Abstract
Sweet potato is grown extensively from tropical to temperate regions and is an important food crop worldwide. In this study, we established detection methods for 17 major sweet potato viruses using single and multiplex RT-PCR assays. To investigate the current incidence of viral diseases, we collected 154 samples of various sweet potato cultivars showing virus-like symptoms from 40 fields in 10 Korean regions, and analyzed them by RT-PCR using specific primers for each of the 17 viruses. Of the 17 possible viruses, we detected eight in our samples. Sweet potato feathery mottle virus (SPFMV) and sweet potato virus C (SPVC) were most commonly detected, infecting approximately 87% and 85% of samples, respectively. Furthermore, Sweet potato symptomless virus 1 (SPSMV-1), Sweet potato virus G (SPVG), Sweet potato leaf curl virus (SPLCV), Sweet potato virus 2 ( SPV2), Sweet potato chlorotic fleck virus (SPCFV), and Sweet potato latent virus (SPLV) were detected in 67%, 58%, 47%, 41%, 31%, and 20% of samples, respectively. This study presents the first documented occurrence of four viruses (SPVC, SPV2, SPCFV, and SPSMV-1) in Korea. Based on the results of our survey, we developed multiplex RT-PCR assays for simple and simultaneous detection of the eight sweet potato viruses we recorded.
Keywords: disease incidence, sweet potato viruses, multiplex RT-PCR
Sweet potato (Ipomea batatas L.), of the family Convolvulaceae, is grown extensively from tropical to temperate regions and is an important food crop worldwide. According to FAOSTAT data, there are 8 million ha of sweet potato in cultivation worldwide, with approximately 103 million metric tons produced in 2012. Major sweet potato-producing nations include China, Tanzania, Nigeria, Uganda, and Indonesia; production in China alone is around 73 million metric tons, or more than 70% of the worldwide total. Korea currently produces approximately 260,000 metric tons from 19,000 ha, with production increasing every year.
Viral diseases of sweet potato have become widespread, causing serious crop losses around the world. In total, more than 30 viruses have now been reported to infect sweet potato (Brunt et al., 1996; Clark et al., 2012). Among these, 23 have been assigned a formal taxonomic position by the International Committee on Taxonomy of Viruses (ICTV) (Table 1). The number continues to increase as virus detection methods are improved. Only a few of the viruses are considered to be of major economic importance. The most severe disease in sweet potato is caused by coinfection with the whitefly-transmitted Sweet potato chlorotic stunt virus (SPCSV) and the aphid-transmitted Sweet potato feathery mottle virus (SPFMV), which results in the synergistic sweet potato virus disease (SPVD) (Gibson et al., 1998; Karyeija et al., 2000; Mukasa et al., 2006). Synergism has also been observed between SPCSV and the possibly whitefly-transmitted Sweet potato mild mottle virus (SPMMV) (Gutiérrez et al., 2003; Hahn, 1979). SPCSV caused synergistic diseases in sweet potato with many other sweet potato viruses (Untiveros et al., 2007).
Table 1.
Major viruses infecting sweet potato
| Virus name | Abb. | Family (genus) | Transmission |
|---|---|---|---|
| Sweet potato feathery mottle virus | SPFMV | Potyviridae (Potyvirus) | Aphid (non-persistent) |
| Sweet potato virus G | SPVG | Potyviridae (Potyvirus) | Aphid (non-persistent) |
| Sweet potato latent virus | SPLV | Potyviridae (Potyvirus) | Aphid (non-persistent) |
| Sweet potato leaf curl virusa | SPLCV | Geminiviridae (Begomovirus) | Whitefly (persistent) |
| Sweet potato mild speckling virus | SPMSV | Potyviridae (Potyvirus) | Aphid (non-persistent) |
| Sweet potato mild mottle virus | SPMMV | Potyviridae (Ipovirus) | Whitefly (persistent) |
| Sweet potato chlorotic stunt virus | SPCSV | Closteroviridae (Crinivirus) | Whitefly(non-persistent) |
| Sweet potato collusive virus | SPCV | Caulimoviridae(Cavemovirus) | * |
| Sweet potato virus 2 | SPV2 | Potyviridae (Potyvirus) | Aphid (non-persistent) |
| Sweet potato virus C | SPVC | Potyviridae (Potyvirus) | Aphid (non-persistent) |
| Sweet potato symptomless virus 1 | SPSMV-1 | Geminiviridae (Mastrevirus) | * |
| Sweet potato chlorotic fleck virus | SPCFV | Betaflexiviridae (Carlavirus) | * |
| Sweet potato vein clearing virus | SPVCV | Caulimoviridae (Solendovirus) | * |
| Sweet potato pakakuy virus | SPPV | Caulimoviridae (Badnavirus) | * |
| Sweet potato C6 virus | SPC6V | Betaflexiviridae (Carlavirus) | * |
| Sweet potato leaf speckling virus | SPLSV | Luteoviridae (Polerovirus) | * |
| Cucumber mosaic virus | CMV | Bromoviridae (Cucumovirus) | Aphid (non-persistent) |
Sweet potato leaf curl virus has been classified into seven species: Sweet potato leaf curl virus (SPLCV), Ipomoea yellow vein virus (IYVV), Sweet potato leaf curl Georgia virus (SPLCGoV), Sweet potato leaf curl China virus (SPLCV-CN), Sweet potato leaf curl Lanzarote virus (SPLCLaV), Sweet potato leaf curl Canary virus (SPLCCaV), and Sweet potato leaf curl Spain virus (SPLCESV) by ICTV.
Not reported.
In Korea, SPFMV, Sweet potato virus G (SPVG), and Sweet potato latent virus (SPLV), all belonging to the family Potyviridae, and Sweet potato leaf curl virus (SPLCV), a member of the Geminiviridae, have been detected (Kwak et al., 2006). Our previous nationwide survey revealed that, in 2003, about 73% of samples were infected with at least one of these four viruses (Kwak et al., 2006). SPFMV and SPVG were especially prevalent (40% and 16%, respectively), and coinfection with SPFMV and SPVG was detected in 11% of diseased sweet potatoes. Although SPCSV was reported by Yun et al. (2002), it has not subsequently been detected in Korea.
In the present study, we established detection methods for 17 major sweet potato viruses using reverse transcription-polymerase chain reaction (RT-PCR), and investigated the current incidence of viral disease in Korean sweet potatoes by analyzing 154 samples of various cultivars showing virus-like symptoms collected from 40 fields in 10 regions. Finally, we developed multiplex RT-PCR methods for simple and simultaneous detection of the eight sweet potato viruses known to occur in Korea.
Materials and Methods
Survey and sample collection
In 2012, we carried out a survey of sweet potato viruses in 40 seedling-cultured fields of sweet potatoes in 10 regions of 5 different Korean provinces (Fig. 1). We collected samples of 154 sweet potato leaves (including petiole and stem) showing virus-like symptoms. The collected samples were treated with insecticides to remove potential insect vectors and maintained in pots in a greenhouse at 20–25°C (Table 2). All samples were inspected for disease symptoms for at least 30 days and analyzed for virus infection by RT-PCR.
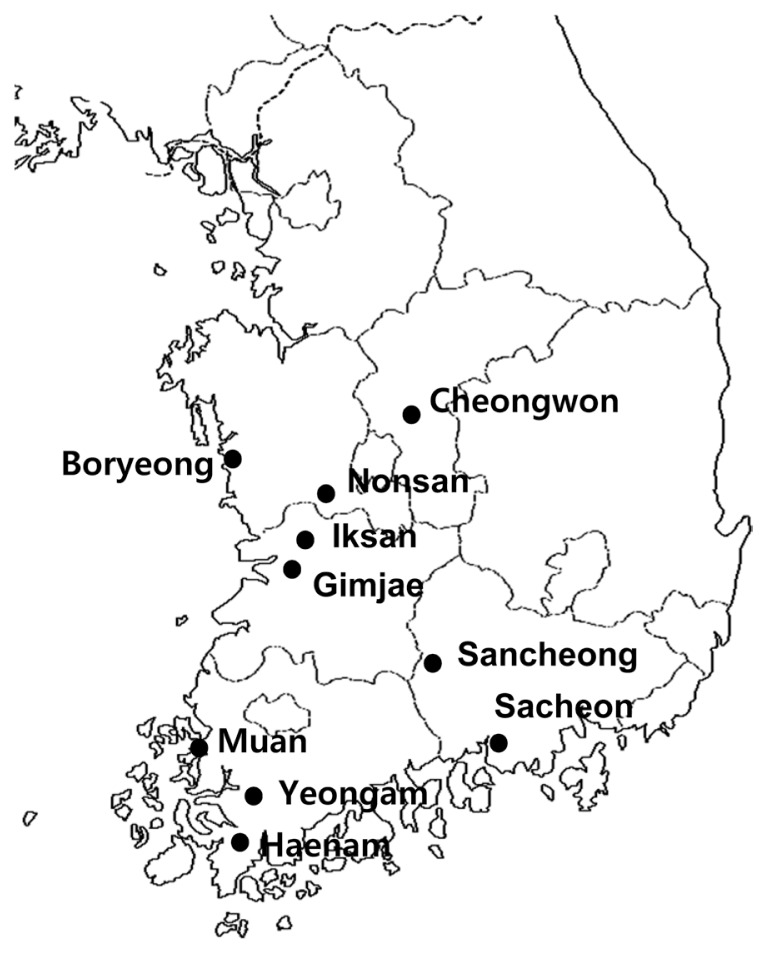
Fig. 1.
Geographic location of the sweet potato fields surveyed in 2012. A total of 154 sweet potato specimens were collected from 40 cultivated fields in 10 regions of 5 Korean provinces, as indicated.
Table 2.
Legend of sweet potato specimens analyzed in this study
| Province | Regionsa | No. of fields investigated | No. of samplesb collected |
|---|---|---|---|
| Jeollanam-do | Haenam | 8 | 40 |
| Yeongam | 4 | 10 | |
| Muan | 7 | 37 | |
|
| |||
| Jeollabuk-do | Iksan | 3 | 16 |
| Gimje | 6 | 14 | |
|
| |||
| Gyeongsangnam-do | Sancheong | 2 | 13 |
| Sacheon | 1 | 4 | |
|
| |||
| Chungcheongbuk-do | Cheongwon | 2 | 7 |
|
| |||
| Chungcheongnam-do | Nonsan | 2 | 3 |
| Boryeong | 5 | 10 | |
|
| |||
| Total | 10 | 40 | 154 |
Regions and fields selected for sample collection were within major Korean sweet potato production areas in 2012.
Samples of sweet potato leaves (including petiole and stem) showing virus-like symptoms were collected.
Total nucleic acids extraction and virus detection by RT-PCR
We extracted total nucleic acids from the infected leaf, petiole and stem samples using the Viral gene-spin™ viral DNA/RNA extraction kit (iNtRON, Korea), according to the manufacturer’s instructions. Typical RT-PCR assays were carried out using the primers shown in Table 3 in a two-step procedure using AMV reverse transcriptase (Promega, USA) for RT and Go-taq polymerase (Promega, USA) for PCR, as described by Kwak et al. (2013). Multiplex RT-PCR assays were performed using two-step RT-PCR or one-step RT-PCR. In the case of two-step RT-PCR, RT reactions were carried out at 42°C for 30 min in a final 5 μl reaction obtained by combining 0.5 μl total RNA (approx. 0.5 μg), 0.5 μl of a mixture of equal amounts of 32 μM reverse primers for four viruses, 1 μl 5 × RT buffer, 0.5 μl 2.5 μM dNTP, 0.1 μl BSA (10 mg/ml), 8 U RNase inhibitor, 0.5 U AMV reverse transcriptase (Promega, USA), and sufficient dH2O to bring the total to 5 μl. RT reactions were terminated by heating at 95°C for 5 min. When RT was completed, we added 20 μl of a solution comprising 0.5 μl of a mixture of equal amounts of 32 μM forward primers for four viruses, 5 μl 5 × PCR buffer, 2.5 μL 25 mM MgCl2, 0.4 μl BSA (10 mg/ml), 1 U Go-Taq DNA polymerase (Promega, USA), and dH2O to the tube containing RT products. PCR was performed in a thermal cycler (Bio-Rad, USA) with the following conditions: pre-denaturing step at 94°C for 5 min; 35 cycles of a denaturing step at 94°C for 30 s; an annealing step at 55°C for 30 s; an extension step at 72°C for 60 s; and a final extension at 72°C for 10 min. One-step RT-PCR was performed in a 20 μl reaction containing total RNA (approx. 0.5 μg), a mixture of equal amounts of 32 μM forward and reverse primers for four viruses, and RT-PCR master mix (GenetBio, Korea) under the following conditions: 30 min at 50°C; 10 min at 94°C; and 35 cycles consisting of 20 s at 94°C, 30 s at 55°C, 60 s at 72°C, and 5 min at 72°C. PCR products were analyzed by electrophoresis on a 1.5% agarose gel at 100 V for 90 min, stained with ethidium bromide, and DNA bands were visualized using a UV transilluminator.
Table 3.
Primers used in multiplex and single RT-PCR assays for the detection of major sweet potato-infecting viruses
| Virus | Primera | Sequence (5′→3′) | Loci | Size (nt) |
|---|---|---|---|---|
| Multiplex RT-PCR primer set 1 | ||||
| SPLCV | SPLC-u1 | TCTGCCGTCGATCTGGAACTC | 2315–2335 | 507 |
| SPLC-d1 | GTGCCCGCCTTTGGTGGAC | 2821–2803 | ||
| SPFMV | SPFMV 1-F | TACACACTGCTAAAACTAGG | 9073–9092 | 356 |
| SPFMV 1-R | AGTTCATCATAACCCCATGA | 9428–9409 | ||
| SPVG | SPG 3-F | CAATGCCAAATGGAAGAATAG | 9945–9965 | 286 |
| SPG 3-R | GCATGATCCAATAGAGGTTTTA | 10230–10209 | ||
| SPLV | SPLV 1-F | GGAGTCAGTTCAATCAATGGTA | 9340–9361 | 184 |
| SPLV 1-R | AGTGGCTTTATTGGGTATGAT′ | 9523–9503 | ||
|
| ||||
| Multiplex RT-PCR primer set 2 | ||||
| SPCFV | SPCFV 2F | AGCTGCTCAAACAAGCAAGAGG | 8526–8547 | 579 |
| SPCFV 2R | GCTCAAAAGTACTTTAAAACATGC | 9104–9081 | ||
| SPVC | SPVC-F | ATTCTTGAATGGGATAGATCACATG | 9353–9377 | 447 |
| SPVC-R | AGCTTCACGAAGCGCAGC | 9799–9782 | ||
| SPV2 | SPV2-F1 | ATGTGTTGAACCATCAGCTGAA | 9414–9435 | 369 |
| SPV2-R1 | GTAACTTGCCTTGGGCTACG | 9782–9763 | ||
| SPSMV-1 | SPSMV-1 F1 | ACCGTGTATTTGATGACGATGTAC | 352–375 | 230 |
| SPSMV-1 R | GGGAAGTTCTGGTAGAACGTATC | 581–559 | ||
|
| ||||
| Single RT-PCR primersb | ||||
| SPMMV | SPMMV 3-F | CCGCGCCAACAA AGGAACTA | 9842–9861 | 298 |
| SPMMV 3-R | TTGATGGGGTAATAAAGCACT | 10140–10120 | ||
| SPCSV | SPCSV-uni-f1 | GGGAAGAMGAGAYATGGAGTTAA | 4484–4506 | 583 |
| SPCSV-uni-r1 | CCTTGTTACAAAGAGCGTTCCT | 5066–5045 | ||
| SPMSV | SPMSV 1-F | GCCAAAACCAACAAGCATCA | 105–124 | 275 |
| SPMSV 1-R | ATTCGCATTTCCTCATCATCT | 380–360 | ||
| SPCV | SPCV F | AGGAAATCCCAGTATTATTCAAC | 4267–4289 | 922 |
| SPCV R | ATTTCTAATTTGGTTTACTAATCC | 5188–5165 | ||
| SPVCV | SPVCV-F | ATCCATTGCCAAATAAGATATTAAGA | 5844–5870 | 308 |
| SPVCV-R | CTTCTTAAGCAATGTTTCATGCTC | 6151–6128 | ||
| SPPV | SPPV-F | ATGAGGAGAA(C)CAGGGGCC | 1486–1503 | 722 |
| SPPV-R | CCAACG(A)TTTGGAGTGTTGGAT | 2207–2187 | ||
| SPC6V | SPC6V-F1 | AAAAGCTTGTTGGCAATTTGTG | 6804–6825 | 590 |
| SPC6V-R1 | TTGGCATTTCGATTGTCCC | 7393–7376 | ||
| SPLSV | SPLSV-F | ATGAGTACGGTCGTGGTTAGAAAC | 1–24 | 612 |
| SPLSV-R | CTACCTATTTGGGTTCTGGAAGG | 612–590 | ||
| CMV | CMV-DP u | CGTCGTGGTTCCCGCTCCG | 1309–1327 | 474 |
| CMV-DP d2 | AGCGCGCATCGCCGAAAGAT | 1782–1763 | ||
Primers were designed based on the nucleotide sequences of sweet potato viruses registered in Genbank.
RT-PCR conditions are as follows ; 30 min at 42°C, 5 min at 94°C, and 35 cycles consisting of 20 s at 94°C, 30 s at 55°C, 60 s at 72°C, and 5 min at 72°C.
Primer design
Specific primers for the detection of sweet potato viruses were designed based on the reported nucleotide sequences of various sweet potato virus isolates retrieved from the GenBank of the National Center for Biotechnology Information (Table 3). In particular, the primers for multiplex RT-PCR were designed to have similar melting temperatures and selected so as not to produce nonspecific bands. Two sets of the multiplex detection primers and the expected sizes of the amplicons are listed in Table 3.
Results
Incidence of viral disease in Korean sweet potatoes in 2012
To investigate the current incidence of viral disease in Korean sweet potatoes, we collected 154 samples from various seedling-cultured sweet potato cultivars in 10 regions of 5 Korean provinces in 2012. We observed various representative symptoms on the infected sweet potato samples, including vein clearing, vein banding, chlorotic local lesions, purpling, malformation, and leaf curling (Fig. 2). We analyzed samples by RT-PCR using specific primers designed to detect 17 major viruses infecting sweet potato (Table 3). In case of SPLCV, universal primers were used to detect seven SPLCV species (Table 1). Among the 17 viruses tested, we detected 8, including 4 viruses not previously detected in Korea (Sweet potato virus C [SPVC], Sweet potato virus 2 [SPV2], Sweet potato chlorotic fleck virus [SPCFV], and Sweet potato symptomless virus 1 [SPSMV-1]; Table 4). SPFMV and SPVC were detected most often, in about 87% and 85% of samples, respectively, including in mixed infections with other viruses (Table 4). Furthermore, SPSMV-1, SPVG, SPLCV, SPV2, SPCFV, and SPLV were detected in 67%, 58%, 47%, 41%, 31%, and 20% of the collected samples, respectively (Table 4). While SPFMV, SPVC, SPVG, SPV2, and SPLCV were detected in all surveyed areas, SPLV and SPSMV-1 were not detected in Boryeong and Sancheong, respectively, and SPCFV was not detected in Nonsan or Boryeong (Table 4). All samples but one were infected with at least one of the eight viruses (Table 5). Only 3.9% of the samples were singly infected with one of the eight viruses, while 95.5% of the samples were found in mixed infections (Table 5). The total rates of double, triple, quadruple, quintuple, sextuple, septuple, and octuple infections were 8.4%, 12.3%, 20.8%, 32.5%, 19.5%, 1.3%, and 0.6%, respectively. The most prevalent infection type was quintuple infection with SPFMV, SPVC, and other viruses (Table 5).
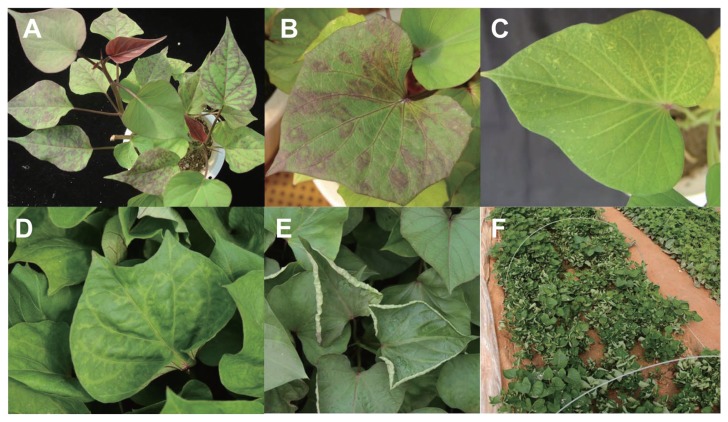
Fig. 2.
Representative virus symptoms observed in sweet potato: (A) vein banding and purpling; (B) chlorotic local lesions and purpling; (C) vein clearing and mottle; (D) vein clearing and leaf malformation; (E) leaf curling; (F) leaf curling in a cultivated field.
Table 4.
Incidence of sweet potato viruses in samples of seedling-cultured sweet potatoes collected in 2012
| Province/Region | No. of samples | No. of virus detections by RT-PCR a | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| SPFMV | SPVC | SPVG | SPV2 | SPLV | SPCFV | SPLCV | SPSMV-1 | |||
| Jeollanam-do | Haenam | 40 | 40 | 40 | 17 | 17 | 8 | 17 | 20 | 29 |
| Yeongam | 10 | 10 | 10 | 7 | 8 | 2 | 1 | 3 | 10 | |
| Muan | 37 | 25 | 27 | 13 | 3 | 2 | 12 | 22 | 34 | |
|
| ||||||||||
| Jeollabuk-do | Iksan | 16 | 13 | 7 | 10 | 7 | 1 | 1 | 6 | 6 |
| Gimje | 14 | 10 | 11 | 8 | 4 | 6 | 3 | 7 | 11 | |
|
| ||||||||||
| Gyeongsangnam-do | Sancheong | 13 | 13 | 13 | 12 | 8 | 6 | 9 | 4 | 0 |
| Sacheon | 4 | 4 | 4 | 4 | 2 | 3 | 2 | 2 | 1 | |
|
| ||||||||||
| Chungcheongbuk-do | Cheongwon | 7 | 7 | 6 | 6 | 4 | 1 | 2 | 5 | 5 |
|
| ||||||||||
| Chungcheongnam-do | Nonsan | 3 | 3 | 3 | 3 | 2 | 1 | 0 | 1 | 2 |
| Boryeong | 10 | 9 | 10 | 9 | 8 | 0 | 0 | 3 | 5 | |
|
| ||||||||||
| Total | 10 | 154 | 134 | 131 | 89 | 63 | 30 | 47 | 73 | 103 |
|
| ||||||||||
| Ratio (%) | 87 | 85 | 58 | 41 | 20 | 31 | 47 | 67 | ||
Including single and mixed infections.
Table 5.
Mixed infection types of eight sweet potato viruses
| Mixed infection type | Detected viruses | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| SPFMV | SPVC | SPVG | SPV2 | SPLV | SPCFV | SPLCV | SPSMV-1 | Subtotal | Total (%) | |
| Octuple | + | + | + | + | + | + | + | + | 1 | 1 (0.6) |
|
| ||||||||||
| Septuple | + | + | + | + | + | + | + | − | 1 | 2 (1.3) |
| + | + | + | − | + | + | + | + | 1 | ||
|
| ||||||||||
| Sextuple | + | + | + | + | + | + | − | − | 4 | 30 (19.5) |
| + | + | + | + | + | − | + | − | 6 | ||
| + | + | + | + | + | − | − | + | 3 | ||
| + | + | + | + | − | + | + | − | 1 | ||
| + | + | + | + | − | − | + | + | 6 | ||
| + | + | + | − | + | + | + | − | 2 | ||
| + | + | + | − | + | − | + | + | 3 | ||
| + | + | + | − | − | + | + | + | 1 | ||
| + | + | − | + | − | + | + | + | 2 | ||
| + | + | − | − | + | + | + | + | 2 | ||
|
| ||||||||||
| Quintuple | + | + | + | + | + | − | + | − | 4 | 50 (32.5) |
| + | + | + | + | − | + | − | − | 3 | ||
| + | + | + | + | − | − | + | − | 6 | ||
| + | + | + | + | − | − | − | + | 12 | ||
| + | + | + | − | + | + | − | − | 1 | ||
| + | + | + | − | − | + | + | − | 1 | ||
| + | + | + | − | − | − | + | + | 8 | ||
| + | + | − | + | − | + | − | + | 2 | ||
| + | + | − | − | − | + | + | + | 13 | ||
|
| ||||||||||
| Quadruple | + | + | + | + | − | − | − | 6 | 32 (20.8) | |
| + | + | + | − | − | + | − | − | 1 | ||
| + | + | + | − | − | − | + | − | 2 | ||
| + | + | + | − | − | − | − | + | 7 | ||
| + | + | − | + | − | − | + | − | 1 | ||
| + | + | − | + | − | − | − | + | 2 | ||
| + | + | − | − | − | + | − | + | 7 | ||
| + | + | − | − | − | − | + | + | 2 | ||
| + | − | + | + | − | − | − | + | 1 | ||
| + | − | + | − | + | + | − | − | 1 | ||
| + | − | + | − | + | − | + | − | 1 | ||
| − | + | − | − | − | + | + | + | 1 | ||
|
| ||||||||||
| Triple | + | + | + | − | − | − | − | − | 2 | 19 (12.3) |
| + | + | − | + | − | − | − | − | 2 | ||
| + | + | − | − | − | − | + | − | 1 | ||
| + | + | − | − | − | − | − | + | 7 | ||
| + | − | − | − | − | − | + | + | 1 | ||
| − | + | − | − | − | − | + | + | 4 | ||
| − | + | + | − | − | − | − | + | 1 | ||
| − | − | − | − | − | + | + | + | 1 | ||
|
| ||||||||||
| Double | + | − | + | − | − | − | − | − | 1 | 13 (8.4) |
| + | − | − | − | − | − | + | − | 1 | ||
| + | − | − | − | − | − | − | + | 4 | ||
| − | + | − | − | − | − | − | + | 2 | ||
| − | − | + | − | − | − | − | + | 2 | ||
| − | − | − | − | − | + | − | + | 1 | ||
| − | − | − | − | − | − | + | + | 2 | ||
|
| ||||||||||
| Single | + | − | − | − | − | − | − | − | 1 | 6 (3.9) |
| − | − | − | − | − | − | + | − | 1 | ||
| − | − | − | − | − | − | − | + | 4 | ||
|
| ||||||||||
| No detection | − | − | − | − | − | − | − | − | 1 | 1 (0.6) |
|
| ||||||||||
| Total | 154 | |||||||||
Development of multiplex RT-PCR assays for simultaneous detection of sweet potato viruses
For simultaneous detection of sweet potato-infecting viruses occurring in Korea, we developed two multiplex RT-PCR assays for the eight major viruses including SPLCV, SPFMV, SPVG, and SPLV in one set, and SPCFV, SPVC, SPV2, and SPSMV-1 in another (Table 3). The primer sets of the assays were designed to generate different sizes of amplicons for the target viruses (Table 3). Each primer pair was highly specific and did not cross-amplify to other viral or sweet potato nucleic acids (Figs. 3 and 4, lanes 1–4). The specificity of each primer pair was confirmed by sequencing the RT-PCR products. Simultaneous detection of all possible combinations among SPLCV, SPFMV, SPVG, and SPLV was performed by multiplex RT-PCR using the singly or mixed-infection field samples (Fig. 3). Likewise, we also performed simultaneous detection of all possible combinations among SPCFV, SPVC, SPV2, and SPSMV-1 by multiplex RT-PCR (Fig. 4). Various field samples of virus-infected sweet potatoes were subjected to the multiplex RT-PCR assays developed in this study, resulting in reliable and sensitive detection and identification of major sweet potato viruses (data not shown).
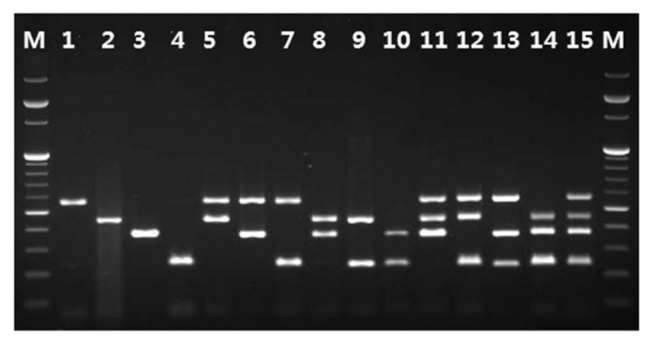
Fig. 3.
Simultaneous detection of all possible combinations among SPLCV, SPFMV, SPVG, and SPLV by multiplex RT-PCR. Lane M: 100 bp ladder; Lane 1: SPLV; Lane 2: SPVG; Lane 3: SPFMV; Lane 4: SPLCV; Lane 5: SPLV and SPVG; Lane 6: SPLV and SPFMV; Lane 7: SPLV and SPLCV; Lane 8: SPVG and SPFMV; Lane 9: SPVG and SPLCV; Lane 10: SPFMV and SPLCV; Lane 11: SPLV, SPVG, and SPFMV; Lane 12: SPLV, SPVG, and SPLCV; Lane 13: SPLV, SPFMV, and SPLCV; Lane 14: SPVG, SPFMV, and SPLCV; Lane 15: SPLV, SPVG, SPFMV, and SPLCV.
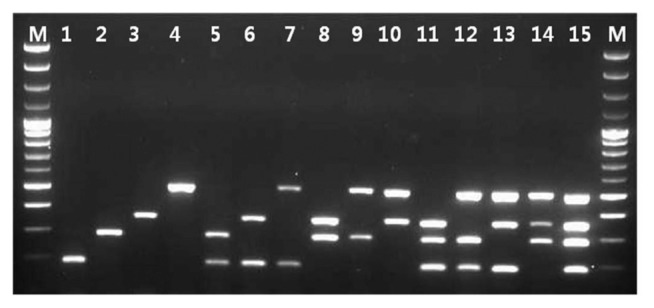
Fig. 4.
Simultaneous detection of all possible combinations among SPCFV, SPVC, SPV2, and SPSMV-1 by multiplex RT-PCR. Lane M: 100 bp ladder; Lane 1: SPCFV; Lane 2: SPVC; Lane 3: SPV2; Lane 4: SPSMV-1; Lane 5: SPCFV and SPVC; Lane 6: SPCFV and SPV2; Lane 7: SPCFV and SPSMV-1; Lane 8: SPVC and SPV2; Lane 9: SPVC and SPSMV-1; Lane 10: SPV2 and SPSMV-1; Lane 11: SPCFV, SPVC, and SPV2; Lane 12: SPCFV, SPVC, and SPSMV-1; Lane 13: SPCFV, SPV2, and SPSMV-1; Lane 14: SPVC, SPV2, and SPSMV-1; Lane 15: SPCFV, SPVC, SPV2, and SPSMV-1.
Discussion
In Korea, SPFMV, SPLV, SPVG, and SPLCV have previously been reported to infect sweet potato (Kwak et al., 2006). In this study, a nationwide survey was performed to investigate the current incidence of viral diseases in Korean sweet potatoes. To this end, we analyzed 154 samples of sweet potatoes showing virus-like symptoms by single or multiplex RT-PCR assays to detect 17 major viruses. Our survey revealed that eight viruses (including four that were previously undocumented: SPVC, SPV2, SPSMV-1, and SPCFV) infect sweet potatoes in Korea. However, other nine viruses (SPCSV, SPMSV, SPMMV, SPCV, SPVCV, SPPV, SPC6V, SPLSV, and CMV) were not detected in our survey. Although the primers used in this study were designed based on the highly conserved regions of each target virus, we could not excluded the possibility that the primers failed to amplify the targets efficiently because positive controls for RT-PCR detection of these viruses were not available in this study. Therefore, we note that our survey results did not conclude that the examined samples were not infected with the undetected viruses.
Six viruses (SPFMV, SPVC, SPVG, SPV2, SPLV, and SPMSV) are members of the genus Potyvirus, in the family Potyviridae (Adams et al., 2011; Clark et al., 2012). Among these potyviruses, five viruses but SPMSV had a high incidence (96%, 148/154 samples collected) in Korea, and SPFMV was especially prevalent. SPFMV has been divided into four representative strains: Russet Crack (RC), Ordinary (O), East Africa (EA), and Common (C) (Abad et al., 1992; Kreuze et al., 2000; Kwak et al., 2007). However, because SPFMV strain C has a relatively low homology with the other SPFMV strains, it was reclassified as a new species, SPVC, by ICTV in 2010. A previous study showed that Korean SPFMV isolates are similar to the RC and O strains, but did not detect any EA-like isolates (Kwak et al., 2007). The EA strain has been detected exclusively in East Africa as a distinct group (Kreuze et al., 2000; Mukasa et al., 2003). However, it was also found in Peru (Untiveros et al., 2008), Vietnam (Ha et al., 2008), Easter Island (Rännäli et al., 2009) and recently in China (Qin et al., 2013). The other potyviruses infecting sweet potato (SPVG, SPLV, and SPV2) have been poorly studied; however, their complete genome sequences were recently reported and compared to SPFMV (Li et al., 2012a; Rodriguez Pardina et al., 2012; Wang et al., 2013). Similar to SPFMV, SPLV was reported to cause synergistic disease when coinfected with SPCSV (Untiveros et al., 2007). SPV2 was first isolated from Taiwan and Nigeria (Rossel et al., 1988) and has distinct biological, serological, and genetic characteristics from SPFMV (Ateka et al., 2007; Li et al., 2012).
Begomoviruses infecting sweet potato are widely distributed worldwide. Twelve sweet potato viruses belonging to the genus Begomovirus in the family Geminiviridae have been reported (Clark et al., 2012), of which seven have been accepted as virus species by the ICTV (Table 1). Incidence of SPLCV in Korea increased markedly from 5% in 2003 to 47% in 2012 (Kwak et al., 2006). This study provides the first record of SPSMV-1 (of the genus Mastrevirus in the family Geminiviridae) in Korea; we detected it frequently (~67%) in most areas, with the exception of Sancheong, in Gyeongsangnam-do. Several SPSMV-1 isolates have been identified from Peru, Tanzania, and China, showing 100% homology in the CP-MP region (Clark et al., 2012; Kreuze et al., 2009). SPCFV (a member of the genus Carlavirus in the family Betaflexiviridae) was first detected in sweet potato showing fine chlorotic spots in Peru. It has since been detected in several countries of South America, Asia, and South Africa and the complete genome sequence of an isolate from Uganda has been characterized (Aritua et al., 2007). Untiveros et al. showed that SPCFV could cause a synergistic disease when sweet potatoes were coinfected with SPCSV. The present study provides the first report of SPCFV in Korea, and we found that it occurred in ~31% of samples collected in most areas except Chungcheongnam-do (Table 4).
Most of the samples were in mixed infections with at least two of the eight viruses (Table 5). The single and mixed infection rates were 3.9% and 95.5%, respectively. The most prevalent infection type (32.5%) was quintuple infection with SPFMV, SPVC, and other viruses. We did not detect single infection of SPVC, SPVG, SPV2, SPLV, or SPCFV (Table 5).
In comparison, among 179 sweet potato samples surveyed in 2003, 73% were infected with SPFMV, SPGV, SPLV, and SPLCV, whereas 27% remained unidentified despite showing virus-like symptoms. However, of the samples collected in 2012, all but one was infected with one or several of the eight aforementioned viruses. In particular, we confirmed that six out of seven samples, which were thought to be virus-free, were actually infected with new viruses (data not shown). Although we detected some differences in virus type and in the degree of multiple infection among sample locations, overall infection rates were consistently high. Consequently, it is essential to test for at least these eight viruses in order to produce virus-free sweet potato seedlings in Korea.
Recently, viral diseases have caused severe damage to sweet potato crops. Thus, detection and identification of viral pathogens is vital for producing virus-free sweet potato seedlings and preventing the spread of viruses. Besides, because most of sweet potatoes were infected with a different combination of viruses, multiplex RT-PCR methods for simultaneous detection of several viruses in one reaction have been developed (Li et al., 2012b; Opiyo et al., 2010; Rukarwa et al., 2010). For rapid, simple, and simultaneous detection of sweet potato-infecting viruses in Korea, we developed two multiplex RT-PCR assays for eight major viruses, including SPLCV, SPFMV, SPVG, and SPLV in one set, and SPCFV, SPVC, SPV2, and SPSMV-1 in another set. We performed these multiplex RT-PCR methods via two-step or one-step reactions and applied them to a variety of sweet potato samples. In case of primer set 1 designed for detecting SPLCV, SPFMV, SPVG and SPLV, two-step multiplex RT-PCR performed well but unfortunately one-step multiplex RT-PCR did not work. Also, in case of primer set 2 designed for detecting SPCFV, SPVC, SPV2 and SPSMV-1, both one-step and two-step multiplex RT-PCRs were successfully. To evaluate the specificity of the assays, we confirmed the amplified products by sequencing. The multiplex RT-PCR assays developed in this study will provide a rapid, convenient, and cost-efficient method for the detection of multiple viruses in sweet potato.
Acknowledgments
This work was supported by a grant from the Agenda Program (PJ006519) of the Rural Development Administration, the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries (iPET: No. 110034-05-3-HD110), and the Ministry of Food, Agriculture, Forestry, and Fisheries, Republic of Korea.
References
- Abad JA, Conkling MA, Moyer JW. Comparison of the capsid protein cistron from serologically distinct strains of Sweet potato feathery mottle virus (SPFMV) Arch Virol. 1992;126:147–157. doi: 10.1007/BF01309691. [DOI] [PubMed] [Google Scholar]
- Adams MJ, Zerbini FM, French R, Rabenstein F, Stenger DC, Valkonen JPT. Virus Taxonomy. In: King AMQ, Lefkowitz E, Adams MJ, Carstens EB, editors. 9th Report of the International Committee for Taxonomy of Viruses. Elsevier Academic Press; San Diego: 2011. pp. 1069–1089. [Google Scholar]
- Aritua V, Barg E, Gibson RW, Adipala E, Vetten HJ. Sequence analysis of the entire RNA genome of Sweet potato chlorotic fleck virus reveals that it belongs to a distinct carlavirus species. Arch Virol. 2007;152:813–818. doi: 10.1007/s00705-006-0891-z. [DOI] [PubMed] [Google Scholar]
- Ateka EM, Barg E, Njeru RW, Thompson G, Vetten HJ. Biological and molecular variability among geographically diverse isolate of Sweet potato virus 2. Arch Virol. 2007;152:479–488. doi: 10.1007/s00705-006-0879-8. [DOI] [PubMed] [Google Scholar]
- Brunt AA, Crabtree K, Dallwitz MJ, Gibbs AJ, Watson L. Viruses of Plants Descriptions and Lists from the VIDE Database. CAB International; Wallingford, UK: 1996. [Google Scholar]
- Clark CA, Davis JA, Abad JA, Cuellar WJ, Fuentes S, Kreuze JF, Gibson RW, Mukasa SB, Tugume AK, Tairo F, Valkonen JPT. Sweetpotato viruses: 15 years of progress on understanding and managing complex diseases. Plant Dis. 2012;96:168–185. doi: 10.1094/PDIS-07-11-0550. [DOI] [PubMed] [Google Scholar]
- Gibson RW, Mpembe I, Alicai T, Carey EE, Mwanga ROM, Seal SE, Vetten HJ. Symptoms, aetiology and serological analysis of sweet potato virus disease in Uganda. Plant Pathol. 1998;47:95–102. [Google Scholar]
- Gutierrez DL, Fuentes S, Salazar LF. Sweet potato virus disease (SPVD): Distribution, incidence, and effect on sweet potato yield in Peru. Plant Dis. 2003;87:297–302. doi: 10.1094/PDIS.2003.87.3.297. [DOI] [PubMed] [Google Scholar]
- Ha C, Revill P, Harding RM, Vu M, Dale JL. Identification and sequence analysis of potyviruses infecting crops in Vietnam. Arch Virol. 2008;153:45–60. doi: 10.1007/s00705-007-1067-1. [DOI] [PubMed] [Google Scholar]
- Hahn SK. Effect of virus (SPDV) on growth and yield of sweet potato. Exp Agric. 1979;15:253–256. [Google Scholar]
- Karyeija RF, Kreuze JF, Gibson RW, Valkonen JPT. Synergistic interactions of a potyvirus and a phloem-limited crinivirus in sweetpotato cultivars. Virology. 2000;269:26–36. doi: 10.1006/viro.1999.0169. [DOI] [PubMed] [Google Scholar]
- Kreuze JF, Karyeija RF, Gibson RW, Valkonen JPT. Comparisons of coat protein gene sequences show that East African isolates of Sweet potato feathery mottle virus form a genetically distinct group. Arch Virol. 2000;145:567–574. doi: 10.1007/s007050050047. [DOI] [PubMed] [Google Scholar]
- Kreuze JF, Perez A, Untiveros M, Quispe D, Fuentes S, Barker I, Simon R. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: A generic method for discovery and sequencing of viruses. Virology. 2009;388:1–7. doi: 10.1016/j.virol.2009.03.024. [DOI] [PubMed] [Google Scholar]
- Kwak HR, Kim MK, Jung MN, Lee SH, Park JW, Kim KH, Choi HS. Virus diseases incidences of sweet potato in Korea. Plant Pathol J. 2006;22:239–247. [Google Scholar]
- Kwak HR, Kim MK, Jung MN, Lee SH, Park JW, Kim KH, Ko SJ, Choi HS. Genetic diversity of Sweet potato feathery mottle virus from sweet potatoes in Korea. Plant Pathol J. 2007;23:13–21. [Google Scholar]
- Kwak HR, Kim MK, Nam M, Kim JS, Kim KH, Cha B, Choi HS. Genetic compositions of Broad bean wilt virus 2 infecting red pepper in Korea. Plant Pathol J. 2013;29:274–284. doi: 10.5423/PPJ.OA.12.2012.0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Xu D, Abad J, Li R. Phyogenetic relationships of closely related potyviruses infecting sweet potato determined by genomic characterization of Sweet potato virus G and Sweet potato virus 2. Virus Genes. 2012a;45:118–125. doi: 10.1007/s11262-012-0749-2. [DOI] [PubMed] [Google Scholar]
- Li F, Zuo R, Abad J, Xu D, Bao G, Li R. Simultaneous detection and differentiation of four closely related sweet potato potyviruses by a multiplex one-step RT-PCR. Journal of Virological Methods. 2012b;186:161–166. doi: 10.1016/j.jviromet.2012.07.021. [DOI] [PubMed] [Google Scholar]
- Mukasa SB, Rubaihayo PR, Valkonen JPT. Sequence variability within the 3′-proximal part of the Sweet potato mild mottle virus genome. Arch Virol. 2003;148:487–496. doi: 10.1007/s00705-002-0930-3. [DOI] [PubMed] [Google Scholar]
- Mukasa SB, Rubaihayo PR, Valkonen JPT. Interactions between a crinivirus, an ipomovirus and a potyvirus in coinfected sweetpotato plants. Plant Pathol. 2006;55:458–467. [Google Scholar]
- Opiyo S, Ateka E, Owuor P, Manguro L, Miano D. Development of a multiplex PCR technique for simultaneous detection of Sweet potato feathery mottle virus and Sweet potato chlorotic stunt virus. Journal of Plant Pathology. 2010;92:363–366. [Google Scholar]
- Qin Y, Zhang Z, Qiao Q, Zhang D, Tian Y, Wang Y. Molecular variability of Sweet potato chlorotic stunt virus (SPCSV) and five potyviruses infecting sweet potato in China. Arch Virol. 2013;158:491–495. doi: 10.1007/s00705-012-1503-8. [DOI] [PubMed] [Google Scholar]
- Rännäli M, Czekaj V, Jones RAC, Fletcher JD, Davis RI, Mu L, Valkonen JPT. Molecular characterization of Sweet potato feathery mottle virus (SPFMV) isolates from Easter Island, French Polynesia, New Zealand and southern Africa. Plant Dis. 2009;93:933–939. doi: 10.1094/PDIS-93-9-0933. [DOI] [PubMed] [Google Scholar]
- Rodriguez Pardina PE, Bejerman N, Luque AV, Di Feo L. Complete nucleotide sequence of an Argentinean isolate of Sweet potato virus G. Virus Genes. 2012;45:593–595. doi: 10.1007/s11262-012-0784-z. [DOI] [PubMed] [Google Scholar]
- Rossel HW, Thottappilly G. Complex virus diseases of sweet potato. Exploration, maintenance, and utilization of sweet potato genetic resources; Report of 1st Sweet Potato Planning Conference; 1987; Lima, Peru: International Potato Centre; 1988. [Google Scholar]
- Rukarwa RJ, Mashingaidze AB, Kyamanywa S, Mukasa SB. Detection and elimination of sweetpotato viruses. Afr Crop Sci J. 2010;18:223–233. [Google Scholar]
- Untiveros M, Fuentes S, Salazar LF. Synergistic interaction of Sweet potato chlorotic stunt virus (Crinivirus) with carla-, cucumo-, ipomo-, and potyviruses infecting sweet potato. Plant Dis. 2007;91:669–676. doi: 10.1094/PDIS-91-6-0669. [DOI] [PubMed] [Google Scholar]
- Untiveros M, Fuentes S, Kreuze J. Molecular variability of sweet potato feathery mottle virus and other potyviruses infecting sweet potato in Peru. Arch Virol. 2008;153:473–483. doi: 10.1007/s00705-007-0019-0. [DOI] [PubMed] [Google Scholar]
- Wang M, Abad J, Fuentes S, Li R. Complete genome sequence of the original Taiwanese isolate of Sweet potato latent virus and its relationship to other potyviruses infecting sweet potato. Arch Virol. 2013;158:2189–2192. doi: 10.1007/s00705-013-1705-8. [DOI] [PubMed] [Google Scholar]
- Yun WS, Lee YH, Kim KH. First report of Sweet potato latent virus and Sweet potato chlorotic stunt virus isolated from sweet potato in Korea. Plant Pathol J. 2002;18:126–129. [Google Scholar]