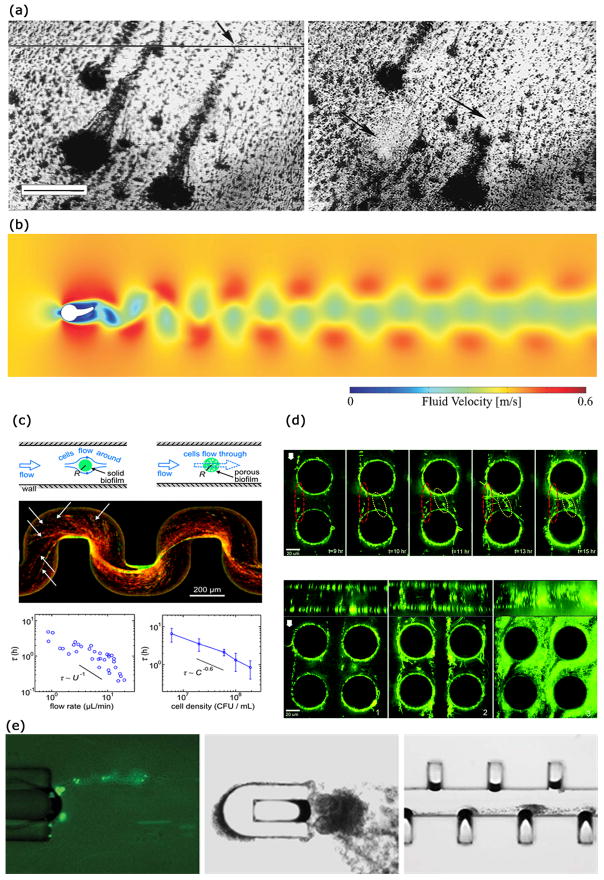
Biofilm streamers have been observed to form in turbulent flow conditions [99, 121], although several recent reports have demonstrated streamer formation in low Reynolds number conditions (Re < 1) [122–124]. Streamer formation in turbulent flow situations represents a more classical aspect of streamer related research (Fig. 5(a)) [99, 121, 125–131]. Streamers forming in such conditions can be observed in nature [125, 132] as well as in industrial situations [133]. Interestingly, some studies observed formation of streamers exclusively in turbulent flow regimes [121, 127] and not in the laminar flow conditions with moderate values of Reynolds number. The length scale of the streamers has been often reported to be on the order of 1 mm, where they begin to shed vortices (Fig. 5(b)) and cause significant pressure drops [121, 126, 131]. The mechanism that has been proposed for such observation of streamers in turbulent flows is as follows. In the initial stage, surface-hugging biofilms and discrete microcolonies form that are firmly attached to the surface [126]. The background turbulent transport results in a larger pressure on the upstream side of the microcolonies and a smaller pressure on the downstream section [126]. This triggers the formation of a wake region in the downstream section of the microcolonies [126]. Turbulent transport is characterized by high shear stresses and these forces cause a stream-lined growth as cells divide and multiply. Thus a preferential accumulation and growth of biomass occurs in the downstream section (wake region) [127]. In the presence of large pressure or form drag (caused by the pressure difference in the upstream and the downstream sides), as well as the large shear stress, this biomass starts to elongate and form a streamer [126]. Since streamers are composed of bacteria connected through EPS filaments, they behave as viscoelastic polymer materials. Formation of biofilm streamers in turbulent flows have also been used to elucidate the rheology of biofilms [130, 134].
FIG. 5.
Streamer formation under different hydrodynamic conditions. (a) Streamer formation by mixed population biofilms under turbulent flow conditions (Re ~ 1000). Images were taken after 7 days of growth. Scale bar is 500 μm. (b) Velocity field generated due to a streamer attached to a cylinder (Re ~ 100). Vortex shedding can induce pressure variations in the conduit. (c) Streamer formation by Pseudomonas aeruginosa in a microfluidic channel with curved sections. Streamers have a porous morphology and lead to catastrophic clogging. (d) Biofilm streamer formation by green fluorescent P. fluorescens in a creeping flow around micro-pillars. Streamer morphology is a function of the imposed flow rate. Streamer growth depends on hydrodynamic conditions as well as simulated pore-structure. Scale bar is 20 μm. (e) Biofilm streamer formation by E. coli due to vortical flows near a horse-shoe structure in a microfluidic device. The cavity in the horse-shoe structure is occupied by an air-bubble approximately 60 μm in width which is oscillating due to acoustic streaming. Images adapted and reproduced with permission from ref. 121, 131, 135, 123 and 124, respectively.
Streamers formed in low Reynolds number systems are relevant to a wide variety of scenarios including clogging of biomedical devices such as heart stents, biofilms in soil, and filtration systems [122, 123, 135, 136]. Biofilm streamers can occur in various membrane based systems such as between spacers in nanofiltration and reverse osmosis equipment leading to pressure drops and loss of process efficiency [137, 138].
Recently, multiple researchers have reported formation of biofilm streamers in microfluidic systems with characteristic Reynolds number much smaller than unity [122–124, 135, 137, 139]. Rusconi et al. [122] investigated the effect of channel curvature and channel geometry on the biofilm formation by Pseudomonas aeruginosa. They found that the streamers can form in curved sections of microchannels, and streamer morphology is related to motility of P. aeruginosa. The investigators reported that due to secondary flows in their device streamers formed only in the mid-section of the microfluidic device [140]. Drescher et al. [135] reported that biofilm streamers can lead to catastrophic disruption of flow in microfluidic systems (Fig. 5(c)). The same group later also showed that S. auerus biofilms formed faster in a microfluidic channel when the channel walls were coated with human blood plasma [141]. Valiei et al. [123] used a microfluidic device with an array of micro-pillars to simulate a porous media structure and studied biofilm formation in this device as a function of fluid flow rate. They found that in a certain flow regime, the bacterium Pseudomonas florescens formed streamers generating a web-like network between different pillars (Fig. 5(d)). Interestingly, streamers were observed to be distributed throughout the cross-section of the device despite the presence of secondary flows. Hassanpourfard et al. [142] provide detailed experimental protocols to study biofilm streamers in microfluidic device with micro-pillars. Marty et al. [137] used micro-pillars to simulate flow through membranes and observed streamer formation by Escherichia coli in non-nutritive conditions. Yazdi and Ardekani [124] showed that Escherichia coli streamers can form in vortical flows originating from an oscillating bubble (Fig. 5(e)). The correlation between the flow structures and formation of biofilm streamers in such low Re situations is not yet well understood. Das and Kumar [143] have recently proposed that these streamers form a highly viscous liquid state of the intrinsically viscoelastic biofilms. They based their conjecture on the observation that the time-scale of biofilm streamer formation typically far exceeds the viscoelastic relaxation time scales of biofilms. Biofilms are known to behave as viscoelastic liquids and hence at time-scales much larger than the viscoelastic relaxation time scale, the viscous behavior dominates [104].
Biofilm streamers represent a challenging scientific problem and here LOC devices can play an important role. Streamers, whether they form in high or low Reynolds number situations, typically are slender bodies with length scales that are compatible with LOC systems. LOC devices not only enable direct visualization of streamer formation, but they can allow for creation of various micro-models of complex habitats. Direct visualization of streamer formation and its impact on the surrounding flow field can lead to experimental data that can be used to calibrate various theoretical and numerical models. On the other hand, complex micro-models can allow us to explore the role of environmental conditions on formation of streamers.
V. QUORUM SENSING
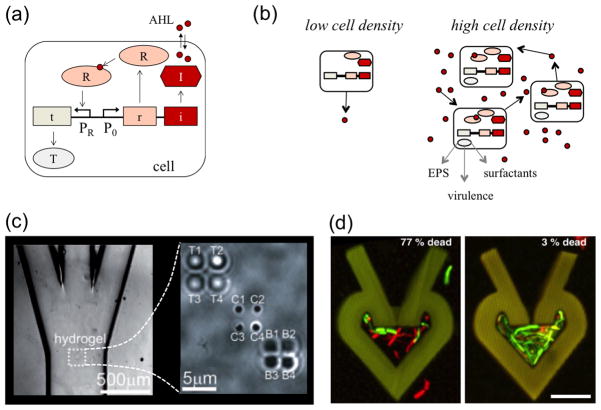
From a biological perspective, biofilm formation is fundamentally a group behavior, whereby bacteria produce and secrete a number of shared resources such as EPS. Not surprisingly, a number of different cell behaviors associated with biofilm formation are regulated through cell-cell communication in the form of quorum sensing. Quorum sensing (QS) merits significant attention in biofilm studies due to its importance in regulating an array of relevant functions [144], but also due to the interplay between quorum sensing dynamics, microenvironment properties, and hydrodynamics. In general, QS systems are comprised of a signal synthase, a signal receptor, and a gene regulatory circuit controlling production of the synthase and receptor (Fig. 6(a)). Gram negative bacterial quorum sensing systems, for example, are typically based on signaling with acyl homoserine lactones (AHL), which can diffuse through cell membranes. Typically, each cell produces a basal level of AHL, and when the population is high enough, AHL concentrations within the cell cross a threshold required for activation of target genes (Fig. 6(b)) [145].
FIG. 6.
(a) Overview of quorum sensing. QS systems are comprised of a signal synthase (I), a signal receptor (R), and a gene regulatory circuit controlling production of the synthase and receptor. Target gene products (T) are expressed when signal concentrations are high enough to activate the receptor. (b) At low local cell densities, cells secrete basal levels of quorum sensing molecules. When local cell density is high, basal levels of quorum sensing molecules accumulate and can lead to sufficient concentrations for activating expression of several genes. These activated genes regulate a variety of products and processes, including EPS secretion, virulence factor production, and surfactant secretion. (c) To study the effects of transport on cell-cell communication, Timp et al. [159] used optical traps to position “transmitter” and “receiver” cells in a hydrogel. This hydrogel was situated in a microfluidic channel, which enabled quantification of the effect of different media flow rates on interaction between the transmitter and receiver cells. (d) To study the effects of confinement on density dependent behavior, Connell et al. [165] optically cross-linked bovine serum albumin to trap cells in microcavities. P. aeruginosa was trapped in different microcavities and exposed for two hours to gentamycin. Dead cells appear red and live cells appear green. The higher density population on the right exhibited far greater survival. Images adapted and reproduced with permission from ref. [159, 165].
One important biofilm process regulated by cell-cell communication is the secretion of structural materials such as extracellular polymeric substances. In many Streptococcus species [146], for example, quorum sensing induces the secretion of DNA, which provides adhesive support to the biofilm [147]. Indeed, the fact that DNA degrading enzymes can cause biofilm dispersal highlights the fact that DNA contributes to the structural integrity and mechanical strength of biofilms [148]. Secretion of EPS components is also upregulated by quorum sensing in P. aeruginosa [149]. However, in this case, quorum sensing induces expression of the pel genes, which catalyze synthesis of a glucose rich exopolysaccharide that forms a key component of the biofilm matrix [149]. In contrast to Streptococcus and Pseudomonas species, other bacteria, such as V. cholera, use quorum sensing to repress EPS secretion. These differing regulation strategies suggest that the benefits of EPS secretion are context dependent. It has been argued that repressing EPS secretion can be beneficial in some cases, due to energy savings and to the facilitation of dispersal [150]. For instance, in the plant pathogen Pantoea stewartii, quorum sensing regulated repression of EPS secretion is necessary for proper maturation of the spatial structure of the biofilm [151].
In addition to regulating EPS secretion, quorum sensing often regulates surfactant secretion. The controlled secretion of surfactants directs the formation of mature biofilm structures as well as biofilm detachment. For example, the P. aeruginosa Rhl quorum sensing system controls secretion of rhamnolipids [152]. Rhamnolipids are associated with the formation of channel structures that facilitate nutrient exchange within the biofilm [153] but also mediate biofilm detachment [154]. Similarly, a recent study revealed that, in Staphylococcus aureus, the Agr quorum sensing system regulates the expression of phenol-soluble modulin surfactant peptides, which play a key role in the formation of channels within the biofilm and in preserving the typical detachment phenotype. Surfactants can also play a role in surface modification and colony spread [155]. In Serratia liquefaciens, quorum sensing controls production of the surfactant serrawettin, which helps to wet dry surfaces to facilitate swarming [156].
Given the importance of intercellular signaling in regulating not only key biofilm formation determinants but also important genes such as virulence factors, it is important to understand the influence of the environmental context on quorum sensing. In particular, the fluidic properties and the spatial structure of the habitat play critical roles in shaping quorum sensing dynamics. As illustrated by the examples below, LOC devices offer a powerful approach for defining the cells microenvironment and quantitatively probing the complex interplay between quorum sensing and biofilm formation.
A. Effect of Hydrodynamics
Since quorum sensing typically involves transport of molecular signals between cells, hydrodynamics play an essential role in determining quorum sensing dynamics. In fact, fundamental questions have been raised over the extent to which cells truly use quorum sensing to sense local population density as opposed to sensing environmental properties such as the local diffusion rate [157, 158]. Several recent studies have applied LOC devices to investigate these questions.
Biofilm mimics have been created to characterize communication between cells as a function of fluidic flow properties. For example, Timp et al. [159] used optical traps to position engineered cells within a microfluidic channel (Fig. 6(c)). The positioned cells were then fixed into place with a hydrogel that mimicked a biofilm extracellular matrix, and media was flowed at different rates through the microfluidic channel. At low flow rates, diffusion dominated transport led to efficient communication between transmitter and receiver cells. On the other hand, at high flow rates, convection dominated transport resulted in an eventual breakdown in communication. Thus, the density of bacteria that defines a critical “quorum” is a function of the hydrodynamics of the surrounding environment.
In another study, Luo et al. [160] developed biofilm mimics by synthesizing chitosan membranes to partition microfluidic channels and subsequently polymerizing a mixture of alginate and cells on either side of the membrane. Using this approach, they characterized transmission of the AI-2 quorum sensing signal under different flow rates. Specifically, they embedded AI-2 transmitter cells that produce AI-2 in alginate on one side of the chitosan membrane and AI-2 receiver cells that fluoresce in response to AI-2 in alginate on the other side. They then flowed fresh media through the channel at different rates. They found that increased flow rates delayed induction of the receiver cells. In fact, the time to reach half-maximal fluorescence induction increased linearly with flow rate.
In a third study, Meyer et al. characterized the quorum sensing performance of Pseudomonas putida under different flow conditions in a simple microfluidic chamber [161]. They found that, when media without AHL was flowed, the flow suppressed quorum sensing induction. Interestingly, although quorum sensing is thought of as coordinating behavior at the population level, significant quorum sensing response variability was observed among different colonies and among different cells within each colony, both with and without flow. This variability is caused by a combination of heterogeneity in the microenvironment as well as stochastic noise [162, 163] and has important evolutionary implications [164].
While the above studies illustrate how flow properties have a key impact on quorum sensing performance, recent work by Drescher et al. shows how quorum sensing determined cell phenotypes can have a strong influence on flow properties [135]. They examined streamer formation for different P. aeruginosa knockout strains, including lasR [135]. LasR is the receptor protein for the Las quorum sensing system, which regulates several processes including EPS secretion [149]. LasR knockout strains exhibited significantly delayed clogging compared to wild type cells, and the duration of clogging was significantly shorter [135].
B. Effect of Confinement
The characteristics of the confining space around cells, such as volume, geometry, and surface properties play a significant role in shaping quorum sensing and biofilm dynamics. Historically, such confinement effects have been difficult to probe, due to lack of control over empirical conditions at relevant scales. However, the application of recent micro and nanofabrication techniques has largely overcome this obstacle, enabling a number of studies of confinement phenomena. For example, Connell et al. trapped bacteria in picoliter-scale microcavities in order to probe quorum sensing dynamics for small population sizes and small volumes (Fig. 6(d)) [165]. These microcavities were produced by cross linking bovine serum albumin, and the resulting structures allowed for the exchange of nutrients and waste products. Bacteria entered the microcavities through small lumens at ambient temperature. However, once the temperature was raised to 37°C, the BSA polymer expanded to seal the lumens, thus trapping the bacteria. The resulting ability to capture small numbers of cells in small volumes enabled cell density to be varied independently from cell population size. Capitalizing on this feature, the authors showed that population size, population density, and external fluid flow rates all play a role in shaping quorum sensing dynamics. For example, they demonstrated that as few as 150 cells can exhibit quorum sensing, as long as the cells are confined to preserve a high cell density. This led to the demonstration that even small bacterial communities can exhibit properties such as antibiotic resistance that are typical of biofilms [165].
In a related study, Boedicker et al. trapped small numbers of P. aeruginosa cells in resin microwells [166]. The trapped cells expressed a fluorescent protein upon quorum sensing induction, which enabled quantification of quorum sensing dynamics. Although multiple cell division events were typically required for triggering quorum sensing gene expression, the authors observed a few cases of single cells expressing quorum sensing controlled genes. Also, in accord with the observations of Meyer et al. [161], significant variability was observed in quorum sensing induction.
C. Lab-on-a-chip Opportunities
As exemplified by the above studies of hydrodynamics and confinement effects, LOC devices have enabled studies and characterizations of quorum sensing behavior and dynamics that would be difficult or impossible through conventional approaches [135, 159–161]. Studies of this nature have been essential in addressing the question of whether cell-cell communication systems truly implement quorum sensing, whereby cells measure their local population density [167], or rather perform “diffusion sensing”, whereby cells measure the transport properties of their environment [157]. The findings that convection dominated transport can disrupt cell-cell communication [159, 160] and the demonstration that even a single, confined cell can activate communication controlled genes have drawn attention to the importance of local environment considerations in cell-cell communication. Recently, the concept of “efficiency sensing” has been proposed to reconcile the different viewpoints of cell-cell communication versus response to environmental conditions. Efficiency sensing holds that cells produce and sense autoinducers in order to estimate the effectiveness of producing extracellular goods, which is a combined function of cell density, hydrodynamic properties, and spatial cell distribution [157]. Due to the multiple factors associated with experimentally probing these fundamental hypotheses, LOC devices are expected to continue to play an essential role due to their capability of precisely controlling hydrodynamic properties, nutrient conditions, and confinement geometry.
A wealth of additional opportunities lies in the investigation of phenomena at the single cell level and at the cell community level. In spite of the potential coordination of behavior that cell-cell communication can enable, expression in individual cells can vary significantly. This variation can have important biological consequences and can, for instance, enable bet-hedging behavior that helps to stabilize the overall population [168]. While some studies have quantified heterogeneity in gene expression within populations [161, 166], much remains to be explored with regards to characterization of noise sources, dynamics, and their ramifications. Setups that facilitate tracking of individual cell function will likely be of particular relevance to these studies [169, 170].
Besides investigating the importance of single cells, investigating interspecies interactions is also of key importance. In nature, species rarely occur in isolation from other species. Thus, examining the role of intercellular communication within the broader context of mixed communities will be of key importance. For example, different species can “eavesdrop” on one another through chemical cues and correspondingly regulate competitiveness factors [171]. The work of Kim et al. exemplifies a step in the direction of quantifying interactions between different cell types [172]. They created a device that enabled three species to be grown in a spatially separated manner while allowing interaction between the species [172]. Specifically, bacteria were seeded in different wells, and each well exchanged small molecules with a microfluidic “communication channel” through a polycarbonate membrane interface. By quantifying strain growth for different spacings of wells and different combinations of community members, Kim et al. [172] were able to characterize the interactions between community members and investigate the importance of spatial structure in maintaining the community.
Like the phenomena of near surface motility and streamer formation, cell-cell communication is affected by hydrodynamics, cell phenotype, and the geometry of the surrounding environment. LOC devices will continue to play an integral role in the study of these phenomena by offering control over the various influencing factors. We now proceed to discuss the study of biofilm formation on patterned surfaces, which is also benefited by the employment of LOC devices.
VI. BIOFILMS ON PATTERNED SURFACES
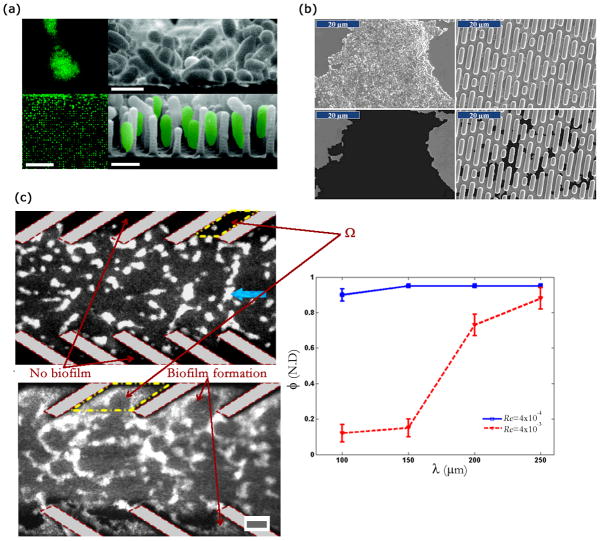
Bacterial attachment to surfaces is a complex process involving physical and chemical events that span a wide range of length and time scales. Bacteria are capable of attaching to a very wide variety of materials that include glass, various metals, and polymer based materials. Recent review articles have detailed the various processes involved in bacteria-surface interaction [61, 173]. On the applied side, the goal usually is to develop surfaces that can resist biofilm formation. Formation of biofilms on a surface can often result in loss of functionality and hence surfaces that can resist biofilm formation are of considerable interest to several industrial sectors. These include marine and biomedical applications. Various strategies have been investigated for such applications including development of various coatings [174] and patterned surfaces. Patterned surfaces can be created by a deliberate design of the surface topography and some commonly used engineered topographies involve the usage of array of posts or holes [175]. Use of an array of micro/nano-scale posts as an engineered topography was investigated by Hochbaum and Aizenberg [176]. They found that P. aeruginosa can modulate its attachment morphology based on presence of topographies (Fig. 7(a)). In another work, Graham et al. observed that cell attachment was strongly dependent upon topographical features under both static and microfluidic flow conditions [177].
FIG. 7.
Effect of patterned topographies on the biofilm formation (a) Adhesion of P. aeruginosa on structures and unstructured substrates. A stark difference in adhesion morphology is observed when the substrate topography is unstructured (top two images) as opposed to micro-textured (bottom two images). Scale bar is 10 μm in left sub-figures and 1 μm in right two sub-figures. (b) SEM images of S. aureus surface coverage on smooth and Sharklet AF surfaces. Bacterial coverage decreases substantially in the latter case. (c) Biofilm formation by fluorescent Shewanella oneidensis on the PDMS substrate flanked by biomimetic baffles. Flow of fluid is from right to left. Secondary flows are produced inside the baffles, which prevent the biofilm formation inside those regions. Prevention of biofilm formation correlates to the baffle pitch and imposed flow rate. The scale bar is 60 μm. Images adapted and reproduced with permission from ref. [176, 181, 184].
Engineered topographies that are biomimetic in design have also been used in the context of understanding and controlling biofilm formation on surfaces [178–180]. Some of these biomimetic designs are inspired by marine organisms whose surface features (e.g. shark-skin, invertebrate shells) are believed to be defense mechanisms against biofouling by the naturally occurring diverse microbial population. Chung et al. [181] demonstrated significantly delayed biofilm formation by Staphylococcus aureus on a topography inspired by certain surface patterns present on shark-skin (Fig. 7(b)). This topography (also known as the Sharklet AF topography) consists of 2 μm wide rectangular ribs spaced in a periodic pattern. These topographies could delay biofilm formation by the microbes for several days as opposed to an unpatterned surface. Similar reduction in settlement of zoospores of macroalga Ulva were demonstrated on these moulded biomimetic topographies [182]. The reduction in surface coverage was later shown to be correlated to an empirically derived engineered roughness index [183]. Surface patterns used to affect biofilm formation have typically employed topographies with length scales in the micro and nano-scale regimes. Use of biomimetic structures in the mesoscopic length scale (length scale ~ 10–100 μm) was investigated by Kumar et al. [184]. The investigators designed baffles in a microfluidic device inspired by shark-skin topography. These baffled structures, were shown to create secondary flows in the presence of an externally applied pressure gradient (Fig. 7(c)). The presence of secondary flow structures affected mass transport across the separating streamline. When fluid flow velocity scale exceeded the velocity scale of bacterial motility, biofilm formation was delayed in the baffled region. Such research bears strong promise in biofouling contexts, where there is a constant flow of fluid (e.g. pipes).
VII. PERSPECTIVE AND FUTURE DIRECTIONS
Scientific appreciation for surface associated bacterial colonies started in the mid twentieth century [185]. Since the early days of biofilm research, substantial effort has been invested in understanding the biological and genetic alterations in microbial cells living in the community structure of a biofilm. In this regard, numerous biofilm assays and screening techniques have been developed to monitor the phenotypic variation of a wide variety of bacteria throughout the processes associated with the formation, growth, and detachment of the biofilms [1, 186, 187]. Despite the abundance of microbiological investigations, substantial lacunae in interdisciplinary research in this topic remain [5, 94, 173]. This disparity, which in the recent years has attracted researchers, has led to new appreciation for the role of the environmental conditions and transport phenomena vis-à-vis biofilm formation. Interestingly, LOC systems afford researchers numerous advantages for investigating the role of the environment on bacterial dynamics and biofilm maturation and growth.
The combination of synthetic biology with cutting edge LOC techniques offers great potential for teasing apart the different interwoven roles of fluidics, microenvironment, cell-cell communication, and gene regulation. The use of forward engineered synthetic gene networks that are more decoupled from native host machinery can facilitate analysis. For example, the study by Timp et al. [159] exemplifies the power of decoupling quorum sensing from its downstream regulatory functions to facilitate cleaner analysis of the interplay between microenvironment properties, fluid flow characteristics, population structure, and quorum sensing function.
In reciprocation, sophisticated LOC devices will also aid biofilm engineering. A growing number of synthetic biology efforts are focusing on either engineering cells for operation within a biofilm context or manipulating biofilms [188–190]. LOC devices can aid the characterization and optimization of future pursuits in this area by providing control over cell confinement, hydrodynamics, and inducer concentrations. An excellent example is the recent construction of a synthetic biofilm consortia in which “disperser” cells can be added to initial colonizer cells to enable removal of engineered biofilms [191]. In this system, disperser cells produce a quorum sensing signal that diffuses to neighboring colonizer cells and activates expression of the biofilm dispersing protein BcdAE50Q. Once disperser cells displace the initial colonizers, the inducer isopropyl-β-D-thiogalactopyranoside (IPTG) can be added to activate Hha13D6 biofilm dispersing signal in order to remove the disperser cells. Characterization and optimization of the system was facilitated by a microfluidic device that allowed control over nutrient flow, introduction of different inducers, and introduction of different cell types [191].
The use of LOC systems extends beyond the fundamental investigations and the development of test beds. These systems can also enable building of application-oriented miniaturized systems. Miniaturized microbial fuel cells that can convert electrochemical energy to electrical energy have already been implemented [192, 193], exhibiting a promising development in utilization of biofilms in practical applications.
There exist several open questions regarding biofilms spanning different time and length scales. For example, the role of background flow field and viscoelasticity of EPS in the interaction of bacterial cells with each other [194, 195] and with nearby surfaces [196–198] is largely unexplored. Advances in LOC technology can help unravel the fundamental physical mechanisms governing the complex interplay of bacterial biofilms and the surrounding environment. However, there are some limitations associated with the LOC approach which necessitate further research. For instance, when the microfluidic chambers are employed to study the behavior and morphology of biofilms in a long time scale, clogging problems might arise due to excessive biofilm growth. This problem is intensified in high aspect-ratio microchannels where the lateral confinement might alter the evolution of the biofilms compared to their natural habitats. Moreover, despite the effort conducted to reproduce the in vivo conditions, current microfluidic devices can mimic the host environments of the bacterial biofilms only to a certain extent. Future LOC investigations need to be directed towards developing sophisticated miniaturized devices capable of replicating the complex dynamics of host-biofilm chemical and physical interactions.
To conclude, biofilms, despite decades of prior research, represent an exciting frontier in interdisciplinary research today. Collaborative effort between engineers, physicists, chemists and biologists is needed to further our understanding of this social form of bacterial living.
Supplementary Material
Acknowledgments
This work was partially supported by the National Science Foundation under grant CBET-1150348-CAREER (AMA), and the Indiana Clinical and Translational Sciences Institute under grant TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, and Clinical and Translational Sciences Award program (AMA). DK acknowledges support from the Independent Research and Development Program of the Johns Hopkins University Applied Physics Laboratory.
References
- 1.O’Toole G, Kaplan HB, Kolter R. Annual Review of Microbiology. 2000;54:49. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 2.Drenkard E. Microbes and Infection. 2003;5:1213. doi: 10.1016/j.micinf.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Davies DG. Society For General Microbiology Symposium. Cambridge: Cambridge University Press; 2000. pp. 37–52. [Google Scholar]
- 4.Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Journal of bacteriology. 1999;181:5993. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong GCL, O’Toole GA. MRS Bulletin. 2011;36:339. doi: 10.1557/mrs.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fairbrother L, Etschmann B, Brugger J, Shapter J, Southam G, Reith F. Environmental Science & Technology. 2013;47:2628. doi: 10.1021/es302381d. [DOI] [PubMed] [Google Scholar]
- 7.Wuertz S, Bishop PL, Wilderer PA. Biofilms in wastewater treatment: An interdisciplinary approach. IWA Publishing; London: 2003. [Google Scholar]
- 8.Prince RC. Trends In Biotechnology. 1997;15:158. [Google Scholar]
- 9.Picioreanu C, van Loosdrecht MCM, Heijnen JJ. In: Community Structure and Co-operation in Biofilms. Allison DG, Gilbert P, Lappin-Scott HM, Wilson M, editors. Society for General Microbiology; 2000. pp. 129–166. [Google Scholar]
- 10.Picioreanu C, van Loosdrecht MCM, Heijnen JJ. Biotechnology and Bioengineering. 2000;69:504. doi: 10.1002/1097-0290(20000905)69:5<504::aid-bit5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 11.Cisneros LH, Cortez R, Dombrowski C, Goldstein RE, Kessler JO. Experiments In Fluids. 2007;43:737. [Google Scholar]
- 12.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Nature. 2002;415:545. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 13.Vertes A, Hitchins V, Phillips KS. Analytical Chemistry. 2012;84:3858. doi: 10.1021/ac2029997. [DOI] [PubMed] [Google Scholar]
- 14.Neethirajan S, Karig D, Kumar A, Mukherjee PP, Retterer ST, Doktycz MJ. Encyclopedia of Nanotechnology. Springer; 2012. pp. 213–219. [Google Scholar]
- 15.Benoit MR, Conant CG, Ionescu-Zanetti C, Schwartz M, Matin A. Applied and Environmental Microbiology. 2010;76:4136. doi: 10.1128/AEM.03065-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer MT, Roy V, Bentley WE, Ghodssi R. Sensors, 2010 IEEE. IEEE; 2010. pp. 2291–2294. [Google Scholar]
- 17.Kim J, Park H-D, Chung S. Molecules. 2012;17:9818. doi: 10.3390/molecules17089818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosier AP, Kaloyeros AE, Cady NC. Journal of Microbiological Methods. 2012;91:198. doi: 10.1016/j.mimet.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Bray D. Cell movements: from molecules to motility. 2. Garland Pub; New York: 2001. [Google Scholar]
- 20.Ottemann KM, Miller JF. Molecular Microbiology. 1997;24:1109. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 21.Harshey RM. Annual Review of Microbiology. 2003;57:249. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 22.Lappin-Scott HM, Costerton JW. Microbial Biofilms. Vol. 5. Cambridge University Press; Cambridge, England: 2003. [Google Scholar]
- 23.Fux CA, Costerton JW, Stewart PS, Stoodley P. Trends in Microbiology. 2005;13:34. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Kolter R, Greenberg EP. Nature. 2006;441:300. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 25.Heijnen JJ, Vanloosdrecht MCM, Mulder R, Weltevrede R, Mulder A. Water Science and Technology. 1993;27:253. [Google Scholar]
- 26.Characklis WG, Nevimons MJ, Picologlou BF. Heat Transfer Engineering. 1981;3:23. [Google Scholar]
- 27.dos Santos VAPM, Yakimov MM, Timmis KN, Golyshin PN. Microbial Biodegradation: Genomics and Molecular Biology. Horizon Scientific Press; 2008. pp. 269–296. [Google Scholar]
- 28.Purcell EM. American Journal of Physics. 1977;45:3. [Google Scholar]
- 29.Berg HC. E. coli in Motion. Springer; New York: 2004. [Google Scholar]
- 30.Lauga E, Powers TR. Reports on Progress in Physics. 2009;72:096601. [Google Scholar]
- 31.Blair DF. Annual Review of Microbiology. 1995;49:489. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 32.Brennen C, Winet H. Annual Review of Fluid Mechanics. 1977;9:339. [Google Scholar]
- 33.Turner L, Ryu WS, Berg HC. Journal of Bacteriology. 2000;182:2793. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell JG. American Naturalist. 2002;160:727. doi: 10.1086/343874. [DOI] [PubMed] [Google Scholar]
- 35.Frymier PD, Ford RM, Berg HC, Cummings PT. Proceedings of the National Academy of Sciences. 1995;92:6195. doi: 10.1073/pnas.92.13.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vigeant MA-S, Ford RM, Wagner M, Tamm LK. Applied and Environmental Microbiology. 2002;68:2794. doi: 10.1128/AEM.68.6.2794-2801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berg HC. Current Biology. 2008;18:R689. doi: 10.1016/j.cub.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 38.Lighthill J. SIAM Review. 1976;18:161. [Google Scholar]
- 39.Berg HC, Brown DA. Nature. 1972;239:500. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 40.Berg HC, Turner L. Biophysical Journal. 1990;58:919. doi: 10.1016/S0006-3495(90)82436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauga E, DiLuzio WR, Whitesides GM, Stone HA. Biophysical Journal. 2006;90:400. doi: 10.1529/biophysj.105.069401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DiLuzio WR, Turner L, Mayer M, Garstecki P, Weibel DB, Berg HC, Whitesides GM. Nature. 2005;435:1271. doi: 10.1038/nature03660. [DOI] [PubMed] [Google Scholar]
- 43.de Kerchove AJ, Elimelech M. Environmental Science and Technology. 2008;42:4371. doi: 10.1021/es703028u. [DOI] [PubMed] [Google Scholar]
- 44.Ramia M, Tullock DL, Phan-Thien N. Biophysical journal. 1993;65:755. doi: 10.1016/S0006-3495(93)81129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shum H, Gaffney EA, Smith DJ. Proceedings of the Royal Society A-Mathematical Physical and Engineering Sciences. 2010;466:1725. [Google Scholar]
- 46.Frymier PD, Ford RM. AIChE Journal. 1997;43:1341. [Google Scholar]
- 47.Biondi SA, Quinn JA, Goldfine H. AIChE Journal. 1998;44:1923. [Google Scholar]
- 48.Giacche D, Ishikawa T, Yamaguchi T. Physical Review E. 2010;82:056309. doi: 10.1103/PhysRevE.82.056309. [DOI] [PubMed] [Google Scholar]
- 49.Taylor BL, Koshland DE. Journal of Bacteriology. 1974;119:640. doi: 10.1128/jb.119.2.640-642.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shigematsu M, Meno Y, Misumi H, Amako K. Microbiology and Immunology. 1995;39:741. doi: 10.1111/j.1348-0421.1995.tb03260.x. [DOI] [PubMed] [Google Scholar]
- 51.McCarter LL. Microbiology and Molecular Biology Reviews. 2001;65:445. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goto T, Nakata K, Baba K, Nishimura M, Magariyama Y. Biophysical Journal. 2005;89:3771. doi: 10.1529/biophysj.105.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie L, Altindal T, Chattopadhyay S, Wu X-L. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2246. doi: 10.1073/pnas.1011953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stocker R. Proceedings of the National Academy of Sciences. 2011;108:2635. doi: 10.1073/pnas.1019199108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magariyama Y, Ichiba M, Nakata K, Baba K, Ohtani T, Kudo S, Goto T. Biophysical Journal. 2005;88:3648. doi: 10.1529/biophysj.104.054049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kudo S, Imai N, Nishitoba M, Sugiyama S, Magariyama Y. FEMS Microbiology Letters. 2005;242:221. doi: 10.1016/j.femsle.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 57.Li G, Tang JX. Biophysical Journal. 2006;91:2726. doi: 10.1529/biophysj.106.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C. Embo Journal. 2009;28:1208. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magariyama Y, Masuda S, Takano Y, Ohtani T, Kudo S. FEMS Microbiology Letters. 2001;205:343. doi: 10.1111/j.1574-6968.2001.tb10970.x. [DOI] [PubMed] [Google Scholar]
- 60.Nakai T, Kikuda M, Kuroda Y, Goto T. Journal of Biomechanical Science and Engineering. 2009;4:2. [Google Scholar]
- 61.Tuson HH, Weibel DB. Soft Matter. 2013;9:4368. doi: 10.1039/C3SM27705D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Houry A, Briandet R, Aymerich S, Gohar M. Microbiology. 2010;156:1009. doi: 10.1099/mic.0.034827-0. [DOI] [PubMed] [Google Scholar]
- 63.Merritt PM, Danhorn T, Fuqua C. Journal of Bacteriology. 2007;189:8005. doi: 10.1128/JB.00566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guttenplan SB, Kearns DB. Fems Microbiology Reviews. 2013;37:849. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Journal of Bacteriology. 2002;184:1140. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. FEMS Immunology and Medical Microbiology. 2010;59:253. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- 67.Boles BR, Horswill AR. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12848. doi: 10.1073/pnas.1210297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Houry A, Gohar M, Deschamps J, Tischenko E, Aymerich S, Gruss A, Briandet R. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13088. doi: 10.1073/pnas.1200791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R. Molecular Microbiology. 2001;39:223. doi: 10.1046/j.1365-2958.2001.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burrows LL. Annual Review of Microbiology. 2012;66:493. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 71.Skerker JM, Berg HC. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6901. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maier B, Potter L, So M, Seifert HS, Sheetz MP. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16012. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Biais N, Ladoux B, Higashi D, So M, Sheetz M. Plos Biology. 2008;6:907. doi: 10.1371/journal.pbio.0060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li G, Brown PJB, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. Molecular Microbiology. 2012;83:41. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holz C, Opitz D, Greune L, Kurre R, Koomey M, Schmidt MA, Maier B. Physical Review Letters. 2010;104:178104. doi: 10.1103/PhysRevLett.104.178104. [DOI] [PubMed] [Google Scholar]
- 76.Maier B, Koomey M, Sheetz MP. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10961. doi: 10.1073/pnas.0402305101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghosh R, Kumar A, Vaziri A. 2014 arXiv preprint arXiv:1409.5472.
- 78.Gibiansky ML, Conrad JC, Jin F, Gordon VD, Motto DA, Mathewson MA, Stopka WG, Zelasko DC, Shrout JD, Wong GCL. Science. 2010;330:197. doi: 10.1126/science.1194238. [DOI] [PubMed] [Google Scholar]
- 79.Conrad JC, Gibiansky ML, Jin F, Gordon VD, Motto DA, Mathewson MA, Stopka WG, Zelasko DC, Shrout JD, Wong GCL. Biophysical journal. 2011;100:1608. doi: 10.1016/j.bpj.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jin F, Conrad JC, Gibiansky ML, Wong GCL. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12617. doi: 10.1073/pnas.1105073108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Toole GA, Kolter R. Molecular Microbiology. 1998;30:295. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 82.Pratt LA, Kolter R. Molecular Microbiology. 1998;30:285. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 83.Li Y, Hao G, Galvani CD, Meng Y, De la Fuente L, Hoch HC, Burr TJ. Microbiology. 2007;153:719. doi: 10.1099/mic.0.2006/002311-0. [DOI] [PubMed] [Google Scholar]
- 84.Chiang P, Burrows LL. Journal of Bacteriology. 2003;185:2374. doi: 10.1128/JB.185.7.2374-2378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Persat A, Stone HA, Gitai Z. Nature Communications. 2014;5:3824. doi: 10.1038/ncomms4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li G, Tam L-K, Tang JX. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18355. doi: 10.1073/pnas.0807305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De La Fuente L, Montanes E, Meng Y, Li Y, Burr TJ, Hoch HC, Wu M. Applied and Environmental Microbiology. 2007;73:2690. doi: 10.1128/AEM.02649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wright E, Neethirajan S, Warriner K, Retterer S, Srijanto B. Lab on a Chip. 2014;14:938. doi: 10.1039/c3lc51138c. [DOI] [PubMed] [Google Scholar]
- 89.De La Fuente L, Burr TJ, Hoch HC. Applied and Environmental Microbiology. 2008;74:5579. doi: 10.1128/AEM.00995-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cruz LF, Cobine PA, De La Fuente L. Applied and Environmental Microbiology. 2012;78:1321. doi: 10.1128/AEM.06501-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bahar O, De La Fuente L, Burdman S. FEMS Microbiology Letters. 2010;312:33. doi: 10.1111/j.1574-6968.2010.02094.x. [DOI] [PubMed] [Google Scholar]
- 92.Flemming H-C, Wingender J. Nature Reviews Microbiology. 2010;8:623. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 93.Donlan RM. Emerging Infectious Diseases. 2002;8:881. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilking JN, Angelini TE, Seminara A, Brenner MP, Weitz DA. Mrs Bulletin. 2011;36:385. [Google Scholar]
- 95.Di Stefano A, D’Aurizio E, Trubiani O, Grande R, Di Campli E, Di Giulio M, Di Bartolomeo S, Sozio P, Iannitelli A, Nostro A, Cellini L. Microbial Biotechnology. 2009;2:634. doi: 10.1111/j.1751-7915.2009.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Davies D. Nature Reviews Drug Discovery. 2003;2:114. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 97.Flemming HC, Wingender J, Mayer C, Korstgens V, Borchard W. In: Community Structure and Cooperation in Biofilms. Allison D, Gilbert P, Lappin-Scott HM, Wilson M, editors. Cambridge University Press; 2000. pp. 87–106. [Google Scholar]
- 98.Rupp CJ, Fux CA, Stoodley P. Applied and Environmental Microbiology. 2005;71:2175. doi: 10.1128/AEM.71.4.2175-2178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stoodley P, Cargo R, Rupp CJ, Wilson S, Klapper I. Journal of Industrial Microbiology and Biotechnology. 2002;29:361. doi: 10.1038/sj.jim.7000282. [DOI] [PubMed] [Google Scholar]
- 100.Vinogradov AM, Winston M, Rupp CJ, Stoodley P. Biofilms. 2004;1:49. doi: 10.1103/PhysRevLett.93.098102. [DOI] [PubMed] [Google Scholar]
- 101.Pavlovsky L, Younger JG, Solomon MJ. Soft Matter. 2013;9:122. doi: 10.1039/C2SM27005F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lieleg O, Caldara M, Baumgaertel R, Ribbeck K. Soft Matter. 2011;7:3307. doi: 10.1039/c0sm01467b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rühs PA, Boeni L, Fuller GG, Inglis RF, Fischer P. Plos One. 2013;8:e78524. doi: 10.1371/journal.pone.0078524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shaw T, Winston M, Rupp CJ, Klapper I, Stoodley P. Physical Review Letters. 2004;93:098102. doi: 10.1103/PhysRevLett.93.098102. [DOI] [PubMed] [Google Scholar]
- 105.Wloka M, Rehage H, Flemming HC, Wingender J. Colloid and Polymer Science. 2004;282:1067. [Google Scholar]
- 106.Branda SS, Vik A, Friedman L, Kolter R. Trends in Microbiology. 2005;13:20. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 107.Hohne DN, Younger JG, Solomon MJ. Langmuir. 2009;25:7743. doi: 10.1021/la803413x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jones WL, Sutton MP, McKittrick L, Stewart PS. Biofouling. 2011;27:207. doi: 10.1080/08927014.2011.554977. [DOI] [PubMed] [Google Scholar]
- 109.Aggarwal S, Hozalski RM. Langmuir. 2012;28:2812. doi: 10.1021/la204342q. [DOI] [PubMed] [Google Scholar]
- 110.Rogers SS, van der Walle C, Waigh TA. Langmuir. 2008;24:13549. doi: 10.1021/la802442d. [DOI] [PubMed] [Google Scholar]
- 111.Galy O, Latour-Lambert P, Zrelli K, Ghigo J-M, Beloin C, Henry N. Biophysical Journal. 2012;103:1400. doi: 10.1016/j.bpj.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lau PCY, Dutcher JR, Beveridge TJ, Lam JS. Biophysical Journal. 2009;96:2935. doi: 10.1016/j.bpj.2008.12.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Squires TM, Mason TG. Annual Review of Fluid Mechanics. 2010;42:413. [Google Scholar]
- 114.Crocker JC, Valentine MT, Weeks ER, Gisler T, Kaplan PD, Yodh AG, Weitz DA. Physical Review Letters. 2000;85:888. doi: 10.1103/PhysRevLett.85.888. [DOI] [PubMed] [Google Scholar]
- 115.Gazuz I, Puertas AM, Voigtmann T, Fuchs M. Physical Review Letters. 2009;102:248302. doi: 10.1103/PhysRevLett.102.248302. [DOI] [PubMed] [Google Scholar]
- 116.Hu X, Boukany PE, Hemminger OL, Lee LJ. Macromolecular Materials and Engineering. 2011;296:308. [Google Scholar]
- 117.Brust M, Schaefer C, Doerr R, Pan L, Garcia M, Arratia PE, Wagner C. Physical Review Letters. 2013;110:078305. doi: 10.1103/PhysRevLett.110.078305. [DOI] [PubMed] [Google Scholar]
- 118.Preira P, Valignat M-P, Bico J, Théodoly O. Biomicrofluidics. 2013;7:024111. doi: 10.1063/1.4802272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pan L, Arratia PE. Microfluidics and Nanofluidics. 2013;14:885. [Google Scholar]
- 120.Drost S, Westerweel J. Journal of Rheology. 2013;57:1787. [Google Scholar]
- 121.Stoodley P, Lewandowski Z, Boyle JD, Lappin-Scott HM. Biotechnology and Bioengineering. 1998;57:536. doi: 10.1002/(sici)1097-0290(19980305)57:5<536::aid-bit5>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 122.Rusconi R, Lecuyer S, Guglielmini L, Stone HA. Journal of the Royal Society Interface. 2010;7:1293. doi: 10.1098/rsif.2010.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Valiei A, Kumar A, Mukherjee PP, Liu Y, Thundat T. Lab on a Chip. 2012;12:5133–5137. doi: 10.1039/c2lc40815e. [DOI] [PubMed] [Google Scholar]
- 124.Yazdi S, Ardekani AM. Biomicrofluidics. 2012;6:044114. doi: 10.1063/1.4771407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hallberg KB, Coupland K, Kimura S, Johnson DB. Applied and Environmental Microbiology. 2006;72:2022. doi: 10.1128/AEM.72.3.2022-2030.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lewandowski Z, Stoodley P. Water Science and Technology. 1995;32:19. [Google Scholar]
- 127.Stoodley P, Dodds I, Boyle JD, Lappin-Scott HM. Journal of Applied Microbiology. 1999;85:19s. doi: 10.1111/j.1365-2672.1998.tb05279.x. [DOI] [PubMed] [Google Scholar]
- 128.Stoodley P, Dodds I, De Beer D, Scott HL, Boyle JD. Biofouling. 2005;21:161. doi: 10.1080/08927010500375524. [DOI] [PubMed] [Google Scholar]
- 129.Stoodley P, Lewandowski Z, Boyle JD, Lappin-Scott HM. Environmental Microbiology. 1999;1:447. doi: 10.1046/j.1462-2920.1999.00055.x. [DOI] [PubMed] [Google Scholar]
- 130.Stoodley P, Lewandowski Z, Boyle JD, Lappin-Scott HM. Biotechnology and Bioengineering. 1999;65:83. [PubMed] [Google Scholar]
- 131.Taherzadeh D, Picioreanu C, Kuttler U, Simone A, Wall WA, Horn H. Biotechnology and Bioengineering. 2010;105:600. doi: 10.1002/bit.22551. [DOI] [PubMed] [Google Scholar]
- 132.Meyer-Dombard DR, Swingley W, Raymond J, Havig J, Shock EL, Summons RE. Environmental Microbiology. 2011;13:2216. doi: 10.1111/j.1462-2920.2011.02476.x. [DOI] [PubMed] [Google Scholar]
- 133.Barton AF, Wallis MR, Sargison JE, Buia A, Walker GJ. Journal of Hydraulic Engineering-Asce. 2008;134:852. [Google Scholar]
- 134.Aravas N, Laspidou CS. Biotechnology and Bioengineering. 2008;101:196. doi: 10.1002/bit.21865. [DOI] [PubMed] [Google Scholar]
- 135.Drescher K, Shen Y, Bassler BL, Stone HA. Proceedings of the National Academy of Sciences. 2013;110:4345–4350. doi: 10.1073/pnas.1300321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Durham WM, Tranzer O, Leombruni A, Stocker R. Physics of Fluids. 2012;24:091107. [Google Scholar]
- 137.Marty A, Roques C, Causserand C, Bacchin P. Biofouling. 2012;28:551. doi: 10.1080/08927014.2012.695351. [DOI] [PubMed] [Google Scholar]
- 138.Suwarno SR, Chen X, Chong TH, Puspitasari VL, McDougald D, Cohen Y, Rice SA, Fane AG. Journal of Membrane Science. 2012;405:219. [Google Scholar]
- 139.Weaver WM, Milisavljevic V, Miller JF, Di Carlo D. Applied and Environmental Microbiology. 2012;78:5890. doi: 10.1128/AEM.01139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rusconi R, Lecuyer S, Autrusson N, Guglielmini L, Stone HA. Biophysical Journal. 2011;100:1392. doi: 10.1016/j.bpj.2011.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim MK, Drescher K, Pak OS, Bassler BL, Stone HA. New Journal of Physics. 2014;16:065024. doi: 10.1088/1367-2630/16/6/065024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hassanpourfard M, Sun X, Valiei A, Mukherjee P, Thundat T, Liu Y, Kumar A. JoVE (Journal of Visualized Experiments) 2014:e51732. doi: 10.3791/51732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Das S, Kumar A. 2013 arXiv preprint arXiv:1312.6056. [Google Scholar]
- 144.Parsek MR, Greenberg EP. Trends in microbiology. 2005;13:27. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 145.Taga ME, Bassler BL. Proc Natl Acad Sci U S A 100 Suppl. 2003;2:14549. doi: 10.1073/pnas.1934514100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Spoering AL, Gilmore MS. Current opinion in microbiology. 2006;9:133. doi: 10.1016/j.mib.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 147.Dominiak DM, Nielsen JL, Nielsen PH. Environmental Microbiology. 2011;13:710. doi: 10.1111/j.1462-2920.2010.02375.x. [DOI] [PubMed] [Google Scholar]
- 148.Jakubovics NS, Shields RC, Rajarajan N, Burgess JG. Letters in Applied Microbiology. 2013;57:467. doi: 10.1111/lam.12134. [DOI] [PubMed] [Google Scholar]
- 149.Sakuragi Y, Kolter R. Journal of bacteriology. 2007;189:5383. doi: 10.1128/JB.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nadell CD, Xavier JB, Levin SA, Foster KR. PLoS Biol. 2008;6:e14. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Koutsoudis MD, Tsaltas D, Minogue TD, von Bodman SB. Proceedings of the National Academy of Sciences. 2006;103:5983. doi: 10.1073/pnas.0509860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pearson JP, Pesci EC, Iglewski BH. Journal of bacteriology. 1997;179:5756. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, Williams P. Molecular microbiology. 2003;50:29. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 154.Boles BR, Thoendel M, Singh PK. Molecular microbiology. 2005;57:1210. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- 155.Daniels R, Vanderleyden J, Michiels J. FEMS Microbiology Reviews. 2004;28:261. doi: 10.1016/j.femsre.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 156.Eberl L, Molin S, Givskov M. Journal of bacteriology. 1999;181:1703. doi: 10.1128/jb.181.6.1703-1712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hense BA, Kuttler C, Müller J, Rothballer M, Hartmann A, Kreft J-U. Nature Reviews Microbiology. 2007;5:230. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- 158.Redfield RJ. Trends in microbiology. 2002;10:365. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 159.Timp W, Mirsaidov U, Matsudaira P, Timp G. Lab on a Chip. 2009;9:925. doi: 10.1039/b810157d. [DOI] [PubMed] [Google Scholar]
- 160.Luo X, Wu H-C, Tsao C-Y, Cheng Y, Betz J, Payne GF, Rubloff GW, Bentley WE. Biomaterials. 2012;33:5136. doi: 10.1016/j.biomaterials.2012.03.037. [DOI] [PubMed] [Google Scholar]
- 161.Meyer A, Megerle JA, Kuttler C, Müller J, Aguilar C, Eberl L, Hense BA, Rädler JO. Physical Biology. 2012;9:026007. doi: 10.1088/1478-3975/9/2/026007. [DOI] [PubMed] [Google Scholar]
- 162.Balázsi G, van Oudenaarden A, Collins JJ. Cell. 2011;144:910. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Simpson ML, Cox CD, Allen MS, McCollum JM, Dar RD, Karig DK, Cooke JF. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. 2009;1:214. doi: 10.1002/wnan.22. [DOI] [PubMed] [Google Scholar]
- 164.Pai A, Tanouchi Y, You L. Proceedings of the National Academy of Sciences. 2012;109:19810. doi: 10.1073/pnas.1211072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Connell JL, Wessel AK, Parsek MR, Ellington AD, Whiteley M, Shear JB. MBio. 2010;1:e00202. doi: 10.1128/mBio.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Boedicker JQ, Vincent ME, Ismagilov RF. Angewandte Chemie-International Edition. 2009;48:5908. doi: 10.1002/anie.200901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fuqua WC, Winans SC, Greenberg EP. Journal of bacteriology. 1994;176:269. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Davidson CJ, Surette MG. Annual Review of Genetics. 2008;42:253. doi: 10.1146/annurev.genet.42.110807.091601. [DOI] [PubMed] [Google Scholar]
- 169.Cho H, Jonsson H, Campbell K, Melke P, Williams JW, Jedynak B, Stevens AM, Groisman A, Levchenko A. PLoS Biol. 2007;5:e302. doi: 10.1371/journal.pbio.0050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Volfson D, Cookson S, Hasty J, Tsimring LS. Proceedings of the National Academy of Sciences. 2008;105:15346. doi: 10.1073/pnas.0706805105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chandler JR, Heilmann S, Mittler JE, Greenberg EP. The ISME journal. 2012;6:2219. doi: 10.1038/ismej.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Proceedings of the National Academy of Sciences. 2008;105:18188. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Renner LD, Weibel DB. MRS Bulletin. 2011;36:347. doi: 10.1557/mrs.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Khoo X, Grinstaff MW. Mrs Bulletin. 2011;36:357. [Google Scholar]
- 175.Carman ML, Estes TG, Feinberg AW, Schumacher JF, Wilkerson W, Wilson LH, Callow ME, Callow JA, Brennan AB. Biofouling. 2006;22:11. doi: 10.1080/08927010500484854. [DOI] [PubMed] [Google Scholar]
- 176.Hochbaum AI, Aizenberg J. Nano Letters. 2010;10:3717. doi: 10.1021/nl102290k. [DOI] [PubMed] [Google Scholar]
- 177.Graham MV, Mosier AP, Kiehl TR, Kaloyeros AE, Cady NC. Soft Matter. 2013;9:6235. [Google Scholar]
- 178.Bhushan B. Philosophical Transactions of the Royal Society a-Mathematical Physical and Engineering Sciences. 2009;367:1445. doi: 10.1098/rsta.2009.0011. [DOI] [PubMed] [Google Scholar]
- 179.Ralston E, Swain G. Bioinspiration & Biomimetics. 2009;4:015007. doi: 10.1088/1748-3182/4/1/015007. [DOI] [PubMed] [Google Scholar]
- 180.Scardino AJ, de Nys R. Biofouling. 2011;27:73. doi: 10.1080/08927014.2010.536837. [DOI] [PubMed] [Google Scholar]
- 181.Chung KK, Schumacher JF, Sampson EM, Burne RA, Antonelli PJ, Brennana AB. Biointerphases. 2007;2:89. doi: 10.1116/1.2751405. [DOI] [PubMed] [Google Scholar]
- 182.Schumacher JF, Carman ML, Estes TG, Feinberg AW, Wilson LH, Callow ME, Callow JA, Finlay JA, Brennan AB. Biofouling. 2007;23:55. doi: 10.1080/08927010601136957. [DOI] [PubMed] [Google Scholar]
- 183.Callow JA, Callow ME. Nature Communications. 2011;2:244. doi: 10.1038/ncomms1251. [DOI] [PubMed] [Google Scholar]
- 184.Kumar A, Karig D, Acharya R, Neethirajan S, Mukherjee PP, Retterer S, Doktycz MJ. Microfluidics and Nanofluidics. 2013;14:895. [Google Scholar]
- 185.Zobell CE. Journal of Bacteriology. 1943;46:39. doi: 10.1128/jb.46.1.39-56.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.O’Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. Biofilms. 1999;310:91. [Google Scholar]
- 187.Hall-Stoodley L, Costerton JW, Stoodley P. Nature Reviews Microbiology. 2004;2:95. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 188.Lu TK, Collins JJ. Proceedings of the National Academy of Sciences. 2007;104:11197. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Brenner K, You L, Arnold FH. Trends in biotechnology. 2008;26:483. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 190.Brenner K, Arnold FH. PLoS One. 2011;6:e16791. doi: 10.1371/journal.pone.0016791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hong SH, Hegde M, Kim J, Wang X, Jayaraman A, Wood TK. Nature communications. 2012;3:613. doi: 10.1038/ncomms1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Qian F, Baum M, Gu Q, Morse DE. Lab on a Chip. 2009;9:3076. doi: 10.1039/b910586g. [DOI] [PubMed] [Google Scholar]
- 193.Li Z, Zhang Y, LeDuc PR, Gregory KB. Biotechnology and Bioengineering. 2011;108:2061. doi: 10.1002/bit.23156. [DOI] [PubMed] [Google Scholar]
- 194.Ardekani AM, Gore E. Physical Review E. 2012;85:056309. doi: 10.1103/PhysRevE.85.056309. [DOI] [PubMed] [Google Scholar]
- 195.Li G-J, Ardekani AM. Physical Review E. 2014;90:013010. doi: 10.1103/PhysRevE.90.013010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, Luijten E, Parsek MR, Wong GCL. Nature. 2013;497:388. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rusconi R, Guasto JS, Stocker R. Nature Physics. 2014;10:212. [Google Scholar]
- 198.Li G-J, Karimi A, Ardekani AM. Rheologica Acta. 2014;53:911–926. doi: 10.1007/s00397-014-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.