Abstract
Background and Question
The harvesting of medicinal plants from wild sources is escalating in many parts of the world, compromising the long-term survival of natural populations of medicinally important plants and sustainability of sources of raw material to meet pharmaceutical industry needs. Although protected areas are considered to play a central role in conservation of plant genetic resources, the effectiveness of protected areas for maintaining medicinal plant populations subject to intense harvesting pressure remain largely unknown. We conducted genetic and demographic studies of Nothapodytes nimmoniana Graham, one of the extensively harvested medicinal plant species in the Western Ghats biodiversity hotspot, India to assess the effectiveness of protected areas in long-term maintenance of economically important plant species.
Methodology/Principal Findings
The analysis of adults and seedlings of N. nimmoniana in four protected and four non-protected areas using 7 nuclear microsatellite loci revealed that populations that are distributed within protected areas are subject to lower levels of harvesting and maintain higher genetic diversity (He = 0.816, Ho = 0.607, A = 18.857) than populations in adjoining non-protected areas (He = 0.781, Ho = 0.511, A = 15.571). Furthermore, seedlings in protected areas had significantly higher observed heterozygosity (Ho = 0.630) and private alleles as compared to seedlings in adjoining non-protected areas (Ho = 0.426). Most populations revealed signatures of recent genetic bottleneck. The prediction of long-term maintenance of genetic diversity using BOTTLESIM indicated that current population sizes of the species are not sufficient to maintain 90% of present genetic diversity for next 100 years.
Conclusions/Significance
Overall, these results highlight the need for establishing more protected areas encompassing a large number of adult plants in the Western Ghats to conserve genetic diversity of economically and medicinally important plant species.
Introduction
The harvesting of medicinal plants from wild sources to meet pharmaceutical industry needs [1]–[3] may reduce populations of many plant species to below minimum viable population sizes, leading to eventual extinction of numerous medicinally important plant species [2], [4]. The long-term survival of these species will largely depend on the effectiveness of protected areas in sustaining viable populations that may serve as genetic stocks to aid replenishing dwindling populations in harvested areas [5]–[9]. Although protected areas may play a central role in conservation of biological diversity and genetic resources [10], [11], their effectiveness in preventing genetic erosion of many species remain largely unknown. Several studies have focused on assessing the effectiveness of protected areas in conserving genetic resources of Non Timber Forest Products (NTFP) in the Western Ghats of India [9], [12]–[15]. These studies have revealed that some plant species including bamboos and rattans harbor higher genetic diversity in protected areas than in non-protected areas or at peripheral regions of the protected areas [13]–[15]. However, the effectiveness of protected areas in conserving medicinal plants remains unknown. Comparative studies of medicinal plants in protected and non-protected areas provide ideal means to evaluate the effectiveness of protected areas in maintenance of genetic diversity and long-term viability of medicinally important plant populations [14].
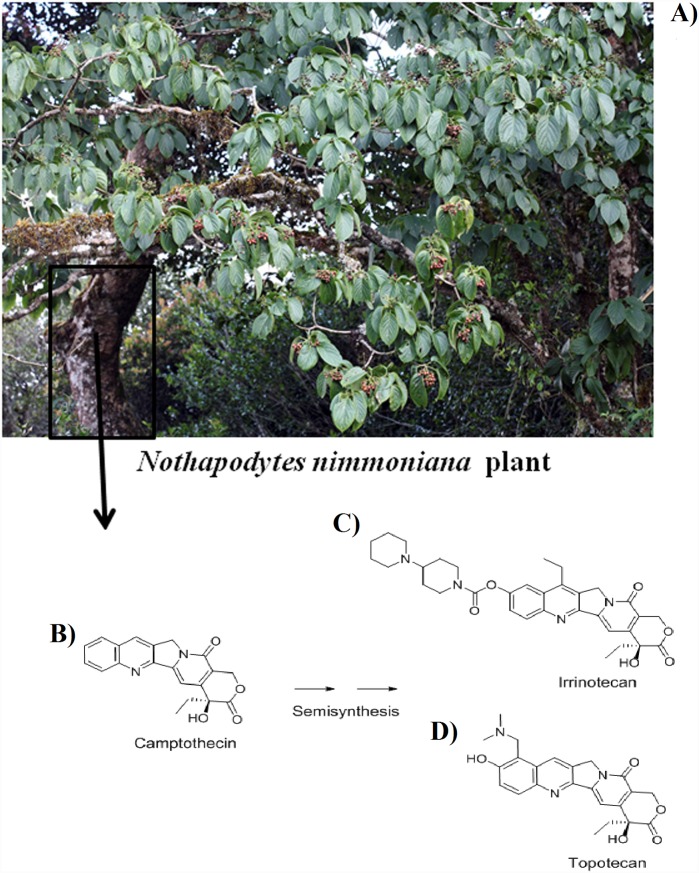
In recent years, Nothapodytes nimmoniana (Graham) Mabb., one of the medicinally important tree species distributed in the Western Ghats has become a major source of DNA topoisomerase inhibiting anti-cancer drug, Camptothecine (CPT) (Fig. 1), one of the alkaloids sought after by the pharmaceutical industries around the world [16]. The global demand for CPT exceeding an annual market value of over US$ 4 billion [17] led to a large-scale exploitation of the species from its wild habitats in the region resulting in an estimated loss of 20% of N. nimmoniana populations in the Western Ghats [18], [19]. Consequently, N. nimmoniana has been declared as an endangered/vulnerable plant species [18], [20].
Figure 1. Image of A) N. nimmoniana plant and chemical structure of B) Camptothecine extracted from wood of N. nimmoniana C) Irinotecan and D) Topotecan, two clinically used drugs synthesized from Camptothecine as a precursor.
We conducted genetic and demographic studies of N. nimmoniana populations in protected and non-protected areas in the central Western Ghats of southern India to assess the genetic and demographic effects of harvesting and evaluate the effectiveness of protected areas in the maintenance of long-term viability of N. nimmoniana. The specific objectives of our study were to 1) assess the genetic structure and diversity of N. nimmoniana populations in protected and non-protected areas, 2) investigate any evidence for genetic bottlenecks of populations and 3) analyze demographic data to predict future population sizes to evaluate long-term viability of N. nimmoniana populations in the Western Ghats.
Materials and Methods
Ethics Statement
The field work and tissue sample collection of Nothopodytes nimmoniana was carried out in the central Western Ghats regions of Karnataka, with permission from the Karnataka Forest Department. Tissue sampling was carried out under the supervision of forest officers and used solely for scientific research. The sampling was non-invasive with no impact on the natural growth or regeneration of N. nimmoniana populations in the wild.
Study sites and design
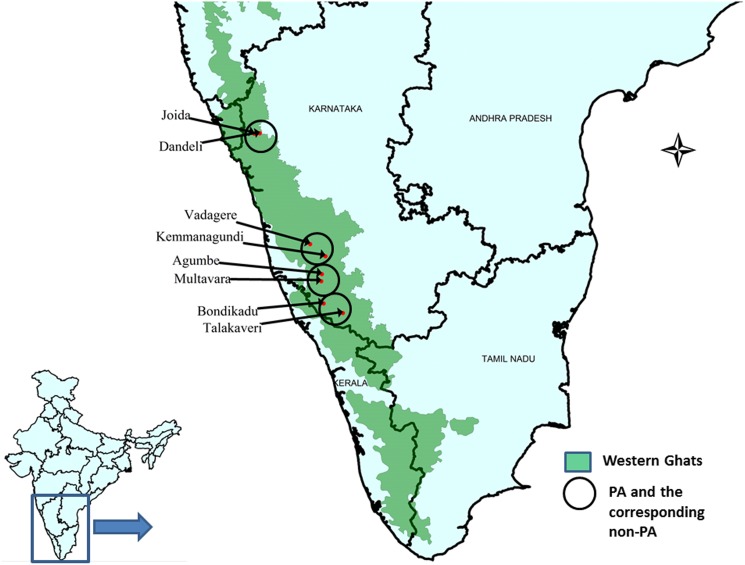
This study was conducted in the Western Ghats, India, one of the 32-biodiversity hotspots of the world [21]. The Western Ghats includes 34 national parks and wildlife sanctuaries covering an area of about 7300 km2 [22]. The distribution map of N. nimmoniana was overlaid with protected area map of the Western Ghats to identify populations that are distributed within protected areas (PAs), and based upon the availability of N. nimmoniana within the PA network of the Western Ghats, four study sites namely: Dandeli Wildlife Sanctuary (established in the year 1974), Talakaveri wild life sanctuary (established in the year 1987), Kudremukh wild life sanctuary (established in the year 1987) and Agumbe medicinal plants protected area (established in the year 1999) were selected for the present study (Fig. 2, Table 1). Similarly, four populations located adjacent to protected areas were also selected and treated as non-protected areas (NPAs) (Fig. 2, Table 1).
Figure 2. Map showing the distribution and sampling locations of Nothapodytes nimmoniana in the Western Ghats, India.
Table 1. Location, geographical coordinates, sample size (N) and private alleles (AP) in eight populations of Nothapodytes nimmoniana.
| Population Name | Status | Latitude | Longitude | Sample size (N) | AP | |
| Adults | Seedlings | |||||
| Dandeli | PA | 15°11′59.1″ | 74°35′59.1″ | 17 | 20 | 11 |
| Talakaveri | PA | 12°24′0″ | 75°30′0″ | 20 | 20 | 13 |
| Kemmangudi | PA | 13°20′59.1″ | 75°27′0″ | 13 | 8 | 6 |
| Agumbe | PA | +−13°3′0″ | 75°5′27.6″ | 13 | 0 | 6 |
| Joida | NPA | 15°10′9.5″ | 74°29′4.2″ | 19 | 12 | 5 |
| Multavara | NPA | 14°22′11.1″ | 75°0′0″ | 11 | 9 | 6 |
| Vadagere | NPA | 12°26′59.1″ | 75°54′0″ | 13 | 0 | 3 |
| Bondikadu | NPA | 12°50′59.1″ | 75°50′59.1″ | 20 | 3 | 7 |
| Over all | PA+NPA | 126 | 72 | 57 | ||
Note: PA = Protected area, NPA = Non-protected area.
Study species
Nothapodytes nimmoniana (Graham) Mabb. (Icacinaceae), (Fig. 1A) formerly known as Nothapodytes foetida (Wight) Sleumer and Mappia foetida Meirs is a medicinally important tree species naturally distributed in many parts of the Western Ghats in South India, some parts of Assam, Himalayan foothills, Sri Lanka, Burma and Thailand [19]. The bark of the stem is one of the richest sources of the anti-cancer compound, Camptothecine (Fig. 1B) [16], [23]. The two clinically used drugs, Irinotecan (Fig. 1C) and Topotecan (Fig. 1D), are currently semi-synthesized using natural Camptothecine as a precursor [24]. These trees are harvested by felling the trunk. Although stumps of cut trees often coppice, the continuous harvesting negatively impact the long term survival and reproduction. The extensive harvesting of N. nimmoniana led to severe reduction in population sizes [19] and currently classified as a ‘vulnerable/endangered’ species [18]. Although N. nimmoniana is a dioecious species, some individuals are polygamous with male, female and bisexual flowers [25].
Population structure
In each of the four PAs and their adjoining non-PAs, 10 quadrats (10 m×10 m) were laid out randomly and data on the number of trees per quadrat, girth of all individuals above 10 cm dbh, and number of harvested or coppicing individuals were recorded. As a measure of regeneration, the number of seedlings and saplings (<1 m height) in each quadrat was recorded. For each quadrat, the number of regenerants (saplings and seedlings) was divided by the number of adults to obtain an index of the regeneration per adult. The number of harvested adults of N. nimmoniana in each quadrat was recorded and expressed as a percentage of the total number of adults harvested for each quadrat [11], [26]. The differences in the various parameters across PA and the non-PA were analyzed using Student’s t-test. The girth-class distributions of adult individuals in each site were determined. The frequency distribution of the girth-class of adults across the PA and non-PA was statistically evaluated using the nonparametric Kolmogorov–Smirnov test [27].
Genetic diversity
DNA extraction and amplification protocol
Leaf samples were collected from 15–20 adults as well as seedlings selected randomly from four PAs and their adjoining Non-PAs. In each site, the area sampled was approximately 0.5 km2. Leaf samples were air-dried and preserved in silica gel until the extraction of total genomic DNA using a modified CTAB (cetryl trimethyl ammonium bromide) protocol [28] at the Conservation Genetics Laboratory at ATREE. The extracted DNA was visualized on ethidium bromide stained 0.8% agarose gels and quantified by measuring the absorption at 260 nm using a spectrophotometer. The DNA was diluted to a concentration of 10 ng/µl and used for PCR amplification. The seven microsatellite primer pairs [29], which gave consistent amplification with good level of polymorphism was selected for genotyping. The PCR amplifications for genotyping were carried out in an Eppendorf Mastercycler Gradient, (Eppendorf) thermal cycler. The PCR amplification was carried out in a 25-µl-volume reaction mixture containing 25 ng template DNA, 2.5 µl 10× reaction buffer containing 15 µM MgCl2, 3 µM of each dNTP, 0.25 µM each forward and reverse primer and 0.5 unit Taq DNA polymerase (Sigma). The thermal cycling parameters were: 94°C for 3 min, 35 cycles of 94°C for 40 s, 58–62°C for 40 s, 72°C for 60 s, followed by a final extension of 5 min at 72°C. The amplified samples were prepared for genotyping by mixing 10 µL of deionized formamide, 0.1 µL of 35–500 bp internal size standards (Tamara GeneScan-500, Applied Biosystems) and 1 µL of PCR product. The mixture was denatured at 95°C for 2 min and immediately placed on ice for a minimum of 5 min and electrophoresed on an ABI PRISM 310 Genetic Analyser (Applied Biosystems) to detect fragment sizes. The electrophorograms were analyzed with the GeneScan 3.7 and GenoTyper 3.7 software programs (Applied Biosystems).
Data analysis
Microsatellite based genetic diversity measures
The following genetic diversity measures were calculated for all eight populations and also separately for adults and seedlings: Allelic distribution, private alleles (alleles unique to a population and not shared with other populations), the mean number of alleles per locus (averaged across 7 loci) and per population were calculated using GENEPOP 3.2a [30]. A standardized estimate of allelic richness per locus (averaged across 7 loci) and per population adjusted to the sample size [31]–[32] was calculated using the program FSTAT 2.9.3 [33]. ARLEQUIN version 3.1 [34] was used to test for heterozygote deficiency at each microsatellite locus for each population using a Hardy-Weinberg test based on Markov Chain iterations [35]. The pairwise test for linkage disequilibrium for each pair of loci for each allele in each population and Wrights F-statistics (Fis and Fst) were calculated using the program GENEPOP 3.2a [30].
Population genetic structure
We calculated the total genetic differentiation among population as Fst: which assumes an Infinite Allele Model (IAM) [36] by computing theta (θ) [37] and Rst: which assumes a Stepwise Mutation Model (SMM) [38] by computing Rho which adjusts for differences in sample size and allele size variances among loci using the software program FSTAT version 2.9.32 [33]. The proportions of genetic variation partitioned among populations and among groups of populations [39] were quantified using analyses of molecular variance (AMOVA) as implemented in ARLEQUIN V3.1 [34], and the statistical significance was tested with 10000 permutations. A model based Bayesian clustering method as implemented in STRUCTURE version 2.3.3 [40] was used as an alternative approach to examine the spatial genetic structure. The programme was run without prior population information under the admixture model (individuals may have mixed ancestry) and correlated allele frequency. Length of the burn-in was 100 000 and the number of MCMC replications after the burn-in was 1 000 000. Twenty independent chains were run for each K from K = 1 to K = 15. The method of Evanno et al [41] was used to find the most likely value of K by plotting log probability (L(K)) and ΔK of the data over multiple runs and as implemented in STRUCTURE HARVESTER [42]. CLUMPP v.1.1.1 [43] software (resolves the label switching and compute average admixture co-efficient) was used to align the repetitions for each K, using G’ (10 000 repeats). The output from CLUMPP was used for ancestry analysis.
Population bottleneck test and simulation of loss of diversity
We assumed that, if extensive harvesting of trees in recent years has affected the populations, they should show signatures of recent genetic bottlenecks between adults and seedlings. We also expect that, the populations outside the PAs should experience higher signatures of genetic bottleneck than populations within PA network, as populations within PAs receive more protection and less harvesting than NPAs. We used following approaches to test these predictions: Populations that experienced a bottleneck often exhibit a reduction of allele number and heterozygosity at polymorphic loci, with allele numbers being reduced at a higher rate than heterozygosity [44], [45]. Thus, observed heterozygosity is higher than that expected based on allele numbers assuming mutation drift equilibrium [46]. We tested for a population bottleneck using BOTTLENECK version 1.2.0.2 [46], [47], which is based on the assumption that populations that may have undergone severe size reductions will show an excess of heterozygotes relative to allelic diversity. The BOTTLENECK analysis was run using the stepwise mutation model (SMM). The significance of genetic diversity excess (H e>H eq) was tested using Wilcoxon signed-rank tests and sign test [46] based on 5000 replications. A population bottleneck is also expected to change the allele frequency distribution [46], which could be detected as a shift in the mode of the allele frequency distribution. The mode-shift indicator test was performed to test [46], if the allele frequency distribution pattern shows a departure from approximately L-shaped (as expected under HWE) distribution.
To assess whether the current population size of N. nimmoniana is sufficient to maintain 90% of present day observed genetic variation over the next 100 years as a measure of long-term genetic viability of the species [44], we simulated future population parameters using the software program BOTTLESIM version 2.6 [48]. Based upon the present genetic diversity and population size, BOTTLESIM simulates future population genetic parameters (observed number of alleles [OA]) under different population bottleneck scenarios. Genetic diversity estimates over 100 years were simulated when retaining 100, 90, 75, 50, and 25 percent of the current population size. We performed 1,000 iterations with constant life history parameters (lifespan = 50 years, age at maturity = 10 years, completely overlapping generations, random mating, dioecious reproduction, and sex ratio of F: M: 1∶1).
Results
Population structure and harvesting pressure
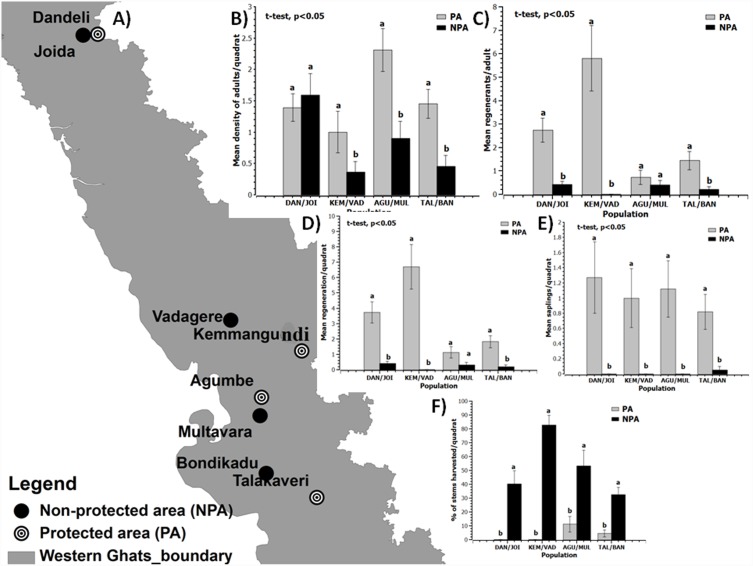
The mean density of adults between populations of PAs and NPAs were significantly different (t-test, P<0.05; Fig. 3B, S1 Table in S1 File). In general, populations in PAs showed higher mean density of adults than populations in the adjoining NPAs. The measure of reproductive turnover (mean regenerants per adult) was also higher in populations of the protected areas (t-test, P<0.05; Fig. 3C). We also found significant difference in overall proportion of reproductive individuals between PA and NPA populations (t-test, p<0.05, S1 Table in S1 File), with higher regeneration per quadrat and mean number of saplings per quadrat from populations of PA (Fig. 3D and 3E). The percentage of harvested adults per quadrat was higher in populations from NPA (t-test, p<0.05; Fig. 3F) than PA populations. Generally we observed that more adults were harvested (>40% of adults) from population of NPA and less than 5% adults were harvested from populations of PA (Fig. 3F) indicating N. nimmoniana populations outside PA are experiencing more threat and harvesting pressure.
Figure 3. Demographic parameters of Nothapodytes nimmoniana in four protected and adjacent non-protected areas in the central Western Ghats, India.
A) Western Ghats map showing sampling locations B) Mean density of adults per quadrat C) Mean regenerants per adult D) Mean regeneration per quadrat E) Mean saplings per quadrat F) Harvesting index. Note: Error bars are standard error. For each parameter, dissimilar letters above the bars are significantly different at 0.05 levels (t-test). PA, protected area; NPA, non-protected area; WG, Western Ghats; KEM, Kemmangundi; MUL, Multavara; TALA, Talakaveri; BAN, Bondikadu; DAN, Dandeli; JOID, Joida; AGU, Agumbe; VADE, Vadagere.
Genetic diversity and population genetic structure
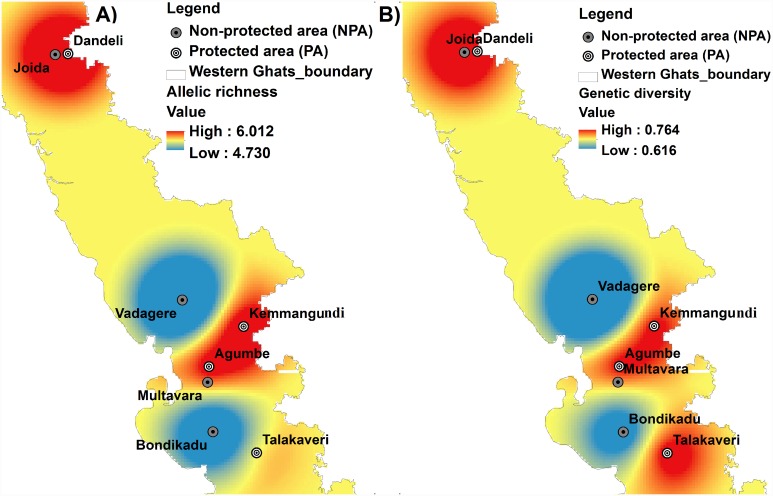
All seven-microsatellite loci were polymorphic in all studied populations. A total of 156 alleles with an average of 22.29 alleles per locus (A) were found at the species level (Table 2). The mean HE and HO across all loci were 0.805 and 0.567 respectively (Table 2). The mean expected heterozygosity (HE) was generally higher than mean observed heterozygosity (Ho) across all loci (Table 3). The locus NNM6a showed highest level of polymorphism with 5–18 alleles per population and expected heterozygosity ranged from 0.576–0.915 followed by the locus NN46 (A = 5–16 and He = 0.725–0.910). The locus NN54 had lowest level of polymorphism with 3–8 alleles per population and expected heterozygosity (HE) ranged from 0.394–0.779 followed by the locus NNT4 (A = 3–10 and HE = 0.504–0.790). The allelic diversity parameters (allelic richness (AR) and mean observed number of alleles (A) per locus (averaged over 7 loci) were highest for the population Dandeli (AR = 6.018 and A = 11.857±4.180) followed by Talakaveri (AR = 5.476 and A = 10.714±4.309), Joida (AR = 5.999 and A = 10.286±3.817) and the lowest values were found in populations Bondikadu (AR = 4.974 and A = 8.143±4.298) and Vadagere (AR = 4.727 and A = 8.571±2.226) (Table 3 and Fig. 4A). Many alleles were exclusive to specific populations, and these alleles were referred to as private alleles (AP). The two of the populations, namely Dandeli and Talakaveri had highest number of private alleles with 11 and 13 private alleles respectively (Table 1). The Nei’s gene diversity across all loci was highest in Joida (0.7633±0.087) and Dandeli (0.7461±0.102) populations. The lowest gene diversity values were observed in Vadagere (0.6158±0.192) and Bondikadu (0.6714±0.134) (Fig. 4B).
Table 2. Summary of genetic variation at seven microsatellite loci scored for eight Nothapodytes nimmoniana populations in the Western Ghats, India: expected and observed heterozygosity (HO and HE), and observed number of alleles per polymorphic locus (A), Weir & Cockerham (1984) estimates of FIT, FST, FIS and RST.
| Locus | HO | HE | A | FIS | FIT | FST | RST |
| NN54 | 0.592* | 0.757 | 12 | 0.098 | 0.215 | 0.130 | 0.113 |
| NNM14 | 0.489* | 0.798 | 22 | 0.342 | 0.409 | 0.102 | 0.124 |
| NNM6a | 0.429* | 0.925 | 39 | 0.489 | 0.560 | 0.139 | 0.154 |
| Nn46 | 0.642* | 0.849 | 31 | 0.200 | 0.250 | 0.062 | 0.067 |
| Nn52 | 0.659* | 0.694 | 18 | 0.016 | 0.127 | 0.112 | 0.003 |
| NNT4 | 0.546* | 0.751 | 13 | 0.182 | 0.276 | 0.115 | 0.045 |
| NN50 | 0.612* | 0.859 | 21 | 0.234 | 0.295 | 0.079 | 0.098 |
| Over all loci | 0.567 * | 0.805 | 22.286 | 0.233 | 0.313 | 0.105 | 0.096 |
*indicates significant deviations of HE from HWE (at 0.05 significance level).
Table 3. Summary of genetic diversity parameters of Nothapodytes nimmoniana in four protected (PA) and adjacent non-protected areas (NPAs) in the central Western Ghats, India.
| Populationgeneticparameter | Protectionstatus | Sampletype | KEM/VAD(Mean±SD) | TALA/BON(Mean±SD) | AGU/MUL(Mean±SD) | DAND/JOI(Mean±SD) | all PA/NPA(Mean±SD) |
| A | PA | Adults | 6.714 (1.799) | 8.857 (3.625) | 8.000 (2.449) | 7.429 (2.149) | 15.286 (5.28) |
| Seedlings | 5.714 (1.976) | 8.143 (2.160) | - | 9.714 (3.352) | 14.429 (6.10) | ||
| Overall | 8.571 (2.225) | 10.714 (4.309) | 8.000 (2.449) | 11.857 (4.180) | 18.857 (8.72) | ||
| NPA | Adults | 8.571 (2.22) | 8.143 (4.298) | 6.857 (2.478) | 9.000 (2.64)a | 15.571 (6.26)a | |
| Seedlings | - | - | 5.143 (1.773) | 5.71 (2.936)b | 8.860 (4.53)b | ||
| Overall | 8.571 (2.226) | 8.143 (4.298) | 8.429 (2.226) | 10.286 (3.817) | 15.571 (6.26) | ||
| H0 | PA | Adults | 0.308 (0.133) | 0.593 (0.162) | 0.495 (0.188) | 0.411 (0.165) | 0.592 (0.12)a |
| Seedlings | 0.357 (0.222) | 0.600 (0.197) | - | 0.632 (0.239) | 0.630 (0.087)a | ||
| Overall | 0.326 (0.144) | 0.696 (0.162)a | 0.495 (0.188)a | 0.524 (0.194) | 0.607 (0.100)a | ||
| NPA | Adults | 0.327 (0.144) | 0.360 (0.220) | 0.481 (0.146)a | 0.556 (0.143)b | 0.490 (0.11)b | |
| Seedlings | - | - | 0.349 (0.268)b | 0.309 (0.214)c | 0.426 (0.200)b | ||
| Overall | 0.327 (0.144)b | 0.360 (0.220)b | 0.421 (0.173) | 0.461 (0.170) | 0.511 (0.111)b | ||
| HE | PA | Adults | 0.710 (0.102) | 0.798 (0.105) | 0.807 (0.096) | 0.782 (0.077) | 0.829 (0.057) |
| Seedlings | 0.696 (0.197) | 0.742 (0.098) | - | 0.802 (0.075) | 0.798 (0.075) | ||
| Overall | 0.739 (0.125) | 0.775 (0.098) | - | 0.806 (0.069) | 0.816 (0.006) | ||
| NPA | Adults | 0.739 (0.124) | 0.717 (0.140) | 0.793 (0.034) | 0.785 (0.097) | 0.511 (0.114) | |
| Seedlings | - | - | 0.645 (0.251) | 0.751 (0.133) | 0.783 (0.085) | ||
| Overall | - | - | 0.746 (0.071) | 0.789 (0.082) | 0.781 (0.094) | ||
| FIS | PA | Adults | 0.397 | 0.342 | 0.397 | 0.297 | 0.209 (0.385) |
| Seedlings | 0.599 | 0.504 | - | 0.217 | 0.205 (0.327)a | ||
| Overall | 0.564 | 0.233 | 0.397 | 0.353 | 0.207 (0.358)a | ||
| NPA | Adults | 0.342 | 0.503 | 0.577 | 0.502 | 0.236 (0.346)a | |
| Seedlings | - | - | 0.474 | 0.195 | 0.452 (0.325)b | ||
| Overall | 0.342 | 0.503 | 0.441 | 0.420 | 0.304 (0.350)b | ||
| FST | PA | Adults | 0.102 | 0.084 | 0.072 | 0.091 | 0.088 (0.010) |
| Seedlings | 0.090 | 0.084 | - | 0.091 | 0.087 (0.026) | ||
| Overall | 0.087 | 0.083 | - | 0.077 | 0.088 (0.029) | ||
| NPA | Adults | 0.099 | 0.093 | 0.082 | 0.067 | 0.092 (0.033) | |
| Seedlings | - | - | 0.072 | 0.078 | 0.075 (0.029) | ||
| Overall | - | - | 0.078 | 0.065 | 0.088 (0.033) |
Note: A = mean observed number of alleles, HO = observed heterozygosity, HE = expected heterozygosity, FIS and FST = population specific fixation indices, − = data not present. Disimilar letters (a and b) above the parameter values indicates t-test significant at p<0.05.
Figure 4. Map showing genetic diversity parameters at sampling locations of Nothapodytes nimmoniana from Western Ghats, India A) Nei’s gene diversity B) Allelic richness.
Note: Regions represented in dark red indicate areas of high genetic diversity and allelic richness.
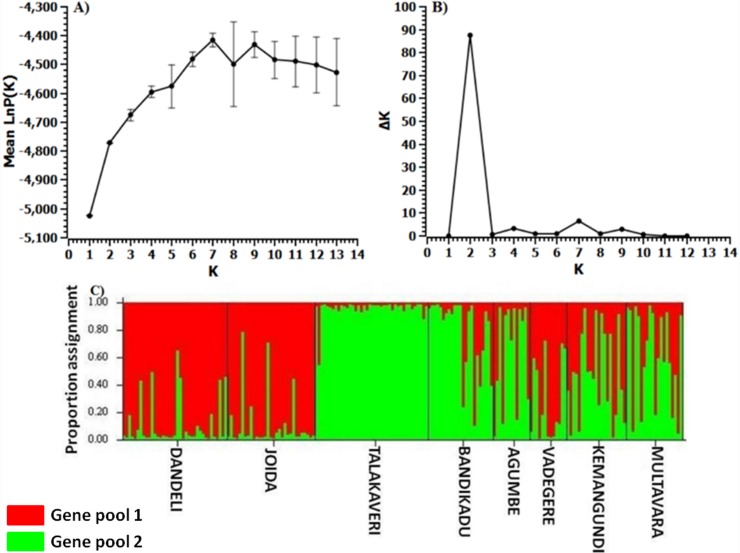
The FST estimate, θ averaged over seven loci was 0.105 (Table 2). The FST values between populations ranged from 0.065 to 0.099 (Table 3), indicating a low but significant level of genetic differentiation of populations. The overall FIS and FIT values were 0.233 and 0.313 respectively indicating loss of heterozygosity both at the population and at meta-population levels (Table 2). The FIS values among populations ranged between 0.233 and 0.564. All populations had positive values of inbreeding (Table 3). The results of AMOVA indicated that most of the variations were within individuals (56.4%) followed by within and among populations (Table 4). The Bayesian clustering method revealed two optimum numbers of genetic clusters (K = 2; Fig. 5A, 5B and 5C) and geographically close populations grouped together corresponding to two genetically distinct clusters (Fig. 5C and Table 5) further supporting significant level of differentiation and genetic structure among populations.
Table 4. Analysis of molecular variance (AMOVA) for 195 individuals of Nothapodytes nimmoniana in protected (PA) and non-protected (NPA) areas.
| Source of variation | d.f. | Sum ofsquares | Variance | % of totalvariance | F-statistics | p-value |
| Among groups (PA*NPA) | 1 | 8.167 | −0.057 | −1.96 | FCT = −0.012 | 0.973 |
| Among populationswithin groups | 6 | 102.886 | 0.284 | 9.72 | FSC = 0.095 | 0.000 |
| Among individualswithin populations | 189 | 707.686 | 1.056 | 35.87 | FIS = 0.389 | 0.000 |
| Within individuals | 197 | 324.5 | 1.647 | 56.35 | FIT = 0.436 | 0.000 |
| Total | 393 | 1143.239 | 2.923 |
Figure 5. Results of Bayesian model based clustering method (STRUCTURE) analysis.
a) Posterior probability of the data LnP(D) (±SD) against the number of K clusters, and increase of LnP(D) given K, calculated as (LnP(D)k–LnP(D)k–1) b) Delta K values from the mean log-likelihood probabilities from STRUCTURE runs where inferred clusters (K) ranged from 1 to 14 (c) Estimates of the proportion of ancestry, Q, in each of K = 2 clusters for 198 Nothapodytes nimmoniana individuals using the model-based cluster analysis, STRUCTURE (version 2.1). Note: A single vertical line represents each individual with estimated membership in each cluster denoted by the different colors. The analysis was based on seven SSR loci and used admixture model of ancestry. Individuals are separated based on their population and black vertical lines in the bar chart are population identifiers. Populations are ordered as per their population name.
Table 5. Membership of each pre-defined population in each of the two (K = 2) clusters generated by CLUMPP based on the results of STRUCTURE v2.2 analysis of SSR data.
| Population name | Q value (proportion of individual ancestry In each cluster) K = 2 | |
| Cluster 1 | Cluster 2 | |
| Dandeli | 0.886 (0.176) | 0.114 (0.176) |
| Joida | 0.895 (0.194) | 0.105 (0.194) |
| Talakaveri | 0.040 (0.071) | 0.960 (0.071) |
| Bondikadu | 0.209 (0.272) | 0.791 (0.272) |
| Agumbe | 0.359 (0.377) | 0.641 (0.377) |
| Vadagere | 0.710 (0.296) | 0.290 (0.296) |
| Kemmangundi | 0.515 (0.318) | 0.485 (0.318) |
| Multavara | 0.376 (0.343) | 0.624 (0.343) |
| Over all | 0.501 (0.409) | 0.499 (0.409) |
Populations contributing >50% of their ancestry to a single cluster are highlighted in bold.
Genetic diversity in adults and seedlings
The comparison of genetic diversity parameters between adults and seedlings at the overall population level revealed no significant differences (A, HO and HE). However, the comparison of genetic diversity parameters between adults and seedlings within individual populations revealed significant decrease in genetic diversity parameters (A and HO) in seedlings as compared to adults in some of the populations (Table 3). Both adults and seedlings showed positive values of FIS (Table 3).
Effect of harvesting on genetic diversity
Genetic diversity varied between populations from PA and NPA sites. Observed heterozygosity (HO) for populations of PA was 0.607, significantly higher than (P<0.05) the 0.511 in populations of NPA. Populations from PA also had highest number of private alleles (Table 1). There was no significant difference in mean HE and observed number of alleles, indicating that differences among PA and NPA populations are due to loss of rare alleles resulting from genetic bottleneck and genetic drift. There was also a significant difference in genetic diversity measures between life history stages (adults and seedlings) from PA and NPA’s. We observed that the adults and seedlings from PA’s had significantly (P<0.05) higher observed number of alleles (A) and highest observed heterozygosity compared to NPA populations (Table 3). We also found significant decrease in genetic diversity measures (A, HE, HO) from adults to seedlings (Table 3) in populations of NPA indicating populations from NPA experience loss of diversity due to harvesting pressure than PAs. The AMOVA results showed that most molecular variance was found within individuals (56.4%) of N. nimmoniana followed by among individuals within populations (36.4%) and among individuals within groups (9.2%) (Table 4). The variance among groups (PA and NPA) was very low and negative (−2%). The recorded FSC and FIT values of 0.095 and 0.436 indicate a moderate level of spatial isolation. Finally, populations from NPAs showed significantly high level of inbreeding than PA populations and inbreeding level in seedlings were higher than in adults (Table 3) indicating a decrease in effective population sizes in NPAs.
The population bottleneck test and prediction of future population size through simulations
The population bottleneck analyses of individual populations with adults and seedlings combined together detected evidence for recent population bottlenecks in five out of eight populations (Wilcoxon’s and Sign test; P>0.05 and 0.01) indicating a deviation from mutation drift equilibrium (Table 6). However, mode shift test did not reveal distortion in allele frequency as compared to normal L-shaped distribution. The population bottleneck analysis between generations (adults and seedlings) showed that out of five populations examined, seedlings from two populations and adults from four populations exhibited excess or deficiency of heterozygotes (Wilcoxon sign rank test (0.05>p<0.01) and sign test (0.05>p<0.01) (Table 6) suggesting a deviation from mutation drift equilibrium. However, allele frequency distribution revealed a normal L-shaped distribution in both adults and seedlings suggesting mutation drift equilibrium. Irrespective of protected status, populations of both PA and NPA showed evidence of recent bottleneck in N. nimmoniana.
Table 6. Analysis of historical and recent genetic bottleneck based on stepwise mutation model (SMM) of microsatellite evolution and Mode shift test for allele frequency distribution.
| Population Name | Sign test (two tailed p-value) | Wilcoxon signed rank test (two tailed p-value | ||||
| SMM | SMM | |||||
| A | S | A+S | A | S | A+S | |
| Dandeli (PA) | 0.002** | 0.022* | 0.002** | 0.008** | 0.016* | 0.008** |
| Joida (NPA) | 0.020* | 0.616 | 0.113 | 0.055* | 0.937 | 0.040* |
| Talakaveri (PA) | 0.124 | 0.022* | 0.002** | 0.375 | 0.015* | 0.008** |
| Bondikadu (NPA) | 0.291 | 0.088 | 0.020* | 0.110 | 0.078 | 0.020* |
| Agumbe (PA) | 0.022* | - | 0.020* | 0.040* | - | 0.039* |
| Vadagere (NPA) | 0.020* | - | 0.021* | 0.040* | - | 0.040* |
| Kemmangundi (PA) | 0.650 | 0.102 | 0.289 | 0.578 | 0.297 | 0.297 |
| Multavara (NPA) | 0.248 | - | 0.097 | 0.375 | - | 0.039* |
Note: – = data absent. *P, <0.05; **P, <0.01. A = adults, S = seedlings.
The significance of gene diversity excess (He. Heq) is an indication of recent effective population size reductions (bottlenecks). The significance was tested using Sign test and Wilcoxon signed ranks test [44] based on 5000 replications.
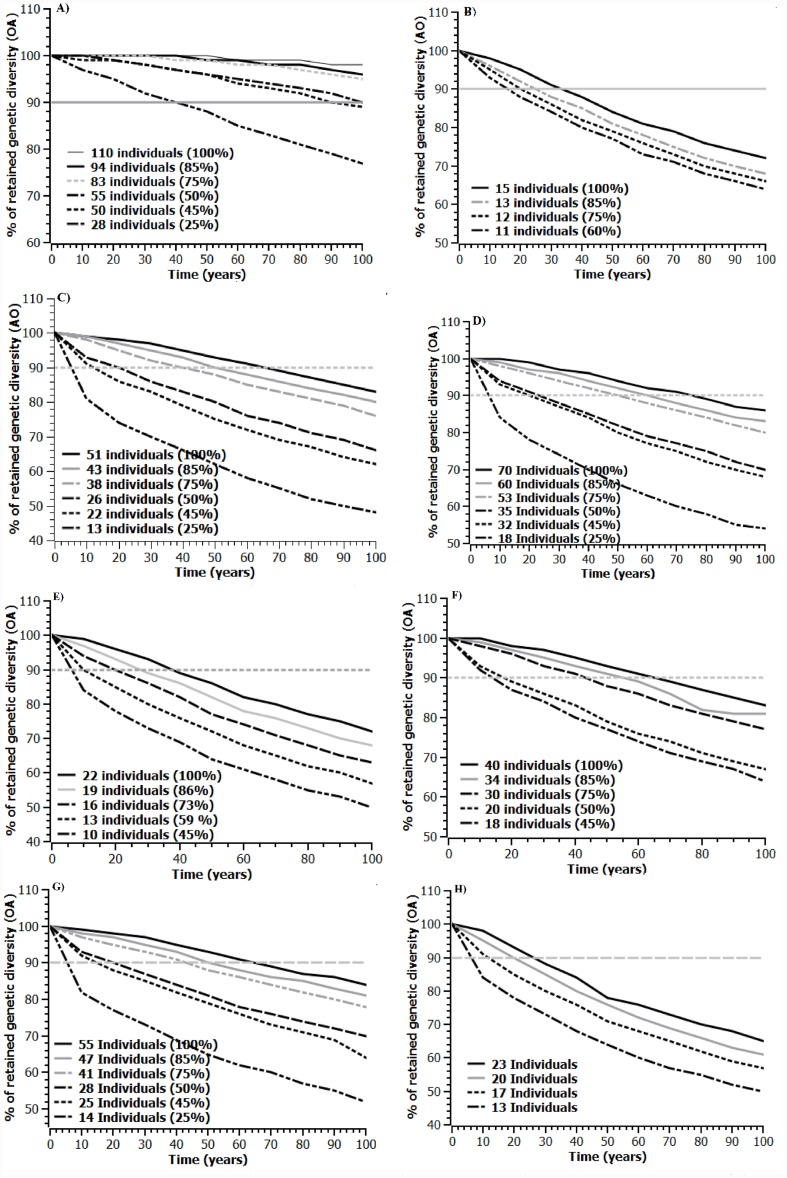
The BOTTLESIM based prediction of future genetic diversity with varying starting population sizes, and assuming current population size as the maximum (100%), the projected genetic diversity continued to decrease over time (Fig. 6). The observed allele diversity is expected to decrease at a higher rate than observed heterozygosity. With the exception of Agumbe population [for both PA and NPA] all other populations showed decline of genetic diversity beyond 90% of current genetic diversity values even at the retention of 100% of individuals (Fig. 6A to H). The populations in NPA are predicted to lose allelic diversity faster than PA populations. The overall results clearly showed that current population size of N. nimmoniana in Western Ghats is not sufficient to maintain present levels of genetic diversity for the next 100 years.
Figure 6. Predicted genetic diversity in eight populations of Nothapodytes nimmoniana sampled from protected (PA) and non-protected (NPA) areas of Western Ghats, India over next 100 years using BOTTLESIM.
The current population size is unable to maintain 90% of current genetic diversity over the next 100 years in almost all sampled populations. A) Agumbe (PA) B)Vadagere (NPA) C)Dandeli (PA) D) Joida (NPA) E) Kemmangundi (PA) F) Multavara (NPA) G) Talakaveri (PA) and H) Bondikadu (NPA). Note: The observed numbers of alleles (OA) were projected to decline (Sex ratio 1∶1 F:M).
Discussion
Genetic diversity
At the species level, pattern of genetic diversity observed within N. nimmoniana is similar to other tropical tree species with similar life history and ecological traits [49]–[54] but higher than other tropical tree species with different life history or ecological traits [55]–[57]. Although populations are sparse and patchy, N. nimmoniana is widely distributed throughout Western Ghats in peninsular India and expected to maintain moderate to high level of genetic diversity. The mating system of N. nimmoniana is mixed, which consists predominantly outcrossing mating system with some selfing [19], [58] and seeds are dispersed by small birds [59]. The genetic diversity in N. nimmoniana (A = 22.28, AR = 4.108, HO = 0.567, HE = 0.805) is comparable to other tropical tree species with mixed mating system and bird dispersed seeds [50], [52], [54]. However, the genetic diversity values of N. nimmoniana are lower than those reported in other microsatellite based studies of tropical tree species [49], [51], [60]–[62].
The comparison of observed (HO) and expected (HE) heterozygosity levels within populations of N. nimmoniana showed significant excess of homozygosity. This could be attributable to non-random mating with high gene exchange among related individuals and selfing due to the mixed mating system of the species. It is possible that reduction in population size due to habitat fragmentation or harvesting may have increased selfing rates and/or mating among closely related individuals resulting in a higher proportion of homozygous individuals in populations.
Genetic structure
The values of inter-population genetic differentiation based on both measures were relatively low (FST = 0.105 RST = 0.096), similar to each other and significantly different from zero suggesting low level of population differentiation with significant amount of gene flow among populations (Nm = 2.125). This observation agrees well with corresponding values for woody perennial species with outcrossing or mixed mating system that maintain most of their variation within populations [63]. In N. nimmoniana, approximately 56% of total genetic variation was found within individuals followed by within populations (36%). This observation is similar to the values of ISSR marker based genetic variation detected in individuals distributed over a broad geographical area [64]. However, the genetic diversity levels detected in N. nimmoniana was higher than corresponding values reported in other microsatellite based analyses of natural populations of tropical tree species [49], [60]–[62], but similar to the values observed in Milicia excelsa [52]. However, historical rates of gene flow among populations estimated based on genetic structure can be confounded by several variables and must be interpreted with caution [65]–[67].
The species level inbreeding coefficient (FIS) indicates that alleles within populations were not united at random and that mating between close relatives may play an important role in determining the genetic structure of N. nimmoniana. At the population level, positive and significant FIS values (Table 3) were detected in all eight populations examined. The significantly higher FIS in NPA populations (Table 3) could be attributable to high harvesting pressure as expected in tree populations experiencing high level of harvesting and fragmentation leading to reduction in population size and the isolation of trees [54], [68], [69].
The results of Bayesian model based clustering method indicated geographical structure with clustering of populations into two major groups based on their geographical location. The neighbouring populations grouped together forming two separate and genetically distinct groups of populations (Fig. 5B and 5C). These results further suggest that genetic structuring of populations is low and weak. Several studies on spatial genetic structure of tropical and temperate species also reported that a weak or low genetic structure could be attributable to extensive gene flow among populations through seeds and pollen [69]–[71]. The structured distribution of two gene pools with predominant distribution of the gene pool 1 in the north and abundance of gene pool 2 in the South could be attributable to limited dispersal of seeds between north and south populations. In addition, several factors including habitat fragmentation and associated increase in selfing and inbreeding may also contribute to the genetic differentiation through genetic drift. Anthropogenic effects may also have contributed to the reduction of seed dispersal via birds leading to genetic structuring of N. nimmoniana [52], [69].
Impact of harvesting on population structure
The size class structure of N. nimmoniana plants in PA and NPA populations were different, and PA populations maintained a higher proportion of larger, potentially older and reproductive plants than NPA populations. Furthermore we found a significant difference in density of adults and proportion of reproductive individuals in PA and NPA populations (P<0.05, Fig. 3B, C, D and E). Significantly, higher proportions of adults were harvested from NPAs as compared to PAs indicating that NPA populations suffer higher harvesting pressure as expected (Fig. 3F). This contributes to significant decline in demographic and reproductive parameters in NPA populations. Our results suggest that reproductive output changes with demographic shift in harvested populations. Populations with highest number of adults harvested had significantly low number of juveniles and saplings as compared to populations that were protected from harvesting. This effect could be attributable to shift in age class structure with harvesting, where non-protected populations continue to experience lower population sizes with fewer reproductive individuals leading to a decline in reproductive output and lower population growth rates.
Adults of N. nimmoniana in each population maintained significantly higher genetic diversity (A and HO) than the seedlings of the same population. This is because adult plants represent pre-harvesting generation with genetically diverse individuals resulting from larger effective population sizes. Recent human induced disturbance activities such as harvesting and fragmentation may have reduced the effective population sizes contributing to lowered genetic diversity over generations. Over harvesting leads to drastic reduction in population sizes and increased inbreeding, which can further reduce the genetic diversity in seedlings. This scenario was supported by the fact that there was significant increase in inbreeding values in seedlings as compared to adults in many populations (Table 3). Similar observations have been reported in several studies where selective logging and overharvesting of tropical tree species led to decrease in genetic diversity and increased inbreeding levels in seedlings [69], [72]. The altered age class distribution in NPA populations of N. nimmoniana due to harvesting may cause evolutionary effects compromising the long-term survival of the species. Documented changes in decline of demographic and reproductive parameters and genetic diversity measures in N. nimmoniana populations highlight the possible negative evolutionary consequences of harvesting. Although the short term demographic effects may be of immediate conservation importance [73], the selective harvesting of mature individuals may have profound long-term evolutionary impacts through the reduction of genetic diversity.
Genetic diversity in protected and non-protected populations
This study indicates that populations in PAs harbor significantly higher level of genetic diversity (HO, AP, AR and gene diversity) than NPA populations. Although other measures of genetic diversity (A and HE), did not differ significantly between two types of populations, the greater HO, AP, AR and gene diversity indicates that allele frequencies are higher in populations in PA’s. These differences are due to difference in harvesting pressure between populations of PA and NPA. On average, more than 45% of adults were extracted from populations of NPA as opposed to less than 5% of adults extracted from PA. This change in demographic decline might have contributed to uneven distribution of alleles in NPA populations. We also observed that the genetic diversity parameters among adults and seedlings differed between populations of PA and NPAs. There was significant decrease in genetic diversity parameters (HO and A) from adults to seedlings within populations of NPAs (Table 3), which is consistent with the general effects of harvesting of natural populations [18], [19]. As populations become small, rare alleles tend to be lost through the effect of genetic drift leading to the erosion of genetic diversity, which is often depicted in seedlings. Overall, our results demonstrate the impact of harvesting on genetic diversity of plants species and highlight the importance of PA network in conservation and management of economically and medicinally important plant species subject to harvesting [13].
Population bottlenecks and predictions of future population sizes
The population bottleneck analyses using excess heterozygosity method (Sign test and Wilcoxon signed rank test) detected evidence of recent genetic bottlenecks in most of the N. nimmoniana populations sampled in the present study, but no difference between PA and NPA were found. The bottleneck analyses results were consistent among both adult and seedlings (Table 6). This suggests that irrespective of protected status of populations, N. nimmoniana populations experienced recent genetic bottlenecks, which is consistent with recent reports of demographic decline of N. nimmoniana [18], [19]. However, the allele frequency based analytical method did not reveal any signatures of bottleneck, suggesting that allele frequency based methods of population bottleneck analyses are not sensitive in detecting recent population bottlenecks. The excess in heterozygosity based methods are known to be more powerful in detecting recent genetic bottleneck signatures as compared to allele frequency distortion methods [46]. Occurrence of large number of rare alleles in a population may alter the distortion of allele frequency due to population bottlenecks and mask the genetic signature of recent bottleneck events. Our results revealed that most of the populations experienced significant decrease in heterozygosity parameters than observed number of alleles, reducing the sensitivity of detection of distortions in allele frequency distributions. The demographic data revealed a decline in N. nimmoniana populations in the wild, which is consistent with the detection of genetic bottlenecks in the present study. The reported 20% decline of N. nimmoniana in recent years may have contributed to reduction of effective population sizes throughout its distribution range leaving genetic signatures of population bottleneck. The excess heterozygosity method can detect population bottlenecks as recent as 6 to 120 years depending on the generation length of a given species [44], [74].
Our simulation study revealed a faster decline of observed number of alleles (OA) than HO and is likely a result of existing low allelic diversity or recent genetic bottleneck. Similar results have been reported for other endangered species [75]. The BOTTLESIM simulation predicted a future decline in genetic diversity in most of the populations analysed (Fig. 6A to 6H). Interestingly, we observed that the current population size of N. nimmoniana in the Western Ghats is not sufficient to maintain present observed levels of genetic diversity over the period of next 100 years. This could be attributable to large scale harvesting of wild populations of N. nimmoniana. After the discovery of camptothecine in N. nimmoniana, the species have been largely exploited from the wild leading to reported 20% decline in natural populations of N. nimmoniana in the Western Ghats [18], [19]. The demographic decline coupled with recent genetic bottleneck events may contribute to further decline in genetic diversity in the future. Simulation results also predicted that populations of NPA lose diversity at a higher rate than populations from PAs. This may be due to variation in harvesting pressure, where over 45% of adults from NPAs have been harvested, but about only 5% of adults in PAs are known to have harvested. However, most of the populations are predicted to lose their diversity during next 100 years (Fig. 6A to 6H). Thus, measures to increase population sizes in PAs are needed to mitigate negative evolutionary consequences ensuring long term survival of N. nimmoniana.
Conservation implications
Our results based on N. nimmoniana highlight the effectiveness of protected areas in conserving genetic diversity of economically and medicinally important plant species. We observed that the populations from PAs had significantly high genetic diversity than populations in NPAs. It was further supported by simulation analysis where NPAs are predicted to lose genetic diversity faster than PA populations in future. PAs were also effective in preventing harvesting pressure on populations as evidenced by harvesting of over 45% from NPAs as compared to about 5% adults harvested from PAs.
The long-term persistence of population depends on population size of a species. A population with large number of individuals is considered to have more genetic diversity, which increases their ability to adapt to changing environmental conditions [76]. On the other hand, reduction in population sizes leads to loss of genetic diversity and allelic richness, inbreeding and increased extinction risk [45]. In the present study, results of genetic bottleneck analysis showed that most of the populations have gone through a phase of recent genetic bottleneck indicating recent reduction in effective population size. The simulation analysis also indicated that the current population size of N. nimmoniana in Western Ghats is not sufficient to maintain 90% of present genetic diversity over the next 100 years. If population sizes of N. nimmoniana further continue to decline in the wild, most populations may lose nearly 50% of present genetic diversity during the next 100 years. The above results are consistent with recent demographic decline of N. nimmoniana in its natural habitat. The reduction in its habitat and population size may have led to genetic bottleneck and further loss in genetic diversity. The conservation strategy should be oriented towards protecting natural populations of N. nimmoniana in Western Ghats from further overexploitation to sustain the long-term survival of the species. As adults are known to maintain more genetic diversity, we recommend that conservation efforts geared toward protection of adult plants in each population should be implemented to protect the reproductive fitness and evolutionary potential of the species [77]. Overall these results highlight the need for establishing more protected areas (PAs) in Western Ghats to conserve genetic diversity of economically and medicinally important plant species.
Supporting Information
Supplementary Table. S1 Table. Summary of t-test statistics for demographic parameters collected for four protected (PA) and non-protected (NPA) N. nimmoniana populations from Western Ghats.
(DOC)
Acknowledgments
Authors thank Naveen Kumar L and Sumangala R. C for assistance in the lab. We thank Dr. Edivani Franceschinelli and an anonymous reviewer for valuable comments that improved the manuscript. Authors thank the Karnataka Forest Department for permission to undertake the fieldwork in the Western Ghats, India.
Funding Statement
The work was supported by the following: Department of Biotechnology, India; NSERC-Canada; and Canadian Commonwealth Scholarships Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nantel P, Gagnon D, Nault AA (1996) Population viability analysis of American ginseng and wild leek harvested in stochastic environments. Conservation Biology 10:608–621. [Google Scholar]
- 2.Sheldon JW, Balick MJ, Laird SA (1997) Medicinal plants: can utilization and conservation coexist? Advances in Economic Botany, vol. 12. 104p, New York Botanical Garden, Bronx, New York, USA.
- 3. Laurance WF (1999) Reflections on the tropical deforestation crisis. Biological Conservation 91:109–117. [Google Scholar]
- 4. Pearce F (1997) Herbal cures means plants suffer. New Scientist 15:6. [Google Scholar]
- 5. Ghimire KB, Pimbert MP Eds., Social Change and Conservation: Environmental Politics and Impacts of National Parks and Protected Areas (Earthscan, London, 1997).
- 6. Rodrigues ASL, Andelman SJ, Bakarr MI, Boitani L, Brooks TM, et al. (2004) Effectiveness of the global protected area network in representing species diversity. Nature 428:640–643. [DOI] [PubMed] [Google Scholar]
- 7. Ehrlich PR, Pringle RM (2008) Where does biodiversity go from here? A grim business-as-usual forecast and a hopeful portfolio of partial solutions. Proceedings of the National Academy of Sciences USA 105:11579–11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thapa S, Chapman DS (2009) Impacts of resource extraction on forest structure and diversity in Bardia National Park, Nepal. Forest Ecology and Management 259:641–649. [Google Scholar]
- 9. Leroux SJ, Krawchuk MA, Schmiegelow F, Cumming SG, Lisgo K, et al. (2010) Global protected areas and IUCN designations: Do the categories match the conditions? Biological Conservation. 143:609–616. [Google Scholar]
- 10. Bruner AG, Gullison RE, Rice RE, Da Fonseca GAB (2001) Effectiveness of parks in protecting tropical biodiversity. Science 291:125–128. [DOI] [PubMed] [Google Scholar]
- 11. Uma Shaanker R, Ganeshaiah KN, Krishnan S, Ramya R, Meera C, et al. (2004) Livelihood gains and ecological costs of non-timber forest product dependence: assessing the roles of dependence, ecological knowledge and market structure in three contrasting human and ecological settings in South India. Environmental Conservation 31:242–253. [Google Scholar]
- 12.Nageswara Rao M, Uma Shaanker R, Ganeshaiah KN (2001) Mapping genetic diversity of sandal (Santalum album L.) in South India: Lessons for in-situ conservation of sandal genetic resources. In: Uma Shaanker R, Ganeshaiah KN, Bawa KS. (eds) Forest genetic resources: Status, threats and conservation strategies. Oxford and IBH Publishing Company Private Limited, New Delhi, India, 49–67.
- 13. Ramesha BT, Ravikanth G, Nageswara Rao M, Ganeshaiah KN, Uma Shaanker R (2007) Genetic structure of rattan, Calamus thwaitesii in core, buffer and peripheral regions of three protected areas at central Western Ghats, India: Do protected areas serve as refugia for genetic resources of economically important plants? Journal of Genetics 86:9–18. [DOI] [PubMed] [Google Scholar]
- 14.Ravikanth G, Nageswara Rao M, Ganeshaiah KN, Uma Shaanker R (2009) Impacts of harvesting on genetic diversity of NTFP species: Implications for conservation. In: Uma Shaanker R, Joseph GC, Hiremath AJ (eds.) Management, utilization, and conservation of non-timber forest products in the South Asia region. Universities Press, Bangalore, India, 53–63.
- 15. Nageswara Rao M, Ravikanth G, Ganeshaiah KN, Uma Shaanker R (2010) Role of protected area in conserving the population and genetic structure of economically important bamboo species, In: Nageswara Rao M, Soneji JR (eds) Tree and forest biodiversity. Bioremediation, Biodiversity and Bioavailability 4:69–76. [Google Scholar]
- 16.Uma Shaanker R, Ramesha BT, Ravikanth G, Gunaga R, Vasudeva R, et al. (2008) Chemical Profiling of Nothapodytes nimmoniana for Camptothecine, an Important Anticancer Alkaloid: Toward the Development of a Sustainable Production System. Pp 197–213. In Bioactive Molecules and Medicinal Plants (ed. Ramawat KG, Merillon JM) Springer Publishing, UK.
- 17. Raskin I, Ribnicky DM, Momarnytsky S, Ilic N, Poulev A, et al. (2002) Plants and human health in the twenty first century, Trends In Biotechnology. 20:522–531. [DOI] [PubMed] [Google Scholar]
- 18.Kumar R, Ved DK (2000) 100 Red listed medicinal plants of conservation concern in Southern India, foundation for revitalization of local health traditions, Bangalore, 261–263.
- 19. Gowda HHC, Vasudeva R, Mathachen GP, Shaanker UR, Ganeshaiah KN (2002) Breeding types in Nothapodytes nimmoniana Graham. Current Science 83:1077–1078. [Google Scholar]
- 20. Ved DK (1997) Trade in medicinal plants: the state of our ignorance. Amruth 1:2–8. [Google Scholar]
- 21.Mittermeier RA, Patricio RG, Hoffman M, Pilgrim J, Brooks T, et al. (2005) Hotspots revisited: earth’s biologically richest and most endangered terrestrial ecoregions. Conservation International. 392 p. [Google Scholar]
- 22.NWDC (National Wildlife Database Centre). 1999 Wild Life Institute of India, the report. Dehra Dun, India.
- 23. Ramesha BT, Amna T, Ravikanth G, Gunaga RP, Vasudeva R, et al. (2008) Prospecting for Camptothecinees from Nothapodytes nimmoniana in theWestern Ghats, South India: Identification of High-Yielding Sources of Camptothecine and New Families of Camptothecinees. Journal of Chromatographic Science 46:362–368. [DOI] [PubMed] [Google Scholar]
- 24. Yamazaki Y, Urano A, Sudo H, Kitajima M, Takayama H, et al. (2003) Metabolite profiling of alkaloids and strictosidine synthase activity in Camptothecine producing plants. Phytochemistry 62:461–470. [DOI] [PubMed] [Google Scholar]
- 25. Sharma MV, Uma Shaanker R, Leather SR, Vasudeva R, Shivanna KR (2011) Floral resources, pollinators and fruiting in a threatened tropical deciduous tree. Journal of Plant Ecology 4(4):259–267. [Google Scholar]
- 26. Ganeshaiah KN, Uma Shaanker R, Murali KS, Uma Shanker A, Bawa KS (1998) Extraction of non-timber forest products in the forest of Biligiri Rangan Hills. 5. Influence of dispersal mode on species response to anthropogenic pressures. Economic Botany 52:316–319. [Google Scholar]
- 27.Siegel S, Castellann JJ (1988) Nonparametric statistics for the behaviour sciences. McGraw-Hill, New York.
- 28. Doyle JJ, Doyle JS (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phyotochemical Bulletin 19:11–15. [Google Scholar]
- 29. Ramesha BT, Srirama R, Ravikanth G, Ravishankar KV, Dayanandan S, et al. (2008) Development of polymorphic microsatellite loci in Nothapodytes nimmoniana, a medicinally important tree from the Western Ghats, India. Molecular Ecology Resources 8(1):365–367. [DOI] [PubMed] [Google Scholar]
- 30. Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity 86:248–249. [Google Scholar]
- 31. Leberg PL (2002) Estimating allelic richness: effects of sample size and bottlenecks. Molecular Ecology 11:2445–2449. [DOI] [PubMed] [Google Scholar]
- 32. El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L) Skeels] endemic to Morocco. Theoretical Applied Genetics 92:832–839. [DOI] [PubMed] [Google Scholar]
- 33. Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet J (1995) FSTAT v-1.2. A computer program to calculate F-statistics. Journal of Heredity 86:485–6. [Google Scholar]
- 34. Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 35. Guo SW, Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportions for multiple alleles. Biometrics 48:361–372. [PubMed] [Google Scholar]
- 36. Kimura M, Crow J (1964) The number of alleles that can be maintained in finite populations. Genetics 49:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- 38. Kimura M, Otha T (1978) Stepwise mutation model and distribution of allelic frequencies in finite populations. Proceedings of National Academy of Science. USA 75:2868–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics. 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Evanno G, Regnaut S, Goudet J (2007) Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14:2611–2620. [DOI] [PubMed] [Google Scholar]
- 42. Earl DA, Von Holdt BM (2011) Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4:359–361. [Google Scholar]
- 43. Jakobsson M, Rosenberg NA (2007) CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. [DOI] [PubMed] [Google Scholar]
- 44. Luikart G, Cornuet JM (1998) Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biology 12:228–237. [Google Scholar]
- 45.Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge (UK), Cambridge University Press.
- 46. Cornuet JM. Luikart (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Piry S, Luikart G, Cornuet JM (1999) bottleneck: a computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity 90:502–503. [Google Scholar]
- 48. Kuo CH, Janzen FJ (2003) BOTTLESIM: a bottleneck simulation program for long-lived species with overlapping generations. Molecular Ecology Notes 3:669–673. [Google Scholar]
- 49. Lemes MR, Gribel R, Proctor J, Grattapaglia D (2003) Population genetic structure of mahogany (Swietenia macrophylla King, Meliaceae) across the Brazilian Amazon based on variation at microsatellite loci, implications for conservation. Molecular Ecology 12:2845–2883. [DOI] [PubMed] [Google Scholar]
- 50. Novick RR, Dick CD, Lemes MR, Navarro C, Caccon A, et al. (2003) Genetic structure of Mesoamerican population of big-leaf mahogany (Swietenia macrophylla) inferred from microsatellite analysis. Molecular Ecology 12:2885–2893. [DOI] [PubMed] [Google Scholar]
- 51. Collevatti RG, Grattapaglia D, Hay JD (2003) Evidence for multiple lineages of Caryocar brasiliense populations in the Brazilian Cerrado based on the analysis of chloroplast DNA sequence and microsatellite haplotype variation. Molecular Ecology 12:105–115. [DOI] [PubMed] [Google Scholar]
- 52.Christine O, Sokpon N, Khasa DP (2009) Genetic diversity and population structure of a threatened African tree species, Milicia excelsa, using nuclear microsatellites DNA markers. International Journal of Forestry Research Article ID 210179, 8 pages.
- 53. Ng KKS, Lee SL, Saw LG, Plotkin JB, Koh CL (2006) Spatial structure and genetic diversity of three tropical tree species with different habitat preferences within a natural forest. Tree Genetics and Genome 2:121–131. [Google Scholar]
- 54. Andrianoelina O, Favreau B, Ramamonjisoa L, Bouvet JM (2009) Small effect of fragmentation on the genetic diversity of Dalbergia monticola, an endangered tree species of the eastern forest of Madagascar, detected by chloroplast and nuclear microsatellites. Annals of Botany 104:1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zucchi MI, Rosana PVB, Pinheiro JB, Chaves LJ, Coelho ASG, et al. (2003) Genetic structure and gene flow in Eugenia dysenterica DC in the Brazilian Cerrado utilizing SSR markers. Genetics and Molecular Biology 26(4):449–457. [Google Scholar]
- 56. Dick CW, Hardy OJ, Jones FA, Petit RA (2008) Spatial scales of pollen and seed-mediated gene flow in tropical rain forest trees. Tropical Plant Biology 1:20–33. [Google Scholar]
- 57. Guidugli MC, Accoroni G, Mestriner MA, Betioli Contel EA, Martinez CA, et al. (2010) Genetic characterization of 12 heterologous microsatellite markers for the giant tropical tree Cariniana legalis (Lecythidaceae). Genetics and Molecular Biology 33(1):131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sharma MV, Shaanker RU, Vasudeva R, Shivanna KR (2010) Functional dioecy in Nothapodytes nimmoniana, a distylous species in the Western Ghats. Current Science 99:1444–1449. [Google Scholar]
- 59. Tadwalkar MD, Joglekar AM, Mhaskar M, Kanade RB, Chavan B, et al. (2012) Dispersal modes of woody species from the northern Western Ghats, India. Tropical Ecology 53:53–67. [Google Scholar]
- 60. Aldrich PR, Hamrick JL, Chavarriaga P, Kochert G (1998) Microsatellite analysis of demographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera . Molecular Ecology 7:933–944. [DOI] [PubMed] [Google Scholar]
- 61. Dayanandan S, Dole J, Bawa KS, Kesseli R (1999) Population structure delineated with microsatellite markers in fragmented populations of a tropical tree, Carapa guianensis (Meliaceae). Molecular Ecology 8:1585–1592. [DOI] [PubMed] [Google Scholar]
- 62. White CM, Boshier DH, Powell W (1999) Genetic variation within a fragmented population of Swietenia humilis Zucc. Molecular Ecology 11:1899–1910. [DOI] [PubMed] [Google Scholar]
- 63. Hamrick JL, Godt MJ (1996) Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London Series B 351:1291–1298. [Google Scholar]
- 64. Abdul Kareem VK, Rajasekharan PE, Mini S, Kumar VT (2011) Genetic diversity and structure of the threatened anti-cancerous plant Nothapodytes nimmoniana as revealed by ISSR analysis. Plant Genetic Resources 9(4):506–514. [Google Scholar]
- 65. Brossart JL, Prowell DP (1998) Genetic estimates of population structure and gene flow: limitations, lessons, and new directions. Trends in Ecology and Evolution 13:202–206. [DOI] [PubMed] [Google Scholar]
- 66. Sork VL, Nason J, Campbell DR, Fernandez JF (1999) Landscape approaches to historical and contemporary gene flow in plants. Trends in Ecology and Evolution 14:219–223s. [DOI] [PubMed] [Google Scholar]
- 67. Whitlock MC, Mccauley DE (1999) Indirect measures of gene flow and migration: FST ≠ 1/(4Nm+1) . Heredity 82:117–125. [DOI] [PubMed] [Google Scholar]
- 68.Savolainen O, Kuittinen H (2000) Small population processes. In: Young A, Boshier D, Boyle T. eds. Forest conservation genetics, principles and practice. Wallingford: CSIRO Publishing, 91–100.
- 69.Shivaprakash KN (2011) Population genetics of vulnerable and endemic tree species Myristica malabarica and evaluation of ecological niche models (ENMs). Master Thesis submitted to University of Agricultural Sciences, Bangalore, India.
- 70. Loiselle BA, Sork VL, Nason J, Graham C (1995) Comparison of genetic variation in bird-dispersed shrubs of a tropical wet forest. Biotropica 27:487–494. [Google Scholar]
- 71. Chung M, Chung MY, Oh G, Epperson B (2000) Spatial genetic structure in a Neolitsea sericea population (Lauraceae). Heredity 85:490–497. [DOI] [PubMed] [Google Scholar]
- 72. Lee CT, Wickneswari R, Mahani MC, Zakri AH. 2002 Effect of selective logging on the genetic diversity of Scaphium macropodum . Biological Conservation 104:107–118. [Google Scholar]
- 73. Schemske DW, Husband BC, Ruckelshaus MH, Goodwillie C, Parker IM, et al. (1994) Evaluating approaches to the conservation of rare and endangered plants. Ecology 75:584–606. [Google Scholar]
- 74. Hundertmark KJ, Van Daele LJ (2010) Founder effect and bottleneck signatures in an introduced, insular population of elk. Conservation Genetics 11:139–147. [Google Scholar]
- 75. Lippe CA, Dumont P, Bernatchez L (2006) High genetic diversity and no inbreeding in the endangered copper redhorse, Moxostoma hubbsi (Catostomidae, Pisces): the positive sides of a long generation time. Molecular Ecology 15:1769–1780. [DOI] [PubMed] [Google Scholar]
- 76. Vrijenhoek RC, Douglas ME, Meffe GK (1985) Conservation genetics of endangered fish populations in Arizona. Science 229:400–402. [DOI] [PubMed] [Google Scholar]
- 77. Cruse-Sanders JM, Hamrick JL (2004) Spatial and genetic structure within populations of wild American ginseng (Panax quinquefolius L., Araliaceae). Journal of Heredity 95:309–321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table. S1 Table. Summary of t-test statistics for demographic parameters collected for four protected (PA) and non-protected (NPA) N. nimmoniana populations from Western Ghats.
(DOC)