Abstract
Canine parvovirus (CPV) is an enteric pathogen causing hemorrhagic enteritis in pups of 3–6 months of age and is mainly transmitted via feco-oral route. In the present study, a total of 85 animals rectal swabs suspected of CPV were tested using a PCR, nested PCR and a newly designed differential PCR. Using PCR 7 (8.23 %) animals were positive whereas 39 (45.88 %) were positive by using nested PCR and 40 (47.05 %) were positive for either one or more than one antigenic types of CPV using differential PCR. Using differential PCR it was found that CPV-2a and CPV-2b were the most prevailing antigenic types. Also it was found that dogs that were vaccinated too yielded positive CPV indicating a possible presence of additional CPV antigenic types. Thus, the primers used in differential PCR can be used in a single PCR reaction to detect various antigenic types of CPV.
Keywords: Antigenic characterization, Canine parvovirus, Differential PCR, Nested PCR
Introduction
Canine parvovirus (CPV) belongs to the genus Parvovirus under the family Parvoviridae. It has a linear single stranded (SS) negative sense DNA. It is an important cause of severe enteritis and systemic disease in dogs throughout the world. The virus replicates autonomously and is genetically related to feline panleukopenia virus (FPLV), mink enteritis virus (MEV) and blue fox parvovirus (BFPV) [22].
CPV was first identified in 1978 and referred to as CPV-2 after distinguishing it from CPV-1 [4]. There are various antigenic variants of CPV 2 that have replaced the original CPV-2 and currently there are three main antigenic variants i.e. 2a, 2b and 2c circulating in the dog population worldwide [7]. Regarding CPV-2c, it was previously designated as CPVGlu-426 mutant that emerged in Italy initially [3] but now has been reported from many countries. CPV-2 causes hemorrhagic gastroenteritis and myocarditis in dogs and it spreads rapidly in the domestic as well as in the wild population of canines. The replication of virus takes place in the villus epithelium of the small intestine that are rapidly dividing and the virus is shed in large quantity in the feces particularly 4–7 days post infection [12]. Infected feces serve as a source of infection. There are a number of methods that are used to diagnose CPV viz., virus isolation using cell culture, haemagglutination (HA), haemagglutination inhibition (HI), electron microscopy (EM), indirect fluorescent test (IFT), enzyme linked immunosorbent assay (ELISA) etc. [20]. Besides these, polymerase chain reaction (PCR) and nested-PCR (NPCR) could be used for its detection as these too have reported high sensitivity and specificity [15, 31].
The genome of CPV is about 5.3 Kb and has two open reading frames (ORF) 1 and 2. ORF1 encodes for two non-structural (NS) proteins; NS1 and NS2, that are translated by alternative splicing of the transcribed mRNA and ORF2 encodes two capsid proteins; VP1 and VP2 [35] having about 10 copies of VP1 and 60–70 copies of VP2 [10]. VP2 plays an important role in the determination of antigenicity and host range of CPV [25] and thus, mutations affecting VP2 are mainly responsible for the evolution of different antigenic variants [20]. So, the continuous emergence of newer strains of CPV is an ever growing concern among dog owners, breeders and veterinarians around the world. Conventional vaccines against CPV include an inactivated and a modified live virus vaccine but often there are reports of vaccination failure due to introduction of newer antigenic variants [34]. Thus, early detection along with the knowledge of genetic variations of VP2 could be of immense help in identifying emerging CPV strains so that this knowledge could be used for the development of vaccine. Keeping in view the above points this study was undertaken to develop a differential PCR that could be used in identification of the CPV antigenic types and also to study the prevalence of CPV.
Materials and methods
Sample collection
Rectal swabs (n = 85) in phosphate buffer saline (pH = 7.2) were collected from dogs exhibiting clinical signs of CPV viz., gastroenteritis, hemorrhagic enteritis etc. from the small animal veterinary clinics, Guru Angad Dev Veterinary and Animal Sciences University, Ludhiana, Punjab. The study was done from June, 2013 till January, 2014. Samples along with the history viz. breed, age, sex and vaccination status of the animal were collected.
DNA extraction
The swabs were squeezed and the liquid was boiled in a water bath for 10 min. The supernatant was collected by centrifugation at 9,390×g for 10 min and was used as a template DNA in the subsequent PCR reactions and was stored at −20 °C till further use.
Extraction of DNA from vaccine
The DNA was extracted from two commercially available vaccines viz., Nobivac DHPPi and Megavac-6 using DNA extraction kit (MoBio Laboratories Inc., Carlsbad, CA) following manufacturer’s instructions with slight modification.
PCR
The primers for PCR were as per Mizak and Rzezutka [19]. The PCR reaction was set up by adding 15 µl of the template DNA, 5.0 µl of 10 × PCR buffer (with 15 mM MgCl2), 1.0 µl of forward and reverse primer (25 pm/µl) each, 1.0 µl of dNTPs mix (10 mM each), 0.5 µl of MgCl2 (50 mM), 1 U Taq DNA polymerase and the reaction was made up to 50 µl using nuclease free water. The reaction was put in a thermocycler (Veriti®, Life Technologies, USA) with 35 cycles of denaturation at 94 °C for 60 s, annealing at 55 °C for 60 s, elongation at 72 °C for 150 s and a final elongation at 72 °C for 10 min.
Nested PCR
The primers used for NPCR were as per Mizak and Rzezutka [19] following the same conditions for NPCR as of PCR. NPCR reaction was set up by adding 5 µl of the PCR product (from above reaction), 2.5 µl of 10 × PCR buffer (with 15 mM MgCl2), 1.0 µl each of forward and reverse primer (25 pm/µl), 1.0 µl of dNTPs (10 mM each), 0.5 µl MgCl2 (50 mM), 1 U Taq DNA polymerase and the final volume was made up to 25 µl by adding nuclease free water. In both the PCR and nested PCR rectal swab from a healthy dog was used as a negative control and a DNA from a vaccine was used as a positive control.
Primers for differential PCR
The primer pairs for the differential PCR for the four antigenic types viz CPV-2, CPV-2a, CPV-2b and CPV-2c (Table 1) were designed using Primer3 software from the NCBI website [28].
Table 1.
Primers for the differential PCR
| S. no. | Antigenic Type | Primer | Sequence | Accession no. | Position in genome | Annealing temperature (°C) | Product size (bp) | |
|---|---|---|---|---|---|---|---|---|
| 1. | 2 | CPV-2GM F | 5′-CTGCTACTCAGCCACCAACT-3′ | EU659116.1 | Whole genome of CPV | 2,981–3,000 | 59 | 719 |
| CPV-2GM R | 5′-AGGTGTTTCTCCTGTTGTGGT-3′ | 3,699–3,679 | ||||||
| 2. | 2a | CPV-2aGM F | 5′-AGAGCATTGGGCTTACCACC-3′ | EU310373.2 | 3,427–3,446 | 60 | 379 | |
| CPV-2aGM R | 5′-ATCTTCCTGTATCTTGATGTGCT-3′ | 3,804–3,782 | ||||||
| 3. | 2b | CPV-2bGM F | 5′-TGTATTGCTACCAACAGATCCA-3′ | JQ743893.1 | VP2 gene of CPV | 1,284–1,305 | 59 | 178 |
| CPV-2bGM R | 5′TGGTGCATTTACATGAAGTCTTGG-3′ | 1,461–1,438 | ||||||
| 4. | 2c | CPV-2cGM F | 5′-GTGGTTCTGGGGGTGTGG-3′ | JF414822.1 | 98–115 | 60 | 470 | |
| CPV-2cGM R | 5′-AGCTGCTGGAGTAAATGGCA-3′ | 567–548 |
Differential PCR
For identifying various antigenic types’ individual PCR reaction was set for the identification of individual antigenic type. The PCR reaction was made by adding 5 µl of the template DNA, 2.5 µl of 10 × PCR buffer (with 15 mM MgCl2), 1.0 µl of forward and reverse primers (25 pm/µl) each, 1.0 µl of dNTPs mix (10 mM each), 0.5 µl of MgCl2 (50 mM), 1 U Taq DNA polymerase and the final volume 25 µl was made by adding nuclease free water. For the differential PCRs the conditions were unaltered and were same as of PCR except for the variation in the annealing temperature (Table 1).
Visualization of PCR, NPCR and differential PCR products
PCR, NPCR and differential PCR products (10 µl) were run using 1 % agarose at 5 volts/cm with Gene Ruler ladder plus 100 bp (New England Biolabs, USA). The gel was visualized and photographed using Gel documentation system (AlphaImager, USA).
Statistical analysis
The results of NPCR and differential PCR were compared using Kappa statistics. The results were interpreted as per the set parameters of the test [6].
Results
In the present study, out of 85 samples from dogs exhibiting clinical signs of CPV 7 (8.23 %) were positive by PCR yielding a product size of 1,198 bp, 39 (45.88 %) samples were positive with nested-PCR yielding a product size of 548 bp (Fig. 1) and 40 (47.05 %) samples were found positive for either one or more than one antigenic type of CPV using differential PCR (Table 2).
Fig. 1.
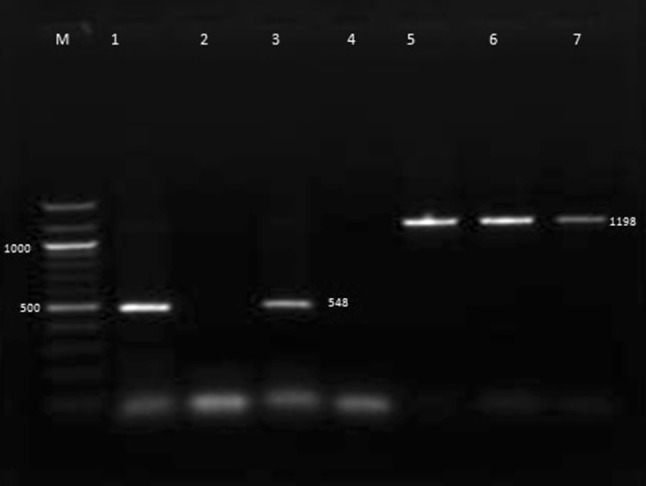
PCR and Nested PCR for Canine Parvovirus. Lane M Gene Ruler 100 bp plus, Lane 1 positive control, Lane 2 negative sample by nested-PCR, Lane 3 positive sample by nested-PCR, Lane 4 negative control, Lane 5 positive control, Lanes 6 and 7 positive samples by PCR
Table 2.
Description of samples positive by PCR, Nested PCR and Differential PCR
| S. no. | Sample | PCR | NPCR | Differential PCR | Age (months) | Sex | Breed | Vaccination status | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 2a | 2b | 2c | Either of 2, 2a, 2b or 2c | ||||||||
| 1. | P1 | − | + | + | + | − | − | + | 4 | F | GSD | + |
| 2. | P2 | − | + | − | − | − | − | − | 3.5 | M | St. Bernard | + |
| 3. | P5 | − | + | − | − | − | − | − | 1.5 | M | ND | + |
| 4. | P9 | − | + | − | − | − | − | − | 5 | M | ND | + |
| 5. | P10 | − | + | − | + | + | − | + | 2 | F | Labrador | + |
| 6. | P12 | − | + | − | + | − | − | + | 2 | M | Labrador | + |
| 7. | P13 | − | + | − | − | − | − | − | 1.5 | M | Rottweiler | + |
| 8. | P14 | − | + | − | − | − | − | − | 2.5 | M | GSD | + |
| 9. | P15 | − | + | − | + | − | − | + | 3 | F | Cocker spaniel | + |
| 10. | P16 | − | − | − | + | + | − | + | 3 | M | Rottweiler | − |
| 11. | P17 | − | − | − | + | − | − | + | 1 | F | St.Bernard | − |
| 12. | P18 | − | + | − | − | − | − | − | 3.5 | F | Labrador | − |
| 13. | P19 | − | + | − | − | − | − | − | 4.5 | M | Rottweiler | + |
| 14. | P20 | − | + | − | − | − | − | − | 3 | M | Labrador | + |
| 15. | P21 | − | + | − | − | − | − | − | 4 | M | Pom | + |
| 16. | P22 | − | − | − | + | − | − | + | 1.5 | F | Labrador | − |
| 17. | P23 | − | − | − | + | + | + | + | 2.5 | M | GSD | + |
| 18. | P26 | − | + | − | − | − | − | − | 2.5 | M | ND | − |
| 19. | P28 | − | + | − | − | − | − | − | 2 | M | Rottweiler | + |
| 20. | P29 | − | + | − | − | − | − | − | 2 | M | ND | − |
| 21. | P30 | − | − | − | − | + | − | + | 1 | F | Rottweiler | − |
| 22. | P31 | − | + | − | − | − | − | − | 1.5 | M | Rottweiler | − |
| 23. | P32 | + | + | − | + | + | + | + | 2 | F | Labrador | + |
| 24. | P34 | + | + | + | + | + | + | + | 4.5 | M | ND | − |
| 25. | P35 | + | + | + | + | + | + | + | 3 | F | Dalmatian | − |
| 26. | P36 | − | + | − | − | − | − | 2 | M | Pug | + | |
| 27. | P37 | − | − | − | − | + | − | + | 5 | M | ND | − |
| 28. | P39 | − | + | + | + | + | + | + | 1.5 | M | Labrador | − |
| 29. | P40 | − | − | − | − | + | − | + | 2.5 | F | Labrador | − |
| 30. | P41 | − | + | − | + | + | − | + | 5 | F | St.Bernard | − |
| 31. | P42 | − | − | − | + | + | − | + | 3 | M | Labrador | + |
| 32. | P43 | − | + | − | + | + | + | + | 4.5 | F | Labrador | − |
| 33. | P44 | − | + | − | − | − | − | − | 6 | F | Labrador | + |
| 34. | P45 | + | + | + | − | + | + | + | 3 | M | Labrador | − |
| 35. | P46 | + | + | + | + | + | + | + | 1.5 | M | ND | − |
| 36. | P47 | + | + | + | + | + | + | + | 3 | F | St.Bernard | − |
| 37. | P48 | − | − | + | − | + | + | + | 2 | F | Labrador | − |
| 38. | P49 | + | + | + | + | + | + | + | 3 | F | Labrador | − |
| 39. | P50 | − | + | + | + | + | + | + | 6 | F | Labrador | + |
| 40. | P56 | − | + | − | − | − | − | − | 8 | F | Pug | + |
| 41. | P61 | − | + | − | − | − | − | − | 10 | M | Mongrel | − |
| 42. | P64 | − | − | − | + | − | + | + | 18 | M | GSD | + |
| 43. | P65 | − | − | − | − | − | + | + | 4.5 | M | Dash hound | + |
| 44. | P68 | − | − | − | + | − | − | + | 2.5 | M | Rottweiler | + |
| 45. | P69 | − | − | − | + | − | + | + | 3 | M | Labrador | + |
| 46. | P71 | − | − | − | − | + | + | + | 2 | F | Sharpie | − |
| 47. | P72 | − | − | − | − | + | + | + | 2.5 | F | GSD | − |
| 48. | P73 | − | − | − | + | + | − | + | 3 | M | Dash hound | − |
| 49. | P74 | − | − | + | + | + | + | + | 6 | M | GSD | − |
| 50. | P75 | − | + | + | − | + | + | + | 10 | M | GSD | − |
| 51. | P76 | − | − | + | + | − | + | + | 4 | M | ND | − |
| 52. | P79 | − | + | + | − | − | + | + | 4 | M | GSD | − |
| 53. | P80 | − | + | − | − | − | − | − | 6 | M | GSD | − |
| 54. | P81 | − | − | − | + | − | − | + | 2 | M | Labrador | − |
| 55. | P82 | − | + | + | + | − | + | + | 5 | F | GSD | − |
| 56. | P83 | − | + | + | + | − | + | + | 3 | M | ND | − |
| 57. | P84 | − | + | − | + | − | − | + | 1.5 | M | ND | − |
| 58. | P85 | − | + | + | + | − | − | + | 1 | F | Pom | − |
| Total | 7 | 39 | 17 | 30 | 24 | 23 | 40 | |||||
(−) Negative/(+) Positive by PCR, Nested PCR and Differential PCR; M Male; F Female; GSD German shepherd dog; ND Non descript; Pom Pomeranian
When the age wise status among positive by NPCR was evaluated, it was found that out of 39, 22 (56.41 %) were 0–3 months of age, 14 (35.89 %) were 3–6 months of age, 1 (2.56 %) of 6–9 months of age and 2 (5.12 %) were of 9–12 months of age. Similarly using differential PCR it was found that out of 40, 27 (67.5 %) were 0–3 months of age, 11(27.5 %) were 3–6 months of age and 2 (5.0 %) were more than 9 months of age.
When the sex wise status among positive by NPCR was evaluated, it was found that out of 39, 24 (61.53 %) were male and 15 (38.46 %) were female. Using differential PCR it was found that out of 40, 21(52.5 %) were male and 19 (47.5 %) were female.
When the breed wise prevalence among positive by NPCR was evaluated it was found that out of 39, 11 (28.20 %) were of Labrador breed, 6 (15.38 %) were of German Shepherd breed, 4 (10.25 %) were of Rottweiler breed, 3 (7.69 %) were of Saint Bernard breed, 2 (5.12 %) each of Pomeranian and Pug breed, one (2.56 %) each of Cocker Spaniel, Dalmatian and Mongrel breed and 8 (20.51 %) were of non-descript type. When the breed wise prevalence among positive using differential PCR was evaluated it was found that out of 40, 14 (35.0 %) were of Labrador breed, 8 (20.0 %) were of German Shepherd breed, 3 (7.5 %) were of Rottweiler breed, 3 (7.5 %) were of Saint Bernard breed, 2 (5.0 %) were of Dash hound breed, 1 (2.5 %) each of Pomeranian, Sharpie, Cocker Spaniel and Dalmatian and 6 (15.0 %) were of non-descript type.
When the vaccination status of the positive by NPCR was evaluated it was found that out of 39 positive 18 (46.15 %) were vaccinated and 21 (53.84 %) were not vaccinated. Using differential PCR it was found that out of 40 positive 12 (30.0 %) were vaccinated and 28 (70.0 %) were not vaccinated.
In the present study, the antigenic characterization of CPV was also done using the designed primers for identification of CPV-2, CPV-2a, CPV-2b and CPV-2c individually (Fig. 2). Out of 40 positive samples using differential PCR, 7 (17.5 %) were found positive for CPV-2a, 3 (7.5 %) were found positive for CPV-2b and 1 (2.5 %) was found positive for CPV-2c. Thus, CPV-2a was found most prevailing antigenic type followed by CPV-2b. When we examined for the presence of more than one antigenic type in a sample it was found that 2 (5.0 %) samples had both CPV-2 and CPV-2a, CPV-2a and CPV-2c and CPV-2b and CPV-2c, 1 (2.5 %) had CPV-2 and CPV-2c, 5 (12.5 %) had CPV-2a and CPV-2b, 3 (7.5 %) had CPV-2a, CPV-2b and CPV-2c, CPV-2, CPV-2b and CPV-2c, CPV-2, CPV-2a and CPV-2c and 8 (20.0 %) had all the four antigenic types viz., CPV-2, CPV-2a, CPV-2b and CPV-2c (Table 2).
Fig. 2.
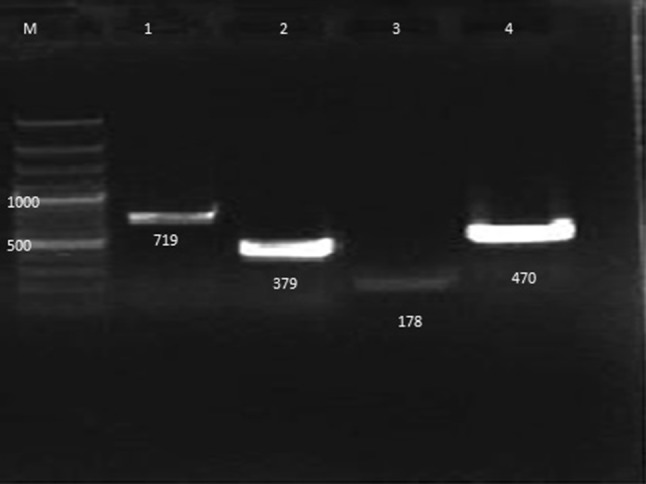
Sample positive for CPV-2, CPV2a, CPV-2b and CPV-2c. Lane M Gene Ruler 100 bp plus, Lane 1 Sample positive for CPV-2 (719 bp), Lane 2 Sample positive for CPV-2a (379 bp), Lane 3 Sample positive for CPV-2b (178 bp), Lane 4 Sample positive for CPV-2c (470 bp)
When the results of NPCR and differential PCR were compared using kappa statistics, it was observed that the kappa value was 0.1282 indicating that there is slight agreement between NPCR and differential PCR.
Discussion
Canine parvovirus has emerged as one of the most important disease of pups in recent times. Although adult dogs show less severe symptoms of disease, they serve as source of infection. Also, due to its immunosuppressive nature it reduces animal’s ability to fight against infections [16] making the animal prone to various infectious diseases. Various methods have been used by earlier workers for its diagnosis that includes virus isolation in cell culture (Madin Darby Canine Kidney, Crandle Feline Kidney, A-72 cell lines), HA, HI, Electron Microscopy, IFT, ELISA and PCR [20]. Virus isolation is very good and has high specificity but it requires availability of cell cultures as well as expert technical staff. Besides, it is time consuming (5–10 days), expensive and laborious method. HA and HI tests are simple inexpensive and easy to perform but require RBC’s of porcine origin afresh every time. Moreover, the presence of non-specific agglutinin in feces makes HA test less reliable for CPV diagnosis [20]. Electron Microscopy (EM) is also a good method but it requires virus to be concentrated. However, sensitivity of these traditional diagnostic methods, has been proven to be inferior to molecular assays and thus, PCR has been used for the detection of CPV-2 in fecal samples with high sensitivity and specificity [31].
Out of 85 samples from dogs exhibiting clinical signs of CPV, 7 were positive by PCR and 39 were positive with NPCR indicating increased sensitivity of nested PCR. The above results are similar to the earlier finding of Mochizuki et al., Hirasawa et al., Sakulwira et al. and Schmitz et al. [11, 20, 29, 30] who stated that nested PCR being more sensitive than conventional PCR. Our results are too in tandem with that of the earlier reports indicating increased number of positive cases when PCR product was used in nested PCR. The possible reason for this is that with the use of NPCR we increase the concentration of PCR product so much so that we are able to visualize it on an agarose gel. Another reason for this could be that the samples containing very few virus particles might be harbouring inhibitory substances as reported by Kumar et al. [15] leading to absence of visualization of the amplified product after a PCR which could have been resolved using a NPCR leading to visualization of NPCR product in an agarose gel. Thus, NPCR can help in early, sensitive and accurate diagnosis of CPV infection in dogs.
The additional use of newly designed differential PCR 40 samples were found positive indicating increased sensitivity of differential PCR over NPCR. It was observed that, there were 21 samples that were positive by both NPCR and differential PCR but there were 19 samples that were negative by NPCR but were positive for either of the antigenic types using differential PCR indicating increased sensitivity of differential PCR.
Further it was also observed that some of the samples (n = 18) that were positive by nested-PCR were found negative by differential PCR. The plausible reason for this might be due to the prevalence of certain other antigenic types besides those that were tested in the present study yielding positive using nested PCR but negative using differential PCR, but further more intensive sequencing studies are required to conclusively prove this statement. Though there have been reports indicative of circulation of newer strains of CPV from India [33].
Maximum pups affected by CPV as indicated by NPCR and differential PCR were of 0–3 months of age followed by 3–6 months substantiating the already established fact that the infection caused by CPV is more severe in young animals [1, 2, 9, 18, 32].
The percent positivity in term of sex indicated that male dogs were more affected than female dogs by CPV as indicated using NPCR and differential PCR both. The above observations are also in tandem to the similar observed reports indicating that the prevalence of CPV infection higher in males when compared with females [13, 14, 33].
Breed wise comparison indicated that German shepherd and Labrador breeds of dogs were mostly affected by CPV as detected by NPCR and differential PCR. These observations too are on the earlier reported facts in which Kumar et al. [15] and Singh et al. [32] reported that in India GSD breed followed by Labrador and Pomeranian breeds of dogs are most predisposed for CPV.
Some of the dogs which were positive for CPV in NPCR and dPCR were vaccinated for CPV which indicates that may be vaccination of pups against CPV is not conferring immunity against the disease. This may be due to the mismatching of vaccine strain and the CPV strain causing infection in dogs. This is also due to the reason that the vaccine is effective against the strain of CPV present in the vaccine and not against the other antigenic strains of CPV that may cause infection and even mortality in dogs [17]. As reported [21] there might be strain variation in vaccine and the prevalent CPV strains in the field condition as most of the vaccines used in India are based on the strain that was isolated about 25–30 years ago.
It was observed that after the emergence of CPV-2, its two more mutants namely CPV-2a and CPV-2b evolved around the world and later, CPV type 2c (CPV-2c) was mostly reported [23]. CPV-2 was reported for the first time in India in 1982 [27] and since then, a large number of outbreaks involving different variants of CPV (2, 2a, 2b and 2c) have been reported from different parts of India in both vaccinated and non-vaccinated dogs [2, 21, 26]. In the present study using differential PCR CPV-2a and CPV-2b were found to be the most prevailing antigenic types and among these two, 2a was more prevalent. Similar to our study, Chinchkar et al. [5] also examined 27 isolates of CPV and indicated that they belonged to CPV-2a and four belonged to CPV-2b. In another study, Perez et al. [24] reported increased CPV-2a variant in a dog population in Uruguay where originally CPV-2c was prevalent indicating shifting trend of antigenic strains in CPV. Similarly in an epidemiological survey for CPV in different European countries CPV-2a was found predominant followed by CPV-2c and CPV-2b [8].
It can be concluded from the above study that canine parvovirus is prevalent in Punjab and the most prevailing antigenic types are CPV-2a and CPV-2b. The differential PCR was found to be highly efficient and quick as it was using only one cycle of PCR for detecting positive CPV. The results (age wise, breed wise, sex wise prevalence etc.) with differential PCR were similar to that of NPCR. The differential PCR developed in the study could be used in tandem with the already established NPCR.
Acknowledgments
The authors are thankful to the University Grant Commission (UGC), New Delhi for the financial support and Director of Research, GADVASU for providing the research facilities.
Conflict of interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Banja BK, Sahoo N, Panda HK, Ray SK, Das PK. Epizootiological status of canine viral haemorrhagic gastroenteritis in Bhubaneswar city. Indian Vet J. 2002;79:850–851. [Google Scholar]
- 2.Biswas S, Das PJ, Ghosh SK, Pradhan NR. Detection of canine parvovirus (CPV) DNA by polymerase chain reaction and its prevalence in dogs in and around Kolkata, West Bengal. Indian J Anim Sci. 2006;76:324–325. [Google Scholar]
- 3.Buonavoglia C, Martella V, Pratelli A, Tempesta M, Cavalli A, Buonavoglia D, Bozzo G, Ella G, Decaro N, Carmichael L. Evidence for evolution of canine parvovirus type 2 in Italy. J Gen Virol. 2001;82:3021–3025. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- 4.Carmichael LE, Schlafler DH, Hashimoto A. Minute virus of canines (MVC, canine parvovirus type-1): pathogenicity for pups and seroprevalence estimate. J Vet Diagn Investig. 1994;6:165–174. doi: 10.1177/104063879400600206. [DOI] [PubMed] [Google Scholar]
- 5.Chinchkar SR, Subramaniam BM, Rao NH, Rangarajan PN, Thiagarajan D, Srinivasan VA. Analysis of VP2 gene sequences of canine parvovirus isolates in India. Arch Virol. 2006;151:1881–1887. doi: 10.1007/s00705-006-0753-8. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- 7.Decaro N, Elia G, Desario C, Roperto S, Martella V, Campolo M, Lorusso A, Cavalli A, Buonavoglia C. A minor groove binder probe real time PCR assay for discrimination between type-2 based vaccines and field strains of canine parvovirus. J Virol Methods. 2006;136:65–70. doi: 10.1016/j.jviromet.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decaro N, Desario C, Amorisco F, Losurdo M, Elia G, Parisi A, Ventrella G, Martella V, Buonavoglia C. Detection of a canine parvovirus type 2c with a non-coding mutation and its implications for molecular characterisation. Vet J. 2013;196:555–557. doi: 10.1016/j.tvjl.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 9.Deepa PM, Saseendrannath MR. Serological studies on canine parvoviral infection. Indian Vet J. 2000;79:643–644. [Google Scholar]
- 10.Gupta PK, Rai A, Rai N, Raunt AA, Chauhan S. Cloning of canine parvovirus VP2 gene and its use as DNA vaccine in dogs. Curr Sci India. 2005;88:778–782. [Google Scholar]
- 11.Hirasawa T, Kaneshige T, Mikazuki K. Sensitive detection of canine parvovirus DNA by the nested polymerase chain reaction. Vet Microbiol. 1994;41:135–145. doi: 10.1016/0378-1135(94)90143-0. [DOI] [PubMed] [Google Scholar]
- 12.Hoelzer K, Shackelton LA, Holmes EC, Parrish CR. Within-host genetic diversity of endemic and emerging parvoviruses of dogs and cats. J Virol. 2008;82:11096–11105. doi: 10.1128/JVI.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houston DM, Ribble CS, Head LL. Risk factors associated with Parvovirus enteritis in dogs: 283 cases (1982–1991) J Am Vet Med Assoc. 1996;208:542–546. [PubMed] [Google Scholar]
- 14.Khan AM, Rabbni M, Muhammad K, Murtaza N, Nazir J. Int J Agric Biol. 2006;8:898. [Google Scholar]
- 15.Kumar M, Chidri S, Nandi S. A sensitive method to detect canine parvoviral DNA in faecal samples by nested polymerase chain reaction. Indian J Biotechnol. 2011;10:183–187. [Google Scholar]
- 16.Legendre AM. Parvovirus in dog. In: Ettinger SJ, Feldman EC, editors. Textbook of veterinary internal medicine, diseases of the dog and cat. Vth ed. vol II. Philadelphia: W B Saunders Co.;2000. p 1958.
- 17.Martella V, Cavalli A, Decaro N, Elia G, Desario C, Campolo M, Bozzo G, Tarsitano E, Buonavoglia C. Immunogenicity of an intranasally administered modified live canine parvovirus type 2b vaccine in pups with maternally derived antibodies. Clin Vaccine Immunol. 2005;12:1243–1245. doi: 10.1128/CDLI.12.10.1243-1245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCaw DL, Hoskins JD. Canine viral enteritis. In: Greene CE, editor. Infectious diseases of the dog and cat. 3rd ed. St. Louis: Saunders Elsevier; 2006. p 63.
- 19.Mizak B, Rzezutka A. Application of nested PCR for detection of canine parvovirus in faeces. Bull Vet Inst Pulawy. 1999;43:19–24. [Google Scholar]
- 20.Mochizuki M, San Gabriel MC, Nakatani H, Yoshida M. Comparison of polymerase chain reaction with virus isolation and haemagglutination assays for the detection of canine parvoviruses in faecal specimens. Res Vet Sci. 1993;55:60–63. doi: 10.1016/0034-5288(93)90035-E. [DOI] [PubMed] [Google Scholar]
- 21.Nandi S, Chidri S, Kumar M, Chauhan RS. Occurrence of canine parvovirus type 2c in the dogs with haemorrhagic enteritis in India. Res Vet Sci. 2010;88:169–171. doi: 10.1016/j.rvsc.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 22.OzKul A, Keles I, Karaoglu T, Cabalar M, Burgu I. Detection and RFLP analysis of canine parvovirus (CPV) DNA by polymerase chain reaction (PCR) in a dog. Turk J Vet Anim Sci. 2002;26:1201–1203. [Google Scholar]
- 23.Parrish CR. Host range relationship and the evolution of canine parvovirus. Vet Microbiol. 1999;69:29–40. doi: 10.1016/S0378-1135(99)00084-X. [DOI] [PubMed] [Google Scholar]
- 24.Pérez R, Bianchi P, Calleros L, Francia L, Hernández M, Maya L, Panzera Y, Sosa K, Zoller S. Recent spreading of a divergent canine parvovirus type 2a (CPV-2a) strain in a CPV-2c homogenous population. Vet Microbiol. 2012;155:214–219. doi: 10.1016/j.vetmic.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 25.Phromnoi S, Sinsiri R, Sirinarumitr T. Expression of recombinant VP2 protein of canine parvovirus in Escherichia coli. Kasetsart J. 2010;44:870–878. [Google Scholar]
- 26.Phukan A, Deka D, Boro PK. Occurrence of canine parvovirus infection in and around Guwahati. Indian J Anim Sci. 2004;74:930–931. [Google Scholar]
- 27.Ramadass P, Khader TGA. Diagnosis of canine parvovirus infection by agar gel precipitation test and fluorescent antibody techniques. Cheiron. 1982;11:323–325. [Google Scholar]
- 28.Rozen S, Skaletsky HJ. Primer 3 on the www for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa: Humana Press; 2007. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 29.Sakulwira K, Oraveerakul K, Poovorawan Y. Detection and genotyping of canine parvovirus in enteric dogs by PCR and RFLP. Sci Asia. 2001;27:143–147. doi: 10.2306/scienceasia1513-1874.2001.27.143. [DOI] [Google Scholar]
- 30.Schmitz S, Coenen C, Matthias K, Heinz-Jurgen T, Neiger R. Comparison of three rapid commercial canine parvovirus antigen detection tests with electron microscopy and polymerase chain reaction. J Vet Diagn Investig. 2009;21:344–345. doi: 10.1177/104063870902100306. [DOI] [PubMed] [Google Scholar]
- 31.Schunck B, Kra W, Truyen U. A simple touch-down polymerase chain reaction for the detection of canine parvovirus and feline panleukopenia virus in feces. J Virol Methods. 1995;55:427–433. doi: 10.1016/0166-0934(95)00069-3. [DOI] [PubMed] [Google Scholar]
- 32.Singh D, Verma AK, Kumar A, Srivastava M, Singh SK, Tripathi AK, Srivastava A, Ahmed I. Detection of canine parvovirus by polymerase chain reaction assay and its prevalence in dogs in and around Mathura, Uttar Pradesh, India. Am J Biochem Mol Biol. doi: 10.3923/ajbmb.2013.
- 33.Srinivas VMV, Mukhopadhyay HK, Thanislass J, Antony PX, Pillai RM. Molecular epidemiology of canine parvovirus in southern India. Vet World. 2013;16:744–749. doi: 10.14202/vetworld.2013.744-749. [DOI] [Google Scholar]
- 34.Truyen U. Emergence and recent evolution of canine parvovirus. Vet Microbiol. 1999;69:47–50. doi: 10.1016/S0378-1135(99)00086-3. [DOI] [PubMed] [Google Scholar]
- 35.Ying H, Runxi X, Manfu Z. Expression or subcellular targeting of virus capsid proteins with cloning genome of a canine parvovirus from China. Res Vet Sci. 2009;87:239–241. doi: 10.1016/j.rvsc.2009.03.001. [DOI] [PubMed] [Google Scholar]


