Abstract
Several EEG parameters are potential endophenotypes for different psychiatric disorders. The present study consists of a comprehensive behavioral- and molecular-genetic analysis of such parameters in a large community sample (N = 4,026) of adolescent twins and their parents, genotyped for 527,829 single nucleotide polymorphisms (SNPs). Biometric heritability estimates ranged from .49 to .85, with a median of .78. The additive effect of all SNPs (SNP heritability) varied across electrodes. Although individual SNPs were not significantly associated with EEG parameters, several genes were associated with delta power. We also obtained an association between the GABRA2 gene and beta power (p < .014), consistent with findings reported by others, although this did not survive Bonferroni correction. If EEG parameters conform to a largely polygenic model of inheritance, larger sample sizes will be required to detect individual variants reliably.
Keywords: EEG, endophenotypes, genome-wide association study, molecular genetics, heritability, GCTA, gene-based tests
Since Berger (1929) first documented the alpha brain rhythm, the electroencephalogram (EEG) has been widely used in a variety of clinical and research contexts. Distinct parameters of the EEG signal have proven diagnostic of certain neurological disorders such as epilepsy, or states of arousal such as sleep or response to anesthesia. EEG activity is often measured in terms of the amplitude of oscillations at differing frequencies, which have typically been grouped into different bands, most commonly delta (0.1 – 4 Hz), theta (4 – 8 Hz), alpha (8 – 13 Hz), and beta (13 – 30 Hz). Changes in the amplitude of activity within specific bands are robustly found to be associated with variations in overall arousal level as well as with different cognitive or perceptual processes. EEG characteristics of neuropsychiatric disorders are subtle, typically consisting primarily of quantitative changes in the amplitude of activity occurring at particular frequencies or over specific scalp locations corresponding to differing areas of cortex. Nevertheless, EEG recording is attractive for use in psychiatry (McLoughlin, Makeig, & Tsuang, 2013), particularly as a means of identifying neurobiological indicators that link psychiatric disorders to genetic risk factors (i.e. endophenotypes; Iacono & Malone, 2011).
The present study is an investigation of the heritable and molecular-genetic basis of several parameters of EEG activity in a large, population-based sample, motivated in part by notions that EEG measures may serve as endophenotypes for different psychiatric disorders. We begin by reviewing evidence that EEG parameters satisfy the criteria for an endophenotype that we have recently articulated (Iacono & Malone, 2011). Because an endophenotype can shed light on the neural and psychological processes implicated in its associated disorder, we then briefly summarize the correlates of different parameters of EEG activity. We conclude the introduction by summarizing extant genetic research on EEG parameters before describing the current study.
EEG parameters as endophenotypes
Endophenotypes are laboratory-based measures of endogenous characteristics of an individual that reflect genetic risk for a specific disorder or spectrum of disorders. One essential criterion for an endophenotype is strong psychometric properties, such as reliability of measurement, and existing research indicates that EEG parameters qualify. Split-half reliability estimates have been found uniformly to approach 1 for measures of EEG power in different frequency ranges (Kondacs & Szabo, 1999), a measure of the energy in a signal, and within-session correlations are relatively robust to the amount of contamination by eye movement activity (Gasser, Bacher, & Steinberg, 1985). Test-retest stability estimates are typically greater than .80 over two- to four-week (Gudmundsson, Runarsson, Sigurdsson, Eiriksdottir, & Johnsen, 2007; Salinsky, Oken, & Morehead, 1991), and have been reported to exceed .70, at least for alpha, beta, and theta power, over two to five years (Kondacs & Szabo, 1999). Stability estimates for delta power are typically smaller (Gasser et al., 1985; Kondacs & Szabo, 1999; Pollock, Schneider, & Lyness, 1991). In addition, individual differences in developmental trajectories have been reported that distinguish children with autism from normally developing children, and it has been proposed that growth curve parameters might constitute promising endophenotypes for autism spectrum disorders (Tierney, Gabard-Durnam, Vogel-Farley, Tager-Flusberg, & Nelson, 2012). Individual differences in EEG activity are highly heritable (Enoch et al., 2008; D. J. Smit, Posthuma, Boomsma, & Geus, 2005; Tang et al., 2007; van Beijsterveldt, Molenaar, de Geus, & Boomsma, 1996), with published estimates of heritability on the order of .80. Heritable individual differences are particularly evident for alpha power and alpha peak frequency, becoming somewhat less prominent toward either end of the power spectrum from the alpha band.
There is mounting evidence to suggest that EEG characteristics can serve as candidate endophenotypes for disorders characterized by disinhibitory psychopathology, such as alcohol abuse and dependence as well as other forms of substance abuse, antisocial behavior, and disruptive disorders (e.g., conduct disorder and ADHD). The most common finding among alcohol-dependent individuals is increased beta power. This has most often been found in studies of individuals in treatment or in active withdrawal (Costa & Bauer, 1997; Herning et al., 1997; Saletu-Zyhlarz et al., 2004). Greater beta power in such individuals has been found to predict whether they continue in treatment or relapse following it (Bauer, 2001; Saletu-Zyhlarz et al., 2004; Winterer et al., 1998). At least one study has reported elevated beta power in non-treatment seeking individuals (Rangaswamy et al., 2002). Other studies conducted with non-treatment samples have reported increases in lower-frequency activity in substance abusers (e.g., theta and delta activity). Increased theta activity in particular has been observed in individuals with various forms of disinhibitory psychopathology: alcohol dependence (Rangaswamy et al., 2003), heavy marijuana use (Struve et al., 1999), antisocial behavior (Ehlers, Phillips, Gizer, Gilder, & Wilhelmsen, 2010), and ADHD (Barry, Clarke, & Johnstone, 2003). Increases in delta power have been reported in community volunteers who were methamphetamine (Newton et al., 2004) or MDMA users (Herning, Better, Tate, & Cadet, 2005). Reductions in alpha power have also been reported among alcohol-dependent individuals, particularly those with comorbid anxiety disorders (Enoch et al., 1995; Enoch et al., 1999) and antisocial behavior (Ducci et al., 2009).
An additional criterion of an endophenotype is that it should be present in individuals whose genetic makeup is similar to those with the manifest disorder even if the disorder is absent. Studies of beta power in first-degree relatives of alcohol-dependent individuals have yielded mixed results. Increased beta power has been observed in high-risk offspring (Bauer & Hesselbrock, 1993; Rangaswamy et al., 2004) as well as other relatives of alcoholics (Pollock, Earleywine, & Gabrielli, 1995), although this is sometimes limited to subjects of one gender (Propping, Kruger, & Mark, 1981), to subjects with comorbid antisocial behavior (Bauer & Hesselbrock, 1993), or to one way of measuring power (Finn & Justus, 1999). There have also been reports of null findings (Cohen, Porjesz, & Begleiter, 1991; Kaplan, Hesselbrock, O’Connor, & DePalma, 1988; Pollock et al., 1983), and reports of reduced alpha power rather than increased beta power (Ehlers & Schuckit, 1991; Finn & Justus, 1999). Alcohol ingestion has been found to result in greater beta power increases in high-risk individuals relative to low-risk individuals (Ehlers & Schuckit, 1990), supporting the notion that there is a familial component to increased beta activity, albeit one that may interact with exposure to alcohol.
The disorder in the spectrum of disinhibitory psychopathology in relation to which EEG activity is most often studied is ADHD. The most robust finding is of increased theta-band activity, as indicated above, along with reduced alpha or beta activity (Barry et al., 2003; Rommelse, Geurts, Franke, Buitelaar, & Hartman, 2011). These are sometimes combined into a theta/beta or theta/alpha ratio to serve as a neurobiological marker of the disorder (Snyder & Hall, 2006). Reduced beta power has also been found to correlate with behavior problems and hyperactivity in children (Deckel, Hesselbrock, & Bauer, 1996) and with a latent externalizing dimension, including substance abuse symptoms, in a community sample of adolescents (Gilmore, Malone, & Iacono, 2010). Thus, although the exact nature of EEG characteristics in disinhibitory psychopathology remains to be better delineated, especially the conditions governing the direction of change in beta activity (increased or decreased), these findings in aggregate suggest that beta EEG power has promise as an endophenotype for such disorders.
EEG has been studied in relation to several other disorders. For instance, increases in both alpha and beta power have been reported among individuals with depressive disorders (Knott, Mahoney, Kennedy, & Evans, 2001; Pollock & Schneider, 1990). Greater alpha power in the right hemisphere relative to the left, a measure with high internal consistency (Allen, Urry, Hitt, & Coan, 2004), seems to be a robust indicator of depression (Henriques & Davidson, 1990, 1991; D. J. Smit, Posthuma, Boomsma, & De Geus, 2007). Indeed, some have considered it a candidate endophenotype for the disorder (Stewart, Bismark, Towers, Coan, & Allen, 2010). Inter-hemispheric differences in power have also been observed among patients with bipolar disorder (Clementz, Sponheim, Iacono, & Beiser, 1994; Koek et al., 1999). A combination of increased slow-wave (delta and theta) power and decreased alpha power has been found in bipolar patients (Clementz et al., 1994) and even more reliably in patients with schizophrenia (Begic et al., 2011; Clementz et al., 1994; Harris, Melkonian, Williams, & Gordon, 2006; Sponheim, Clementz, Iacono, & Beiser, 1994, 2000; Sponheim, Iacono, Thuras, Nugent, & Beiser, 2003). However, EEG abnormalities are more often reported for schizophrenic patients than their unaffected relatives, suggesting that they may reflect disease status, illness progression, or long-term medication effects (Ranlund et al., 2014; Venables, Bernat, & Sponheim, 2009), rather than endophenotypic properties.
Changes in EEG activity have also been reported in patients with neurodegenerative conditions, such as Alzheimer’s disease, which is characterized by a slowing of the dominant (alpha) frequency as well as increases in low-frequency power (Petit, Gagnon, Fantini, Ferini-Strambi, & Montplaisir, 2004; Stam, 2005). The anomalies in this case appear to reflect the disease process rather than a genetically influenced vulnerability. However, understanding the molecular-genetic influences on the EEG parameters themselves is relevant.
Neurochemical and psychological correlates of EEG parameters
A potentially important benefit of endophenotypes is that they can convey information about specific neurobiological and pathophysiological processes involved in the particular disorder with which they are associated (Iacono & Malone, 2011). EEG activity is intimately related to the organism’s level of arousal and attention, both of which are directly modulated by cholinergic pathways in the ascending reticular formation (Steriade, Gloor, Llinas, Lopes de Silva, & Mesulam, 1990). For instance, decreased discharge levels of cholinergic projections to basal forebrain neurons results in large EEG slow waves, such as those observed during sleep. Noradrenergic modulation complements cholinergic activity during waking states (Steriade et al., 1990). In general, the specific form of EEG activity, particularly the magnitude of activity and the frequency of EEG oscillations, reflects the balance between excitatory and inhibitory neuronal interactions, both within and between populations of neurons (David & Friston, 2003). Different frequencies of oscillations in turn are consistently associated with particular cognitive and perceptual states. The alpha rhythm, for example, originates in multiple thalamocortical projections (Lopes da Silva, Vos, Mooibroek, & Van Rotterdam, 1980), and it is typically observed during periods when an individual is relaxed but alert. It is most prominent when measured over the visual cortex in an individual with eyes closed; it thus appears to reflect an intrinsic rhythm that is greatest in amplitude in the absence of sensory load and in a state of inactivity (Goldman, Stern, Engel, & Cohen, 2002). It has been proposed that alpha activity (increased synchronization) serves a control function, inhibiting activity in particular neural circuits, which facilitates in turn the appropriate timing of cortical processes (Klimesch, Sauseng, & Hanslmayr, 2007) as well as protecting against intrusion by irrelevant information into selective attention and short-term memory (Foxe & Snyder, 2011; Payne, Guillory, & Sekuler, 2013).
Some individuals exhibit very low levels of alpha activity. Recent studies have indicated that such low-voltage alpha (LVA) is associated with anxiety, including anxiety comorbid with alcoholism (Enoch et al., 1999). There is also indirect evidence of an association between LVA and variance common to antisocial behavior and alcoholism (Ducci et al., 2009). Robust individual differences are observed for the dominant (or peak) frequency of the alpha rhythm, which declines with aging (Obrist, 1979) and neurodegenerative diseases such as Alzheimer’s (Stam, 2005). Individual differences in alpha peak frequency have been reported to be associated with working memory (Clark et al., 2004) and intelligence (Grandy et al., 2013), although not in all studies (Posthuma, Neale, Boomsma, & de Geus, 2001). Like alpha power, alpha peak frequency is highly heritable, with estimates of approximately .80 (Posthuma et al., 2001; C. M. Smit, Wright, Hansell, Geffen, & Martin, 2006). Although there is a modest (inverse) phenotypic correlation between alpha power and peak frequency, bivariate biometric models fit to twin data indicate that they are genetically independent (C. M. Smit et al., 2006).
Whereas alpha activity is largely characteristic of the organism at rest, beta activity is associated with active mental states related to emotional or cognitive processing and even anxious arousal (Ray & Cole, 1985). Early preclinical research found that increased beta activity was observed in cats in hunting situations, which was interpreted as reflecting a state of expectancy and inhibition of movement (Steriade et al., 1990). More recent research with humans has corroborated the role of ‘sensorimotor’ beta activity in motor inhibition by demonstrating that initiating movement is robustly accompanied by decreases in beta power (Engel & Fries, 2010). Benzodiazepines, which bind specifically to GABA receptors (Tallman & Gallager, 1985), produce increases in beta power (Domino, French, Pohorecki, Galus, & Pandit, 1989), which has been interpreted as evidence for a role of GABA in the production of beta activity. This hypothesis is supported by findings from empirical work and computational modeling approaches (Muthukumaraswamy, Edden, Jones, Swettenham, & Singh, 2009; Whittington, Traub, Kopell, Ermentrout, & Buhl, 2000).
Theta-band activity dominates the EEG signal recorded from the hippocampus of most non-human mammals during wakefulness, and intracranial recordings suggest that theta activity in humans is modulated by behavior in similar ways (Steriade et al., 1990). Hippocampal theta activity, which is thought to reflect an “on-line” state of the hippocampus (Buzsaki, 2002), appears to be related to long-term potentiation and thus to memory consolidation (Steriade et al., 1990). Theta oscillations have also been observed in prefrontal cortex (PFC) (Benchenane, Tiesinga, & Battaglia, 2011). In rodents, coherent theta oscillations between hippocampus and PFC are critical for spatial working memory and encoding relevant information for long-term storage (Benchenane et al., 2010). In humans, theta oscillations in the prefrontal cortex have been linked to working memory performance (Tesche & Karhu, 2000), and theta activity in the resting EEG has been reported to be associated with reward sensitivity and decision-making that is compromised by preference for immediate reward (Massar, Kenemans, & Schutter, 2013; Schutter & Van Honk, 2005).
Candidate gene findings
There have been relatively few studies of genetic variants in relation to EEG parameters, and only one finding that has been replicated to date. There have been reports of associations between variants in the HTR3B gene, which codes for a serotonin receptor, and reduced alpha power, as well as alcohol dependence with comorbid antisocial behavior (Ducci et al., 2009), between the Val158/Met polymorphism of the catechol-O-methyltransferase gene (COMT) and low-frequency power (delta and theta) in schizophrenic patients (Venables et al., 2009) and slower alpha peak frequency in normal controls (Bodenmann et al., 2009), as well as reports of associations between asymmetry in EEG activity and candidate genes (Bismark et al., 2010; Bulgin et al., 2008). The ε4 allele of the apolipoprotein E (APOE) gene, which confers risk for Alzheimer’s dementia (Elias-Sonnenschein, Viechtbauer, Ramakers, Verhey, & Visser, 2011), has been found to be associated with reduced alpha power (Ponomareva, Korovaitseva, & Rogaev, 2008). Most importantly, this association has been observed in healthy young adults (Lee et al., 2012). A similar association has been reported (Ponomareva et al., 2013) between reduced alpha power and a variant of the CLU gene (encoding glycoprotein clusterin, or apolipoprotein J), which also confers risk for cognitive impairment in later life (Golenkina et al., 2010; Harold et al., 2009; Lambert et al., 2009). Although it was once thought that LVA was due to a single dominant gene (Anokhin et al., 1992; Vogel, 1986), this hypothesis has not been borne out. Recently, associations have been reported in women between LVA and the Val158/Met polymorphism in COMT (Enoch, Xu, Ferro, Harris, & Goldman, 2003) as well as a polymorphism in the GABAB receptor gene, independent of gender (Winterer et al., 2003). Other candidate gene findings exist (Loo et al., 2010), although involving a different type of polymorphism than a SNP, which is the focus of the present investigation.
Genome-wide studies
Several studies have taken the approach of examining the entire genome, rather than focusing on a single candidate gene. Linkage analysis uses markers consisting of polymorphisms varying either in sequence or size in samples comprising families. If a marker is coinherited with a trait, the two are said to be linked, and the gene that influences the trait is thought to be located near the marker. However, such markers tend to be widely spaced, and linkage analysis can only identify a chromosomal region, or “hot spot,” and not individual variants. An early analysis found linkage between beta power and a region of chromosome 4 (Porjesz et al., 2002). Subsequent analysis of individual SNPs to determine the source of this signal obtained significant associations between beta activity and SNPs in the gene that encodes the GABA α2 receptor subunit (GABRA2) (Edenberg et al., 2004). A recent case-control study of alcohol dependence obtained significant associations between beta activity and several GABRA2 SNPs (Lydall et al., 2011). Although characterizing EEGs as dominated by beta or not, rather than evaluating quantitative measures of power, this study broadly corroborated the beta power-GABRA2 association. Another genome-wide linkage scan reported an area of linkage between alpha and beta power and a region of chromosome 5. Follow-up association analysis focused on the gene coding for corticotropin releasing hormone binding protein (CRHBP) obtained a significant association with alpha EEG power as well as alcoholism and anxiety (Enoch et al., 2008).
GWAS represents a potentially powerful genome-wide approach. Because it is atheoretical, it is immune to any claims of cherry picking that can be made about candidate gene studies. The genotyping arrays contain many more variants than can be examined in linkage analysis, facilitating the identification of specific causal variants, and judicious selection of markers that “tag” others makes it possible to obtain much more comprehensive coverage of the genome. Moreover, EEG rhythms appear to reflect common characteristics of the mammalian brain, which suggests that a “common disease [phenotype]–common variant” model of inheritance is particularly appropriate. Genotyping arrays used in GWAS are designed to assess such common genetic variants. However, only one GWAS of resting EEG parameters has been conducted to date, which did not demonstrate significant associations with variants in any of the genes reviewed above that have been reported to be associated with EEG parameters. It demonstrated instead an association between theta power and several SNPs in the SGIP1 gene, involved in neurotransmission through synaptic vesicle formation (Hodgkinson et al., 2010). This finding was replicated in the same study and related to alcoholism as well. However, a subsequent study failed to replicate either finding (Derringer et al., 2011). Thus, our understanding of the molecular-genetic basis of EEG parameters remains extremely limited.
The current study
The present investigation consisted of an analysis of EEG parameters in a large population-based sample of adolescent and adult participants from three independent cohorts of the Minnesota Center for Twin and Family Research (MCTFR) who had been genotyped for 527,829 SNPs. The analysis plan for the GWASs in this special issue is described in depth in Iacono, Malone, Vaidyanathan, and Vrieze (2014). In brief, we examined the power of the EEG signal in four frequency ranges as well as the peak frequency of the alpha rhythm using a four-pronged approach: estimate the heritability of each EEG parameter using twin and twin-family biometric models; estimate the total genetic variance in each parameter accounted for by all SNPs in aggregate by means of genome-wide complex trait analysis (GCTA; Yang, Lee, Goddard, & Visscher, 2011); assess associations between each individual SNP and EEG parameters in a GWAS; and assess associations between individual genes and EEG parameters by aggregating the effect of all SNPs in a gene using VEGAS, a versatile gene-based test for association studies (Liu et al., 2010). Analyses of individual SNPs and genes comprised both purely atheoretical analyses of the whole genome as well as more targeted analyses of candidate genetic variants.
Method
Participants
As described in Iacono et al. (2014), the sample is a subset of the larger sample in a recent family-based GWAS of substance abuse and related psychopathology conducted at the Minnesota Center for Twin and Family Research (MCTFR) (McGue et al., 2013; Miller et al., 2012). Participants for the present investigation are from the older and younger cohorts and enrichment samples of the Minnesota Twin Family Study (MTFS; Iacono, Carlson, Taylor, Elkins, & McGue, 1999; Keyes et al., 2009; McGue et al., 2013). The sample for this investigation comprised all twins who completed their age-17 laboratory assessment of twins and all parents who had completed an identical laboratory assessment. (See Iacono et al., 2014 for further details.) Participants in MCTFR studies gave written informed consent or assent, if under the age of 18, to participate in the initial study as well as to allow data used in GWASs to be placed in a public repository to be shared with other researchers.
The sample is broadly representative ethnically of the state of Minnesota during the relevant birth years; it is thus predominantly Caucasian (96%). To avoid population stratification of allele frequencies, which confounds genetic analyses, we limited this investigation to Caucasian subjects, based on self-reported ethnicity corroborated by principal component analysis (PCA) of genotype data (Iacono et al., 2014; Miller et al., 2012).
EEG recording
Participants sat in a comfortable chair with neck support in a darkened room and were instructed to close their eyes and relax as completely as possible. EEG data were collected continuously for 5 min. Data were collected over the course of more than 20 years using two different systems. For participants in the MTFS older and younger cohorts (71% of the sample), EEG was recorded via Grass Neurodata 12 systems (128-Hz sampling rate, passband from 1 to 30 Hz with a rolloff of 6 dB). Hardware constraints limited the number of signals recorded. EEG was recorded from the bipolar derivations O1–P7 and O2–P8 and from Cz referenced to linked earlobes. Eye blinks and other eye movements were recorded by means of a transverse electrode arrangement, with one superior to the eye and one on the outer canthus. For participants in the ES sample (with the exception of 14 subjects), a Biosemi ActiveTwo system was used to collect continuously recorded EEG data with a sampling rate of 1024 Hz. ActiveTwo amplifiers are DC-coupled, and signals were lowpass-filtered using a digital 5th-order Bessel anti-aliasing sinc filter with a cutoff frequency (3-dB attenuation) of 205 Hz. ActiveTwo signals are monopolar.
EEG processing
Data processing was conducted in MATLAB® (2009a, The Mathworks, Natick, MA) using identical methods for both systems, based on functions in the Psychophysiology Toolbox (http://sourceforge.net/projects/psychophys/) and custom scripts. Biosemi data were transformed to be comparable to the original data. Data were downsampled using the Matlab resample function. The signal from Cz was referenced to an algebraic average of the two ears, whereas the signals from O1 and O2 were referenced to P7 and P8, respectively. All signals were filtered with finite impulse response (FIR) filters and a Kaiser window as implemented in the firfilt plugin to EEGLAB (Delorme & Makeig, 2004): a highpass filter with a half-amplitude cutoff frequency of 1 Hz (1-Hz transition band, filter order of 1286, ripple of .0001) and subsequently a lowpass FIR filter with a half-amplitude cutoff frequency of 30 Hz (transition band of 20 Hz, filter order of 24, ripple of .0001). A transverse EOG lead was created from the appropriate pair of monopolar electrodes.
Notes recorded when the data were originally collected guided us in visually identifying problematic data that might need to be excluded. In addition, segments containing transient artifacts and excessively small or large voltage deflections were tagged for exclusion by computer algorithm. We used a robust version of Mahalanobis distance from the robustbase package (Rousseeuw et al., 2011) in the R statistical programming environment (R Development Core Team, 2010) to identify multivariate outliers with respect to all three leads and EEG power in all bands as well as total power. Data for which the robust Mahalanobis distance fell in the upper 10th percentile of the cumulative chi-square distribution were flagged for review and excluded if visual inspection of the raw data indicated that the data were contaminated by noise (e.g., high-frequency noise) or artifacts (e.g., electrocardiogram) or if the EEG showed clear signs of drowsiness or sleep (e.g., approximately 6-Hz theta bursts, sharp vertex waves). Individual recording sites were excluded from analyses if fewer than 45 artifact-free sweeps were available.
We excluded 68 subjects for use of alcohol, marijuana or other illicit drug the day of the assessment; a history of serious head injury; neurological disorders; medication likely to affect psychophysiological responses; not refraining from taking medication for ADHD, such as methylphenidate, the day of their assessment, as was requested of all adolescent participants. An additional 110 subjects were excluded for recording problems that made the data unusable.
In addition, we compared all subjects who reported sleeping or were noted to have fallen asleep by the experimenter (n = 252) to the remaining subjects with respect to different measures of EEG power in a series of univariate analyses using lme4 in R to conduct a linear mixed model (LMM) with a random intercept at the family level to accommodate the dependency of EEG measures within families. Age, gender, age cohort, and recording system were included as covariates (see below). Eleven of the 12 LMMs yielded significant effects, with a clear trend: subjects observed to fall asleep had greater low frequency power (theta and delta, t-statistics ranging from 2.64 to 9.28 across electrodes and the two bands) and less high frequency power (alpha and beta, t-statistics ranging from −0.90 to −5.08 across electrodes and bands). Only beta power at Cz did not show a significant effect, although the direction of effect was the same as the direction in the occipital-parietal locations. In addition, peak alpha frequency at the occipital sites (described below) was slower among those reported to sleep (t-statistics of −2.62 and −2.99). As a result, these subjects were excluded. The final sample consisted of 4,026 individuals, 2,383 adolescents (1,153 males) and 1,643 adults (903 males) from 1,613 families. The majority of families were MZ twin families (1038, or 64%). The mean age was 17.7 (range, 16.6–20.0) for adolescent participants and 45.1 (range, 29.6–65.3) for the parents.
Continuous EEG data were divided into 2-s half-overlapping segments (i.e., Welch’s method). For each 2-s segment, the mean voltage was subtracted from the data and the resulting mean-centered data were tapered with a 50% Hanning window. The Fast Fourier Transform, as implemented in Matlab, was used to obtain spectral power estimates, which were averaged across all segments. Mean power estimates were calculated for each of four frequency bands: delta (0.5 to 3.5 Hz), theta (4 to 7.5 Hz), alpha (8 to 12.5 Hz), and beta (13 to 30 Hz). The natural logarithm of mean power in each band was used in analyses (cf. Pivik et al., 1993; Pizzagalli, 2007). In addition, we examined the peak (dominant) frequency in the alpha band. For this purpose we padded the (tapered) data with 0s to a series length of 1024, which yielded a frequency resolution of 0.125 Hz, rather than the 0.5-Hz resolution of the original data. We used “gravity frequency” (see Klimesch, 1999 for a discussion), which involves weighting power estimates at each spectral frequency within the alpha band by its corresponding frequency, represented in the formula Σ(a(f) × f)/Σ a(f) where a(f) represents power at a given frequency, f. This weighted frequency measure is particularly advantageous when the power spectrum consists of multiple peaks in the alpha band (Klimesch, 1999). We calculated peak frequency between 7 and 14 Hz.
Molecular-genetic data
As is common, we used PCA as implemented in EIGENSTRAT (Price et al., 2006) to identify the major dimensions of genetic variation in our sample of Caucasian subjects, and the 10 components (PCs) accounting for the most variance were included as covariates in our GWAS to control for any remaining population stratification (cf. Price et al., 2006). Genomic inflation statistics from genome-wide analyses were subsequently examined for evidence of meaningful residual population stratification.
Phenotypes
We analyzed log-transformed power in each of the four frequency bands as well as total power at Cz. Bipolar leads act as a spatial high pass filter and minimize contributions from far sources, which makes our two bipolar leads particularly useful for measuring alpha activity arising from occipital-parietal areas. Log-transformed alpha power estimates and peak alpha frequency estimates from the bipolar leads were highly correlated (r = .94 for both). We therefore averaged them into a single measure of alpha power and alpha peak frequency, respectively. There were 3,966 subjects with occipital-parietal data and 3,948 with data from Cz. We used as covariates in all analyses generation (parent or adolescent twin), gender, and chronological age. We also included a dummy variable representing recording system (Biosemi or Grass) and the 10 PCs from EIGENSTRAT as covariates (cf. Iacono et al., 2014).
Statistical analyses
Biometric heritability
The amount of heritable variance in each measure was estimated using standard biometric approaches to modeling twin-family data (Neale, Boker, Xie, & Maes, 2003). All measures were adjusted for the effects of covariates (chronological age, gender, generation cohort, recording system, and the 10 EIGENSTRAT PCs), and biometric models fit to the residuals. In typical biometric models three latent variables account for the variance in each phenotype: additive genes (A), common or shared environment (C), and unique or unshared environment (E). The correlations among family members with respect to the three latent variables determine the within-family phenotypic correlations. For instance, the family environment is shared equally by all family members, so the correlation for C is 1, whereas DZ twins and parents and offspring share half their genes, so the genetic correlation is 0.5 in these pairs but 1 in MZ twins. E is by definition unique to each individual. Models were fit to data from four-member families as well as based only on twins, the latter to facilitate comparisons between the present results and published data, which is predominantly based on twin data. Additional detail concerning our model-fitting approach is provided in Iacono et al. (Iacono et al., 2014). As noted in that paper, we also evaluated possible dominance (D) effects, and fit and report the results of ADE model fitting in the twin sample where appropriate. Because variances tended to differ between males and females as well as between adolescents and adults (see Table 2 in Iacono et al., 2014, this issue), dummy variables representing these two characteristics were included as (scalar) moderators of phenotype variance in our biometric models.
SNP heritability
In addition, we estimated the proportion of variance in each EEG parameter accounted for by the combined additive effect of all Illumina markers (and those in linkage disequilibrium, or LD, with them) using GCTA (Yang et al., 2011). GCTA estimates the degree of phenotypic similarity among genetically unrelated individuals, which is then assumed due to the specific genetic variants they share. Genotypic similarity is represented in the form of a genetic relatedness matrix (GRM), which resembles a correlation matrix representing pairwise genetic similarity. In samples consisting of closely related individuals, Yang and colleagues (Yang, Lee, Goddard, & Visscher, 2013) have recommended filtering the sample by means of several thresholds of genetic relatedness in order to look for consistency across the resulting estimates. We used thresholds of .025, .05, and .10, which remove all but distant relatives. The same covariates were used as in all other analyses (age, gender, generation, recording system, and the 10 PCs from EIGENSTRAT). Because LD can bias SNP heritability estimates upward (Speed, Hemani, Johnson, & Balding, 2012), we repeated the three analyses after weighting SNPs by local LD patterns. It has been recommended more recently when the sample consists of closely related individuals to estimate the total genetic variance while modeling the environmental influences family members share (the C latent variable in biometric models) (Yang et al., 2013). This produces an estimate of genetic influence unconfounded by shared environmental influence. In addition to this, we conducted the same analysis without modeling shared environmental influences (i.e., without any threshold of genetic relatedness). These two analyses, one that models C and one that does not, allowed us to assess the influence of shared environmental effects by comparing the two estimates.
SNP effects: Genome-wide scan
Our sample consists of individuals nested within families, which creates a correlation that violates the assumption of independent residuals in regression analyses. We used Rapid Feasible Generalized Least Squares (RFGLS; Li, Basu, Miller, Iacono, & McGue, 2011), a computationally efficient form of generalized least squares, to account for this source of dependency. Correlations are estimated separately for MZ and DZ twin families; the 65 stepparents in the present sample (61 of them male) were treated as independent observations. Additive SNP effects were modeled, with each SNP represented as a count of the number of minor alleles. The conventional threshold for genome-wide significance of 5 × 10−8 was used to evaluate the significance of each SNP.
SNP effects: Candidate SNPs
Subsequent to this genome-wide scan, we examined associations with two targeted sets of candidate SNPs: those implicated in previous genetic studies of EEG (18 EEG-specific candidate SNPs) and those implicated in recent meta-analyses of disorders and traits that are themselves associated with the endophenotypes examined in this special issue (1,180 endophenotype-general candidate SNPs). The papers we consulted for SNPs were identified through MEDLINE and are listed in the Supplementary Material. SNPs not on the Illumina array were imputed using minimac (Howie, Fuchsberger, Stephens, Marchini, & Abecasis, 2012) with reference CEU haplotypes from 1000 Genomes (2/2012) after having been prephased with BEAGLE (Browning & Browning, 2009). Imputation produces an allele dosage for each SNP, which is a weighted sum of the minor allele frequency (0, 1, or 2) and the posterior probability that the imputed SNP belongs to each of these frequency categories. The sum of minor allele dosages constituted the independent variable in these analyses.
Gene effects: Genome-wide scan
We used VEGAS (Liu et al., 2010) to conduct gene-based tests of all 17,601 genes identified by VEGAS, to parallel our genome-wide scan of individual SNPs. VEGAS aggregates the effects of all SNPs within a gene by converting the p-values for each SNP into a chi-squared statistic and summing these into a single score, which is adjusted for LD between the SNPs (see Iacono et al., 2014). In order to capture regulatory SNPs and those in LD with SNPs in the gene proper, VEGAS includes all SNPs within 50 kilobases of each end of the gene. Because the p-values were produced by RFGLS, they accurately reflect the clustered nature of our sample. The null distribution of the test statistic in the presence of LD is determined using Monte Carlo methods and the LD structure of a reference sample from HapMap, for which purpose we used the CEU sample of Caucasians. A Bonferroni-corrected threshold of 2.84 × 10−6 was used to determine statistical significance.
Gene effects: Candidate genes
We evaluated three sets of candidate genes. The first comprised a small number of candidate genes (n = 16) previously reported to be associated specifically with EEG measures (EEG-specific candidate genes). (One of the 14 genes in this set has been reported to be associated with three different EEG parameters. We treated each as independent associations.) One of the candidate genes was APOE. Because the APOE ε4 risk allele is defined by two SNPs, we did not use VEGAS, which aggregates over SNPs within a gene and flanking it, to evaluate its role. Although not on the Illumina array, these two SNPs could be imputed relatively accurately with reference to haplotypes in the 1000 Genomes reference panel, with imputation r2 values of .875 and .911. We created a dummy variable coding for risk, consisting of either the ε3/ε4 or ε4/ε4 haplotypes, and examined its effect on both measures of alpha power in RFGLS analyses using the same covariates as in all other analyses.
The second candidate set consisted of 204 candidate genes that are likely relevant to understanding genetic influences on all of the endophenotypes examined in this special issue because they are part of the major neurotransmitter and neuromodulator systems systems (dopamine, noradrenaline, acetylcholine, GABA, glutamate, and serotonin), they are involved in metabolizing nicotine and alcohol, or they are part of the endogenous cannabinoid and opioid systems. We consulted the NeuroSNP database to identify relevant genes {Saccone, 2009 #558}. The third set of candidate genes comprised autosomal 92 genes found to be associated with one or more endophenotypes for schizophrenia by the Consortium on the Genetics of Schizophrenia (Greenwood et al., 2011).
Results
Table 2 in Iacono et al. (2014) in this issue presents descriptive statistics for all measures. The distribution of each measure, after it had been adjusted for all model covariates, is characterized graphically in Figures S1 to S7. These indicate that the assumption of normality required by regression models is reasonably well met. Correlations among the measures are provided in Table S3 of the supplement to Vrieze et al. (2014) in this issue and are illustrated graphically in the form of a heatmap in Figure 2 in Iacono et al. (2014).
Heritability from biometric models
Family correlations from RFGLS, which have been adjusted for effects on mean levels of the various model covariates, are presented in Table 1. The median correlation was .80 for MZ twin pairs, .39 for DZ pairs, and .27 for parents and offspring. The mother-father correlation was essentially 0. Overall, the pattern of correlations is consistent with substantial additive genetic influence and negligible shared environmental influence (Iacono et al., 2014). Table 2 presents estimates of the proportion of genetic and environmental variance in each EEG measure from ACE models, along with confidence intervals. These are estimated from data from all four family members as well as from MZ and DZ twins only. Heritability estimates were uniformly large, ranging from .49, for delta power, to .85, for beta power, with a median of .78. Estimates were very similar for family-based models and those based only on twins, especially for the higher frequencies and total power. The remaining variance was due to unique environmental influence; point estimates of common environmental influence were 0 with two exceptions: alpha power for both the occipital measure and Cz in the twin model. The upper limit of 95% confidence intervals around C was not greater than .02 for estimates from family data, although it ranged as high as .20 for estimates from twin data.
Table 1.
Family correlations
| Mother-Father | Mother-Offspring | Father-Offspring | MZ Twins | DZ Twins | |
|---|---|---|---|---|---|
| Alpha power, occipital | .003 | .275 | .272 | .796 | .419 |
| Alpha power, Cz | .041 | .270 | .298 | .848 | .449 |
| Beta power, Cz | −.030 | .329 | .216 | .855 | .388 |
| Theta Power, Cz | −.083 | .227 | .132 | .735 | .365 |
| Delta power, Cz | −.119 | .214 | .068 | .561 | .237 |
| Total power, Cz | −.058 | .282 | .183 | .782 | .379 |
| Alpha peak frequency | .071 | .316 | .292 | .836 | .412 |
Note: Alpha peak frequency is derived from the occipital-parietal locations. All correlations are from RFGLS and are adjusted for the covariates in our model: chronological age, generation, gender, recording system (Grass or Biosemi), and the first 10 PCs from EIGENSTRAT.
Table 2.
ACE parameter estimates
| Measure | Data | A | C | E | |
|---|---|---|---|---|---|
| O1O2 | Alpha power | Family | .781 (.751–.807) | .000 (.000–.016) | .219 (.193–.249) |
| Twin | .772 (.616–.816) | .021 (.000–.173) | .207 (.184–.234) | ||
| Alpha frequency | Family | .826 (.799–.847) | .000 (.000–.019) | .174 (.153–.198) | |
| Twin | .836 (.692–.855) | .000 (.000–.143) | .164 (.145–.185) | ||
|
|
|||||
| Cz | Alpha power | Family | .838 (.814–.858) | .000 (.000–.014) | .162 (.142–.185) |
| Twin | .799 (.649–.863) | .047 (.000–.196) | .154 (.136–.174) | ||
| Beta power | Family | .848 (.826–.867) | .000 (.000–.011) | .152 (.133–.174) | |
| Twin | .853 (.757–.870) | .000 (.000–.095) | .147 (.130–.166) | ||
| Theta power | Family | .690 (.648–.728) | .000 (.000–.011) | .310 (.272–.352) | |
| Twin | .733 (.572–.763) | .000 (.000–.155) | .267 (.237–.300) | ||
| Delta power | Family | .488 (.432–.540) | .000 (.000–.014) | .512 (.460–.568) | |
| Twin | .558 (.423–.605) | .000 (.000–.120) | .442 (.395–.493) | ||
| Total power | Family | .757 (.722–.787) | .000 (.000–.012) | .243 (.213–.278) | |
| Twin | .782 (.638–.806) | .000 (.000–.140) | .218 (.184–.246) | ||
Note: A represents additive genetic influence, C represents shared environmental influence, and E represents unique, or unshared, environmental influence. Estimates are standardized, and they therefore give the proportion of total phenotypic variance accounted for by each latent variable. Ninety-five percent confidence intervals are provided in parentheses. Data indicates whether analyses were based on the entire family or only on the MZ and DZ twins. All power measures are log-transformed. O1O2 indicates the average of O1–P7 and O2–P8.
SNP heritability
SNP heritability estimates are presented in Table 3. Values are provided for each threshold of genetic relatedness used to create subsamples of unrelated individuals, and are based on unweighted SNPs as well as SNPs that had been weighted SNPs by local LD patterns to attenuate the effects on SNP heritability estimates of high levels of LD with causal SNPs. Because estimates are based on unrelated individuals, standard errors are quite large. Our interest is in patterns in the point estimates. As expected, the weighted estimates tend to have smaller point estimates and larger SEs, both of which are arguably appropriate (Speed et al., 2012). The occipital-parietal measures of alpha power and peak frequency produced median point estimates of .45 and .48, respectively, across the two methods and three thresholds of genetic relatedness, which were consistently larger than the Cz estimates, including the estimate for alpha power at Cz. SNP heritability estimates for alpha and beta power at Cz were relatively modest, with median values of approximately .20. Estimates for the other parameters tended to be quite small, although those for delta varied somewhat across thresholds and methods.
Table 3.
GCTA estimates of SNP heritability
| Measure | SNPs | Threshold of Genetic Relatedness
|
GCTA-Family | ||||
|---|---|---|---|---|---|---|---|
| .025 | .05 | .10 | None | ||||
| O1O2 | Alpha power | Unweighted | .578 (.192) | .483 (.186) | .481 (.185) | .774 (.014) | .774 (.036) |
| Weighted | .420 (.244) | .407 (.237) | .373 (.235) | ||||
| Alpha frequency | Unweighted | .584 (.185) | .676 (.179) | .649 (.178) | .824 (.011) | .823 (.034) | |
| Weighted | .306 (.243) | .383 (.236) | .367 (.235) | ||||
|
|
|||||||
| Cz | Alpha power | Unweighted | .308 (.191) | .316 (.185) | .296 (.184) | .849 (.009) | .849 (.033) |
| Weighted | .124 (.243) | .139 (.236) | .143 (.236) | ||||
| Beta power | Unweighted | .220 (.187) | .205 (.183) | .248 (.182) | .872 (.008) | .872 (.033) | |
| Weighted | .144 (.240) | .113 (.236) | .174 (.235) | ||||
| Theta power | Unweighted | .048 (.191) | .115 (.188) | .117 (.185) | .726 (.017) | .725 (.038) | |
| Weighted | .000 (.239) | .029 (.236) | .036 (.233) | ||||
| Delta power | Unweighted | .122 (.187) | .123 (.185) | .167 (.181) | .528 (.021) | .528 (.045) | |
| Weighted | .079 (.241) | .292 (.239) | .303 (.235) | ||||
| Total power | Unweighted | .089 (.191) | .157 (.187) | .139 (.184) | .793 (.013) | .793 (.035) | |
| Weighted | .000 (.239) | .024 (.235) | .039 (.233) | ||||
Note: Following the recommendation of Yang and colleagues (Yang et al., 2013), we used several different thresholds on the genetic relatedness matrix (GRM) to create a sample of genetically unrelated individuals. Weighted estimates (and SEs) are based on weighting SNPs by local LD patterns to avoid inflating SNP heritability estimates (Speed et al., 2012). Unweighted estimates are based on the unweighted, measured SNPs. Estimates based on the whole sample obtained by not imposing a threshold of genetic relatedness are in the column “None.” “GCTA-Family” refers to the method that uses all individuals in estimating genetic influence while modeling, and thus controlling statistically, shared environmental influence (Yang et al., 2013). O1O2 refers to the average of O1–P7 and O2–P8.
Finally, we include estimates based on the entire sample, whether by not imposing a threshold of genetic relatedness or by modeling shared environmental influences. These were virtually identical, suggesting that shared environmental influences were unimportant overall (an inference confirmed by GCTA estimates of shared environmental influence). Estimates also closely approximated the biometric heritability estimates.
SNP effects: Genome-wide scan
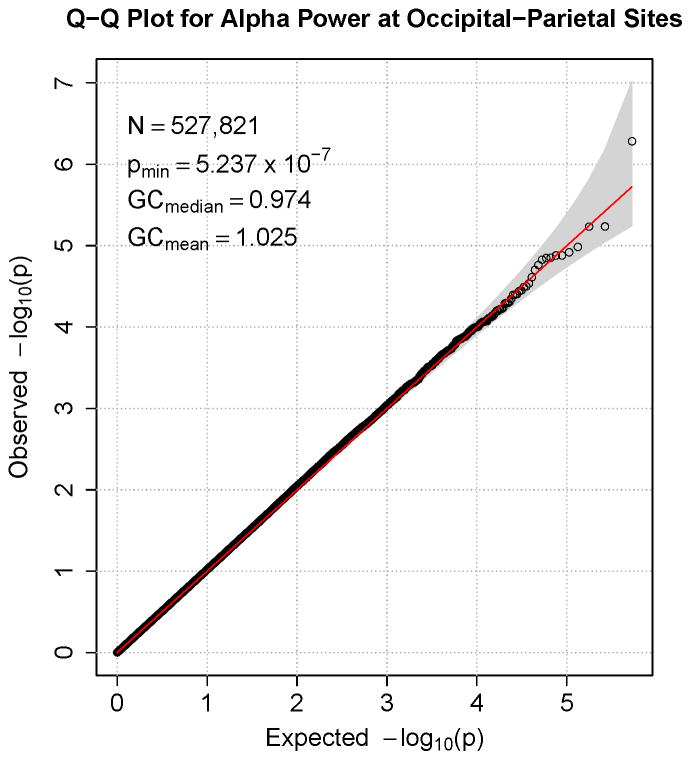
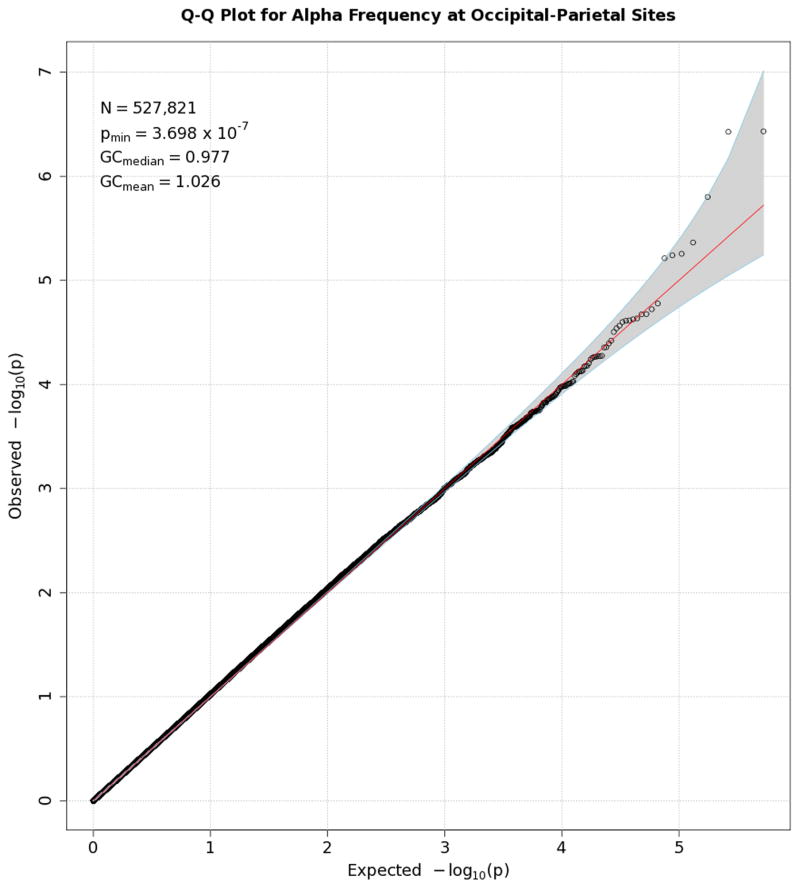
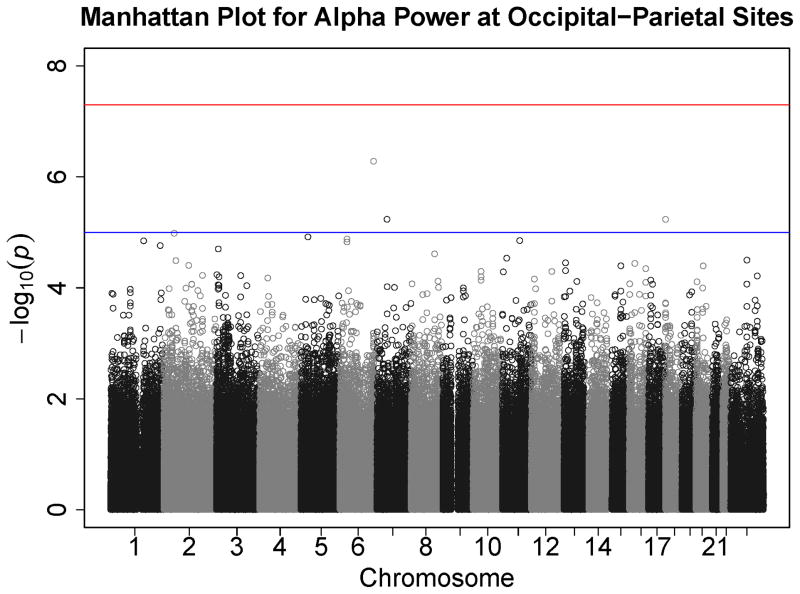
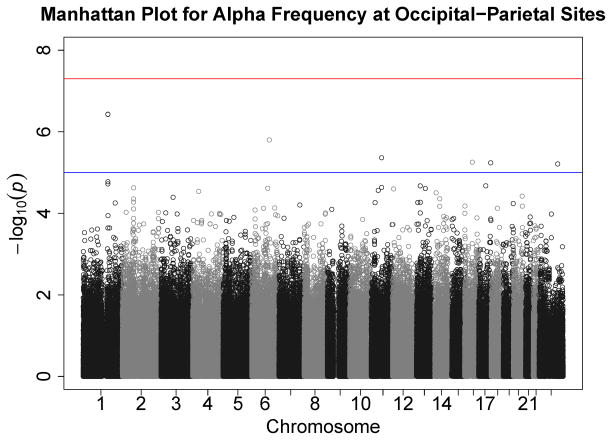
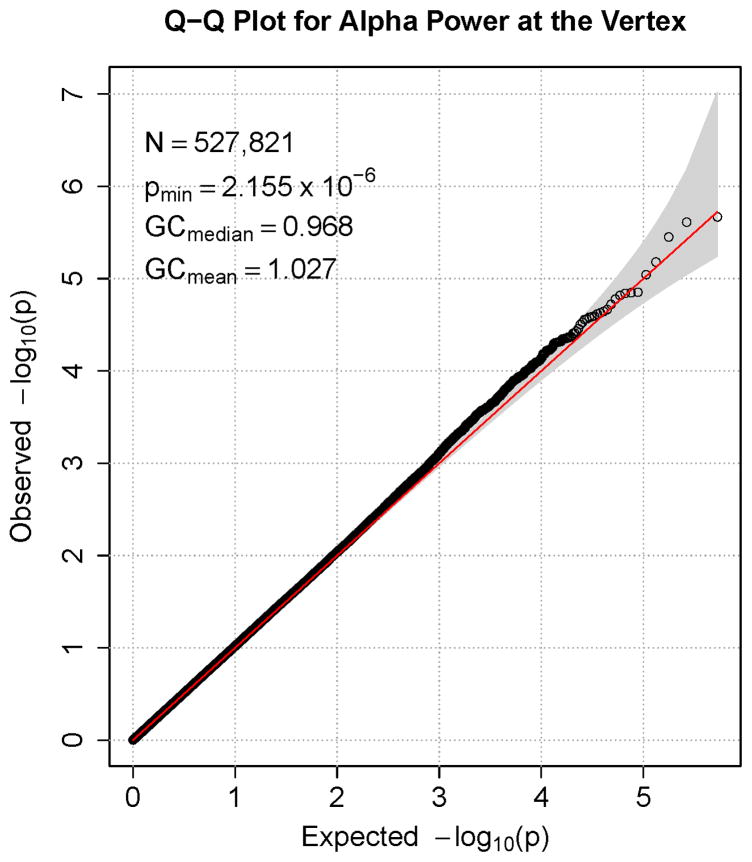
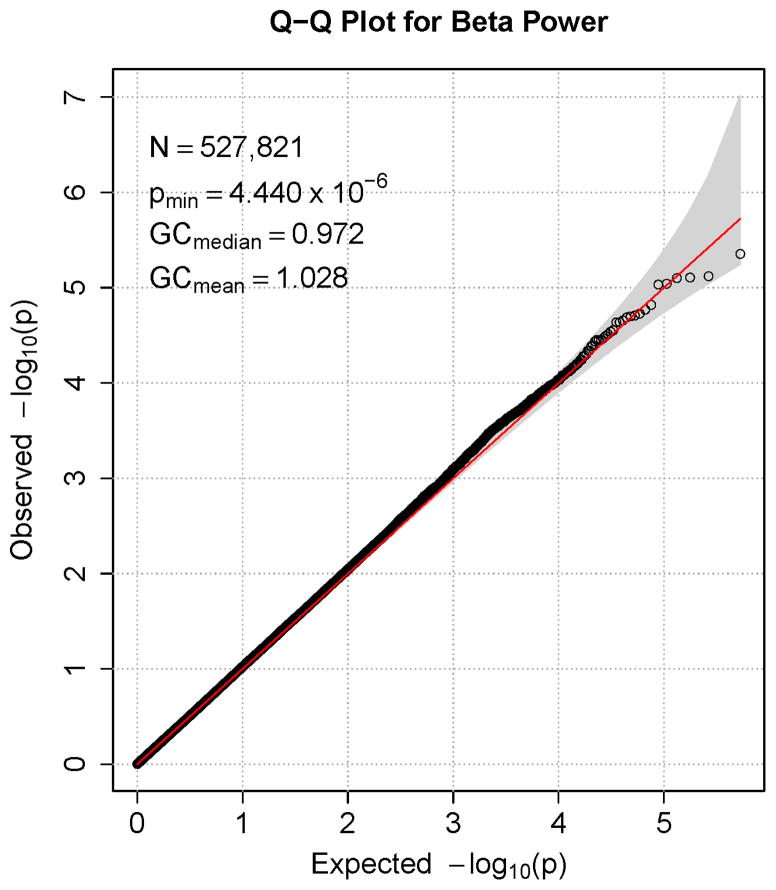
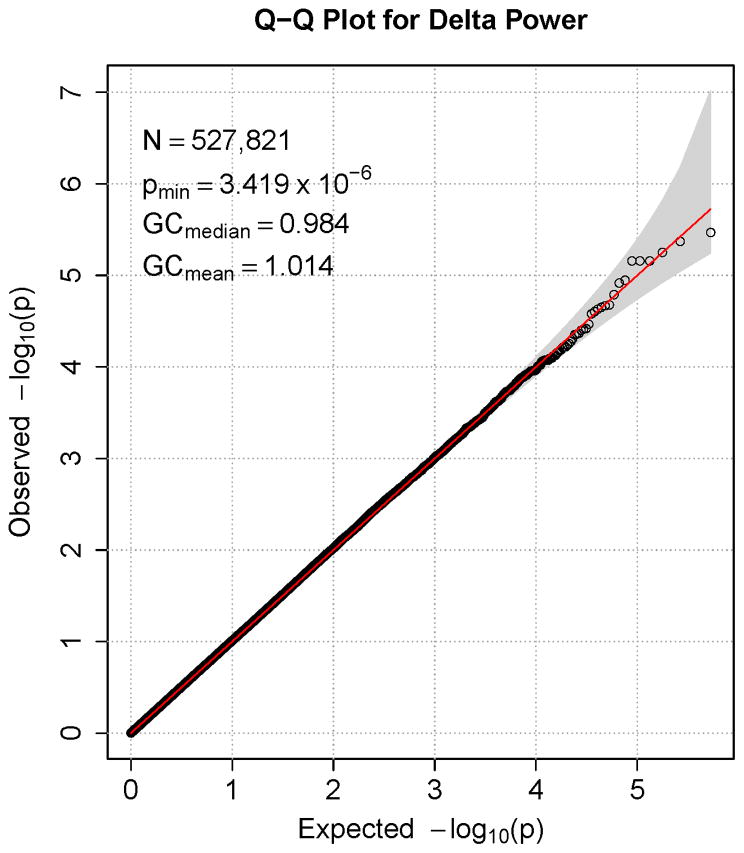
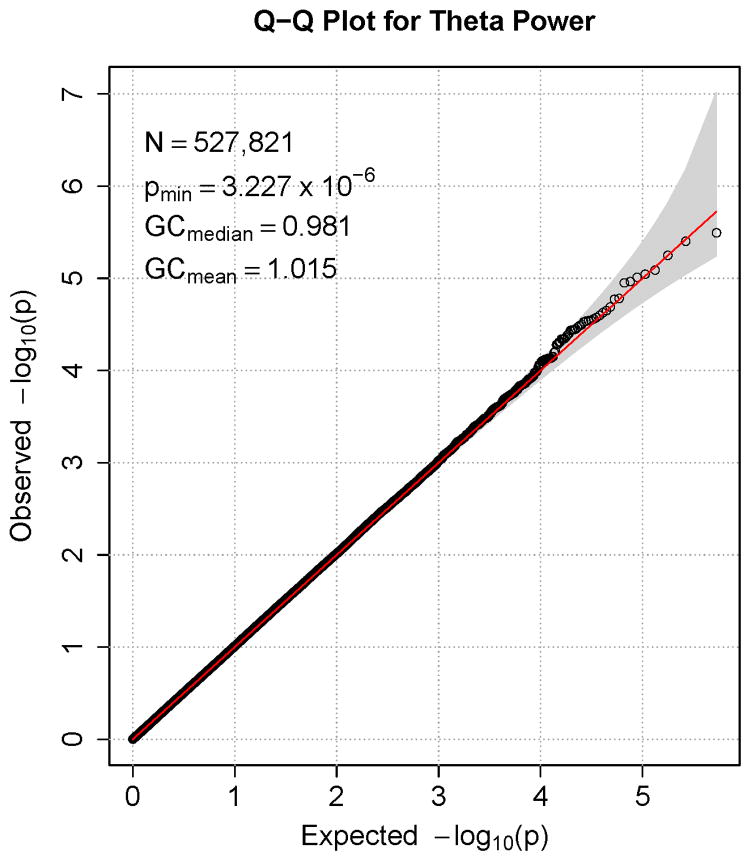
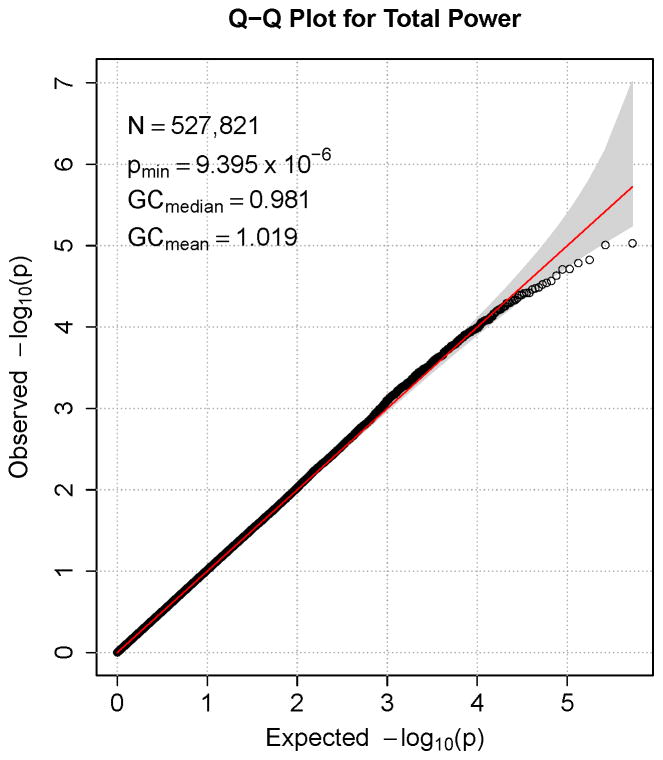
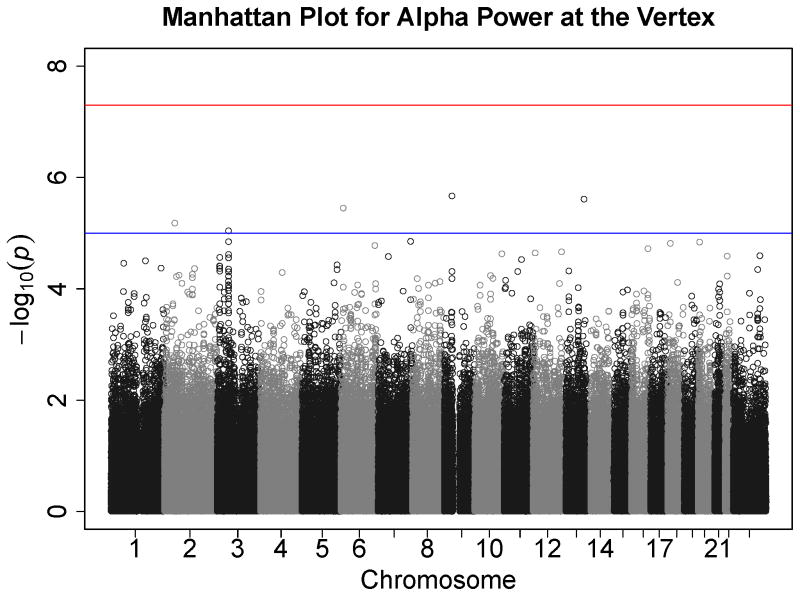
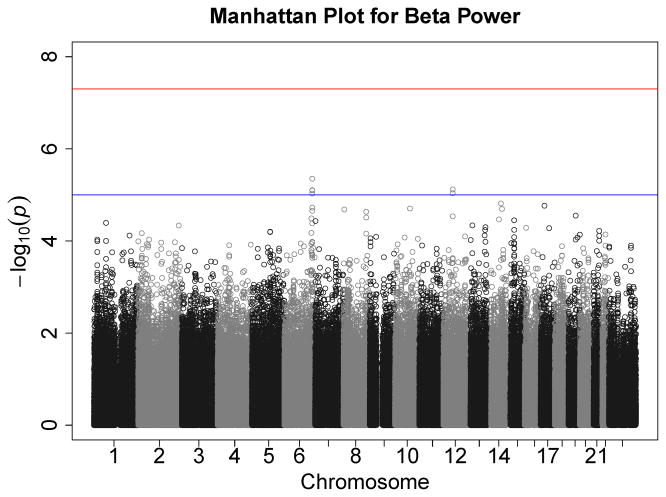
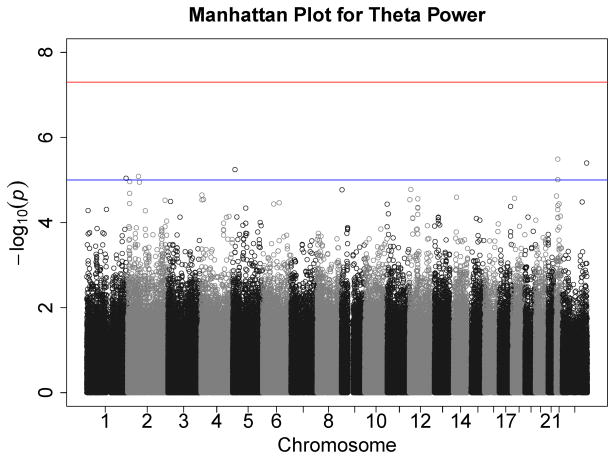
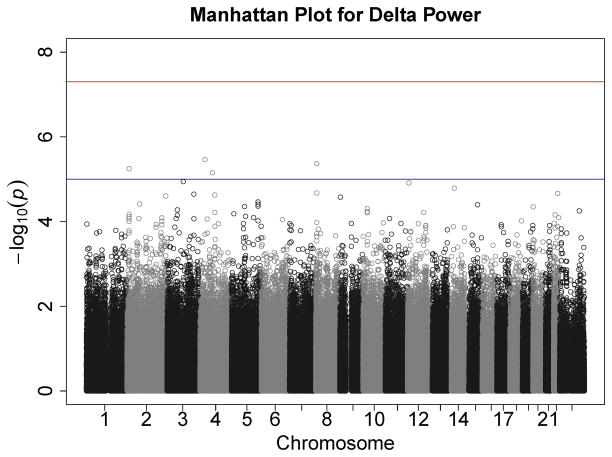
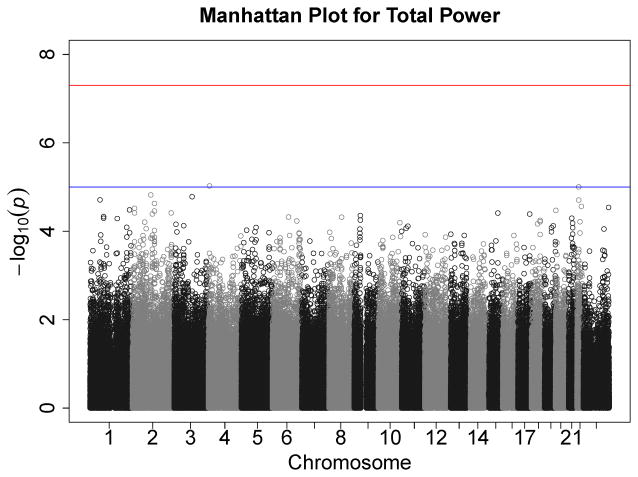
Figures 1–7 consist of Q-Q plots for each of the EEG measures of observed p-values against expected p-values under the null distribution, using the additive inverse of log10(p) to emphasize small p-values. There is no evidence in these plots of inflation that might be due to population stratification in allele frequencies unrelated to outcome measure. Genomic inflation factors associated with each analysis are close to 1, ranging from .97 to .99, confirming that the observed values conform to our expectation. In addition, however, none of the p-values approaches the significance threshold of 5 × 10−8. Figures 8–14 give the corresponding Manhattan plots, which order values of −log10(p) by the location of the SNP associated with it on each chromosome. These are unremarkable and do not reveal any obvious local signal. Results for all SNPs associated with p-values less than 10−4 are documented in Tables S1–S7.
Figure 1.
Q-Q plot for SNP associations with alpha power at occipital-parietal leads. The 45° line gives the expected value under the null distribution. The area shaded in gray corresponds to the 95% acceptance region. Median and mean genomic control values are given in the inset in the upper left. N refers to the number of SNPs, which is 8 fewer than the number of SNPs on the array because there was no variation for 8 SNPs in this sample. Q-Q plots in GWAS give the observed p-values against the expected p-values under the null distribution of no association, although the additive inverse of the common log of p-values (−log10[p]) is used in order to emphasize small p-values. Because the vast majority of SNPs are not expected to be associated with a given phenotype, observed p-values should conform closely to their expected values, falling on or very close to the 45° line. The gray region in each plot depicts the 95% confidence region (null acceptance region).
Figure 7.
Q-Q plot for SNP associations with alpha peak frequency at occipital-parietal leads. The 45° line gives the expected value under the null distribution. The area shaded in gray corresponds to the 95% acceptance region. Median and mean genomic control values are given in the inset in the upper left. N refers to the number of SNPs, which is 8 fewer than the number of SNPs on the array because there was no variation for 8 SNPs in this sample. Q-Q plots in GWAS give the observed p-values against the expected p-values under the null distribution of no association, although the additive inverse of the common log of p-values (−log10[p]) is used in order to emphasize small p-values. Because the vast majority of SNPs are not expected to be associated with a given phenotype, observed p-values should conform closely to their expected values, falling on or very close to the 45° line. The gray region in each plot depicts the 95% confidence region (null acceptance region).
Figure 8.
Manhattan plot of individual SNP associations with alpha power at occipital-parietal leads. Manhattan plots also depict the distribution of −log10(p) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p-values. The horizontal line at 7.3 indicates the genome-wide significance level (5E-08). The horizontal line at 5 indicates E-05, which is sometimes used to indicate “suggestive” significance.
Figure 14.
Manhattan plot of individual SNP associations with alpha peak frequency at occipital-parietal leads. Manhattan plots also depict the distribution of −log10(p) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p-values. The horizontal line at 7.3 indicates the genome-wide significance level (5E-08). The horizontal line at 5 indicates E-05, which is sometimes used to indicate “suggestive” significance.
SNP effects: Candidate SNPs
We specifically examined 18 SNPs identified by Hodgkinson and colleagues (2010); none of these EEG-specific candidate SNPs was significant (all p-values > .40) (Table S8). Regression coefficients and p-values for SNPs in our set of endophenotype-general candidate SNPs, selected because they are associated with relevant phenotypes, are presented in Tables S9–S15. None of the 1,180 SNPs in this set was associated with a p-value that survives Bonferroni correction (α = .05/1180 = 4.24 × 10−5). The smallest p-value for each EEG measure ranged from 1.71 × 10−3 to 6.94 × 10−4.
Gene effects: Genome-wide scan
Our gene-based test of 17,601 genes on autosomal chromosomes yielded two genes associated with delta power that survive Bonferroni correction (α = 2.84 × 10−6): DEFA4 and DEFA6 on chromosome 8 (both p-values < 1 × 10−6). Both encode defensins, a family of peptides thought to be involved in host defense. DEFA4 encodes a protein expressed in neutrophils (leukocytes) (i.e., in the blood) that inhibits corticotropin stimulated corticosterone production, and DEFA6 encodes a protein that is highly expressed in the bowel. These two genes are located in close proximity to each other. We followed up this finding by using RFGLS to analyze SNPs imputed with reference to 1000 Genomes to provide denser coverage of this region than the Illumina array provided. Results are plotted in Figure S8 in the supplement to this article. None of the imputed SNPs yielded a genome-wide significant p-value. The smallest p-values largely fall between the two genes and appear to represent a single signal.
Gene effects: candidate genes
RFGLS analyses of APOE ε4 risk status indicated that although risk was associated with reduced alpha power at both occipital-parietal and vertex sites, neither association was close to significant (both p-values greater than .45). VEGAS results for the remaining 15 genes (gene–EEG parameter associations) in our set of EEG-specific candidate genes are provided in Table 4. None survived correction for the number of tests (α = 3.12 × 10−3). Two associations were nominally significant (p < .05): between GABRA2 and beta power, and between CRH-BP and alpha power at Cz (although this was not observed for occipital-parietal alpha power). As in Derringer et al. (2011), we did not obtain a significant association between SGIP1 and theta power.
Table 4.
Associations with EEG-specific candidate genes implicated in previous research
| EEG measure | Gene | Chr | N SNPs | test-statistic | p-value | Source |
|---|---|---|---|---|---|---|
| Beta | GABRA2 | 4 | 18 | 70.46 | .014 | 6,11 |
| Alpha, occipital | CRH-BP | 5 | 33 | 71.63 | .036 | 7 |
| Alpha, Cz | CRH-BP | 5 | 33 | 42.95 | .212 | 7 |
| Theta | CHRM2* | 7 | 61 | 116.55 | .055 | 9 |
| Theta | GRM8* | 7 | 236 | 321.06 | .140 | 4 |
| Alpha, Cz | UGDH | 4 | 31 | 46.28 | .157 | 8 |
| Alpha, occipital | UGDH | 4 | 31 | 44.95 | .174 | 8 |
| Theta | SGIP1 | 1 | 99 | 131.95 | .199 | 8 |
| Theta | HTR7* | 10 | 35 | 34.59 | .413 | 14 |
| Theta | KCNJ6* | 21 | 126 | 128.12 | .442 | 10 |
| Alpha, occipital | HTR1A | 5 | 9 | 7.74 | .447 | 1 |
| Alpha, Cz | HTR1A | 5 | 9 | 4.68 | .694 | 1 |
| Alpha, occipital | CLU | 8 | 36 | 32.28 | .468 | 12 |
| Alpha, Cz | CLU | 8 | 36 | 24.99 | .677 | 12 |
| Alpha, occipital | HTR3B | 11 | 34 | 29.95 | .466 | 5 |
| Alpha, Cz | HTR3B | 11 | 34 | 20.66 | .740 | 5 |
| Theta | BDNF | 11 | 30 | 18.19 | .702 | 3 |
| Theta | COMT | 22 | 55 | 38.10 | .720 | 13 |
| Alpha frequency | COMT | 22 | 55 | 33.32 | .815 | 2 |
| Delta | COMT | 22 | 55 | 28.52 | .887 | 13 |
Note: Chr is the chromosome on which each gene is located. N SNPs gives the number of SNPs considered part of that gene by VEGAS (those within 50 kilobases of each end of the gene). Each test has 1 df. Source gives the paper reporting the relevant previous results (see below). Candidate genes for event-related theta activity (as opposed to resting theta activity) are indicated by an asterisk.
Bismark, A. W., Moreno, F. A., Stewart, J. L., Towers, D. N., Coan, J. A., Oas, J., … Allen, J. J. (2010). Polymorphisms of the HTR1a allele are linked to frontal brain electrical asymmetry. Biological Psychology, 83, 153–158. doi: 10.1016/j.biopsycho.2009.12.002
Bodenmann, S., Rusterholz, T., Durr, R., Stoll, C., Bachmann, V., Geissler, E., … Landolt, H. P. (2009). The functional Val158Met polymorphism of COMT predicts interindividual differences in brain alpha oscillations in young men. Journal of Neuroscience, 29, 10855–10862. doi: 29/35/10855 [pii]10.1523/JNEUROSCI.1427-09.2009
Bulgin, N. L., Strauss, J. S., King, N. A., Shaikh, S. A., George, C. J., Fox, N. A., … Kennedy, J. L. (2008). Association study of theta EEG asymmetry and brain-derived neurotrophic factor gene variants in childhood-onset mood disorder. Neuromolecular Medicine, 10, 343–355. doi: 10.1007/s12017-008-8038-x
Chen, A. C., Tang, Y., Rangaswamy, M., Wang, J. C., Almasy, L., Foroud, T., … Porjesz, B. (2009). Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 150B, 359–368. doi: 10.1002/ajmg.b.30818
Ducci, F., Enoch, M. A., Yuan, Q., Shen, P. H., White, K. V., Hodgkinson, C., … Goldman, D. (2009). HTR3B is associated with alcoholism with antisocial behavior and alpha EEG power--an intermediate phenotype for alcoholism and co-morbid behaviors. Alcohol, 43, 73–84. doi: S0741-8329(08)00334-0 [pii]10.1016/j.alcohol.2008.09.005
Edenberg, H. J., Dick, D. M., Xuei, X., Tian, H., Almasy, L., Bauer, L. O., … Begleiter, H. (2004). Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. American Journal of Human Genetics, 74, 705–714.
Enoch, M. A., Shen, P. H., Ducci, F., Yuan, Q., Liu, J., White, K. V., … Goldman, D. (2008). Common genetic origins for EEG, alcoholism and anxiety: the role of CRH-BP. PLoS One, 3, e3620. doi: 10.1371/journal.pone.0003620
Hodgkinson, C. A., Enoch, M. A., Srivastava, V., Cummins-Oman, J. S., Ferrier, C., Iarikova, P., … Goldman, D. (2010). Genome-wide association identifies candidate genes that influence the human electroencephalogram. Proceedings of the National Academy of Sciences of the United States of America, 107, 8695–8700. doi: 0908134107 [pii]10.1073/pnas.0908134107
Jones, K. A., Porjesz, B., Almasy, L., Bierut, L., Dick, D., Goate, A., … Begleiter, H. (2006). A cholinergic receptor gene (CHRM2) affects event-related oscillations. Behavior Genetics, 36, 627–639.
Kang, S. J., Rangaswamy, M., Manz, N., Wang, J. C., Wetherill, L., Hinrichs, T., … Dick, D. (2012). Family-based genome-wide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes, Brain and Behavior, 11, 712–719.
Lydall, G. J., Saini, J., Ruparelia, K., Montagnese, S., McQuillin, A., Guerrini, I., … Gurling, H. M. (2011). Genetic association study of GABRA2 single nucleotide polymorphisms and electroencephalography in alcohol dependence. Neuroscience Letters, 500, 162–166. doi: S0304-3940(11)00879-2 [pii] 10.1016/j.neulet.2011.05.240
Ponomareva, N. V., Andreeva, T., Protasova, M., Shagam, L., Malina, D., Goltsov, A., … Rogaev, E. I. (2013). Age-dependent effect of Alzheimer’s risk variant of CLU on EEG alpha rhythm in non-demented adults. Front Aging Neurosci, 5, 86. doi: 10.3389/fnagi.2013.00086
Venables, N. C., Bernat, E. M., & Sponheim, S. R. (2009). Genetic and disorder-specific aspects of resting state EEG abnormalities in schizophrenia. Schizophrenia Bulletin, 35, 826–839. doi: sbn021 [pii]10.1093/schbul/sbn021
Zlojutro, M., Manz, N., Rangaswamy, M., Xuei, X., Flury-Wetherill, L., Koller, D., … Kuperman, S. (2011). Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 156, 44–58.
The association between beta power and the GABRA2 gene produced the smallest p-value. Because there is stronger prior evidence for this association than the other candidate genes (other than possibly APOE ε4), we elected to follow it up. In order to compare the present results to published findings, it was necessary to impute SNPs in the gene with reference to 1000 Genomes because these SNPs were not on the Illumina genotyping array. This also provided finer coverage of the gene. Results are plotted in the top panel of Figure 15, which indicates that a large segment at the 3′ untranslated region (UTR) and the region immediately flanking it yielded somewhat elevated and relatively homogeneous –log10(p), although none was genome-wide significant. Table 5 shows p-values from the two previous studies of this gene (Edenberg et al., 2004; Lydall et al., 2011) and the present study. Although all SNPs were imputed in our data, with a single exception imputation r2 values uniformly approached 1. Ten SNPs with nominally significant associations (p < .05) in previous studies and the present one are highlighted. In all, 18 SNPs in our data were associated with beta power at this level of significance. Substantial pairwise LD characterizes this region of the chromosome. Table 5 therefore includes the median LD statistic (r2) for each SNP in relation to all others. These values tend to be quite close to 1 for the majority of SNPs, suggesting a high level of dependency among them, and therefore that the nominally significant p-values represent a single signal.
Figure 15.
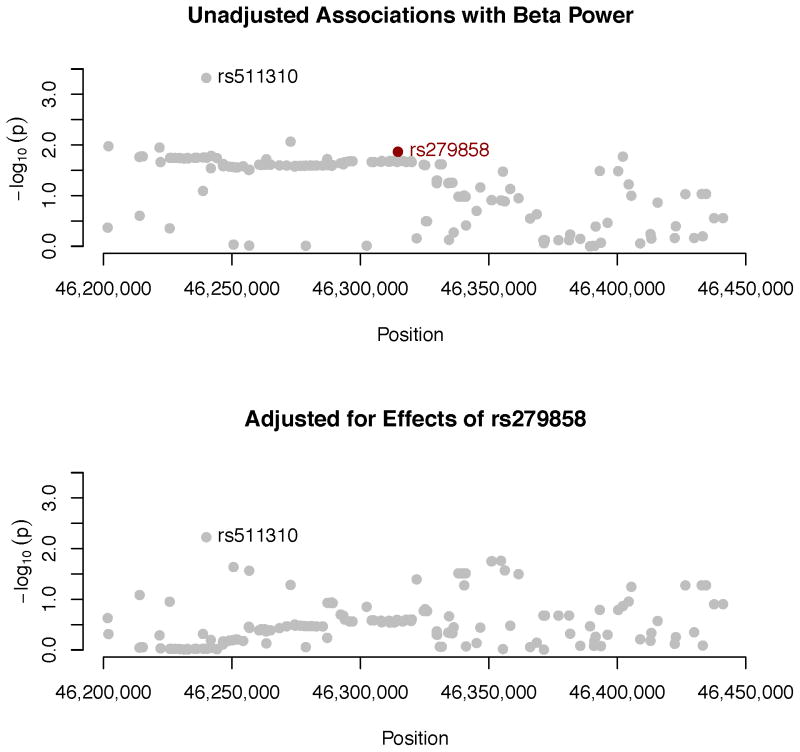
Associations between individual SNPs within or near GABRA2 and beta power. SNPs were imputed with reference to 1000 Genomes haplotypes (see text for additional detail). p-values produced by RFGLS analyses of the dosages for these imputed SNPs are expressed as −log10(p). SNP positions are from GRCh37 build 37 (hg19). Raw p-values are plotted in the upper panel. rs279858, which tags a haplotype associated with problematic alcohol use (Enoch, 2008), is indicated in black. The lower panel gives results are from a follow-up analysis that adjusts for effects of this SNP as an additional covariate, to determine to what extent the elevated p-values in the upper panel likely represent a single signal.
Table 5.
Concordance between present and previous associations between beta power and GABRA2
| SNP | Position | COGA
|
Lydall | MCTFR
|
LD r2 | |||
|---|---|---|---|---|---|---|---|---|
| AUD | Beta | t-statistic | p-value | impute r2 | ||||
| rs561779 | 46238778 | .048 | .044 | −2.37 | .018 | .999 | .870 | |
| rs572227 | 46251393 | .038 | .019 | −2.20 | .028 | 1.000 | .872 | |
| rs573400 | 46252066 | .062 | .270 | −2.20 | .028 | 1.000 | .872 | |
| rs481311 | 46254382 | .076 | .170 | −2.22 | .027 | .999 | .867 | |
| rs532780 | 46261366 | .079 | .016 | −2.25 | .025 | .998 | .872 | |
| rs548583 | 46263344 | .012 | .028 | .018 | −2.26 | .024 | .998 | .872 |
| rs496650 | 46264385 | .054 | .750 | −2.25 | .024 | .998 | .872 | |
| rs540363 | 46274246 | .044 | .490 | −2.22 | .026 | .998 | .872 | |
| rs526752 | 46276629 | .120 | .070 | −2.23 | .026 | .998 | .870 | |
| rs530329 | 46281119 | .034 | .048 | −2.23 | .026 | 1.000 | .872 | |
| rs483160 | 46287075 | .150 | .036 | −2.35 | .019 | .986 | .900 | |
| rs279871 | 46305733 | <.001 | .049 | .039 | ||||
| rs279867 | 46308303 | .240 | .050 | −2.32 | .021 | 1.000 | .903 | |
| rs279866 | 46309764 | .029 | .037 | |||||
| rs279863 | 46313022 | .017 | .011 | .027 | −2.32 | .021 | 1.000 | .872 |
| rs279861 | 46313325 | .037 | .045 | −2.32 | .021 | 1.000 | .872 | |
| rs279858 | 46314593 | .009 | .220 | .064 | −2.47 | .014 | .960 | .903 |
| rs175931 | 46316323 | .100 | .071 | −2.31 | .021 | 1.000 | .903 | |
| rs279843 | 46325204 | .049 | .300 | −2.24 | .025 | .990 | .840 | |
| rs279846 | 46329886 | .017 | .012 | −1.96 | .050 | .990 | .721 | |
| rs279826 | 46334209 | .001 | .250 | −1.91 | .057 | .995 | .747 | |
| rs279828 | 46334810 | .009 | .020 | −1.91 | .056 | .996 | .781 | |
| rs279834 | 46338299 | .015 | .027 | −1.62 | .104 | .999 | .777 | |
| rs279836 | 46339070 | .007 | .007 | .064 | .837 | |||
| rs279837 | 46339323 | .035 | .064 | −1.62 | .105 | 1.000 | .840 | |
| rs279841 | 46340763 | .038 | .018 | .018 | −1.62 | .105 | 1.000 | .843 |
| rs189957 | 46346679 | .053 | .270 | −1.82 | .069 | .998 | .781 | |
| rs1442061 | 46371220 | .370 | .240 | 0.31 | .756 | .997 | .064 | |
| rs1442062 | 46377076 | .220 | .130 | 0.31 | .756 | .999 | .060 | |
| rs3756007 | 46391064 | .990 | .980 | −0.02 | .984 | .963 | .088 | |
| rs894269 | 46393612 | .097 | .840 | 0.19 | .849 | .776 | .014 | |
| rs1545234 | 46404413 | .410 | .620 | −1.88 | .060 | .968 | .015 | |
Note: AUD indicates alcohol dependence. Lydall gives the p-values for beta power from Table 3 in Lydall et al. (2011). t-statistic is the test statistic for association with beta power in the MCTFR sample, p-value its corresponding significance level, and “impute r2” is the imputation accuracy. SNPs in the table are all SNPs reported in Edenberg et al. (2004) that were also analyzed by Lydall et al. or here. Base pair locations were converted from Edenberg et al. to the GRCh37 (hg19) assembly using the lift Over software utility from the UCSC Genome Bioinformatics website. “LD r2” gives the median LD between each SNP and all others (in the HapMap CEU population), as determined by the program WGA Viewer (Ge et al., 2008). The majority of SNPs are characterized by strong LD. SNPs significantly associated with beta activity in Edenberg et al. (2004) and in the present investigation are shaded gray.
GABRA2 contains two large haplotype blocks, and a synonymous SNP in the exon of the GABRA2 gene, rs279858, tags a haplotype that is correlated with severity of alcoholism and is itself associated with heavy drinking (Enoch, 2008). This SNP was not on the Illumina array but could be imputed successfully (with an r2 of .960), and the risk allele, G, was associated with reduced beta power in our sample, t = −2.46, p = .014. To assess the degree to which there might be an additional signal in GABRA2, we repeated our analysis of imputed SNPs, this time including each individual’s dosage for rs279858 as a covariate. Adjusting for this SNP substantially reduced the magnitude of associations between the other SNPs in the gene and beta power, presented graphically in the bottom panel of Figure 15. rs511310, which previously had yielded the smallest p-value and which is not in strong LD with the other SNPs (median across all SNPs in the gene, 0.09) remained the SNP with the strongest effect.
Analyses of the 204 genes in our set of endophenotype-general candidate genes produced one association that survived Bonferroni correction (α = 2.45 × 10−4): GABRA1, which encodes the α1 receptor for GABA, was associated with delta power, p = 2.33 × 10−4. GABRA1 has been associated with several drinking-related behaviors and consequences in the COGA sample (Dick, Plunkett, et al., 2006). Association results for all EEG measures are presented in Tables S16 to S22. Analyses of 92 genes in our set of schizophrenia endophenotype candidate genes are presented in Tables S23 to S28. None of the genes was associated with any EEG endophenotype at a level that survived Bonferroni correction (α = 5.43 × 10−4).
Discussion
Biometric and SNP heritability of EEG parameters
Biometric model-fitting analyses indicated that a substantial proportion of the variation in the different EEG measures examined in this study reflects heritable individual differences, with a median heritability estimate of .78. Measures of alpha and beta power as well as alpha peak frequency were particularly heritable, with estimates of additive genetic influence ranging from .79 to .85. These results, derived from fitting biometric models to both family and twin data, are consistent with the substantial heritability estimates obtained in previous research (Enoch et al., 2008; D. J. Smit et al., 2005; Tang et al., 2007; van Beijsterveldt et al., 1996), particularly for the higher frequencies.
GCTA analyses indicated that, although point estimates varied somewhat across endophenotypes, all SNPs on the Illumina array in aggregate (as well as those in LD with the Illumina markers) accounted for a substantial proportion of the variance only in occipital-parietal measures (alpha power and alpha peak frequency). SNP heritability estimates were smaller on the whole for power measures from Cz, especially in the lower frequencies. The large standard errors associated with these estimates make definitive inferences impossible. However, the heritability estimates provide a rough idea of the degree to which common variants influence each endophenotype. In general, they seem to influence alpha power and peak frequency at occipital-parietal sites to a greater degree than the measures from the vertex. Of the measures from Cz, common variants may influence alpha and beta power somewhat more than measures of power in the lowest frequencies, especially the theta band. This difference between higher and lower frequencies may reflect the fact that activity in higher frequencies predominate in the resting EEG, whereas lower-frequency activity is more vulnerable to artifacts and can reflect drowsiness, which may have escaped our data-cleaning procedure. In general, the SNP heritability estimates from measures derived from the Cz electrode fell short of the biometric estimates, thus indicating that genetic effects other than those captured by the common SNPs on the Illumina chip contribute to the observed biometric heritability.
Analysis of individual variants
Despite the findings that the various measures are likely substantially influenced by additive genetic effects, analyses of individual SNPs did not yield any evidence of genome-wide significant associations. Gene-based tests of all autosomal genes, which aggregate over all SNPs in a gene rather than examining each SNP by itself, did, yielding evidence of association between delta power and two genes. However, they are most highly expressed outside the brain, and the proximity of the two genes to each other suggests that this represents one signal, which is likely to be a false positive. Our analysis of 204 candidate genes selected because they are likely relevant to understanding EEG measures or related phenotypes also yielded an association between delta power and GABRA1, a gene that encodes the α1 subunit of the GABA receptor. GABRA1 has been associated with measures of alcohol consumption (Dick, Plunkett, et al., 2006). However, evidence of GABA system involvement in EEG is limited to high frequencies (Whittington et al., 2000). In addition, given the number of tests across different EEG measures, this one finding must be treated cautiously.
Our analyses of EEG-specific candidate genes corroborated Derringer et al. (2011) in failing to replicate the finding of an association between SGIP1 and theta power (Hodgkinson et al., 2010). Perhaps our most compelling gene-related finding was an association between beta power and the GABRA2 gene, which has been reported previously by two independent groups (Edenberg et al., 2004; Lydall et al., 2011; Rangaswamy et al., 2002). We specifically found an association between the G allele of rs279858, a synonymous exonic variant in GABRA2, and reduced beta power. This allele tags a haplotype that confers risk for heavy drinking and an early onset of alcohol dependence (Enoch, 2008). Our finding is thus consistent with evidence that beta power is reduced in alcohol dependence and related disinhibitory psychopathology. As others have indicated, GABRA2 is a plausible candidate for association with beta power in particular. GABRA2 encodes the α2 subunit of the GABAA receptor, and inhibitory GABAergic interneurons are centrally involved in producing high frequency oscillations, particularly those in the beta and gamma range. The specific frequency of oscillations in neuronal networks is dependent on the magnitude and kinetics of inhibitory synaptic potentials between interneurons mediated specifically by GABAA receptors (Whittington et al., 2000). Of course, this is not the only source of beta activity; coherent oscillations of excitatory neurons represent another important mechanism (Whittington et al., 2000). Nevertheless, GABAergic activity is clearly particularly important for “inhibition-based” high-frequency rhythms. In addition, a number of studies have reported associations between the GABRA2 gene and externalizing spectrum disorders, such as heavy drinking or alcohol or drug dependence (Agrawal et al., 2006; Bauer et al., 2007; Covault, Gelernter, Hesselbrock, Nellissery, & Kranzler, 2004; Drgon, D’Addario, & Uhl, 2006; Edenberg et al., 2004; Enoch, Schwartz, Albaugh, Virkkunen, & Goldman, 2006; Fehr et al., 2006; Soyka et al., 2008), perhaps with anxiety (Enoch et al., 2006), and antisocial personality (Dick, Agrawal, et al., 2006; Dick, Bierut, et al., 2006), or related personality traits (Villafuerte, Strumba, Stoltenberg, Zucker, & Burmeister, 2013). GABRA2 has been implicated in schizophrenia as well (Volk et al., 2002). These disorders are typically characterized by altered magnitude of EEG beta activity. However, associations between GABA system genes and alcohol use and abuse have not been replicated in the MCTFR data (Irons et al., 2014).
Limitations
The most obvious limitation of the present study is that only a few EEG electrodes were included, providing sparse coverage of the scalp. In particular, we did not have electrodes over frontal brain regions, which may have led us to fail to detect some associations, including our failure to replicate the finding of an association between SGIP1 and theta activity obtained in the only other GWAS of EEG parameters (Hodgkinson et al., 2010). Our sample also comprised two different age cohorts: individuals in late adolescence and adults, who ranged considerably in age but were primarily middle-aged. This was required in order to have as large a sample as possible. Although our analyses accounted for differences between age cohorts in mean levels of EEG parameters, it may be that the genetic influences expressed during these different developmental periods are different. Even with the combined age cohorts, our sample was small by current standards (although not by the standards in place when we began this investigation). In addition, to the degree that dominance effects influence these EEG parameters, we will have overestimated the magnitude of additive genetic influence in our biometric model-fitting analyses (i.e., the narrow sense heritability of each endophenotype). However, the evidence for such effects in ADE twin models was extremely weak.
Conclusions
Null findings are now the norm in individual GWASs, and they are often taken as an indication of polygenic inheritance; common phenotypes are due to a large number of genes, each with very small effect, such that individual GWASs are underpowered to detect them. Thus, individual GWASs may not be well suited for discovery. Larger samples obtained through meta-analysis are likely required, as demonstrated recently for volume of the hippocampus (Stein et al., 2012). An implication of this is that one promise of endophenotypes — that they might help identify genes for psychiatric disorders — cannot yet be meaningfully assessed, and it may never be meaningfully assessed. In the present investigation we obtained several findings that were significant even with Bonferroni correction of (endophenotype-wide) Type I error rate. By contrast, a GWAS of behavioral phenotypes in a sample that is essentially a superset of the sample used here and that is approximately twice as large, did not obtain any genome-wide significant findings (McGue et al., 2013). Does this imply that the EEG endophenotypes we examined are better situated to detect individual variants? This is impossible to know without replication and meta-analysis, as well as a better understanding of the variants we did identify. However, the cost of collecting most psychophysiological measures may prohibit the kinds of sample sizes required, including for replication studies (de Geus, 2010)
At the same time, GCTA results indicate that common variants account for varying degrees of the variance in these EEG endophenotypes. For some endophenotypes, SNP heritability estimates were greater than half the heritability estimate. Thus, there appears to be substantial additive genetic variance related to EEG measures, which is consonant with our phenotypic heritability estimates. However, identifying specific genetic variants or genes remains a challenge. More sophisticated analytic methods might be useful.
Such methods may have to reflect a somewhat different conceptual model as well. Two things are typically absent in GWA studies: the environment and development. The development of ocular dominance columns and normal vision depends critically on the experience of patterned light (Wiesel & Hubel, 1965). Although it would of course be extremely unlikely for an infant not to have this form of experience, it is nevertheless necessary. It is a truism that genes are expressed in particular environments over the course of development, yet this is not reflected in the GWAS design. Progress in understanding the genetic influences on complex traits may require analytic approaches that account for this interplay.
A strength of this investigation, and the others in this special issue, is that our sample comprises families, which allowed us to estimate the heritability of EEG parameters by means of biometric models and test for specific genetic variants in the same sample. Heritability estimates were substantial, and SNP heritability estimates indicated that as much as half the heritable variance in EEG endophenotypes is likely accounted for by the common variants on our genotyping array. However, we identified few specific variants, whether SNPs or genes. If EEG parameters are in fact a result of the additive effects of many genes each with very small influence, understanding the genetics of such traits is likely to continue to present interesting challenges, both methodologically and conceptually.
Supplementary Material
Figure 2.
Q-Q plot for SNP associations with alpha power at Cz. The 45° line gives the expected value under the null distribution. The area shaded in gray corresponds to the 95% acceptance region. Median and mean genomic control values are given in the inset in the upper left. N refers to the number of SNPs, which is 8 fewer than the number of SNPs on the array because there was no variation for 8 SNPs in this sample. Q-Q plots in GWAS give the observed p-values against the expected p-values under the null distribution of no association, although the additive inverse of the common log of p-values (−log10[p]) is used in order to emphasize small p-values. Because the vast majority of SNPs are not expected to be associated with a given phenotype, observed p-values should conform closely to their expected values, falling on or very close to the 45° line. The gray region in each plot depicts the 95% confidence region (null acceptance region).
Figure 3.
Q-Q plot for SNP associations with beta power at Cz. The 45° line gives the expected value under the null distribution. The area shaded in gray corresponds to the 95% acceptance region. Median and mean genomic control values are given in the inset in the upper left. N refers to the number of SNPs, which is 8 fewer than the number of SNPs on the array because there was no variation for 8 SNPs in this sample. Q-Q plots in GWAS give the observed p-values against the expected p-values under the null distribution of no association, although the additive inverse of the common log of p-values (−log10[p]) is used in order to emphasize small p-values. Because the vast majority of SNPs are not expected to be associated with a given phenotype, observed p-values should conform closely to their expected values, falling on or very close to the 45° line. The gray region in each plot depicts the 95% confidence region (null acceptance region).
Figure 4.
Q-Q plot for SNP associations with theta power at Cz. The 45° line gives the expected value under the null distribution. The area shaded in gray corresponds to the 95% acceptance region. Median and mean genomic control values are given in the inset in the upper left. N refers to the number of SNPs, which is 8 fewer than the number of SNPs on the array because there was no variation for 8 SNPs in this sample. Q-Q plots in GWAS give the observed p-values against the expected p-values under the null distribution of no association, although the additive inverse of the common log of p-values (−log10[p]) is used in order to emphasize small p-values. Because the vast majority of SNPs are not expected to be associated with a given phenotype, observed p-values should conform closely to their expected values, falling on or very close to the 45° line. The gray region in each plot depicts the 95% confidence region (null acceptance region).
Figure 5.
Q-Q plot for SNP associations with delta power at Cz. The 45° line gives the expected value under the null distribution. The area shaded in gray corresponds to the 95% acceptance region. Median and mean genomic control values are given in the inset in the upper left. N refers to the number of SNPs, which is 8 fewer than the number of SNPs on the array because there was no variation for 8 SNPs in this sample. Q-Q plots in GWAS give the observed p-values against the expected p-values under the null distribution of no association, although the additive inverse of the common log of p-values (−log10[p]) is used in order to emphasize small p-values. Because the vast majority of SNPs are not expected to be associated with a given phenotype, observed p-values should conform closely to their expected values, falling on or very close to the 45° line. The gray region in each plot depicts the 95% confidence region (null acceptance region).
Figure 6.
Q-Q plot for SNP associations with total power at Cz. The 45° line gives the expected value under the null distribution. The area shaded in gray corresponds to the 95% acceptance region. Median and mean genomic control values are given in the inset in the upper left. N refers to the number of SNPs, which is 8 fewer than the number of SNPs on the array because there was no variation for 8 SNPs in this sample. Q-Q plots in GWAS give the observed p-values against the expected p-values under the null distribution of no association, although the additive inverse of the common log of p-values (−log10[p]) is used in order to emphasize small p-values. Because the vast majority of SNPs are not expected to be associated with a given phenotype, observed p-values should conform closely to their expected values, falling on or very close to the 45° line. The gray region in each plot depicts the 95% confidence region (null acceptance region).
Figure 9.
Manhattan plot of individual SNP associations with alpha power at Cz. Manhattan plots also depict the distribution of −log10(p) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p-values. The horizontal line at 7.3 indicates the genome-wide significance level (5E-08). The horizontal line at 5 indicates E-05, which is sometimes used to indicate “suggestive” significance.
Figure 10.
Manhattan plot of individual SNP associations with beta power at Cz. Manhattan plots also depict the distribution of −log10(p) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p-values. The horizontal line at 7.3 indicates the genome-wide significance level (5E-08). The horizontal line at 5 indicates E-05, which is sometimes used to indicate “suggestive” significance.
Figure 11.
Manhattan plot of individual SNP associations with theta power at Cz. Manhattan plots also depict the distribution of −log10(p) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p-values. The horizontal line at 7.3 indicates the genome-wide significance level (5E-08). The horizontal line at 5 indicates E-05, which is sometimes used to indicate “suggestive” significance.
Figure 12.
Manhattan plot of individual SNP associations with delta power at Cz. Manhattan plots also depict the distribution of −log10(p) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p-values. The horizontal line at 7.3 indicates the genome-wide significance level (5E-08). The horizontal line at 5 indicates E-05, which is sometimes used to indicate “suggestive” significance.
Figure 13.
Manhattan plot of individual SNP associations with total power at Cz. Manhattan plots also depict the distribution of −log10(p) but are ordered by SNP location on a chromosome, which provides information about the location of any SNPs associated with small p-values. The horizontal line at 7.3 indicates the genome-wide significance level (5E-08). The horizontal line at 5 indicates E-05, which is sometimes used to indicate “suggestive” significance.
Acknowledgments
We are grateful to Jon Klaphake, Dragana Vidovic, Jennifer Donnelly, and Micah Hammer and for their assistance with organizing, processing, and screening the data. This research was supported by NIH grants DA 024417, DA 05147, AA 09367, DA 13240, and DA 036216.
References
- Agrawal A, Edenberg HJ, Foroud T, Bierut LJ, Dunne G, Hinrichs AL, Dick DM. Association of GABRA2 with drug dependence in the collaborative study of the genetics of alcoholism sample. Behavior Genetics. 2006;36(5):640–650. doi: 10.1007/s10519-006-9069-4. [DOI] [PubMed] [Google Scholar]
- Allen JJ, Urry HL, Hitt SK, Coan JA. The stability of resting frontal electroencephalographic asymmetry in depression. Psychophysiology. 2004;41(2):269–280. doi: 10.1111/j.1469-8986.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- Anokhin AP. Genetic psychophysiology: Advances, problems, and future directions. International Journal of Psychophysiology. 2014;93(2):173–197. doi: 10.1016/j.ijpsycho.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokhin AP, Steinlein O, Fischer C, Mao Y, Vogt P, Schalt E, Vogel F. A genetic study of the human low-voltage electroencephalogram. Human Genetics. 1992;90(1–2):99–112. doi: 10.1007/BF00210751. [DOI] [PubMed] [Google Scholar]
- Barry RJ, Clarke AR, Johnstone SJ. A review of electrophysiology in attention-deficit/hyperactivity disorder: I. Qualitative and quantitative electroencephalography. Clinical Neurophysiology. 2003;114(2):171–183. doi: 10.1016/s1388-2457(02)00362-0. S1388245702003620 [pii] [DOI] [PubMed] [Google Scholar]
- Bauer LO. Predicting relapse to alcohol and drug abuse via quantitative electroencephalography. Neuropsychopharmacology. 2001;25(3):332–340. doi: 10.1016/S0893-133X(01)00236-6. S0893-133X(01)00236-6 [pii] [DOI] [PubMed] [Google Scholar]
- Bauer LO, Covault J, Harel O, Das S, Gelernter J, Anton R, Kranzler HR. Variation in GABRA2 predicts drinking behavior in project MATCH subjects. Alcoholism, Clinical and Experimental Research. 2007;31(11):1780–1787. doi: 10.1111/j.1530-0277.2007.00517.x. ACER517 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer LO, Hesselbrock VM. EEG, autonomic and subjective correlates of the risk for alcoholism. Journal of Studies on Alcohol. 1993;54(5):577–589. doi: 10.15288/jsa.1993.54.577. [DOI] [PubMed] [Google Scholar]
- Begic D, Popovic-Knapic V, Grubisin J, Kosanovic-Rajacic B, Filipcic I, Telarovic I, Jakovljevic M. Quantitative electroencephalography in schizophrenia and depression. Psychiatria Danubina. 2011;23(4):355–362. [PubMed] [Google Scholar]
- Benchenane K, Peyrache A, Khamassi M, Tierney PL, Gioanni Y, Battaglia FP, Wiener SI. Coherent theta oscillations and reorganization of spike timing in the hippocampal- prefrontal network upon learning. Neuron. 2010;66(6):921–936. doi: 10.1016/j.neuron.2010.05.013. S0896-6273(10)00381-8 [pii] [DOI] [PubMed] [Google Scholar]
- Benchenane K, Tiesinga PH, Battaglia FP. Oscillations in the prefrontal cortex: a gateway to memory and attention. Current Opinion in Neurobiology. 2011;21(3):475–485. doi: 10.1016/j.conb.2011.01.004. S0959-4388(11)00008-0 [pii] [DOI] [PubMed] [Google Scholar]
- Berger H. Über das Elektrenkephalogramm des Menschen. Archiv für Psychiatrie und Nervenkrankheiten. 1929;87:527–570. [Google Scholar]
- Bismark AW, Moreno FA, Stewart JL, Towers DN, Coan JA, Oas J, Allen JJ. Polymorphisms of the HTR1a allele are linked to frontal brain electrical asymmetry. Biological Psychology. 2010;83(2):153–158. doi: 10.1016/j.biopsycho.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmann S, Rusterholz T, Durr R, Stoll C, Bachmann V, Geissler E, Landolt HP. The functional Val158Met polymorphism of COMT predicts interindividual differences in brain alpha oscillations in young men. Journal of Neuroscience. 2009;29(35):10855–10862. doi: 10.1523/JNEUROSCI.1427-09.2009. 29/35/10855 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. American Journal of Human Genetics. 2009;84(2):210–223. doi: 10.1016/j.ajhg.2009.01.005. S0002-9297(09)00012-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgin NL, Strauss JS, King NA, Shaikh SA, George CJ, Fox NA, Kennedy JL. Association study of theta EEG asymmetry and brain-derived neurotrophic factor gene variants in childhood-onset mood disorder. Neuromolecular Medicine. 2008;10(4):343–355. doi: 10.1007/s12017-008-8038-x. [DOI] [PubMed] [Google Scholar]
- Buzsaki G. Theta oscillations in the hippocampus. Neuron. 2002;33(3):325–340. doi: 10.1016/s0896-6273(02)00586-x. S089662730200586X [pii] [DOI] [PubMed] [Google Scholar]
- Clark RC, Veltmeyer MD, Hamilton RJ, Simms E, Paul R, Hermens D, Gordon E. Spontaneous alpha peak frequency predicts working memory performance across the age span. International Journal of Psychophysiology. 2004;53(1):1–9. doi: 10.1016/j.ijpsycho.2003.12.011. S0167876003002782 [pii] [DOI] [PubMed] [Google Scholar]
- Clementz BA, Sponheim SR, Iacono WG, Beiser M. Resting EEG in first-episode schizophrenia patients, bipolar psychosis patients, and their first-degree relatives. Psychophysiology. 1994;31(5):486–494. doi: 10.1111/j.1469-8986.1994.tb01052.x. [DOI] [PubMed] [Google Scholar]
- Cohen HL, Porjesz B, Begleiter H. EEG characteristics in males at risk for alcoholism. Alcoholism, Clinical and Experimental Research. 1991;15(5):858–861. doi: 10.1111/j.1530-0277.1991.tb00613.x. [DOI] [PubMed] [Google Scholar]
- Costa L, Bauer L. Quantitative electroencephalographic differences associated with alcohol, cocaine, heroin and dual-substance dependence. Drug and Alcohol Dependence. 1997;46(1–2):87–93. doi: 10.1016/s0376-8716(97)00058-6. S0376-8716(97)00058-6 [pii] [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2004;129B(1):104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- David O, Friston KJ. A neural mass model for MEG/EEG: coupling and neuronal dynamics. Neuroimage. 2003;20(3):1743–1755. doi: 10.1016/j.neuroimage.2003.07.015. S1053811903004579 [pii] [DOI] [PubMed] [Google Scholar]
- de Geus EJ. From genotype to EEG endophenotype: a route for post-genomic understanding of complex psychiatric disease? Genome Medicine. 2010;2(9):1–4. doi: 10.1186/gm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckel AW, Hesselbrock V, Bauer L. Antisocial personality disorder, childhood delinquency, and frontal brain functioning: EEG and neuropsychological findings. Journal of Clinical Psychology. 1996;52(6):639–650. doi: 10.1002/(SICI)1097-4679(199611)52:6<639::AID-JCLP6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Delorme A, Makeig S. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. Journal of Neuroscience Methods. 2004;134(1):9–21. doi: 10.1016/j.jneumeth.2003.10.009. S0165027003003479 [pii] [DOI] [PubMed] [Google Scholar]
- Derringer J, Krueger RF, Manz N, Porjesz B, Almasy L, Bookman E, Bierut LJ. Nonreplication of an association of SGIP1 SNPs with alcohol dependence and resting theta EEG power. Psychiatric Genetics. 2011;21(5):265–266. doi: 10.1097/YPG.0b013e32834371fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Agrawal A, Schuckit MA, Bierut L, Hinrichs A, Fox L, Begleiter H. Marital status, alcohol dependence, and GABRA2: evidence for gene-environment correlation and interaction. Journal of Studies on Alcohol. 2006;67(2):185–194. doi: 10.15288/jsa.2006.67.185. [DOI] [PubMed] [Google Scholar]
- Dick DM, Bierut L, Hinrichs A, Fox L, Bucholz KK, Kramer J, Foroud T. The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behavior Genetics. 2006;36(4):577–590. doi: 10.1007/s10519-005-9041-8. [DOI] [PubMed] [Google Scholar]
- Dick DM, Plunkett J, Wetherill LF, Xuei XL, Goate A, Hesselbrock V, Foroud T. Association between GABRA1 and drinking behaviors in the collaborative study on the genetics of alcoholism sample. Alcoholism-Clinical and Experimental Research. 2006;30(7):1101–1110. doi: 10.1111/j.1530-0277.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- Domino EF, French J, Pohorecki R, Galus CF, Pandit SK. Further observations on the effects of subhypnotic doses of midazolam in normal volunteers. Psychopharmacology Bulletin. 1989;25(3):460. [PubMed] [Google Scholar]
- Drgon T, D’Addario C, Uhl GR. Linkage disequilibrium, haplotype and association studies of a chromosome 4 GABA receptor gene cluster: candidate gene variants for addictions. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2006;141B(8):854–860. doi: 10.1002/ajmg.b.30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducci F, Enoch MA, Yuan Q, Shen PH, White KV, Hodgkinson C, Goldman D. HTR3B is associated with alcoholism with antisocial behavior and alpha EEG power--an intermediate phenotype for alcoholism and co-morbid behaviors. Alcohol. 2009;43(1):73–84. doi: 10.1016/j.alcohol.2008.09.005. S0741-8329(08)00334-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. American Journal of Human Genetics. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Phillips E, Gizer IR, Gilder DA, Wilhelmsen KC. EEG spectral phenotypes: heritability and association with marijuana and alcohol dependence in an American Indian community study. Drug and Alcohol Dependence. 2010;106(2–3):101–110. doi: 10.1016/j.drugalcdep.2009.07.024. S0376-8716(09)00317-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Schuckit MA. EEG fast frequency activity in the sons of alcoholics. Biological Psychiatry. 1990;27(6):631–641. doi: 10.1016/0006-3223(90)90531-6. 0006-3223(90)90531-6 [pii] [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Schuckit MA. Evaluation of EEG alpha activity in sons of alcoholics. Neuropsychopharmacology. 1991;4(3):199–205. [PubMed] [Google Scholar]
- Elias-Sonnenschein LS, Viechtbauer W, Ramakers IH, Verhey FR, Visser PJ. Predictive value of APOE-epsilon4 allele for progression from MCI to AD-type dementia: a meta-analysis. Journal of Neurology, Neurosurgery and Psychiatry. 2011;82(10):1149–1156. doi: 10.1136/jnnp.2010.231555. jnnp.2010.231555 [pii] [DOI] [PubMed] [Google Scholar]
- Engel AK, Fries P. Beta-band oscillations--signalling the status quo? Current Opinion in Neurobiology. 2010;20(2):156–165. doi: 10.1016/j.conb.2010.02.015. S0959-4388(10)00039-5 [pii] [DOI] [PubMed] [Google Scholar]
- Enoch MA. The role of GABAA receptors in the development of alcoholism. Pharmacology Biochemistry and Behavior. 2008;90(1):95–104. doi: 10.1016/j.pbb.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Rohrbaugh JW, Davis EZ, Harris CR, Ellingson RJ, Andreason P, et al. Relationship of genetically transmitted alpha EEG traits to anxiety disorders and alcoholism. American Journal of Medical Genetics. 1995;60(5):400–408. doi: 10.1002/ajmg.1320600510. [DOI] [PubMed] [Google Scholar]
- Enoch MA, Schwartz L, Albaugh B, Virkkunen M, Goldman D. Dimensional anxiety mediates linkage of GABRA2 haplotypes with alcoholism. American Journal of Medical Genetics B: Neuropsychiatric Genetics. 2006;141B(6):599–607. doi: 10.1002/ajmg.b.30336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Shen PH, Ducci F, Yuan Q, Liu J, White KV, Goldman D. Common genetic origins for EEG, alcoholism and anxiety: the role of CRH-BP. PloS One. 2008;3(10):e3620. doi: 10.1371/journal.pone.0003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, White KV, Harris CR, Robin RW, Ross J, Rohrbaugh JW, Goldman D. Association of low-voltage alpha EEG with a subtype of alcohol use disorders. Alcoholism, Clinical and Experimental Research. 1999;23(8):1312–1319. 00000374-199908000-00004 [pii] [PubMed] [Google Scholar]
- Enoch MA, Xu K, Ferro E, Harris CR, Goldman D. Genetic origins of anxiety in women: a role for a functional catechol-O-methyltransferase polymorphism. Psychiatric Genetics. 2003;13(1):33–41. doi: 10.1097/01.ypg.0000054709.85338.c3. [DOI] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatric Genetics. 2006;16(1):9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. 00041444-200602000-00003 [pii] [DOI] [PubMed] [Google Scholar]
- Finn PR, Justus A. Reduced EEG alpha power in the male and female offspring of alcoholics. Alcoholism, Clinical and Experimental Research. 1999;23(2):256–262. 00000374-199902000-00010 [pii] [PubMed] [Google Scholar]
- Foxe JJ, Snyder AC. The Role of Alpha-Band Brain Oscillations as a Sensory Suppression Mechanism during Selective Attention. Frontiers in Psychology. 2011;2:154. doi: 10.3389/fpsyg.2011.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser T, Bacher P, Steinberg H. Test-retest reliability of spectral parameters of the EEG. Electroencephalography and Clinical Neurophysiology. 1985;60(4):312–319. doi: 10.1016/0013-4694(85)90005-7. [DOI] [PubMed] [Google Scholar]
- Ge D, Zhang K, Need AC, Martin O, Fellay J, Urban TJ, Goldstein DB. WGAViewer: software for genomic annotation of whole genome association studies. Genome Research. 2008;18(4):640–643. doi: 10.1101/gr.071571.107. gr.071571.107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore CS, Malone SM, Iacono WG. Brain electrophysiological endophenotypes for externalizing psychopathology: a multivariate approach. Behavior Genetics. 2010;40(2):186–200. doi: 10.1007/s10519-010-9343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RI, Stern JM, Engel J, Jr, Cohen MS. Simultaneous EEG and fMRI of the alpha rhythm. Neuroreport. 2002;13(18):2487–2492. doi: 10.1097/01.wnr.0000047685.08940.d0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golenkina SA, Gol’tsov A, Kuznetsova IL, Grigorenko AP, Andreeva TV, Reshetov DA, Rogaev EI. Analysis of clusterin gene (CLU/APOJ) polymorphism in Alzheimer’s disease patients and in normal cohorts from Russian populations. Molekuliarnaia Biologiia. 2010;44(4):620–626. [PubMed] [Google Scholar]
- Grandy TH, Werkle-Bergner M, Chicherio C, Lovden M, Schmiedek F, Lindenberger U. Individual alpha peak frequency is related to latent factors of general cognitive abilities. Neuroimage. 2013;79:10–18. doi: 10.1016/j.neuroimage.2013.04.059. S1053-8119(13)00406-0 [pii] [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ, Hardiman G. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. American Journal of Psychiatry. 2011;168(9):930–946. doi: 10.1176/appi.ajp.2011.10050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson S, Runarsson TP, Sigurdsson S, Eiriksdottir G, Johnsen K. Reliability of quantitative EEG features. Clinical Neurophysiology. 2007;118(10):2162–2171. doi: 10.1016/j.clinph.2007.06.018. S1388-2457(07)00299-4 [pii] [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nature Genetics. 2009;41(10):1088–1093. doi: 10.1038/ng.440. ng.440 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Melkonian D, Williams L, Gordon E. Dynamic spectral analysis findings in first episode and chronic schizophrenia. International Journal of Neuroscience. 2006;116(3):223–246. doi: 10.1080/00207450500402977. [DOI] [PubMed] [Google Scholar]
- Henriques JB, Davidson RJ. Regional brain electrical asymmetries discriminate between previously depressed and healthy control subjects. Journal of Abnormal Psychology. 1990;99(1):22–31. doi: 10.1037//0021-843x.99.1.22. [DOI] [PubMed] [Google Scholar]
- Henriques JB, Davidson RJ. Left frontal hypoactivation in depression. Journal of Abnormal Psychology. 1991;100(4):535–545. doi: 10.1037//0021-843x.100.4.535. [DOI] [PubMed] [Google Scholar]
- Herning RI, Better W, Tate K, Cadet JL. Neuropsychiatric alterations in MDMA users: preliminary findings. Annals of the New York Academy of Sciences. 2005;1053:20–27. doi: 10.1196/annals.1344.003. 1053/1/20 [pii] [DOI] [PubMed] [Google Scholar]
- Herning RI, Guo X, Better WE, Weinhold LL, Lange WR, Cadet JL, Gorelick DA. Neurophysiological signs of cocaine dependence: increased electroencephalogram beta during withdrawal. Biological Psychiatry. 1997;41(11):1087–1094. doi: 10.1016/S0006-3223(96)00258-2. S0006-3223(96)00258-2 [pii] [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Enoch MA, Srivastava V, Cummins-Oman JS, Ferrier C, Iarikova P, Goldman D. Genome-wide association identifies candidate genes that influence the human electroencephalogram. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(19):8695–8700. doi: 10.1073/pnas.0908134107. 0908134107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature Genetics. 2012;44(8):955–959. doi: 10.1038/ng.2354. ng.2354 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Carlson SR, Taylor J, Elkins IJ, McGue M. Behavioral disinhibition and the development of substance use disorders: Findings from the Minnesota Twin Family Study. Development and Psychopathology. 1999;11:869–900. doi: 10.1017/s0954579499002369. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Malone SM. Developmental endophenotypes: Indexing genetic risk for substance abuse with the P300 brain event-related potential. Child Development Perspectives. 2011;5(4):239–247. doi: 10.1111/j.1750-8606.2011.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, Vaidyanathan U, Vrieze SI. Genome wide scans of genetic variants for psychophysiological endophenotypes: A methodological overview. Psychophysiology. 2014 doi: 10.1111/psyp.12343. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons DE, Iacono WG, Oetting WS, Kirkpatrick RM, Vrieze SI, Miller MB, McGue M. Gamma-aminobutyric acid system genes -- No evidence for a role in alcohol use and abuse in a community-based sample. Alcoholism: Clinical and Experimental Research. 2014;38(4):938–947. doi: 10.1111/acer.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan RF, Hesselbrock VM, O’Connor S, DePalma N. Behavioral and EEG responses to alcohol in nonalcoholic men with a family history of alcoholism. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 1988;12(6):873–885. doi: 10.1016/0278-5846(88)90083-8. [DOI] [PubMed] [Google Scholar]
- Keyes MA, Malone SM, Elkins IJ, Legrand LN, McGue M, Iacono WG. The enrichment study of the Minnesota twin family study: increasing the yield of twin families at high risk for externalizing psychopathology. Twin Research and Human Genetics. 2009;12(5):489–501. doi: 10.1375/twin.12.5.489. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimesch W. EEG alpha and theta oscillations reflect cognitive and memory performance: a review and analysis. Brain Research: Brain Research Reviews. 1999;29(2–3):169–195. doi: 10.1016/s0165-0173(98)00056-3. [DOI] [PubMed] [Google Scholar]
- Klimesch W, Sauseng P, Hanslmayr S. EEG alpha oscillations: the inhibition-timing hypothesis. Brain Res Rev. 2007;53(1):63–88. doi: 10.1016/j.brainresrev.2006.06.003. S0165-0173(06)00083-X [pii] [DOI] [PubMed] [Google Scholar]
- Knott V, Mahoney C, Kennedy S, Evans K. EEG power, frequency, asymmetry and coherence in male depression. Psychiatry Research. 2001;106(2):123–140. doi: 10.1016/s0925-4927(00)00080-9. [DOI] [PubMed] [Google Scholar]
- Koek RJ, Yerevanian BI, Tachiki KH, Smith JC, Alcock J, Kopelowicz A. Hemispheric asymmetry in depression and mania. A longitudinal QEEG study in bipolar disorder. Journal of Affective Disorders. 1999;53(2):109–122. doi: 10.1016/s0165-0327(98)00171-2. [DOI] [PubMed] [Google Scholar]
- Kondacs A, Szabo M. Long-term intra-individual variability of the background EEG in normals. Clinical Neurophysiology. 1999;110(10):1708–1716. doi: 10.1016/s1388-2457(99)00122-4. S1388-2457(99)00122-4 [pii] [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nature Genetics. 2009;41(10):1094–1099. doi: 10.1038/ng.439. ng.439 [pii] [DOI] [PubMed] [Google Scholar]
- Lee TW, Yu YW, Hong CJ, Tsai SJ, Wu HC, Chen TJ. The influence of apolipoprotein E Epsilon4 polymorphism on qEEG profiles in healthy young females: a resting EEG study. Brain Topography. 2012;25(4):431–442. doi: 10.1007/s10548-012-0229-y. [DOI] [PubMed] [Google Scholar]
- Li X, Basu S, Miller MB, Iacono WG, McGue M. A rapid generalized least squares model for a genome-wide quantitative trait association analysis in families. Human Heredity. 2011;71(1):67–82. doi: 10.1159/000324839. 000324839 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Mcrae AF, Nyholt DR, Medland SE, Wray NR, Brown KM. A versatile gene-based test for genome-wide association studies. American Journal of Human Genetics. 2010;87(1):139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo SK, Hale ST, Hanada G, Macion J, Shrestha A, McGough JJ, Smalley SL. Familial clustering and DRD4 effects on electroencephalogram measures in multiplex families with attention deficit/hyperactivity disorder. Journal of the American Academy of Child and Adolescent Psychiatry. 2010;49(4):368–377. S0890-8567(10)00076-6 [pii] [PMC free article] [PubMed] [Google Scholar]
- Lopes da Silva FH, Vos JE, Mooibroek J, Van Rotterdam A. Relative contributions of intracortical and thalamo-cortical processes in the generation of alpha rhythms, revealed by partial coherence analysis. Electroencephalography and Clinical Neurophysiology. 1980;50(5–6):449–456. doi: 10.1016/0013-4694(80)90011-5. [DOI] [PubMed] [Google Scholar]
- Lydall GJ, Saini J, Ruparelia K, Montagnese S, McQuillin A, Guerrini I, Gurling HM. Genetic association study of GABRA2 single nucleotide polymorphisms and electroencephalography in alcohol dependence. Neuroscience Letters. 2011;500(3):162–166. doi: 10.1016/j.neulet.2011.05.240. S0304-3940(11)00879-2 [pii] [DOI] [PubMed] [Google Scholar]
- Massar SA, Kenemans JL, Schutter DJ. Resting-state EEG theta activity and risk learning: sensitivity to reward or punishment? International Journal of Psychophysiology. 2013 doi: 10.1016/j.ijpsycho.2013.10.013. S0167-8760(13)00303-6 [pii] [DOI] [PubMed] [Google Scholar]
- McGue M, Zhang Y, Miller MB, Basu S, Vrieze S, Hicks B, Iacono WG. A genome-wide association study of behavioral disinhibition. Behavior Genetics. 2013;43:363–373. doi: 10.1007/s10519-013-9606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin G, Makeig S, Tsuang MT. In search of biomarkers in psychiatry: EEG-based measures of brain function. American Journal of Medical Genetics B: Neuropsychiatric Genetics. 2013 doi: 10.1002/ajmg.b.32208. [DOI] [PubMed] [Google Scholar]
- Miller MB, Basu S, Cunningham J, Eskin E, Malone SM, Oetting WS, McGue M. The Minnesota Center for Twin and Family Research genome-wide association study. Twin Research and Human Genetics. 2012;15(6):767–774. doi: 10.1017/thg.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukumaraswamy SD, Edden RA, Jones DK, Swettenham JB, Singh KD. Resting GABA concentration predicts peak gamma frequency and fMRI amplitude in response to visual stimulation in humans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(20):8356–8361. doi: 10.1073/pnas.0900728106. 0900728106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. Mx: Statistical modeling. 6. Richmond, VA: Department of Psychiatry, Virginia Commonwealth University; 2003. [Google Scholar]
- Newton TF, Kalechstein AD, Hardy DJ, Cook IA, Nestor L, Ling W, Leuchter AF. Association between quantitative EEG and neurocognition in methamphetamine-dependent volunteers. Clinical Neurophysiology. 2004;115(1):194–198. doi: 10.1016/s1388-2457(03)00314-6. [DOI] [PubMed] [Google Scholar]
- Obrist WD. Brain function in old age. Springer; 1979. Electroencephalographic changes in normal aging and dementia; pp. 102–111. [Google Scholar]
- Payne L, Guillory S, Sekuler R. Attention-modulated alpha-band oscillations protect against intrusion of irrelevant information. Journal of Cognitive Neuroscience. 2013;25(9):1463–1476. doi: 10.1162/jocn_a_00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit D, Gagnon JF, Fantini ML, Ferini-Strambi L, Montplaisir J. Sleep and quantitative EEG in neurodegenerative disorders. Journal of Psychosomatic Research. 2004;56(5):487–496. doi: 10.1016/j.jpsychores.2004.02.001. S0022399904000182 [pii] [DOI] [PubMed] [Google Scholar]
- Pivik RT, Broughton RJ, Coppola R, Davidson RJ, Fox N, Nuwer MR. Guidelines for the recording and quantitative analysis of electroencephalographic activity in research contexts. Psychophysiology. 1993;30(6):547–558. doi: 10.1111/j.1469-8986.1993.tb02081.x. [DOI] [PubMed] [Google Scholar]
- Pizzagalli DA. Electroencephalography and high-density electrophysiological source localization. In: Cacioppo JT, Tassinary LG, Berntson GG, editors. Handbook of psychophysiology. 3. Cambridge, UK: Cambridge University Press; 2007. pp. 56–84. [Google Scholar]
- Pollock VE, Earleywine M, Gabrielli WF. Personality and EEG beta in older adults with alcoholic relatives. Alcoholism, Clinical and Experimental Research. 1995;19(1):37–43. doi: 10.1111/j.1530-0277.1995.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Pollock VE, Schneider LS. Quantitative, waking EEG research on depression. Biological Psychiatry. 1990;27(7):757–780. doi: 10.1016/0006-3223(90)90591-o. 0006-3223(90)90591-O [pii] [DOI] [PubMed] [Google Scholar]
- Pollock VE, Schneider LS, Lyness SA. Reliability of topographic quantitative EEG amplitude in healthy late-middle-aged and elderly subjects. Electroencephalography and Clinical Neurophysiology. 1991;79(1):20–26. doi: 10.1016/0013-4694(91)90152-t. [DOI] [PubMed] [Google Scholar]
- Pollock VE, Volavka J, Goodwin DW, Mednick SA, Gabrielli WF, Knop J, Schulsinger F. The EEG after alcohol administration in men at risk for alcoholism. Archives of General Psychiatry. 1983;40(8):857–861. doi: 10.1001/archpsyc.1983.01790070047006. [DOI] [PubMed] [Google Scholar]
- Ponomareva NV, Andreeva T, Protasova M, Shagam L, Malina D, Goltsov A, Rogaev EI. Age-dependent effect of Alzheimer’s risk variant of CLU on EEG alpha rhythm in non-demented adults. Frontiers in Aging Neuroscience. 2013;5:86. doi: 10.3389/fnagi.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomareva NV, Korovaitseva GI, Rogaev EI. EEG alterations in non-demented individuals related to apolipoprotein E genotype and to risk of Alzheimer disease. Neurobiology of Aging. 2008;29(6):819–827. doi: 10.1016/j.neurobiolaging.2006.12.019. S0197-4580(07)00006-1 [pii] [DOI] [PubMed] [Google Scholar]
- Porjesz B, Almasy L, Edenberg HJ, Wang KM, Chorlian DB, Foroud T, Begleiter H. Linkage disequilibrium between the beta frequency of the human EEG and a GABA(A) receptor gene locus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(6):3729–3733. doi: 10.1073/pnas.052716399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma D, Neale MC, Boomsma DI, de Geus EJ. Are smarter brains running faster? Heritability of alpha peak frequency, IQ, and their interrelation. Behavior Genetics. 2001;31(6):567–579. doi: 10.1023/a:1013345411774. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Propping P, Kruger J, Mark N. Genetic disposition to alcoholism. An EEG study in alcoholics and their relatives. Human Genetics. 1981;59(1):51–59. doi: 10.1007/BF00278854. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R: Foundation for Statistical Computing; Vienna, Austria: 2010. Retrieved from . http://www.R-project.org. [Google Scholar]
- Rangaswamy M, Porjesz B, Chorlian DB, Choi K, Jones KA, Wang K, Begleiter H. Theta power in the EEG of alcoholics. Alcoholism, Clinical and Experimental Research. 2003;27(4):607–615. doi: 10.1097/01.ALC.0000060523.95470.8F. [DOI] [PubMed] [Google Scholar]
- Rangaswamy M, Porjesz B, Chorlian DB, Wang K, Jones KA, Bauer LO, Begleiter H. Beta power in the EEG of alcoholics. Biological Psychiatry. 2002;52(8):831–842. doi: 10.1016/s0006-3223(02)01362-8. S0006322302013628 [pii] [DOI] [PubMed] [Google Scholar]
- Rangaswamy M, Porjesz B, Chorlian DB, Wang K, Jones KA, Kuperman S, Begleiter H. Resting EEG in offspring of male alcoholics: beta frequencies. International Journal of Psychophysiology. 2004;51(3):239–251. doi: 10.1016/j.ijpsycho.2003.09.003. S0167876003002186 [pii] [DOI] [PubMed] [Google Scholar]
- Ranlund S, Nottage J, Shaikh M, Dutt A, Constante M, Walshe M, Bramon E. Resting EEG in psychosis and at-risk populations - A possible endophenotype? Schizophrenia Research. 2014 doi: 10.1016/j.schres.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray WJ, Cole HW. EEG alpha activity reflects attentional demands, and beta activity reflects emotional and cognitive processes. Science. 1985;228(4700):750–752. doi: 10.1126/science.3992243. [DOI] [PubMed] [Google Scholar]
- Rommelse NNJ, Geurts HM, Franke B, Buitelaar JK, Hartman CA. A review on cognitive and brain endophenotypes that may be common in autism spectrum disorder and attention-deficit/hyperactivity disorder and facilitate the search for pleiotropic genes. Neuroscience and Biobehavioral Reviews. 2011;35(6):1363–1396. doi: 10.1016/j.neubiorev.2011.02.015. [DOI] [PubMed] [Google Scholar]
- Rousseeuw P, Crous C, Todorov V, Ruckstuhl A, Salibian-Barrera M, Verbeke T, Maechler M. robustbase: Basic Robust Statistics (Version R package version 0.7-3) 2011 Retrieved from http://CRAN.R-project.org/package=robustbase.
- Saletu-Zyhlarz GM, Arnold O, Anderer P, Oberndorfer S, Walter H, Lesch OM, Saletu B. Differences in brain function between relapsing and abstaining alcohol-dependent patients, evaluated by EEG mapping. Alcohol and Alcoholism. 2004;39(3):233–240. doi: 10.1093/alcalc/agh041. [DOI] [PubMed] [Google Scholar]
- Salinsky MC, Oken BS, Morehead L. Test-retest reliability in EEG frequency analysis. Electroencephalography and Clinical Neurophysiology. 1991;79(5):382–392. doi: 10.1016/0013-4694(91)90203-g. [DOI] [PubMed] [Google Scholar]
- Schutter DJLG, Van Honk J. Electrophysiological ratio markers for the balance between reward and punishment. Cognitive Brain Research. 2005;24(3):685–690. doi: 10.1016/j.cogbrainres.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Smit CM, Wright MJ, Hansell NK, Geffen GM, Martin NG. Genetic variation of individual alpha frequency (IAF) and alpha power in a large adolescent twin sample. International Journal of Psychophysiology. 2006;61(2):235–243. doi: 10.1016/j.ijpsycho.2005.10.004. S0167-8760(05)00261-8 [pii] [DOI] [PubMed] [Google Scholar]
- Smit DJ, Posthuma D, Boomsma DI, De Geus EJ. The relation between frontal EEG asymmetry and the risk for anxiety and depression. Biological Psychology. 2007;74(1):26–33. doi: 10.1016/j.biopsycho.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Smit DJ, Posthuma D, Boomsma DI, Geus EJ. Heritability of background EEG across the power spectrum. Psychophysiology. 2005;42(6):691–697. doi: 10.1111/j.1469-8986.2005.00352.x. [DOI] [PubMed] [Google Scholar]
- Snyder SM, Hall JR. A meta-analysis of quantitative EEG power associated with attention-deficit hyperactivity disorder. Journal of Clinical Neurophysiology. 2006;23(5):441–456. doi: 10.1097/01.wnp.0000221363.12503.78. [DOI] [PubMed] [Google Scholar]
- Soyka M, Preuss UW, Hesselbrock V, Zill P, Koller G, Bondy B. GABA-A2 receptor subunit gene (GABRA2) polymorphisms and risk for alcohol dependence. Journal of Psychiatric Research. 2008;42(3):184–191. doi: 10.1016/j.jpsychires.2006.11.006. S0022-3956(06)00244-5 [pii] [DOI] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. American Journal of Human Genetics. 2012;91(6):1011–1021. doi: 10.1016/j.ajhg.2012.10.010. S0002-9297(12)00533-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponheim SR, Clementz BA, Iacono WG, Beiser M. Resting EEG in first-episode and chronic schizophrenia. Psychophysiology. 1994;31(1):37–43. doi: 10.1111/j.1469-8986.1994.tb01023.x. [DOI] [PubMed] [Google Scholar]
- Sponheim SR, Clementz BA, Iacono WG, Beiser M. Clinical and biological concomitants of resting state EEG power abnormalities in schizophrenia. Biological Psychiatry. 2000;48(11):1088–1097. doi: 10.1016/s0006-3223(00)00907-0. [DOI] [PubMed] [Google Scholar]
- Sponheim SR, Iacono WG, Thuras PD, Nugent SM, Beiser M. Sensitivity and specificity of select biological indices in characterizing psychotic patients and their relatives. Schizophrenia Research. 2003;63(1–2):27–38. doi: 10.1016/s0920-9964(02)00385-7. [DOI] [PubMed] [Google Scholar]
- Stam CJ. Dementia and EEG. In: Niedermeyer E, Lopes da Silva FH, editors. Electroencephalography: basic principles, clinical applications, and related fields. Lippincott Williams & Wilkins; 2005. pp. 375–393. [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Thompson PM. Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics. 2012;44(5):552–561. doi: 10.1038/ng.2250. ng.2250 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steriade M, Gloor P, Llinas RR, Lopes de Silva FH, Mesulam MM. Basic mechanisms of cerebral rhythmic activities. Electroencephalography and Clinical Neurophysiology. 1990;76(6):481–508. doi: 10.1016/0013-4694(90)90001-z. [DOI] [PubMed] [Google Scholar]
- Stewart JL, Bismark AW, Towers DN, Coan JA, Allen JJ. Resting frontal EEG asymmetry as an endophenotype for depression risk: sex-specific patterns of frontal brain asymmetry. Journal of Abnormal Psychology. 2010;119(3):502–512. doi: 10.1037/a0019196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve FA, Straumanis JJ, Patrick G, Leavitt J, Manno JE, Manno BR. Topographic quantitative EEG sequelae of chronic marihuana use: a replication using medically and psychiatrically screened normal subjects. Drug and Alcohol Dependence. 1999;56(3):167–179. doi: 10.1016/s0376-8716(99)00029-0. [DOI] [PubMed] [Google Scholar]
- Tallman JF, Gallager DW. The GABA-ergic system: a locus of benzodiazepine action. Annual Review of Neuroscience. 1985;8:21–44. doi: 10.1146/annurev.ne.08.030185.000321. [DOI] [PubMed] [Google Scholar]
- Tang Y, Chorlian DB, Rangaswamy M, Porjesz B, Bauer L, Kuperman S, Begleiter H. Genetic influences on bipolar EEG power spectra. International Journal of Psychophysiology. 2007;65(1):2–9. doi: 10.1016/j.ijpsycho.2007.02.004. S0167-8760(07)00038-4 [pii] [DOI] [PubMed] [Google Scholar]
- Tesche CD, Karhu J. Theta oscillations index human hippocampal activation during a working memory task. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(2):919–924. doi: 10.1073/pnas.97.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney AL, Gabard-Durnam L, Vogel-Farley V, Tager-Flusberg H, Nelson CA. Developmental trajectories of resting EEG power: an endophenotype of autism spectrum disorder. PloS One. 2012;7(6):e39127. doi: 10.1371/journal.pone.0039127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beijsterveldt CE, Molenaar PC, de Geus EJ, Boomsma DI. Heritability of human brain functioning as assessed by electroencephalography. American Journal of Human Genetics. 1996;58(3):562–573. [PMC free article] [PubMed] [Google Scholar]
- Venables NC, Bernat EM, Sponheim SR. Genetic and disorder-specific aspects of resting state EEG abnormalities in schizophrenia. Schizophrenia Bulletin. 2009;35(4):826–839. doi: 10.1093/schbul/sbn021. sbn021 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villafuerte S, Strumba V, Stoltenberg SF, Zucker RA, Burmeister M. Impulsiveness mediates the association between GABRA2 SNPs and lifetime alcohol problems. Genes, Brain and Behavior. 2013 doi: 10.1111/gbb.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F. Principles and significance of genetically-induced variability of the normal human EEG. EEG-EMG Zeitschrift für Elektroenzephalographie, Elektromyographie und Verwandte Gebiete. 1986;17(4):173–188. [PubMed] [Google Scholar]
- Volk DW, Pierri JN, Fritschy JM, Auh S, Sampson AR, Lewis DA. Reciprocal alterations in pre- and postsynaptic inhibitory markers at chandelier cell inputs to pyramidal neurons in schizophrenia. Cerebral Cortex. 2002;12(10):1063–1070. doi: 10.1093/cercor/12.10.1063. [DOI] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Pankratz N, Vaidyanathan U, Miller MB, Kang HM, Iacono WG. Genetic associations of nonsynonymous exonic variants with psychophysiological endophenotypes. Psychophysiology. 2014 doi: 10.1111/psyp.12349. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington MA, Traub RD, Kopell N, Ermentrout B, Buhl EH. Inhibition-based rhythms: experimental and mathematical observations on network dynamics. International Journal of Psychophysiology. 2000;38(3):315–336. doi: 10.1016/s0167-8760(00)00173-2. S0167876000001732 [pii] [DOI] [PubMed] [Google Scholar]
- Wiesel TN, Hubel DH. Comparison of the effects of unilateral and bilateral eye closure on cortical unit responses in kittens. Journal of Neurophysiology. 1965;28(6):1029–1040. doi: 10.1152/jn.1965.28.6.1029. [DOI] [PubMed] [Google Scholar]
- Winterer G, Kloppel B, Heinz A, Ziller M, Dufeu P, Schmidt LG, Herrmann WM. Quantitative EEG (QEEG) predicts relapse in patients with chronic alcoholism and points to a frontally pronounced cerebral disturbance. Psychiatry Research. 1998;78(1–2):101–113. doi: 10.1016/s0165-1781(97)00148-0. S0165-1781(97)00148-0 [pii] [DOI] [PubMed] [Google Scholar]
- Winterer G, Mahlberg R, Smolka MN, Samochowiec J, Ziller M, Rommelspacher HP, Sander T. Association analysis of exonic variants of the GABA(B)-receptor gene and alpha electroencephalogram voltage in normal subjects and alcohol-dependent patients. Behavior Genetics. 2003;33(1):7–15. doi: 10.1023/a:1021043315012. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. S0002-9297(10)00598-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. Genome-wide complex trait analysis (GCTA): methods, data analyses, and interpretations. Genome-Wide Association Studies and Genomic Prediction. 2013 Jun 13;1019:215–236. doi: 10.1007/978-1-62703-447-0_9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.