Abstract
West Nile virus (WNV) is an increasing public health concern in Europe with numerous human cases. A total of 23,029 female mosquitoes were tested for a variety of mosquito-borne flaviviruses and orthobunyaviruses supposedly endemic in Southern Transdanubia, Hungary, in the frames of a large-scale surveillance between 2011 and 2013. WNV nucleic acid was detected in a single pool containing Uranotaenia unguiculata mosquitoes. Sequence- and phylogenetic analyses for two different regions (NS5 and E) of the viral genome showed that the novel Hungarian WNV strain was different from other previously described WNV lineages. These findings may indicate the presence of a putative, novel lineage of WNV in Europe. Our results also indicate that U. unguiculata mosquito may become relevant species as a potential vector for West Nile virus in Europe.
Keywords: West Nile virus, Lineage, Hungary, Uranotaenia unguiculata
Mosquito-borne viruses (MBV) represent a considerable healthcare problem worldwide, causing numerous infections both in humans and animals. MBVs belong to a variety of virus families; most prominent members of MBVs are those within the Flaviviridae (e.g. West Nile virus, Dengue virus, Usutu virus), Bunyaviridae (e.g. Tahyna virus), and Togaviridae (e.g. Sindbis virus). In many parts of Europe, the greatest medical and veterinary burden is attributed to flaviviruses, and particularly West Nile virus (WNV) [5]. WNV has been subdivided into at least eight distinct lineages (WNV-1 to WNV-8): WNV-1 and WNV-2 strains have been identified most frequently in human and animal diseases, while strains within WNV-3 to WNV-8 have been detected sporadically during MBV surveillance in parts of Europe, Asia, and Africa. WNV-3 is represented by strain Rabensburg 97–103 isolated from Culex pipiens in Czech Republic, WNV-4 is represented by strain LEIVKrnd88-190 isolated from Dermacentor marginatus ticks in the Caucasus, whereas WNV-5 (strain 804994) was identified in India. Representative strains of other putative lineages include the strain KUN MP502-66 from Malaysia (WNV-6), Dak Ar D 5443 from Senegal (WNV-7) and HU2925/06 from Spain (WNV-8) [3]. In the past decade, WNV-2 has emerged in Europe and caused outbreaks in Hungary, Austria, Greece, Italy, Romania, Serbia and Croatia, and now has become endemic in many regions across Europe. Until now, only WNV-1 and WNV-2 have been described in Hungary, causing infections and excess mortality in birds, horses and humans. WNV-2, since its first detection in 2004, has caused sporadic infections until 2008, when, after its explosive spread, 22 human and 37 animal cases were diagnosed with neuroinvasive disease in the country [1]. In this study we report the description of a novel putative lineage of WNV (WNV-9) which was identified during a 3 year surveillance of MBVs conducted in Southwest Hungary.
To identify MBVs in Southwest Hungary we collected 23,029 female mosquitoes, between 2011 and 2013. Mosquitoes were collected with the human-landing collection method and updraft box traps equipped with white light attractant [4]. A total of 25 mosquito species were identified according to the taxonomic keys of Becker et al. [2]. Specimens were grouped by species, collection site and date, and finally pooled with a maximum size of 50 individuals. With this approach a total of 586 pools were generated. Mosquitoes were homogenized in 600 μl 1 × PBS with a 3 mm metal bead using Qiagen® TissueLyser. After centrifugation at 21,500 × g for 10 min, RNA was extracted from 200 μl of supernatants using DiaExtract Total RNA Isolation Kit (DIAGON Ltd., Hungary) following the manufacturer’s recommendations. Samples were tested by nested reverse transcription-PCR assays. These included a screening panel using both universal and virus specific primers targeting a variety of mosquito-borne flaviviruses and orthobunyaviruses supposed to be endemic in the study area [6–8].
In 31.5 % (185 pools) of the total 586 tested pools, a strong 250 nucleotide-long specific signal was identified by nested RT-PCR with universal flavivirus primers. A representative positive pool from each tested mosquito genera, i.e. Aedes sp., Anopheles sp. and Uranotaenia sp. was sequenced (BigDye Terminator v1.1 Cycle Sequencing Kit, ABI Prism 310 DNA Sequencer—Applied Biosystems). Insect-specific flaviviruses were identified in all of the tested mosquito genera except Uranotaenia sp. In a single pool of Uranotaenia unguiculata females, sequencing of the 250 bp amplicon revealed WNV. This pool was collected in a marshland near the city of Pécs (latitude: latitude 46° 3′23.48″N; longitude 18° 9′23.97″E) containing 32 individuals of U. unguiculata. These mosquitoes were collected in August 2011 using updraft box traps equipped with white light attractant. Thus far, West Nile-like virus (so-called “Volgograd virus”) was detected only in U. unguiculata mosquitoes in Russia (A.E. Platonov, unpub. data, GenBank accession number: FJ159131). Because the short fragment of NS5 gene was not suitable for classification of the Hungarian U. unguiculata WNV strain, a longer region of the same gene (875 bp) and the envelope glycoprotein (E) gene (865 bp) were sequenced (GenBank accession numbers: KJ592829 and KJ592830). In the phylogenetic tree, the Hungarian WNV sequence (WNV/189Uu/2011) clustered with other WNV strains, but unambiguously formed a separate monophyletic branch (Fig. 1). The NS5 gene-based analyses showed that the Hungarian novel WNV strain shared the greatest similarity with representatives of WNV-4 (up to 82 %, phylogenetic distance: 0.413) and the putative lineage 8 strain (83 %, phylogenetic distance: 0.360), originating from Russia and Spain, respectively. Similarly, the E gene-based analyses indicated that the novel Hungarian WNV strain was different from the WNV strain that derived from the same mosquito species (U. unguiculata) in Volgograd, Russia, which rather clustered together with WNV-4.
Fig. 1.
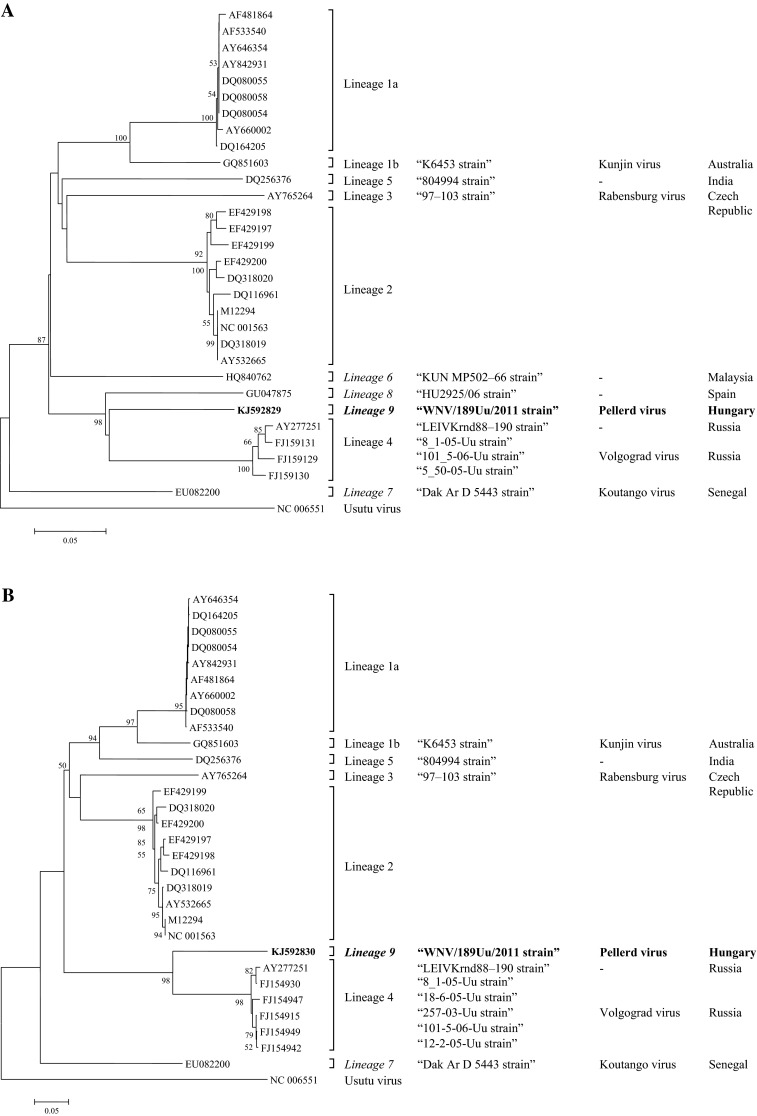
Phylogenetic trees were constructed with MEGA v5.0 software using neighbor-joining algorithm with Maximum Composite Likelihood parameter model based on nucleic acid sequences of a 875 bp long region of the non-structural gene (NS5) (a) and a 865 bp sequence of the envelope glycoprotein gene (E) (b). The Hungarian WNV strain (WNV/189Uu/2011, KJ592829 and KJ592830) detected in this study was marked in bold face. Number of bootstrap replications was 1,000, and the analysis was performed rooted using Usutu virus as outgroup
In order to isolate WNV/189/Uu/2011 strain, 50 μl of mosquito supernatant was filtered through 0.22 μm sterile filter, then preheated inoculum (37 °C) was injected intracranially into 5 days old CD-1® suckling mice. Mice remained asymptomatic even after 10 days postinfection. Euthanized animals were dissected, and then infectivity was verified by RT-PCR. Viral nucleic acid was not detected in the supernatant of homogenized mouse brain and lung tissues. Additionally, 100 μl of filtered (0.22 μm) mosquito pool supernatant was inoculated onto subconfluent monolayer culture of Vero E6 cells (CRL-1586), then cells were incubated for 10 days at 37 °C humidified air supplied with 5 % carbon dioxide. After three blind passages neither cytopathic effect nor positive RT-PCR was observed. All kinds of manipulation with potentially infectious materials were carried out in a BSL-3 facility of the University of Pécs.
In the present study, 23,029 female mosquitoes were screened for a variety of MBVs using both virus-specific and universal primers. In the sample set a WNV, named “Pellerd virus” (WNV/189Uu/2011), was detected form a single U. unguiculata mosquito pool. Sequence- and phylogenetic analyses for two different regions of the viral genome showed that the novel Hungarian WNV strain was different from other previously described WNV lineages, and formed a separate monophyletic branch. Although, the sequence was related to WNV-4 and putative WNV-8, we assumed that strain WNV/189/Uu/2011 could be a new putative lineage of WNV (68–78 % homology, phylogenetic distance: 0.265–0.584). Even though, U. unguiculata is a frequent species throughout the Mediterranean region, this mosquito was also described in several European countries including Hungary [2].
In conclusion, we described a putative new lineage, lineage 9, of WNV from an uncommon mosquito species in Hungary. Our results also indicate that U. unguiculata mosquito may become relevant species as a potential vector for West Nile virus in Europe.
Acknowledgments
Research activity of Gábor Kemenesi and Ferenc Jakab was supported by the TÁMOP 4.2.4. A/2-11-1-2012 0001—National Excellence Program Elaborating and operating an inland student and researcher personal support system. The project was subsidized by the European Union and co-financed by the European Social Fund. The study was also partially supported by the European Union FP7-261504 EDENext Project (www.edenext.eu) and is catalogued by the EDENext Steering Committee as EDENext227. The contents of this paper are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission. This study was supported by the Hungarian Scientific Research Fund (OTKA; PD77977) project. Krisztián Bányai was supported by the “Momentum program”.
References
- 1.Bakonyi T, Ferenczi E, Erdélyi K, Kutasi O, Csörgő T, Seidel B, Weissenböck H, Bruggeri K, Bánc E, Nowotny N. Explosive spread of a neuroinvasive lineage 2 West Nile virus in Central Europe, 2008/2009. Vet Microbiol. 2013;165:61–70. doi: 10.1016/j.vetmic.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Becker N, Petric D, Zgomba M, Boase C, Madon M, Dahl C, Kaiser A. Mosquitoes and their control. New York: Kluwer Academic/Plenum Publisher; 2003. [Google Scholar]
- 3.Donadieu E, Bahuon C, Lowenski S, Zientara S, Coulpier M, Lecollinet S. Differential virulence and pathogenesis of West Nile viruses. Viruses. 2013;11:2856–2880. doi: 10.3390/v5112856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall-Mendelin S, Ritchie SA, Johansen CA, Zborowski P, Cortise G, Dandridgef S, Halla RA, Hurk AF. Exploiting mosquito sugar feeding to detect mosquito-borne pathogens. PNAS. 2010;107:11255–11259. doi: 10.1073/pnas.1002040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hubálek Z. Mosquito-borne viruses in Europe. Parasitol Res. 2008;103:29–43. doi: 10.1007/s00436-008-1064-7. [DOI] [PubMed] [Google Scholar]
- 6.Hubálek Z, Rudolf I, Bakonyi T, Kazdová K, Halouzka J, Sebesta O, Sikutová S, Juricová Z, Nowotny N. Mosquito (Diptera: Culicidae) surveillance for arboviruses in an area endemic for West Nile (Lineage Rabensburg) and Tahyna viruses in Central Europe. J Med Entomol. 2010;47:466–472. doi: 10.1603/ME09219. [DOI] [PubMed] [Google Scholar]
- 7.Scaramozzino N, Crance JM, Jouan A, DeBriel DA, Stoll F, Garin D. Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-pcr assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J Clin Microbiol. 2001;39:1922–1927. doi: 10.1128/JCM.39.5.1922-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weissenböck H, Bakonyi T, Chvala S, Nowotny N. Experimental Usutu virus infection of suckling mice causes neuronal and glial cell apoptosis and demyelination. Acta Neuropathol. 2004;108:453–460. doi: 10.1007/s00401-004-0916-1. [DOI] [PubMed] [Google Scholar]


