Abstract
Background
Mesothelin, previously shown to be expressed in triple negative breast cancer (TNBC), is a potential therapeutic target and prognostic marker in breast cancer.
Methods
We analyzed clinical data from two cohorts comprising of 141 patients treated between 2009 and 2011 at our institution (discovery cohort) and 844 patients from The Cancer Genome Atlas (TCGA) (validation cohort). Mesothelin expression was quantified by immunohistochemistry (IHC) or by RNA transcript levels as measured by whole-transcriptome sequencing in the discovery and validation cohorts respectively.
Results
In the discovery cohort, the median follow up was 3.55 years. Univariate analyses demonstrated that tumor size (hazard ratio (HR) =1.30, 95% confidence interval (CI) 1.11–1.51), positive (+) axillary lymph nodes (HR=3.34; 95% CI 1.51–7.39), and mesothelin expression (HR = 2.03; 95% CI 1.10–3.74) were associated with overall and disease-specific survival. We used a Cox-proportional hazard (Cox-PH) model to adjust for the two independent predictors of survival, namely (+) axilla lymph nodes and tumor size, and we found a significant association between mesothelin expression and overall and disease-specific survival in the discovery cohort (HR = 3.06, 95% CI 1.40–6.68). Using the TCGA dataset, we confirmed that, over a median follow-up of 16.0 months, patients with mesothelin-expressing tumors had poorer overall survival (HR=1.46; 95% CI 1.05–2.03). On Cox-PH multivariate analysis, mesothelin-positivity was an independent predictor of worse survival, after adjusting for (+) axillary lymph nodes and tumor size (HR = 1.69; 95%CI 1.17–2.42).
Conclusions
Our results suggest that mesothelin is a prognostic breast tumor marker whose expression is highly enriched in TNBC tumors, especially in African American women. As there is no existing targeted therapy for TNBC, mesothelin may be a promising drug target for TNBC. Future work is needed to evaluate the efficacy of mesothelin directed targeted therapy in the treatment of breast cancer.
Keywords: Mesothelin, breast cancer subtype, basal tumor subtype, triple negative breast cancer (TNBC), tumor marker, breast cancer outcome, targeted therapy
Introduction
Mesothelin is a 40 kDa glycophosphatidylinositol-anchored cell membrane glycoprotein encoded by the 2138 bp mesothelin gene which was first identified as a cell surface antigen recognized by the mouse K1 monoclonal antibody [1]. Mesothelin was subsequently cloned by Chang and Pastan, who were the first to show its expression on the surface of human ovarian carcinoma cells [2]. Mesothelin is initially synthesized as a 69kDa precursor protein, which is subsequently cleaved post-translationally into a 40 kDa membrane-bound C-terminal fragment, mesothelin, and a 31 kDa N-terminal soluble secreted protein fragment, megakaryocyte potentiating factor (hMPF) [3].
Mesothelin is expressed in the lining of the peritoneum, pleura and pericardium [1]. The biological function of mesothelin is unknown but mice with homozygous null mutation showed no detectable anatomic, developmental or reproductive defects indicating that mesothelin is not likely to be an essential protein in mice [4]. Mesothelin appears to be involved in cell adhesion via its interaction with CA125 and has been proposed to play a role in cancer progression [5].
Only recently has mesothelin been identified as a tumor antigen in breast cancer [6–8], in part because tumors of the most common breast cancer subtype, i.e. luminal A, rarely express mesothelin. In contrast, mesothelin is expressed in nearly half of all tumors belonging to the less common breast cancer subtype, basal or triple negative breast cancer (TNBC) [8]. This skewed expression pattern of mesothelin suggests that mesothelin may be a unique therapeutic target in TNBC. As numerous targeted therapeutic strategies directed against mesothelin have been developed for the treatment of malignancies such as mesothelioma, ovarian and some biliary and pancreatic carcinoma (summarized in a recent review [9]), these strategies, which include mesothelin-specific immune toxins, monoclonal antibodies, antibody-drug conjugates, tumor vaccines and cell-based immunotherapies, may be adopted as novel treatment strategies for TNBC. However, prior to adopting these mesothelin directed targeted therapy for clinical use, the mechanistic role of mesothelin in breast cancer pathogenesis has to be better understood.
The association between mesothelin expression in tumor cells and unfavorable clinical outcome has been reported in several gastrointestinal malignancy including biliary adenocarcinoma [10], and gastric carcinoma [11]. However, there has also been conflicting results that demonstrated that mesothelin expression was associated with prolonged survival in patients with advanced stage in epithelial ovarian carcinomas [12]. As for breast cancer, efforts to elucidate its prognostic significance have likely been dampened due to conflicting results from two studies aimed at evaluating the association between mesothelin expression and clinical outcomes in breast cancer [13, 14].
We therefore performed this study using data from two breast cancer patient cohorts comprising of patients treated at a single institution (n=141, discovery cohort) and patients from a multi-center cohort (n=844, validation cohort) obtained from The Cancer Genome Atlas (TCGA) to further clarify the equivocal status of mesothelin as a bona fide prognostic tumor marker in breast cancer. Our results demonstrate that mesothelin is indeed a prognostic tumor marker in breast cancer. Our findings support the need for further research to elucidate the mechanistic role of mesothelin in breast cancer progression and to evaluate the efficacy of some of the established mesothelin-targeted therapies in the treatment of breast cancer, in particular, TNBC.
Materials and Methods
Patients and tissue specimens
The discovery cohort is comprised of 141 patients. After obtaining approval from our Institutional Review Board, we identified all consecutive cases of TNBC (n=70) treated between 2009 and 2011 at our breast center located in a tertiary medical center. In addition, we included an additional 71 consecutively-treated patients who were diagnosed with receptor positive breast cancer (defined below) within the same study period. The majority of the patients in the discovery cohort (n=134) were diagnosed with primary operable breast cancer except for seven patients who had undergone surgical resection of their in-breast or chest wall recurrence in the same study period. These seven patients were initially diagnosed with their primary breast cancer between 2001 and 2008. All patients were treated according to the National Comprehensive Cancer Network (NCCN) guidelines. All patients included in the study were reviewed to ensure that there was sufficient archival tumor tissue for additional IHC studies.
Immunohistochemistry staining Protocol
Estrogen (ER), progesterone (PR), and Her-2/neu (Her2) receptor expression were evaluated by standard immunohistochemistry (IHC) staining techniques on formalin-fixed paraffin-embedded (FFPE) breast cancer tissue samples for all participants as part of standard pathology evaluation at our institution. The FDA-approved PharmDx ER and PR test kits (DAKO, Carpinteria,CA) and HercepTest (DAKO, Carpinteria, CA) were used to evaluate ER, PR and Her2 expression following manufacturer’s guidelines. The tests were reported as negative if the Allred score was 2 or less for ER and PR and 0 or 1+ for Her2. Fluorescent in situ hybridization (FISH) was performed using PathVision HER2 DNA probe kit (Abbott, Late County, IL) on all TNBC to confirm Her2 receptor status. FISH analysis was also performed to confirm the Her2 status in all Her2 2+ tumors as determined by IHC.
Expression of mesothelin was evaluated on FFPE tissue sections by IHC staining using a mouse monoclonal antibody specific for mesothelin (clone 5B2, 1:100, Thermo Scientific MS-1320) using a fully-automated Leica Bond™ Polymer Refine Detection System. Slides were pre-treated with Bond ER2 solution for 20 minutes at 100°C. Mesothelioma tissue was used as positive control.
All IHC-stained tumor sections were evaluated and scored by a board-certified pathologist who was blinded to the clinical characteristics and outcome of the corresponding patient. Because mesothelin staining is heterogeneous within and across tumor sections, the H-score, defined as the product of % positive tumor cells and IHC staining intensity (1, 2, or 3 with 3 being the most intense), was determined as the mean score from three separate high power fields.
Statistical analyses
For the discovery cohort, clinical characteristics, including African-American (AA) race, age at diagnosis (defined as the date of initial breast cancer surgery), tumor subtypes according to receptor status, tumor size (as continuous variable or as categorical variable by TNM classification), and number of involved or (+) axillary lymph nodes (as ordinal or as categorical variable by TNM classification). For this study, we classified tumor subtypes into three groups: Group 1 is comprised of ER+ breast cancer which expresses either ER or PR and lacks Her2 expression (n=34); Group 2 is comprised of Her2+ breast cancer which expresses Her2 as determined by IHC and/or fluorescence in situ hybridization with or without expression of ER or PR (n=37); and Group 3 is comprised of TNBC which lacks expression of ER, PR, or Her2 (n=70). In addition, we also stratified the cohort into mesothelin (−) vs. (+) tumors and evaluated the association of mesothelin expression with clinical and pathological characteristics using either a two-tailed student’s t-test for continuous covariates (i.e. age and tumor size) or a two-tailed Fisher’s Exact Test for discrete or ordinal covariates (ie. histologic types, and number of (+) axillary lymph nodes (0 vs. ≥1). Univariate and multivariate analyses were performed to assess both overall and disease-specific survival outcomes using the Kaplan-Meier and Cox-proportional hazards models, respectively. For the discovery cohort, the overall and disease-specific survival outcomes were identical as all deaths were attributed to disease progression.
For the TCGA validation cohort, we acquired the data on total mesothelin mRNA expression levels from breast cancer patients who had participated in the TCGA study. These expression data were retrieved from publicly available datasets after obtaining approval from the TCGA study group. All data files were stored in secure servers as per TCGA data handling requirements. We derived a threshold for mesothelin expression based on the observed mRNA expression levels in tumors as compared to matched, normal breast tissue mesothelin levels (when available). We downloaded the RNA-Seqv2 rsem estimated raw counts for genes and isoforms from solid tumor and adjacent non-tumor samples. Transcripts from the UCSC Known Genes Table, Refseq, Wega, and Ensemble and predictions were used and scores (number of tags in each transcript) were obtained from each sample [15]. We normalized both gene and isoform level estimated raw counts using the bioconductor R software package edgeR [16]. Scores were normalized with respect to total tags in the sample as well as total tags in the chromosome. Only the tags that overlap with transcripts were used in counting total tags. If the same tag is mapped to several places, a ratio is taken in counting. These estimation counts were obtained by TCGA using methods as published [15] . We retained only those samples with available mesothelin expression data (n=844) for further analysis.
We assessed the distribution of mesothelin expression scores (ranging from 0 to 994.5) and noted that the distribution of mesothelin expression was non-normal, precluding the use of the expression scores directly as a continuous variable. After normalizing the mesothelin mRNA expression levels, a mesothelin normalized mRNA expression score was derived for each patient based on total normalized read counts. We dichotomized the mesothelin expression covariate as either positive or negative. To determine a threshold for mesothelin mRNA expression positivity, we compared the distribution of mesothelin expression scores in normal versus tumor breast tissues to arrive at a cutoff threshold. As mesothelin mRNA is expressed in normal breast tissues albeit at low levels, we used a threshold such that 95% of all matching tumor and normal tissues had a tumor mesothelin mRNA level greater than the matching normal sample. This cutoff, which is equivalent to a normalized mesothelin expression score of (1.64), was retained to dichotomize the mesothelin expression variable as mesothelin-positive or negative tumors.
Results
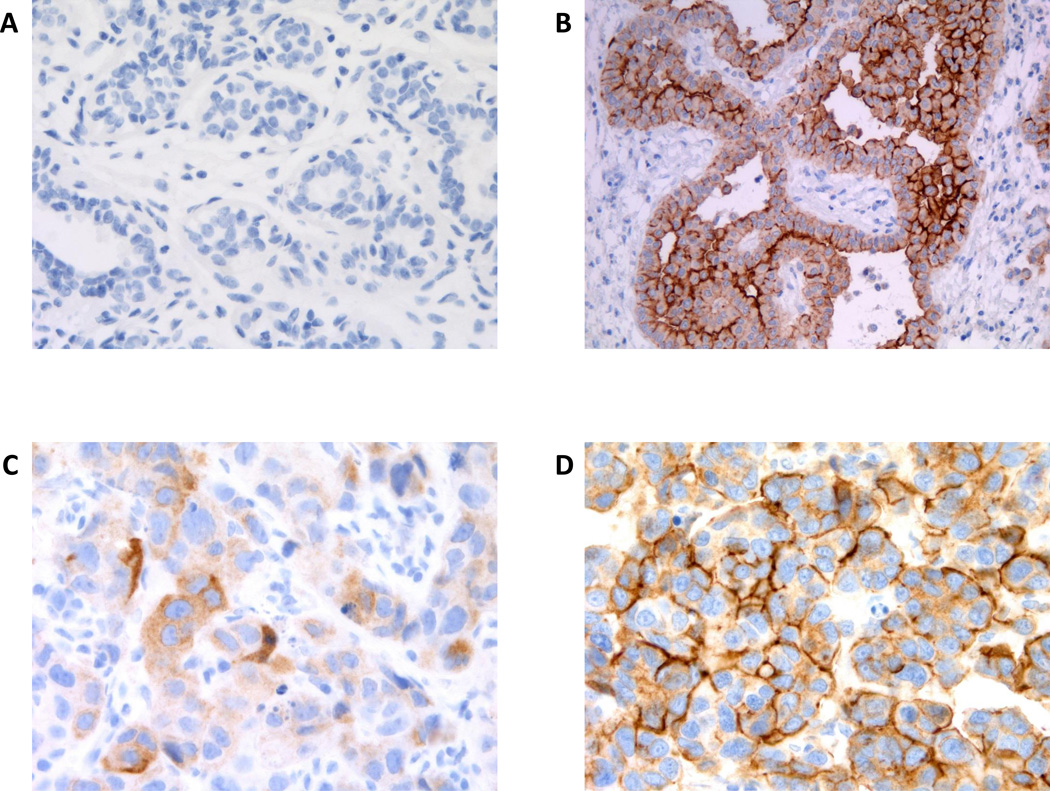
For our discovery cohort, mesothelin expression was quantified by IHC. Representative IHC staining results were shown in Fig. 1. Mesothelin was not detected in normal breast tissue by IHC (Fig. 1A) while mesothelin was highly expressed in mesothelioma tissue, a positive control (Fig. 1B). Staining pattern was predominantly membranous but cytoplasmic staining was also observed in breast tumor sections with heterogeneity noted within and across tumor samples. Representative IHC staining patterns with associated H-scores of 40 and 200 were shown in Fig. 1C and D respectively.
Figure 1. Expression of mesothelin in Mesothelioma and Breast Cancer Tissues.
The expression of mesothelin as assessed by Immunohistochemistry (IHC) and representative IHC staining results in various tissues were shown. A. Normal breast tissue; B. mesothelioma; C. 2+ cytoplasmic staining intensity, overall H-score = 40; D. 3+ membranous staining intensity, overall H-score = 200.
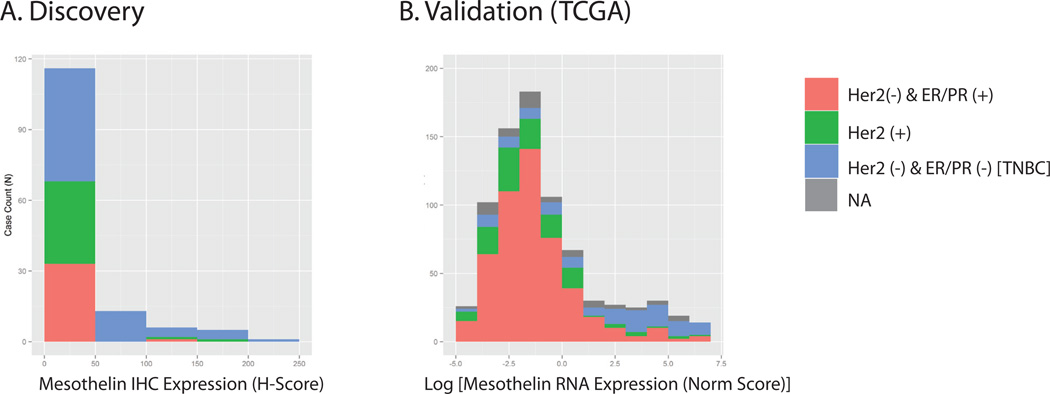
For the discovery cohort, we noted that mesothelin H-scores (ranging from 0 – 225) was not normally-distributed (skewness = 2.36 and kurtosis = 5.12) with a significant number of samples showing H-scores under one (mean = 22.68 and median = 0). The distribution of H-scores according to tumor subtypes was shown in Fig. 2A. The majority of tumor tissues have H-score <10. In addition, most tumors that were mesothelin (+) belonged to a specific tumor subtype, i.e. TNBC. To determine a method for dichotomizing the H-score, we visualized the histogram of H-scores and noted a natural distribution of patients either with ≥10 or <10 H-scores. We therefore used H-score of 10 as the threshold to dichotomize tumor samples into mesothelin positive vs. negative when H-score was ≥ 10 or H-score <10 respectively. Of note, the same H-score threshold was used in a recent study [14].
Figure 2. A and B. Distribution of tumor mesothelin status stratified by receptor subtypes in the two study cohorts.
Histogram plots illustrating the stratified distribution of tumor mesothlin expression levels/scores in the two cohorts, Discovery (A) and TCGA (B) cohorts by the three receptor subtypes (as described in Methods). Note that NA denotes samples whereby receptor subtype information was not available. Also note that the mesothelin expression level (based on RNA sequencing data obtained for the TCGA dataset) was normalized and a natural log transform is shown (B).
For the TCGA (validation) cohort, we used total mRNA expression of all mesothelin transcripts from the TCGA whole-transcriptome sequencing dataset to evaluate mesothelin expression. The distribution of mesothelin RNA expression values in the TCGA cohort also failed to follow a normal distribution, with the majority of the tumors showing very low or non-detectable expression (Fig. 2B). Similar to the discovery cohort, the majority of tumor tissues which expressed mesothelin were of the TNBC subtype (Fig. 2B). In both the discovery and validation cohort datasets, our empirically determined threshold, which was based on mesothelin expression levels as visualized by kernel density plot, provided a clear cutoff for dichotomizing tumors [14]. Subsequent univariate and multivariate analyses to assess association of mesothelin positivity with survival outcomes would use this binary classification to dichotomize tumors as either mesothelin (−) vs. (+).
The clinical characteristics of both study cohorts as stratified by mesothelin positivity were summarized in Table 1. For the discovery cohort, we noted that the mesothelin (+) subgroup was significantly enriched for patients of African American (AA) race (p = 0.02). Most importantly, we noted that 63% of TNBC samples (44 of 70) expressed mesothelin with H-scores ranging from 10 to 225 (mean = 22.68). In contrast, mesothelin expression was only observed in 1 out of 34 ER+ tumors (3%) and 5 out of 37 Her2+ tumors (14%) (Table 1).
Table 1. Clinical characteristics of the two breast cancer patient cohorts stratified by mesothelin expression status.
Clinical characteristics for both cohorts are shown for all patients included in our analyses. Patients with missing covariates were excluded from multivariate analyses and model search but were included in univariate analyses to identify significant covariates as stratified by mesothelin positivity.
| Discovery Cohort | TCGA Cohort | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Overall | Mesothelin (−)* | Mesothelin (+) | p value | Overall | Mesothelin (−) | Mesothelin (+) | p value | ||
| Patients (N) | 141 | 91 | 50 | 844 | 666 | 178 | |||
| Race | non-AA | 61 | 46 | 15 | 2.14E-02 | 645 | 525 | 120 | 4.84E-07 |
| AA | 80 | 45 | 35 | 70 | 37 | 33 | |||
| NA | 0 | 0 | 0 | 139 | 114 | 25 | |||
| Age at diagnosis (years) | mean ± sd | 55.63±13.93 | 57.13±13.95 | 52.90±13.61 | 0.07 | 58.35±13.21 | 58.65±13.34 | 57.26±12.69 | 0.31 |
| ≤50 | 56 | 31 | 25 | 254 | 195 | 59 | |||
| >50 | 85 | 60 | 25 | 590 | 471 | 119 | |||
| Tumor size (cm) | mean ± sd | 2.32±1.80 | 2.22±1.83 | 2.50±1.74 | 0.31 | 0.12 | |||
| T1 | <2 | 69 | 48 | 21 | 228 | 187 | 41 | ||
| T2 | 2–5 | 55 | 31 | 24 | 495 | 380 | 115 | ||
| T3 | >5 | 10 | 7 | 3 | 95 | 80 | 15 | ||
| NA | 7 | 5 | 2 | 26 | 19 | 7 | |||
| Stage | I | 47 | 33 | 14 | 0.53 | 148 | 119 | 29 | 0.87 |
| II | 62 | 37 | 25 | 503 | 394 | 109 | |||
| III | 21 | 14 | 7 | 193 | 153 | 40 | |||
| NA | 11 | 7 | 4 | 0 | 0 | 0 | |||
| (+) axilla lymph node(s) | 0 | 79 | 47 | 32 | 0.13 | 363 | 274 | 89 | 0.09 |
| ≥1 | 52 | 38 | 14 | 356 | 288 | 68 | |||
| NA | 10 | 6 | 4 | 125 | 104 | 21 | |||
| Receptor subtype | ER/PR+ & Her2- | 34 | 33 | 1 | 6.66E-12 | 527 | 463 | 64 | 1.07E-31 |
| Any Her2+ | 37 | 32 | 5 | 144 | 126 | 18 | |||
| TNBC | 70 | 26 | 44 | 116 | 40 | 76 | |||
| NA | 0 | 0 | 0 | 57 | 37 | 20 | |||
| Length of Follow Up (days) | Mean ± sd | 1206.65±571.79 | 1286.65±597.34 | 1058.45±493.19 | 1.36E-02 | 851.92±571.79 | 834.869±597.34 | 915.78±493.19 | 0.15 |
| Median | 1294.5 | 1390 | 1135 | 479 | 456.5 | 581.5 | |||
| Clinical Outcome | NED | 110 | 74 | 36 | 4.96E-03 | 586 | 472 | 114 | 8.08E-03 |
| AWD | 8 | 8 | 0 | 18 | 15 | 3 | |||
| DOO | 0 | 0 | 0 | 45 | 33 | 12 | |||
| DOD | 22 | 9 | 13 | 34 | 19 | 15 | |||
| NA | 0 | 0 | 1 | 161 | 127 | 34 | |||
Mesothelin expression is defined by IHC staining (discovery cohort) or mRNA expression levels (TCGA validation cohort) as described in the methods
NED = no evidence of disease; AWD = alive with disease; DOD = died of disease; DOO = Died of Other causes; AA = African American
For the TCGA validation cohort, we again noted a significantly higher proportion of women with AA descent whose tumors were mesothelin (+) (p = 4.84 E-07) (Table 1). We also observed a significant enrichment for mesothelin (+) expression in 66% TNBC tumors (76 of 116) as compared to all other receptor subtypes combined (12%) (p = 1.07 E-31).
For the discovery cohort, clinical and pathological characteristics that correlated with worse prognosis included tumor size (both as continuous variable in cm and as categorical variable in T stage), (+) axilla nodes, and (+) mesothelin expression with HR = 1.30, 95% CI 1.11–1.51; HR = 3.34, 95% CI 1.51–7.39; and HR = 2.03, 95% CI 1.10–3.74 respectively. Of note, our data did not show that breast cancer subtype was significantly associated with breast cancer outcomes although there was a trend towards worse prognosis for TNBC tumor subtype (HR = 2.35, CI 0.95–5.93) (Table 2A).
Table 2.
Results from univariate Kaplan Meier survival analysis showing three-year survival estimates for either the Discovery or TCGA cohorts based on base-line survival estimates along with estimated HRs.
| A Univariate Survival Characteristics Based on 3-year Estimated Overall Survival from Kaplan Meier Estimates in the Discovery Cohort | |||||||
|---|---|---|---|---|---|---|---|
| Discovery Cohort | 3-yr Est Survival | Hazard Ratio | |||||
| Variable | Factor | 3-yr Est Survival |
95% Confidence Interval |
Factor | Hazard Ratio |
95% Confidence Interval |
P-value; Logrank test |
| Race | AA | 0.821 | 0.74–0.91 | AA:nonAA | 1.803 | 0.32–4.44 | 2.00E-01 |
| non-AA | 0.891 | 0.81–0.98 | |||||
| Age at diagnosis | >50 | 0.871 | 0.80–0.95 | >50: ≤50 | 0.630 | 0.27–1.45 | 2.78E-01 |
| ≤50 | 0.820 | 0.72–0.93 | |||||
| Tumor size (cm) | T2+T3 (>=2cm) | 0.833 | 0.74–0.93 | T2+T3: T1 | 1.445 | 0.73–2.83 | 2.84E-01 |
| T1 (<2cm) | 0.917 | 0.84–0.98 | Continuous | 1.300 | 1.11–1.51 | 4.58E-04 | |
| Tumor stage (cm) | I | 0.976 | 0.93–1.00 | II:I | 4.736 | 0.57–39.35 | 1.50E-01 |
| II | 0.913 | 0.84–0.99 | III:I | 21.294 | 2.68–168.69 | 3.78E-03 | |
| III | 0.592 | 0.41–0.86 | |||||
| Involved axilla node(s) | >=1 | 0.762 | 0.65–0.90 | ≥1:0 | 3.342 | 1.51–7.39 | 2.86E-03 |
| 0 | 0.945 | 0.89–1.00 | |||||
| Breast cancer subtypes | TNBC | 0.802 | 0.71–0.91 | TNBC: ER+/PR+/Her2+ | 2.347 | 0.95–5.93 | 6.57E-02 |
| ER+/PR+ or Her2+ | 0.895 | 0.82–0.97 | |||||
| Mesothelin# | mesothelin(+) | 0.772 | 0.66–0.91 | mesothelin(+): mesothelin(−) | 2.031 | 1.10–3.74 | 2.32E-02 |
| mesothelin(−) | 0.893 | 0.83–0.96 | |||||
| B Univariate Analyses to Evaluate Association Between Clinical Covariates and Estimated Overall Survival from Kaplan Meier Estimates in the TCGA Cohort | |||||||
|---|---|---|---|---|---|---|---|
| TCGA Cohort | 3-yr Est Survival | Hazard Ratio | |||||
| Variable | Factor | 3-yr Est Survival |
95% Confidence Interval |
Factor | Hazard Ratio |
95% Confidence Interval |
P-value; Logrank test |
| Race | AA | 0.89 | 0.80–0.99 | AA:nonAA | 0.954 | 0.50–1.81 | 0.89 |
| non-AA | 0.93 | 0.90–0.96 | |||||
| Age at diagnosis | >50 | 0.90 | 0.86–0.94 | >50: ≤50 | 1.302 | 0.80–2.09 | 0.28 |
| ≤50 | 0.94 | 0.91–0.99 | |||||
| Tumor size$ | T1 (<2cm) | 0.94 | 0.89–0.98 | T2:T1 | 1.202 | 0.70–2.07 | 0.51 |
| T2 (2–5cm) | 0.94 | 0.90–0.97 | T3:T1 | 1.013 | 0.48–2.14 | 0.97 | |
| T3 (>5 cm) | 0.87 | 0.78–0.98 | |||||
| Tumor stage (cm) | I | 0.97 | 0.92–1.00 | II:I | 1.301 | 0.68–2.47 | 0.05 |
| II | 0.93 | 0.89–0.97 | III:I | 2.116 | 1.06–4.21 | 0.03 | |
| III | 0.82 | 0.74–0.90 | |||||
| (+) axilla lymph node(s) | ≥1 | 0.89 | 0.85–0.94 | ≥1:0 | 1.604 | 1.18–2.35 | 3.86E-03 |
| 0 | 0.96 | 0.92–0.99 | |||||
| Tumor Receptor Subtypes | TNBC | 0.85 | 0.77–0.95 | TNBC:ER/PR (+) | 2.186 | 1.13–4.22 | 2.85E-03 |
| Her2+ | 0.85 | 0.76–0.96 | Her2 (+):ER/PR (+) | 2.528 | 1.38–4.65 | 0.02 | |
| ER/PR+& Hert2(−) | 0.96 | 0.93–0.98 | |||||
| Mesothelin# | mesothelin(+) | 0.93 | 0.90–0.96 | mesothelin(+): mesothelin(−) | 1.463 | 1.05–2.03 | 0.02 |
| mesothelin(−) | 0.86 | 0.79–0.93 | |||||
AA: African American
mesothelin positivity is based on IHC Score dichotimization as described in the methods
AA: African American;
Quantiative tumor size (in cm) is not available
mesothelin positivity is based on mRNA transcript level-derived dichomization as described in the methods
For the TCGA cohort, the clinical variables that correlated with worse prognosis included tumor size (as categorized by TMN staging classification), (+) axillary lymph nodes, tumor subtypes, specifically TNBC and Her2+ tumor subtypes, and (+) mesothelin expression with HR = 2.12, 95% CI 1.06–4.21; HR=1.60, 95% CI 1.18–2.35; HR=2.19, 95% CI 1.13–4.22; HR=2.53, 95% CI 1.38–4.65; and HR 1.46, 95% CI 1.05–2.03 respectively (Table 2B).
To further evaluate the prognostic significance of mesothelin expression, Kaplan-Meier analyses were performed to correlate mesothelin expression with overall survival for patients in both study cohorts. As shown in Fig. 3, mesothelin expression is significantly associated with worse survival outcome in both Discovery and TCGA patient cohorts.
Figure 3. A and B Kaplan Meier plots showing breast cancer patient survival probabilities stratified by mesothelin expression positivity in the Discovery (A) and TCGA (B) cohorts.
Patient survival rates are shown for all patients in either the Discovery (A) or TCGA (B) cohorts as a function of the survival time (in days). Probabilities are based on those derived from the Kaplan Meier survival estimates using only those patients included in the analyses as described.
When multivariate analyses were performed using data from the discovery cohort taking into consideration of all identified significant covariates from Table 2A, we identified three independent prognostic factors that predicted outcome. These prognostic factors were: 1) (+) axilla lymph nodes, 2) tumor size (in cm) and 3) mesothelin (+) disease with HR=1.13, 95% CI 1.05–1.21; HR=1.37, 95% CI 1.13–1.67; and HR=3.06, 95% CI 1.40–6.68 respectively (Tables 3A). Of note, we were not able to demonstrate that the following covariates, namely, women with AA race and tumor subtypes, were independent prognostic factors in the discovery cohort. To illustrate this further, we stratified our data by either AA race or tumor subtype. Importantly, the three prognostic variables remained statistically significant upon stratification by AA race (Table 3A), suggesting that despite the association between mesothelin (+) tumors and AA race, (+) mesothelin expression remained an independent predictor of clinical outcome (HR = 3.14, 95% CI 1.24–6.17) (Table 3A). In contrast, when we stratified the data by tumor subtypes, mesothelin was no longer significantly associated with outcomes (HR=1.63, 95% CI 0.65–4.10) suggesting a strong association between mesothelin expression and tumor subtypes, specifically TNBC.
Table 3.
Results from multivariate Cox proportional hazard analysis of the Discovery and TCGA datasets using the multivariate model identified in the discovery cohort based on those covariates that are independently associated with survival were summarized in Table 3A and 3B.
| A. Multivariate Cox Proportional Hazardz Analysis using data from the discovery cohort | |||
|---|---|---|---|
| Penn Study Cohort | |||
| Forward stepwise parsimonious model | HR | 95% CI | p value |
| Involved axilla node(s) | 1.13 | 1.05–1.21 | 1.13E-03 |
| Mesothelin(+):(−) | 3.06 | 1.40–6.68 | 5.01E-03 |
| Tumor size (cm) | 1.37 | 1.13–1.67 | 1.44E-03 |
| Stratified by AA Race | |||
| Involved axilla node(s) | 1.13 | 1.01–4.60 | 4.69E-02 |
| Mesothelin(+):(−) | 3.14 | 1.24–6.17 | 6.14E-03 |
| Tumor size (cm) | 1.35 | 1.15–1.68 | 5.78E-04 |
| Stratified by Receptor Type | |||
| Involved axilla node(s) | 2.42 | 1.13–5.19 | 2.27E-02 |
| Mesothelin(+):(−) | 1.63 | 0.65–4.10 | 3.00E-01 |
| Tumor size (cm) | 1.43 | 1.17–1.75 | 4.46E-04 |
| B. Multivariate Cox Proportional Hazardz Analysis using data from the TCGA cohort | |||
|---|---|---|---|
| TCGA cohort | |||
| Based on model in discovery cohort | HR | 95% CI | p value |
| Involved axilla node(s) | 1.10 | 1.06–1.13 | 2.53E-03 |
| Mesothelin(+):(−) | 1.69 | 1.17–2.42 | 1.24E-02 |
| Tumor size (T2:T1) | 0.95 | 0.53–1.7 | 7.84E-01 |
| Tumor size (T3:T1 | 0.53 | 0.22–1.26 | 2.67E-01 |
| Stratified by AA Race | |||
| Involved axilla node(s) | 1.08 | 1.04–1.11 | 2.65E-03 |
| Mesothelin(+):(−) | 1.72 | 1.19–2.46 | 1.71E-02 |
| Stratified by Receptor Type | |||
| Involved axilla node(s) | 1.09 | 1.04–1.12 | 5.94E-03 |
| Mesothelin(+):(−) | 1.20 | 0.75–1.89 | 5.99E-01 |
When multivariate analyses were performed using data from the TCGA cohort taking into consideration of the significant covariates identified in the Discovery cohort, we identified two independent prognostic factors that predicted outcome. These factors were: 1) (+) axilla nodes and 2) mesothelin (+) tumors with HR = 1.10, 95% CI 1.06–1.13; and 1.69, 95% CI 1.17–2.42. Interestingly, tumor size as categorized by T stage was no longer associated with survival in this analysis (Table 3B). Again, the association between clinical outcome and (+) axilla nodes and (+) mesothelin expression was preserved upon stratification by AA race, indicating that these two prognostic factors were independent of AA race. As was observed with the discovery cohort, the significant association between mesothelin expression and outcome no longer persisted upon stratification by tumor subtypes, suggesting a very strong correlation between mesothelin (+) tumors and the TNBC tumor subtype. Involved axilla nodes remained an independent prognostic variable regardless of tumor subtypes, HR=1.09, 95% CI 1.04–1.12 Table 3B).
Discussion
TNBC enjoys no benefit from targeted therapy directed against estrogen, progesterone, or Her-2/neu receptors. Therefore, the search for a molecular therapeutic target for TNBC is ongoing. We reported earlier that mesothelin is a promising molecule that may serve as a tumor marker for TNBC especially since the majority of TNBC express mesothelin [8]. Two studies [13][14] have reported on the prognostic value of mesothelin expression in breast cancer but only one showed a significant correlation with clinical outcome [13]. The divergent results of these two studies may be attributable to differences in study design and differences in clinical demographics of the patient cohorts.
The study that did demonstrate a significant prognostic significance of mesothelin was a retrospective analysis involving 182 breast cancer patients. The study included tumors of all receptor subtypes. Results from that study demonstrated that aside from nodal status, tumor size, and Her2+ disease, mesothelin expression was significantly associated with poor prognosis. However, a recent retrospective study, which restricted their analyses to patients diagnosed with TNBC subtype only (n=109), did not demonstrate a significant correlation between mesothelin expression and poor outcome. In that TNBC study, although a statistical significance was not reached, a trend towards worse prognosis was noted in patients with mesothelin (+) tumors (HR=1.02, 95% CI 0.35 – 2.98). The inability to demonstrate a significant association between mesothelin expression and poor outcome in the TNBC study may be due to an inadequate sample size even though the study included 109 TNBC patients. Extrapolating from our data and using the expected HR of 1.69 (Table 3B) derived from the TCGA study cohort dataset, we estimated that a sample size of at least 208 TNBC patients will be needed to achieve sufficient power (0.8) to test the prognostic value of mesothelin within this specific tumor subtype.
Given that another retrospective study may share intrinsic limitations including inadequate sample size and selection bias, to overcome these potential limitations, we included in our study two independent patient cohorts. The smaller study cohort was comprised of patients treated at a single institution and served as a discovery cohort while the second larger cohort was comprised of patients participating in a multi-institution clinical trial for The Cancer Genome Atlas project (TCGA) and served as a validation cohort. Our results demonstrated that mesothelin is indeed a prognostic breast tumor marker and is significantly associated with poorer overall and disease-specific survival in both patient cohorts. In addition, we observed in both study cohorts that mesothelin expression is primarily observed in TNBC.
Nevertheless, our study has limitations. Despite the large sample size provided by the TCGA cohort, we could not demonstrate that the association between mesothelin expression and worse outcome is independent of tumor receptor subtypes. As mentioned earlier, we would need a cohort that has at least 208 patients diagnosed with TNBC to elucidate the prognostic value of mesothelin expression in this specific tumor subtype. Due to the strong correlation between tumor mesothelin positivity and receptor status (~60% of all TNBC tumors expressed mesothelin), we estimate that we will require a sample size twice that of the TCGA cohort to be sufficiently powered to dissect the prognostic value of mesothelin independent of receptor subtypes. Indeed, future mechanistic studies will help address the biology of the association between mesothelin expression and receptor status.
Another notable difference between our study and the TNBC study [14] was that we found a higher proportion of TNBC tumors that were mesothelin (+) (~60%) as compared to 34% as reported in the TNBC study. This difference may be attributed to the difference in methodology of the two studies. As opposed to using tissue microarrays for IHC analyses in the TNBC study, we performed all of our IHC staining using whole tumor tissue sections. The use of tissue microarrays may have resulted in a lower rate of detection of mesothelin (+) tumors since mesothelin expression was noted to be heterogeneous within and across tumor samples when we performed our IHC analyses.
Taken together, our results demonstrated that mesothelin positivity was significantly associated with TNBC and poor clinical outcome. Our results support future work to dissect the mechanistic role of mesothelin in breast cancer pathogenesis. Our results also provide the rationale to adapt existing mesothelin targeted therapies intended for other malignancies as novel treatment strategies for TNBC.
Acknowledgements
This research was, in part, funded by the NCI Cancer Center Support Grant (2-P30-CA-016520-35) (J. Tchou), the Linda and Paul Richardson Breast Cancer Research Funds (J. Tchou), the Breast Cancer Immunotherapy Funds (J. Tchou), the 2013 Exceptional Project Award from the Breast Cancer Alliance (J. Tchou), the Pennsylvania Department of Health grant #0972501 (C.H. June), Fundacion Alonso Martin Escudero (A. Perales-Puchalt), and RO1s CA178687 (J. Conejo-Garcia), CA157664 (J. Conejo-Garcia) and CA124515 (J. Conejo-Garcia). We thank the Bioinformatics Facility at The Wistar Institute for help with analyses of TCGA datasets. Y.R. Li is supported by the Paul and Daisy Soros Fellowship for New Americans and the NIH F30 Individual NRSA Training Grant.
Footnotes
Conflict of Interest
All authors have declared no conflict of interest.
References
- 1.Chang K, Pai LH, Pass H, et al. Monoclonal antibody K1 reacts with epithelial mesothelioma but not with lung adenocarcinoma. Am J Surg Pathol. 1992;16:259–268. doi: 10.1097/00000478-199203000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci U S A. 1996;93:136–140. doi: 10.1073/pnas.93.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kojima T, Oh-eda M, Hattori K, et al. Molecular cloning and expression of megakaryocyte potentiating factor cDNA. J Biol Chem. 1995;270:21984–21990. doi: 10.1074/jbc.270.37.21984. [DOI] [PubMed] [Google Scholar]
- 4.Bera TK, Pastan I. Mesothelin is not required for normal mouse development or reproduction. Mol Cell Biol. 2000;20:2902–2906. doi: 10.1128/mcb.20.8.2902-2906.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rump A, Morikawa Y, Tanaka M, et al. Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J Biol Chem. 2004;279:9190–9198. doi: 10.1074/jbc.M312372200. [DOI] [PubMed] [Google Scholar]
- 6.Argani P, Iacobuzio-Donahue C, Ryu B, et al. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE) Clin Cancer Res. 2001;7:3862–3868. [PubMed] [Google Scholar]
- 7.Ordonez NG. Value of mesothelin immunostaining in the diagnosis of mesothelioma. Mod Pathol. 2003;16:192–197. doi: 10.1097/01.MP.0000056981.16578.C3. [DOI] [PubMed] [Google Scholar]
- 8.Tchou J, Wang LC, Selven B, et al. Mesothelin, a novel immunotherapy target for triple negative breast cancer. Breast Cancer Res Treat. 2012;133:799–804. doi: 10.1007/s10549-012-2018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly RJ, Sharon E, Pastan I, Hassan R. Mesothelin-targeted agents in clinical trials and in preclinical development. Mol Cancer Ther. 2012;11:517–525. doi: 10.1158/1535-7163.MCT-11-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawamata F, Kamachi H, Einama T, et al. Intracellular localization of mesothelin predicts patient prognosis of extrahepatic bile duct cancer. Int J Oncol. 2012;41:2109–2118. doi: 10.3892/ijo.2012.1662. [DOI] [PubMed] [Google Scholar]
- 11.Einama T, Homma S, Kamachi H, et al. Luminal membrane expression of mesothelin is a prominent poor prognostic factor for gastric cancer. Br J Cancer. 2012;107:137–142. doi: 10.1038/bjc.2012.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yen MJ, Hsu CY, Mao TL, et al. Diffuse mesothelin expression correlates with prolonged patient survival in ovarian serous carcinoma. Clin Cancer Res. 2006;12:827–831. doi: 10.1158/1078-0432.CCR-05-1397. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Niu Z, Zhang L, et al. Clinicopathological significance of mesothelin expression in invasive breast cancer. J Int Med Res. 2012;40:909–916. doi: 10.1177/147323001204000309. [DOI] [PubMed] [Google Scholar]
- 14.Parinyanitikul N, Blumenschein GR, Wu Y, et al. Mesothelin expression and survival outcomes in triple receptor negative breast cancer. Clin Breast Cancer. 2013;13:378–384. doi: 10.1016/j.clbc.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]