Abstract
Iris yellow spot virus (IYSV) is a plant pathogenic virus which has been reported to continuously occur in onion bulbs, allium field crops, seed crops, lisianthus, and irises. In South Korea, IYSV is a “controlled” virus that has not been reported, and inspection is performed when crops of the genus Iris are imported into South Korea. In this study, reverse-transcription polymerase chain reaction (RT-PCR) and nested PCR inspection methods, which can detect IYSV, from imported crops of the genus Iris at quarantine sites, were developed. In addition, a modified positive plasmid, which can be used as a positive control during inspection, was developed. This modified plasmid can facilitate a more accurate inspection by enabling the examination of a laboratory contamination in an inspection system. The inspection methods that were developed in this study are expected to contribute, through the prompt and accurate inspection of IYSV at quarantine sites to the plant quarantine in South Korea.
Keywords: Iris yellow spot virus, nested PCR, RT-PCR, quarantine
Iris yellow spot virus (IYSV) is a plant pathogenic virus, which is classified as Group V negative-sense ssRNA viruses, genus Tospovirus (Cortês et al., 1998), and is known to be spread by Frankliniella fusca and Thrips tabaci (Srinivasan et al., 2012). In the United Kingdom, the western region of the United States, southwest Germany, Réunion Island, and India, IYSV is reported to occur continuously in onion bulbs, allium field crops, seed crops, lisianthus, and irises, and, thus, it is thus considered an economically important plant disease (Bag et al., 2012; Krauthausen et al., 2012; Mumford et al., 2008; Ravi et al., 2006; Robène-Soustrade et al., 2006). In South Korea, IYSV has not been reported and is a “controlled” virus that is inspected when crops of the genus Iris are imported into the country (Animal, Plant, and Fisheries Quarantine and Inspection Agency, 2013). According to the Pest Information System of the Animal and Plant Quarantine Agency, for the crops of the genus Iris that have been imported via cargo, mail, and carry-on baggage, a total of 385 cases of import inspections have been performed during the last five years. In South Korea, an enzyme-linked immunosorbent assay (ELISA) has been used as the plant quarantine inspection method for a long time, but the low detection sensitivity, and false positive reactions have arisen (Caruso et al., 2003; Priou et al., 2006), so, ELISA has gradually been replaced by nucleic acid inspection methods (Kim et al., 2005; Lee et al., 2011a). The PCR diagnosis systems with the same conditions, which can be used at quarantine sites, have been developed (Lee et al., 2013a, b, and c; Lee and Shin, 2014). On the other hand, as IYSV is a quarantine virus that is not common in South Korea, it is not possible to use an infected sample as the positive control for inspection. Thus, there is a need to develop a positive control that enables contamination verification during the PCR diagnosis (Lee et al., 2013a and b). Therefore, in this study, the RT-PCR and nested PCR inspection methods, which can promptly and accurately diagnose IYSV at quarantine sites, were developed, and a system that is capable of prompt and accurate diagnosis of IYSV at quarantine sites was developed through the development of a genetically modified positive control, facilitating the contamination verification system of a PCR inspection.
For the experiment, IYSV-infected samples were purchased from Adgen (Scotland, UK), through the approval of prohibited goods import, and the cDNAs of the reference virus strains (Tomato spotted wilt virus and Tobacco rattle virus), which were to be used for the selection of species-specific primers, were purchased from a domestic enterprise (Bione, Korea).
To design the primers for diagnosis so that they are species-specific to IYSV, the gene sequences of the RNA segment S (NCBI accession number AF001387), which performs the coding of nonstructural protein and nucleoprotein genes among the linear RNA (total of 17.2 kb) of IYSV, and the reference virus strains such as Tomato spotted wilt virus (D00645), Impatiens necrotic spot virus (X66972), Peanut yellow spot virus (AF013994), Tomato chlorotic spot virus (AF282982), and Groundnut ringspot virus (AF513219) were obtained from the National Center for Biotechnology Information (NCBI). Using the DNAMAN software package version 6.0 (Lynnon Biosoft, Canada), the species-specific sequences were sought at 51–59°C as the annealing temperatures, which was the same condition in the method described in a previous paper (Lee and Shin, 2014). The manufactured primers for the specific detection of IYSV were six forward primers and six reverse primers (for total of 12 primers) (Supplementary Fig. S1); The synthesis of oligonucleotides was requested from a domestic company (Bioneer, Korea); and twenty-four sets that are capable of the PCR amplification of 135–807 bp were obteined through the combination of the manufactured primers (Supplementary Table 1).
The RNA extraction from the samples was performed by using an RNeasy®Plant mini-kit (Qiagen, Netherlands), and the genomic DNA extraction from the iris rhizome was performed by using a DNeasy®Plant mini-kit (Qiagen) and following its protocol. On the other hand, the RT-PCR, nested PCR, and electrophoresis processes were performed by using the same methods as those used in a previous paper (Lee and Shin, 2014). To select the optimal combinations, which were to be used for the PCR diagnosis of IYSV, from among the species-specific primer combinations, the specific reaction of IYSV (i.e., the target virus) and the non-specific reactions to the reference virus strains and the genomic DNA of the iris rhizome, were tested. For the selected PCR primer combinations, the sensitivity was tested by diluting tenfold the RNA of IYSV (down to 10−7) and, then selecting two optimal RT-PCR primer sets for IYSV. Nested PCR primer combinations were designed based on the location where the final selected RT-PCR primer sets acted, and the nested PCR primers were finally selected by the reaction of the template and product, which had been obtained by the 100-fold dilution of the RT-PCR product. In addition, to examine the annealing temperature reaction range of the two, final selected RT-PCR primer sets, RT-PCR was performed at the annealing temperatures of 51, 53, 55, 57, and 59°C.
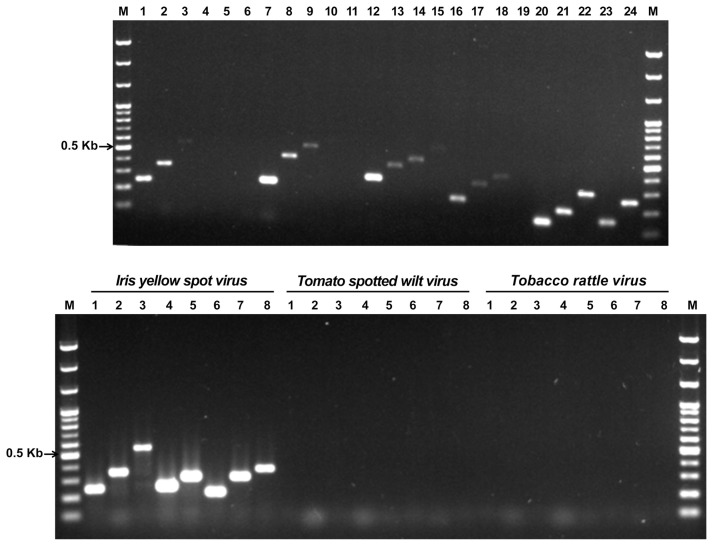
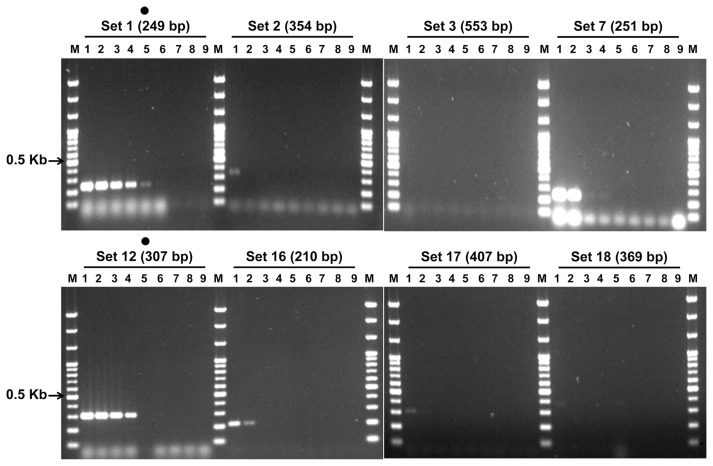
In the first selection of the primer sets for IYSV diagnosis, among the twenty-four sets of combinations, sets 1, 2, 3, 7, 12, 16, 17, and 18 formed specific bands and were selected (Fig. 1A). These eight PCR primer sets, did not show any reaction to either reference virus of Tomato spotted wilt virus or Tobacco rattle virus, and they also did not show a specific reaction to the genomic DNA of the iris rhizome. Thus, the non-specific reaction was not observed (Fig. 1B and Supplementary Fig. S2). The sensitivity experiment of these eight selected PCR primer sets indicated that sets 1 and 12 had the highest sensitivity at 10−4 and 10−3, respectively, and that the remaining sets had a detection sensitivity of less than 10−2. Thus, sets 1 and 12 were finally selected (Fig. 2). For the nested PCR primer sets of sets 1 and 12, which had been selected as the RT-PCR primer sets, one and two combinations were designed (data not shown). The nested PCR experiment indicated that the bands were formed in all the three lanes. Between the two nested PCR primer combinations of RT-PCR primer set 12, a final selection was made based on the length. The final selected RT-PCR primer combinations 1 and 12 can amplify 249 and 307 bp, respectively, and the nested PCR products of sets 1 and 12 can amplify 146 and 210 bp, respectively (Table 1). On the other hand, for all the RT-PCR primer sets, specific bands were observed at the 51–59°C, the annealing temperatures (Supplementary Fig. S3).
Fig. 1.
First and second selections of primer sets for the IYSV detection. Panel A. First selection of specific primer sets. Lane M, 100 bp step DNA Ladder maker (Genepia, Korea); lanes 1–24, IYSV-specific primer sets 1–24; dot, selected primer set. Panel B. Second selection of non-specific primer sets. lane number, primer set number; TSWV, Tomato spotted wilt virus; TRV, Tobacco rattle virus.
Fig. 2.
Ten-fold diluted sensitivity test of Lane M, 100 bp step DNA Ladder maker (Genepia, Korea); lane 1, undiluted RNA temlpate; lane 2, 10−1; lane 3, 10−2; lane 4, 10−3; lane 5, 10−4; lane 6, 10−5; lane 7, 10−6; lane 8, 10−7; lane 9, negative control; dot, selected primer set.
Table 1.
Selected primer sets for RT-PCR and nested PCR of IYSV
| Primer | Sequence (5′→3′) | Length (nt) | Product (nt) | ||
|---|---|---|---|---|---|
|
| |||||
| Set | PCR | Name | |||
| 1 | RT | IYS_N10 | TCTATCTTTCTTGGAGGGAG | 20 | 249 |
| IYS_C60 | TCAACTTACGAGCAACTCTG | 20 | |||
| Nested | IYS_N20 | ATCAATGAAGCAGCACCC | 18 | 146 | |
| IYS_C60 | TCAACTTACGAGCAACTCTG | 20 | |||
| 12 | RT | IYS_N30 | GCTCGTAAGTTGAGAATCTGC | 21 | 307 |
| IYS_C40 | TGGACATTCAGGAGGTTG | 18 | |||
| Nested | IYS_N40 | TGCCATCAAAGTGAGAGG | 18 | 210 | |
| IYS_C40 | TGGACATTCAGGAGGTTG | 18 | |||
Lastly, to develop a PCR diagnosis system that could be used at quarantine sites, a genetically modified positive control for IYSV that can detect contamination was developed. The 807 bp PCR product was amplified by using IYSV-N10 and C10 primer, which included all of the sections where the selected primer sets had acted. Cloning was performed by using a pGEM®-T easy vector (Promega, USA) as the insert DNA (Lee et al., 2013a). A total of five clones were taken, and pure isolation was done. Then, sequencing was conducted, and one clone, which corresponded to the binding sites of the primers developed in this study, was selected. For the selected clone, six nucleotides were inserted into the amplification part of the nested primer set by using a site-directed mutagenesis kit (Enzynomics, Korea) (Ramsdell, 1995). These six nucleotides are the sequences with which the restriction enzyme can react. The EZchange™ site-directed mutagenesis kit uses a three-stage reaction. In this reaction, a new mutation-linear plasmid is amplified by the PCR method, which uses plasmid DNA, as the template, and the primer with a mutation. Then, the original plasmid is removed; and transformation is performed by simultaneously conducting an end kination and ligation. In this study, the restriction enzyme Xho I (CTCGAG) was inserted. To check the insertion of the Xho I site in the plasmid, a nested PCR was performed three times, and the reaction of the restriction enzyme Xho I was examined. The results indicated that two bands were formed in all the cases, confirming the insertion of the six sequences (Supplementary Fig. S4).
According to the detailed execution method for a precise laboratory inspection, which is regulated by the Animal and Plant Quarantine Agency (subparagraph 107), IYSV is inspected when the genus Iris (Iris species) is imported. With the recent implementation of the import and export liberalization policy, the importing of irises and other crops is expected to increase. Also, due to the global report of the occurrence of IYSV in onion bulbs, allium field crops, seed crops, and Eustoma grandiflorum (Bag et al., 2012; Krauthausen et al., 2012; Mumford et al., 2008; Ravi et al., 2006; Robène-Soustrade et al., 2006), prompt quarantine inspection of crops is required so that any crops that could be infected by IYSV can be quarantined. In the case of IYSV and based on the ELISA inspection, there have thus far been no quarantine cases of disposal or return, but, according to the Disease and Insect Pest Information System of the Animal and Plant Quarantine Agency, plant pathogens, such as Botrytis cinerea, Fusarium solani, Penicillium expansum, Aspergillus niger, Penicillium cyclopium, Fusarium moniliforme, Fusarium oxysporum, Rhizopus stolonifer, Alternaria alternata, Iris mild mosaic virus, and Penicillium hirsutum were detected in the rhizomes and seedlings of irises, the seeds of blue-eyed grass, and the seedlings of blue flag. On the other hand, the PCR diagnosis systems, which had been developed for the diagnosis of seed-transmitted quarantine viruses in imported seeds, have been gradually applied to frontline quarantine sites from 2007, and as a result, the detection record improved significantly from 78 cases in 2006 to 145 cases in 2007. Also, as a result, Cherry leaf roll virus, a seed-transmitted quarantine virus that had not been detected before 2006, was detected. In the case of Wheat streak mosaic virus, which is known to be transmitted from corn- and wheat-seeds, three cases (1,836 kg) were detected at frontline quarantine sites. The contaminated items were discarded or returned. These actions occurred for a year after the RT-PCR and nested PCR inspection methods were developed (Lee et al., 2013a). In addition, the RT-PCR and nested PCR diagnosis systems for the four species (Tomato black ring virus, Arabis mosaic virus, Cherry leaf roll virus and Grapevine fanleaf virus) that belong to Nepovirus were reported (Lee et al., 2013c), and according to the Pest Information System of the Animal and Plant Quarantine Agency, the detection records that are relevant to the four viruses showed 27 cases for the last three years (2010–2012). As shown above, virus detection has continuously increased at the sites due to the development of the single-condition RT-PCR and nested PCR quarantine diagnosis system, and, thus, quarantine safety during the import and export distribution has been secured in South Korea. Therefore, it is expected that the PCR diagnosis system for IYSV developed in this study will make a significant contribution to the frontline quarantine sites.
Acknowledgments
This research was supported by the 2011 R&D fund of the Animal and Plant Quarantine Agency.
References
- Animal, Plant and Fisheries Quarantine and Inspection Agency. List of plant quarantine viruses in Korea in newly revised in 2013. Res Plant Dis. 2013;19:65–75. [Google Scholar]
- Bag S, Singh J, Davis RM, Chounet W, Pappu HR. Iris yellow spot virus in onion in Nevada and northern California. Plant Dis. 2009;93:674. doi: 10.1094/PDIS-93-6-0674C. [DOI] [PubMed] [Google Scholar]
- Caruso P, Bertolini E, Cambra M, López MM. A new and sensitive co-operational polymerase chain reaction for rapid detection of Ralstonia solanacearum in water. J. Microbiol Methods. 2003;55:257–272. doi: 10.1016/s0167-7012(03)00161-1. [DOI] [PubMed] [Google Scholar]
- Cortês I, Livieratos IC, Derks A, Peters D, Kormelink R. Molecular and serological characterization of Iris yellow spot virus, a new and distinct Tospovirus species. Virology. 1998;88:1276–1282. doi: 10.1094/PHYTO.1998.88.12.1276. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Park S, Yie SW, Kim KH. RT-PCR detection of dsRNA Mycoviruses infecting Pleurotus ostreatus and Agaricus blazei Murrill. Plant Pathol. J. 2005;21:343–348. [Google Scholar]
- Krauthausen HJ, Leinhos GME, Müller J, Radtke PC, Jehle JA. Identification and incidence of Iris yellow spot virus in Allium field crops in Southwest Germany. Eur. J. Plant Pathol. 2012;134:345–356. [Google Scholar]
- Lee JS, Cho WK, Lee SH, Choi HS, Kim KH. Development of RT-PCR based method for detecting five non-reported quarantine plant viruses infecting the family Cucurbitaceae or Solanaceae. Plant Pathol. J. 2011a;27:93–97. [Google Scholar]
- Lee S, Kang EH, Chu YM, Shin YG, Ahn TY. Development of PCR diagnosis system for plant quarantine seed-borne Wheat streak mosaic virus. Kor. J Microbiol. 2013a;49:112–117. [Google Scholar]
- Lee S, Kang EH, Heo NY, Kim SM, Kim YJ, Shin YG. Detection of Carnation necrotic fleck virus and Carnation ringspot virus using RT-PCR. Res. Plant Dis. 2013b;19:36–44. [Google Scholar]
- Lee S, Kang EH, Shin YG, Lee SH. Development of RT-PCR and nested PCR for detecting four quarantine plant viruses belonging to Nepovirus. Res. Plant Dis. 2013c;19:220–225. [Google Scholar]
- Lee S, Shin YG. Development and practical use of RT-PCR for seed-transmitted Prune dwarf virus in quarantine. Plant Pathol J. 2014 doi: 10.5423/PPJ.NT.10.2013.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Cho WK, Choi HS, Kim KH. RT-PCR detection of five quarantine plant RNA viruses belonging to Poty and Tospoviruses. Plant Pathol. J. 2011b;27:291–296. [Google Scholar]
- Mumford RA, Glover R, Daly M, Nixon T, Harju V, Skelton A. Iris yellow spot virus (IYSV) infecting Lisianthus (Eustoma grandiflorum) in the UK: first finding and detection by real-time PCR. New Dis. Rep. 2008;16:6. [Google Scholar]
- Ramsdell DC, Gillett JM, Bird GW. Susceptibility of American grapevine scion cultivars and French hybrid rootstock and scion cultivars to infection by peach rosette mosaic nepovirus. Plant Dis. 1995;79:154–157. [Google Scholar]
- Ravi KS, Kitkaru AS, Winter S. Iris yellow spot virus in onions: a new tospovirus record from India. Plant Pathol. 2006;55:288. [Google Scholar]
- Robène-Soustrade I, Laurent P, Gagnevin L, Jouen E, Pruvost O. Specific detection of Xanthomonas axonopodis pv. dieffenbachiae in Anthurium (Anthurium andreanum) tissues by nested PCR. Appl. Environ. Microbiol. 2006;72:1072–1078. doi: 10.1128/AEM.72.2.1072-1078.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan B, Sundaraj S, Pappu HR, Diffie S, Riley DG, Gitaitis RD. Transmission of Iris yellow spot virus by Frankliniella fusca and Thrips tabaci (Thysanoptera: Thripidae) J. Econ. Entomol. 2012;105:40–47. doi: 10.1603/ec11094. [DOI] [PubMed] [Google Scholar]
- Priou S, Gutarra L, Aley P. An improved enrichment broth for the sensitive detection of Ralstonia solanacearum (biovars 1 and 2A) in soil using DAS-ELISA. Plant Pathol. J. 2006;55:36–45. [Google Scholar]