Abstract
Autotrophic ammonia oxidation is performed by two distinct groups of microorganisms: ammonia-oxidising archaea (AOA) and ammonia-oxidising bacteria (AOB). AOA outnumber their bacterial counterparts in many soils, at times by several orders of magnitude, but relatively little is known of their physiology due to the lack of cultivated isolates. Although a number of AOA have been cultivated from soil, Nitrososphaera viennensis was the sole terrestrial AOA in pure culture and requires pyruvate for growth in the laboratory. Here, we describe isolation in pure culture and characterisation of two acidophilic terrestrial AOA representing the Candidatus genus Nitrosotalea and their responses to organic acids. Interestingly, despite their close phylogenetic relatedness, the two Nitrosotalea strains exhibited differences in physiological features, including specific growth rate, temperature preference and to an extent, response to organic compounds. In contrast to N. viennensis, both Nitrosotalea isolates were inhibited by pyruvate but their growth yield increased in the presence of oxaloacetate. This study demonstrates physiological diversity within AOA species and between different AOA genera. Different preferences for organic compounds potentially influence the favoured localisation of ammonia oxidisers within the soil and the structure of ammonia-oxidising communities in terrestrial ecosystems.
Keywords: ammonia-oxidising archaea, acidic soil, low pH, mixotrophy, autotrophy, nitrification
Introduction
Ammonia oxidation is the first and rate-limiting step of nitrification. While ammonia-oxidising bacteria (AOB) have been cultivated for more than a century, the first ammonia-oxidising archaeon (AOA) Nitrosopumilus maritimus was not isolated until 2005 (Könneke et al., 2005). Since then, 27 different cultivated AOA strains have been reported (Supporting information, Table S1), spanning six proposed genera and originating from a diverse range of aquatic and terrestrial habitats. However, only two pure cultures have been obtained so far, and Nitrososphaera viennensis is the only representative from soil (Tourna et al., 2011). This lack of pure cultures currently limits studies of comparative physiology within AOA and between AOA and AOB.
Ammonia oxidisers, whether bacterial or archaeal, are generally considered autotrophs. While AOB fix CO2 via the Calvin–Benson–Bassham cycle (Arp et al., 2007), AOA are thought to assimilate 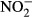 via a modified version of the 3-hydroxypropionate-4-hydroxybutyrate pathway (Berg et al., 2010). However, many autotrophs are able to supplement their inorganic carbon metabolism by organic compounds that are either pathway intermediates or otherwise readily incorporated into their metabolic pathways. The capacity for mixotrophy has been proposed as an important attribute for soil AOA (Prosser & Nicol, 2012). While this trait may provide a competitive advantage to AOA over AOB in the environment, the hypothesis is almost exclusively fuelled by observations of N. viennensis. Pyruvate enables N. viennensis to grow approximately 10 times faster than in inorganic medium alone, with pyruvate-C assimilated into biomass (Tourna et al., 2011). Furthermore, the marine archaeon N. maritimus can grow autotrophically, but growth is stimulated by the presence of pyruvate and α-ketoglutarate (Stahl & de la Torre, 2012). AOA are thought to operate an incomplete tricarboxylic acid (TCA) cycle in which pyruvate and α-ketoglutarate are intermediates (Walker et al., 2010).
via a modified version of the 3-hydroxypropionate-4-hydroxybutyrate pathway (Berg et al., 2010). However, many autotrophs are able to supplement their inorganic carbon metabolism by organic compounds that are either pathway intermediates or otherwise readily incorporated into their metabolic pathways. The capacity for mixotrophy has been proposed as an important attribute for soil AOA (Prosser & Nicol, 2012). While this trait may provide a competitive advantage to AOA over AOB in the environment, the hypothesis is almost exclusively fuelled by observations of N. viennensis. Pyruvate enables N. viennensis to grow approximately 10 times faster than in inorganic medium alone, with pyruvate-C assimilated into biomass (Tourna et al., 2011). Furthermore, the marine archaeon N. maritimus can grow autotrophically, but growth is stimulated by the presence of pyruvate and α-ketoglutarate (Stahl & de la Torre, 2012). AOA are thought to operate an incomplete tricarboxylic acid (TCA) cycle in which pyruvate and α-ketoglutarate are intermediates (Walker et al., 2010).
Nitrosotalea devanaterra is an obligately acidophilic ammonia oxidiser that grows by converting ammonia into nitrite within the pH range 4.0–5.5 (Lehtovirta-Morley et al., 2011). Nitrosotalea is an abundant, globally distributed genus of AOA found in acidic soils (Gubry-Rangin et al., 2011). Thirty per cent of the world's soils are considered acidic (pH < 5.5; von Uexküll & Mutert, 1995), and ammonia oxidation in low pH soils is dominated by AOA, rather than AOB (Gubry-Rangin et al., 2010; Lehtovirta-Morley et al., 2011). It is therefore of great interest to identify factors affecting the physiology and activity of N. devanaterra-like strains. Nitrosotalea devanaterra is the first and, currently, the only cultivated obligately acidophilic ammonia oxidiser and the model organism for studying ammonia oxidation in low pH ecosystems (Lehtovirta-Morley et al., 2011).
pH plays a major role in cell physiology and moderates the effect of organic compounds. Organic acids can become toxic under acidic conditions, as small protonated acids can pass readily through the cell membrane (Alexander et al., 1987; Kitko et al., 2009). Upon entering the cytoplasm, dissociation of the proton results in acidification of the cytoplasm and loss of proton motive force (Salmond et al., 1984). The inability of the cell to control transmembrane transport of organic acids can render acidophilic organisms vulnerable. The capability to consume organic compounds may alleviate the toxic effects of these compounds and thus make organisms less susceptible to toxicity of organic acids. This has been suggested as a reason why the most extreme acidophiles are heterotrophs; for example, the acidophile Picrophilus sp. which grows at an optimum pH of 1 is an obligate heterotroph (Baker-Austin & Dopson, 2007). Conversely, autotrophic acidophiles are generally adapted to oligotrophic environments and fail to respond rapidly to organic acids (Ñancucheo & Johnson, 2010). Toxicity of organic acids has been reported in several autotrophic acidophiles, including several species of Acidithiobacillus and Leptospirillum ferriphilum (Alexander et al., 1987; Aston et al., 2009; Ñancucheo & Johnson, 2010). In addition, organic acids are inhibitory to some acidophilic heterotrophs; Acidiphilium strains typically grown with low concentration of organics are inhibited by lactate and succinate (Kishimoto et al., 1990).
Here, we report isolation of two strains of acidophilic soil AOA belonging to the Nitrosotalea genus. Nitrosotalea devanaterra Nd1 was isolated from a previously described enrichment culture originating from an acidic agricultural soil in Scotland (Lehtovirta-Morley et al., 2011), while Nitrosotalea sp. Nd2 was cultivated from an acidic paddy field soil in China. The effects of pH and temperature were investigated on the two strains. In addition, the capacity for mixotrophy was examined by supplying cultures with intermediates of the TCA cycle at micromolar concentrations.
Materials and methods
Enrichment culture conditions
Nitrosotalea devanaterra Nd1 was originally enriched from an agricultural soil (pH 4.5) sampled from the Craibstone Estate, Scotland's Rural College (SRUC), Aberdeen, as previously described (Lehtovirta-Morley et al., 2011). The field site is subject to an 8-year crop rotation cycle as described in detail by Kemp et al. (1992) and experiences a mean annual temperature of 8.3 °C. Enrichment cultures for additional acidophilic ammonia oxidisers were established using an acid sulphate paddy soil (pH 4.7) from Taishan County, Guangdong Province in Southern China (latitude/longitude: 22.11 N/112.81 E) and experiences a mean annual temperature of 21.8 °C. Double rice cropping has been practiced since 2003 where urea is used as a nitrogen fertilizer, with the paddy field receiving four applications per year, averaging 150 kg N ha−1 for early rice and 150 kg N ha−1 for later rice.
Enrichment cultures were established in triplicate as previously described (Lehtovirta-Morley et al., 2011). Briefly, 1% soil inoculum was introduced into sterile mineral salts medium (pH 4.7) containing 500 μM NH4Cl, and enrichment cultures were incubated in darkness at 28 and 37 °C without shaking. Growth was monitored by measuring nitrite production. Both cultures were maintained routinely by transferring 2% mid-exponential culture into fresh mineral salts medium at pH 5.0. Nitrosotalea devanaterra Nd1 was maintained at 25 °C and Nitrosotalea sp. Nd2 at 37 °C.
The sensitivity of culturable bacterial contaminants to antibiotics in initial enrichment cultures was tested by adding sterile paper discs containing antibiotics to cultures spread-plated on 10% tryptone soy agar (TSA) plates. Antibiotics (ampicillin, carbenicillin, cephalexin, mecillinam, bacitracin, gentamycin, kanamycin, clindamycin, chloramphenicol, tetracycline, erythromycin, nalidixic acid) representing a range of modes of action (e.g. cell wall synthesis, translation and DNA replication) were applied as 50 μg antibiotic disc−1, except nalidixic acid which was used at a concentration of 15 μg disc−1. Inhibitory antibiotics identified from these initial screens were added to sterile liquid medium (50 mg L−1 for each antibiotic except nalidixic acid which was used at a concentration of 15 mg L−1) before inoculating with the enrichment cultures. Co-cultivated bacteria were successfully removed from the Nitrosotalea sp. Nd2 culture by application of 50 mg L−1 kanamycin. However, contaminants could not be eliminated from the N. devanaterra Nd1 culture with individual or combinations of antibiotics. Bacteria were therefore excluded on the basis of their comparatively larger cell size by filtration (0.45 μm pore-size) of the N. devanaterra Nd1 enrichment culture directly into medium containing 50 mg L−1 kanamycin. Neither kanamycin treatment nor filtration alone was sufficient to remove all bacteria. Purity was confirmed and monitored by plating on 10% TSA, microscopy and a lack of PCR amplification of bacterial 16S rRNA genes.
All physiology experiments were conducted in triplicate at 25 °C for N. devanaterra Nd1 and at 37 °C for Nitrosotalea sp. Nd2 unless specified otherwise. The effects of selected organic acids were determined in the presence of 100 μM of each compound. pH of the medium was adjusted to 5.0 after the addition of organic acids and was monitored throughout the experiments. Stimulation and inhibition by organic acids were expressed as the percentage increase/decrease in total nitrite concentration or specific growth rate and calculated as (treatment − inorganic control)/inorganic control. Specific growth rates was calculated by fitting a slope according to the equation: μ = (lnN1 − lnN0)/(t1 − t0), where μ represents the specific growth rate, (lnN1 − lnN0) the change in natural logarithms of nitrite concentration and (t1 − t0) the change in time. At least four time points were used for each growth rate calculation.
Colorimetric determination of inorganic nitrogen
Ammonia and nitrite were measured colorimetrically using a 96-well plate format. Ammonia concentration was determined by the indophenol method described by Kandeler & Gerber (1988). Nitrite was measured by diazotising and coupling with Griess reagent (Shinn, 1941). Duplicate standards ranged between 50 and 500 μM NH4Cl and 0.781 and 50 μM NaNO2 for ammonia and nitrite assays, respectively. Absorbance was recorded at 620 and 540 nm, respectively, using an Infinite F50® microplate reader (Labtech, Uckfield, UK).
Nucleic acid extraction and PCR amplification
DNA was extracted according to Tourna et al. (2010) with modifications described by Lehtovirta-Morley et al. (2011). Briefly, cultures were centrifuged, and pellets were bead-beaten in SDS-based buffer and phenol : chloroform : isoamyl alcohol mixture (25 : 24 : 1) After treating the aqueous phase with chloroform : isoamyl alcohol (24 : 1), DNA was precipitated in the presence of linear acrylamide and PEG6000. DNA was washed with 70% ethanol, dried and re-suspended in dH2O.
Bacteria-specific PCR was performed to monitor purity of cultures using two different primer sets targeting the bacterial 16S rRNA gene. Primers 27F and a revised version of 1492R were used together, and the second primer set consisted of P1 and P2 (Table 1). Archaeal amoA and 16S rRNA genes were amplified for sequencing and phylogenetic analysis from Nitrosotalea sp. Nd2 using primer sets 23F and 616R, and A109F and 1492R, respectively (Table 1). The composition of PCR reagent mixtures and cycling conditions have been described previously (Nicol et al., 2008). PCR-amplified Nitrosotalea sp. Nd2 16S RNA and amoA gene sequences were sequenced along both strands and have been deposited in GenBank with accession numbers KJ540205 and KJ540206, respectively. Nitrosotalea devanaterra Nd1 16S rRNA gene and amoA gene sequences used for the phylogenetic analysis were sequenced previously (Lehtovirta-Morley et al., 2011) and have accession numbers JN227488 and JN227489, respectively.
Table 1.
Primers used in this study
| Primer | Target organism | Gene | Sequence 5′-3′ | Reference |
|---|---|---|---|---|
| P1 | Bacteria | 16S rRNA | CCTACGGGAGGCAGCAG | Muyzer et al. (1993) |
| P2 | Bacteria | 16S rRNA | ATTACCGCGGCTGCTGG | Muyzer et al. (1993) |
| 27F | Bacteria | 16S rRNA | AGAGTTTGATCCTGGCTCAG | Lane (1991) |
| 1492R | Universal | 16S rRNA | GYYACCTTGTTACGACCT | Nicol et al. (2008) |
| A109F | Archaea | 16S rRNA | ACKGCTCAGTAACACGT | Großkopf et al. (1998) |
| 23F | Archaea | amoA | ATGGTCTGGCTWAGACG | Tourna et al. (2008) |
| 616R | Archaea | amoA | GCCATCCATCTGTATGTCCA | Tourna et al. (2008) |
Phylogenetic analysis
Nitrosotalea 16S rRNA gene and amoA gene sequences were aligned against sequences from cultured organisms and selected environmental clones using ClustalW implemented in bioedit Sequence Alignment Editor (Hall, 1999) before removing regions of ambiguous alignment. Phylogenetic analyses were performed using General Time Reversible-corrected maximum-likelihood (phyml; Guindon & Gascuel, 2003), parsimony (mega5; Tamura et al., 2011) and Tamura's three-parameter pairwise distance analysis (mega5) for 16S rRNA gene analysis, and Jones–Taylor–Thornton (JTT)-corrected maximum-likelihood (phyml), parsimony (mega5) and JTT-corrected pairwise distance (mega5) analyses of inferred translated amino acid sequences for amoA genes. Where appropriate, analyses used estimated variable sites only with gamma-distributed site variation, and bootstrap support for all methods was calculated 1000 times.
Cell enumeration
Total cell concentration was determined in 1 mL samples, fixed by addition of formaldehyde (final concentration 5%; v/v) and stored at 4 °C until enumeration. Thirty microlitres of 200 μg mL−1 DAPI (4,6-diamidino-2-phenylindole) was added to 1 mL of fixed cells, and samples were incubated for 5 min in the dark. Stained cells were placed onto a Cyclopore 0.22-μm pore-size black polycarbonate filter (Sigma-Aldrich, Gillingham, UK) by applying a vacuum in a standard filtration set-up and dried filters were mounted on glass slides with immersion oil and a cover slip. Cells were imaged using an Olympus BX61 fluorescence microscope equipped with a U-MWU2 fluorescence mirror unit (Olympus, Southend-on-Sea, UK). Automated cell counts were performed using imagej software (Schneider et al., 2012). Five fields of view were counted on each filter. Depending on the growth stage, this equated to 10–500 cells per sample.
Electron microscopy
Cells for both scanning and transmission electron microscopy (TEM) were harvested from an exponential culture and fixed in 4% (v/v) glutaraldehyde in phosphate-buffered saline. For scanning electron microscopy, cells were allowed to adhere to glass coverslips coated with poly-l-lysine. Slides were then rinsed with dH2O and postfixed with 1% OsO4. After washing with dH2O, specimens were dehydrated through a graded series of increasing ethanol concentrations (from 50% to 100%) and critical-point dried with liquid CO2. Samples were sputtered with Au and examined using an Eva MA 10 scanning electron microscope (Carl Zeiss, Cambridge, UK).
TEM was performed by postfixing cells with 1% OsO4, followed by dehydration via a graded ethanol series. Cells were transferred to propylene oxide and embedded in resin. Ultrathin sections were generated using a Leica UC6 ultramicrotome (Leica, Milton Keynes, UK), stained with uranyl acetate and lead citrate and examined using a JEM-1400 Plus transmission electron microscope (JEOL, Welwyn Garden City, UK) equipped with an UltraVUE camera (AMT, Bury St. Edmunds, UK).
Prediction of permeability properties of organic acids
Permeability of compounds was estimated both by molecular polar surface area and the predicted concentration of protonated form of each compound. Molecular polar surface area was calculated for the studied organic acids as described by Ertl et al. (2000). This method is based on tabulated contributions of atoms to the polar surface area based on their bonding patterns. This value was only calculated for the form of compounds where all carboxyl groups are protonated. The threshold for permeability is considered < 60 Å2. If any carboxyl groups are in their dissociated form, the molecule will have a negative charge and is impermeable. Concentrations of protonated and nonprotonated forms of the compounds were calculated as follows: for pyruvate, using Henderson–Hasselbach equation pH = pKa + log10([A−]/[HA]); for all other compounds except citrate using equation αH2A = [H+]2/([H+]2 + [H+]Ka1 + Ka1Ka2), and for citrate using equation αH3A = [H+]3/([H+]3 + [H+]2Ka1 + [H+]Ka1Ka2 + Ka1Ka2Ka3), where Ka values are dissociation constants, and αH2A and αH3A are alpha fractions containing the protonated, that is the permeable form of the compound. It is necessary to use different equations because pyruvate has one, citrate has three, and all other compounds have two carboxyl groups.
Statistical analyses
Statistical tests were performed in sigmaplot v12 (Systat Software). Normality and homogeneity of variance were tested with Kolmogorov–Smirnov and Levene's tests, respectively. Raw data (nitrite yield, growth rate and cell counts) were subjected to anova followed by Holm–Sidak test.
Results
Isolation of AOA
Nitrosotalea devanaterra Nd1 was maintained in an enrichment culture containing Burkholderia sp. for 3 years. Bacterial contaminants were removed by filtration of the culture through 0.45-μm pore-sized filters into medium containing 50 mg L−1 kanamycin, but ammonia oxidation and growth were not detected in this pure culture of N. devanaterra in mineral salts medium. A number of other antibiotics were tested but were not suitable. Nalidixic acid, tetracycline and chloramphenicol inhibited many strains of co-cultivated bacteria but inhibited nitrification. In addition, ampicillin, mecillinam and carbenicillin inhibited a few strains of bacteria on plates, but not in liquid culture, either alone or in combinations with other antibiotics. Attempts to restore growth of N. devanaterra Nd1 were made by supplementing medium with an array of organic compounds, including organic acids, amino acids, yeast extract and vitamins, but none enabled growth (Table S2). However, both filtered ‘spent’ medium (obtained after incubation of the mixed culture for 2 days and passed through a 0.22-μm filter) or supplementation of fresh medium with 80 mg L−1 casamino acids restored growth and ammonia oxidation activity by N. devanaterra Nd1. Although this initially suggested that N. devanaterra Nd1 may depend on organic supplements for growth, after approximately 10 subcultures, N. devanaterra Nd1 was able to grow autotrophically in unsupplemented mineral salts medium, using ammonia as its sole energy source and bicarbonate as the carbon source.
Nitrosotalea sp. Nd2 was enriched from a Chinese acidic paddy field soil by cultivation at 37 °C. A highly enriched AOA culture was obtained after several months of serial transfer using the same approach as that for N. devanaterra Nd1. Isolation was accomplished by addition of 50 mg L−1 kanamycin to inhibit the co-cultured bacteria and, unlike N. devanaterra, Nitrosotalea sp. Nd2 was able to grow autotrophically immediately after isolation. Conversion of ammonia to nitrite was stoichiometric in both isolates of Nitrosotalea (Fig. 1).
Fig 1.
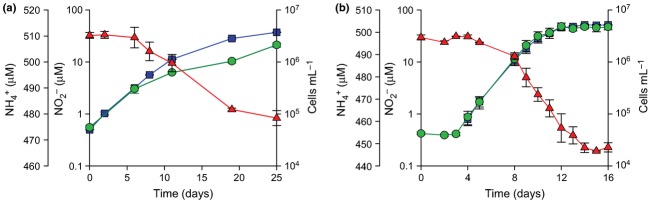
Growth and stoichiometric ammonia oxidation to nitrite by two Nitrosotalea isolates. (a) Nitrosotalea devanaterra Nd1, (b) Nitrosotalea sp. Nd2. Red triangles = [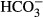 ], blue squares = [
], blue squares = [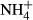 ], green circles = cell abundance. Error bars represent the standard error of the mean of triplicate cultures. Nitrite was below the detection limit until day 4 in Nitrosotalea sp. Nd2 cultures.
], green circles = cell abundance. Error bars represent the standard error of the mean of triplicate cultures. Nitrite was below the detection limit until day 4 in Nitrosotalea sp. Nd2 cultures.
Phylogeny
Despite originating from different soils separated by considerable geographical distance, the two strains share 98.9% 16S rRNA gene and 95.1% amoA gene sequence identity. The novel AOA isolate from paddy field soil is phylogenetically affiliated with the Nitrosotalea cluster (Fig. 2, Fig. S2; Pester et al., 2012), which is an abundant taxon in acidic soils globally (Gubry-Rangin et al., 2011). Although the two strains have a greater 16S rRNA gene similarity than the recommended species cut-off of 97.5% at the 16S rRNA gene level (Konstantinidis & Tiedje, 2005), the name Nitrosotalea sp. Nd2 is proposed for this organism, in contrast to the type strain N. devanaterra Nd1, due to moderate differences amoA gene sequence identity and a lack of genome data for comparison.
Fig 2.
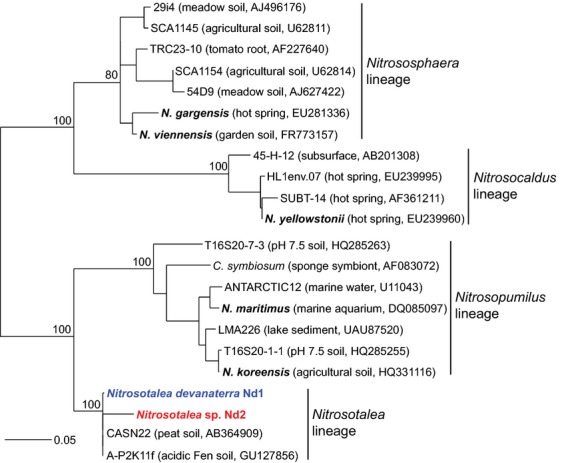
Maximum-likelihood phylogenetic analysis of 16S rRNA genes of Nitrosotalea devanaterra Nd1 and Nitrosotalea sp. Nd2 with sequences from other cultivated AOA (names in bold) and cloned environmental sequences placed with in four major AOA lineages as described by Pester et al. (2012) (assuming genera-level congruence between 16S rRNA gene- and amoA gene-defined phylogenies). Analyses were performed on 597 unambiguously aligned positions and values at major nodes represent the most conservative bootstrap support from three methods of analysis (ML, parsimony and distance). The scale bar represents 0.05 changes per nucleotide position.
Morphology
Both N. devanaterra isolates are morphologically indistinguishable and are small rods, typically 0.5–1 μm in length (Fig. 3). Both display a slightly angular appearance with electron-dense poles. Interestingly, during exponential growth, N. devanaterra cells occasionally became very elongated, reaching up to 2 μm in length (Fig. 3a). Intracellular compartmentalisation was evident in TEM. However, the membrane-bound intracellular compartment previously reported in Nitrosopumilus-like species (Jung et al., 2011; Yan et al., 2012) was not visible in the Nitrosotalea strains. Both strains contained apparent subcellular particles and inclusion bodies (indicated by arrows in Fig. 3), which may be storage granules.
Fig 3.
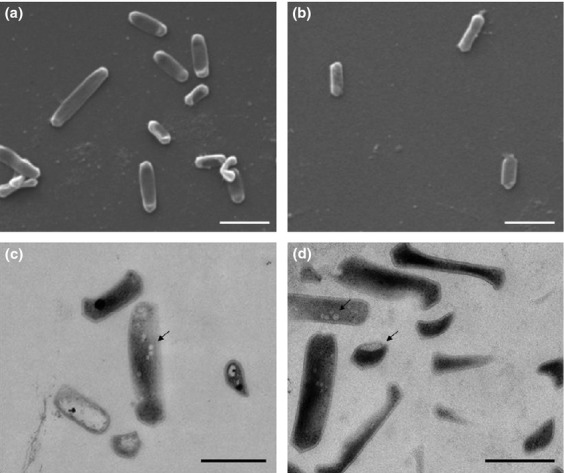
Scanning (a and b) and transmission (c and d) electron micrographs of Nitrosotalea devanaterra Nd1 (a and c) and Nitrosotalea sp. Nd2 (b and d). Arrows indicate subcellular particles. Scale bars are 1 μm (a and b) and 0.5 μm (c and d).
Physiology of AOA isolates
Despite being closely related, the two Nitrosotalea isolates exhibit different physiologies. Specific growth rates of N. devanaterra Nd1 and Nitrosotalea sp. Nd2 were strikingly different; Nitrosotalea sp. Nd2 grows approximately twice as fast as N. devanaterra Nd1 [μ ≈ 0.60 (SE 0.01) day−1 and 0.29 (SE 0.01) day−1, respectively]. Cell yields of N. devanaterra Nd1 and Nitrosotalea sp. Nd2 cultures were similar, with 6.45 × 104 (SE 5.1 × 102) cells (μM 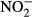 )−1 and 7.0 × 104 (SE 8.2 × 103 cells (μM
)−1 and 7.0 × 104 (SE 8.2 × 103 cells (μM 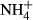 )−1, respectively. Cell activities were also similar: 71.8 (SE 4.3) amol
)−1, respectively. Cell activities were also similar: 71.8 (SE 4.3) amol 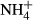 cell−1 h−1 for N. devanaterra Nd1 and 64.5 (SE 4.4) amol
cell−1 h−1 for N. devanaterra Nd1 and 64.5 (SE 4.4) amol 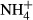 cell−1 h−1 for Nitrosotalea sp. Nd2. Assuming a similar protein content to that of N. maritimus, based on their similar size and morphology, these activities can be alternatively expressed as approximately 7.0 (SE 0.42) and 6.3 (SE 0.43) μmol
cell−1 h−1 for Nitrosotalea sp. Nd2. Assuming a similar protein content to that of N. maritimus, based on their similar size and morphology, these activities can be alternatively expressed as approximately 7.0 (SE 0.42) and 6.3 (SE 0.43) μmol 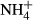 mg protein−1 h−1 for N. devanaterra Nd1 and Nitrosotalea sp. Nd2, respectively. Activity and yield of N. devanaterra Nd1 were different in pure cultures and enrichments, most likely due to different enumeration methods (qPCR vs. direct cell counts). Discrepancy between the two methods has been previously reported in a pure culture of N. maritimus (Nakagawa & Stahl, 2013). Alternatively, it is possible that this difference is a result from the lack of bacterial metabolites after isolation. Growth of both cultures was affected by nitrite accumulation and subsequent toxicity from nitrous acid, and growth terminated before all ammonium was depleted. Maximum nitrite concentration at the onset of stationary phase was strongly pH-dependent, as previously reported for the enrichment culture of N. devanaterra Nd1 (Lehtovirta-Morley et al., 2011), and N. devanaterra Nd1 and Nitrosotalea sp. Nd2 were inhibited at 0.91–3.50 μM and 1.61–5.70 μM HNO2, respectively (assumed from the Henderson–Hasselbach equation). Under typical growth conditions (pH 5), final nitrite concentration was ∼50 μM and the highest values recorded were 49.0 (SE 1.2) μM for N. devanaterra Nd1 and 54.3 (SE 0.1) μM for Nitrosotalea sp. Nd2. The highest nitrite concentrations in inorganically grown cultures were 101 (SE 0.9) μM for N. devanaterra Nd1 (pH 5.6) and 112 (SE 0.8) μM for Nitrosotalea sp. Nd2 (pH 6.1).
mg protein−1 h−1 for N. devanaterra Nd1 and Nitrosotalea sp. Nd2, respectively. Activity and yield of N. devanaterra Nd1 were different in pure cultures and enrichments, most likely due to different enumeration methods (qPCR vs. direct cell counts). Discrepancy between the two methods has been previously reported in a pure culture of N. maritimus (Nakagawa & Stahl, 2013). Alternatively, it is possible that this difference is a result from the lack of bacterial metabolites after isolation. Growth of both cultures was affected by nitrite accumulation and subsequent toxicity from nitrous acid, and growth terminated before all ammonium was depleted. Maximum nitrite concentration at the onset of stationary phase was strongly pH-dependent, as previously reported for the enrichment culture of N. devanaterra Nd1 (Lehtovirta-Morley et al., 2011), and N. devanaterra Nd1 and Nitrosotalea sp. Nd2 were inhibited at 0.91–3.50 μM and 1.61–5.70 μM HNO2, respectively (assumed from the Henderson–Hasselbach equation). Under typical growth conditions (pH 5), final nitrite concentration was ∼50 μM and the highest values recorded were 49.0 (SE 1.2) μM for N. devanaterra Nd1 and 54.3 (SE 0.1) μM for Nitrosotalea sp. Nd2. The highest nitrite concentrations in inorganically grown cultures were 101 (SE 0.9) μM for N. devanaterra Nd1 (pH 5.6) and 112 (SE 0.8) μM for Nitrosotalea sp. Nd2 (pH 6.1).
Temperature preference differed between the Nitrosotalea strains (Fig. 4a). Optimal growth of N. devanaterra Nd1 was observed at 25 °C [μ = 0.29 (SE 0.01) day−1] and growth occurred within the range of 20–30 °C. In contrast, Nitrosotalea sp. Nd2 grew optimally at 35 °C [0.60 (SE 0.01) day−1], a temperature that completely inhibited growth of N. devanaterra Nd1. In addition, Nitrosotalea sp. Nd2 had a wider temperature range (20–42 °C) than N. devanaterra Nd1. Interestingly, the specific growth rate of Nitrosotalea sp. Nd2 was lower at 25 °C and comparable to that of N. devanaterra Nd1 under the same conditions. Preference for low pH is the defining feature of currently recognised members of the genus Nitrosotalea. Nitrosotalea sp. Nd2 is an obligate acidophile with a slightly broader pH range for growth (4.0–6.1; Fig. 4b) than N. devanaterra Nd1 (4.2–5.6).
Fig 4.
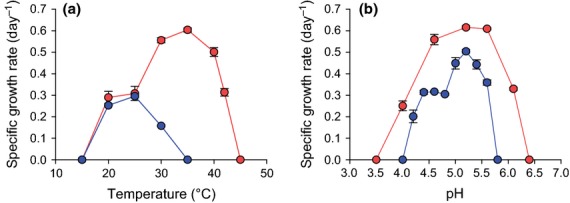
The effect of temperature (a) and pH (b) on the specific growth rate of Nitrosotalea devanaterra isolates. In a, the left y-axis represents N. devanaterra Nd1, the right y-axis Nitrosotalea sp. Nd2. Blue circles = N. devanaterra Nd1, red circles = Nitrosotalea sp. Nd2. Error bars represent the standard error of the mean of triplicate cultures.
Effect of organic acids on acidophilic ammonia oxidisers
Surprisingly, many TCA cycle intermediates inhibited N. devanaterra Nd1 and Nitrosotalea sp. Nd2. Pyruvate (100 μM) decreased the cell yield of both organisms (Fig. 5 and Fig. S2) and reduced the final nitrite concentration, being 26% lower in N. devanaterra Nd1 cultures [22.0 (SE 1.2) vs. 36.4 (SE 0.4) μM]. In addition, cell yield of N. devanaterra Nd1 was reduced (45%) in the presence of pyruvate (Fig. S2a). Exposure of Nitrosotalea sp. Nd2 to pyruvate reduced nitrite and cell yields to 24% and 34%, respectively. Specific growth rate of N. devanaterra Nd1 was reduced by pyruvate from 0.29 (SE 0.01) day−1 to 0.12 (SE 0.01) day−1. Growth rate of Nitrosotalea sp. Nd2 was not reduced significantly in the presence of pyruvate.
Fig 5.
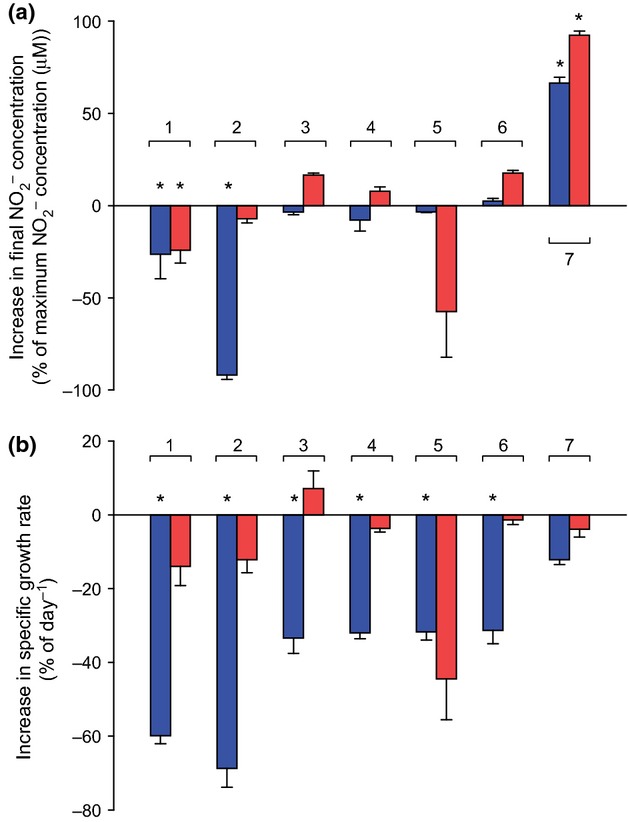
Effects of addition of 100 μM TCA cycle intermediates on nitrite yield (a) and specific growth rate (b) of Nitrosotalea isolates expressed as a percentage compared with inorganically grown control. Blue bars = Nitrosotalea devanaterra Nd1, red bars = Nitrosotalea sp. Nd2. 1 = pyruvate, 2 = citrate, 3 = α-ketoglutarate, 4 = succinate, 5 = fumarate, 6 = malate, 7 = oxaloacetate. Error bars represent the standard error of the mean of triplicate cultures. Asterisk indicates statistically significant difference to inorganically grown control (P < 0.05).
The Nitrosotalea strains responded differently to many organic acids, underlining their distinct physiologies. Apart from oxaloacetate, all the tested TCA cycle intermediates significantly decreased the specific growth rate of N. devanaterra Nd1 (Fig. 5b), while none of the tested compounds had a significant effect on the specific growth rate of Nitrosotalea sp. Nd2. Citrate strongly inhibited N. devanaterra Nd1, resulting in decreases to 69% and 93% in nitrite and biomass yield, but had no effect on Nitrosotalea sp. Nd2. In contrast, fumarate inhibited Nitrosotalea sp. Nd2 but not N. devanaterra Nd1.
Oxaloacetate was the only tested compound to stimulate both Nitrosotalea strains significantly. While specific growth rates remained unchanged, both nitrite and cell yields increased in the presence of oxaloacetate. Cell abundance was 60% and 117% higher for N. devanaterra Nd1 and Nitrosotalea sp. Nd2, respectively, while maximum nitrite concentrations were 66% and 92% greater. Generally the yield of acidophilic AOA is dictated by pH and the consequent concentration of nitrous acid. However, the apparent advantage granted by oxaloacetate may not be due to buffering effects or pH changes. The final pH was 0.09 and 0.16 units higher for N. devanaterra Nd1 and Nitrosotalea sp. Nd2 cultures, respectively, in oxaloacetate-supplemented cultures compared with the control. The final concentration of nitrous acid was 3.63 (SE ± 0.07) and 6.26 (SE 0.07 μM) for N. devanaterra Nd1 and Nitrosotalea sp. Nd2, respectively, which indicates slightly higher tolerance than in inorganically grown cultures.
Although oxaloacetate increased nitrite and cell concentrations, proportional changes were similar (Fig. S2b) and yield expressed as cells generated (1 μM 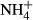 produced)−1 was not significantly different between any of the treatments with organic acids or the control. This suggests that under the tested conditions, Nitrosotalea strains derive their energy for growth by oxidising ammonia rather than organic compounds.
produced)−1 was not significantly different between any of the treatments with organic acids or the control. This suggests that under the tested conditions, Nitrosotalea strains derive their energy for growth by oxidising ammonia rather than organic compounds.
Membrane permeability of organic acids could play a critical role in their toxicity in low pH. Permeability estimated as the molecular polar surface area was calculated for the studied compounds (Table S3). Only fully protonated compounds can be permeable, so concentration of this form was also calculated for each compound. Only pyruvate was highly permeable, with a value of 54.4 Å2 (< 60 Å2 is considered a threshold for permeability).
Discussion
This study describes isolation and characterisation of two acidophilic AOA: N. devanaterra Nd1 and Nitrosotalea sp. Nd2. Interestingly, while the two AOA isolates are very closely related in terms of 16S rRNA gene and amoA gene phylogenies, their physiologies differed with respect to optimal growth temperature, specific growth rate and response to organic acids. 16S rRNA gene similarity is used as the foundation in taxonomy and molecular ecology studies. This therefore highlights the dangers in assuming strong links between 16S rRNA gene and functional gene phylogeny and function at the levels of sequence resolution achievable using such genes and typically employed in molecular studies. Despite high identity in 16S rRNA gene and amoA genes, these two strains differed in physiological characteristics that are likely to be of ecological importance. In addition, it is interesting that such (phylogenetically) closely related strains were isolated from ecosystems separated by a vast distance. This may indicate the selection by the growth medium and cultivation conditions employed for N. devanaterra-like phylotypes rather than other ammonia oxidisers present in acidic soils.
Contrasting temperature preferences of the Nitrosotalea isolates may reflect the origins of the organisms; the mean annual temperature of 21.8 °C in the Chinese paddy field soil was considerably higher than that of the Scottish agricultural soil (8.3 °C). The optimal growth temperature (35 °C) of Nitrosotalea sp. Nd2 is also similar to cultivated Nitrososphaera gargensis and N. viennensis (Hatzenpichler et al., 2008; Tourna et al., 2011).
Although cell yields of the Nitrosotalea strains were similar to each other, they were substantially lower than typically observed for AOA [4 × 105 cells (μM 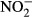 )−1 for N. maritimus]. Furthermore, cell activities of both Nitrosotalea were approximately eightfold lower than values reported for N. maritimus (51.9 μmol
)−1 for N. maritimus]. Furthermore, cell activities of both Nitrosotalea were approximately eightfold lower than values reported for N. maritimus (51.9 μmol 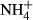 mg protein−1 h−1). This difference may reflect a higher energy expenditure required for intracellular pH regulation in acidic conditions. Interestingly, the specific growth rate of Nitrosotalea sp. Nd2 is comparable to that of N. maritimus (maximum reported μ = 0.64 day−1; Martens-Habbena et al., 2009) and considerably higher than that of N. devanaterra Nd1.
mg protein−1 h−1). This difference may reflect a higher energy expenditure required for intracellular pH regulation in acidic conditions. Interestingly, the specific growth rate of Nitrosotalea sp. Nd2 is comparable to that of N. maritimus (maximum reported μ = 0.64 day−1; Martens-Habbena et al., 2009) and considerably higher than that of N. devanaterra Nd1.
In contrast to neutrophilic AOA, growth of both Nitrosotalea isolates was inhibited by nitrite accumulation and ceased prior to substrate depletion. As maximum nitrite yield in Nitrosotalea cultures depended strongly on the cultivation pH, nitrous acid rather than nitrite is presumably mediating this toxicity. The amount of nitrous acid accumulated by the Nitrosotalea strains is equivalent to > 13 mM nitrite at pH 7.5. Little is known about nitrite/nitrous acid toxicity in other AOA, but it is plausible that Nitrosotalea strains are no more sensitive to nitrous acid than any neutrophilic AOA. In terms of ecology, this suggests that AOA must maintain a tight partnership with nitrite oxidisers in acidic soils. Although nitrite rarely accumulates in soils (Paul & Clarke, 1989), nitrite oxidisers may play a more crucial role for ammonia oxidisers in acidic rather than neutral soils. In soil microcosms where N. devanaterra drives nitrification, close to 100 μg nitrate–N g−1 soil was accumulated without apparent toxicity (Lehtovirta-Morley et al., 2011), indicating that nitrate is likely to be less toxic than nitrite is in these ecosystems.
Inhibition of growth by organic acids may be caused by interference with intracellular pH regulation. Small, nonpolar molecules can readily penetrate membranes when protonated, and dissociation within the cytoplasm leads to acidification and uncoupling of energy-generating mechanisms (Baker-Austin & Dopson, 2007). Molecular polar surface area was used as a predictor of membrane permeability (Ertl et al., 2000). Pyruvate was the only highly membrane-permeable compound in this study (Table S3). However, in a neutral pH environment, metabolism of pyruvate may necessitate active transport, as a very small proportion will exist in the membrane-permeable, protonated form. For example, even at the highest reported concentration (10 mM) for N. viennensis, only 0.1 μM pyruvate is protonated. In contrast, 0.32 μM pyruvate was present in the protonated form in the experiments with both Nitrosotalea strains, equivalent to ∼32 mM pyruvate in the typical growth conditions of N. viennensis.
The concentrations of organic acids tested in this study were micromolar to mimic concentrations typical of rhizosphere soil (Jones, 1998). In soil ecosystems, readily assimilable organic compounds are in their highest concentrations in the rhizosphere (Cieśliński et al., 1998). Although N. viennensis grows in medium containing a wide range of pyruvate concentrations (0.001–10 mM), stimulation was greatest at 100 μM and did not increase at higher pyruvate concentrations. No information has been released about the stimulatory concentrations of organic acids in N. maritimus. If mixotrophy is a genuine metabolic feature of AOA, these data indicate that there is variation in preference for the organic substrates utilised. AOA belonging to Nitrososphaera and Nitrosopumilus genera may preferentially reside in the rhizospheric soil, whereas the opposite would be true for species belonging to Nitrosotalea. Alternatively, in soil Nitrosotalea strains may benefit from proximity of heterotrophs which rapidly degrade inhibitory organic acids. It remains to be seen whether AOA belonging to Nitrosopumilus and Nitrososphaera can successfully compete against heterotrophs for organics in soil. Results from previously published in situ studies are contradictory and thaumarchaeal/AOA abundance has been reported to be both higher (Ochsenreiter et al., 2003; Dias et al., 2012) and lower (Herrmann et al., 2008) in bulk soil than in the rhizosphere. The differential preferences of ammonia oxidisers to organic compounds may contribute to this discrepancy and also shape ammonia oxidiser community structures.
In conclusion, this study demonstrates that Nitrosotalea strains can be isolated in pure culture and even closely related AOA strains do not display identical physiology Furthermore, although pyruvate is stimulatory to N. viennensis and N. maritimus, it is slightly inhibitory to Nitrosotalea strains. Capacity for mixotrophy, or the lack of it, may be an important contributor to niche differentiation of AOA.
Acknowledgments
The authors would like to thank Dr Martin Könneke (University of Bremen) for advice and helpful discussions on AOA carbon metabolism, and Mr Kevin MacKenzie and Ms Gillian Milne (University of Aberdeen) for technical support with scanning and transmission electron microscopy. This work was financially supported by Natural Environmental Research Council (standard grant NE/I027835/1), the Royal Society (International Exchange grant IE111001) and the National Science Foundation of China (grant 81101283).
Authors' contribution
L.E.L.-M. and C.G. contributed equally to this work.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Effects of addition of 100 μM TCA cycle intermediates on cell abundance (a) and yield normalised against 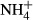 production (b).
production (b).
Maximum-likelihood phylogenetic analysis of inferred AmoA amino acid sequences translated from amoA gene sequences of Nitrosotalea devanaterra Nd1 and Nitrosotalea sp.
AOA cultures reported in literature.
Table S2. Capacity of organic compounds to restore growth in the pure culture of Nitrosotalea devanaterra.
Table S3. Properties of the examined organic acids.
Supplementary
References
- Alexander B, Leach S. Ingledew WJ. The relationship between chemiosmotic parameters and sensitivity to anions and organic acids in the acidophile Thiobacillus ferrooxidans. J Gen Microbiol. 1987;133:1171–1179. [Google Scholar]
- Arp DJ, Chain PSG. Klotz MG. The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu Rev Microbiol. 2007;61:503–528. doi: 10.1146/annurev.micro.61.080706.093449. [DOI] [PubMed] [Google Scholar]
- Aston JE, Apel WA, Lee BD. Peyton BM. Toxicity of select organic acids to the slightly thermophilic acidophile Acidithiobacillus caldus. Environ Toxicol Chem. 2009;28:279–286. doi: 10.1897/08-277.1. [DOI] [PubMed] [Google Scholar]
- Baker-Austin C. Dopson M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007;15:165–171. doi: 10.1016/j.tim.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, Alber BE. Fuchs G. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- Cieśliński G, Van Rees KCJ, Szmigielska AM, Krishnamurti GSR. Huang PM. Low-molecular-weight organic acids in rhizosphere soils of durum wheat and their effect on cadmium bioaccumulation. Plant Soil. 1998;203:109–117. [Google Scholar]
- Dias ACF, Hoogwout EF, Pereira e Silva MDC, Salles JF, van Overbeek LS. van Elsas JD. Potato cultivar type affects the structure of ammonia oxidizer communities in field soil under potato beyond the rhizosphere. Soil Biol Biochem. 2012;50:85–95. [Google Scholar]
- Ertl P, Rohde B. Selzer P. Fast calculation of molecular polar surface area as a sum of fragment based contributions and its application to the prediction of drug transport properties. J Med Chem. 2000;43:3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- Grosßkopf R, Stubner S. Liesack W. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl Environ Microbiol. 1998;64:4983–4989. doi: 10.1128/aem.64.12.4983-4989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubry-Rangin C, Nicol GW. Prosser JI. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol. 2010;74:566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, Schloter M, Griffiths RI, Prosser JI. Nicol GW. Niche specialization of terrestrial archaeal ammonia oxidizers. P Natl Acad Sci USA. 2011;108:21206–21211. doi: 10.1073/pnas.1109000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. Gascuel O. phyml: a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall TA. bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H. Wagner M. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. P Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann M, Saunders AM. Schramm A. Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl Environ Microbiol. 2008;74:3279–3283. doi: 10.1128/AEM.02802-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL. Organic acids in the rhizosphere – a critical review. Plant Soil. 1998;205:25–44. [Google Scholar]
- Jung MY, Park SJ, Min D, Kim JS, Rijpstra WIC, Sinninghe Damste JS, Kim GJ, Madsen EL. Rhee SK. Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl Environ Microbiol. 2011;77:8635–8647. doi: 10.1128/AEM.05787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeler E. Gerber H. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol Fertil Soils. 1988;6:68–72. [Google Scholar]
- Kemp JS, Paterson E, Gammack SM, Cresser MS. Killham K. Leaching of genetically modified Pseudomonas fluorescens through organic soils: influence of temperature, soil pH, and roots. Biol Fertil Soils. 1992;13:218–224. [Google Scholar]
- Kishimoto N, Inagaki K, Sugio T. Tano T. Growth inhibition of Acidiphilium species by organic acids contained in yeast extract. J Ferment Bioeng. 1990;70:7–10. [Google Scholar]
- Kitko RD, Cleeton RL, Armentrout EI, Lee GE, Noguchi K, Berkmen MB, Jones BD. Slonczewski JL. Cytoplasmic acidification and the benzoate transcriptome in Bacillus subtilis. PLoS One. 2009;4:e8255. doi: 10.1371/journal.pone.0008255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB. Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT. Tiedje JM. Genomic insights that advance the species definition for prokaryotes. P Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Goodfellow M, editor; Stackebrandt E, editor. Nucleic Acid Techniques in Bacterial Systematics. New York, NY: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI. Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. P Natl Acad Sci USA. 2011;108:15892–15897. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR. Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Muyzer G, De Waal EC. Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T. Stahl DA. Transcriptional response of the archaeal ammonia oxidizer Nitrosopumilus maritimus to low and environmentally relevant ammonia concentrations. Appl Environ Microbiol. 2013;79:6911–6916. doi: 10.1128/AEM.02028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ñancucheo I. Johnson DB. Production of glycolic acid by chemolithotrophic iron- and sulfur-oxidizing bacteria and its role in delineating and sustaining acidophilic sulfide mineral-oxidizing consortia. Appl Environ Microbiol. 2010;76:461–467. doi: 10.1128/AEM.01832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol GW, Leininger S, Schleper C. Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L. Schleper C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol. 2003;5:787–797. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- Paul EA. Clarke FE. Soil Microbiology and Biochemistry. San Diego: Academic Press; 1989. [Google Scholar]
- Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A. Wagner M. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol. 2012;14:525–539. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser JI. Nicol GW. Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 2012;20:523–531. doi: 10.1016/j.tim.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Salmond CV, Kroll RG. Booth IR. The effect of food preservatives on pH homeostasis in Escherichia coli. J Gen Microbiol. 1984;130:2845–2850. doi: 10.1099/00221287-130-11-2845. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS. Eliceiri KW. NIH Image to imagej: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinn MB. Colorimetric method for determination of nitrite. Ind Eng Chem. 1941;13:33–35. [Google Scholar]
- Stahl DA. de la Torre JR. Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol. 2012;66:83–101. doi: 10.1146/annurev-micro-092611-150128. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M. Kumar S. mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourna M, Freitag TE, Nicol GW. Prosser JI. Growth, activity and temperature responses of ammonia oxidising archaea and bacteria in soil microcosms. Environ Microbiol. 2008;10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- Tourna M, Freitag TE, Nicol GW. Prosser JI. Stable isotope probing analysis of interactions between ammonia oxidizers. Appl Environ Microbiol. 2010;76:2468–2477. doi: 10.1128/AEM.01964-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourna M, Stieglmeier M, Spang A, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. P Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Uexküll HR. Mutert E. Global extent, development and economic impact of acid soils. In: Probert ME, editor; Date RA, Grundon NJ, Raymet GE, editors. Plant–Soil Interactions at Low pH: Principles and Management. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1995. pp. 5–19. [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, et al. The Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine Archaea. P Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Haaijer SC, Op den Camp HJ, van Niftrik L, Stahl DA, Könneke M, Rush D, Sinninghe Damsté JS, Hu YY. Jetten MS. Mimicking the oxygen minimum zones: stimulating interaction of aerobic archaeal and anaerobic bacterial ammonia oxidizers in a laboratory-scale model system. Environ Microbiol. 2012;14:3146–3158. doi: 10.1111/j.1462-2920.2012.02894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of addition of 100 μM TCA cycle intermediates on cell abundance (a) and yield normalised against 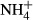 production (b).
production (b).
Maximum-likelihood phylogenetic analysis of inferred AmoA amino acid sequences translated from amoA gene sequences of Nitrosotalea devanaterra Nd1 and Nitrosotalea sp.
AOA cultures reported in literature.
Table S2. Capacity of organic compounds to restore growth in the pure culture of Nitrosotalea devanaterra.
Table S3. Properties of the examined organic acids.
Supplementary


