Abstract
Dinoflagellates are a major cause of harmful algal blooms (HABs), with consequences for coastal marine ecosystem functioning and services. Alexandrium fundyense (previously Alexandrium tamarense) is one of the most abundant and widespread toxigenic species in the temperate Northern and Southern Hemisphere and produces paralytic shellfish poisoning toxins as well as lytic allelochemical substances. These bioactive compounds may support the success of A. fundyense and its ability to form blooms. Here we investigate the impact of grazing on monoclonal and mixed set-ups of highly (Alex2) and moderately (Alex4) allelochemically active A. fundyense strains and a non-allelochemically active conspecific (Alex5) by the heterotrophic dinoflagellate Polykrikos kofoidii. While Alex4 and particularly Alex5 were strongly grazed by P. kofoidii when offered alone, both strains grew well in the mixed assemblages (Alex4 + Alex5 and Alex2 + Alex5). Hence, the allelochemical active strains facilitated growth of the non-active strain by protecting the population as a whole against grazing. Based on our results, we argue that facilitation among clonal lineages within a species may partly explain the high genotypic and phenotypic diversity of Alexandrium populations. Populations of Alexandrium may comprise multiple cooperative traits that act in concert with intraspecific facilitation, and hence promote the success of this notorious HAB species.
Keywords: Alexandrium fundyense, grazing protection, harmful algal blooms, phenotypic diversity, allele-specific quantitative PCR, associational resistance
1. Introduction
Marine phytoplankton account for approximately half of the global annual net primary production [1]. Their high biomass turnover rate and conversion of light energy, CO2 and inorganic nutrients into organic material drive the marine pelagic ecosystem. Like many aquatic microorganisms, phytoplankton can have large population sizes, and typically have high rates of predominantly asexual reproduction. Furthermore, phytoplankton live in a rather open and seemingly homogeneous pelagic habitat. Yet, communities may also exhibit a patchy distribution even on small geographical scales, may show horizontal organization in thin layers and seem to be, at least partly, organized as metapopulations [2–5]. Molecular data show that populations of plankton species are temporally and spatially distributed based on historical, ecological and local oceanographic conditions [6–9]. Furthermore, distinct phytoplankton populations have high genetic diversity [9–13] and typically comprise a variety of genotypes and phenotypes [7,14,15].
Some phytoplankton species have the ability to produce toxins, and their proliferation leads to harmful algal blooms (HABs). These HABs can have major implications for marine ecosystems, causing mortality of fishes and other marine life, and threaten human health through accumulation of toxins in the food chain [16,17]. Among the notorious HAB formers, some dinoflagellates are found to produce a variety of potent bioactive substances of which paralytic shellfish poisoning toxins (PSTs) are most common [17]. Additionally, many dinoflagellates have the ability to produce and release allelochemical compounds of poorly characterized chemical nature that exert negative effects on a range of unicellular eukaryotic organisms [18–20]. Intracellular PSTs and the extracellular allelochemicals have been shown to provide cells with protection against grazers [19,21–23] and competitors under certain conditions [24]. Since grazing can remove up to 50% of gross biomass production [25], production and release of bioactive compounds may add to the ecological success of dinoflagellates [20,26,27].
Understanding the evolution of phenotypic traits, such as the production of PSTs or allelochemicals, and their variation in genotypically diverse dinoflagellate populations [7,14] is a challenge, as only some individuals carry the costs, whereas the benefits are shared within the whole population. Thus, the production of toxins and allelochemicals provide an advantage to non-producers (i.e. cheaters), particularly in spatially unstructured populations [28]. Within a structured population of closely related individuals, however, cooperative traits can be favoured as a public good and facilitate the success of the entire population [29–32]. This principle of facilitation [33,34] has been shown in populations of various organisms, including bacteria [35], toxigenic cyanobacteria [36], amoeba [37] and yeast [38]. Little is yet known about intraspecific facilitation in populations of planktonic algae. Among flagellated algae many are known to produce extracellular toxins [20,27,39–41], and, depending on the environmental conditions, these toxins might serve as a direct benefit at the cell level or as an exploitable public good for the whole population [42].
Here, we assess whether facilitation may occur within phenotypically diverse populations of the common toxigenic bloom forming dinoflagellate Alexandrium fundyense (previously Alexandrium tamarense [43]) [7,14], and whether production of allelochemical substances by some individuals in an experimental population provides benefits to non-allelochemical producing individuals and thereby facilitate the success of multiple strains. To answer this question, we investigated whether Alexandrium strains with intermediate and high allelochemical activity can protect a non-allelochemical producing conspecific against the heterotrophic protist Polykrikos kofoidii. In this study, we applied for the first time, to our knowledge, allele-specific quantitative PCR (asqPCR) in a marine microalgae. The method proved to be highly reliable in mixtures of known cell ratios of clonal assemblies and thus allows assessing the relative cell numbers with high precision in time course experiments to obtain insights into intraspecific processes. We show that the allelochemically non-active strain is protected by the active strains. Our results thereby demonstrate intraspecific facilitation, which may partly explain the high genotypic and phenotypic diversity that is often observed in marine dinoflagellate populations. Hence, intraspecific facilitation might be another, yet often overseen mechanism that can explain the occurrence of exploitable public goods in genotypically diverse populations.
2. Material and methods
(a). Algal cultures
Three clonal strains of A. fundyense (group I) were isolated in May 2004 from the North Sea coast of Scotland [21] and grown in K-medium [44] prepared with sterile filtered north seawater (salinity 33), pH adjusted to 8.0 at 15°C, with an incident light intensity of 150 µmol photons m–2 s–1, provided by cool white fluorescent lamps at a 14 L : 10 D photocycle. All three strains, Alex2, Alex4 and Alex5 are morphologically indistinguishable, have a similar cell size and produce PSTs. Shortly after their isolation, Alex2 and Alex4 were shown to produce allelochemically active compounds. Alex2 was highly allelochemically active (lytic), Alex4 was moderately active (lytic), and Alex5 was non-active (non-lytic) [21]. The culture of P. kofoidii was established in 2009 also from coastal waters off Scotland [45]. The culture was routinely held in 63 ml culture flasks on a slow rotating plankton wheel (1 r.p.m.) at 15°C and low light (10–20 µmol photons m–2 s–1) and fed with Lingulodinium polyedrum (CCMP 1738). A dense subculture used for experiment inoculation was starved for approximately 1 day so that no food algae were present (i.e. Polykrikos cells contained no visible food vacuoles, see the electronic supplementary material, figure S1), and Polykrikos had also not yet started massive gamete formation [44].
(b). Experimental design and set-up
Before starting the experiment, each Alexandrium strain was treated by a multi-antibiotic cocktail (50 µg ml–1 ampicillin, 3.3 µg ml–1 gentamycin, 25 µg ml–1 streptomycin, 1 µg ml–1 chloramphenicol and 10 µg ml–1 ciprofloxacin) for 5 days in order to reduce background bacterial numbers [46], without affecting the dinoflagellate nor its allelochemical properties. Small sub-samples were stained with acridine orange and checked by epifluorescence microscopy, confirming the removal of bacterial contamination [46]. After a 1 : 10 dilution in new antibiotic free media, all three strains grew in the exponential phase for 5 days. Thereafter, monoclonal cultures were grown for scaling up from 500 ml to a final 5000 ml in serial batch cultures and kept in exponential growth to ensure a similar physiological status of the strains in the experiment. Cells in exponential growth phase were washed three times with sterile filtered K-medium over a 10 µm pore size sieve in order to remove extracellular allelochemical compounds. Thus, any observed allelochemical mediated effect is assumed to be a result of the allelochemicals accumulated during the course of the experiment.
The experiment was performed in a temperature- and light-controlled culture room on a slowly rotating plankton wheel with a speed of 1 r.p.m. allowing homogeneous mixing, but with minimal turbulence. The three strains of Alexandrium were grown in monoclonal cultures with starting cell densities of 500 cells ml–1. Additionally, Alex5 was grown in two distinct two-strain mixtures with Alex2 (i.e. Alex2 + Alex5) and with Alex4 (i.e. Alex4 + Alex5) to a final concentration of 1000 cells ml–1 (i.e. 500 cells ml–1 per strain). To test for a potential impact of the higher cell densities in the mixed cultures on the ability of Polykrikos to exert control over the Alexandrium population compared to the monoclonal culture experiments, additional experiments with monoclonal cultures of Alex5 with 1000 cells ml–1 were performed (see the electronic supplementary material, figure S2). All experimental Alexandrium cultures were grown in triplicate with and without adding Polykrikos cells (20 cells ml–1). The experiment started in completely filled 1000 ml Schott flasks, and 500 ml was harvested after 24 h. The remaining cultures were further incubated in 500 ml flasks and 250 ml was harvested at day 2. Again, the remaining culture was incubated in 250 ml flasks and 125 ml was harvested at day 3. The remaining cultures incubated in 125 ml flasks was harvested on day 4. The harvested samples were divided for cell counts of Alexandrium and Polykrikos, and for DNA extraction and subsequent asqPCR.
(c). Counting procedure
Lugol's fixed (1% final concentration) Alexandrium cells were counted after sedimentation of 3 × 1 ml aliquots using an inverted microscope. All or at least 300 cells in each 1 ml aliquot were counted. For counting Polykrikos, 10 ml samples were fixed with a mixture of formalin (1% final concentration), and Lugol's iodine solution (0.3% final concentration) and settled in 10 ml settling chambers. Whole chambers were counted. For each sample, Polykrikos was scored as either containing food particles in their food vacuoles or without visible food vacuoles, in order to estimate the proportion of active grazers (see the electronic supplementary material, figure S1).
(d). Allele-specific quantitative PCR
The three haploid Alexandrium strains used in this study were genotyped at 18 microsatellite loci [47,48] (see also the electronic supplementary material, table S1) and were found to carry differently sized alleles at certain microsatellite loci. Hence, strain-specific amplicons derived by PCR from a mixed DNA template, such as those that were derived from mixed culture experiments, could be distinguished and relatively quantified by asqPCR [49]. Genomic DNA extractions were performed with a DNeasy plant mini Kit (Qiagen, Hilden, Germany) with slight modifications of the manufacturer's instructions. Cells (approx. 50–75 × 103) from experimental cultures were harvested in a 50 ml reaction tube and centrifuged at 3000g for 5 min. Cells were resuspended after addition of 400 µl buffer AP1 and the suspension was transferred to a microcentrifuge tube, into which a mixture of approximately 300 µl of 1 mm and 0.3–0.6 mm-sized glass beads was added beforehand. Contents of the tube were then mixed by vortexing and incubated for 15 min at 95°C (ThermoMixer, Eppendorf, Hamburg, Germany). Remaining intact cells were disrupted in a TissueLyser (Qiagen) for 2 × 1 min at 20 Hz. Afterwards, 4 µl RNaseA stock solution (100 mg ml−1, Qiagen) was added and the sample was incubated at 65°C for 15 min. The following steps for DNA purification have been performed according to the manufacturer's instructions (Qiagen).
PCR reactions were carried out with a Type-it Microsatellite PCR Kit (Qiagen, Hilden, Germany) with 25 µl reaction containing 1 µl (10 ng) template DNA, 12.5 µl 2 × Type-it Multiplex PCR Master Mix (including Taq polymerase and reaction buffer), and 0.2 µl (0.2 µMol, final conc.) of each primer per reaction (Atama15; Fwd: CCACATGCTCAACATTCACGTATACAG, Rev: GTATTTGCTCATATGGCTTGG [48]. For fragment analysis purposes, the forward primer was labelled with the fluorescent dye 6-FAM. For better resolution in subsequent fragment analysis, 2.5 µl of Q-Solution (5×) were added to the reaction mix. After the initial denaturation (95°C, 5 min), 35 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 1 min and elongation at 70°C for 30 s were carried out, followed by a final extension at 60°C for 30 min in a Gradient Mastercycler (Eppendorf, Hamburg, Germany). Sizing of amplified microsatellite alleles was carried out with GeneMapper v. 3.7/4.0 (Applied Biosystems, Darmstadt, Germany) after capillary electrophoresis on a 3130xl Genetic Analyzer (Applied Biosystems).
Cell numbers of the different Alexandrium strains in mixed cultures were calculated from the peak area under the specific allele peak, i.e. the sum of fluorescence signal from a strain specific allele. Total peak area was calculated for each sample as the sum of the peak area values of the two differently sized microsatellite alleles, each representative for a specific strain. The estimate of the relative abundance of a strain was then calculated as the proportion of the peak area of the specific allele of a strain from the total peak area (i.e. the sum of peak areas of alleles from both strains). This relative abundance estimation for both strains in the mixed assemblages was then converted to cell numbers by multiplication of strain specific relative abundance estimates with total cell numbers obtained by mircoscopical cell counts from the respective sample. The asqPCR assays were validated with standard curves derived from DNA mixes; i.e. from mixed samples with relative contributions of one of the two strains to the mixed population from 0, 20, 40, 50, 60, 80 and 100%. The linear regression estimated for these mixed populations of the combinations Alex2 + Alex5 and Alex4 + Alex5 showed that the relative contribution of the peak area of the allele of one strain was directly proportional to the actual proportion of cells of the respective allele in the mixture. The slope and the regression coefficient of strain Alex5 were 1.057 and r2 = 0.96 and 1.001 and r2 = 0.995 for mixed populations with Alex2 and Alex4, respectively (see the electronic supplementary material, figure S3). Cell numbers of strains from experimental samples are presented as mean values of triplicate cultures and their standard deviation.
(e). Statistical analysis
The population growth in replicated culture set-ups was calculated from day 1 to day 4 in all cases but the set-up with Alex5 with grazer, for which the experiment lasted only until day 3 when all cells were grazed. Growth rate (µ) was calculated by fitting an exponential function through all replicate cell counts in the respective time periods according to
| 3.1c |
where A refers to the cell density, A1 to cell density at the day 1, and t to the time of the experiment.
Differences in Alexandrium growth between treatments were tested using three-way ANOVA with strain and mix versus mono culture and/or grazing treatment as fixed factors, followed by post-hoc comparison of the means using Tukey's HSD [49] in Statistica v.6 (StatSoft, Hamburg, Germany). Differences of Polykrikos growth and grazing was either tested with a one-way ANOVA followed by post-hoc comparison of the means using Tukey's HSD, or using a t-test [49] and were carried out in Sigmaplot v. 12 (Systat Software, Erkrath, Germany). Normality was tested according to Shapiro–Wilk [50].
3. Results
(a). Growth in monoclonal and mixed cultures without grazer
All Alexandrium cultures started with exponential growth after day 1. The attained growth rates of all three strains in the monoclonal cultures were not significantly different and were not significantly different within the mix culture set-ups (table 1 and figure 1a). Growth rates of Alex5 in cultures starting with 500 or with 1000 cells ml–1 were not significantly different, with 0.41 ± 0.06 d–1 and 0.37 ± 0.02 d–1, respectively (t-test, t4 = 1.169; p = 0.307; see also the electronic supplementary material, figure S2).
Table 1.
Summary of three-factorial ANOVA results on the effect of the treatments (grazing, Alexandrium strain identity and culture form) on clonal growth rates. (d.f., degrees of freedom; MQ, mean square; F, test statistic; p, level of significance.)
| d.f. | MQ | F | p | |
|---|---|---|---|---|
| grazing (yes/no) | 1 | 2.366040 | 271.1199 | <0.000001 |
| strain (Alex2, Alex4, Alex5) | 2 | 0.918121 | 105.2057 | <0.000001 |
| culture form (monoclonal culture/mixed culture) | 1 | 2.648118 | 303.4428 | <0.000001 |
| grazing × strain | 2 | 0.956716 | 109.6282 | <0.000001 |
| grazing × culture form | 1 | 3.086409 | 353.6656 | <0.000001 |
| strain × culture form | 2 | 0.718774 | 82.3630 | <0.000001 |
| grazing × strain × culture form | 2 | 0.728015 | 83.4219 | <0.000001 |
| 30 | 0.008727 |
Figure 1.
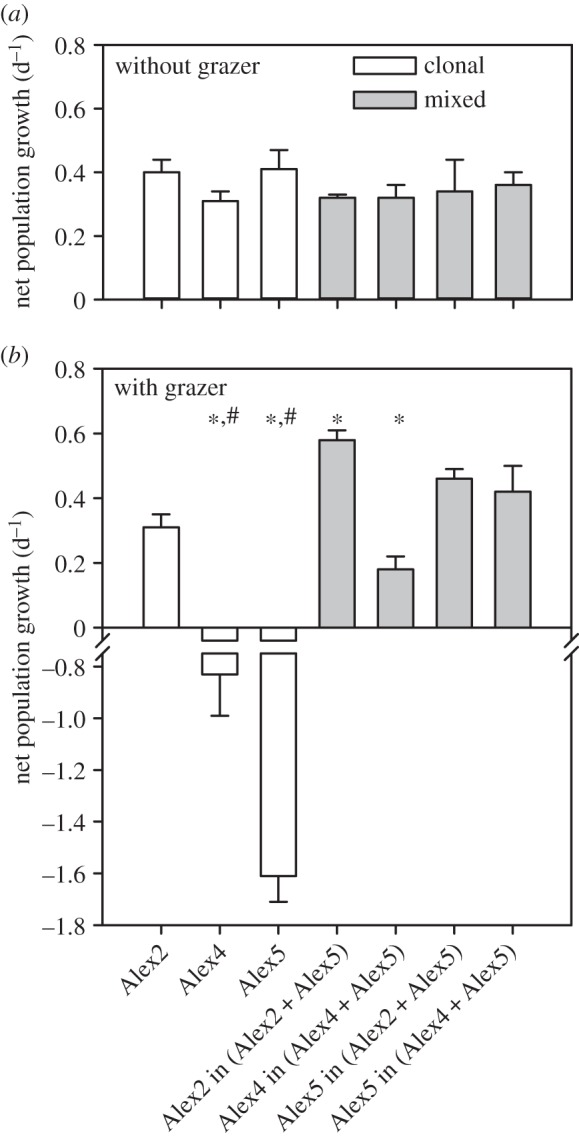
Overview of Alexandrium fundyense growth rates. (a) Monoclonal and mixed assemblages without Polykrikos kofoidii. (b) Monoclonal and mixed assemblages with P. kofoidii grazer. Values for mixed assemblages show growth rates of individual clones with the mixtures Alex2 + Alex5, and Alex4 + Alex5. Asterisks indicate significant differences between clonal and mixed assemblages, and hashtags indicate significant differences between set-ups without and with grazers (Tukey HSD post-hoc test, p < 0.05). Error bars indicate standard deviation (n = 3).
(b). Grazing impact on Alexandrium in monoclonal and mixed cultures
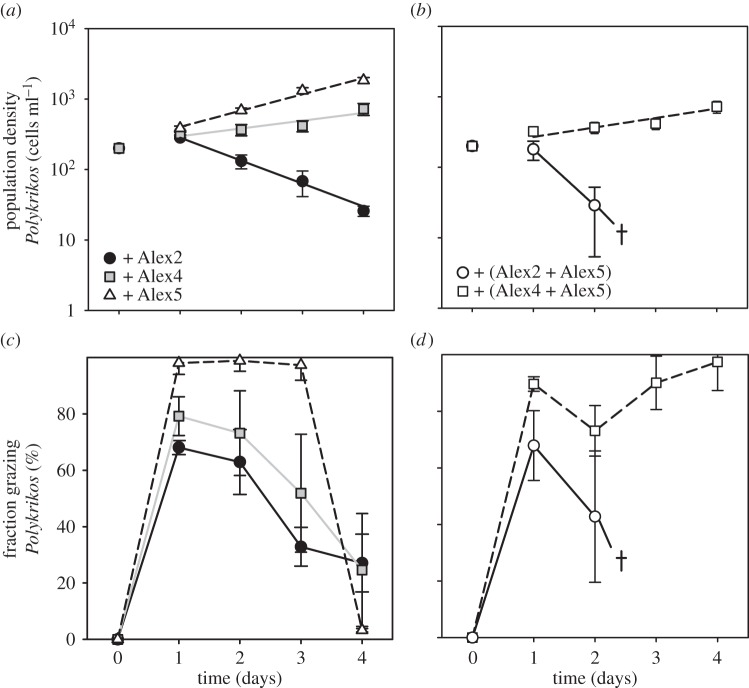
In the monoclonal cultures, net population growth of both Alex4 and Alex5 became negative after addition of Polykrikos, whereas growth of Alex2 remained unaffected compared to the control (table 1 and figures 1b and 2a,b). All strains showed a higher growth rate in the mixed cultures Alex4 + Alex5 and Alex2 + Alex5 as compared to their respective monoclonal cultures under grazing pressure (table 1 and figure 1b). Growth rates of Alex5 and Alex2 in the mixed culture Alex2 + Alex5 together with Polykrikos appear to be slightly increased, although this was statistically not significant compared with the growth rates in the mixed culture without grazer. Within the other mixed culture with Polykrikos (i.e. Alex4 + Alex5), Alex5 showed no change in growth rate, while growth of Alex4 seemed to have reduced (table 1 and figure 1).
Figure 2.

Growth of Alexandrium fundyense strains when exposed to Polykrikos kofoidii. (a) Growth rates in the monoclonal set-up with the lytic Alex2, the moderate-lytic Alex4 and the non-lytic Alex5. (b) Growth rates of individual strains in the mixed assemblages. Error bars indicate standard deviation (n = 3).
(c). Alexandrium impact on the grazer
In all experimental set-ups where Polykrikos was added, initial grazing was documented and in more than 60% Polykrikos cells at least one visible food particle at day 1 was observed (figure 3c,d). After day 1, 60–95% of Polykrikos cells had grazed on Alexandrium in all cultures. In monoclonal cultures, however, the number of Polykrikos cells with visible food vacuoles on day 1 was significantly lower when fed on the lytic strains Alex2 and Alex4 when compared with the non-lytic strain Alex5 (figure 3c,d). After day 1, the number of Polykrikos cells with visible food vacuoles declined when fed on monoclonal cultures of Alex2 and Alex4, as well as on the mixture Alex2 + Alex5, reaching 0–30% by the end of the experiment (figure 3c,d). When grown on the mixture Alex4 + Alex5, a majority of Polykrikos cells possessed food vacuoles until the end of the experiment. When Polykrikos was fed on different Alex5 concentrations, its growth rate was higher when more food was provided (t-test, t4 = 6.707, p = 0.003), with µ = 0.39 ± 0.04 and µ = 0.58 ± 0.03 for 500 and 1000 cell ml−1, respectively. Polykrikos grazed down all cells of the Alex5 cultures and became fully starved at the end of the experiment (figure 3c,d), independent of the initial Alex5 cell densities (data not shown).
Figure 3.
Growth and grazing of Polykrikos kofoidii in monoclonal and mixed assemblages of Alexandrium fundyense. (a) Growth of P. kofoidii grazing on monoclonal set-ups with lytic Alex2, the moderate-lytic Alex4 and the non-lytic Alex5. (b) Growth of P. kofoidii grazing on mixed assemblages. (c) Percentage of P. kofoidii cells with food particles when fed on monoclonal cultures, or (d) mixed assemblages. Error bars indicate standard deviation (n = 3).
Initial growth of Polykrikos (i.e. from day 0 to day 1) was positive, except when fed on the mixed culture Alex2 + Alex5 (figure 3a,b). Subsequent growth rates of Polykrikos depended on the Alexandrium strain being fed with (one-way ANOVA, F2,6 = 245, p < 0.001), showing highest growth rates when fed on monoclonal cultures of Alex5 (µ = 0.39 ± 0.04), intermediate growth rates when fed on Alex4 (µ = 0.27 ± 0.07), and negative growth rates when fed on Alex2 (µ = –0.57 ± 0.06). When Alexandrium clones were provided in a mixture, growth of Polykrikos, when compared to the monoclonal culture of Alex5 (1000 cells ml–1; µ = 0.58 ± 0.03), was reduced on Alex4 + Alex5 as a food source (µ = 0.26 ± 0.06, t-test, t4 = 8.904, p < 0.001) and even became negative when offered Alex2 + Alex5 (µ = –1.27 ± 0.09, t-test, t4 = 8.205, p = 0.001).
4. Discussion
In our experiments, all three Alexandrium strains exhibited comparable growth rates when grown in monoclonal cultures without a grazer. The strains differ significantly in their lytic activity under the chosen culture conditions, which according to our current understanding, is best explained by distinct levels of production and release of allelochemical active substances [21]. This shows that for the three strains used herein, potential cost of allelochemical production does not lead to a reduced growth rate. Furthermore, growth rates of all strains remained unaltered in mixed cultures, proving that allelochemicals produced by the lytic strains did not have intraspecific inhibitory (or supportive) effects as they do not affect growth of the non-lytic strain. While in monoclonal cultures the net population growth of the most lytic strain, Alex2, was not affected by Polykrikos, growth of both Alex4 and Alex5 decreased as a result of grazing by Polykrikos (figures 1b and 2a). Interestingly, net population growth of all three strains was positive when grown with the other Alexandrium strain in mixed cultures (i.e. Alex2 + Alex5 and Alex4 + Alex5) with Polykrikos (figures 1 and 2b). The slightly higher growth rate of the lytic strain Alex2 in the presence of Polykrikos and Alex5 indicates a benefit not only for the non-lytic Alex5 but perhaps also for the lytic strain Alex2. Whereas the beneficial effect for Alex5 can be explained by reduction of grazing owing to effects of allelochemicals produced by Alex2, a beneficial effect for Alex2 is more difficult to explain and needs to be confirmed in additional experiments. Increased growth solely related to mixotrophy is unlikely, as Alex2 does not benefit in terms of growth from lysed grazers in the monoclonal grazing set-up, indicating that other mutual processes might be involved in the mixed cultures. An additional hint for synergistic effects in the mixed set-ups is that Polykrikos was slightly more affected in the mixed culture Alex2 + Alex5, indicated by the extinction of all Polykrikos cells before the third day of the experiment (cf. figure 3c,d).
The increase in growth rate of Alex4 in the mixed culture Alex4 + Alex5 with grazers when compared with that in the monoculture with grazers seems to be an indirect positive effect of the presence of Alex5. The grazing pressure of Polykrikos on the lytic Alex4 in the mixed culture could be lower simply because Alex5 was available as alternative food source. As a consequence, population densities of Alex4 could remain relatively high allowing a sufficient production of allelochemicals for protection against Polykrikos, although Polykrikos did not show a reduced growth in the Alex4 + Alex5 mixed culture compared to the monoclonal culture of Alex4 (figure 2). Yet, growth of Polykrikos in mixed culture with Alex4 is reduced compared with the growth when Alex5 is provided as food alone, when its growth was highest, demonstrating the effect of the allelochemical substances as a grazer deterrent (figure 3a).
Polykrikos grew well on monoclonal cultures of Alex5. The mixture Alex2 + Alex5 reduced its growth when compared with growth on Alex5 alone. Although the mixed assemblage started at a higher cell density (with 1000 versus 500 cells ml−1 in the monoclonal cultures), it is unlikely that this initial density difference negatively affected Polykrikos growth, as Polykrikos grew better on a higher Alex5 population density (see the electronic supplementary material, figure S2). Hence, a detrimental effect of allelochemicals produced by Alex2 and Alex4 on Polykrikos is the most likely cause for its reduced growth in the presence of allelochemically active Alexandrium.
The view that allelochemicals play an important ecological role is widely shared (see reviews by [20,26,40,41,51]). It may be argued that other competing species might also benefit from reducing grazer fitness [52]. However, it has been shown that the same allelochemicals strongly affect growth of many phytoplankton species that are potential resource competitors [24,53–55]. Therefore, any indirect beneficial effects for competitors (e.g. through release from grazing pressure) might be reduced or excluded as long as their growth is also affected by the allelochemicals. Our results clearly demonstrate that allelochemicals can protect both the producers as well as a non-producing conspecific against grazing. The observed facilitation by grazing protection might resemble associational resistance [56,57], described for terrestrial plant communities [58–60], as well as among macroalgal species and their epiphytes [61–63]. Such associational resistance may also occur in pelagic microalgae populations even at the intraspecific level, within assemblages of sufficiently high population densities. Our results indicate that facilitation plays a role in phytoplankton populations and we show that benefits are shared between producer and non-producer strains.
It is as challenging to understand what drives the evolution of a trait such as production and release of allelochemical substances that benefit the producer as well as the population as a whole, as to understand how these traits are maintained in the population [64]. What is the selective advantage of allelochemical substances for the producer, i.e. how can this trait evolve and how can it be maintained in phenotypically diverse populations under natural selection? According to theory, selection for a public good might take place when costs versus benefits for the producer are relatively low and the relatedness between producer and non-producer is relatively high [31,38]. Positive population effects of allelochemicals might result at the cellular level (private good) and higher structural levels (populations) could benefit indirectly [65]. Before allelochemicals start serving as a public good, a high relative abundance of producers is required in order to support the entire population. Consequently, the allelopathic phenotype as a trait might be maintained by frequency-dependent selection. Indeed, in a natural population of A. fundyense, only two out of 88 clonal isolates were non-producers, whereas all others were allelochemically active, though the allelochemical potency was normally distributed and varied widely [14].
In general, intraspecific genetic and phenotypic diversity is discussed to have an important impact on evolutionary and ecological process and hence the population's dynamics and success [66–68]. A high variability in phenotypic traits involved in interactions among individuals of different clonal linages within a population may allow mutualistic intraspecific facilitation in various ways, and thereby promote the overall success of Alexandrium. If cooperative traits governing intraspecific interactions are common in mixed Alexandrium populations, the high phenotypic and genotypic diversity of these populations may be explained for example, by compatibility among these beneficial phenotypic traits in different strains. Such alternative traits may include for instance, chain formation, swimming speed, nutrient uptake capabilities, intrinsic growth rate and PST content [22,23,69–71]. Indeed, the non-lytic strain Alex5 in our experiment contained the highest amount of PSTs (data not shown), a trait that potentially allows protection against grazing by copepods [22,23,69]. It is conceivable that with a higher genotypic diversity, more cooperative traits can be provided that benefit the entire population.
The observed high genotypic diversity of phytoplankton populations [9–13] may be sustained by mutualistic interactions of cooperative traits. Yet, with respect to the functioning of extracellular allelochemical substances, the seemingly homogeneous or ephemeral spatial distribution pattern of marine phytoplankton populations and their typical low population densities may contradict with (or limit) the effectiveness of beneficial interactions derived from laboratory experiments with high cell concentration [64]. The functioning of allelochemical mediated facilitation in natural populations will thus depend on the degree of spatial dispersal, i.e. the local accumulation of a population, as well as on the rate at which extracellular allelochemicals are produced and excreted, and on the rate of their diffusion and degradation [51,72]. In dinoflagellates, bloom formation typically occurs at low mixing and water column stratification [26]. Under such conditions, plankton populations are often not homogeneously distributed, but rather show a spatially structured distribution, for instance as patches or thin layers [2–4,72]. Flagellar movement may favour accumulation in patches [72], which is presumably also required for sexual reproduction in the life cycle of Alexandrium [17], as increased encounter rates in patches may allow Alexandrium gametes to find their corresponding mating type [73]. The fine-scale analysis of the spatial distribution of dinoflagellates in the water column indicates that they tend to accumulate. Cooperative traits such as allelochemicals might facilitate the population success within patches of high cell densities.
The conditions in our culture experiment obviously represent a simplification of the natural environment, where Alexandrium populations are composed of a much higher diversity in genotypes and phenotypic traits. We partially accounted for this natural diversity by selecting three strains with distinct allelochemical activities, i.e. one highly active (Alex2), one intermediately active (Alex4) and one non-active (Alex5). Such a range of allelochemical activities is also found in the natural environment, and it is therefore very likely that the observed facilitation by allelochemicals occurs during natural Alexandrium blooms. In our experiment, we worked with cell densities reflecting dense Alexandrium bloom conditions rather than pre-bloom conditions [74,75]. Obviously, production of allelochemicals becomes favourable when sufficient cells are present, and these compounds may thus play a crucial role in the prolongation of HABs [64]. Especially, if phenotypic traits in allelopathic microalgae serve multiple purposes over the course of a bloom, for example by functioning as private versus public good during low and high population densities, respectively [42]. It is conceivable that the transient nature of a selective advantage by allelochemical production leads to an increase in the phenotypic diversity, and its underlying genetic diversity, of natural populations, because the selective advantage of various phenotypes at different stages of population development will be balanced over time.
In this study, by adopting the approach of asqPCR for A. fundyense, we were able to follow the strain-specific responses to grazing in mixed culture set-ups, and we showed that allelochemical active A. fundyense strains can protect a non-lytic conspecific from grazing by P. kofoidii. Our findings are in line with the view that a multitude of hitherto not well-recognized cooperative traits, including allelochemical mediated intraspecific facilitation, may contribute to the high genotypic and phenotypic diversity of Alexandrium populations. Multiple traits potentially lead to mutual facilitation among phenotypically diverse clonal lineages within an Alexandrium population, and thereby further promote the success of these notorious HAB species.
Supplementary Material
Acknowledgements
The authors are grateful to Helmut Hillebrand, Victor Smetacek and Mathias Wegner for their helpful comments on the manuscript.
Data accessibility
Additional experimental data are available at http://dx.doi.org/10.1594/PANGAEA.836236
Funding statement
Financial support was provided by the PACES research program of the Alfred-Wegener-Institute Helmholtz-Zentrum für Polar- und Meeresforschung.
References
- 1.Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJR. 2004. The evolution of modern eukariotic phytoplankton. Science 305, 354–360. ( 10.1126/science.1095964) [DOI] [PubMed] [Google Scholar]
- 2.Durham WM, Stocker R. 2012. Thin phytoplankton layers: characteristics, mechanisms, and consequences. Annu. Rev. Mar. Sci. 4, 177–207. ( 10.1146/annurev-marine-120710-100957) [DOI] [PubMed] [Google Scholar]
- 3.Menden-Deuer S. 2012. Structure-dependent phytoplankton photosynthesis and production rates: implications for the formation, maintenance, and decline of plankton patches. Mar. Ecol. Prog. Ser. 468, 15–30. ( 10.3354/meps09968) [DOI] [Google Scholar]
- 4.Montagnes DJS, Poulton AJ, Shammon TM. 1999. Mesoscale, finescale and microscale distribution of micro- and nanoplankton in the Irish Sea, with emphasis on ciliates and their prey. Mar. Biol. 134, 167–179. ( 10.1007/s002270050535) [DOI] [Google Scholar]
- 5.Rynearson TA, Lin EO, Armbrust EV. 2009. Metapopulation structure in the planktonic diatom Ditylum brightwellii (Bacillariophyceae). Protist 160, 111–121. ( 10.1016/j.protis.2008.10.003) [DOI] [PubMed] [Google Scholar]
- 6.Foissner W. 2006. Biogeography and dispersal of microorganisms: a review emphasizing protists. Acta Protozool. 45, 111–136. [Google Scholar]
- 7.Alpermann TJ, Beszteri B, John U, Tillmann U, Cembella AD. 2009. Implications of life history transitions on the population genetic structure of the toxigenic marine dinoflagellate Alexandrium tamarense. Mol. Ecol. 18, 2122–2133. ( 10.1111/j.1365-294X.2009.04165.x) [DOI] [PubMed] [Google Scholar]
- 8.Nagai S, et al. 2007. Microsatellite markers reveal population genetic structure of the toxic dinoflagellate Alexandrium tamarense (Dinophyceae) in Japanese coastal waters. J. Phycol. 43, 43–54. ( 10.1111/j.1529-8817.2006.00304.x) [DOI] [Google Scholar]
- 9.Tahvanainen P, Alpermann TJ, Figueroa RI, John U, Hakanen P, Nagai S, Blomster J, Kremp A. 2012. Patterns of post-glacial genetic differentiation in marginal populations of a marine microalga. PLoS ONE 7, e53602 ( 10.1371/journal.pone.0053602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans KM, Kühn SF, Hayes PK. 2005. High levels of genetic diversity and low levels of genetic differentiation in North Sea Pseudo-nitzschia pungens (Bacillariophyceae) populations. J. Phycol. 41, 506–514. ( 10.1111/j.1529-8817.2005.00084.x) [DOI] [Google Scholar]
- 11.Logares R, Boltovskoy A, Bensch S, Laybourn-Parry J, Rengefors K. 2009. Genetic diversity patterns in five protist species occurring in lakes. Protist 160, 301–317. ( 10.1016/j.protis.2008.10.004) [DOI] [PubMed] [Google Scholar]
- 12.Iglesias-Rodríguez MD, Schofield OM, Batley J, Medlin LK, Hayes PK. 2006. Intraspecific genetic diversity in the marine coccolithophore Emiliania huxleyi (Prymnesiophyceae): the use of microsatellite analysis in marine phytoplankton population studies. J. Phycol. 42, 526–536. ( 10.1111/j.1529-8817.2006.00231.x) [DOI] [Google Scholar]
- 13.Lowe CD, Montagnes DJS, Martin LE, Watts PC. 2010. High genetic diversity and fine-scale spatial structure in the marine flagellate Oxyrrhis marina (Dinophyceae) uncovered by microsatellite loci. PLoS ONE 5, e15557 ( 10.1371/journal.pone.0015557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alpermann TJ, Tillmann U, Beszteri B, Cembella AD, John U. 2010. Phenotypic variation and genotypic diversity in a planktonic population of the toxigenic marine dinoflagellate Alexandrium tamarense (Dinophyceae). J. Phycol. 46, 18–32. ( 10.1111/j.1529-8817.2009.00767.x) [DOI] [Google Scholar]
- 15.Bachvaroff TR, Adolf JE, Place AR. 2009. Strain variation in Karlodinium veneficum (Dinophyceae): toxin profiles, pigments, and growth characteristics. J. Phycol. 45, 137–153. ( 10.1111/j.1529-8817.2008.00629.x) [DOI] [PubMed] [Google Scholar]
- 16.Hallegraeff GM. 2003. Harmful algal blooms: a global overview. In Manual on harmful marine microalgae (eds Hallegraeff GM, Anderson DM, AD Cembella), pp. 25–49. Paris, France: UNESCO Publishing. [Google Scholar]
- 17.Anderson DM, Alpermann TJ, Cembella AD, Collos Y, Masseret E, Montresor M. 2012. The globally distributed genus Alexandrium: multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 14, 10–35. ( 10.1016/j.hal.2011.10.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma HY, Krock B, Tillmann U, Cembella A. 2009. Preliminary characterization of extracellular allelochemicals of the toxic marine dinoflagellate Alexandrium tamarense using a Rhodomonas salina bioassay. Mar. Drugs 7, 497–522. ( 10.3390/md7040497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tillmann U, John U. 2002. Toxic effects of Alexandrium spp. on heterotrophic dinoflagellates: an allelochemical defence mechanism independent of PSP-toxin content. Mar. Ecol. Prog. Ser. 230, 47–58. ( 10.3354/meps230047) [DOI] [Google Scholar]
- 20.Cembella AD. 2003. Chemical ecology of eukaryotic microalgae in marine ecosystems. Phycologia 42, 420–447. ( 10.2216/i0031-8884-42-4-420.1) [DOI] [Google Scholar]
- 21.Tillmann U, Alpermann T, da Purificação RC, Krock B, Cembella A. 2009. Intra-population clonal variability in allelochemical potency of the toxigenic dinoflagellate Alexandrium tamarense. Harmful Algae 8, 759–769. ( 10.1016/j.hal.2009.03.005) [DOI] [Google Scholar]
- 22.Wohlrab S, Iversen MH, John U. 2010. A molecular and co-evolutionary context for grazer induced toxin production in Alexandrium tamarense. PLoS ONE 5, e15039 ( 10.1371/journal.pone.0015039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selander E, Thor P, Toth G, Pavia H. 2006. Copepods induce paralytic shellfish toxin production in marine dinoflagellates. Proc. R. Soc. B 273, 1673–1680. ( 10.1098/rspb.2006.3502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tillmann U, Hansen PJ. 2009. Allelopathic effects of Alexandrium tamarense on other algae: evidence from mixed growth experiments. Aquat. Microb. Ecol. 57, 101–112. ( 10.3354/ame01329) [DOI] [Google Scholar]
- 25.Landry MR, Calbet A. 2004. Microzooplankton production in the oceans. ICES J. Mar. Sci. 61, 501–507. ( 10.1016/j.icesjms.2004.03.011) [DOI] [Google Scholar]
- 26.Smayda TJ. 2002. Adaptive ecology, growth strategies and the global bloom expansion of dinoflagellates. J. Oceanogr. 58, 281–294. ( 10.1023/A:1015861725470) [DOI] [Google Scholar]
- 27.Granéli E, Hansen PJ. 2006. Allelopathy in harmful microalgae: a mechanism to compete for resources? In Ecology of harmful algae (eds Granéli E, Turner JT.), pp. 189–201. Berlin, Germany: Springer. [Google Scholar]
- 28.Lewis WM., Jr 1986. Evolutionary interpretations of allelochemical interactions in phytoplankton algae. Am. Nat. 127, 184–194. ( 10.1086/284477) [DOI] [Google Scholar]
- 29.Hamilton WD. 1964. The genetical evolution of social behavior I. J. Theor. Biol. 7, 1–16. ( 10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- 30.Hamilton WD. 1964. The genetical evolution of social behaviour II. J. Theor. Biol. 7, 17–52. ( 10.1016/0022-5193(64)90039-6) [DOI] [PubMed] [Google Scholar]
- 31.Xavier JB. 2011. Social interaction in synthetic and natural microbial communities. Mol. Syst. Biol. 7, 483 ( 10.1038/msb.2011.16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkins TJ, Swain PS. 2009. Strategies for cellular decision-making. Mol. Syst. Biol. 5, 326 ( 10.1038/msb.2009.83) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruno JF, Stachowicz JJ, Bertness MD. 2003. Inclusion of facilitation into ecological theory. Trends Ecol. Evol. 18, 119–125. ( 10.1016/S0169-5347(02)00045-9) [DOI] [Google Scholar]
- 34.West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat. Rev. Micro. 4, 597–607. (http://www.nature.com/nrmicro/journal/v4/n8/suppinfo/nrmicro1461_S1.html) [DOI] [PubMed] [Google Scholar]
- 35.Lee HH, Molla MN, Cantor CR, Collins JJ. 2010. Bacterial charity work leads to population-wide resistance. Nature (Lond.) 467, 82–86. ( 10.1038/nature09354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Gremberghe I, Vanormelingen P, Vanelslander B, Van der Gucht K, D'hondt S, De Meester L, Vyverman W. 2009. Genotype-dependent interactions among sympatric Microcystis strains mediated by Daphnia grazing. Oikos 118, 1647–1658. ( 10.1111/j.1600-0706.2009.17538.x) [DOI] [Google Scholar]
- 37.Mehdiabadi NJ, Jack CN, Farnham TT, Platt TG, Kalla SE, Shaulsky G, Queller DC, Strassmann JE. 2006. Social evolution: kin preference in a social microbe. Nature 442, 881–882. (http://www.nature.com/nature/journal/v442/n7105/suppinfo/442881a_S1.html). [DOI] [PubMed] [Google Scholar]
- 38.MacLean RC, Fuentes-Hernandez A, Greig D, Hurst LD, Gudelj I. 2010. A mixture of ‘cheats’ and ‘co-operators’ can enable maximal group benefit. PLoS Biol. 8, e1000486 ( 10.1371/journal.pbio.1000486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poulson KL, Sieg RD, Kubanek J. 2009. Chemical ecology of the marine plankton. Nat. Prod. Rep. 26, 729–745. ( 10.1039/b806214p) [DOI] [PubMed] [Google Scholar]
- 40.Legrand C, Rengefors K, Fistarol GO, Granéli E. 2003. Allelopathy in phytoplankton—biochemical, ecological and evolutionary aspects. Phycologia 42, 406–419. ( 10.2216/i0031-8884-42-4-406.1) [DOI] [Google Scholar]
- 41.Tillmann U, John U, Krock B, Cembella AD. 2008. Allelopathic effects of bioactive compounds produced by harmful algae. In Proc. of the 12th Int. Conf. on Harmful Algae (eds Moestrup Ø, et al.), pp. 12–18. Copenhagen, Denmark: International Society for the Study of Harmful Algae and Intergovernmental Oceanographic Commission of UNESCO. [Google Scholar]
- 42.Driscoll WW, Espinosa NJ, Eldakar OT, Hackett JD. 2012. Allelopathy as an emergent, exploitable public good in the bloo-forming microalga Prymnesium parvum. Evolution 67, 1582–1590. ( 10.1111/evo.12030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.John U, et al. In press. Formal revision of the Alexandrium tamarense species complex (Dinophyceae) taxonomy: the introduction of five species with emphasis on molecular-based (rDNA) classification. Protist. ( 10.1016/j.protis.2014.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keller MD, Selvin RC, Claus W, Guillard RRL. 1987. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23, 633–638. ( 10.1111/j.1529-8817.1987.tb04217.x) [DOI] [Google Scholar]
- 45.Tillmann U, Hoppenrath M. 2013. Life cycle of the pseudocolonial dinoflagellate Polykrikos kofoidii (Gymnodiniales, Dinoflagellata). J. Phycol. 49, 298–317. ( 10.1111/jpy.12037) [DOI] [PubMed] [Google Scholar]
- 46.Jaeckisch N, Yang I, Wohlrab S, Glöckner G, Kroymann J, Vogel H, Cembella A, John U. 2011. Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii. PLoS ONE 6, e28012 ( 10.1371/journal.pone.0028012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alpermann TJ, John U, Medlin LK, Edwards KJ, Hayes PK, Evans KM. 2006. Six new microsatellite markers for the toxic marine dinoflagellate Alexandrium tamarense. Mol. Ecol. Notes 6, 1057–1059. ( 10.1111/j.1471-8286.2006.01432.x) [DOI] [Google Scholar]
- 48.Nagai S, Lian C, Hamaguchi M, Matsuyama Y, Itakaru S, Hogetsu T. 2004. Development of microsatellite markers in the toxic dinoflagellate Alexandrium tamarense (Dinophyceae). Mol. Ecol. Notes 4, 83–85. ( 10.1046/j.1471-8286.2003.00576.x) [DOI] [Google Scholar]
- 49.Meyer JR, Ellner SP, Hairston NG, Jones LE, Yoshida T. 2006. Prey evolution on the time scale of predator–prey dynamics revealed by allele-specific quantitative PCR. Proc. Natl Acad. Sci. USA 103, 10 690–10 695. ( 10.1073/pnas.0600434103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokal RR, Rohlf FJ. 1995. Biometry: the principles and practice of statistics in biological research, 3rd edn, 880 p New York, NY: Freeman. [Google Scholar]
- 51.Prince EK, Myers TL, Kubanek J. 2008. Effects of harmful algal blooms on competitors: allelopathic mechanisms of the red tide dinoflagellate Karenia brevis. Limnol. Oceanogr. 53, 531–541. ( 10.4319/lo.2008.53.2.0531) [DOI] [Google Scholar]
- 52.Flynn KJ. 2008. Attack is not the best form of defense: lessons from harmful algal bloom dynamics. Harmful Algae 8, 129–139. ( 10.1016/j.hal.2008.08.007) [DOI] [Google Scholar]
- 53.Tillmann U, Alpermann T, John U, Cembella A. 2008. Allelochemical interactions and short-term effects of the dinoflagellate Alexandrium on selected photoautotrophic and heterotrophic protists. Harmful Algae 7, 52–64. ( 10.1016/j.hal.2007.05.009) [DOI] [Google Scholar]
- 54.Fistarol GO, Legrand C, Selander E, Hummert C, Stolte W, Granéli E. 2004. Allelopathy in Alexandrium spp.: effect on a natural plankton community and on algal monocultures. Aquat. Microb. Ecol. 35, 45–56. ( 10.3354/ame035045) [DOI] [Google Scholar]
- 55.Weissbach A, Rudstrom M, Olofsson M, Bechemin C, Icely J, Newton A, Tillmann U, Legrand C. 2011. Phytoplankton allelochemical interactions change microbial food web dynamics. Limnol. Oceanogr. 56, 899–909. ( 10.4319/lo.2011.56.3.0899) [DOI] [Google Scholar]
- 56.Tahvanainen JO, Root RB. 1972. The influence of vegetational diversity on the population ecology of a specialized herbivore, Phyllotreta cruciferae (Coleoptera: Chrysomelidae). Oecologia 10, 321–346. ( 10.1007/BF00345736) [DOI] [PubMed] [Google Scholar]
- 57.Barbosa P, Hines J, Kaplan I, Martinson H, Szczepaniec A, Szendrei Z. 2009. Associational resistance and associational susceptibility: having right or wrong neighbors. Annu. Rev. Ecol. Evol. Syst. 40, 1–20. ( 10.1146/annurev.ecolsys.110308.120242) [DOI] [Google Scholar]
- 58.Atsatt PR, O'Dowd DJ. 1976. Plant defense guilds. Science 193, 24–29. ( 10.1126/science.193.4247.24) [DOI] [PubMed] [Google Scholar]
- 59.Hambäck PA, Agren J, Ericson L. 2000. Associational resistance: insect damage to purple loosestrife reduced in thickets of sweet gale. Ecology 81, 1784–1794. ( 10.1890/0012-9658(2000)081[1784:ARIDTP]2.0.CO;2) [DOI] [Google Scholar]
- 60.Hambäck PA, Pettersson J, Ericson L. 2003. Are associational refuges species-specific? Funct. Ecol. 17, 87–93. ( 10.1046/j.1365-2435.2003.00699.x) [DOI] [Google Scholar]
- 61.Wahl M, Hay ME. 1995. Associational resistance and shared doom: effects of epibiosis on herbivory. Oecologia 102, 329–340. ( 10.1007/BF00329800) [DOI] [PubMed] [Google Scholar]
- 62.Karez R, Engelbrecht S, Sommer U. 2000. ‘Co-consumption’ and ‘protective coating’: two new proposed effects of epiphytes on their macroalgal hosts in mesograzer-epiphyte-host interactions. Mar. Ecol.-Prog. Ser. 205, 85–93. ( 10.3354/meps205085) [DOI] [Google Scholar]
- 63.Smith TB, Fong P, Kennison R, Smith J. 2010. Spatial refuges and associational defenses promote harmful blooms of the alga Caulerpa sertularioides onto coral reefs. Oecologia 164, 1039–1048. ( 10.1007/s00442-010-1698-x). [DOI] [PubMed] [Google Scholar]
- 64.Jonsson PR, Pavia H, Toth G. 2009. Formation of harmful algal blooms cannot be explained by allelopathic interactions. Proc. Natl Acad. Sci. USA 106, 11 177–11 182. ( 10.1073/pnas.0900964106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oshaka S. 2012. Emergence, hierarchy and top-down causation in evolutionary biology. Interface Focus 2, 49–54. ( 10.1098/rsfs.2011.0046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bijlsma R, Loeschcke V. 2005. Environmental stress, adaptation and evolution: an overview. J. Evol. Biol. 18, 744–749. ( 10.1111/j.1420-9101.2005.00962.x) [DOI] [PubMed] [Google Scholar]
- 67.Hughes AR, Inouye BD, Johnson MT, Underwood N, Vellend M. 2008. Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623. ( 10.1111/j.1461-0248.2008.01179.x) [DOI] [PubMed] [Google Scholar]
- 68.Bolnick DI, et al. 2011. Why intraspecific trait variation matters in community ecology. Trends Ecol. Evol. 26, 183–192. ( 10.1016/j.tree.2011.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Selander E, Jakobsen HH, Lombard F, Kiørboe T. 2011. Grazer cues induce stealth behavior in marine dinoflagellates. Proc. Natl Acad. Sci. USA 108, 4030–4034. ( 10.1073/pnas.1011870108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van de Waal DB, Eberlein T, John U, Wohlrab S, Rost B. 2013. Impact of elevated pCO2 on PSP toxin production by Alexandrium tamarense. Toxicon 78, 58–67. ( 10.1016/j.toxicon.2013.11.011) [DOI] [PubMed] [Google Scholar]
- 71.Van de Waal DB, Tillmann U, Mingming Z, Koch BP, Rost B, John U. 2013. Nutrient pulse induces dynamic changes in cellular C:N:P, amino acids, and paralytic shellfish poisoning toxins in Alexandrium tamarense. MEPS 493, 57–69. ( 10.3354/meps10532) [DOI] [Google Scholar]
- 72.Durham WM, Climent E, Barry M, De Lillo F, Boffetta G, Cencini M, Stocker R. 2013. Turbulence drives microscale patches of motile phytoplankton. Nat. Commun. 4, 2148 ( 10.1038/ncomms3148) [DOI] [PubMed] [Google Scholar]
- 73.Wyatt T, Jenkinson IR. 1997. Notes on Alexandrium population dynamics. J. Plankton Res. 19, 551–575. ( 10.1093/plankt/19.5.551) [DOI] [Google Scholar]
- 74.McGillicuddy DJ, Jr, Brosnahan ML DAC, Kaefer BA, Manning JP, Martin JL, Pilskaln CH, Townsend DW, Anderson DM. 2014. A red tide of Alexandrium fundyense in the Gulf of Maine. Deep-Sea Res. II 103, 174–184. ( 10.1016/j.dsr2.2013.05.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harwood DT, Boundy M, Selwood AI, van Ginkel R, MacKenzie L, McNabb PS. 2013. Refinement and implementation of the Lawrence method (AOAC 2005.06) in a commercial laboratory: assay performance during an Alexandrium catenella bloom event. Harmful Algae 24, 20–31. ( 10.1016/j.hal.2013.01.003) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional experimental data are available at http://dx.doi.org/10.1594/PANGAEA.836236