Abstract
Mesembryanthemum crystallinum, a halophilic, inducible Crassulacean acid metabolism (CAM) species, was grown at NaCl concentrations of 20 and 400 millimolar in the rooting medium. Plants from the low salinity treatment showed exclusively C3-photosynthetic net CO2 fixation, whereas plants exposed to the high salinity level exhibited net CO2 dark fixation involving CAM. Mesophyll protoplasts, isolated from both tissues, were gently ruptured, and the intracellular localization of enzymes was studied following differential centrifugation and Percoll density gradient centrifugation of protoplast extracts. Both centrifugation techniques resulted in the separation of intact chloroplasts, with up to 90% yield, from other organelles and the nonparticulate fraction of cells. Enzymes were identified by determination of activity and by sodium dodecyl sulfate gel electrophoresis of enzyme protein.
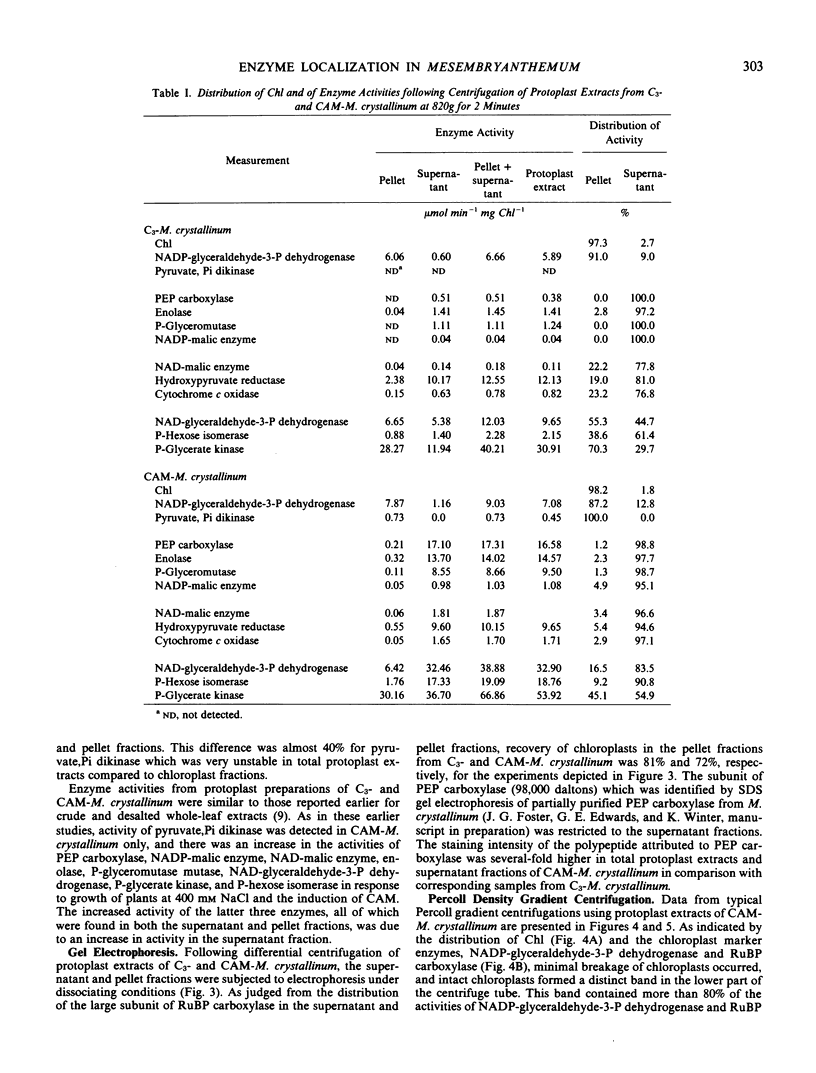
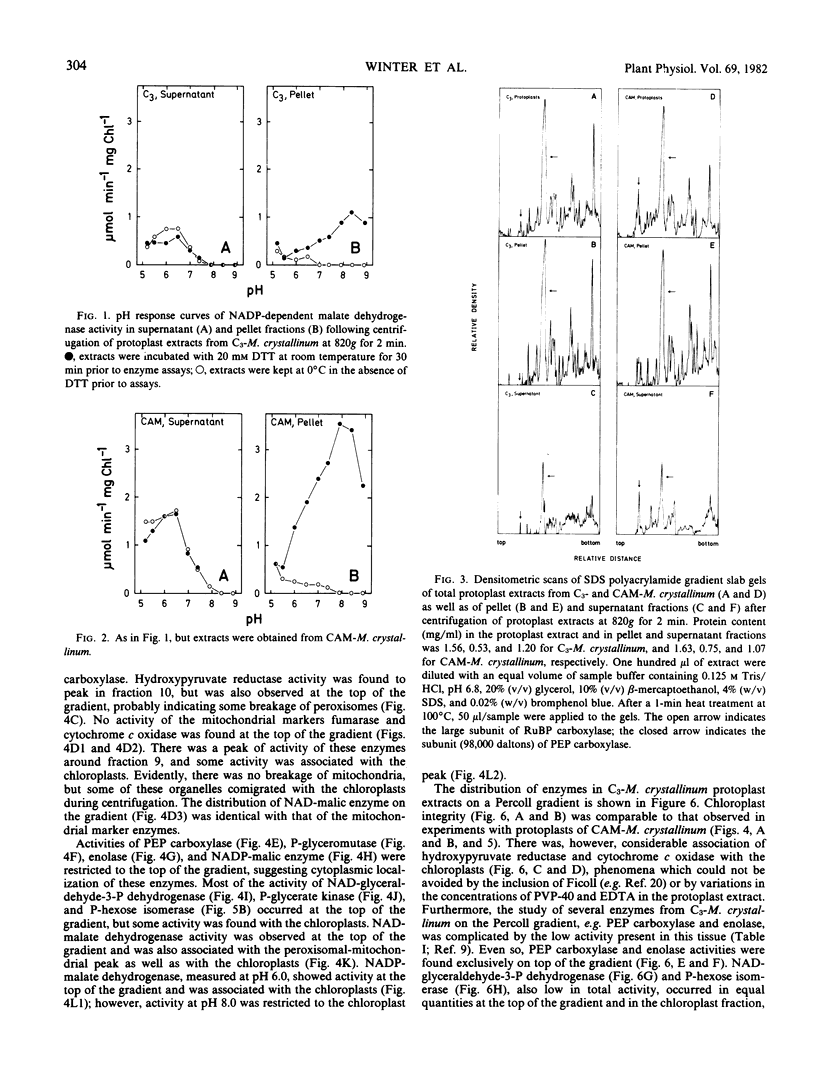
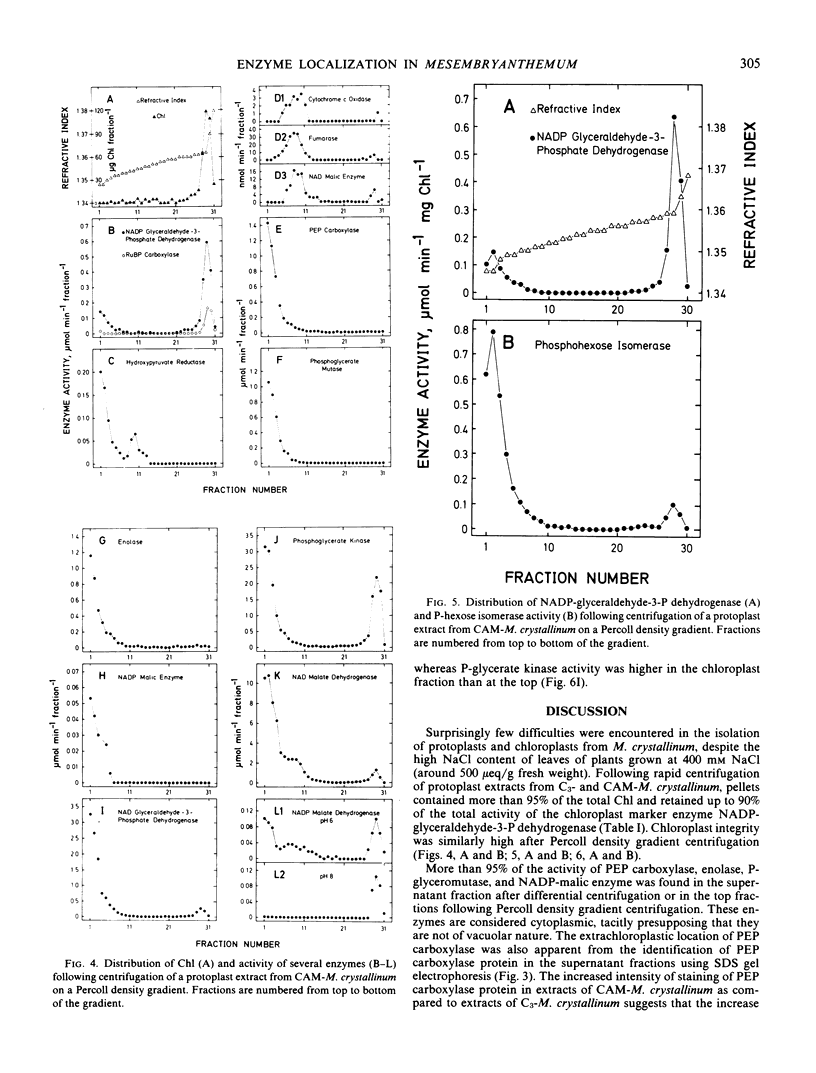
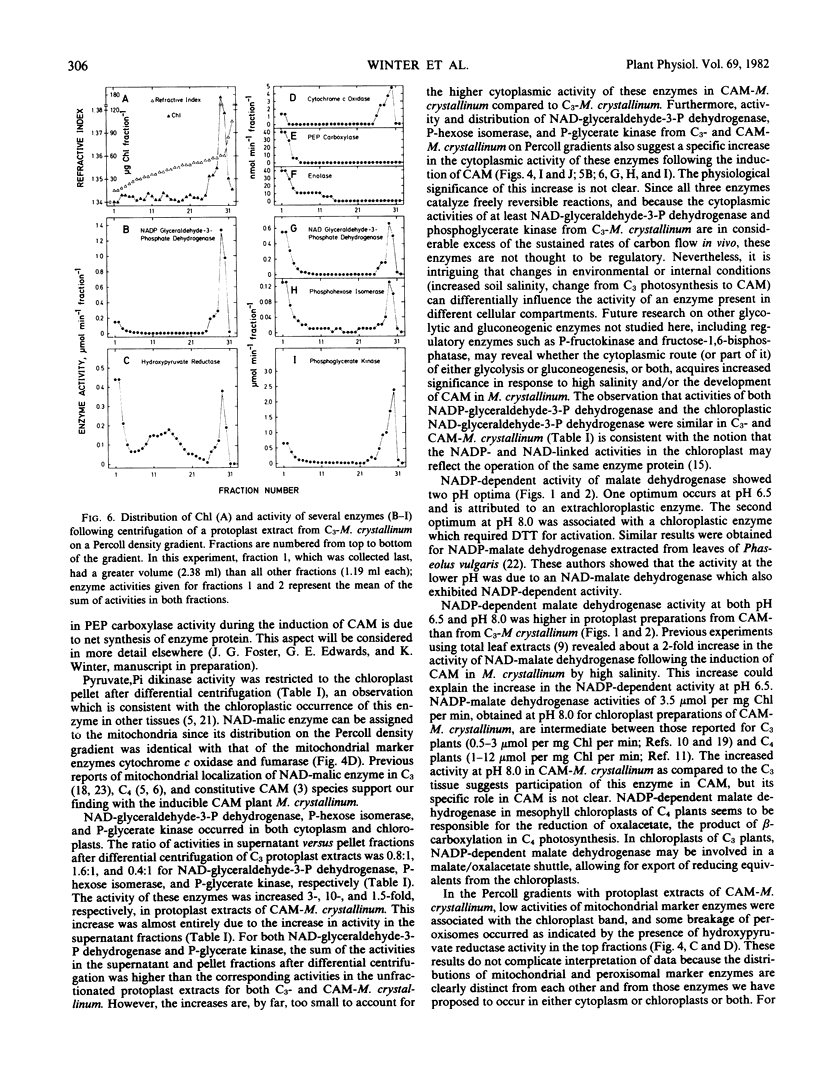
Experiments established the extraorganellar (cytoplasmic) location of phosphoenolpyruvate carboxylase, enolase, phosphoglyceromutase, and NADP-malic enzyme; the mitochondrial location of NAD-malic enzyme; and the chloroplastic location of pyruvate, Pi dikinase. NAD-glyceraldehyde-3-phosphate dehydrogenase, phosphohexose isomerase, and phosphoglycerate kinase were associated with both cytoplasm and chloroplasts. NADP-dependent malate dehydrogenase activity was found in both the chloroplastic and extrachloroplastic fractions; the activity in the chloroplast showed an optimum at pH 8.0 and was dependent upon preincubation of enzyme with dithiothreitol. The extrachloroplastic activity showed an optimum at pH 6.5 and was independent of pretreatment with dithiothreitol. Protoplast extracts of M. crystallinum performing CAM exhibited higher activities (expressed per mg chlorophyll per min) of phosphoenolpyruvate carboxylase, pyruvate, Pi dikinase, NADP-malic enzyme, NAD-malic enzyme, NADP-malate dehydrogenase, enolase, phosphoglyceromutase, NAD-glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, and phosphohexose isomerase than protoplast extracts from M. crystallinum not exhibiting CAM. The increase in total activity of the latter three enzymes following exposure of plants to 400 millimolar NaCl and the development of CAM was due to specific increases in the levels of activity in the cytoplasm.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnon D. I. COPPER ENZYMES IN ISOLATED CHLOROPLASTS. POLYPHENOLOXIDASE IN BETA VULGARIS. Plant Physiol. 1949 Jan;24(1):1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich P. Nicotinamide Adenine Dinucleotide-specific "Malic" Enzyme in Kalanchoë daigremontiana and Other Plants Exhibiting Crassulacean Acid Metabolism. Plant Physiol. 1976 Feb;57(2):310–314. doi: 10.1104/pp.57.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M. D., Slack C. R. NADP-specific malate dehydrogenase and glycerate kinase in leaves and evidence for their location in chloroplasts. Biochem Biophys Res Commun. 1969 Mar 10;34(5):589–593. doi: 10.1016/0006-291x(69)90778-5. [DOI] [PubMed] [Google Scholar]
- Johnson H. S., Hatch M. D. Properties and regulation of leaf nicotinamide-adenine dinucleotide phosphate-malate dehydrogenase and 'malic' enzyme in plants with the C4-dicarboxylic acid pathway of photosynthesis. Biochem J. 1970 Sep;119(2):273–280. doi: 10.1042/bj1190273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H. S. NADP-malate dehydrogenase: photoactivation in leaves of plants with Calvin cycle photosynthesis. Biochem Biophys Res Commun. 1971 May 21;43(4):703–709. doi: 10.1016/0006-291x(71)90672-3. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lorimer G. H., Badger M. R., Andrews T. J. D-Ribulose-1,5-bisphosphate carboxylase-oxygenase. Improved methods for the activation and assay of catalytic activities. Anal Biochem. 1977 Mar;78(1):66–75. doi: 10.1016/0003-2697(77)90009-4. [DOI] [PubMed] [Google Scholar]
- Macrae A. R. Isolation and properties of a 'malic' enzyme from cauliflower bud mitochondria. Biochem J. 1971 May;122(4):495–501. doi: 10.1042/bj1220495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R., Beck E. On the Mechanism of Activation by Light of the NADP-dependent Malate Dehydrogenase in Spinach Chloroplasts. Plant Physiol. 1979 Nov;64(5):744–748. doi: 10.1104/pp.64.5.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding M. H., Schmitt M. R., Ku S. B., Edwards G. E. Intracellular Localization of Some Key Enzymes of Crassulacean Acid Metabolism in Sedum praealtum. Plant Physiol. 1979 Apr;63(4):738–743. doi: 10.1104/pp.63.4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedding R. T., Black M. K., Pap D. Malate Dehydrogenase and NAD Malic Enzyme in the Oxidation of Malate by Sweet Potato Mitochondria. Plant Physiol. 1976 Dec;58(6):740–743. doi: 10.1104/pp.58.6.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K. Day/Night Changes in the Sensitivity of Phosphoenolpyruvate Carboxylase to Malate during Crassulacean Acid Metabolism. Plant Physiol. 1980 May;65(5):792–796. doi: 10.1104/pp.65.5.792. [DOI] [PMC free article] [PubMed] [Google Scholar]


