Abstract
The microorganisms that inhabit the human gastrointestinal tract comprise a complex ecosystem with functions that significantly contribute to our systemic metabolism and have an impact on health and disease. In line with its importance, the human gastrointestinal microbiota has been extensively studied. Despite the fact that a significant part of the intestinal microorganisms has not yet been cultured, presently over 1000 different microbial species that can reside in the human gastrointestinal tract have been identified. This review provides a systematic overview and detailed references of the total of 1057 intestinal species of Eukarya (92), Archaea (8) and Bacteria (957), based on the phylogenetic framework of their small subunit ribosomal RNA gene sequences. Moreover, it unifies knowledge about the prevalence, abundance, stability, physiology, genetics and the association with human health of these gastrointestinal microorganisms, which is currently scattered over a vast amount of literature published in the last 150 years. This detailed physiological and genetic information is expected to be instrumental in advancing our knowledge of the gastrointestinal microbiota. Moreover, it opens avenues for future comparative and functional metagenomic and other high-throughput approaches that need a systematic and physiological basis to have an impact.
Keywords: microbiota, diversity, gut, gastrointestinal, microbiome, function
Introduction – a historical perspective
Human beings, similar to other higher organisms, live in symbiosis with their coevolved microbiota (Bäckhed et al., 2005). The majority of the human microorganisms reside in the gastrointestinal tract, where, besides contributing to the digestion, they perform various other functions that are essential for the human host. These functions include the production of vitamins, education of the immune system, communication with the intestinal cells, and modulation of the host's behavior (Bäckhed et al., 2005; Cryan & Dinan, 2012; Rajilić-Stojanović, 2013). The first report of living creatures in the human gastrointestinal tract dates from 1681 when Antonie van Leeuwenhoek reported a variety of ‘little animals’ in his stool samples and identified what is now thought to be a Giardia spp. when suffering from diarrhea (Dobell, 1932). Almost two centuries passed before the first detailed descriptions of pure cultures of gastrointestinal microorganisms were reported, of which the earliest is most likely the description of the eukaryal intestinal parasite Pentatrichomonas hominis (at the time named Trichomonas hominis), by Casimir Davaine in 1854 (Hemmeter, 1902). Since P. hominis, similar to other intestinal Eukarya, has a very low prevalence, this discovery did not trigger further analysis of the gastrointestinal microbiota. However, intensive studies of the gastrointestinal microbiota followed the first cultivation of the intestinal bacterium, now known as Escherichia coli. From a historical perspective, this and several other events, here termed turning points, can be recognized as having impacted the discovery of the gastrointestinal microbiota constituents. These turning points are evident when the number of described gastrointestinal tract species is considered in view of time (Fig. 1).
Fig 1.
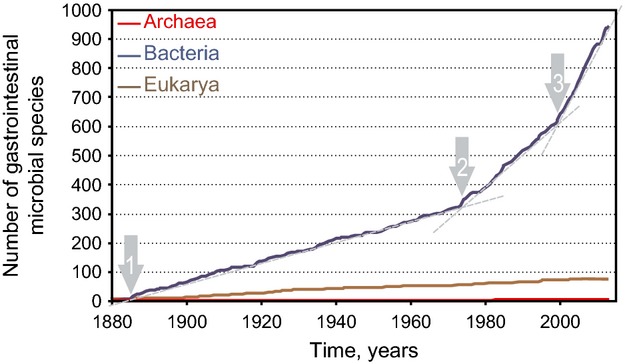
Graphical representation of the cumulative number of cultured species from Bacteria, Archaea and Eukarya from the human gastrointestinal tract as a function of time. The arrows indicate the turning points of the gastrointestinal microbiota research: (1) Isolation of the first gastrointestinal bacterial species, (2) Introduction of strictly anaerobic techniques, and (3) Introduction of molecular techniques in the field of the gastrointestinal microbiota research.
The first turning point (Fig. 1) marks the first description of a gastrointestinal bacterium, which is the isolation of Bacterium coli commune (later renamed to E. coli), by the German pediatrician Teodor Escherich in 1885 (Shulman et al., 2007). The studies that followed shortly thereafter led to the description of representatives of a number of the major gastrointestinal bacterial groups, including the genera Bacteroides, Bifidobacterium, and Bacillus as well as proteolytic cocci (Flügge, 1886; Veillon & Zuber, 1898; Moro, 1900; Tissier, 1900; Passini, 1905; Tissier, 1908; Distaso, 1911). During this period that lasted till the late sixties of the 20th century, Bifidobacterium and Bacteroides spp. were considered to be the dominant groups in the human gastrointestinal tract. Aerobes, referred as coliforms, streptococci and lactobacilli, were found as minor groups, while clostridia, staphylococci and aerobic spore-formers were reported as rare and not always detectable (Haenel, 1970). However, the vast majority of the gastrointestinal microorganisms are now known to be strict anaerobes, and this was for the first time shown in 1931 (Sanborn, 1931). Therefore, the early cultivation studies provided only a partial view of the gastrointestinal microbiota and it enabled isolation of only a minority (10–25%) of the gastrointestinal microorganisms (Finegold, 1969).
The improvements of anaerobic cultivation techniques by Hungate (1969) marked the second turning point in the gastrointestinal microbiota research, approximately 50 years ago (Fig. 1). In this second period of the gastrointestinal microbiota research that lasted from the early seventies till the molecular revolution in the beginning of this century (Fig. 1), it was recognized that the microbiota in the gastrointestinal tract is dominated by bacterial species that belong to the following genera: Bacteroides, Clostridium, Eubacterium, Veillonella, Ruminococcus, Bifidobacterium, Fusobacterium, Lactobacillus, Peptostreptococcus, and Peptococcus (Moore & Holdeman, 1974a). Using strict anaerobic techniques, it was reportedly possible to cultivate up to 88% of the total microscopic counts in fecal samples (Moore & Holdeman, 1974a). However, due to the enormous complexity of the gastrointestinal microbiota, many of the hundreds of isolates were not characterized beyond the genus level (Finegold et al., 1974; Moore & Holdeman, 1974a; Benno et al., 1986). Moreover, as processing of even a single sample yielded an enormous amount of different isolates, it was physically impossible to compare these all and make a full description based on the morphological, biochemical and physiological characteristics that could be determined at that time (Moore & Holdeman, 1974b). Hence, due to these technical limitations, the gastrointestinal microbiota remained only partially characterized.
Finally, the third turning point in the gastrointestinal microbiota research can be ascribed to the incorporation of molecular techniques about a dozen years ago (Fig. 1). These include global and culture-independent studies based on the sequence analysis of the small subunit ribosomal RNA (SSU rRNA) that had provided the molecular basis for microbial taxonomy that is currently used (Woese et al., 1990). However, the complexity of the gastrointestinal tract microbial ecosystem hampers the rapid application of SSU rRNA-based methods as well as (meta)genomics (Zoetendal et al., 2008). Hence, the first gastrointestinal tract study using SSU rRNA sequences dealt with a single adult sample (Wilson & Blitchington, 1996). Subsequent SSU rRNA-based studies in multiple adults demonstrated the individuality, temporal stability and site specificity of the intestinal microbiota with a diversity that was only partially grasped in cultivation-based studies (Zoetendal et al., 1998; Suau et al., 1999; Zoetendal et al., 2001). These novel findings sparked a revival of the scientific interest in the gastrointestinal microbiota that was initially compared to a Renaissance (Tannock, 1999). However, the years that followed showed this to be more of a revolution that incorporated metagenome and whole-genome characterizations (Nelson et al., 2010; Qin et al., 2010; Brown et al., 2013).
When integrated with cultivation-based studies, the analysis of the SSU rRNA gene sequences as phylogenetic markers enabled rapid identification of the new gastrointestinal isolates, and illustrated the need for the reclassification of many species. In addition, the SSU rRNA gene sequences enabled the detection of not yet cultured microorganisms and their phylogenetic positioning. Finally, the research field expanded to another dimension with the application of high-throughput approaches, including next-generation sequencing of the SSU rRNA gene sequences or the entire genomic material (Zoetendal et al., 2008). The latter metagenomic analyses generated a baseline of over 3 million, mainly bacterial, genes present in the human gastrointestinal tract (Qin et al., 2010; Brown et al., 2013) and demonstrated that the majority of the gastrointestinal microorganisms contain genomes that have not yet been characterized (Qin et al., 2010; Le Chatelier et al., 2013).
The present-day view of the gastrointestinal microbiota composition is quite different than prior to the molecular revolution. Most importantly, it is evident that still many of the gastrointestinal microorganisms have not yet been cultured and this in particular concerns phylogenetically distinct bacterial groups belonging to the Firmicutes phylum (Rajilić-Stojanović et al., 2007). Furthermore, several of the bacterial groups that based on cultivation studies had been recognized as dominant gastrointestinal genera, have been reclassified and renamed. Most notably this concerns the Bacteroides spp. that have been reclassified into the genera Alistipes, Prevotella, Paraprevotella, Parabacteroides, and Odoribacter. Moreover, it is evident that different members of the Bacteroidetes phylum and not the Bacteroides genus sensu stricto are dominant in the human gastrointestinal tract. Furthermore, the abundance of the Peptostreptococcus spp. demonstrated in cultivation-based studies, could primarily be attributed to Peptostreptococcocus productus (Holdeman et al., 1976). However, SSU rRNA gene analysis has shown that this species does not belong to the genus Peptostreptococcocus, and the species was reclassified first as Ruminococcus productus (Ezaki et al., 1994) and finally as Blautia producta (Liu et al., 2008). Today, it is clear that Blautia spp., in contrast to Peptostreptococcus spp., form one of the most abundant groups in the human gastrointestinal tract. Many other so-called dominant genera are still in need for major reclassification, and the best example of this is the Clostridium genus, for which a detailed phylogenetic analysis led to a proposed grouping into 19 clusters (Collins et al., 1994). Bacteria belonging to the Clostridium spp. are highly abundant in the adult gastrointestinal tract, and in particular, the members of the species that cluster within the Clostridium cluster IV (C. leptum group, which major constituent is the Ruminococcacea family) and the Clostridium cluster XIVa (C. coccoides group, which resembles the Lachnospiraceae family). Furthermore, the Ruminococcus genus is polyphyletic or paraphyletic and its members cluster within two families — the Ruminococcaceae and Lachnospiraceae. A recent metagenomic study reported that the abundance of Ruminococcus spp. is a driver of one of the proposed enterotype status of the microbiota (Arumugam et al., 2011). However, as the present metagenomic analyses do not provide accurate phylogenetic information, it is unclear which of the two distinct groups of Ruminococcus spp., is the actual driver of this enterotype status. This example illustrates the need for a systematic and detailed presentation of the microbiota analysis in a phylogenetic framework.
The extensive period of the studying of the gastrointestinal microbiota, its complexity and its variation between individuals have generated a massive amount of information, which is scattered in the literature. To unify the knowledge of the gastrointestinal microbiota that has accumulated since its discovery, we have performed a search of the publications covering more than a century (Fig. 1). We found references that link the human gastrointestinal microbiota with a total of 1057 intestinal species belonging to the Eukarya (92), Archaea (8) and Bacteria (957; Fig. 2, Supporting information, Tables S1–S3). These species were analyzed in arb software-based database of the SSU rRNA sequences (Pruesse et al., 2007). The phylogenetic trees presented here were extracted from the reference phylogenetic tree of the silva database (Yarza et al., 2008). From this phylogenetic and literature analysis, it is clear that bacteria that cluster within the phyla Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria, are the most diverse and abundant microorganisms in the adult gastrointestinal tract (Fig. 2). The gastrointestinal microbiota also contains members of the less diverse, although in some cases still abundant, bacterial phyla, including the Verrucomicrobia, Lentisphaerae, Synergistetes, Planctomycetes, Tenericutes and the Deinococcus-Thermus group. In addition to these established phylogenetic groups, the SSU rRNA gene sequences of not yet cultured bacteria that cluster within the TM7 candidate phylum, Melainabacteria and Gemmatimonacetes, can be detected in the human gastrointestinal tract (Fig. 2). Several archaeal species that cluster within two phyla have been detected in the human gastrointestinal tract. The Euryarchaeota include the methanogens that are relatively abundant. Among the Eukarya there are organisms that are highly adapted to the human gastrointestinal tract, such as some Candida spp., while many other eukaryote species can be present at a low abundance and may be passengers. Altogether, our present analysis confirms that the human gastrointestinal microbiota is composed of representatives of all three domains of life — Bacteria, Archaea, and Eukarya.
Fig 2.
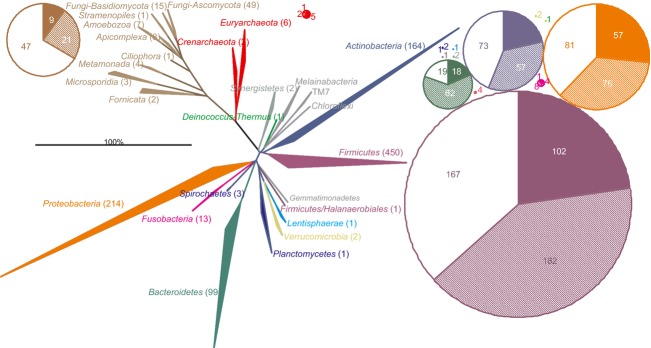
Phylogenetic tree of the human gastrointestinal microbiota. The numbers in parentheses indicate the number of cultured species given per phylum. The pie charts illustrate distribution between the number of species with full genome sequence genome (full sectors), the number of species with partial genome sequence (semi-full sectors) and number of species without any genome sequence (empty sectors) given for Archaea, Eukarya and per phylum for Bacteria. The color code of pies corresponds to the color code of the phylogenetic tree.
The gastrointestinal microbiota research is very dynamic, and in the last decade, 239 novel gastrointestinal tract species have been detected or described, confirming the earlier notion that the majority of the gastrointestinal microorganisms are cultivable but not yet cultured. While traditional cultivation media and strategies are efficient in obtaining novel species within the Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria phyla, for the detection of the gastrointestinal representatives of the phyla Verrucomicrobia and Lentisphaerae, the development of specific media and culturing approaches was needed (Zoetendal et al., 2003; Derrien et al., 2004). This suggests that for the cultivation of the gastrointestinal microorganisms belonging to phyla that lack any cultured representatives from the human intestine (e.g., TM7 candidate phylum or the Oscillospira genus), alternative and creative cultivation approaches should be developed and applied. Some new and promising developments include the use of high-throughput solid phase growth (Ingham et al., 2007), advanced culturing approaches using gnotobiotic mice (Goodman et al., 2011), or gel microdroplet culturing (Fitzsimons et al., 2013). The use of high-throughput culturing systems that employ a large set of growth media coupled to genomic characterization has proven to be very fruitful (Lagier et al., 2012a; Dubourg et al., 2013; Hamad et al., 2013; Pfleiderer et al., 2013). This recent attention for culturing the gastrointestinal microorganisms reflects the perceived need for detailed physiological, ecological and genetic studies. While a variety of functional metagenomics approaches have been described and applied, it is the integration with culturing approaches that is needed to further advance the understanding of the function of the intestinal ecosystem in health and disease. The power of this combination has recently been illustrated with the example of the abundant mucus-utilizing bacterium, Akkermansia muciniphila as a paradigm (Belzer & de Vos, 2012). Currently, the complete genome of at least one strain of 225 gastrointestinal species has been fully sequenced, assembled, and published, while many other genomic sequencing projects are ongoing (Fig. 2, Tables S1–S3). The physiological and genetic characteristics of these currently recognized gastrointestinal species and their association with particular functions of the ecosystem or diseases are systematized in this review that aims to provide the basis for future comparative and functional metagenomic and other high-throughput approaches applied on the gastrointestinal microbiota.
Actinobacteria
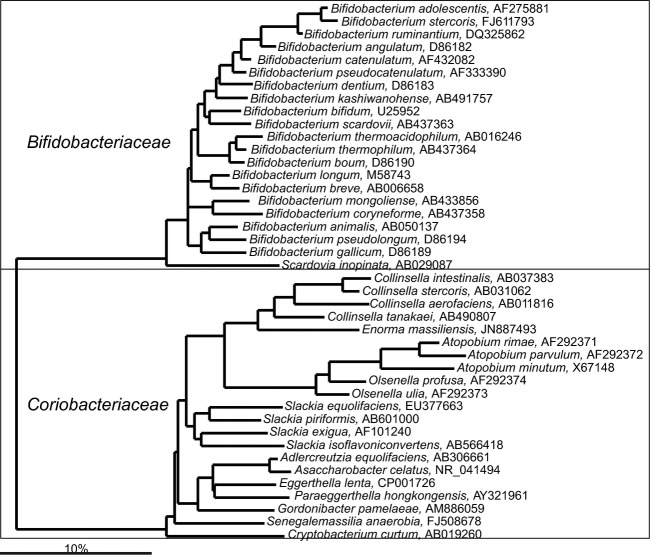
Actinobacteria are common and abundant in the human gastrointestinal tract. They are also known as gram-positive bacteria with a high G + C content in their DNA. As they are particularly difficult to lyse and their SSU RNA needs specific PCR primers to be amplified (Satokari et al., 2001), this group of bacteria is often underrepresented in molecular surveys of the gastrointestinal microbiota (notably in one of the first global studies of the infants' microbiota; Palmer et al., 2007). Members of the orders Bifidobacteriales (in particular Bifidobacterium spp.) and Coriobacteriales (mainly Collinsella spp.) are highly prevalent already since early life, while members of the order Actinobacteriales are human-associated bacteria that are subabundant and only scarcely detected in the gastrointestinal tract.
Actinobacteria-Bifidobacteriales
Bifidobacterium spp. form a dominant fraction of the human gastrointestinal microbiota, particularly in infants (Benno et al., 1984). Bifidobacteria are present in the abundance ranging between 108 and 1010 cells g−1 of intestinal content (Finegold et al., 1974; Moore & Holdeman, 1974a; Tannock, 1995). The majority of Bifidobacterium spp. have been recovered exclusively from human or animal gastrointestinal samples, and for two species (B. minimum and B. subtile) that were isolated from sewage (Scardovi & Trovatelli, 1974), an intestinal origin can be suspected, showing the high adaption of this genus to the gastrointestinal tract. Phylogenetically, Bifidobacterium spp. form a homogenous group, with 20 cultured species linked to the human gastrointestinal tract (Fig. 3). The first Bifidobacterium spp. was recovered from infant feces in 1900 by Henri Tissier, as a part of his PhD thesis work (Tissier, 1900). It was named Bacillus bifidus-communis. Already in 1924, this bacterium was renamed to Bifidobacterium bifidum, but Bifidobacterium was not recognized as an independent genus until 1974 (Biavati et al., 2000). The members of the Bifidobacterium genus are nonmotile, anaerobic or microaerophilic bacteria that produce acetate and lactate as major fermentation products from sugars. The degradation of sugars by these bacteria is performed through a phosphoketolase pathway, also known as the Bifido shunt. Bifidobacterium spp. degrade monosaccharides, galacto-, manno-, and fructo-oligosaccharides, while some strains are able to ferment complex carbohydrates such as starch, arabinogalactan, arabic gum, and gastric mucin (Crociani et al., 1994). As mother milk's contains nondigestible oligosaccharides that can be degraded by Bifidobacterium spp. (Marcobal et al., 2011), this bacterial group is strongly stimulated in breast-fed infants resulting in the dominance of Bifidobacterium spp. in the gastrointestinal microbiota before weaning. The Bifidobacterium spp. are assumed to have a beneficial effect on health (Mitsuoka, 1990) and several members of the Bifidobacterium genus are commercially applied as probiotics. The most relevant observation is that these bacteria have decreased abundance in relation to a number of diseases including vitamin K deficiency (Benno et al., 1985), atopic diseases (Kalliomaki et al., 2001), irritable bowel syndrome (Kerckhoffs et al., 2009; Rajilić-Stojanović et al., 2011), and autism (Wang et al., 2011a). Moreover, Bifidobacterium spp. represent a very stable component of the gastrointestinal microbiota of each person, the composition of which hardly changes throughout years (Rajilić-Stojanović et al., 2013b). Remarkably, a recent study of the microbiota of the Hazda tribe from Tanzania suggested that these adult hunter-gatherers do not carry any Bifidobacterium spp., which was explained by absence of dietary components such as meat and dairy that could support growth of these bacteria (Schnorr et al., 2014).
Fig 3.
Phylogenetic tree of the human gastrointestinal species that belong to the orders of the Bifidobacteriales and Coriobacteriales. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated.
Actinobacteria-Coriobacteriales
Coriobacteriales species constitute a frequently detected group of the gastrointestinal microbiota composed of representatives of 12 different genera (Fig. 3). Collinsella is the most dominant among other members of the order, and a representative of these rod-shaped, nonmotile obligate anaerobes was for the first time detected in human feces in 1935 (Eggerth, 1935). Both cultivation- and molecular-based studies show that Collinsella aerofaciens is a prevalent and an abundant gastrointestinal microorganism (Moore & Holdeman, 1974a; Benno et al., 1986; Kageyama et al., 2000). Four different types of Collinsella aerofaciens were initially recognized and later reclassified into distinct species (Kageyama & Benno, 2000). Collinsella spp. can ferment a wide range of different carbohydrates including complex sugars, such as starch but also glycogen to produce hydrogen gas, ethanol, formate, and lactate (Eggerth, 1935; Kageyama et al., 1999a). Experiments with an in vitro model of the human colon showed that Collinsella spp. along with Bifidobacterium spp. are the major lactose utilizers in the human gastrointestinal microbiota (Kovatcheva-Datchary, 2010). Moreover, Collinsella spp. are capable of deconjugation of bile acids and their abundance shows significant positive correlation with plasma cholesterol levels (Lahti et al., 2013).
Eggerthella are assacharolytic bacteria that produce acids only from glucose, but not from other sugars. The first representative of this bacterial group was isolated in 1935 by Arnold Eggerth (Eggerth, 1935). These bacteria produce formate and lactate. Until now, only Eggerthella lenta and the still not fully characterized Eggerthella sp. YY7918 are associated with the human gastrointestinal tract. Eggerthella lenta has been implied in producing anti-tumor substances that stimulate natural killer cells (Hatta, 1995), while Eggerthella sp. YY7918 has been reported to produce s-equol (Yokoyama & Suzuki, 2008), which has anticarcinogenic properties (Yuan et al., 2007).
Slackia spp. are asaccharolytic bacteria with the common feature of converting dietary isoflavones. These isoflavones have been proposed to prevent hormone-dependent diseases, while their conversion by gastrointestinal bacteria impacts their biological effectiveness. Among the bacterial products, s-equol appears to be the most relevant to human physiology (Yuan et al., 2007). At least two Slackia spp. are capable of equol production from isoflavones (Matthies et al., 2009; Jin et al., 2010), while Adlercreutzia equolifaciens that also belongs to the Coriobacteriales order, is another gastrointestinal species capable to produce equol (Maruo et al., 2008). The ability to produce s-equol is more abundant among microbiota of Asian than the Caucasian subjects (Song et al., 2006) and can be explained by the adaptation of the microbiota to the higher availability of isoflavones — particularly those derived from soy beans.
Atopobium species are anaerobic bacteria that cluster within the Actionobacteria phylum, and, in contrast to the rest of the phylum, contain DNA with a low G + C content. The main product of their metabolism is lactate, which is in line with the previous classification of these bacteria within the Lactobacillus and Streptococcus genera (Collins & Wallbanks, 1992). Based on the literature data, it can be concluded that Atopobium spp. are among the earliest colonizers of the human intestinal tract as they are reported to be present in gastrointestinal contents of 6-week-old infants (Fallani et al., 2011). However, the data on Atopobium quantification are based on the application of a FISH probe for the Atopobium cluster, which in addition to Atopobium, hybridizes to species that belong to the Coriobacterium, Eggerthella and Collinsella genera (Harmsen et al., 2000). Therefore, it is not clear if the Atopobium or the other targeted genera are colonizing the gastrointestinal tract of infants. Bacteria belonging to the Atopobium cluster are significantly associated with the major products of protein fermentation, suggesting that these bacteria are responsible for protein degradation in the gastrointestinal tract (Shen et al., 2010; Thompson-Chagoyan et al., 2011).
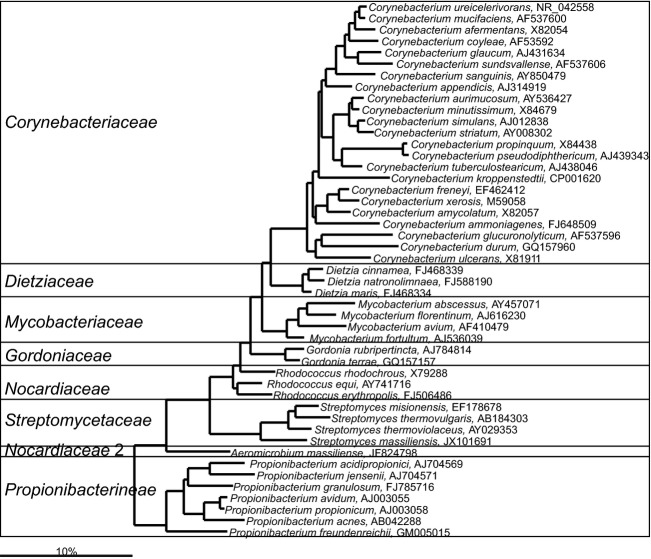
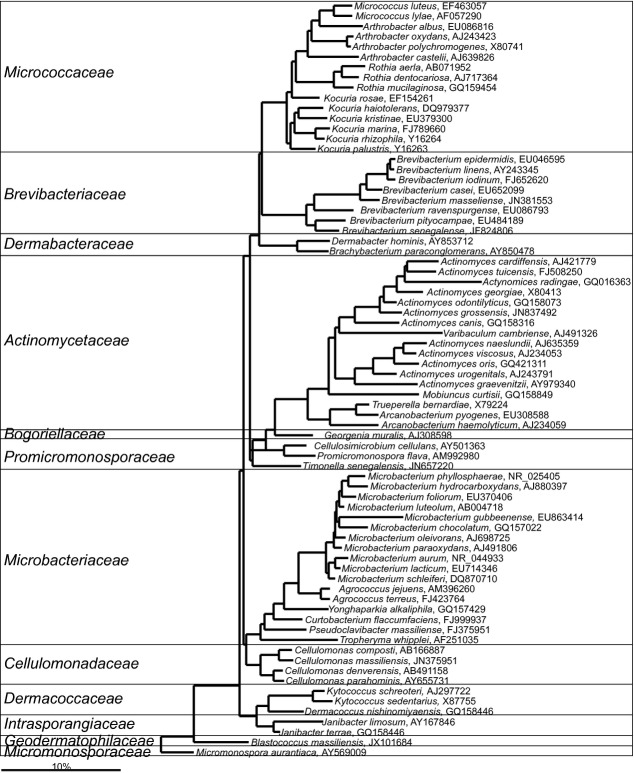
Actinobacteria-Actinomycetales
The Actinobacteria of the human gastrointestinal tract include diverse members of the order Actinomycetales (Figs4 and 5 — for clarity the phylogenetic tree of this numerous order was split into two parts). These bacteria are rarely detected in human gastrointestinal samples, but this is most likely due to their low abundance in the gastrointestinal tract that is in the range from 102 to 103 cells g−1 of feces (Hoyles et al., 2012). Their low abundance can explain the fact that many representatives of this group were detected only in studies that specifically targeted this group of bacteria (Hoyles et al., 2013), or in studies that targeted low abundant bacteria within the gastrointestinal microbiota (Lagier et al., 2012a; Dubourg et al., 2013). Various different Actinomycetales species, of which many are still uncultured, were identified in a molecular study of these specific subcommunity within the gastrointestinal microbiota of healthy humans of different ages, showing a high prevalence of these bacteria (Hoyles et al., 2013). The most diverse and the frequently detected Actinomycetales of the human gastrointestinal tract include Propionibacterium spp. and Corynebacterium spp. (Fig. 4). These bacteria typically colonize the human skin and are found in high abundance in infants that are born using Caesarean section (Dominguez-Bello et al., 2010). In adults, Corynebacterium spp. are more frequently detected in the samples of the upper gastrointestinal tract (Justesen et al., 1984). Cultivation studies have indicated that Propionibacterium spp. are the major proteolytic bacteria in the human intestine (Macfarlane et al., 1986). Propionibacterium spp. are applied as probiotics as they are major vitamin B12 producers and produce propionate from lactate. In vitro experiments showed that the metabolic products of two gastrointestinal Propionibacterium spp. can induce apoptosis of colorectal carcinoma cells (Jan et al., 2002).
Fig 4.
Phylogenetic tree of a fraction of the human gastrointestinal species that belong to the order of the Actinomycetales. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. For the other human gastrointestinal species that cluster within the Actinomycetales see Fig. 5.
Fig 5.
Phylogenetic tree of a fraction of the human gastrointestinal species that belong to the order of the Actinomycetales. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. For the other human gastrointestinal species that cluster within the Actinomycetales see Fig. 4.
Bacteria belonging to the Rhodococcus genus are rarely detected in the human gastrointestinal microbiota but have been found in an extreme abundance (up to 68%) in mucosal biopsies of ulcerative colitis patients (Lepage et al., 2011). Mycobacterium spp., notably Mycobacterium avium, have also been implied in ulcerative colitis and the expression of intestinal cells in inflammatory bowel disease patients appears to have similarities to Mycobacterium infection (Sibartie et al., 2010).
The gastrointestinal Actinomycetales also include Rothia spp. that are frequently detected in the upper gastrointestinal tract (Ou et al., 2009), but rarely in fecal samples. These species contribute to the degradation of gluten (Zamakhchari et al., 2011) and their abundance and activity might be relevant for celiac disease and other conditions related to gluten digestion. Another species of this order — Actinomyces graevenitzii — which is detected in an increased abundance in the small intestine of celiac disease patients, might be a relevant risk factor for the development of this disease (Ou et al., 2009). Various other Actinomyces spp. can be detected in low abundance in the fecal samples of healthy humans (Hoyles et al., 2012; Hoyles et al., 2013), and while the role of these bacteria in the gastrointestinal tract is still to be determined, it is noteworthy that an Actinomyces spp. was detected as colonizer of the infant gastrointestinal tract using sensitive molecular methods already in the first days of life (Favier et al., 2002).
Micrococcus spp. are relatively prevalent (present in 20% of the analyzed subjects) in the samples of the upper gastrointestinal tract in patients predisposed to the development of the small intestinal bacterial overgrowth syndrome (Bouhnik et al., 1999). Although representatives of this genus can be detected in the fecal samples (Finegold et al., 1974), these bacteria typically inhabit human skin. Similar applies to six Kocuria spp., which are human skin and oropharynx mucosa commensals (Savini et al., 2010), although two recent studies have reported presence of Kocuria spp. in gastrointestinal samples (Lagier et al., 2012a; Fitzsimons et al., 2013).
The other members of the Actinomycetales order include the representative species of the following genera: Brevibacterium, Cellulomonas, and Microbacterium. These genera are typically associated with other ecosystems, namely the skin (Brevibacterium), and soil (Cellulomonas and Microbacterium). Nevertheless, most of these bacteria are already recognized as relevant for human health, as many of these species can cause infections of different tissues, particularly in immuno-suppressed patients (Funke et al., 1997). It has been suggested that gastrointestinal tract represents the natural niche of these bacteria (Funke et al., 1997).
Bacteroidetes
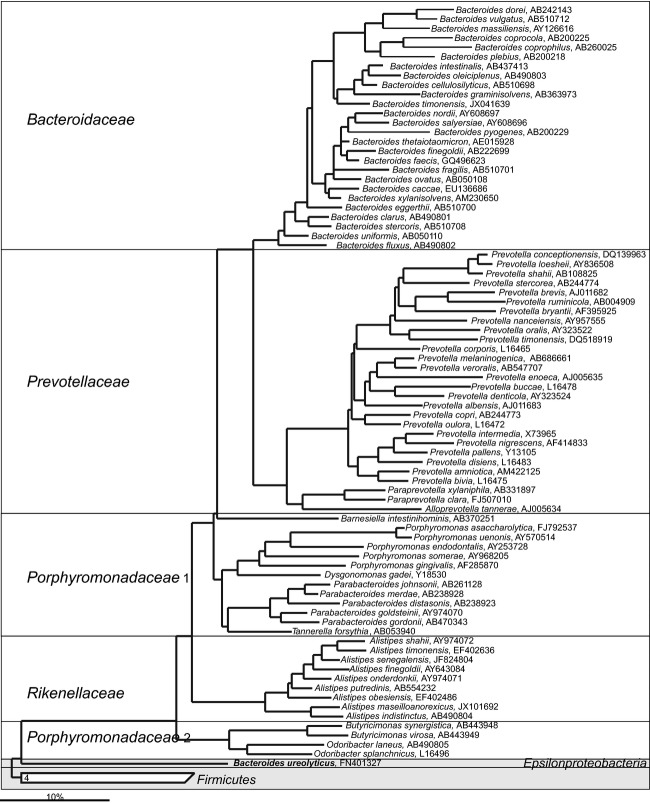
The Gram-negative bacteria that belong to the phylum Bacteroidetes are common, abundant and diverse within the human gastrointestinal tract. The first Bacteroides species — Bacteroides fragilis — was isolated in 1898 as a human pathogen linked to appendicitis among other clinical cases (Veillon & Zuber, 1898). Although some Bacteroides spp. are still considered to be opportunistic pathogens, several decades of research have testified that many Bacteroidetes species are highly adjusted to the gastrointestinal tract, where they live in high abundance (up to 1011 cells g−1 of intestinal material; Eggerth & Gagnon, 1933; Moore & Holdeman, 1974a; Benno et al., 1986). Hence, they perform metabolic conversions that are essential for the host, often related to the degradation of proteins or complex sugar polymers. The colonization of the gastrointestinal tract with the Bacteroidetes is promoted already in infants, as mother milk's nondigestible oligosaccharides support the growth of both Bacteroides and Bifidobacterium spp. (Marcobal et al., 2011). Furthermore, animal model experiments have shown that the colonization of the normal gastrointestinal tract, as illustrated by experiments with pure cultures of Bacteroides spp., is a result of the recognition and selection by the immune system of the host (Rakoff-Nahoum et al., 2004), mediated through the toll-like receptors (Round et al., 2011; Lopez-Siles et al., 2012) and other specific host-microorganism interactions (Hooper et al., 2012).
For a long time, it was thought that the majority of Gram-negative gastrointestinal tract bacteria belonged to the Bacteroides genus, but in recent years many earlier designed Bacteroides spp. were assigned to other genera within the Bacteroidetes phylum. Currently, only four gastrointestinal Bacteroides spp. form deep branches in the phylogenetic tree (Fig. 6), suggesting that these bacteria (B. ureolyticus, B. galacturonicus, B. pectinophilus, and B. coagulans) still should be reclassified to other phylogenetic groups. A similar situation applies to Anaerorhabdus furcosa, which is still classified as a member of the Bacteroidaceae family, but based on its SSU rRNA gene sequence clusters within the Firmicutes phylum. The majority of the gastrointestinal Bacteroidetes spp. belongs to the following bacterial families: Bacteroidaceae, Prevotellaceae, Rikenellaceae, and Porphyromonadaceae (Fig. 6). These bacterial species share the common feature that they produce succinic acid, acetic acid, and in some cases propionic acid, as the major end-products. Species belonging to the genera Alistipes, Bacteroides, Parabacteroides, Prevotella, Paraprevotella, Alloprevotella, Barnesiella, and Tannerella are saccharolytic, while species belonging to Odoribacter and Porphyromonas are predominantly asaccharolytic. Some Bacteroides spp. and Prevotella spp. can degrade complex plant polysaccharides such as starch, cellulose, xylans, and pectins (Wu et al., 1992; Morotomi et al., 2009; Sakamoto & Ohkuma, 2012). The Bacteroidetes species play also an important role in protein metabolism, as some species have proteolytic activity, assigned to the proteases that are linked to the cell wall (Macfarlane et al., 1986; Macfarlane et al., 1988), while some Bacteroides spp. have a potential to utilize urea as a nitrogen source (Yatsunenko et al., 2012). Other important functions of Bacteroides spp. include the deconjugation of bile acids (Narushima et al., 2006) and growth on mucus (Leitch et al., 2007). The Bacteroidetes contribute to the recently proposed classification of the gastrointestinal microbiota into enterotypes (Arumugam et al., 2011). The importance of the Bacteroidetes is further illustrated by the fact that this group is the most stable component of the gastrointestinal microbiota over time in healthy adults (Rajilić-Stojanović et al., 2013b). Anecdotally, a unique case report described the microbiota of a critically ill patient that harbored no Bacteroidetes — this patient passed away soon after sampling (Dubourg et al., 2013).
Fig 6.
Phylogenetic tree of the human gastrointestinal species that belong to the class of the Bacteroidia. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. Deeply rooted Bacteroides spp., which based on the SSU rRNA gene sequence cluster within distant phylogenetic groups are depicted in the gray area.
Because of their broad metabolic potential, the role of the Bacteroidetes in the gastrointestinal microbiota is complex: while the reduced abundance of the Bacteroidetes in some cases is associated with obesity (Ley, 2010) and irritable bowel syndrome (Rajilić-Stojanović et al., 2011), this bacterial group appears to be enriched in patients suffering from type 1 and type 2 diabetes (Larsen et al., 2010). Moreover, Bacteroides spp. in contrast to Prevotella spp. were recently found to be enriched in the metagenomes of subjects with low gene richness that were associated with adiposity, insulin resistance and dyslipidaemia as well as an inflammatory phenotype (Le Chatelier et al., 2013).
Bacteroidetes species that belong to classes Flavobacteriales and Sphingobacteriales are only occasionally detected in the gastrointestinal tract (Fig. 7, Table S1). With an exception of Capnocytophaga spp. and Sphingobacterium spp. that can be detected in the human oral cavity, the other bacteria of this group are typically associated with other ecosystems (primarily soil). There is no data about the role of these bacteria in the gastrointestinal microbiota, but it is noteworthy that several of these bacteria were detected only in the SSU rRNA gene clone libraries of the microbiota of inflammatory bowel disease patients (Frank et al., 2007).
Fig 7.
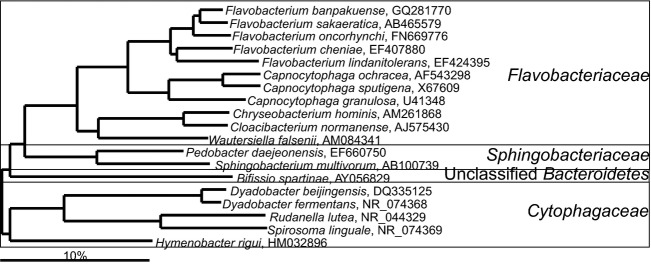
Phylogenetic tree of the human gastrointestinal species that belong to the classes of the Cytophagia and Sphingobacteria. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated.
Firmicutes
Firmicutes are the most diverse and abundant group of the gastrointestinal microbiota, making up over half and in many cases around 80% of the gastrointestinal microbiota of healthy adults. The gastrointestinal Firmicutes are distributed over four classes: Bacilli, Clostridia, Erysipelotrichi, and Negativicutes. Traditionally, this group is considered to include Gram-positive bacteria with a low GC content in their DNA, although recent studies have shown that Gram-positive staining is not a feature of many Firmicutes. This can be illustrated with Faecalibacterium prausnitzii, which is a Gram-negative-staining bacterium, previously classified within the Fusobacteria phylum (Duncan et al., 2002a), novel gastrointestinal isolates such as Christensenella minuta (Morotomi et al., 2012), but also typical Gram-negative bacteria such as members of the Veillonellacea family (Marchandin et al., 2010). While the vast majority of the Firmicutes are indeed low GC content bacteria, this also, is not a common feature of the phylum as seen in an example of Anaerofustis stercorihominis, which DNA has a content of the GC of around 70% (Finegold et al., 2004). Most of the Firmicutes, notably the Clostridium spp. and Bacillus spp., are spore-formers and this property confers special survival value in and beyond the gastrointestinal tract.
The most abundant gastrointestinal microorganisms are members of the class Clostridia and within this class the families Ruminococcaceae and Lachnospiraceae (Tap et al., 2009; Jalanka-Tuovinen et al., 2011). Another diverse group of the Firmicutes is the class Bacilli that includes the genera of Lactobacillus, Enterococcus, and Streptococcus, which are dominant in the upper part of the gastrointestinal tract. In line with its enormous diversity, the Firmicutes in the gastrointestinal tract perform a number of different functions that stretch from health promoting of some probiotic Lactobacillus spp. to pathogenic properties of Clostridium difficile. The vast majority of the currently uncultured gastrointestinal inhabitants belong to the phylum Firmicutes (Rajilić-Stojanović et al., 2007), which illustrates that future research is expected to dramatically expand our knowledge about the functional contribution of this group to the ecosystem and the host.
Bacilli
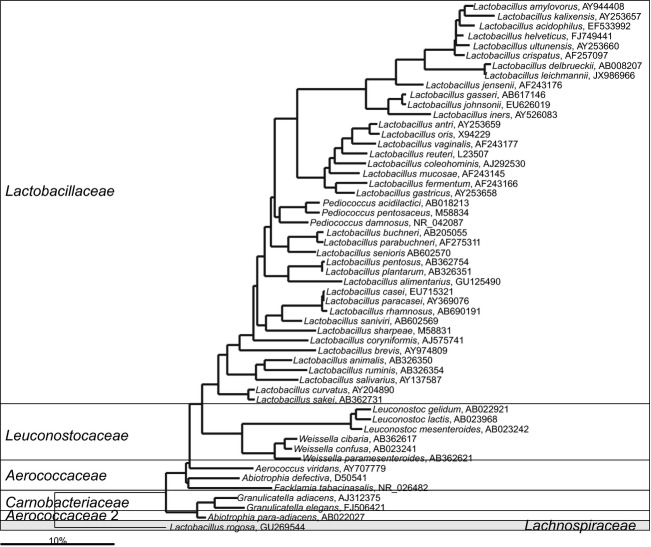
The first representative of the Bacilli class retrieved from the gastrointestinal tract was a member of Lactobacillales order and was isolated in 1900 — Bacillus acidophilus (Moro, 1900). The description of this species is vague, based on the currently accepted standards, and as the original strain was lost, it is not clear if this species is Lactobacillus acidophilus or one of the other five species derived from the so-called L. acidophilus group (Mitsuoka, 1992). Lactobacilli comprise a group of gastrointestinal inhabitants that has received particular scientific attention (Tannock, 2004), mainly because of the health claims proposed by Metchnikoff (1908) in the beginning of the nineteenth century and their later application as probiotics. Although highly important for the health, Lactobacilli are rarely detected as markers of the gastrointestinal microbiota dysbiosis, but such reports exist and include a reduced abundance in patients suffering from inflammatory bowel disease (Keighley et al., 1978; Ott et al., 2004), type 1 diabetes (Murri et al., 2013). This might be related to the fact that lactobacilli are only a minor fraction of the fecal microbiota where they can reach counts of up to 108 cells g−1 (Simon & Gorbach, 1984), and most of the analysis of the gastrointestinal microbiota is based on the use of stool samples. In the small intestine Lactobacillus spp. represent one of the predominant groups obtained by culturing (Reuter, 2001). However, while molecular studies could confirm their presence in the upper intestinal tract, these also showed that the Lactobacilli are quite variable and not as abundant as other gastrointestinal genera at that location, such as Streptococcus and Veillonella (Booijink, 2009; Booijink et al., 2010). This may explain why Lactobacillus spp. should be part of the diet, as consumed probiotic strains of Lactobacillus spp. have a beneficial effect on human health and specific induction of gene expression has been observed in duodenal biopsies after exposure of Lactobacillus plantarum (van Baarlen et al., 2009). Specific media, developed already in the 1950s (Rogosa et al., 1951), enabled the isolation of numerous Lactobacillus spp. Nevertheless, new Lactobacillus spp. from human gastrointestinal tract are still being reported (Roos et al., 2005; Oki et al., 2012), indicating that even the 38 known gastrointestinal Lactobacillus sp. (Fig. 8) are not covering the group's full diversity. Several previously misclassified Lactobacillus spp. have now been reclassified into novel genera, including Weissella, Atopobium, Eggerthia, and Kandleria (Collins & Wallbanks, 1992; Bjorkroth et al., 2002; Salvetti et al., 2011). Currently, only Lactobacillus rogosae is strongly outgrouping from the remaining Lactobacillus spp., although even after exclusion of the strongly outgrouping species, the species show a large degree of the SSU rRNA gene variation and form several groups in the phylogenetic tree (Fig. 8) As Lactobacillus spp. produce lactic acid as the major fermentation production that can be accompanied with ethanol and carbon dioxide in some species and under some conditions, traditionally Lactobacillus spp. are classified into three groups: obligately homofermentative, facultatively homofermentative, and obligately heterofermentative. However, the phylogenetic position of the species does not seem to be related to their fermentation profile.
Fig 8.
Phylogenetic tree of a fraction of the human gastrointestinal species that belong to the order of the Lactobacillales. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. For the other gastrointestinal species that belong to the order of the Lactobacillales see Fig. 9.
In addition to Lactobacillus spp., other related, lactic acid bacteria can be detected in the gastrointestinal tract. Members of the genera Leuconostoc and Weissella used to be considered as occasional and possibly transient members of the gastrointestinal microbiota. However, a recent study showed that Leuconostoc spp. and Weissella spp. are abundant (representing up to 24% of total microbial community) and widely distributed in colonic mucosa after bowel cleansing (Hong et al., 2011). Moreover, Leuconostoc was identified as the most abundant bacterial genus, representing almost a quarter of the total microbial community in a group of meconium samples of newborns, which persisted in the gastrointestinal tract until 7 months of age (Gosalbes et al., 2013). In the same study, Weissella spp. were also detected as the earliest colonizers of the gastrointestinal tract of some newborns. These bacteria utilize simple sugars and their presence in the lower parts of the gastrointestinal tract is dependent on the activity of other gastrointestinal microorganisms that have the ability to degrade complex sugars, resistant to human digestive enzymes.
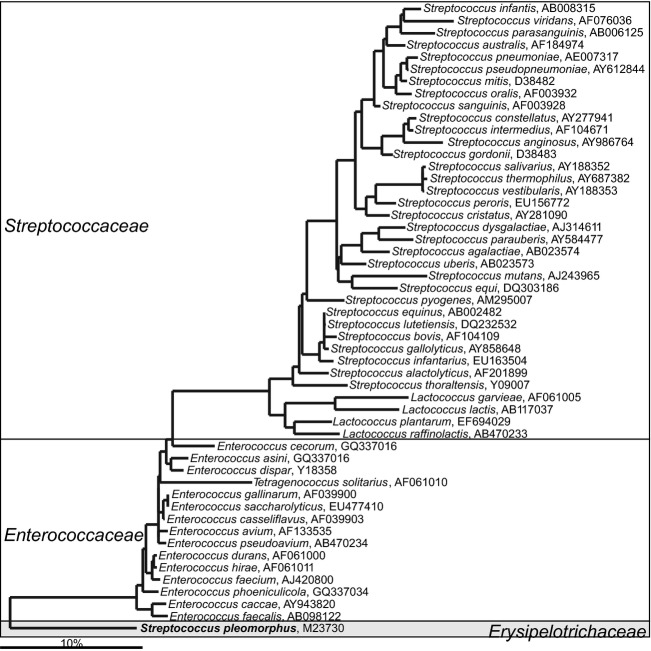
Other relevant gastrointestinal bacteria belonging to the Lactobacillales order include members of the genera Streptococcus and Enterococcus. These two genera have only recently been separated, although the presence of the subgroup within the genus Streptococcus was noticed as late as in the 1930s (Sherman, 1938). They are one of the dominant bacterial fractions in the upper part of the small intestine (Simon & Gorbach, 1986; Reuter, 2001). Forty-six species of these two genera are known to be gastrointestinal inhabitants (Fig. 9). In addition, Streptococcus pleomorphus, which also can be part of the gastrointestinal microbiota, forms a deep branch in the SSU rRNA gene sequence–based phylogenetic tree, suggesting that this species should be reclassified into another genus within the Erysipelotrichaceae family. The ample presence of the Enterococcus and Streptococcus spp. can be explained by the fact that the species are oxygen tolerant and easily cultivable. The oldest isolate of the group, Enterococcus faecalis, was for the first time plated in 1899 from a case of endocarditis (MacCallum & Hastings, 1899), and only 7 years later it was recovered from intestinal samples (Andrewes & Horder, 1906). Both Streptococcus and Enterococcus spp. are among the first established species in the infant's gastrointestinal tract that can be detected already in the first day of life (Solís et al., 2010; Gosalbes et al., 2013). Although this early presence would suggest an important role in the ecosystem, the data on the role of Streptococcus and Enterococcus spp. in human health are conflicting. Enterococcus spp. are widely recognized as opportunistic pathogens, although these species are common, and can even exhibit probiotic properties (Ó Cuív et al., 2013). The abundance of a Streptococcus species is decreased in mucosal biopsies in Crohn's disease patients (Li et al., 2012), while Streptococcus and Enterococcus phylotypes are found to be increased in fecal samples of colorectal cancer patients (Wang et al., 2012). However, in the adult gastrointestinal tract, Streptococcus spp. are particularly abundant in the upper part of the gastrointestinal tract, where they are active in the process of simple sugar fermentation into lactate (Zoetendal et al., 2012). Moreover, they may form a tropic chain with the equally abundant Veillonella spp. that convert the produced lactate into propionate (Zoetendal et al., 2012).
Fig 9.
Phylogenetic tree of the human gastrointestinal species that belong to the families of Streptococcaceae and Enterococcaceae. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. For the other gastrointestinal species that belong to the order of the Lactobacillales see Fig. 8.
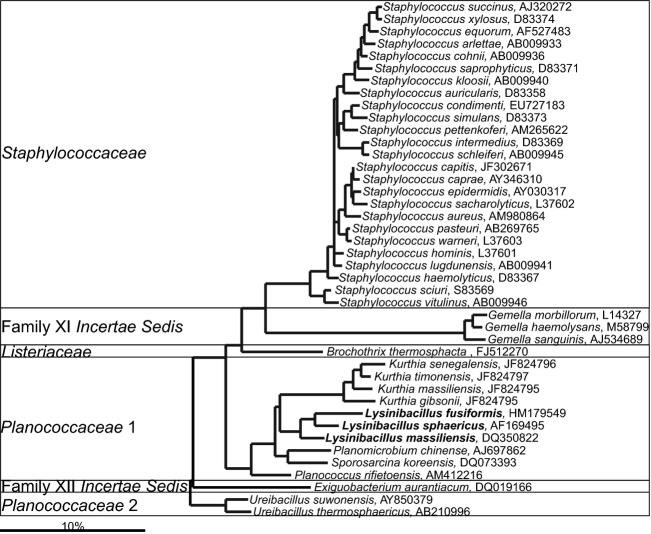
Various members of the Bacillales order can be low-level constituents of the human gastrointestinal microbiota (Figs10 and 11 — for clarity the phylogenetic tree of this numerous order was split into two parts). Among them, a large number of Staphylococcus spp., which typically are associated with the human skin, can be detected in the human gastrointestinal tract (Fig. 11). These bacteria are one of the earliest colonizers of the gastrointestinal tract, particularly in infants that were delivered by cesarean section (Dominguez-Bello et al., 2010). The predominant early colonization with Staphylococcus spp. is, however, coupled with several health risks, as it induces strong stimulation of the immune system, which can be a trigger for the development of asthma and rhinitis in later childhood (Johansson et al., 2012). Furthermore, predominant colonization of the gastrointestinal tract of premature infants with Staphylococcus spp. is associated with fatal sepsis (Madan et al., 2012). An increased abundance of bacteria belonging the Staphylococcus genus, both in the upper and lower gastrointestinal tract, is associated with celiac disease, although the abundance of these bacteria can be depleted with the withdrawal of gluten from the diet of patients (Collado et al., 2009).
Fig 10.
Phylogenetic tree of a fraction of the human gastrointestinal species that belong to the order of the Bacillales. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. The species indicated in bold are based on the SSU rRNA gene sequence clustering within the families that are designated on the figure in contrast to their official classification. For the other gastrointestinal species that belong to the order of the Bacillales see Fig. 11.
Fig 11.
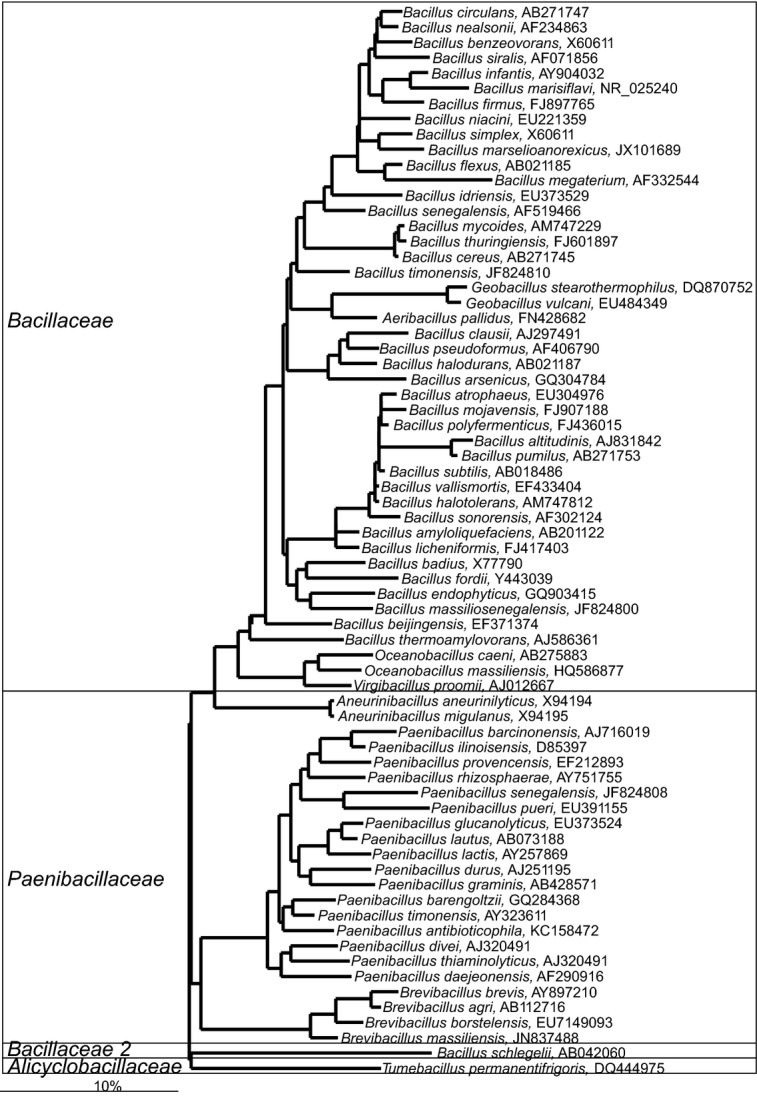
Phylogenetic tree of a fraction of the human gastrointestinal species that belong to the order of the Bacillales. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. For the other gastrointestinal species that belong to the order of the Bacillales order see Fig. 10.
Gemella spp. are abundant in the upper gastrointestinal tract and specially the proximal small intestine (Ou et al., 2009). While their function in the gastrointestinal tract has not been determined, these species can include pathogenic strain that translocate to other organs. For instance, Gemella spp. have been described as likely causing agents of endocarditis, particularly in patients that suffer from gastrointestinal disorders (such as colon cancer; Lopez-Dupla et al., 1996).
Numerous members of the Bacillus and Paenibacillus genera have been detected in the samples of the human gastrointestinal tract. The first representatives of this group of bacteria were isolated in 1919 by Marjorie Batchelor, who reported Bacillus cereus as the most prevalent member of the aerobic sporogenic bacteria in infant feces (Batchelor, 1919). Members of the Bacillus genus were often reported in the older cultivation-based studies, but the vast majority of the species of this genus and related genera were reported only recently (Hoyles et al., 2012; Lagier et al., 2012a; Zoetendal et al., 2012). One of these studies was designed for the targeted cultivation of Bacillus and related species from human samples (Hoyles et al., 2012). It has been shown that Bacillus spp. could be retrieved from all analyzed samples, although these bacteria have very low abundance of 102–104 cells mL−1 of intestinal content. Many of the Bacillus spp. isolated in this study exhibited notable antimicrobial activity. This feature is in line with the use of several Bacillus spp. as potent probiotics with immunomodulatory potential (Duc et al., 2004). Little is known about the function of these bacteria in the ecosystem, but it is noteworthy that two independent studies have shown that members of the Bacillales order, more specifically Aneurinibacillus spp., have an increased abundance in feces of irritable bowel syndrome patients (Krogius-Kurikka et al., 2009; Rajilić-Stojanović et al., 2011), while a significantly higher abundance of Bacillus subtilis was found in the feces of bottle-fed than breast-fed babies (Benno et al., 1984).
Clostridia
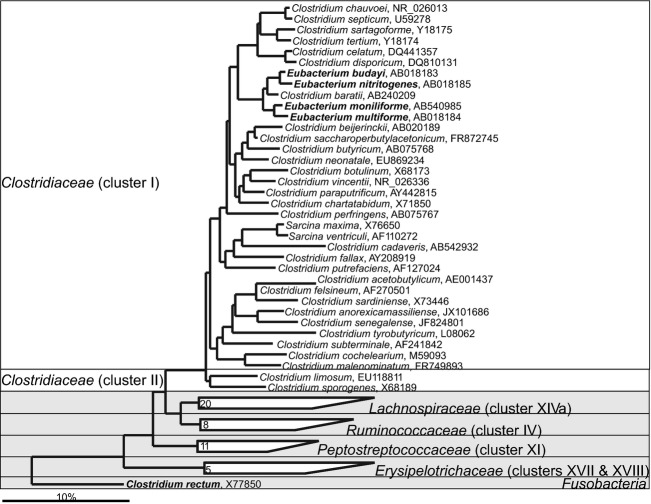
The class Clostridia clusters bacteria that are dominant and frequently detected in the lower gastrointestinal tract that are distributed within the families: Clostridiaceae, Christensenellaceae, Eubacteriaceae, Lachnospiraceae, Peptostreptococcaceae, Ruminococcacea as well as bacteria with an unclear taxonomic status that are classified within Clostridiales Incertae Sedis families XI and XIII (Garrity et al., 2005). Members of the Clostridia class are heterogeneous and many of its members were initially assigned to Clostridium genus and subsequently reclassified into novel genera. The Clostridium sensu stricto — the real Clostridium spp. — are grouped around the type species Clostridium butyricum and belong to the Clostridium cluster I within the Clostridiaceae family (Fig. 12; Stackebrandt et al., 1999). In addition to Clostridium spp., the Clostridiaceae officially groups Sarcina spp., Butyricicoccus pullicaecorum and Lactonifactor longoviformis, Anoxynatronum sibiricum while the latter three, based on their SSU rRNA gene sequence, should be assigned to various other Clostridiales families (Figs13, 14 and 16). The first human gastrointestinal Clostridium isolate, C. perfringens, was recovered in 1905 (Passini, 1905). The same species, previously known as Bacillus aerogenes capsulatus and Clostridium welchii, was earlier isolated from a case of endocarditis (Welch & Nuttall, 1892). Both isolation sites fit the nowadays known properties of C. perfringens, which is a commensal gastrointestinal bacterium that can cause bacteraemia (Petit et al., 1999). Up to now, 72 Clostridium spp. have been detected in the human gastrointestinal samples, of which 30 belong to the Clostridium sensu stricto (Fig. 12). The other Clostridium spp. belong to different families within the Firmicutes phylum, while Clostridium rectum belongs to the Fusobacteria phylum. Members of the Clostridium sensu stricto are generally perceived as pathogenic, although cultivation-based studies show that C. perfringens and other real clostridia can be found in densities of up to 1010 cells g−1 intestinal content of healthy individuals (Finegold et al., 1974), and up 107 cells g−1 intestinal content of healthy infants (Mevissen-Verhage et al., 1987). Still, the presence of these bacteria, notably as seen for C. perfringens in elderly Irish subjects, is interpreted as an indicator of a less healthy microbiota (Lakshminarayanan et al., 2013).
Fig 12.
Phylogenetic tree the human gastrointestinal species that belong to the family of Clostridiaceae. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. The species indicated in bold are based on the SSU rRNA gene sequence clustering within the families that are designated on the figure in contrast to their official classification. Deeply rooted Clostridium spp., which based on the SSU rRNA gene sequence cluster within distant phylogenetic groups, are depicted in the gray area.
Fig 13.
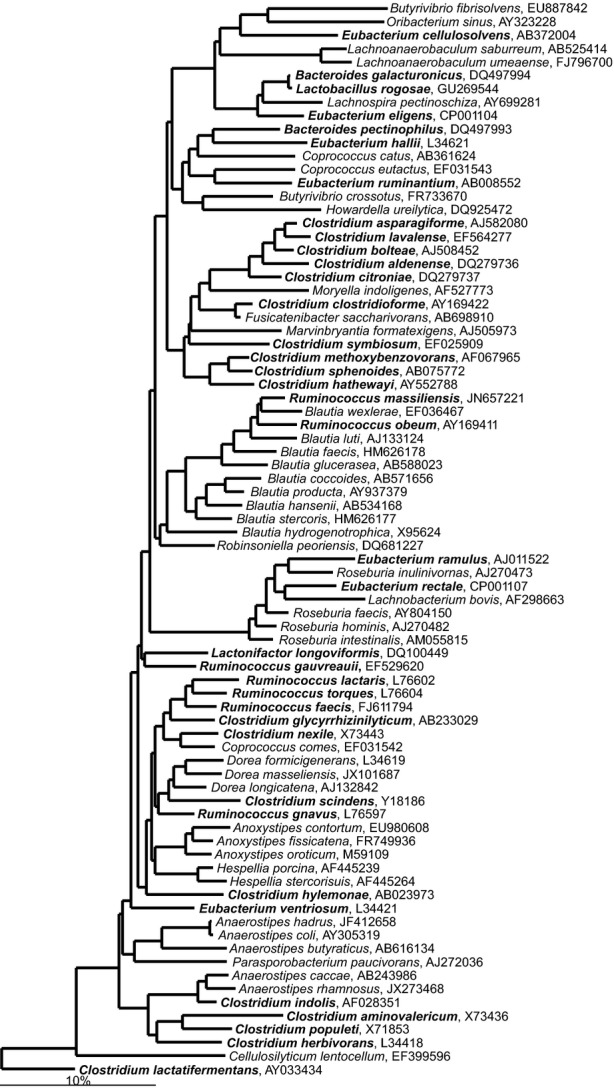
Phylogenetic tree the human gastrointestinal species that belong to the family of the Lachnospiraceae. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. The species indicated in bold are based on the SSU rRNA gene sequence clustering within the families that are designated on the figure in contrast to their official classification.
Fig 14.
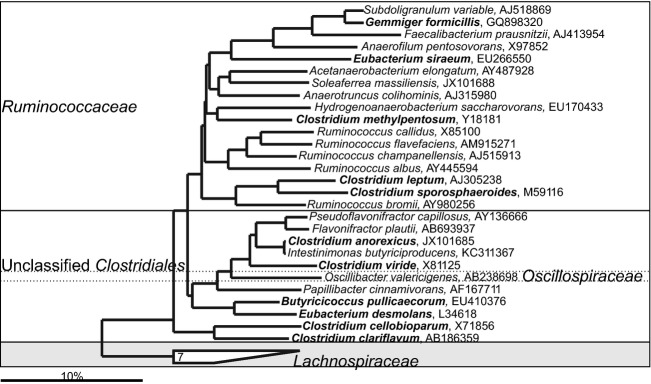
Phylogenetic tree the human gastrointestinal species that belong to the Clostridium cluster IV, most of which belong to the family of the Ruminococcaceae. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. The species indicated in bold are based on the SSU rRNA gene sequence clustering within the families that are designated on the figure in contrast to their official classification. Deeply rooted Ruminococcus spp., which based on the SSU rRNA gene sequence cluster within distant phylogenetic groups, are depicted in the gray area.
Fig 16.
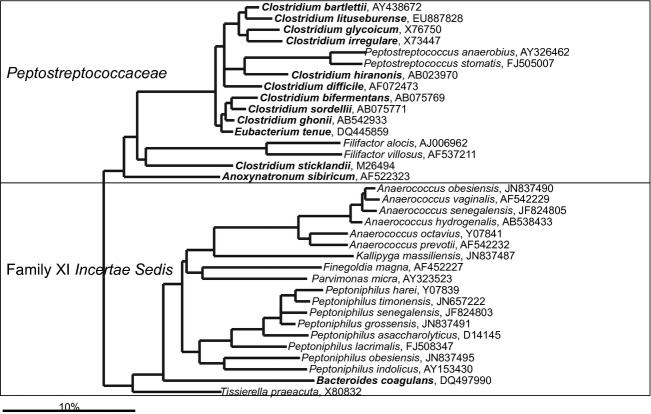
Phylogenetic tree the human gastrointestinal species that belong to families of the Peptostreptococcaceae and Clostridiales Family XI Incertae Sedis. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. The species indicated in bold are based on the SSU rRNA gene sequence clustering within the families that are designated on the figure in contrast to their official classification.
The most abundant and diverse gastrointestinal family is the Lachnospiraceae. This family groups 24 different genera, most of which can be detected in the human gastrointestinal tract. In addition, a number of species that are officially classified into the genera Clostridium, Eubacterium, and Ruminococcus, cluster within the Lachnospiraceae based on their SSU rRNA gene sequence (Fig. 3). Members of the Lachnospiraceae are also among the first to be established in the gastrointestinal tract. A recent study showed that Ruminococcus gnavus is an exclusive representative of this family in 2-months old breast-fed infants, while infants fed with cow-milk based formula have a more diverse Lachnospiraceae community (Tannock et al., 2013). Analysis of the microbiota of children and adults showed that this group of bacteria is predominant in both young children and in adults, which indicates the early establishment of these bacteria (Ringel-Kulka et al., 2013).
Several members of the Lachnospiraceae family are butyrate producers including Anaerostipes spp., Butyrivibrio spp., Coprococcus spp., Roseburia spp., Eubacterium rectale- and Eubacterium hallii-related species. Butyrate can be used as an energy source by the gut epithelial cells, and it has anticarcinogenic and anti-inflammatory properties (Hamer et al., 2008). Furthermore, a recent study shows that butyrate produced by intestinal microorganisms has beneficial effects on glucose and energy homeostasis (De Vadder et al., 2014). The decrease in the relative abundance of the butyrate-producing Lachnospiraceae in the gastrointestinal microbiota is associated with compromised health status of subjects suffering from colorectal cancer (Wang et al., 2012), ulcerative colitis (Rajilić-Stojanović et al., 2013a), type 1 (Murri et al., 2013) and type 2 diabetes (Qin et al., 2012). This bacterial group seems to be stimulated by an omnivore diet, since it is present in lower abundance in vegetarians (Kabeerdoss et al., 2012). This is an intriguing but not yet explained finding, as it could be expected that vegetarian, plant-based diets, which are rich in fibers, would favor butyrate production in the colon and promote health.
The gastrointestinal Lachnospiraceae include Dorea spp., which are the major gas producers in the gastrointestinal tract and its end-products of glucose fermentation include both hydrogen and carbon dioxide (Taras et al., 2002). Dorea spp. were found in an increased in abundance in both pediatric and adult irritable bowel syndrome patients (Rajilić-Stojanović et al., 2011; Saulnier et al., 2011), which probably could explain the symptom of bloating, experienced by the majority of these patients.
Blautia is a recently described bacterial genus that groups several abundant gastrointestinal bacteria that were previously assigned to the Ruminococcus genus – notably those related to Ruminococcus obeum (Fig. 4). The common feature of Blautia spp. is the utilization of hydrogen and carbon dioxide to form acetate (Bernalier et al., 1996). Blautia spp. are among the most abundant members of the entire gastrointestinal tract and can encompass between 2.5% and 16% of the total microbiota (Zoetendal et al., 2002). This abundant bacterial group is significantly depleted in elderly subjects (Hayashi et al., 2003; Biagi et al., 2010) and in mucosal samples of colorectal cancer patients (Chen et al., 2012). In contrast, increased levels of Blautia spp. are observed in irritable bowel syndrome patients (Rajilić-Stojanović et al., x2011), but this could reflect the adaption of the ecosystem to the larger amount of gasses produced by Dorea spp., which can be utilized by Blautia spp.
An interesting group within the Lachnospiraceae family is the misclassified Ruminococcus spp., including R. gnavus, R. torques, R. lactaris, and R. faecis. These bacteria are abundant in the gastrointestinal tract (Holdeman & Moore, 1974), and apparently associated with a number of important metabolic functions. R. torques and other currently uncultured species related to R. torques, are among the most potent mucus utilizes that enable mucus degradation by secretion of several different extracellular glycosidases (Hoskins et al., 1985). Furthermore, the abundance of these bacteria is strongly associated with the level of triglycerides in blood serum (Lahti et al., 2013). Several studies of the microbiota of irritable bowel syndrome patients and controls have shown that organisms related to these misclassified Ruminococcus spp. are significantly elevated in patients (Kassinen et al., 2007; Rajilić-Stojanović et al., 2011; Saulnier et al., 2011). Moreover, the abundance of these bacteria is positively correlated with irritable bowel syndrome symptoms (Malinen et al., 2010) and significantly reduced by probiotics consumption that reduces these symptoms (Lyra et al., 2010).
The Ruminococcaceae family is another relevant group of gastrointestinal bacteria within the Clostridiales order. It includes the true Ruminococcus spp. — members of the Ruminococcus sensu stricto namely R. albus, R. bromii, R. callidus, R. champanellensis, and R. flavefaciens. Several other frequently detected gastrointestinal genera that are recognized members of the Clostridium cluster IV (Clostridium leptum group) are either members or are closely related to this family (Fig. 14). The true Ruminococcus spp. are an abundant fraction of the human gastrointestinal microbiota that can reach densities of up to 1010 cells g−1 of intestinal content (Finegold et al., 1977). Being strictly anaerobic cellulolytic cocci, Ruminococcus spp. were isolated from human gastrointestinal samples only after the improvement of the anaerobic techniques and media for studying rumen anaerobes (Hungate, 1947). The first human gastrointestinal Ruminococcus spp. reported is Ruminococcus bromii isolated in 1972 (Moore et al., 1972). Similar to the other gastrointestinal bacteria, the initially defined Ruminococcus spp. are a heterogeneous group, which based on the SSU rRNA gene sequence clusters within the Ruminococcaceae and Lachnospiraceae families. Recently, five gastrointestinal Ruminococcus spp. were reclassified into Blautia genus, leaving seven others to be reclassified (Liu et al., 2008; Figs9 and 10). Bacteria that belong to the Ruminococcus sensu stricto degrade complex sugars to produce acetate as the major fermentation product. Both in vitro and in vivo studies have shown that R. bromii is the major degrader of the resistant starch in the human gastrointestinal tract (Kovatcheva-Datchary et al., 2009; Walker et al., 2011). Application of the resistant starch in the diet has a wide range of health-promoting effects, suggesting the importance of the metabolic activity of R. bromii for the wellbeing of the host (Higgins & Brown, 2013). The importance of the members of the Ruminococcus sensu stricto for the intestinal health is indicated by their reduced abundance in feces of Crohn's disease (Kang et al., 2010) and ulcerative colitis patients (Rajilić-Stojanović et al., 2013a).
Among the Ruminococcaceae family, Faecalibacterium prausnitzii (previously known as Fusobacterium prausnitzii) is the most prevalent and abundant gastrointestinal microorganism (Holdeman et al., 1976). Faecalibacterium prausnitzii can utilize glucose, fructose, and fructo-oligosacharides, as well as complex molecules such as pectin and N-acetylglucosamine to produce butyrate, formate and lactate (Duncan et al., 2002a; Lopez-Siles et al., 2012). It is one of the major butyrate producers in the gastrointestinal tract, which is a relevant feature because of the health-promoting properties of butyrate. The reduced abundance of this bacterium is detected in association with Crohn's disease (Sokol et al., 2006; Kang et al., 2010) and with colon cancer (Chen et al., 2012). This bacterium is important for the gastrointestinal microbiota homeostasis as it has found to show anti-inflammatory properties in mice (Sokol et al., 2008) and is associated with a range of metabolic processes in the human mucosa (Lepage et al., 2011). Health-promoting properties are also exhibited by B. pullicaecorum, another species with the SSU rRNA gene sequence that is related to the Ruminococcaceae family. This bacterium is significantly reduced in inflammatory bowel disease patients, while its oral administration strengthens the epithelial barrier function in animal models by increasing the trans-epithelial resistance (Eeckhaut et al., 2013).
The first Eubacterium spp. from a human gastrointestinal sample was isolated already in 1908 when Henri Tissier plated Bacillus ventriousus, later renamed into Eubacterium ventriosum (Tissier, 1908). The genus Eubacterium was for a long time recognized as one of the most abundant genera of the human gastrointestinal microbiota, with densities of up to 1010 cells g−1 of intestinal content (Moore & Holdeman, 1974a). However, Eubacterium, similar to Clostridium, is a genus that is very vaguely described. Defined as anaerobic, rod-shaped, Gram-positive bacteria that do not form endospores, Eubacterium genus includes a consortium of distantly related species. Some Eubacterium spp. have been reclassified into novel genera within two bacterial phyla — Actinobacteria and Firmicutes — of which six genera (Dorea, Collinsella, Eggerthella, Flavonifractor, Mogibacterium and Pseuodramibacter) can be members of the gastrointestinal microbiota (Willems & Collins, 1996; Kageyama et al., 1999a, b; Nakazawa et al., 2000; Taras et al., 2002; Carlier et al., 2010). Further reclassification of the genus can be expected, as only four gastrointestinal Eubacterium spp. belong to the Eubacterium sensu stricto (Fig. 5). In a recent study of the gastrointestinal microbiota of centenarians, Eubacterium spp. (notably those related to Eubacterium limosum) were reported as signature bacteria of the long life, being 10-fold increased in centenarians (Biagi et al., 2010). It is known that E. limosum has the ability to transform dietary phytoestrogens into forms that might have a positive impact on health (Clavel et al., 2006; Possemiers & Verstraete, 2009). Furthermore, E. limosum is selectively stimulated by prebiotics that improve the symptoms of inflammatory bowel disease patients (Kanauchi et al., 2005).
Fig 15.
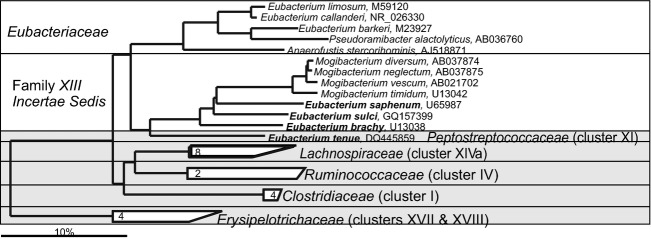
Phylogenetic tree the human gastrointestinal species that belong to the family of the Eubacteriaceae and Clostridiales Family XIII Incertae Sedis. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. The species indicated in bold are based on the SSU rRNA gene sequence clustering within the families that are designated on the figure in contrast to their official classification. Deeply rooted Eubacterium spp., which based on the SSU rRNA gene sequence cluster within distant phylogenetic groups are depicted in the gray area.
Mogibacterium is a genus established by reclassification of the intestinal bacterium – Eubacterium timidum. Mogibacterium spp. are enriched in mucosa-associated microbiota in colon cancer patients but not much is known about this group of bacteria belonging to Clostridium Family XIII Incertae Sedis (Chen et al., 2012).
Prior to the molecular revolution of the gastrointestinal microbiota research (Fig. 1), Peptococcus spp. and Peptostreptococcus spp. were considered as the dominant and abundant in the human gastrointestinal tract (Holdeman et al., 1976). However, the latter research has shown that these two genera are heterogeneous and led to the major reclassification resulting in definition of novel genera that include Anaerococcus, Blautia, Finegoldia, Parvimonas, and Peptoniphilus (Murdoch & Shah, 1999; Ezaki et al., 2001; Tindall & Euzeby, 2006; Liu et al., 2008). All these genera, with exception of Blautia that is a genus in the Lachnospiraceae family, belong to the Clostridiales Family XI Incertae Sedis (Fig. 16). Among this group of bacteria, Peptoniphilus asaccharolyticus is the most frequently detected and also was the first to be cultured and described (Distaso, 1911). Currently, only two true Peptostreptococcus spp. have been recognized as gastrointestinal inhabitants, although a number of other species that are officially classified within different genera (predominantly Clostridium) belong to the Peptostreptococcaceae family according to their SSU rRNA gene sequence (Fig. 6). Members of the Peptostreptococcaceae family are, in principle, associated with compromised health, and the most convincing example of this is Clostridium difficile. Although C. difficile can be present in low numbers in healthy subjects without exhibiting pathogenic properties (Ozaki et al., 2004), many strains are toxin producing and are well-established pathogens that cause severe diarrhea. Furthermore, recent studies have shown that Peptostreptococcus spp. have an increased abundance in association with ulcerative colitis (Rajilić-Stojanović et al., 2013a) and colorectal cancer (Chen et al., 2012; Wang et al., 2012). Based on the SSU rRNA gene sequence, the members of the Peptococcaceae family form two paraphyletic groups within the Firmicutes phylum, of which the group that contains the two human gastrointestinal bacteria is closely related to Negativicutes class and is discussed in the following section.
In addition to the already mentioned species, five other gastrointestinal bacteria that officially belong to the Clostridia class, form distinct branches in the phylogenetic tree (Fig. 17). Two of these gastrointestinal bacteria, namely Catabacter hongkongensis and C. minuta, form a separate cluster within the Clostridiales order of the Clostridia class and are the only cultured representatives of a phylogenetic group that was previously detected only in various molecular studies and was in a previous review designated as uncultured Clostridiales II (Rajilić-Stojanović et al., 2007). These two species are officially assigned to two different families (Catabacteriaceae and Christensenellaceae), although based on the SSU rRNA gene sequence similarity (96.5%), they should be grouped in the same family, and, most likely, in the same genus. Catabacter hongkongensis was isolated in 2007 from a blood sample, although the intestinal origin of the bacterium was suspected (Lau et al., 2007). This bacterium was later isolated from patients with acute appendicitis, but also from other tissues where it was a causative agent of fatal bacteremia (Lau et al., 2012). Christensenella minuta is an intestinal isolate, described in 2012 (Morotomi et al., 2012). Not much is known about the role of this group of strictly anaerobic bacteria in the human gastrointestinal tract, but it is noteworthy that bacteria that belong to the Christensenella/Catabacter group were reported to be dramatically (20-fold) depleted in fecal samples of ulcerative patients relative to controls (Rajilić-Stojanović et al., 2013a) and significantly (fivefold) depleted in fecal samples of patients with postinfectious irritable bowel syndrome (Jalanka-Tuovinen et al., 2013).
Fig 17.
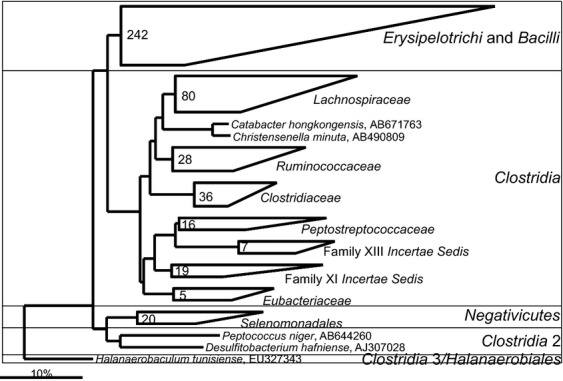
Partially opened phylogenetic tree the human gastrointestinal species that belong to the Firmicutes phylum on which five species that cluster within the three families with low diversity are indicated.
Two members of the Peptococcaceae family were reported as members of the human gastrointestinal microbiota (Fig. 17). These bacteria are officially classified within the Clostridia class and Clostridiales order, although based on the SSU rRNA gene sequence they are closely related to the members of the Selenomonadales order within the Negativicutes class (Fig. 18). Based on their SSU rRNA gene sequence, gastrointestinal members of the Peptococcaceae family will be reclassified either the Negativicutes class or into another novel class, different from Clostridia sensu stricto. Peptococcus niger is the only representative of the genus. This strictly anaerobic bacterium has been isolated from various body sites, while the strain isolated from feces showed an ability to desulfate and perform other chemical transformation of steroid molecules (Van Eldere et al., 1987). This feature of P. niger makes it an important player in enterophatic circulation of various steroid molecules, primarily steroid hormones, which has a major impact on human metabolism. Another member of the Peptococcaceae family, Desulfitobacterium hafniense, has been reported only once as an inhabitant of the human gastrointestinal tract (van de Pas et al., 2001). In contrast to other members of the genus, the human D. hafniense is not able to use chloroethenes or chlorophenol as terminal electron acceptors (Smidt & de Vos, 2004). This bacterium is capable of using sulfite as terminal electron acceptor and hence produces hydrogen sulfide. Hydrogen sulfide is also produced by P. niger (Wilkins et al., 1975), and this property that may be detrimental to health as described below for the Deltaproteobacteria.
Fig 18.
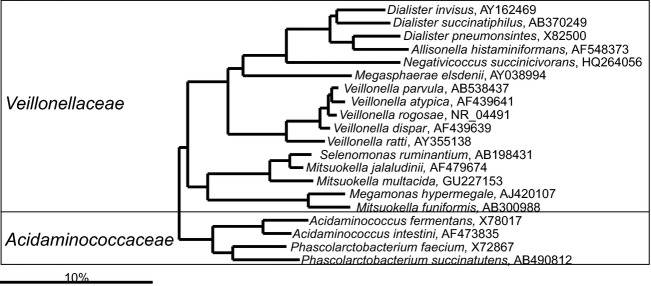
Phylogenetic tree the human gastrointestinal species that belong to the order of the Negativicutes. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated.
Finally, Halanaerobaculum tunisiense has been identified by pyrosequencing of the V6 variable region of the SSU rRNA gene in a recent study that compared efficiency of culturomics and pyrosequencing approach for studying the gastrointestinal microbiota diversity (Lagier et al., 2012a). Halanaerobaculum tunisiense is a recently described anaerobic bacterium that was isolated from hypersaline lake in Tunisia (Hedi et al., 2009). Given the conditions that the isolated strain of this bacterium requires for its growth (the minimal required NaCl concentration of 14%), it is not likely that it represents a member of the gastrointestinal microbiota, although this should be confirmed in further studies.
Negativicutes
The Negativicutes include bacteria that were previously assigned to the Clostridium cluster IX (Collins et al., 1994; Marchandin et al., 2010), distributed within the following genera: Acidamoinococcus, Dialister, Megamonas, Megasphaera, Phascolarctobacterium and Veillonella (Fig. 8). Bacteria of this group used to be classified within the order Clostridiales, although based on their SSU rRNA gene sequence, they are distant from other Clostridiales. Therefore, following the description of a novel gastrointestinal inhabitant — Negativicoccus succinicivorans, the novel class of Negativicutes and novel order of Selenomonadales were introduced to accommodate the Gram-negative staining bacteria within the Firmicutes phylum (Marchandin et al., 2010). The first record of this bacterial group dates from 1898 when Veillon and Zuber isolated Staphylococcus parvulus, which was later reclassified as Veillonella parvula, from infected appendix tissue (Veillon & Zuber, 1898). The Negativicutes are typically isolated from the oral cavity or the proximal small bowel (Rogosa, 1965; Simon & Gorbach, 1986), but representative species from this group can be detected in high abundances even in the lower intestinal tract. In that line V. parvula, can reach densities of up to 1011 cells g−1 of feces (Finegold et al., 1977), while molecular quantification of Phascolarctobacterium spp. showed that these bacteria represent more than 2% of the total fecal microbiota in some subjects (Paliy et al., 2009). Members of the Negativicutes are assacharolytic and utilize end=products of sugar metabolisms of other gastrointestinal bacteria (such as lactate or succinate) to produce propionate, forming an important trophic chain. Propionate is a beneficial product of the gastrointestinal microbiota as it has anti-inflammatory potential, is utilized by adipose tissue and the liver, plays a role in the satiety sensation, influences glucose and energy homeostasis, and improves insulin sensitivity (Vipperla & O'Keefe, 2012; De Vadder et al., 2014). In the upper gastrointestinal tract, Veillonella spp. are an indispensable component of the gastrointestinal microbiota (van den Bogert et al., 2011) where they form a trophic chain with the lactate and acetate-producing Streptococcus spp. (Zoetendal et al., 2012). Currently, there is no evidence about the role of Veillonella spp. in human health, although several studies have shown an increased abundance of Veillonella spp. in fecal samples of irritable bowel patients (Malinen et al., 2005; Tana et al., 2010; Saulnier et al., 2011), which could indicate an increased transit of the ileal microbiota to the lower part of the gastrointestinal tract.
Erysipelotrichi
The Erysipelotrichi constitute a class of bacteria within the Firmicutes phylum that was introduced into bacterial systematics in 2009, to accommodate members of earlier established family Erysipelotrichaceae (Ludwig et al., 2009). The majority of the human gastrointestinal bacteria that based on their SSU rRNA gene sequence cluster within the Erysipelotrichi class are still officially classified within other groups of the Firmicutes (Fig. 9). This indicates that a major revision of this group can be expected in the future. There are several studies that link Erysipelotrichi with compromised health. An increased abundance of Erysipelotrichi in patients suffering from colon cancer was reported (Chen et al., 2012). Animal model experiments have shown that members of this group are increased on high fat, and western type diets (Turnbaugh et al., 2009; Fleissner et al., 2010), while their increased abundance is associated with obesity (Turnbaugh et al., 2006). Furthermore, it has been shown that an increased abundance of Erysipelotrichaceae correlates with choline deficiency-induced fatty liver disease (Spencer et al., 2011), which causes multiple organ dysfunctions. Choline is an important component of our diet, and recently, it was found that choline and phospatidylcholine are converted by the intestinal microbiota to trimethylamine, which is further metabolized to proatherogenic trimethylamine-N-oxide, linking diet and microbiota to cardiovascular disease (Wang et al., 2011b; Koeth et al., 2013).
Tenericutes
Tenericutes is a recently introduced phylum that accommodates the Mollicutes class, which was previously positioned within the Firmicutes phylum. The assignment of the Mollicutes to the novel phylum was supported by the unique properties of these bacteria, in particular the lack of rigid cell walls (Ludwig et al., 2009), although based on the SSU rRNA gene sequence, they are interrelated with the members of the Erysipelotrichi class of the Firmicutes (Fig. 19). The majority of the gastrointestinal Tenericutes are currently uncultured species that are frequently detected in the molecular surveys. Their detection even in the studies of the limited depth suggests a relatively high relative abundance of these bacteria (see for instance, Suau et al., 1999). Concerning their function in the gastrointestinal tract, a recent study has shown a significant positive correlation between the abundance of the Tenericutes and the levels of trimethylamine-N-oxide, which is a metabolite that is believed to accelerate atherosclerosis (Koeth et al., 2013).
Fig 19.
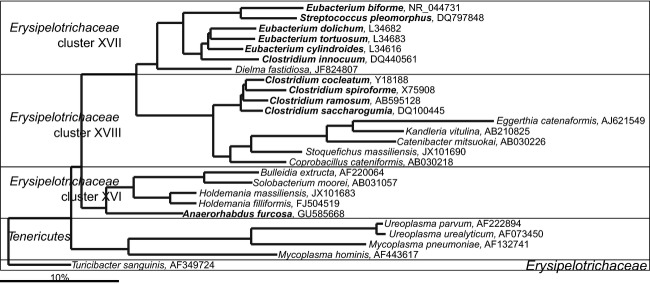
Phylogenetic tree the human gastrointestinal species that belong to the order of the Erysipelotrichi. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species, while the family is divided over Clostridium clusters (Collins et al., 1994). The species indicated in bold are based on the SSU rRNA gene sequence clustering within the Erysipelotrichi in contrast to their official classification.
It has been suggested that members of the Tenericutes are involved in inflammatory bowel disease, as they have the ability to adhere to and to fuse with epithelial and immune system cells, which could explain intracellular epithelial structures in the Crohn's disease patients' biopsies detected by electron microscopy (Roediger & Macfarlane, 2002). A molecular survey has revealed that a cultured representative of this group — the pathogenic Mycoplasma pneumoniae — can be detected in mucosal biopsies of both inflammatory bowel disease patients and healthy controls alike, although Crohn's disease patients had a significantly higher abundance of this bacterium than ulcerative colitis patients and healthy subjects (Chen et al., 2001). The first report of the Mycoplasma in the human gastrointestinal tract dates from 1973 (Bhat et al., 1973), while specific searches for Ureaplasma urealyticum and Mycoplasma hominis revealed the presence of these two species in anal swaps of more than half of the analyzed subjects (Munday et al., 1981).
Fusobacteria
Fusobacteria are another phylum of the frequently detected gastrointestinal bacteria, the majority of which belong to the genus Fusobacterium (Fig. 20). These bacteria are pointed, nonsporulating, Gram-negative, anaerobic bacilli (Knorr, 1922). The first record of a Fusobacterium spp. originates from 1886 when Bacillus fusiforme (now known as Fusobacterium necrophorum) was reported as a pathogen related to appendicitis (Flügge, 1886). Although Fusobacterium spp. can be isolated from gastrointestinal samples of healthy humans in densities of up to 1010 cells g−1 of feces (Benno et al., 1989), this group of bacteria seems to be relevant for intestinal inflammation. Recent studies have shown that the majority of cases of acute appendicitis are associated with a local infection of Fusobacterium spp. (Swidsinski et al., 2011), while the increased abundance of Fusobacterium spp. is associated with ulcerative colitis (Rajilić-Stojanović et al., 2013a) and colorectal cancer (Castellarin et al., 2012; Kostic et al., 2012). The human gastrointestinal tract-associated Fusobacteria also include representatives of the Leptotrichia genus and the misclassified C. rectum (Fig. 20).
Fig 20.
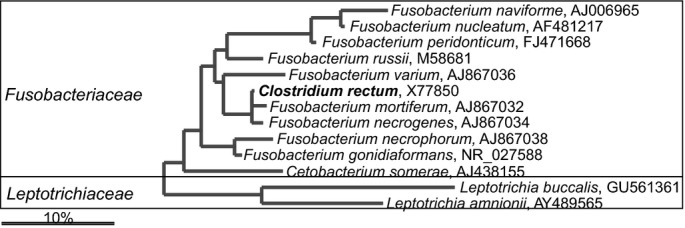
Phylogenetic tree the human gastrointestinal species that belong to the phylum of the Fusobacteria. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. The species indicated in bold are based on the SSU rRNA gene sequence clustering within the families that are designated on the figure in contrast to their official classification.
Proteobacteria
Bacteria belonging to the phylum Proteobacteria are commonly detected in the gastrointestinal samples and this group of the true Gram-negative bacteria is particularly diverse, although not very abundant – typically all Proteobacteria account about 1% of the total microbiota (Holdeman et al., 1976). Members of five different classes of Proteobacteria, namely the Alpha-, Beta-, Gamma-, Delta- and Epsilonproteobacteria can be part of the gastrointestinal microbiota, and among them Enterobacteriaceae within Gammaproteobacteria are the most abundant and the prevalent group.
Alphaproteobacteria
Gemmiger formicilis was the first and the only bacterium from the Alphaproteobacteria class that was associated with the human gastrointestinal tract, throughout the 20th century (Holdeman et al., 1976; Benno et al., 1986; Moore & Moore, 1995; Macfarlane et al., 2004). Ironically, the recent SSU rRNA gene sequence analysis of G. formicilis indicated that this species has been misclassified and belongs to Firmicutes (Clostridia/Clostridiales/Ruminococcaceae) and has almost an identical SSU rRNA gene sequence as Subdoligranulum variabile (98.3% sequence similarity). Nevertheless, other representatives of the true Alphaproteobacteria seem to be a part of the normal gastrointestinal microbiota. In a recent study, four alphaproteobacterial species were cultivated from gastrointestinal samples (Lagier et al., 2012a). Twenty-four other members of this subdivision were detected by retrieving SSU rRNA gene sequences identical to previously cultured bacteria from gastrointestinal samples (Fig. 21, Table S1). AlphapProteobacteria seem to be characteristic for the upper part of the gastrointestinal tract as sequences of these bacteria were detected only in studies where samples from the upper intestine were included (Eckburg et al., 2005; Wang et al., 2005), or when the microbiota of patients with ileal pouch was analyzed (McLaughlin et al., 2010). There are no data that correlate Alphaproteobacteria with any specific function in the gastrointestinal tract or any disease. However, some genera are known to perform specific metabolic transformations — for example, Methylobacterium spp. include bacteria that can oxidize methylamine or methanol to generate energy. Although the metabolism of these compounds has not been studied in the gastrointestinal tract, and the methylotrophic community is not typically associated with the human body, a recent study has demonstrated that methanotrophs are ubiquitous in the human oral microbiota (Hung et al., 2011). Similarly, it can be anticipated that they are present in the upper gastrointestinal tract, where SSU rRNA gene sequences of these bacteria were detected, and where oxygen needed for their metabolic activity is present. Sphingomonas spp. include metabolically versatile aerobic bacteria that can be found in different environments. Studies of these and most other Alphaproteobacteria focus on outbreaks of infections involving these bacteria in immuno-suppressed patients. However, animal models studies have shown that Sphingomonas spp. are important for the development of the immune system of the host, since these species can stimulate the maturation of invariant natural killer T cells (Wingender et al., 2012).
Fig 21.
Phylogenetic tree the human gastrointestinal species that belong to the class of the Alphaproteobacteria class. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated.
Betaproteobacteria
The first bacterium from the Betaproteobacteria class — Alcaligenes faecalis — was isolated from a human fecal sample in 1896 (Petruschky, 1896). This asaccharolytic rod that can utilize urea, a range of amino-acids, and can produce nitric oxide (NO; Denault et al., 1953) has frequently been detected in later studies of the gastrointestinal microbiota. A number of other bacteria within this class have been detected in recent years, showing that Betaproteobacteria are diverse and ubiquitous members of the gastrointestinal microbiota (Fig. 22). Among them, Sutterella and Parrasutterella are the most frequently encountered and the abundant gastrointestinal microorganisms (Nagai et al., 2009; Mukhopadhya et al., 2011). These bacteria are typical for the gastrointestinal microbiota ecosystem, assacharolytic and inactive in classical microbiological tests. Their abundance is increased in autistic children (Williams et al., 2012) and in patients suffering from type 2 diabetes (Larsen et al., 2010). Hence, it is of interest to define which metabolic transformations are catalyzed by these bacteria, as they might be particularly relevant for understanding the global impact of the gastrointestinal microbiota on human health. Alcaligenes faecalis and related bacteria are common gastrointestinal tract inhabitants, notably of the ileum. Alcaligenes spp. may be opportunists that inhabit Peyers' patches and signal to the immune system as shown in model animals (Obata et al., 2010). Moreover, Alcaligenes spp. produce NO, which is an important biological regulator (Culotta & Koshland, 1992; Anderson et al., 1993).
Fig 22.
Phylogenetic tree the human gastrointestinal species that belong to the class of the Betaproteobacteria. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated.
Oxalobacter formigenes is another relevant intestinal inhabitant. It has been isolated in 1985 and described as a unique intestinal bacterium that degrades exclusively oxalate and can reach densities of up to 107 cells g−1 of feces (Allison et al., 1985). Due to its metabolic activity, O. formigenes regulates oxalate concentrations in feces and urine, and indirectly influences the formation of kidney stones (Duncan et al., 2002b). This has led to its application in probiotic formulations. Recently, it has been shown that some strains of another intestinal inhabitant Ancylobacter polymorphus (belonging to the Alphaproteobacteria) can utilize oxalate (Lang et al., 2008), suggesting that there are alternative pathways for oxalate removal from the gastrointestinal tract.
Neisseria spp. are inhabiting mucosal surfaces of the genital, the respiratory and the upper gastrointestinal tract. Neisseria gonorhhoea and Neisseria meningitis are the most studied as they are important pathogens causing gonorrhoea and meningitis. However, the majority of Neisseria spp. are nonpathogenic and their presence in the gastrointestinal tract samples was detected in the early cultivation-based studies (Gray & Shiner, 1967; Bhat et al., 1980). The presence of specific Neisseria spp. in the gastrointestinal tract was only recently reported in a high-throughput culturing study (Lagier et al., 2012a) and emerged from SSU rRNA gene sequencing-based studies (Fig. 22, Table S1). Nonpathogenic Neisseria spp. do not catabolize many carbohydrates, while some species are even asaccharolytic, but they can reduce nitrate. It is known that Neisseria spp. are able to grow on amino acids and can use sulfur directly from sulfate (McDonald & Johnson, 1975), but their function in the gastrointestinal tract has not been exploited.
The human gastrointestinal Betaproteobacteria also include a number of other species within various genera, which are occasionally reported as members of the gastrointestinal microbiota (Fig. 22). Among them, Acidovorax spp. were found to be ubiquitously present in the colonic mucosa (Hong et al., 2011), Burkholderia spp. were found in an increased abundance in hepatic encephalopathy and were linked to poor cognition and inflammation (Bajaj et al., 2012), while Variovorax spp. seem to be particularly abundant in the upper gastrointestinal tract (van den Bogert et al., 2011). There is no available information about association of other species with specific gastrointestinal sites, diseases or functions of the gastrointestinal microbiota.
Gammaproteobacteria
Within the class Gammaproteobacteria, nine different families distributed within six different orders have been detected in the human gastrointestinal samples. Escherichia coli was the first bacterial isolate of the Gammaproteobacteria characterized from human gastrointestinal samples in 1885 (Shulman et al., 2007). Escherichia coli belongs to the Enterobacteriaceae family, which is the most diverse, prevalent and abundant of all gastrointestinal Proteobacteria (Fig. 23). Most of Enterobacteriaceae members are associated with diarrhea (Thielman & Guerrant, 2004), although representatives of this family are not necessarily causing any symptoms and are actually one of the first to be found in the newborn gastrointestinal tract (Favier et al., 2002). The abundance of this bacterial group increases with age, but it remains subdominant and in elderly subjects represents about 1% of the total gastrointestinal microbiota (Hopkins et al., 2001). Escherichia coli is the most prevalent representative of this family that is often the most abundant facultative anaerobe in the gastrointestinal samples. The different strains of E. coli can exhibit different properties, varying from probiotic (Kruis et al., 2004) to pathogenic causing diarrhea or infections on other sites (Ron, 2006). The majority of other Enterobacteriaceae spp. are infrequently isolated from gastrointestinal samples (Bucher & von Graevenitz, 1982; Müller, 1986). In that line, a study of dedicated isolation of the Morganella-Proteus-Providencia group from feces of almost 3000 healthy subjects and patients suffering from enteric diseases, showed that species of this group are subabundant and have joined prevalence between 10% and 20%, depending on the health status (Müller, 1986). Still, the Enterobacteriaceae is one of the most comprehensively described gastrointestinal families, which can be explained by its development as a paradigm for genetic studies and its clinical relevance (Grimont et al., 1981; Hickman-Brenner et al., 1984; Farmer et al., 1985; Hickman-Brenner et al., 1985a, b). A specific case is represented by the so-called adherent-invasive E. coli strains that have been implied in various forms of inflammation in the gastrointestinal tract of human and animal models (Negroni et al., 2012; Chassaing et al., 2014). Phylogenetically, the Enterobacteriaceae is a diverse group and while some genera form separate clusters (e.g., Yersinia), species of other genera are mixed up in the SSU rRNA phylogenetic tree (Fig. 23). The absence of genus-specific SSU rRNA gene sequences for these genera, could explain a recent the finding that sequences assigned to E. coli, Salmonella enterica, Citrobacter koseri, and Enterobacter cancerogenus appear together (Lozupone et al., 2012).
Fig 23.
Phylogenetic tree the human gastrointestinal species that belong to the family of the Enterobacteriaceae. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated.
In addition to the Enterobacteriaceae, representatives of eight other bacterial families with the Gammaproteobacteria class can be detected in the human gastrointestinal tract (Fig. 24). Among these, members of the Moraxellaceae are relatively frequently detected using both cultivation-based and molecular studies. Within this family, Acinetobacter spp. are the most diverse and are frequently detected in infants (Chang et al., 2011; Pandey et al., 2012), with an increased abundance in infants that develop allergy (Nakayama et al., 2011). A recent study has indicated that members of the Gammaproteobacteria and in particular Haemophilus spp. are elevated in irritable bowel syndrome pediatric patients (Saulnier et al., 2011). Members of the same phylogenetic group were found to correlate with irritable bowel syndrome symptom score in an independent study (Rajilić-Stojanović et al., 2011). Haemophilus spp. are relatively frequently detected in the upper parts of the gastrointestinal tract of healthy humans of different ages (Justesen et al., 1984; Ou et al., 2009), but also can be detected inflamed and stool specimens from children with diarrhea, with relatively low prevalence (Mégraud et al., 1988).
Fig 24.
Phylogenetic tree the human gastrointestinal species that belong to the class of the Gammaproteobacteria without Enterobacteriaceae family. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated.
Aeromonas spp. are medically significant as these species are implicated in the development of the gastroenteritis and diarrhea. The role of these bacteria in human health has been a subject of a long-lasting debate resulting in a conclusion that at least four gastrointestinal species (A. caviae, A. hydrophila, A. jandaei, and A. veronii), are pathogenic (Janda & Abbott, 1998). The presence of these species in the gastrointestinal tract is not necessarily inducing any symptoms, although in vitro experiments have indicated that they are cytotoxic and induce lesions in the intestinal mucosa (Pitarangsi et al., 1982).
The Succinivibrionaceae family groups strictly anaerobic bacteria that ferment carbohydrates to produce succinate and acetate. Representatives of three genera of this family can be detected in the human gastrointestinal tract: Anaerobiospirillum spp. are motile, spiral bacteria that are implicated in the development of diarrhea (Malnick, 1997); while Succinatimonas and Succinivibrio representatives are subdominant bacteria that were isolated from healthy humans (Table S1).
Members of the Vibrionaceae are pathogens that cause acute, self-limiting gastroenteritis (Hou et al., 2011). The natural habitat of Vibrio spp. is the aquatic ecosystem, and hence they are only rarely detected in the human gastrointestinal samples — notably after infection that induces diarrhea.
Eight different Pseudomonas spp. can be detected in the human gastrointestinal tract. Among them, Pseudomonas aeruginosa is the most prevalent and was frequently reported as a member of the fecal microbiota of healthy humans (Finegold et al., 1974; Benno et al., 1986). Although a member of the normal gastrointestinal microbiota, Pseudomonas aeruginosa can act as an opportunistic pathogen in critically ill or immuno-suppressed patients and cause sepsis as it can interact with our immune system (Wu et al., 2005) and can disrupt the intestinal epithelial barrier (Zaborina et al., 2006). Pseudomonas fluorescens is a less prevalent member of the genus that has been implicated in the development of Crohn's disease (Wei et al., 2002). A recent study showed that Pseudomonas spp., among other Proteobacteria, have increased abundance in infants with colic (de Weerth et al., 2013).
Until today, only two bacteria from Xanthomonadaceae have been isolated from gastrointestinal samples: Stenotrophomonas maltophilia from a stool and an ileal sample derived from atypical clinical cases (Tamura et al., 1988; Apisarnthanarak et al., 2003), and Lysobacter soli in a recent high-throughput cultivation of the normal gastrointestinal microbiota (Lagier et al., 2012a). In addition, four other related bacterial species were detected based on the SSU rRNA gene sequence. These bacteria were previously isolated from different ecosystems and include Nevskia ramose and Rhodanobacter ginsenosidimutans from soil, Pseudoxanthomonas mexicana from sludge and urine, and Silanimonas lenta from a hot spring. Based on their low prevalence in the gastrointestinal tract, it is most likely that Xanthomonadaceae are transient members of the gastrointestinal microbiota.
Deltaproteobacteria
Sulfate-reducing bacteria that cluster within the δ class of the phylum Proteobacteria inhabit the human gastrointestinal tract where they utilize sulfate that can be diet derived or released from mucins. Human gastrointestinal tract-associated sulfate-reducing bacteria include the acetate-utilizing Desulfobacter spp., the lactate-, and H2-utilizing Desulfovibrio spp., and the propionate-utilizing Desulfobulbus spp. (Gibson et al., 1988). This group of related bacteria has been subject of numerous studies because the end-product of their metabolism — hydrogen sulfide — is a highly toxic compound that inhibits butyrate oxidation within the colonocytes (Attene-Ramos et al., 2006). Hydrogen sulfide overproduction in the gastrointestinal tract has been linked to ulcerative colitis and colon cancer. Although some cultivation, studies showed an association between the presence of sulfate-reducing bacteria and ulcerative colitis, the overproduction of hydrogen sulfide has a stronger correlation with dietary protein (Magee et al., 2000) than with dietary sulfate (Deplancke et al., 2003), challenging the hypothesis that the metabolic activity of sulfate-reducing bacteria is involved in compromised health, at least in healthy subjects. Sulfate-reducing bacteria are normally present in low abundance, of approximately 106–107 cells g−1 (Fite et al., 2004), and although at least three different genera of this group of bacteria can be found in the human gastrointestinal tract, only Desulfovibrio spp. are characterized below the genus level (Fig. 25). In addition, Bilophila wadsworthia is another member of Deltaproteobacteria that is present in approximately half of the studied humans (Baron et al., 1992; Baron, 1997). Bilophila wadsworthia is capable of utilizing taurine, which is released by deconjugation of bile salts or present in the diet, and also generates hydrogen sulfide as the major end-product. Its involvement in promoting colitis via taurine metabolism in mice has recently been established (Devkota et al., 2012).
Fig 25.
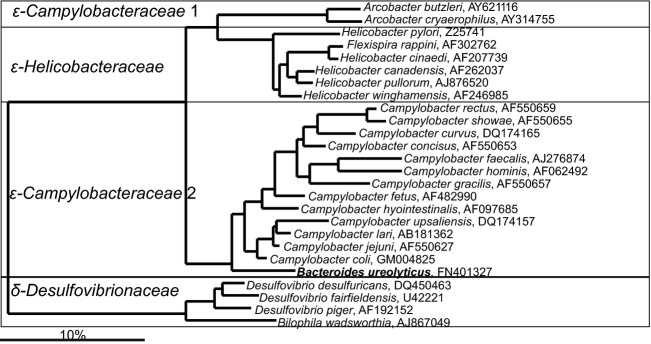
Phylogenetic tree the human gastrointestinal species that belong to the classes of the Deltaproteobacteria and Epsilonproteobacteria. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the family names are indicated. The species indicated in bold is based on the SSU rRNA gene sequence clustering within the indicated family in contrast to its official classification.
Epsilonproteobacteria
The class of Epsilonproteobacteria is represented by two main genera in the human gastrointestinal tract: Campylobacter and Helicobacter. Campylobacter is a genus that groups diverse isolates from mucosal surfaces of gastrointestinal, oral and urogenital tract. The first report of Campylobacter isolation from the human gastrointestinal tract dates from 1946 (Levy, 1946). Campylobacter spp. were originally described as members of genus Vibrio, and reclassified into an independent genus in 1973 (Veron & Chatelain, 1973). Species of this genus are principally considered to be pathogenic organisms involved in diarrheal illness. However, Campylobacter hominis has been isolated from a gastrointestinal sample of a healthy subject (Lawson et al., 2001), while C. concisus is a clinical isolate that also has been recovered from healthy individual (Engberg et al., 2000). Finally, a recent study of the microaerophilic fecal microbiota of children revealed diverse and prevalent colonization of Campylobacter spp. of gastrointestinal tract of both healthy and children suffering from inflammatory bowel diseases (Hansen et al., 2013). The presence of Campylobacter spp. in the gastrointestinal tract can cause watery or bloody diarrhea, whereas it also can be associated with intestinal diseases such as ulcerative colitis (Rajilić-Stojanović et al., 2013a). However, Campylobacter spp. can also remain asymptomatic, suggesting that at least some species of this genus are commercial members of the gastrointestinal microbiota.
The Arcobacter genus was introduced to accommodate an independent cluster identified based on the SSU rRNA gene sequences, of species that were previously classified within the Campylobacter genus (Vandamme et al., 1991). These bacteria are also associated with diarrheal disease, although the prevalence of their isolation, even from clinical samples is very low (Engberg et al., 2000).
The genus Helicobacter has been derived from Campylobacter after reclassification of the latter (Goodwin et al., 1989; Vandamme et al., 1991). Helicobacter spp. are spiral-shaped bacteria that were detected in human gastric mucosa as late as in 1906 (Krienitz, 1906). They received exceptional attention once Helicobacter pylori was discovered to induce the gastric and duodenal ulcers (for recent review see, Fock et al., 2013). Helicobacter spp. are mainly located in the stomach but can be detected in the other gastrointestinal samples of healthy individuals but only when highly sensitive techniques are applied, suggesting that they may lyse and disappear in transit from the stomach (MacKay et al., 2003; Ceelen et al., 2005). Hence, their prevalence is low in samples from the lower gastrointestinal tract (Hansen et al., 2013).
Lentisphaerae
Victivallis vadensis is the only species within the phylum of Lentisphaerae that has been isolated from the gastrointestinal tract (Fig. 26). This species was isolated in 2003, as a bacterium that was able to grow in basal liquid medium containing cellobiose as the sole carbon source, but not on the same medium solidified with agar (Zoetendal et al., 2003). Further studies have confirmed its presence in the gastrointestinal tract of humans (Claesson et al., 2012), indicating the adaptation of this species (and most likely the entire bacterial group) to the gastrointestinal tract conditions. The genome of this bacterium has been sequenced (van Passel et al., 2011), revealing a host of functions, but its activity in the gastrointestinal tract and its impact on the human host remains to be determined.
Fig 26.
Phylogenetic tree the human gastrointestinal species that belong to the different bacterial phyla with limited diversity, and two archaeal phyla. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the phylum names are indicated.
Spirochetes
The first report of the Spirochetes in the human gastrointestinal samples dates from 1923 when a cultivation-based study reported the 28% prevalence of these bacteria in fecal samples of healthy individuals (Parr, 1923). The Spirochetes are established pathogens in veterinary medicine and their pathogenicity in humans has been debated for a long period of time. Two species of the phylum Spirochetes can be detected in the human gastrointestinal samples: Brachyspira pilosicoli and Brachyspira aalborgi (Fig. 26). Their presence in the gastrointestinal tract is termed intestinal spirochetosis, which in clinical cases is associated with abdominal pain and diarrhea. However, a retrospective review of cases diagnosed as intestinal spirochetosis revealed that the presence of Spirochetes in the gastrointestinal biopsies is often asymptomatic and may not have pathological significance (Carr et al., 2010). The question of implication of the Spirochetes in clinical cases might be a question of their density in the gut. These bacteria typically colonize intestinal mucosa where they attach and penetrate short distances into the surface epithelial cells (Harland & Lee, 1967). In clinical cases of spirochetosis they form a dense biofilm that covers the entire colonic surface, as indicated by scanning electron microscopy images (Gad et al., 1977). Another factor might be relevant to the currently undefined role of the Spirochetes in human health. A recent molecular study showed that in addition to the two cultured species, another, currently uncultured Brachyspira spp. is more frequent than the other two Brachyspira spp. in the human colonic biopsies. Although this study confirmed the absence of a correlation between these species and physical complaints, it appeared that B. pilosicoli is associated with intestinal inflammation (Westerman et al., 2012).
Another relevant group of the gastrointestinal Spirochetes are formed by the Treponema spp. Members of the Treponema genus were detected in molecular-based studies of the gut microbiota of five geographically separate rural African and Native American tribes (De Filippo et al., 2010; Tito et al., 2012; Yatsunenko et al., 2012; Ou et al., 2013; Schnorr et al., 2014). Only in one study, a representative of this group was identified at species level. A sequence 99% similar to Treponema berlinense was detected when analyzing the microbiota coprolite (fossilized feces) taken from archaeological site in Mexico (Tito et al., 2012). Treponema spp. are in principle considered to be pathogenic in industrial societies (Giacani & Lukehart, 2014), but its reproducible detection in the gastrointestinal microbiota of isolated rural communities suggest alternative symbiotic roles played by these bacteria. These might include degradation of fiber rich foods and enhancement of anti-inflammatory capability, as suggested by de Fillipo and coauthors, who were the first to detected Treponema spp. in healthy human gut (De Filippo et al., 2010).
Synergistetes
Synergistetes is a recently recognized bacterial phylum that is typically subdominant in the ecosystems where it resides, and its members can be present in abundance of about 0.01% in human fecal samples (Horz et al., 2006). The first attempt to detect this group of bacteria in the human gastrointestinal tract yielded a sole SSU rRNA gene sequence that is identical to that of a later on cultured Cloacibacillus evryensis (Ganesan et al., 2008). Cloacibacillus spp. are amino acid degrading bacteria that use sulfate as terminal electron acceptor and that are capable to use mucin as sole carbon source (Looft et al., 2013). Their presence in the human gastrointestinal microbiota was confirmed by several molecular studies. Furthermore, a recent high-throughput culturomics study retrieved another gastrointestinal Synergistetes bacterium — Pyramidobacter piscolens (Lagier et al., 2012a). Pyramidobacter piscolens was described in 2009, as an asaccharolytic, anaerobic oral isolate capable of hydrogen sulfide production.
Although a minor group, the gastrointestinal Synergistetes might be relevant for the human health, as indicated by their increased abundance in mucosal samples associated with colorectal cancer (Chen et al., 2012). Mucin degradation coupled with hydrogen sulfide produced by these bacteria might be relevant for the colorectal cancer etiology, since the produced metabolite increases mucosal apoptosis, goblet cell depletion, and superficial ulceration (Aslam et al., 1992; Deplancke & Gaskins, 2003).
TM7 candidate phylum
The TM7 phylum represents a recently recognized, widely distributed group of yet uncultured filamentous bacteria (Hugenholtz et al., 2001). These bacteria can be detected in the human oral cavity and the gastrointestinal tract, while a recent study of the microbiota along the gastrointestinal tract has shown that TM7 bacteria are one of the common microorganisms, widely distributed among humans (Stearns et al., 2011). Although the presence of these bacteria is not determinative of the health status, it has been shown that different TM7 bacteria inhabit the gastrointestinal tracts of inflammatory bowel disease patients and healthy controls (Kuehbacher et al., 2008).
Verrucomicrobia
Currently, only two species within the Verrucomicrbia phylum have been detected in the human gastrointestinal tract. Akkermansia muciniphila was described in 2004 as a unique human gastrointestinal bacterium that is able to grow on intestinal mucus as a sole carbon source (Derrien et al., 2004). This bacterium is widely distributed and can be detected in fecal material of humans of all age groups (Collado et al., 2007), and although its abundance varies between subjects, it is probably one of the members of the core microbiota. Recent literature shows that A. muciniphila is important for a healthy host as its decreased abundance is associated with compromised health including acute appendicitis (Swidsinski et al., 2011), ulcerative colitis (Vigsnæs et al., 2012; Rajilić-Stojanović et al., 2013a), autism (Wang et al., 2011a), and atopic diseases (Candela et al., 2012). Finally, the abundance of A. muciniphila is inversely correlated with obesity (Karlsson et al., 2012). A recent study suggests that A. muciniphila plays a pivotal role in obesity as its duodenal delivery regulates fat-mass gain, metabolic endotoxemia, adipose tissue inflammation, and insulin resistance in an animal model experiment (Everard et al., 2013). Given the very short history of the research related to A. muciniphila, the wealth of data that support its beneficial role provides evidence of the remarkable importance of this bacterium. In addition, one other species of the Verrucomicrobia — Prosthecobacter fluviatilis — has been detected in the study of bacteria of a patient with ileal pouch (McLaughlin et al., 2010), while sequences classified within this genus were detected in the study of the microbiota of infants (Palmer et al., 2007).
Other gastrointestinal bacterial phyla
There are a few gastrointestinal bacterial phyla with low diversity that have not yet been discussed, including the Deinococcus-Thermus bacteria, the Melainabacteria the Gemmatimonadetes and the Planctomycetes (Fig. 26). The Deinococcus-Thermus bacteria were for the first time associated with the human gastrointestinal microbiota in 2006, when uncultured phylotypes within this phylum were recovered in a molecular survey of the microbiota of the human stomach (Bik et al., 2006). Several studies have confirmed the presence of these bacteria in the gastrointestinal microbiota, while a recent high-throughput culturing study detected a single cultivated bacterium of this phylum (Lagier et al., 2012a). Currently, no particular function has been assigned to this bacterial group, although there is evidence that these bacteria are active in the distal parts of the gastrointestinal tract (Peris-Bondia et al., 2011).
Molecular studies have retrieved SSU rRNA gene sequences from the gastrointestinal tract of humans and other animals that cluster into a distinct clade, related to, but separate from cultured photosynthetic Cyanobacteria (Ley et al., 2005). A novel name – Melainabacteria — was recently proposed for this group of bacteria (Di Rienzi et al., 2013). Sequences representing these bacteria were detected in the stomach (Andersson et al., 2008), while two independent studies have shown that these bacteria are active in the distal part of the human gastrointestinal tract (Rajilić-Stojanović et al., 2008; Peris-Bondia et al., 2011), which indicates their wide distribution along the gastrointestinal tract. The role of these bacteria in the ecosystem is currently undetermined and although no cultured representatives are available now, the genomes of these bacteria have been sequenced and annotated. Based on the genomic information, it was concluded that these are motile, strictly anaerobic, fermentative bacteria (Di Rienzi et al., 2013).
In addition to the bacterial phyla that are reproducibly detected by many studies of the human gastrointestinal microbiota, some bacterial phyla are only occasionally found. For instance, some studies have reported the presence of the Gemmatimonadetes in the human gastrointestinal tract (Andersson et al., 2008), but even when present, these bacteria constitute an extremely minor fraction, as indicated by the fact that only one of over 15 000 SSU rRNA gene sequences in the study of the microbiota of inflammatory bowel disease patients and controls originated from a Gemmatimonadetes bacterium (Frank et al., 2007). Furthermore, a few pyrosequencing studies have reported Nitrospira bacteria in the human gastrointestinal tract (Hung et al., 2011). It is interesting to note that in a recent study of the microbiota of pediatric irritable bowel syndrome patients only one Nitrospira phylotype was detected by pyrosequencing. However, the further attempts to retrieve this bacterium revealed that the detected sequence was actually retrieved from a novel taxon related to the genus Ruminococcus (Saulnier et al., 2011). This example testifies to the fact that identification of sequences based on very short reads, such as those produced by currently used high-throughput sequencing technologies, are not always reliable (Werner et al., 2012).
The presence of the Planctomycetes in the human gastrointestinal tract was reported only in molecular-based studies (Wilson & Blitchington, 1996; De Filippo et al., 2010; Hong et al., 2011). A sequence of an uncultured bacterium (Gene Bank Accession Number U58225) was reported in one of the first molecular studies of the human gastrointestinal microbiota, and was designated as an uncultured Plantomycetes bacterium (Wilson & Blitchington, 1996). However, the detected SSU rRNA gene sequence was probably retrieved from a representative of a new genus within the Lentisphaerae phylum, since it has the highest similarity (88%) with V. vadensis (that was isolated and described after publication of the molecular study). Nevertheless, Plantomycetes might be a part of the human gastrointestinal microbiota, as a recent review indicated the detection of a diverse community of the Planctomycetes in the human gastrointestinal tract (Lagier et al., 2012b). When analyzing the publicly available data, we found that only one (JQ287572) had high similarity (98%) to the SSU rRNA gene sequence of a cultured bacterium—Schlesneria paludicola. However, based on the characteristics of this bacterium (e.g., it grows in the temperature range 4–32 °C), it is not likely that this bacterium resides in the human gastrointestinal tract.
Archaea
Bacteria are the dominant but not an exclusive component of the human gastrointestinal microbiota. Archaea, primarily the methanogenic ones, can be relatively abundant component of the gastrointestinal microbiota with densities of up to 1010 cells g−1 of feces (Bond et al., 1971; Miller & Wolin, 1986). In total, eight archaeal species have been associated with the human gastrointestinal tract (Fig. 26, Table S2). In an early cultivation study, which dates from 1968, a single methanogenic species, isolated from four of five individuals, was identified as Methanobrevibacter ruminantium (Nottingham & Hungate, 1968). Today, Methanobrevibacter smithii is recognized as the most abundant, and often an exclusive methanogen of human gastrointestinal microbiota (Miller & Wolin, 1986; Dridi et al., 2009), which suggests a possible misidentification of the isolates in the earlier study. In addition to M. smithii, Methanosphaera stadtmaniae is a relatively prevalent, but atypical methanogenic archaea that reduces methanol and that can be found in human feces in low concentrations (Miller & Wolin, 1985; Dridi et al., 2009). Similar to M. stadtmaniae, a recently isolated Methanomassiliicoccus luminyensis can also utilize methanol in the presence of hydrogen, but these two gastrointestinal archaeal species are phylogenetically distant (Dridi et al., 2012). Methanogenic archaea have been extensively studied as the process of methane synthesis from carbon dioxide and hydrogen results in a significant gas removal in the gastrointestinal tract. The role of methanogenic archaea might be particularly relevant for bloating, which is one of the symptoms of irritable bowel syndrome, and a recent study has shown a highly significant (fourfold) reduction of methanogenic archaea in irritable bowel syndrome patients relative to controls (Rajilić-Stojanović et al., 2011). In addition to methanogenic archaea, two cultured species of halophilic archaea have been detected in the study of the microbiota of Korean subjects (Nam et al., 2008), while the presence of low numbers of these organisms was confirmed by the analysis of the colonic mucosa of inflammatory bowel patients (Oxley et al., 2010). In addition to the confirmed presence of the Euryarchaeota phylum members, the human gastrointestinal archaea might also include a number of yet uncultured species within the Thermoplasma and the Crenarchaeota phylum and putative novel orders, as detected in molecular-based studies and recently reviewed (Dridi et al., 2011). The presence of Crenarchaeota phylum representatives was detected by retrieving the specific partial SSU rRNA gene sequences in a study that dates from 2005 (Rieu-Lesme et al., 2005). One of the amplified SSU rRNA gene sequences (AY887079) shows 97% sequence similarity with SSU rRNA gene sequence of Sulfolobus solfataricus, and another (AY887074) 99% sequence similarity with the SSU rRNA gene of Candidatus Nitrososphaera gargensis. It should be noted that both Sulfolobus solfataricus and Nitrososphaera gargensis are hyperthermophilic species, and it is highly unlikely that these species inhabit the gastrointestinal tract of humans. Until cultured representatives or at least the full SSU rRNA gene sequences of the Crenarchaeota species are obtained in the future independent studies, the presence of the this archaeal phylum in the human gastrointestinal tract remains questionable.
Eukarya
Different microeukaryal species can be detected in the human gastrointestinal tract, and although this group of organisms is subdominant, it is widely distributed component of the gastrointestinal microbiota. The first molecular-based study of this component of the gastrointestinal microbiota was published only recently (Scanlan & Marchesi, 2008). Very few other studies have been reported since, and the results of these have been recently summarized (Hamad et al., 2013). The most prevalent human gastrointestinal Eukarya are yeasts, while a number of different microeukaryal intestinal parasites can be detected in the human gastrointestinal samples. These species have been excessively studied by epidemiologists, and although their presence is in most cases the result of an infection with contaminated food or water, some species establish in healthy humans and are probably a part of the normal gastrointestinal microbiota of some humans (Scanlan & Marchesi, 2008).
Fungi
The most prevalent Eukarya in the human gastrointestinal tract are yeast-like fungi and in total 57 intestinal species distributed between the two phyla, Ascomycota and Basidiomycota, have been detected (Fig. 27). The first report of yeasts in the human gastrointestinal tract dates from 1901 when Candida albicans was isolated from feces of patients infected with tropical sprue (Kohlbrugge, 1901). A thorough analysis of the yeasts diversity in the gastrointestinal tract in the early twentieth century showed that yeasts can be detected in about one out of five subjects and that the detected yeast community is similar in healthy subjects and patients suffering from various gastrointestinal disorders (Anderson, 1917). A recent molecular analysis of the fungal diversity allowed for detecting low amounts of fungi in any studied subject (Ott et al., 2008). Among yeasts, Candida spp. are the most prevalent and there is considerable evidence that C. albicans and C. rugosa are part of the normal gastrointestinal microbiota, while other Candida spp. are scarcely detected in gastrointestinal samples (Fig. 27, Table S3). Candida albicans is early established in the ecosystem, as illustrated by the fact that it can be detected in over 95% of 1-month-old infants (Kumamoto & Vinces, 2005). Candida spp. are subdominant in the ecosystem and typically present in densities lower than 106 cells g−1 of intestinal content (Anderson, 1917; Finegold et al., 1974; Finegold et al., 1977). Although the natural environment of Candida spp. is the gastrointestinal tract, where they are either symbiotic or commensal to the human host, under specific circumstances these organisms can cause a variety of candidiasis in different organs of the human body. It has been proposed that in the developed world the increased intake of drugs, processed foods and pollutants can cause overgrowth of Candida spp. and trigger a Candida-associated complex of symptoms (Schulze & Sonnenborn, 2009). This might be relevant for gastrointestinal health, as it has been shown that Crohn's disease patients and their first relatives have a significantly higher abundance of Candida spp. compared to the controls (Standaert-Vitse et al., 2009). Furthermore, there is a considerable overlap between the symptoms of the irritable bowel syndrome, and the symptoms of intestinal candidiasis (Santelmann & Howard, 2005), although the association between the irritable bowel syndrome and Candida spp. has not been adequately studied. Other yeasts in the gastrointestinal tract include several Saccharomyces spp., of which Saccharomyces cerevisiae was the most reproducibly detected in molecular studies (Nam et al., 2008; Ott et al., 2008; Scanlan & Marchesi, 2008). Furthermore, Galactomyces geotrichum has been detected in the human fecal samples using molecular techniques (Scanlan & Marchesi, 2008) and cultivation (Gouba et al., 2013; Hamad et al., 2013). In contrast to Candida spp. that have the gastrointestinal tract as their natural niche, both Saccharomyces spp. and G. geotrichum are yeasts that are used in food production and their detection in fecal samples of humans could be a result of the dietary intake prior to sampling. This hypothesis is supported by the results of the recently published gastrointestinal microbiota analysis of adults on extreme diets, as a number of foodborn microorganisms (including yeasts) were detected in the gastrointestinal tract of subjects on animal-based (meat and cheese rich) diet (David et al., 2014).
Fig 27.
Phylogenetic tree the human gastrointestinal Fungi. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the phylum names are indicated.
Filamentous fungi are another group of Eukarya that can be present in the human gastrointestinal tract. Although they are not widely distributed, their presence was noticed in a number of studies (Finegold et al., 1974; Finegold et al., 1977; Benno et al., 1986). Several Aspergillus spp. and Penicillium spp. have been identified during a 9-year long culture-based monitoring of an immunodeficient child's microbiota (Taylor et al., 1985), while different species of these two genera have been detected in recent years using both cultivation on various media (Gouba et al., 2013), or molecular-based methods (Scanlan & Marchesi, 2008). Many of these species are associated with dietary sources, as they are used as starters for cheese production or ripening, whereas some are just food contaminants. Having in mind their source, and instability (as illustrated in the results of longitudinal follow up of fungal microbiota; Scanlan & Marchesi, 2008), it is likely that these fungi are not a constant and functionally relevant part of the gastrointestinal microbiota.
Eukarya-intestinal parasites
The gastrointestinal tract of humans can be inhabited by a number of different micro-eukaryotes that belong to the phyla: Apicomplexa, Amoebozoa, Ciliophora, Metamonada, Micosporidia, Parabasalia, and Stramenopiles (Fig. 28). Some of these organisms are pathogenic infectious agents, which after ingestion (through contaminated water or food) can cause gastrointestinal symptoms, most frequently diarrhea. Because of their clinical significance, these organisms, which are often referred as intestinal protozoa, are relatively thoroughly studied as infections agents. Little attention has been given to these organisms in terms of their role in the ecosystem, but as several studies have shown that some of these species can be present in the healthy human gastrointestinal tract, it is reasonable to expect that future research will reveal the true role of these organisms in the gastrointestinal microbiota.
Fig 28.
Phylogenetic tree the human gastrointestinal microeukarya. GenBank Accession Numbers of the SSU rRNA gene sequence are provided for each species and the phylum names are indicated.
A number of Eukarya classified within the phylum of Apicomplexa in addition to Micosporidia form a group of intestinal spore-forming protozoa that cause intracellular infections, primarily in the epithelial cells of the gastrointestinal tract. All Apicomplexa species that can be retrieved from the human gastrointestinal tract are considered to be pathogenic (Farthing, 2006). Cryptosporidium parvum is the most widely distributed representative of the group, and it has a high prevalence of infection in children of the developing world (Checkley et al., 1997). Although infections with Cryptosporidium parvum can be asymptomatic, even when it does not cause diarrhea, this organism affects absorption of nutrients and has a negative effect on weight gain of children (Checkley et al., 1997).
Blastocystis spp. (classified within the Stramenopiles) are single-celled protozoan organisms. Blastocystis spp. was for the first time isolated from the human gastrointestinal tract in 1911 and was reported under name Blastocystis enterocola, which was, at the time, designated as yeast (Alexieff, 1911). According to the current convention, all Blastocystis isolated from humans are identified as Blastocystis hominis, although their SSU rRNA gene sequence analysis showed that nine different phylotypes of these organisms can be detected in the human gastrointestinal tract (Arisue et al., 2003; Scanlan & Marchesi, 2008). This ‘within species’ diversity of the B. hominis can explain the fact that the role of Blastocystis in human disease is still not defined (Zierdt, 1991). Longitudinal study of the Eukarya in the human gastrointestinal tract has shown that Blastocystis spp. are stable and frequently detected organisms in healthy subjects (Scanlan & Marchesi, 2008). Although some authors have suggested the link between Blastocystis and intestinal diseases such as diarrhea, irritable bowel syndrome and inflammatory bowel disease, the detection of Blastocystis in 105 patients suffering from various gastrointestinal diseases and 96 healthy controls, indicated that Blastocystis is equally frequent in patients and healthy subjects, although different phylotypes of Blastocystis are associated with different health status (Dogruman-Al et al., 2008). Another recent study could establish a significantly higher incidence of Blastocystis in ulcerative colitis patients, when compared to controls (Cekin et al., 2012).
Neobalantidium coli is the only representative of the Ciliophora phylum. Neobalantidium coli is the largest protozoan parasite that infects humans, but its natural hosts are pigs. Although the organism can reproduce within the intestinal lumen of humans without attacking the tissues and therefore remain asymptomatic, the infection with this species is typically followed by diarrhea and bloody stools (Katz et al., 1982).
The Amoebozoa that can be detected in the human gastrointestinal tract include Endolimax nana and Iodamoeba bütschlii and six Entamoeba spp. (Table S3). The SSU rRNA gene sequence is available only for five Entamoeba spp. (Fig. 28). While most of the intestinal Amoebozoa are nonpathogenic, there is sufficient evidence that Entamoeba histolytica is pathogenic for humans and causes amebiasis — dysentery or amebic colitis with a high mortality rate (Fotedar et al., 2007). Entamoeba histolytica was the first described 1875, although the species name Entamoeba histolytica was assigned later, in 1903 by Fritz Schaudinn (Saklatvala, 1993). There are several studies that show the successful detection of Entamoeba spp. in clinical samples using molecular methods, typically in stool samples taken from patients with diarrhea. Molecular studies of the Eukarya as part of the ecosystem in the human gastrointestinal tract already confirmed the presence of two Entamoeba species: Entamoeba coli (Scanlan & Marchesi, 2008) and Entamoeba hartmanni (Hamad et al., 2013) in the gastrointestinal tract of a healthy man.
Members of the phyla Micosporidia, Parabasalia and Metamonada are micro-eukaryotic organisms that contain flagella and are often commonly termed as flagellates. Micosporidia are obligate intracellular protozoan parasites that spread in the environment via spores. Four species of Micosporidia can infect the human gastrointestinal tract — Enterocytozoon bieneusi (Desportes et al., 1985), Encephalitozoon intestinalis (Weber et al., 1994), Encephalitozoon cuniculi (Franzen et al., 1995), and Retortamonas intestinalis (Jones-Engel et al., 2004; Fig. 28, Table S3). Retortamonas intestinalis, for which SSU rRNA gene sequence is not available, is the oldest isolate of this group that was for the first time cultured in 1879 (Hogue, 1933). Other Micosporidia were isolated from the gastrointestinal tract of a subject infected with HIV (Desportes et al., 1985). The presence of Micosporidia in the gastrointestinal tract is typically associated with diarrhea, mostly in immuno-suppressed patients, although spores of these organisms can be detected in gastrointestinal samples of asymptomatic subjects (Cegielski et al., 1999; Mungthin et al., 2005; Wichro et al., 2005). If asymptomatic, the presence of Micosporidia in the gastrointestinal tract is associated with malnutrition.
Among Parabasalia, two species can be associated with the human gastrointestinal tract. Pentatrichomonas hominis is generally regarded as a harmless commensal organism, although it is occasionally designated as a causal agent of diarrhea. This organism, which actually represents the oldest gastrointestinal isolate retrieved in 1854, has a low prevalence of the human gastrointestinal tract colonization that varies from 0.1% to 30.9% depending on the geographical location (Honigberg, 1990). In 1918 another member of this group, Dientamoeba fragilis was reported as a commensal in the human gastrointestinal tract. However, the latter research has suggested that D. fragilis might be associated with a number of diseases including diarrhea, abdominal pain, anorexia, irritable bowel syndrome or allergic colitis (reviewed in Johnson et al., 2004). The uncertainty of the pathogenicity of D. fragilis might be, similar to B. hominis, due to the presence of different phylotypes, since two different phylotypes of this species, with 2% SSU rRNA gene sequence divergence, have been identified (Johnson & Clark, 2000).
The Metamonada phylum includes Giardia lamblia and two rarely detected and principally nonpathogenic species — Enteromonas homins and R. intestinalis (Katz et al., 1982). Giardia lamblia is the most common flagellate of the human gastrointestinal tract. When ingested, typically via contaminated food or water, G. lamblia attaches to the mucosal surface of the duodenum or jejunum and multiplies by binary fission (Wolfe, 1992). Infection with G. lamblia is termed giardiasis, and although it may remain asymptomatic, giardiasis can be followed by a range of symptoms that include steatorrhoea, diarrhea and weight loss (Wolfe, 1992). It is not clear why some infections are asymptomatic, but already in the 1970s, it was suggested that the symptoms might depend on the relation between the parasite and the enteric gastrointestinal microbiota (Tandon et al., 1977). Postinfective to the giardiasis, patients might develop a range of novel symptoms that resemble those of the irritable bowel syndrome (Hanevik et al., 2009). This condition, which can be developed after infection with other infectious agents, such as Campylobacter spp., is recognized as postinfectious irritable bowel syndrome (Spiller & Garsed, 2009). A recent study showed that the bacterial fraction of the gastrointestinal microbiota of the postinfectious irritable bowel syndrome patients have a distinct composition relative to controls, which most likely reflects a consequence of an intensive interaction between the ecosystem and the infectious agent (Jalanka-Tuovinen et al., 2013). This illustrates that infectious agent, although not true members of the gastrointestinal microbiota should be kept in mind when studying the gastrointestinal microbiota, as their short- and long-term impact on the microbiota composition and function can be profound.
Concluding remarks
The knowledge generated during more than a century of studying the human gastrointestinal microbiota has shown that this ecosystem is indeed a forgotten organ of the human body. The wealth of data about the gastrointestinal microbiota is highly scattered in time as well as in space — a caveat that this review aims to correct in an attempt to systematize the generated knowledge. We have given particular attention to the diversity and the defined functions of the abundant and important microbiota groups. Our inventory reports 1057 cultured gastrointestinal species, while many more are still expected to be cultured. Although cultivation of the gastrointestinal microbiota is laborious, it is an essential step for the detailed physiological and biochemical characterization of the individual gastrointestinal isolates that is needed for the progress of this research field. This has been increasingly recognized and the recent high-throughput culturing studies have proven that cultivation can be used as a powerful methodology in discovery of currently unknown gastrointestinal inhabitants (Lagier et al., 2012a; Dubourg et al., 2013; Pfleiderer et al., 2013). The future cultivation of the remaining majority of the gastrointestinal microbiota is expected to improve our understanding of this ecosystem, while this review can serve as a baseline for the gastrointestinal microbiota diversity and function when the first 1000 intestinal species had been discovered.
Acknowledgments
Part of this work was supported by the Spinoza Award and Gravity Grant of the Netherlands Organization of Scientific Research and the ERC Advanced Grant 250172 (Microbes Inside) of the European Research Council to WMdV. The authors declare no conflict of interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
List of bacterial species that can be members of the human gastrointestinal microbiota, with a reference that provides the link between the species and gastrointestinal tract as ecological niche.
Table S2. List of archaeal species that can be members of the human gastrointestinal microbiota, with a reference that provides the link between the species and gastrointestinal tract as ecological niche.
Table S3. List of eukaryal species that can be members of the human gastrointestinal microbiota, with a reference that provides the link between the species and gastrointestinal tract as ecological niche.
References
- Alexieff A. Sur la nature des formations dites ‘Kystes de Trichomonas intestinalis’. CR Soc Biol. 1911;71:296–298. [Google Scholar]
- Allison MJ, Dawson KA, Mayberry WR. Foss JG. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol. 1985;141:1–7. doi: 10.1007/BF00446731. [DOI] [PubMed] [Google Scholar]
- Anderson HW. Yeast-like fungi of the human intestinal tract. J Infect Dis. 1917;21:341–386. [Google Scholar]
- Anderson IC, Poth M, Homstead J. Burdige D. A comparison of NO and N2O production by the autotrophic nitrifier Nitrosomonas europaea and the heterotrophic nitrifier Alcaligenes faecalis. Appl Environ Microbiol. 1993;59:3525–3533. doi: 10.1128/aem.59.11.3525-3533.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P. Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrewes FW. Horder TJ. A study of the streptococci pathogenic for man. Lancet. 1906;2:708–713. [Google Scholar]
- Apisarnthanarak A, Fraser VJ, Dunne WM, Little JR, Hoppe-Bauer J, Mayfield JL. Polish LB. Stenotrophomonas maltophilia intestinal colonization in hospitalized oncology patients with diarrhea. Clin Infect Dis. 2003;37:1131–1135. doi: 10.1086/378297. [DOI] [PubMed] [Google Scholar]
- Arisue N, Hashimoto T. Yoshikawa H. Sequence heterogeneity of the small subunit ribosomal RNA genes among blastocystis isolates. Parasitology. 2003;161:1–9. doi: 10.1017/s0031182002002640. [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;474:666. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M, Batten JJ, Florin TH, Sidebotham RL. Baron JH. Hydrogen sulphide induced damage to the colonic mucosal barrier in the rat. Gut. 1992;33:S69. [Google Scholar]
- Attene-Ramos MS, Wagner ED, Plewa MJ. Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA. Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bajaj JS, Hylemon PB, Ridlon JM, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675–G685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron EJ. Bilophila wadsworthia: a unique Gram-negative anaerobic rod. Anaerobe. 1997;3:83–86. doi: 10.1006/anae.1997.0075. [DOI] [PubMed] [Google Scholar]
- Baron E, Curren M, Henderson G, et al. Bilophila wadsworthia isolates from clinical specimens. J Clin Microbiol. 1992;30:1882–1884. doi: 10.1128/jcm.30.7.1882-1884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor MD. Aerobic spore-bearing bacteria in the intestinal tract of children. J Bacteriol. 1919;4:23–3415. doi: 10.1128/jb.4.1.23-34.15.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzer C. de Vos WM. Microbes inside – from diversity to function: the case of Akkermansia. ISME J. 2012;6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benno Y, Sawada K. Mitsuoka T. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol Immunol. 1984;28:975–986. doi: 10.1111/j.1348-0421.1984.tb00754.x. [DOI] [PubMed] [Google Scholar]
- Benno Y, Sawada K. Mitsuoka T. The intestinal microflora of infants: fecal flora of infants with vitamin K deficiency. Microbiol Immunol. 1985;29:243–250. doi: 10.1111/j.1348-0421.1985.tb00823.x. [DOI] [PubMed] [Google Scholar]
- Benno Y, Sawada K. Mitsuoka T. Comparison of the fecal microflora in rural Japanese and urban Canadians. Microbiol Immunol. 1986;30:521–532. doi: 10.1111/j.1348-0421.1986.tb02978.x. [DOI] [PubMed] [Google Scholar]
- Benno Y, Endo K, Takeo M, Namba Y, Komori T. Mitsuoka T. Comparison of fecal microflora of elderly persons in rural and urban areas of Japan. Appl Environ Microbiol. 1989;55:1100–1105. doi: 10.1128/aem.55.5.1100-1105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernalier A, Willems A, Leclerc M, Rochet V. Collins MD. Ruminococcus hydrogenotrophicus sp. nov., a new H2/CO2-utilizing acetogenic bacterium isolated from human feces. Arch Microbiol. 1996;166:176–183. doi: 10.1007/s002030050373. [DOI] [PubMed] [Google Scholar]
- Bhat P, Boulter EA, Rajan D, Shanthakumari S, Kapadia CR. Baker SJ. Mycoplasma in the upper gastrointestinal tracts of Southern Indian control subjects and patients with tropical sprue. Am J Clin Pathol. 1973;59:825–828. doi: 10.1093/ajcp/59.6.825. [DOI] [PubMed] [Google Scholar]
- Bhat P, Albert MJ, Rajan D, Ponniah J, Mathan VI. Baker SJ. Bacterial flora of the jejunum: a comparison of luminal aspirate and mucosal biopsy. J Med Microbiol. 1980;13:247–256. doi: 10.1099/00222615-13-2-247. [DOI] [PubMed] [Google Scholar]
- Biagi E, Nylund L, Candela M, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biavati B, Vescovo M, Torriani S. Bottazzi V. Bifidobacteria: history, ecology, physiology and applications. Ann Microbiol. 2000;50:117–131. [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ. Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. P Natl Acad Sci USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkroth KJ, Schillinger U, Geisen R, Weiss N, Hoste B, Holzapfel WH, Korkeala HJ. Vandamme P. Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov., detected in food and clinical samples. Int J Syst Evol Microbiol. 2002;52:141–148. doi: 10.1099/00207713-52-1-141. [DOI] [PubMed] [Google Scholar]
- Bond JH, Jr, Engel RR. Levitt MD. Factors influencing pulmonary methane excretion in man: an indirect method of studying the in situ metabolism of the methane-producing colonic bacteria. J Exp Med. 1971;133:572–588. doi: 10.1084/jem.133.3.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booijink CCGM. Wageningen, the Netherlands: Wageningen University; 2009. Analysis of diversity and function of the human small intestinal microbiota. PhD. [Google Scholar]
- Booijink CCGM, El-Aidy S, Rajilić-Stojanović M, Heilig HG, Troost FJ, Smidt H, Kleerebezem M, de Vos WM. Zoetendal EG. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010;12:3213–3227. doi: 10.1111/j.1462-2920.2010.02294.x. [DOI] [PubMed] [Google Scholar]
- Bouhnik Y, Alain S, Attar A, Flourie B, Raskine L, Sanson-Le Pors MJ. Rambaud J-C. Bacterial populations contaminating the upper gut in patients with small intestinal bacterial overgrowth syndrome. Am J Gastroenterol. 1999;94:1327–1331. doi: 10.1111/j.1572-0241.1999.01016.x. [DOI] [PubMed] [Google Scholar]
- Brown J, de Vos WM, DiStefano PS, Dore J, Huttenhower C, Knight R, Lawley TD, Raes J. Turnbaugh P. Translating the human microbiome. Nat Biotechnol. 2013;31:304–308. doi: 10.1038/nbt.2543. [DOI] [PubMed] [Google Scholar]
- Bucher C. von Graevenitz A. Evaluation of three differential media for detection of Enterobacter agglomeransErwinia herbicola. J Clin Microbiol. 1982;15:1164–1166. doi: 10.1128/jcm.15.6.1164-1166.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela M, Rampelli S, Turroni S, et al. Unbalance of intestinal microbiota in atopic children. BMC Microbiol. 2012;12:95. doi: 10.1186/1471-2180-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier J-P, Bedora-Faure M, K'ouas G, Alauzet C. Mory F. Proposal to unify Clostridium orbiscindens Winter et al. 1991 and Eubacterium plautii (Séguin 1928) Hofstad and Aasjord 1982, with description of Flavonifractor plautii gen. nov., comb. nov., and reassignment of Bacteroides capillosus to Pseudoflavonifractor capillosus gen. nov., comb. nov. Int J Syst Evol Microbiol. 2010;60:585–590. doi: 10.1099/ijs.0.016725-0. [DOI] [PubMed] [Google Scholar]
- Carr NJ, Mahajan H, Tan KL. Sharma R. The histological features of intestinal spirochetosis in a series of 113 patients. Int J Surg Pathol. 2010;18:144–148. doi: 10.1177/1066896908330203. [DOI] [PubMed] [Google Scholar]
- Castellarin M, Warren R, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceelen L, Decostere A, Verschraegen G, Ducatelle R. Haesebrouck F. Prevalence of Helicobacter pullorum among patients with gastrointestinal disease and clinically healthy persons. J Clin Microbiol. 2005;43:2984–2986. doi: 10.1128/JCM.43.6.2984-2986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegielski JP, Ortega YR, McKee S, et al. Cryptosporidium, Enterocytozoon, and Cyclospora infections in pediatric and adult patients with diarrhea in Tanzania. Clin Infect Dis. 1999;28:314–321. doi: 10.1086/515131. [DOI] [PubMed] [Google Scholar]
- Cekin A, Cekin Y, Adakan Y, Tasdemir E, Koclar F. Yolcular B. Blastocystosis in patients with gastrointestinal symptoms: a case–control study. BMC Gastroenterol. 2012;12:122. doi: 10.1186/1471-230X-12-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JY, Shin SM, Chun J, Lee J-H. Seo J-K. Pyrosequencing-based molecular monitoring of the intestinal bacterial colonization in preterm infants. J Pediatr Gastroenterol Nutr. 2011;53:512–519. doi: 10.1097/MPG.0b013e318227e518. [DOI] [PubMed] [Google Scholar]
- Chassaing B, Koren O, Carvalho FA, Ley RE. Gewirtz AT. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2014;63:1069–1080. doi: 10.1136/gutjnl-2013-304909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley W, Gilman RH, Epstein LD, Suarez M, Diaz JF, Cabrera L, Black RE. Sterling CR. Asymptomatic and symptomatic cryptosporidiosis: their acute effect on weight gain in Peruvian children. Am J Epidemiol. 1997;145:156–163. doi: 10.1093/oxfordjournals.aje.a009086. [DOI] [PubMed] [Google Scholar]
- Chen W, Li D, Paulus B, Wilson I. Chadwick V. High prevalence of Mycoplasma pneumoniae in intestinal mucosal biopsies from patients with inflammatory bowel disease and controls. Dig Dis Sci. 2001;46:2529–2535. doi: 10.1023/a:1012352626117. [DOI] [PubMed] [Google Scholar]
- Chen W, Liu F, Ling Z, Tong X. Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Clavel T, Henderson G, Engst W, Doré J. Blaut M. Phylogeny of human intestinal bacteria that activate the dietary lignan secoisolariciresinol diglucoside. FEMS Microbiol Ecol. 2006;55:471–478. doi: 10.1111/j.1574-6941.2005.00057.x. [DOI] [PubMed] [Google Scholar]
- Collado MC, Derrien M, Isolauri E, de Vos WM. Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. 2007;73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Donat E, Ribes-Koninckx C, Calabuig M. Sanz Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J Clin Pathol. 2009;62:264–269. doi: 10.1136/jcp.2008.061366. [DOI] [PubMed] [Google Scholar]
- Collins MD. Wallbanks S. Comparative sequence analyses of the 16S rRNA genes of Lactobacillus minutus Lactobacillus rimae and Streptococcus parvulus: proposal for the creation of a new genus Atopobium. FEMS Microbiol Lett. 1992;74:235–240. doi: 10.1016/0378-1097(92)90435-q. [DOI] [PubMed] [Google Scholar]
- Collins M, Lawson P, Willems A, Cordoba J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H. Farrow J. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- Crociani F, Alessandrini A, Mucci MM. Biavati B. Degradation of complex carbohydrates by Bifidobacterium spp. Int J Food Microbiol. 1994;24:199–210. doi: 10.1016/0168-1605(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Cryan JF. Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- Culotta E. Koshland DE. NO news is good news. Science. 1992;258:1862–1865. doi: 10.1126/science.1361684. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G. Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. P Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F. Mithieux G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- de Weerth C, Fuentes S, Puylaert P. de Vos WM. Intestinal microbiota of infants with colic: development and specific signatures. Pediatrics. 2013;131:550–558. doi: 10.1542/peds.2012-1449. [DOI] [PubMed] [Google Scholar]
- Denault LJ, Cleverdon RC. Kulp WL. Nitrogen compounds utilized by Alcaligenes faecalis. J Bacteriol. 1953;66:465–467. doi: 10.1128/jb.66.4.465-467.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B. Gaskins HR. Hydrogen sulfide induces serum-independent cell cycle entry in nontransformed rat intestinal epithelial cells. FASEB J. 2003;17:1310–1312. doi: 10.1096/fj.02-0883fje. [DOI] [PubMed] [Google Scholar]
- Deplancke B, Finster K, Graham WV, Collier CT, Thurmond JE. Gaskins HR. Gastrointestinal and microbial responses to sulfate-supplemented drinking water in mice. Exp Biol Med. 2003;228:424–433. doi: 10.1177/153537020322800413. [DOI] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM. de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- Desportes I, Charpentier YL, Galian A, Bernard F, Cochand-Priollet B, Lavergne A, Ravisse P. Modigliani R. Occurrence of a new microsporidan: Enterocytozoon bieneusi n. g., n. sp., in the enterocytes of a human patient with AIDS. J Protozool. 1985;32:250–254. doi: 10.1111/j.1550-7408.1985.tb03046.x. [DOI] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B. Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Sharon I, Wrighton KC, et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. eLife. 2013;2:e01102. doi: 10.7554/eLife.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distaso A. Sur les microbes protéolytiques de la flore intestinale de l'homme et des animaux. Zbl Bakt Parasit. 1911;59:97–103. [Google Scholar]
- Dobell C. Antony Van Leeuwenhoek and His Little Animals. New York: Harcourt, Brace & Company; 1932. [Google Scholar]
- Dogruman-Al F, Kustimur S, Yoshikawa H, Tuncer C, Simsek Z, Tanyukse M, ArazI E. Boorom K. Blastocystis subtypes in irritable bowel syndrome and inflammatory bowel disease in Ankara, Turkey. Mem Inst Oswaldo Cruz. 2008;104:724–727. doi: 10.1590/s0074-02762009000500011. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N. Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. P Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B, Henry M, El Khéchine A, Raoult D. Drancourt M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS One. 2009;4:e7063. doi: 10.1371/journal.pone.0007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B, Raoult D. Drancourt M. Archaea as emerging organisms in complex human microbiomes. Anaerobe. 2011;17:56–63. doi: 10.1016/j.anaerobe.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Dridi B, Fardeau M-L, Ollivier B, Raoult D. Drancourt M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol. 2012;62:1902–1907. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- Dubourg G, Lagier JC, Armougom F, Robert C, Hamad I, Brouqui P. Raoult D. The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur J Clin Microbiol Infect Dis. 2013;32:637–645. doi: 10.1007/s10096-012-1787-3. [DOI] [PubMed] [Google Scholar]
- Duc LH, Hong HA, Barbosa TM, Henriques AO. Cutting SM. Characterization of Bacillus probiotics available for human use. Appl Environ Microbiol. 2004;70:2161–2171. doi: 10.1128/AEM.70.4.2161-2171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Hold GL, Harmsen H, Stewart CS. Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol. 2002a;52:2141–2146. doi: 10.1099/00207713-52-6-2141. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Richardson AJ, Kaul P, Holmes RP, Allison MJ. Stewart CS. Oxalobacter formigenes and its potential role in human health. Appl Environ Microbiol. 2002b;68:3841–3847. doi: 10.1128/AEM.68.8.3841-3847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE. Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhaut V, Machiels K, Perrier C, et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut. 2013;62:1745–1752. doi: 10.1136/gutjnl-2012-303611. [DOI] [PubMed] [Google Scholar]
- Eggerth AH. The Gram-positive non-spore-bearing anaerobic bacilli of human feces. J Bacteriol. 1935;30:277–299. doi: 10.1128/jb.30.3.277-299.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggerth AH. Gagnon BH. The bacteroides of human feces. J Bacteriol. 1933;25:389–413. doi: 10.1128/jb.25.4.389-413.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg J, On SLW, Harrington CS. Gerner-Smidt P. Prevalence of Campylobacter Arcobacter Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J Clin Microbiol. 2000;38:286–291. doi: 10.1128/jcm.38.1.286-291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. P Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki T, Li N, Hashimoto Y, Miura H. Yamamoto H. 16S ribosomal DNA sequences of anaerobic cocci and proposal of Ruminococcus hansenii comb. nov. and Ruminococcus productus comb. nov. Int J Syst Bacteriol. 1994;44:130–136. doi: 10.1099/00207713-44-1-130. [DOI] [PubMed] [Google Scholar]
- Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L. Shu S. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol. 2001;51:1521–1528. doi: 10.1099/00207713-51-4-1521. [DOI] [PubMed] [Google Scholar]
- Fallani M, Amarri S, Uusijarvi A, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157:1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- Farmer JJ, 3rd, Fanning GR, Davis BR, O'Hara CM, Riddle C, Hickman-Brenner FW, Asbury MA, Lowery VA. Brenner DJ. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens. J Clin Microbiol. 1985;21:77–81. doi: 10.1128/jcm.21.1.77-81.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farthing MJG. Treatment options for the eradication of intestinal protozoa. Nat Clin Pract Gastroenterol Hepatol. 2006;3:436–445. doi: 10.1038/ncpgasthep0557. [DOI] [PubMed] [Google Scholar]
- Favier CF, Vaughan EE, de Vos WM. Akkermans ADL. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold SM. Intestinal bacteria. The role they play in normal physiology, pathologic physiology, and infection. Calif Med. 1969;110:455–459. [PMC free article] [PubMed] [Google Scholar]
- Finegold SM, Howard RA. Vera LS. Effect of diet on human intestinal fecal flora: comparison of Japanese and American diets. Am J Clin Nutr. 1974;27:1456–1469. doi: 10.1093/ajcn/27.12.1456. [DOI] [PubMed] [Google Scholar]
- Finegold SM, Vera LS, Paul TS, Harvey AE, Shirley ML. Ronald LP. Fecal microbial flora in Seventh Day Adventist populations and control subjects. Am J Clin Nutr. 1977;30:1781–1792. doi: 10.1093/ajcn/30.11.1781. [DOI] [PubMed] [Google Scholar]
- Finegold SM, Lawson PA, Vaisanen ML, Molitoris DR, Song Y, Liu C. Collins MD. Anaerofustis stercorihominis gen. nov., sp. nov., from human faeces. Anaerobe. 2004;10:41–45. doi: 10.1016/j.anaerobe.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Fite A, Macfarlane GT, Cummings JH, Hopkins MJ, Kong SC, Furrie E. Macfarlane S. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut. 2004;53:523–529. doi: 10.1136/gut.2003.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimons MS, Novotny M, Lo C-C, et al. Nearly finished genomes produced using gel microdroplet culturing reveal substantial intraspecies genomic diversity within the human microbiome. Genome Res. 2013;23:878–888. doi: 10.1101/gr.142208.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner CK, Huebel N, Abd El-Bary MM, Loh G, Klaus S. Blaut M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr. 2010;104:919–929. doi: 10.1017/S0007114510001303. [DOI] [PubMed] [Google Scholar]
- Flügge C. Die Mikroorganismen. Vogel, Leipzig: 1886. [Google Scholar]
- Fock KM, Graham DY. Malfertheiner P. Helicobacter pylori research: historical insights and future directions. Nat Rev Gastroenterol Hepatol. 2013;10:495–500. doi: 10.1038/nrgastro.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotedar R, Stark D, Beebe N, Marriott D, Ellis J. Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev. 2007;20:511–532. doi: 10.1128/CMR.00004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N. Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. P Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen C, Schwartz DA, Visvesvara GS, et al. Immunologically confirmed disseminated, asymptomatic Encephalitozoon cuniculi infection of the gastrointestinal tract in a patient with AIDS. Clin Infect Dis. 1995;21:1480–1484. doi: 10.1093/clinids/21.6.1480. [DOI] [PubMed] [Google Scholar]
- Funke G, von Graevenitz A, Clarridge JE. Bernard KA. Clinical microbiology of coryneform bacteria. Clin Microbiol Rev. 1997;10:125–159. doi: 10.1128/cmr.10.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad A, Willén R, Furugård K, Fors B. Hradsky M. Intestinal spirochaetosis as a cause of longstanding diarrhoea. Ups J Med Sci. 1977;82:49–54. doi: 10.3109/03009737709179059. [DOI] [PubMed] [Google Scholar]
- Ganesan A, Chaussonnerie S, Tarrade A, Dauga C, Bouchez T, Pelletier E, Le Paslier D. Sghir A. Cloacibacillus evryensis gen. nov., sp. nov., a novel asaccharolytic, mesophilic, amino-acid-degrading bacterium within the phylum ‘Synergistetes’, isolated from an anaerobic sludge digester. Int J Syst Evol Microbiol. 2008;58:2003–2012. doi: 10.1099/ijs.0.65645-0. [DOI] [PubMed] [Google Scholar]
- Garrity GM, Bell JA. Lilburn T. The revised road map to the manual. In: Garrity GM, editor; Brenner K, Staley JT, editors. Bergey's Manual of Systematic Bacteriology, 2nd edn Part A, Introductory Essays, Vol. 2. New York: Springer; 2005. pp. 159–220. [Google Scholar]
- Giacani L. Lukehart SA. The endemic treponematoses. Clin Microbiol Rev. 2014;27:89–115. doi: 10.1128/CMR.00070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GR, Cummings JH. Macfarlane GT. Use of a three-stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl Environ Microbiol. 1988;54:2750–2755. doi: 10.1128/aem.54.11.2750-2755.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G. Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. P Natl Acad Sci USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin CS, Armstrong JA, Chilvers T, Peters M, Collins MD, Sly L, McConnell M. Harper WES. Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Helicobacter pylori comb. nov, and Helicobacter mustelae comb. nov., respectively. Int J Syst Bacteriol. 1989;39:397–405. [Google Scholar]
- Gosalbes MJ, Llop S, Vallès Y, Moya A, Ballester F. Francino MP. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy. 2013;43:198–211. doi: 10.1111/cea.12063. [DOI] [PubMed] [Google Scholar]
- Gouba N, Raoult D. Drancourt M. Plant and fungal diversity in gut microbiota as revealed by molecular and culture Investigations. PLoS One. 2013;8:e59474. doi: 10.1371/journal.pone.0059474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JD. Shiner M. Influence of gastric pH on gastric and jejunal flora. Gut. 1967;8:574–581. doi: 10.1136/gut.8.6.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont PA, Grimont F, Farmer JJ., 3rd Asbury MA. Cedecea davisae gen. nov., sp. nov. and Cedecea lapagei sp. nov., new Enterobacteriaceae from clinical specimens. Int J Syst Bacteriol. 1981;31:317–326. [Google Scholar]
- Haenel H. Human normal and abnormal gastrointestinal flora. Am J Clin Nutr. 1970;23:1433–1439. doi: 10.1093/ajcn/23.11.1433. [DOI] [PubMed] [Google Scholar]
- Hamad I, Sokhna C, Raoult D. Bittar F. Molecular detection of Eukaryotes in a single human stool sample from Senegal. PLoS One. 2013;7:e40888. doi: 10.1371/journal.pone.0040888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ. Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- Hanevik K, Dizdar V, Langeland N. Hausken T. Development of functional gastrointestinal disorders after Giardia lamblia infection. BMC Gastroenterol. 2009;9:27. doi: 10.1186/1471-230X-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R, Berry SH, Mukhopadhya I, et al. The microaerophilic microbiota of de-novo paediatric inflammatory bowel disease: the BISCUIT study. PLoS One. 2013;8:e58825. doi: 10.1371/journal.pone.0058825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland WA. Lee FD. Intestinal spirochaetosis. Br Med J. 1967;3:718–719. doi: 10.1136/bmj.3.5567.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen HJM, Wildeboer-Veloo ACM, Grijpstra J, Knol J, Degener JE. Welling GW. Development of 16S rRNA-Based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl Environ Microbiol. 2000;66:4523–4527. doi: 10.1128/aem.66.10.4523-4527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M. Antitumor mechanisms of Eubacterium lentum and its components. Asian Pac J Allergy Immunol. 1995;13:129–137. [PubMed] [Google Scholar]
- Hayashi H, Sakamoto M. Benno Y. Molecular analysis of fecal microbiota in elderly individuals using 16S rDNA library and T-RFLP. Microbiol Immunol. 2003;47:557–570. doi: 10.1111/j.1348-0421.2003.tb03418.x. [DOI] [PubMed] [Google Scholar]
- Hedi A, Fardeau M-L, Sadfi N, Boudabous A, Ollivier B. Cayol J-L. Characterization of Halanaerobaculum tunisiense gen. nov., sp. nov., a new halophilic fermentative, strictly anaerobic bacterium isolated from a hypersaline lake in Tunisia. Extremophiles. 2009;13:313–319. doi: 10.1007/s00792-008-0218-y. [DOI] [PubMed] [Google Scholar]
- Hemmeter JC. Diseases of the Intestines, Their Special Pathology. Diagnosis and Treatment. Philadelphia: P. Blakiston's Son & Co; 1902. [Google Scholar]
- Hickman-Brenner FW, Huntley-Carter GP, Saitoh Y, Steigerwalt AG, Farmer JJ., 3rd Brenner DJ. Moellerella wisconsensis, a new genus and species of Enterobacteriaceae found in human stool specimens. J Clin Microbiol. 1984;19:460–463. doi: 10.1128/jcm.19.4.460-463.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman-Brenner FW, Huntley-Carter GP, Fanning GR, Brenner DJ. Farmer JJ., 3rd Koserella trabulsii, a new genus and species of Enterobacteriaceae formerly known as Enteric Group 45. J Clin Microbiol. 1985a;21:39–42. doi: 10.1128/jcm.21.1.39-42.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman-Brenner FW, Vohra MP, Huntley-Carter GP, Fanning GR, Lowery VA, Brenner DJ. Farmer JJ., 3rd Leminorella, a new genus of Enterobacteriaceae: identification of Leminorella grimontii sp. nov. and Leminorella richardii sp. nov. found in clinical specimens. J Clin Microbiol. 1985b;21:234–239. doi: 10.1128/jcm.21.2.234-239.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JA. Brown IL. Resistant starch: a promising dietary agent for the prevention/treatment of inflammatory bowel disease and bowel cancer. Curr Opin Gastroenterol. 2013;29:190–194. doi: 10.1097/MOG.0b013e32835b9aa3. [DOI] [PubMed] [Google Scholar]
- Hogue MJ. A new variety of Retortamonas Embadomonas intestinalis from man. Am J Epidemiol. 1933;18:433–441. [Google Scholar]
- Holdeman LV. Moore WEC. New Genus, Coprococcus, twelve new species, and emended description of four previously described species of bacteria from human feces. Int J Syst Bacteriol. 1974;24:260–277. [Google Scholar]
- Holdeman LV, Good IJ. Moore WEC. Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl Environ Microbiol. 1976;31:359–375. doi: 10.1128/aem.31.3.359-375.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P-Y, Croix JA, Greenberg E, Gaskins HR. Mackie RI. Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS One. 2011;6:e25042. doi: 10.1371/journal.pone.0025042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg BM. Trichomonad found outside the urogenital tract of humans. In: Honigberg BM, editor. Trichomonads Parasitic in Humans. New York: Springer-Verlag; 1990. pp. 342–393. [Google Scholar]
- Hooper LV, Littman DR. Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins MJ, Sharp R. Macfarlane GT. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut. 2001;48:198–205. doi: 10.1136/gut.48.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horz H-P, Citron DM, Warren YA, Goldstein EJC. Conrads G. Synergistes group organisms of human origin. J Clin Microbiol. 2006;44:2914–2920. doi: 10.1128/JCM.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins LC, Agustines M, McKee WB, Boulding ET, Kriaris M. Niedermeyer G. Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest. 1985;75:944–953. doi: 10.1172/JCI111795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou CC, Lai CC, Liu WL, Chao CM, Chiu YH. Hsueh PR. Clinical manifestation and prognostic factors of non-cholerae Vibrio infections. Eur J Clin Microbiol Infect Dis. 2011;30:819–824. doi: 10.1007/s10096-011-1162-9. [DOI] [PubMed] [Google Scholar]
- Hoyles L, Honda H, Logan NA, Halket G, La Ragione RM. McCartney AL. Recognition of greater diversity of Bacillus species and related bacteria in human faeces. Res Microbiol. 2012;163:3–13. doi: 10.1016/j.resmic.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Hoyles L, Clear JA. McCartney AL. Use of denaturing gradient gel electrophoresis to detect Actinobacteria associated with the human faecal microbiota. Anaerobe. 2013;22:90–96. doi: 10.1016/j.anaerobe.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Tyson GW, Webb RI, Wagner AM. Blackall LL. Investigation of candidate division TM7, a recently recognized major lineage of the domain bacteria with no known pure-culture representatives. Appl Environ Microbiol. 2001;67:411–419. doi: 10.1128/AEM.67.1.411-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung W-L, Wade W, Boden R, Kelly D. Wood A. Facultative methylotrophs from the human oral cavity and methylotrophy in strains of Gordonia Leifsonia, and Microbacterium. Arch Microbiol. 2011;193:407–417. doi: 10.1007/s00203-011-0689-6. [DOI] [PubMed] [Google Scholar]
- Hungate RE. Studies on cellulose fermentation. The culture and isolation of cellulose decomposing bacteria from the rumen of cattle. J Bacteriol. 1947;53:631–645. doi: 10.1128/jb.53.5.631-645.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungate RE. Methods in Microbiology. London: Academic Press; 1969. [Google Scholar]
- Ingham CJ, Sprenkels A, Bomer J, Molenaar D, van den Berg A, van Hylckama Vlieg JET. de Vos WM. The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. P Natl Acad Sci USA. 2007;104:18217–18222. doi: 10.1073/pnas.0701693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalanka-Tuovinen J, Salonen A, Nikkilä J, Immonen O, Kekkonen R, Lahti L, Palva A. de Vos WM. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One. 2011;6:e23035. doi: 10.1371/journal.pone.0023035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalanka-Tuovinen J, Salojärvi J. Salonen A, et al. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut. 2013 doi: 10.1136/gutjnl-2013-305994. doi: 10.1136/gutjnl-2013-305994. [DOI] [PubMed] [Google Scholar]
- Jan G, Belzacq AS, Haouzi D, Rouault A, Métivier D, Kroemer G. Brenner C. Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ. 2002;9:179–188. doi: 10.1038/sj.cdd.4400935. [DOI] [PubMed] [Google Scholar]
- Janda JM. Abbott SL. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin Infect Dis. 1998;27:332–344. doi: 10.1086/514652. [DOI] [PubMed] [Google Scholar]
- Jin J-S, Kitahara M, Sakamoto M, Hattori M. Benno Y. Slackia equolifaciens sp. nov., a human intestinal bacterium capable of producing equol. Int J Syst Evol Microbiol. 2010;60:1721–1724. doi: 10.1099/ijs.0.016774-0. [DOI] [PubMed] [Google Scholar]
- Johansson MA, Saghafian-Hedengren S, Haileselassie Y, Roos S, Troye-Blomberg M, Nilsson C. Sverremark-Ekström E. Early-life gut bacteria associate with IL-4−, IL-10− and IFN-γ production at two years of age. PLoS One. 2012;7:e49315. doi: 10.1371/journal.pone.0049315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA. Clark CG. Cryptic genetic diversity in Dientamoeba fragilis. J Clin Microbiol. 2000;38:4653–4654. doi: 10.1128/jcm.38.12.4653-4654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EH, Windsor JJ. Clark CG. Emerging from obscurity: biological, clinical, and diagnostic aspects of Dientamoeba fragilis. Clin Microbiol Rev. 2004;17:553–570. doi: 10.1128/CMR.17.3.553-570.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Engel L, Engel GA, Schillaci MA, Froehlich J, Paputungan U. Kyes RC. Prevalence of enteric parasites in pet macaques in Sulawesi, Indonesia. Am J Primatol. 2004;62:71–82. doi: 10.1002/ajp.20008. [DOI] [PubMed] [Google Scholar]
- Justesen T, Nielsen OH. Krasilnikoff PA. Normal cultivable microflora in upper jejunal fluid in children without gastrointestinal disorders. J Pediatr Gastroenterol Nutr. 1984;3:683–686. doi: 10.1097/00005176-198411000-00007. [DOI] [PubMed] [Google Scholar]
- Kabeerdoss J, Shobana Devi R, Regina Mary R. Ramakrishna BS. Faecal microbiota composition in vegetarians: comparison with omnivores in a cohort of young women in southern India. Br J Nutr. 2012;108:953–957. doi: 10.1017/S0007114511006362. [DOI] [PubMed] [Google Scholar]
- Kageyama A. Benno Y. Emendation of genus Collinsella and proposal of Collinsella stercoris sp. nov. and Collinsella intestinalis sp. nov. Int J Syst Evol Microbiol. 2000;50:1767–1774. doi: 10.1099/00207713-50-5-1767. [DOI] [PubMed] [Google Scholar]
- Kageyama A, Benno Y. Nakase T. Phylogenetic and phenotypic evidence for the transfer of Eubacterium aerofaciens to the genus Collinsella as Collinsella aerofaciens gen. nov., comb. nov. Int J Syst Bacteriol. 1999a;49:557–565. doi: 10.1099/00207713-49-2-557. [DOI] [PubMed] [Google Scholar]
- Kageyama A, Benno Y. Nakase T. Phylogenetic evidence for the transfer of Eubacterium lentum to the genus Eggerthella as Eggerthella lenta gen. nov., comb. nov. Int J Syst Bacteriol. 1999b;49:1725–1732. doi: 10.1099/00207713-49-4-1725. [DOI] [PubMed] [Google Scholar]
- Kageyama A, Sakamoto M. Benno Y. Rapid identification and quantification of Collinsella aerofaciens using PCR. FEMS Microbiol Lett. 2000;183:43–47. doi: 10.1111/j.1574-6968.2000.tb08931.x. [DOI] [PubMed] [Google Scholar]
- Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S. Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- Kanauchi O, Matsumoto Y, Matsumura M, Fukuoka M. Bamba T. The beneficial effects of microflora, especially obligate anaerobes, and their products on the colonic environment in inflammatory bowel disease. Curr Pharm Des. 2005;11:1047–1053. doi: 10.2174/1381612053381675. [DOI] [PubMed] [Google Scholar]
- Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M. McSweeney CS. Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- Karlsson CLJ, Önnerfält J, Xu J, Molin G, Ahrné S. Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity. 2012;20:2257–2261. doi: 10.1038/oby.2012.110. [DOI] [PubMed] [Google Scholar]
- Kassinen A, Krogius-Kurikka L, Mäkivuokko H, Rinttilä T, Pulin L, Corander J, Malinen E, Apajalahti J. Palva A. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Katz M, Despommier DD. Gwadz RW. Parasitic Diseases. New York: Springer-Verlag; 1982. [Google Scholar]
- Keighley MR, Arabi Y, Dimock F, Burdon DW, Allan RN. Alexander-Williams J. Influence of inflammatory bowel disease on intestinal microflora. Gut. 1978;19:1099–1104. doi: 10.1136/gut.19.12.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerckhoffs APM, Samsom M, van der Rest ME, de Vogel J, Knol J, Ben-Amor K. Akkermans LM. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol. 2009;15:2887–2892. doi: 10.3748/wjg.15.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr M. Über die fusispirillare Symbiose, die Gattung Fusobacterium (K. B. Lehman) und Spirillum sputigenum. Die Gattung Fusobacterium. Zbl Bakt Parasit. 1922;87:536–545. [Google Scholar]
- Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlbrugge JHI. Die Atiologie der Aphthae tropicae. In: Cassel CM, editor. Archiv für Schiffs- und Tropen-Hygiene. Vol. 5. Leipzig: Johann Amrosius Barth; 1901. pp. 394–397. [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovatcheva-Datchary P. Wageningen, the Netherlands: University of Wageningen; 2010. Analyzing the functionality of the human intestinal microbiota by stable isotope probing. PhD. [Google Scholar]
- Kovatcheva-Datchary P, Egert M, Maathuis A, Rajilić-Stojanović M, de Graaf AA, Smidt H, de Vos WM. Venema K. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ Microbiol. 2009;11:914–926. doi: 10.1111/j.1462-2920.2008.01815.x. [DOI] [PubMed] [Google Scholar]
- Krienitz W. Über das auftreten von spirochaeten verschiedener form im magenihalt bei carcinoma ventriculi. Dtsch Med Wochenschr. 1906;32:872. [Google Scholar]
- Krogius-Kurikka L, Lyra A, Malinen E, Aarnikunnas J, Tuimala J, Paulin L, Makivuokko H, Kajander K. Palva A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruis W, Frič P, Pokrotnieks J, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004;53:1617–1623. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehbacher T, Rehman A, Lepage P, Hellmig S, Fölsch UR, Schreiber S. Ott SJ. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol. 2008;57:1569–1576. doi: 10.1099/jmm.0.47719-0. [DOI] [PubMed] [Google Scholar]
- Kumamoto CA. Vinces MD. Alternative Candida albicans lifestyles: growth on surfaces. Annu Rev Microbiol. 2005;49:113–133. doi: 10.1146/annurev.micro.59.030804.121034. [DOI] [PubMed] [Google Scholar]
- Lagier JC, Armougom F, Million M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012a;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- Lagier JC, Million M, Hugon P, Armougom F. Raoult D. Human gut microbiota: repertoire and variations. Front Cell Infect Microbiol. 2012b;2:136. doi: 10.3389/fcimb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti L, Salonen A, Kekkonen RA, Salojärvi J, Jalanka-Tuovinen J, Palva A, Orešič M. de Vos WM. Associations between the human intestinal microbiota, Lactobacillus rhamnosus GG and serum lipids indicated by integrated analysis of high-throughput profiling data. PeerJ. 2013;1:e32. doi: 10.7717/peerj.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarayanan B, Harris HMB, Coakley M, et al. Prevalence and characterization of Clostridium perfringens from the faecal microbiota of elderly Irish subjects. J Med Microbiol. 2013;62:457–466. doi: 10.1099/jmm.0.052258-0. [DOI] [PubMed] [Google Scholar]
- Lang E, Swiderski J, Stackebrandt E, Schumann P, Sproer C. Sahin N. Description of Ancylobacter oerskovii sp. nov. and two additional strains of Ancylobacter polymorphus. Int J Syst Evol Microbiol. 2008;58:1997–2002. doi: 10.1099/ijs.0.65666-0. [DOI] [PubMed] [Google Scholar]
- Larsen N, Vogensen FK, van den Berg FW, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, McNabb A, Woo GK, Hoang L, Fung AM, Chung LM, Woo PC. Yuen K-Y. Catabacter hongkongensis gen. nov., sp. nov., isolated from blood cultures of patients from Hong Kong and Canada. J Clin Microbiol. 2007;45:395–401. doi: 10.1128/JCM.01831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Fan RY, Lo H-W, et al. High mortality associated with Catabacter hongkongensis bacteremia. J Clin Microbiol. 2012;50:2239–2243. doi: 10.1128/JCM.00128-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson AJ, On SLW, Logan JMJ. Stanley J. Campylobacter hominis sp. nov., from the human gastrointestinal tract. Int J Syst Evol Microbiol. 2001;51:651–660. doi: 10.1099/00207713-51-2-651. [DOI] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Leitch EC, Walker AW, Duncan SH, Holtrop G. Flint HJ. Selective colonization of insoluble substrates by human faecal bacteria. Environ Microbiol. 2007;9:667–679. doi: 10.1111/j.1462-2920.2006.01186.x. [DOI] [PubMed] [Google Scholar]
- Lepage P, Häsler R, Spehlmann ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Levy AJ. A gastroenteritis outbreak probably due to a bovine strain of Vibrio. Yale J Biol Med. 1946;18:243–247. [PMC free article] [PubMed] [Google Scholar]
- Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD. Gordon JI. Obesity alters gut microbial ecology. P Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang C, Tang C, Li N. Li J. Molecular-phylogenetic characterization of the microbiota in ulcerated and non-ulcerated regions in the patients with Crohn's disease. PLoS One. 2012;7:e34939. doi: 10.1371/journal.pone.0034939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Finegold SM, Song Y. Lawson PA. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2008;58:1896–1902. doi: 10.1099/ijs.0.65208-0. [DOI] [PubMed] [Google Scholar]
- Looft T, Levine UY. Stanton TB. Cloacibacillus porcorum sp. nov., a mucin-degrading bacterium from the swine intestinal tract and emended description of the genus Cloacibacillus. Int J Syst Evol Microbiol. 2013;63:1960–1966. doi: 10.1099/ijs.0.044719-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Dupla M, Creus C, Navarro O. Raga X. Association of Gemella morbillorum endocarditis with adenomatous polyps and carcinoma of the colon: case report and review. Clin Infect Dis. 1996;22:379–380. doi: 10.1093/clinids/22.2.379. [DOI] [PubMed] [Google Scholar]
- Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJM, Garcia-Gil LJ. Flint HJ. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol. 2012;78:420–428. doi: 10.1128/AEM.06858-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK. Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Schleifer K-H, Whitman WB. Revised road map to the phylum Firmicutes. In: De Vos P, dGarrity G, Jones D, et al., editors. Bergey's Manual of Systematic Bacteriology. Vol. 3. New York: Springer-Verlag; 2009. pp. 1–13. The Firmicutes. [Google Scholar]
- Lyra A, Krogius-Kurikka L, Nikkila J, Malinen E, Kajander K, Kurikka K, Korpela R. Palva A. Effect of a multispecies probiotic supplement on quantity of irritable bowel syndrome-related intestinal microbial phylotypes. BMC Gastroenterol. 2010;10:110. doi: 10.1186/1471-230X-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum WG. Hastings TW. A case of acute endocarditis caused by Micrococcus zymogenes (nov. spec.), with a description of the microorganism. J Exp Med. 1899;4:521–534. doi: 10.1084/jem.4.5-6.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, Cummings JH. Allison C. Protein degradation by human intestinal bacteria. J Gen Microbiol. 1986;132:1647–1656. doi: 10.1099/00221287-132-6-1647. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Allison C, Gibson SA. Cummings JH. Contribution of the microflora to proteolysis in the human large intestine. J Appl Bacteriol. 1988;64:37–46. doi: 10.1111/j.1365-2672.1988.tb02427.x. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Furrie E, Cummings JH. Macfarlane GT. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin Infect Dis. 2004;38:1690–1699. doi: 10.1086/420823. [DOI] [PubMed] [Google Scholar]
- MacKay WG, Williams CL, McMillan M, Ndip RN, Shepherd AJ. Weaver LT. Evaluation of protocol using gene capture and PCR for detection of Helicobacter pylori DNA in feces. J Clin Microbiol. 2003;41:4589–4593. doi: 10.1128/JCM.41.10.4589-4593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan JC, Salari RC, Saxena D, et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed. 2012;97:F456–F462. doi: 10.1136/fetalneonatal-2011-301373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee EA, Richardson CJ, Hughes R. Cummings JH. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am J Clin Nutr. 2000;72:1488–1494. doi: 10.1093/ajcn/72.6.1488. [DOI] [PubMed] [Google Scholar]
- Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, Krogius L, Saarela M, Korpela R. Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- Malinen E, Krogius-Kurikka L, Lyra A, Nikkilä J, Jääskeläinen A, Rinttilä T, Vilpponen-Salmela T, von Wright AJ. Palva A. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol. 2010;16:4532–4540. doi: 10.3748/wjg.v16.i36.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnick H. Anaerobiospirillum thomasii sp. nov., an anaerobic spiral bacterium isolated from the feces of cats and dogs and from diarrheal feces of humans, and emendation of the genus Anaerobiospirillum. Int J Syst Bacteriol. 1997;47:381–384. doi: 10.1099/00207713-47-2-381. [DOI] [PubMed] [Google Scholar]
- Marchandin H, Teyssier C, Campos J, Jean-Pierre H, Roger F, Gay B, Carlier J-P. Jumas-Bilak E. Negativicoccus succinicivorans gen. nov., sp. nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales ord. nov. and Acidaminococcaceae fam. nov. in the bacterial phylum Firmicutes. Int J Syst Evol Microbiol. 2010;60:1271–1279. doi: 10.1099/ijs.0.013102-0. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Erica D, Sonnenburg, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo T, Sakamoto M, Ito C, Toda T. Benno Y. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol. 2008;58:1221–1227. doi: 10.1099/ijs.0.65404-0. [DOI] [PubMed] [Google Scholar]
- Matthies A, Blaut M. Braune A. Isolation of a human intestinal bacterium capable of daidzein and genistein conversion. Appl Environ Microbiol. 2009;75:1740–1744. doi: 10.1128/AEM.01795-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald IJ. Johnson KG. Nutritional requirements of some non-pathogenic Neisseria grown in simple synthetic media. Can J Microbiol. 1975;21:1198–1204. doi: 10.1139/m75-179. [DOI] [PubMed] [Google Scholar]
- McLaughlin SD, Walker AW, Churcher C, et al. The bacteriology of pouchitis: a molecular phylogenetic analysis using 16S rRNA gene cloning and sequencing. Ann Surg. 2010;252:90–98. doi: 10.1097/SLA.0b013e3181e3dc8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mégraud F, Bébéar C, Dabernat H. Delmas C. Haemophilus species in the human gastrointestinal tract. Eur J Clin Microbiol Infect Dis. 1988;7:437–438. doi: 10.1007/BF01962361. [DOI] [PubMed] [Google Scholar]
- Metchnikoff E. Etude sur la flore intestinale. Ann Inst Pasteur. 1908;22:929–955. [Google Scholar]
- Mevissen-Verhage EA, Marcelis JH, de Vos MN, Harmsen-van Amerongen WC. Verhoef J. Bifidobacterium, Bacteroides, and Clostridium spp. in fecal samples from breast-fed and bottle-fed infants with and without iron supplement. J Clin Microbiol. 1987;25:285–289. doi: 10.1128/jcm.25.2.285-289.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TL. Wolin MJ. Methanosphaera stadtmanae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen. Arch Microbiol. 1985;141:116–122. doi: 10.1007/BF00423270. [DOI] [PubMed] [Google Scholar]
- Miller TL. Wolin MJ. Methanogens in human and animal tracts. Syst Appl Microbiol. 1986;7:223–229. [Google Scholar]
- Mitsuoka T. Bifidobacteria and their role in human health. J Ind Microbiol Biotechnol. 1990;6:263–267. [Google Scholar]
- Mitsuoka T. The human gastrointestinal tract. In: Wood BJB, editor. The Lactic Acid Bacteria in Health and Disease. Vol. 1. Essex: Elsevier Science Publishers, Ltd; 1992. pp. 69–114. [Google Scholar]
- Moore WEC. Holdeman LV. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974a;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WEC. Holdeman LV. Special problems associated with the isolation and identification of intestinal bacteria in fecal flora studies. Am J Clin Nutr. 1974b;27:1450–1455. doi: 10.1093/ajcn/27.12.1450. [DOI] [PubMed] [Google Scholar]
- Moore WEC. Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WEC, Cato EP. Holdeman LV. Ruminococcus bromii sp. n. and emendation of the description of Ruminococcus Sijpestein. Int J Syst Bacteriol. 1972;22:78–80. [Google Scholar]
- Moro E. Ueber den Bacillus acidophilus. Jahrb Kinderh. 1900;52:38–55. [Google Scholar]
- Morotomi M, Nagai F, Sakon H. Tanaka R. Paraprevotella clara gen. nov., sp. nov. and Paraprevotella xylaniphila sp. nov., members of the family ‘Prevotellaceae’ isolated from human faeces. Int J Syst Evol Microbiol. 2009;59:1895–1900. doi: 10.1099/ijs.0.008169-0. [DOI] [PubMed] [Google Scholar]
- Morotomi M, Nagai F. Watanabe Y. Description of Christensenella minuta gen. nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. nov. Int J Syst Evol Microbiol. 2012;62:144–149. doi: 10.1099/ijs.0.026989-0. [DOI] [PubMed] [Google Scholar]
- Mukhopadhya I, Hansen R, Nicholl CE, et al. A comprehensive evaluation of colonic mucosal isolates of Sutterella wadsworthensis from inflammatory bowel disease. PLoS One. 2011;6:e27076. doi: 10.1371/journal.pone.0027076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller HE. Occurrence and pathogenic role of Morganella–Proteus–Providencia group bacteria in human feces. J Clin Microbiol. 1986;23:404–405. doi: 10.1128/jcm.23.2.404-405.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday PE, Furr PM. Taylor-Robinson D. The prevalence of Ureaplasma urealyticum and Mycoplasma hominis in the cervix and anal canal of women. J Infect. 1981;3:253–257. doi: 10.1016/s0163-4453(81)90888-4. [DOI] [PubMed] [Google Scholar]
- Mungthin M, Subrungruang I, Naaglor T, Aimpun P, Areekul W. Leelayoova S. Spore shedding pattern of Enterocytozoon bieneusi in asymptomatic children. J Med Microbiol. 2005;54:473–476. doi: 10.1099/jmm.0.45832-0. [DOI] [PubMed] [Google Scholar]
- Murdoch DA. Shah HN. Reclassification of Peptostreptococcus magnus (Prevot 1933) Holdeman and Moore 1972 as Finegoldia magna comb. nov. and Peptostreptococcus micros (Prevot 1933) Smith 1957 as Micromonas micros comb. nov. Anaerobe. 1999;5:555–559. [Google Scholar]
- Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones F, Cardona F, Soriguer F. Queipo-Ortuno MI. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case–control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai F, Morotomi M, Sakon H. Tanaka R. Parasutterella excrementihominis gen. nov., sp. nov., a member of the family Alcaligenaceae isolated from human faeces. Int J Syst Evol Microbiol. 2009;59:1793–1797. doi: 10.1099/ijs.0.002519-0. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Kobayashi T, Tanaka S, Korenori Y, Tateyama A, Sakamoto N, Kiyohara C, Shirakawa T. Sonomoto K. Aberrant structures of fecal bacterial community in allergic infants profiled by 16S rRNA gene pyrosequencing. FEMS Immunol Med Microbiol. 2011;63:397–406. doi: 10.1111/j.1574-695X.2011.00872.x. [DOI] [PubMed] [Google Scholar]
- Nakazawa F, Sato M, Poco SE, Hashimura T, Ikeda T, Kalfas S, Sundqvist G. Hoshino E. Description of Mogibacterium pumilum gen. nov., sp. nov. and Mogibacterium vescum gen. nov., sp. nov., and reclassification of Eubacterium timidum (Holdeman et al. 1980) as Mogibacterium timidum gen. nov., comb. nov. Int J Syst Evol Microbiol. 2000;50:679–688. doi: 10.1099/00207713-50-2-679. [DOI] [PubMed] [Google Scholar]
- Nam Y-D, Chang H-W, Kim K-H, et al. Bacterial, archaeal, and eukaryal diversity in the intestines of Korean people. J Microbiol. 2008;46:491–501. doi: 10.1007/s12275-008-0199-7. [DOI] [PubMed] [Google Scholar]
- Narushima S, Itoh K, Miyamoto Y, Park S-H, Nagata K, Kuruma K. Uchida K. Deoxycholic acid formation in gnotobiotic mice associated with human intestinal bacteria. Lipids. 2006;41:835–843. doi: 10.1007/s11745-006-5038-1. [DOI] [PubMed] [Google Scholar]
- Negroni A, Costanzo M, Vitali R, et al. Characterization of adherent-invasive Escherichia coli isolated from pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:913–924. doi: 10.1002/ibd.21899. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinstock GM, Highlander SK, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottingham PM. Hungate RE. Isolation of methanogenic bacteria from feces of man. J Bacteriol. 1968;96:2178–2179. doi: 10.1128/jb.96.6.2178-2179.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ó Cuív P, Klaassens ES, Smith WJ, et al. Draft genome sequence of Enterococcus faecalis PC1.1, a candidate probiotic strain isolated from human feces. Genome Announc. 2013;1:e00160–00112. doi: 10.1128/genomeA.00160-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Goto Y, Kunisawa J, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. P Natl Acad Sci USA. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki K, Kudo Y. Watanabe K. Lactobacillus saniviri sp. nov. and Lactobacillus senioris sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2012;62:601–607. doi: 10.1099/ijs.0.031658-0. [DOI] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, Timmis KN. Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott SJ, Kühbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- Ou G, Hedberg M, Hörstedt P, et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am J Gastroenterol. 2009;104:3058–3067. doi: 10.1038/ajg.2009.524. [DOI] [PubMed] [Google Scholar]
- Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, Gaskins HR. O'Keefe SJ. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98:111–120. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxley AP, Lanfranconi MP, Würdemann D, Ott S, Schreiber S, McGenity TJ, Timmis KN. Nogales B. Halophilic archaea in the human intestinal mucosa. Environ Microbiol. 2010;12:2398–2410. doi: 10.1111/j.1462-2920.2010.02212.x. [DOI] [PubMed] [Google Scholar]
- Ozaki E, Kato H, Kita H, et al. Clostridium difficile colonization in healthy adults: transient colonization and correlation with enterococcal colonization. J Med Microbiol. 2004;53:167–172. doi: 10.1099/jmm.0.05376-0. [DOI] [PubMed] [Google Scholar]
- Paliy O, Kenche H, Abernathy F. Michail S. High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl Environ Microbiol. 2009;75:3572–3579. doi: 10.1128/AEM.02764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, Digiulio DB, Relman DA. Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey PK, Verma P, Kumar H, Bavdekar A, Patole MS. Shouche YS. Comparative analysis of fecal microflora of healthy full-term Indian infants born with different methods of delivery (vaginal vs cesarean): Acinetobacter sp. prevalence in vaginally born infants. J Biosci. 2012;37:989–998. doi: 10.1007/s12038-012-9268-5. [DOI] [PubMed] [Google Scholar]
- Parr LE. Intestinal spirochetosis. J Infect Dis. 1923;33:369–383. [Google Scholar]
- Passini F. Studien uber faulnisserregende anaerobe Bakterien des normalen menschlichen Darmes und ihre Bedeutung. Z Hyg Infektionskr. 1905;49:135. [Google Scholar]
- Peris-Bondia F, Latorre A, Artacho A, Moya A. D'Auria G. The active human gut microbiota differs from the total microbiota. PLoS One. 2011;6:e22448. doi: 10.1371/journal.pone.0022448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit L, Gibert M. Popoff MR. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 1999;7:104. doi: 10.1016/s0966-842x(98)01430-9. [DOI] [PubMed] [Google Scholar]
- Petruschky J. Bacillus faecalis alcaligenes (n.sp.) Zentbl Bakteriol Parasitenk Infektionskr Hyg Abt I. 1896;19:187–191. [Google Scholar]
- Pfleiderer A, Lagier JC, Armougom F, Robert C, Vialettes B. Raoult D. Culturomics identified 11 new bacterial species from a single anorexia nervosa stool sample. Eur J Clin Microbiol Infect Dis. 2013;32:1471–1481. doi: 10.1007/s10096-013-1900-2. [DOI] [PubMed] [Google Scholar]
- Pitarangsi C, Echeverria P, Whitmire R, Tirapat C, Formal S, Dammin GJ. Tingtalapong M. Enteropathogenicity of Aeromonas hydrophila and Plesiomonas shigelloides: prevalence among individuals with and without diarrhea in Thailand. Infect Immun. 1982;35:666–673. doi: 10.1128/iai.35.2.666-673.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemiers S. Verstraete W. Oestrogenicity of prenylflavonoids from hops: activation of pro-oestrogens by intestinal bacteria. Environ Microbiol Rep. 2009;1:100–109. doi: 10.1111/j.1758-2229.2009.00011.x. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J. Glöckner FO. silva: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M. Function of the microbiota. Best Pract Res Clin Gastroenterol. 2013;27:5–16. doi: 10.1016/j.bpg.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Smidt H. de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, de Vos WM. Zoetendal EG. The human intestinal microbiota and its impact on human health. In: Zengler K, editor; Accessing Uncultivated Microorganisms: From the Environment to Organisms and Genomes and Back. Herndon, VA: ASM Press; 2008. pp. 11–32. [Google Scholar]
- Rajilić-Stojanović M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S. de Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Guarner F, Shanahan F. de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013a;19:481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Heilig HG, Tims S, Zoetendal EG. de Vos WM. Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol. 2013b;15:1146–1159. doi: 10.1111/1462-2920.12023. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S. Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Reuter G. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr Issues Intest Microbiol. 2001;2:43–53. [PubMed] [Google Scholar]
- Rieu-Lesme F, Delbès C. Sollelis L. Recovery of partial 16S rDNA sequences suggests the presence of Crenarchaeota in the human digestive ecosystem. Curr Microbiol. 2005;51:317–321. doi: 10.1007/s00284-005-0036-8. [DOI] [PubMed] [Google Scholar]
- Ringel-Kulka T, Cheng J, Ringel Y, Salojärvi J, Carroll I, Palva A, de Vos WM. Satokari R. Intestinal microbiota in healthy U.S. young children and adults – a high throughput microarray analysis. PLoS One. 2013;8:e64315. doi: 10.1371/journal.pone.0064315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger WEW. Macfarlane GT. A role for intestinal mycoplasmas in the aetiology of Crohn's disease? J Appl Microbiol. 2002;92:377–381. doi: 10.1046/j.1365-2672.2002.01531.x. [DOI] [PubMed] [Google Scholar]
- Rogosa M. The Genus Veillonella IV. Serological groupings, and genus and species emendations. J Bacteriol. 1965;90:704–709. doi: 10.1128/jb.90.3.704-709.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogosa M, Mitchell JA. Wiseman RF. A selective medium for the isolation and enumeration of oral and fecal lactobacilli. J Bacteroil. 1951;62:132–133. doi: 10.1128/jb.62.1.132-133.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron EZ. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr Opin Microbiol. 2006;9:28. doi: 10.1016/j.mib.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Roos S, Engstrand L. Jonsson H. Lactobacillus gastricus sp. nov., Lactobacillus antri sp. nov., Lactobacillus kalixensis sp. nov. and Lactobacillus ultunensis sp. nov., isolated from human stomach mucosa. Int J Syst Evol Microbiol. 2005;55:77–82. doi: 10.1099/ijs.0.63083-0. [DOI] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA. Mazmanian SK. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M, Ohkuma M. Reclassification of Xylanibacter oryzae Ueki et al. 2006 as Prevotella oryzae comb. nov., with an emended description of the genus Prevotella. Int J Syst Evol Microbiol. 2012;62:2637–2642. doi: 10.1099/ijs.0.038638-0. [DOI] [PubMed] [Google Scholar]
- Saklatvala T. Milestones in parasitology. Parasitol Today. 1993;9:347–348. [Google Scholar]
- Salvetti E, Felis GE, Dellaglio F, Castioni A, Torriani S. Lawson PA. Reclassification of Lactobacillus catenaformis (Eggerth 1935) Moore and Holdeman 1970 and Lactobacillus vitulinus Sharpe et al. 1973 as Eggerthia catenaformis gen. nov., comb. nov. and Kandleria vitulina gen. nov., comb. nov., respectively. Int J Syst Evol Microbiol. 2011;61:2520–2524. doi: 10.1099/ijs.0.029231-0. [DOI] [PubMed] [Google Scholar]
- Sanborn AG. The faecal flora of adults, with particular attention to individual differences and their relationship to the effects of various diets. J Infect Dis. 1931;48:541–569. [Google Scholar]
- Santelmann H. Howard JM. Yeast metabolic products, yeast antigens and yeasts as possible triggers for irritable bowel syndrome. Eur J Gastroenterol Hapatol. 2005;17:21–26. doi: 10.1097/00042737-200501000-00005. [DOI] [PubMed] [Google Scholar]
- Satokari RM, Vaughan EE, Akkermans ADL, Saarela M. de Vos WM. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67:504–513. doi: 10.1128/AEM.67.2.504-513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier DM, Riehle K, Mistretta T-A, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savini V, Catavitello C, Masciarelli G, Astolfi D, Balbinot A, Bianco A, Febbo F, D'Amario C. D'Antonio D. Drug sensitivity and clinical impact of members of the genus Kocuria. J Med Microbiol. 2010;59:1395–1402. doi: 10.1099/jmm.0.021709-0. [DOI] [PubMed] [Google Scholar]
- Scanlan PD. Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2:1183–1193. doi: 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- Scardovi V. Trovatelli LD. Bifidobacterium animalis (Mitsuoka) comb. nov. and the ‘minimum’ and ‘subtile’ groups of new bifidobacteria found in sewage. Int J Syst Evol Microbiol. 1974;24:21–28. [Google Scholar]
- Schnorr SL, Candela M, Rampelli S, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J. Sonnenborn U. Yeasts in the gut: from commensals to infectious agents. Dtsch Arztebl Int. 2009;106:837–842. doi: 10.3238/arztebl.2009.0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Chen YA. Tuohy KM. A comparative in vitro investigation into the effects of cooked meats on the human faecal microbiota. Anaerobe. 2010;16:572–577. doi: 10.1016/j.anaerobe.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Sherman JM. The enterococci and related streptococci. J Bacteriol. 1938;35:81–93. doi: 10.1128/jb.35.2.81-93.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman ST, Friedmann HC. Sims RH. Theodor Escherich: the first pediatric infectious diseases physician? Clin Infect Dis. 2007;45:1025–1029. doi: 10.1086/521946. [DOI] [PubMed] [Google Scholar]
- Sibartie S, Scully P, Keohane J, O'Neill S, O'Mahony J, O'Hanlon D, Kirwan WO, O'Mahony L. Shanahan F. Mycobacterium avium subsp. paratuberculosis (MAP) as a modifying factor in Crohn's disease. Inflamm Bowel Dis. 2010;16:296–304. doi: 10.1002/ibd.21052. [DOI] [PubMed] [Google Scholar]
- Simon GL. Gorbach SL. Intestinal flora in health and disease. Gastroenterology. 1984;86:174–193. [PubMed] [Google Scholar]
- Simon GL. Gorbach SL. The human intestinal microflora. Dig Dis Sci. 1986;31:147S–162S. doi: 10.1007/BF01295996. [DOI] [PubMed] [Google Scholar]
- Smidt H. de Vos WM. Anaerobic microbial dehalogenation. Annu Rev Microbiol. 2004;58:43–73. doi: 10.1146/annurev.micro.58.030603.123600. [DOI] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, Podglajen I, Marteau P. Doré J. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. P Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís G, de los Reyes-Gavilan CG, Fernández N, Margolles A. Gueimonde M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. 2010;16:307–310. doi: 10.1016/j.anaerobe.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Song KB, Atkinson C, Frankenfeld CL, Jokela T, Wähälä K, Thomas WK. Lampe JW. Prevalence of daidzein-metabolizing phenotypes differs between Caucasian and Korean American women and girls. J Nutr. 2006;136:1347–1351. doi: 10.1093/jn/136.5.1347. [DOI] [PubMed] [Google Scholar]
- Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH. Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller R. Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology. 2009;136:1979–1988. doi: 10.1053/j.gastro.2009.02.074. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Kramer I, Swiderski J. Hippe H. Phylogenetic basis for a taxonomic dissection of the genus Clostridium. FEMS Immunol Med Microbiol. 1999;24:253–258. doi: 10.1111/j.1574-695X.1999.tb01291.x. [DOI] [PubMed] [Google Scholar]
- Standaert-Vitse A, Sendi B, Joossens M, et al. Candida albicans colonization and ASCA in familial Crohn's disease. Am J Gastroenterol. 2009;104:1745–1753. doi: 10.1038/ajg.2009.225. [DOI] [PubMed] [Google Scholar]
- Stearns JC, Lynch MDJ, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G. Neufelda JD. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suau A, Bonnet R, Sutren M, Godon J-J, Gibson GR, Collins MD. Doré J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Dörffel Y, Loening-Baucke V, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- Tamura H, Yamashita S, Kusano N, Suzuki C, Yamaguchi Y, Tanigawa K, Masuhara M, Okita K. Murakami F. Fulminant hepatitis complicated by small intestine infection and massive hemorrhage. J Gastroenterol. 1988;33:412–418. doi: 10.1007/s005350050105. [DOI] [PubMed] [Google Scholar]
- Tana C, Umesaki Y, Imaoka A, Handa T, Kanazawa M. Fukudo S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–519. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- Tandon BN, Tandon RK, Satpathy BK. Shriniwas Mechanism of malabsorption in giardiasis: a study of bacterial flora and bile salt deconjugation in upper jejunum. Gut. 1977;18:176–181. doi: 10.1136/gut.18.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW. Normal Microflora: An Introduction to Microbes Inhabiting the Human Body. London: Charpman & Hill; 1995. [Google Scholar]
- Tannock GW. Analysis of the intestinal microflora: a renaissance. Antonie Van Leeuwenhoek. 1999;76:265–278. [PubMed] [Google Scholar]
- Tannock GW. A special fondness for lactobacilli. Appl Environ Microbiol. 2004;70:3189–3194. doi: 10.1128/AEM.70.6.3189-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW, Lawley B, Munro K, et al. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl Environ Microbiol. 2013;79:3040–3048. doi: 10.1128/AEM.03910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap J, Mondot S, Levenez F, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- Taras D, Simmering R, Collins MD, Lawson PA. Blaut M. Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2002;52:423–428. doi: 10.1099/00207713-52-2-423. [DOI] [PubMed] [Google Scholar]
- Taylor GR, Kropp KD. Molina TC. Nine-year microflora study of an isolator-maintained immunodeficient child. Appl Environ Microbiol. 1985;50:1349–1356. doi: 10.1128/aem.50.6.1349-1356.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielman NM. Guerrant RL. Clinical practice: acute infectious diarrhea. N Engl J Med. 2004;350:38–47. doi: 10.1056/NEJMcp031534. [DOI] [PubMed] [Google Scholar]
- Thompson-Chagoyan OC, Fallani M, Maldonado J, Vieites JM, Khanna S, Edwards C, Doré J. Gil A. Faecal microbiota and short-chain fatty acid levels in faeces from infants with cow's milk protein allergy. Int Arch Allergy Immunol. 2011;156:325–332. doi: 10.1159/000323893. [DOI] [PubMed] [Google Scholar]
- Tindall BJ. Euzeby JP. Proposal of Parvimonas gen. nov. and Quatrionicoccus gen. nov. as replacements for the illegitimate, prokaryotic, generic names Micromonas Murdoch and Shah 2000 and Quadricoccus Maszenan et al. 2002, respectively. Int J Syst Evol Microbiol. 2006;56:2711–2713. doi: 10.1099/ijs.0.64338-0. [DOI] [PubMed] [Google Scholar]
- Tissier MH. Recherches sur la flore intestinale normale et pathologique du nourrisson. Paris: Medicine. Université de Paris; 1900. [Google Scholar]
- Tissier MH. Recherches sur la flore intestinale normale des enfants ages dún an a cin. ans. Ann Inst Pasteur. 1908;22:189–207. [Google Scholar]
- Tito RY, Knights D, Metcalf J, et al. Insights from characterizing extinct human gut microbiomes. PLoS One. 2012;7:e51146. doi: 10.1371/journal.pone.0051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER. Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R. Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baarlen P, Troost FJ, van Hemert S, van der Meer C, de Vos WM, de Groot PJ, Hooiveld GJ, Brummer R-JM. Kleerebezem M. Differential NF-κB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. P Natl Acad Sci USA. 2009;106:2371–2376. doi: 10.1073/pnas.0809919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pas BA, Harmsen HJ, Raangs GC, de Vos WM, Schraa G. Stams AJ. A Desulfitobacterium strain isolated from human feces that does not dechlorinate chloroethenes or chlorophenols. Arch Microbiol. 2001;176:391–392. doi: 10.1007/s002030100276. [DOI] [PubMed] [Google Scholar]
- van den Bogert B, de Vos WM, Zoetendal EG. Kleerebezem M. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol. 2011;77:2071–2080. doi: 10.1128/AEM.02477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eldere JR, De Pauw G. Eyssen HJ. Steroid sulfatase activity in a Peptococcus niger strain from the human intestinal microflora. Appl Environ Microbiol. 1987;53:1655–1660. doi: 10.1128/aem.53.7.1655-1660.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Passel MWJ, Kant R, Palva A, et al. Genome sequence of Victivallis vadensis ATCC BAA-548, an anaerobic bacterium from the phylum Lentisphaerae, isolated from the human gastrointestinal tract. J Bacteriol. 2011;193:2373–2374. doi: 10.1128/JB.00271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R. De Ley J. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol. 1991;41:88–103. doi: 10.1099/00207713-41-1-88. [DOI] [PubMed] [Google Scholar]
- Veillon A. Zuber A. Recherches sur quelques microbes strictement anaerobies et leur role en pathologie. Arch Med Exp Anat Pathol. 1898;10:517–545. [Google Scholar]
- Veron M. Chatelain R. Taxonomic study of the genus Campylobacter Sebald and Veron and designation of the neotype strain for the type species Campylobacter fetus (Smith and Taylor) Sebald and Veron. Int J Syst Bacteriol. 1973;23:122–134. [Google Scholar]
- Vigsnæs LK, Brynskov J, Steenholdt C, Wilcks A. Licht TR. Gram-negative bacteria account for main differences between faecal microbiota from patients with ulcerative colitis and healthy controls. Benef Microbes. 2012;3:287–297. doi: 10.3920/BM2012.0018. [DOI] [PubMed] [Google Scholar]
- Vipperla K. O'Keefe SJ. The microbiota and its metabolites in colonic mucosal health and cancer risk. Nutr Clin Pract. 2012;27:624–635. doi: 10.1177/0884533612452012. [DOI] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Ahrne S, Jeppsson B. Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54:219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT. Conlon MA. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microbiol. 2011a;77:6718–6721. doi: 10.1128/AEM.05212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011b;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S. Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber R, Sauer B, Spycher MA, Deplazes P, Keller R, Ammann R, Briner J. Lüthy R. Detection of Septata intestinalis in stool specimens and coprodiagnostic monitoring of successful treatment with albendazole. Clin Infect Dis. 1994;19:342–345. doi: 10.1093/clinids/19.2.342. [DOI] [PubMed] [Google Scholar]
- Wei B, Huang T, Dalwadi H, Sutton CL, Bruckner D. Braun J. Pseudomonas fluorescens encodes the Crohn's disease-associated I2 sequence and T-Cell superantigen. Infect Immun. 2002;70:6567–6575. doi: 10.1128/IAI.70.12.6567-6575.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch WH. Nuttall GHF. A gas producing bacillus (Bacillus aerogenes capsulatus, nov. spec.) capable of rapid development in the blood vessels after death. Bull Johns Hopkins Hosp. 1892;3:81. [Google Scholar]
- Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, Angenent LT, Knight R. Ley RE. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2012;6:94–103. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerman LJ, Stel HV, Schipper MEI, et al. Development of a real-time PCR for identification of Brachyspira species in human colonic biopsies. PLoS One. 2012;7:e52281. doi: 10.1371/journal.pone.0052281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichro E, Hoelzl D, Krause R, Bertha G, Reinthaler F. Wenisch C. Microsporidiosis in travel-associated chronic diarrhea in immune-competent patients. Am J Trop Med Hyg. 2005;73:285–287. [PubMed] [Google Scholar]
- Wilkins TD, Moore WEC, West SEH. Holdeman LV. Peptococcus niger (Hall) Kluyver and van Niel 1936: emendation of description and designation of neotype strain. Int J Syst Bacteriol. 1975;25:47–49. [Google Scholar]
- Willems A. Collins MD. Phylogenetic relationships of the genera Acetobacterium and Eubacterium sensu stricto and reclassification of Eubacterium alactolyticum as Pseudoramibacter alactolyticus gen. nov., comb. nov. Int J Syst Bacteriol. 1996;46:1083–1087. doi: 10.1099/00207713-46-4-1083. [DOI] [PubMed] [Google Scholar]
- Williams BL, Hornig M, Parekh T. Lipkin WI. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. mBio. 2012;3:e00261–00211. doi: 10.1128/mBio.00261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KH. Blitchington RB. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender G, Stepniak D, Krebs P, Lin L, McBride S, Wei B, Braun J, Mazmanian SK. Kronenberg M. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143:418–428. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O. Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. P Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS. Giardiasis. Clin Microbiol Rev. 1992;5:93–100. doi: 10.1128/cmr.5.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-C, Johnson JL, Moore WEC. Moore LVH. Emended descriptions of Prevotella denticola Prevotella loescheii Prevotella veroralis, and Prevotella melaninogenica. Int J Syst Bacteriol. 1992;42:536–541. doi: 10.1099/00207713-42-4-536. [DOI] [PubMed] [Google Scholar]
- Wu L, Estrada O, Zaborina O, et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;309:774–777. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer K-H, Ludwig W, Glöckner FO. Rosselló-Móra R. The all-species living tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S. Suzuki T. Isolation and characterization of a novel equol-producing bacterium from human feces. Biosci Biotechnol Biochem. 2008;72:2660–2666. doi: 10.1271/bbb.80329. [DOI] [PubMed] [Google Scholar]
- Yuan J-P, Wang J-H. Liu X. Metabolism of dietary soy isoflavones to equol by human intestinal microflora – implications for health. Mol Nutr Food Res. 2007;51:765–781. doi: 10.1002/mnfr.200600262. [DOI] [PubMed] [Google Scholar]
- Zaborina O, Kohler J, Wang Y, Bethel C, Shevchenko O, Wu L, Turner J. Alverdy J. Identification of multi-drug resistant Pseudomonas aeruginosa clinical isolates that are highly disruptive to the intestinal epithelial barrier. Ann Clin Microbiol Antimicrob. 2006;5:14. doi: 10.1186/1476-0711-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamakhchari M, Wei G, Dewhirst F, Lee J, Schuppan D, Oppenheim FG. Helmerhorst EJ. Identification of Rothia bacteria as gluten-degrading natural colonizers ofthe upper gastro-intestinal tract. PLoS One. 2011;6:e24455. doi: 10.1371/journal.pone.0024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierdt CH. Blastocystis hominis–past and future. Clin Microbiol Rev. 1991;4:61–79. doi: 10.1128/cmr.4.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermans AD. de Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermans AD, Akkermans-van Vliet WM, de Visser AJGM. de Vos WM. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb Ecol Health Dis. 2001;13:129–134. [Google Scholar]
- Zoetendal EG, Ben-Amor K, Harmsen HJM, Schut F, Akkermans ADL. de Vos WM. Quantification of uncultured Ruminococcus obeum-like bacteria in human fecal samples by fluorescent In situ hybridization and flow cytometry using 16S rRNA-targeted probes. Appl Environ Microbiol. 2002;68:4225–4232. doi: 10.1128/AEM.68.9.4225-4232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Plugge CM, Akkermans ADL. de Vos WM. Victivallis vadensis gen. nov., sp. nov., a sugar-fermenting anaerobe from human faeces. Int J Syst Evol Microbiol. 2003;53:211–215. doi: 10.1099/ijs.0.02362-0. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Rajilić-Stojanović M, de Vos M. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Raes J, van den Bogert B, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of bacterial species that can be members of the human gastrointestinal microbiota, with a reference that provides the link between the species and gastrointestinal tract as ecological niche.
Table S2. List of archaeal species that can be members of the human gastrointestinal microbiota, with a reference that provides the link between the species and gastrointestinal tract as ecological niche.
Table S3. List of eukaryal species that can be members of the human gastrointestinal microbiota, with a reference that provides the link between the species and gastrointestinal tract as ecological niche.