Abstract
Bradyrhizobium sp. DOA9, a non-photosynthetic bacterial strain originally isolated from the root nodules of the legume Aeschynomene americana, is a divergent nod-containing strain. It exhibits a broad host range, being able to colonize and efficiently nodulate the roots of most plants from the Dalbergioid, Millettioid, and Robinioid tribes (7 species of Papilionoideae). In all cases, nodulation was determinate. The morphology and size of DOA9 bacteroids isolated from the nodules of various species of Papilionoideae were indistinguishable from the free-living form. However, they were spherical in Arachis hypogaea nodules. GusA-tagged DOA9 also colonized rice roots as endophytes. Since broad-host-range legume symbionts often carry multiple replicons in their genome, we analyzed the replicons for symbiosis genes by electrophoresis. DOA9 carried two replicons, a chromosome (cDOA9) and single megaplasmid (pDOA9) larger than 352 kb. The genes for nodulation (nodA, B, C) and nitrogen fixation (nifH) were localized on the megaplasmid. Southern blot hybridization revealed two copies of nodA on the megaplasmid, single copies of nodB and C on the megaplasmid, and one copy each of nifH on the chromosome and megaplasmid. These results suggested that Bradyrhizobium sp. DOA9 may have the unusual combination of a broad host range, bacteroid differentiation, and symbiosis-mediating replicons.
Keywords: Bradyrhizobium, Aeschynomene americana, broad host range, megaplasmid
The genus Aeschynomene belongs to the Dalbergioid clade of the subfamily Papilionoideae of the family Fabaceae (19). Aeschynomene species establish a symbiotic relationship with bacteria of the genus Bradyrhizobium (19, 27). They have been classified into one of three cross-inoculation (CI) groups: CI group 1 species, such as A. americana and A. elaphroxylon, are only nodulated on their roots by non-photosynthetic bradyrhizobia. CI group 2 species, such as A. afraspera and A. nilotica, are nodulated on their roots and stems by both non-photosynthetic and photosynthetic bradyrhizobia. CI group 3 species, such as A. indica and A. sensitiva, are nodulated on their roots and stems by photosynthetic bradyrhizobia. CI group 3 species are nodulated by strains BTAi1 and ORS278 in a Nod factor–independent manner (27). However, A. indica (CI group 3) was recently found to be nodulated by a non-photosynthetic strain isolated from A. americana (CI group 1). This strain could nodulate several CI groups of Aeschynomene, as well as the peanut and mung bean (29).
Bradyrhizobium sp. DOA9 (Alphaproteobacteria) was isolated from the root nodules of A. americana in Thailand. This strain was assigned to B. yuanmingense based on its phenotypic characteristics and multilocus sequences analysis of the 16S rRNA gene and housekeeping genes (dnaK, recA, and glnB). DOA9 was called a divergent nod-containing strain based on Southern blot hybridization with the nod gene (nodA, B, and C) probes. In addition, on the basis of nifH sequence similarities, DOA9 was placed in a cluster of non-photosynthetic bradyrhizobial strains that are able to fix nitrogen in the free-living form. Moreover, it can nodulate a wide range of legumes, including Macroptilium atropurpureum, Arachis hypogaea, Vigna radiata, and Aeschynomene afraspera (29). Since information regarding this strain is limited, we investigated the root colonization, infection, and nodulation efficiency of DOA9 in several species from the legume subfamilies Papilionoideae and Mimosoideae, as well as rice. We also examined bacteroids and nodule development. Previous studies demonstrated that symbionts with a broad host range generally carried multiple replicons in their genome (12, 36, 42); therefore, we examined the replicon structure, localization, and copy number of symbiosis genes.
Materials and Methods
Bacterial strains and growth conditions
Bradyrhizobium sp. DOA9, B. diazoefficiens USDA110, and Mesorhizobium loti MAFF303099 were cultured at 28°C in HM medium supplemented with l-arabinose (8). GUS-tagged DOA9 was cultured in HM supplemented with streptomycin (200 μg mL−1) (29).
Plant growth and inoculation
Peanut (Arachis hypogaea), mung bean (Vigna radiata), soybean (Glycine max cv. SJ5), siratro (Macroptilium atropurpureum), Sesbania rostrata, lupin (Lupinus polyphyllus), Desmodium sp., alfalfa (Medicago sativa), Medicago truncatula, and Acacia mangium seeds were sterilized as described previously (37). Aeschynomene americana (a local Thai variety), A. afraspera, A. evenia (provided by Eric Giraud), and A. indica (ecotype Tottori, Japan; ecotype Tomeshi, Japan; and a local Thai variety) were sterilized by incubating in concentrated sulfuric acid for 30 min. The seeds of Indigofera tinctoria, Lotus japonicus, Stylosanthes hamata, Crotalaria juncea, Lespedeza sp., Leucaena leucocephala, Mimosa pudica, Neptunia natans, and Samanea saman were sterilized by incubating in concentrated sulfuric acid for 10 min. The seeds of rice (Oryza sativa ssp. indica) were sterilized as described previously (26). All seeds were washed with sterilized water and then soaked in sterilized water overnight at ambient temperature. All seeds were germinated on sterilized 0.8% (w/v) water agar for 1 to 2 d at 28°C in the dark. The germinated seeds were transferred onto Hoagland’s agar (15). Bradyrhizobium sp. DOA9 was washed with 0.85% NaCl, and optical density at 600 nm (OD600) was adjusted to 1 with sterilized water, corresponding to approximately 109 cells mL−1. Each seedling was then inoculated with 100 μL of the bacterial culture. All plants were grown at 25°C under a 16 h light/8 h dark cycle at a light intensity of 639 μE m−2 s−1 for 7 to 14 d (43). These samples were used for microscopic observations. The symbiotic abilities of DOA9 were determined in Leonard’s jars containing sterilized vermiculite and inoculated with 1 mL of bacterial culture, as described above. N-free Hoagland’s solution was added to each jar as required. Plants were harvested after 35 d, used in an analysis of nitrogenase activity by the Acetylene Reduction Assay, and the number of nodules was scored. The dry weights of plants were determined after drying at 70°C for 72 h.
Microscopic observation of root colonization and nodulation
Root colonization and nodulation were revealed by GUS staining. Samples inoculated with GUS-tagged DOA9 were immersed in GUS assay solution (40 mL 20 mg mL−1 X-Gluc in N,N-dimethyl-formamide, 20 mg SDS, 2 mL methanol, 0.2 mL 1 M sodium phosphate buffer, and 7.76 mL distilled water) in a vacuum for 120 min and left in this solution for 12 h at 28°C. GUS staining was observed under a light microscope.
Differential interference contrast and fluorescence microscope observations of isolated bacteroids
Nodules were mashed in bacteroid extraction buffer (125 mM KCl, 50 mM Sodium succinate, 50 mM TES buffer, pH 7.0, with 1% (w/v) BSA) (24). To remove plant cell debris, the suspension was centrifuged at 100×g at 4°C for 10 min. To precipitate the bacteroids, the supernatant was centrifuged at 3,000×g at 4°C for 10 min. The precipitate was observed using Differential Interference Contrast (DIC) microscopy. Regarding fluorescence microscopy, the bacteroid fraction was first stained with 4′,6-diamidino-2-phenylindole (DAPI; 50 μg mL−1) and then with propidium iodide (PI; 2 μg mL−1).
DNA extraction and megaplasmid detection
Bradyrhizobium sp. DOA9 total DNA was prepared as described previously (23). Megaplasmids were isolated by electrophoresis (2, 40) with the modifications described here. Bacteria were cultured on HM broth medium (37) with 0.05% (w/v) l-arabinose, 0.05% (w/v) yeast extract for B. diazoefficiens USDA110 and M. loti MAFF303099, and no l-arabinose for Bradyrhizobium strain DOA9, to reduce the production of polysaccharides. USDA110 and DOA9 were cultured at 30°C for 3 to 5 d on a rotary shaker at 200 rpm until the late-log phase was reached. A total of 1% (v/v) of these pre-cultures was then inoculated into new tubes containing HM broth medium. The cultures were incubated for 3 d until the exponential growth phase (the mid-log phase) was reached. MAFF303099 was incubated for 2 d, inoculated into a fresh tube containing HM medium, and then cultured under the same conditions for 24–36 h. Cell pellets were harvested by centrifugation at 3,000×g for 10 min. The cells were resuspended in 0.85% (w/v) NaCl to OD600=1. They were harvested from 1 mL of a cell suspension and washed with M9 salts (34) containing 0.5 M NaCl, and then with 1 mL 0.1% (w/v) Sarcosyl. The supernatant was removed immediately and the sediment was resuspended in 50 μL of lysis buffer (1 mg mL−1 lysozyme, 1 mg mL−1 RNase A [33], 0.1% (w/v) bromophenol blue in Trisborate buffer (pH 8.2, 89 mM Tris base, 12.5 mM disodium EDTA, and 8.9 mM boric acid [10], and 40% [v/v] glycerol).
Before sample loading, electrophoresis was performed on a 0.7% agarose gel, which leveled off with 0.5×TBE buffer (4°C) until it touched the gel. The wells were then filled with 50 μL Sodium Dodecyl Sulfate (SDS; 10% [w/v]) mixed with xylene cyanol (1 mg mL−1). The current was run for 10–15 min at 100 V with reversed polarity until SDS was 1 cm above the wells. After that, 50 μL of sample mixed with lysis buffer was directly loaded and left for 15 min before 15 μL of Proteinase K (5 mg mL−1) in 40% (w/v) glycerol was overlaid. The wells were sealed with melted agarose gel, and 0.5×TBE buffer was then added to cover the gel. After 1 h, electrophoresis was carried out in a cold chamber at 4°C. The current was run at 10 mA for 14 h and at 50 mA for a further 10 h. The DNA in the gel was stained for 30 min in ethidium bromide (0.5 μg mL−1) and washed with distilled water before being viewed under UV light.
Southern blot hybridization
The megaplasmid and chromosomal DNAs separated on the gel were used for the Southern blot hybridization of nodulation (nod) and nitrogen fixation (nifH) genes as described (29). Briefly, probes for nodA (550 bp), nodB (530 bp), and nodC (1 kb) were obtained through PCR amplification using the genomic DNA of B. yuanmingense SUTN6-2, B. canariense SUTN7-2, and B. diazoefficiens USDA110, respectively. DNA fragments of the respective strains were amplified with the primer pairs nodAYF46/nodAYR595 (nodA), nodB26/nodB625 (nodB), and nodC195/nodCI (nodC) (29). The probe for nifH was derived from B. yuanmingense using nifHF/nifHI primer pairs (18). DNA probes were labeled overnight at 37°C by random priming, and hybridized with the Digoxigenin (DIG) High Prime DNA Labeling and Detection Starter Kit I (Roche, Switzerland). DNA was capillary-transferred to a Hybond-N+ nylon membrane (Amersham, Cardiff, UK) as described previously (34). Low-stringency conditions were used for hybridization: membranes were hybridized at 40°C (nodA and nodC genes) or 42°C (nodB and nifH) for 18 h and then washed twice in 2× SSC + 0.1% (w/v) SDS at 25°C for 15 min and in 0.5× SSC + 0.1% (w/v) SDS at 62°C for 15 min.
DOA9 genomic DNA was digested with EcoRI, EcoRV, HindIII, or NotI to evaluate the copy numbers of nodB, nodC, and nifH; with EcoRI, HindIII, BglII, or NotI for nodA. Fragments were separated on 1% (w/v) agarose gels before hybridization as described above.
Results and Discussion
Nodulation of the subfamily Papilionoideae
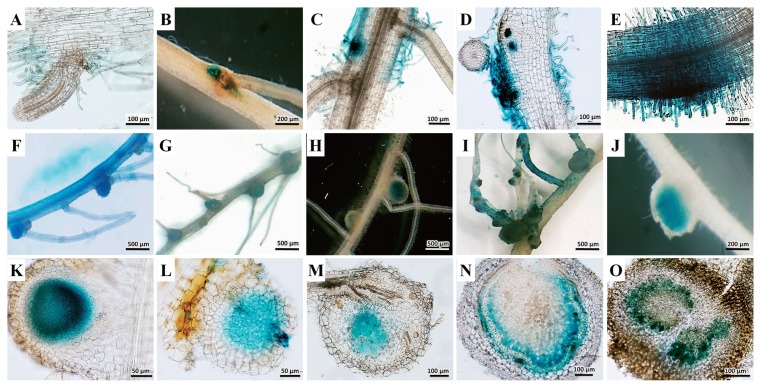
We tested 19 species and 3 varieties of the ecotypes of A. indica (Table 1). DOA9 was able to nodulate the roots of 15 species. Seven of the tested species were effective in terms of nitrogen-fixing efficiency, as indicated by a significant increase in the plant dry weight when inoculated with DOA9 (39) (Table 1 and Table S1 in the supplemental material). An ineffective symbiotic was detected in DOA9 with S. hamata, M. atropurpureum, Lepedeza sp., L. japonicus, L. leucocephala, and S. saman. This strain colonized the lateral root (Fig. 1A–D) and also the root surface (Fig. 1E). In addition, nodulation was related to the colonization sites (Fig. 1F–I). All nodules were determinate (Fig. 1F–J). A thin section of nodules in this group showed achynomenoid types (Fig. 1K–O) (38). DOA9 nodulated both genistoids, but only induced effective nodules on C. juncea. Among the Dalbergioids, DOA9 induced effective nodules on A. americana, A. afraspera, and A. hypogaea. Nodules in this group were clearly of the aeschynomenoid type (Fig. 1K–L), which were formed via the crack infection pathway (Fig. 1F and G) (38). This route of infection bypassed some of the complex processes involved in infection via root hairs, which depend on the Nod-factor structure (14). DOA9 induced nodules on all the Millettioids tested, except for G. max, even though soybean-nodulating bacteria were distributed among three genera (6, 7). Rhizobia so far known to nodulate Millettioids are all classical rhizobia, mainly slow-growing (“brady-”) types (38). Of the two Robinioids, only L. japonicus was nodulated; however, these nodules were ineffective (see Fig. S1 and Table S1). This strain could nodulate the model legume L. japonicus. L. japonicus may have permitted intercellular infection of the cortex (22). However, DOA9 could not nodulate S. rostrata, even though Sesbania accepted infection via the crack entry mode (14). Among the Inverted Repeat–Lacking Clade (IRLC), only M. sativa was nodulated; however, these nodules were ineffective. These Papilionoideae tribes are generally nodulated with fast-growing α-rhizobia (Alphaproteobacteria) and typically show a high degree of specificity between symbiotic partners (38).
Table 1.
Nodulation by Bradyrhizobium sp. DOA9 and bacteroid morphology in various legumes
| Plants | Nodulationa | Nodule type | Bacteroidc |
|---|---|---|---|
| Papilionoideae | |||
| Genistoids | |||
| Lupinus polyphyllus | (+) | determinate | unswollen |
| Crotalaria juncea | + | determinate | unswollen |
| Dalbergioids | |||
| Aeschynomene americana (a local Thai variety) | + | determinate | unswollen |
| Aeschynomene indica (ecotype Tottori, Japan) | − | − | − |
| Aeschynomene indica (ecotype Tomeshi, Japan) | − | − | − |
| Aeschynomene indica (a local Thai variety) | − | − | − |
| Aeschynomene afraspera | + | determinate | unswollen |
| Aeschynomene evenia | − | − | − |
| Arachis hypogaea | + | determinate | swollen |
| Stylosanthes hamata | (+) | determinate | swollen |
| Millettioids | |||
| Glycine max (cv. SJ5) | − | − | − |
| Macroptilium atropurpureum | (+) | determinate | unswollen |
| Vigna radiata | + | determinate | unswollen |
| Desmodium sp. | + | determinate | unswollen |
| Lespedeza sp. | (+) | determinate | unswollen |
| Indigofera tinctoria | + | determinate | unswollen |
| Robinioids | |||
| Lotus japonicus (ecotype Miyagi, Japan) | (+) | determinate | unswollen |
| Sesbania rostrata | − | − | − |
| IRLC (inverted repeat–lacking clade) | |||
| Medicago truncatula | − | − | − |
| Medicago sativa | (+) | determinate | unswollen |
| Trifolium repens (white clover) | − | − | − |
| Mimosoideae | |||
| Mimoseae | |||
| Mimosa pudica | − | − | − |
| Leucaena leucocephala | (+) | determinate | unswollen |
| Neptunia natans | − | − | − |
| Ingeae | |||
| Samanea saman | (+) | determinate | unswollen |
+, positive with effective nodules; (+), positive with ineffective nodules; −, negative.
Fig. 1.
Examples of (A–E) root colonization, (F–J) nodule morphology, and (K–O) thin sections of nodules inoculated with GUS-tagged DOA9 in various legumes. (A, F, and K); Aeschynomene americana, (B, G, and L); Stylosanthes hamate, (C, H, and M); Indigofera tinctoria, (D, I, and N) Macroptilium atropurpureum, and (E, J, and O) Lotus japonicus.
Nodulation of the subfamily Mimosoideae
We tested 4 species (Table 1). Although DOA9 nodulated L. leucocephala and S. saman, these nodules were not effective. This limited result may have been due to host range specificity, as species of the tribe Mimoseae may be nodulated by β-rhizobia (Betaproteobacteria) (3, 11).
Bradyrhizobium sp. DOA9 is able to nodulate several species of legumes (Table 1) (29). In addition, A. americana plants are commonly nodulated by Bradyrhizobium spp. of the cowpea miscellaneous group (41). These findings high-lighted the broad host range characteristics of these bacteria. DOA9 also nodulated many of the Dalbergioids, Millettioids, and Robinioids. DOA9 has been shown to contain divergent nod-genes (29), which may facilitate the broad host range nodulation ability. Moreover, the ability of DOA9 to invade roots via the cracks through which lateral roots emerge (29) may allow it to infect a wide variety of legumes; however, we found no infection threads in any sample. These results indicated that DOA9 showed a broad host range.
Root colonization and infection of rice
DOA9 colonized the roots and infected the tissues of rice. The expression of GUS was indicated by blue staining (Fig. 2). At 1 d after inoculation, the histochemical staining of β-galactosidase activity revealed the strong colonization of the root cap (Fig. 2A). At 5 d, this strain colonized the entire root (Fig. 2B) and intercellular cells (Fig. 2C and D), as shown in blue. However, we did not measure ARA in rice and its efficiency could not be decided based on plant growth. This should be further analyzed using the Kjeldahl method for the total nitrogen content and compared between inoculated and un-inoculated rice. Previous studies reported that photosynthetic Bradyrhizobium strains induced N2-fixing nodules on the stems and roots of the genus Aeschynomene (28), and also formed a natural endophytic association with the wild rice species Oryza breviligulata (5), which grows in association with several aquatic legumes. However, no studies have yet been published on non-photosynthetic bradyrhizobial strains in endophytic association with rice. Therefore, it was interesting that the non-photosynthetic DOA9 could colonize and infect rice. In Thailand, the semi-aquatic A. americana frequently grows in association with rice. The genes in DOA9 that are involved in rice infection, which may also play a role in the early interaction between A. americana and DOA9, should be examined in more detail. The findings of such a study may reveal a common determinant in the symbiotic interaction between rhizobia and ancestor plants before the symbiotic relationship evolved.
Fig. 2.
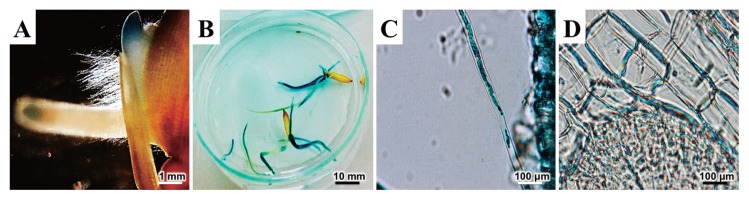
Light microscopic observations of Oryza sativa ssp. indica seedlings inoculated with GUS-tagged Bradyrhizobium sp. DOA9. (A) 1 d after inoculation. (B) 5 d after inoculation. (C) Close-up view of root hair and (D) close-up view of root tissue 5 days after inoculation.
Microscopic observation of isolated bacteroids
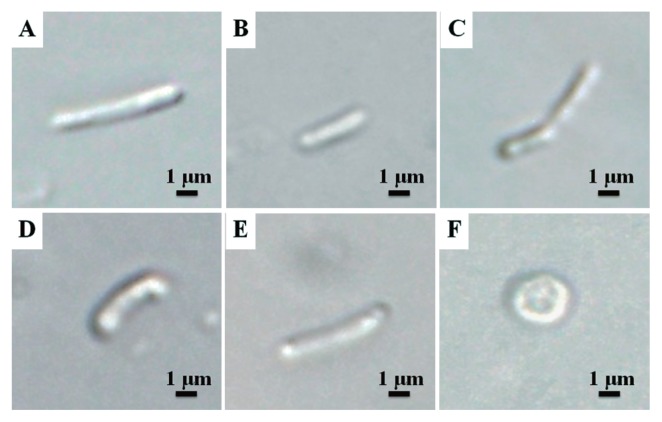
Bacteroids isolated from the various species of Papilionoideae were indistinguishable from the free-living form (Figs. 3, and 4). No significant differences were observed in the shape or size of bacteroids between this group and the free-living form (Fig. 3A–E). However, bacteroids from A. hypogaea were spherical (Figs. 3F, 4B, 4F, 4J). It is widely accepted that the size and shape of N2-fixing bacteroids vary widely and are controlled by the legume host rather than by the rhizobial genotype (31). The bacteroids isolated from the nodules of four out of five Papilionoideae species were not swollen and were the same size (2–4 μm) as the free-living form (Fig. 3A–E).
Fig. 3.
Examples of Differential Interference Contrast (DIC) views of Bradyrhizobium sp. DOA9 bacteroids. (A) Free-living. (B–F) Bacteroids isolated from (B) Indigofera tinctoria, (C) Macroptilium atropurpureum, (D) Aeschynomene americana, (E) Aeschynomene afraspera, and (F) Arachis hypogaea.
Fig. 4.
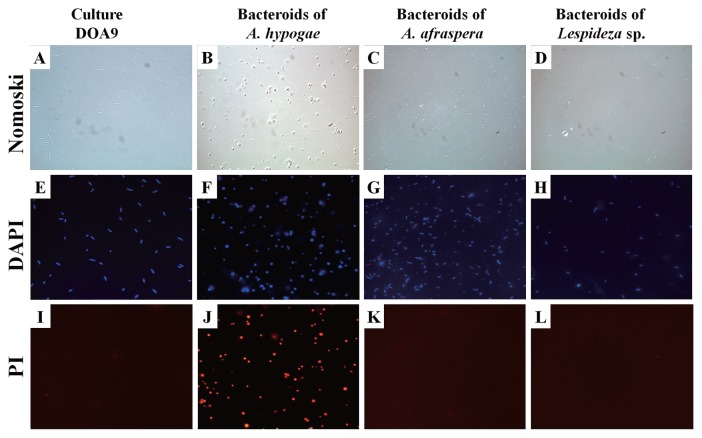
Examples of shapes of (left-hand column) free-living DOA9 bacteria and (other columns) resident bacteroids isolated from (B, F, J) Arachis hypogaea, (C, G, K) Aeschynomene afraspera, and (D, H, L) Lespedeza sp. nodules. DIC, differential interference contrast microscopy; DAPI, fluorescence microscopy with 4′,6-diamidino-2-phenylindole stain; PI, fluorescence microscopy with propidium iodide stain.
Furthermore, PI only stained the bacteroids from A. hypogaea (Fig. 4J). In addition, no colony appeared when the bacteroid fraction prepared from the nodules of A. hypogaea was plated on agar media. Although further quantitative analyses are needed, this result suggests that bacteroids in the nodules of A. hypogaea lost their productivity. These characteristics, the absence of PI staining, and the ability to form colonies on agar plates place DOA9 in the group that continues to reproduce after leaving the nodules (25). However, the bacteroids isolated from A. hypogaea were spherical and non-reproductive after leaving the nodules. Since the morphology of bacteroids (which is determined by the host legume) has been linked to reproductive viability (25), the host species can have implications for rhizobial evolution (32).
Megaplasmid detection and hybridization
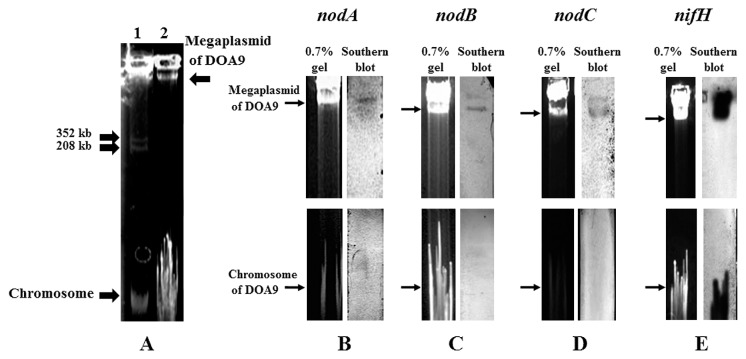
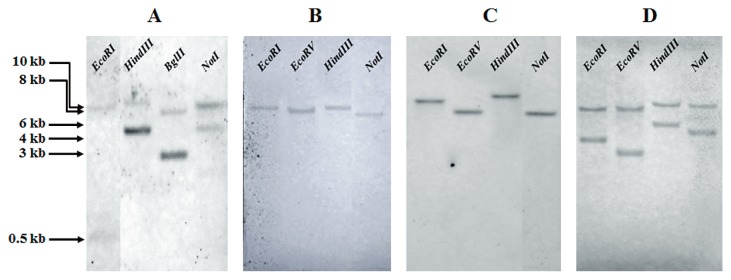
We successfully extracted both chromosomal and megaplasmid DNAs. The megaplasmid size was estimated from the plasmid profile of M. loti MAFF303099, which contained megaplasmids of 208 and 352 kb in size (16) (Fig. 5A lane 1). DOA9 revealed one megaplasmid, which was larger than 352 kb (Fig. 5A lane 2). The megaplasmid hybridized with all the probes for nodA, nodB, nodC, and nifH (Fig. 5B–E). The nifH probe also hybridized with the chromosomal DNAs (Fig. 5E).
Fig. 5.
Determination of Bradyrhizobium sp. DOA9 replicons and symbiosis genes. (A) Megaplasmid profiles: lane 1, Mesorhizobium loti MAFF303099 (208 and 352 kb); lane 2, DOA9; (B–E) Southern blot hybridization signals of nodulation and N-fixation genes on the megaplasmid and chromosome of DOA9 under low stringency conditions: (B) nodA; (C) nodB; (D) nodC; (E) nifH.
Broad-host-range symbionts generally carry multiple replicons on their genome. Sinorhizobium sp. NGR234, which has an extremely broad host range, has three replicons: a symbiosis plasmid, megaplasmid, and chromosome (12); S. fredii USDA257 has a chromosome and plasmid (36); S. fredii HH103 has one chromosome and five plasmids (42); DOA9 has a chromosome (cDOA9) and megaplasmid (pDOA9) larger than 352 kb. A large number of the natural isolates of Rhizobium spp. have been shown to carry various large plasmids (21). Genes for a few functions have been detected on these large plasmids, particularly the nod and nif genes of fast-growing rhizobia. In contrast, the published genomes of Bradyrhizobium spp. are composed of only a chromosome, except for Bradyrhizobium sp. BTAi1, which also harbors a plasmid of 228 kb (9). There have not yet been any studies published on symbiosis genes localized on the plasmids of Bradyrhizobium (21). The symbiotic islands of B. diazoefficiens USDA110 have been located on a chromosome, while the symbiosis genes of Sinorhizobium sp. were located on plasmids (13). This characteristic may contribute to the broad host range of these rhizobia. It will be interesting to find out whether symbiosis genes on plasmid have extended their host range.
Copy number
At least two fragments were detected using the probe for nodA (Fig. 6A). This result suggested that nodA on the megaplasmid was a single or two copies. Regarding nodB and nodC, a single fragment was detected on each lane, suggesting that these two genes may be a single copy on the megaplasmid (Fig. 6B, and C). At least two fragments were detectable when probed by nifH. The result supported the two nifH genes being separately located on the megaplasmid and chromosome (Figs. 5E, and 6D).
Fig. 6.
Determination of copy numbers of (A) nodA, (B) nodB, (C) nodC, and (D) nifH by Southern blot hybridization. Bradyrhizobium sp. DOA9 genomic DNA was digested with the restriction enzymes shown, and the blot was hybridized with probes for nodA (from B. yuanmingense SUTN6-2), nodB (from B. canariense SUTN7-2), nodC (from B. japonicum USDA110), and nifH (from B. yuanmingense SUTN6-2).
DOA9 has at least two copies of nifH, one on the megaplasmid and one on the chromosome. Two copies of nifH were also shown to be present in photosynthetic Bradyrhizobium ORS278 and BTAi1 (30), and one copy in B. diazoefficiens USDA110 (17), in all cases on the chromosome. We already sequenced nodA of DOA9 (Accession No. DF 820426.1), which was the corresponding fragment of the nodA probe used in this study, and no restriction sites of EcoRI, HindIII, BglII, or NotI were identified; therefore, the hybridization data probed by nodA suggested that DOA9 may have at least two copies of nodA on the megaplasmid. An increase in the copy number of the symbiotic region has been indicated to promote the plant phenotype. The inoculation of alfalfa with a moderate increase in the copy number of the symbiotic region of S. meliloti resulted in enhanced plant growth (4).
Therefore, this is the first study on the symbiosis genes of Bradyrhizobium sp. located on a megaplasmid. A detailed investigation of both replicons of DOA9 is warranted. Previous studies on the host range of rhizobia have focused on Sinorhizobium sp. strain NGR234, S. fredii USDA257, and S. fredii HH103. Since the genome sequences of these three strains share a high degree of synteny (35, 36, 42), the host range of rhizobia may be related to the number of specialized protein secretion systems they carry (35). Therefore, a future study is needed to determine and compare the protein secretion systems of DOA9.
Conclusion
Since many of the photosynthetic bradyrhizobial (PB) strains have been reported to be naturally endophytic with rice (5), we concluded that rice may have evolved approximately 120 million years before legumes (1, 20). Thus, PB strains may also be an ancestor of non-photosynthetic bradyrhizobia (41). In the present study, DOA9 exhibited the characteristic of a broad host range in legume and rice infections. Therefore, DOA9 may represent the interval of evolution in the nod factor-independent symbiotic system, but may have later lost photosynthetic activity. Moreover, DOA9 is a divergent nod-containing strain rendering the incomplete production of nod factors. This may also support DOA9 associating with host plants in a nod-independent manner since and during its evolution. To more clearly understand the evolution of this group, whole genome assessments, symbiotic genes comparisons and disruptions, as well as protein secretion systems and lipopolysaccharide structures should be explored in more detail.
Supplementary Information
Acknowledgements
This work was supported by Suranaree University of Technology; by the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission; and by a JSPS Ronpaku scholarship. Thanks are extended to Dr. Issra Pramoolsook for advice and comments on the manuscript.
References
- 1.Allen KD. Assaying gene content in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:9568–9572. doi: 10.1073/pnas.142126599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argandoña M, Martínez Checa F, Llamas I, Quesada E, Moral A. Megaplasmids in Gram negative, moderately halophilic bacteria. FEMS Microbiol Lett. 2003;227:81–86. doi: 10.1016/S0378-1097(03)00651-7. [DOI] [PubMed] [Google Scholar]
- 3.Barrett CF, Parker MA. Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica. Appl. Environ. Microbiol. 2006;72:1198–1206. doi: 10.1128/AEM.72.2.1198-1206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castillo M, Flores M, Mavingui P, Martínez-Romero E, Palacios R, Hernández G. Increase in alfalfa nodulation, nitrogen fixation, and plant growth by specific DNA amplification in Sinorhizobium meliloti. Appl Environ Microbiol. 1999;65:2716–2722. doi: 10.1128/aem.65.6.2716-2722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaintreuil C, Giraud E, Prin Y, Lorquin J, Bâ A, Gillis M, de Lajudie P, Dreyfus B. Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl Environ Microbiol. 2000;66:5437–5447. doi: 10.1128/aem.66.12.5437-5447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen WX, Yan GH, Li JL. Numerical taxonomic study of fast-growing soybean rhizobia and a proposal that Rhizobium fredii be assigned to Sinorhizobium gen. nov. Int J Syst Bacteriol. 1988;38:392–397. [Google Scholar]
- 7.Chen W, Wang E, Wang S, Li Y, Chen X, Li Y. Characteristics of Rhizobium tianshanense sp. nov., a moderately and slowly growing root nodule bacterium isolated from an arid saline environment in Xinjiang, People’s Republic of China. Int J Syst Bacteriol. 1995;45:153–159. doi: 10.1099/00207713-45-1-153. [DOI] [PubMed] [Google Scholar]
- 8.Cole MA, Elkan GH. Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrob Agents Chemother. 1973;4:248–253. doi: 10.1128/aac.4.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cytryn EJ, Jitacksorn S, Giraud E, Sadowsky MJ. Insights learned from pBTAi1, a 229-kb accessory plasmid from Bradyrhizobium sp. strain BTAi1 and prevalence of accessory plasmids in other Bradyrhizobium sp. strains. The ISME journal. 2008;2:158–170. doi: 10.1038/ismej.2007.105. [DOI] [PubMed] [Google Scholar]
- 10.Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978;1:584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- 11.Elliott GN, Chen WM, Chou JH, Wang HC, Sheu SY, Perin L, Reis VM, Moulin L, Simon MF, Bontemps C. Burkholderia phymatum is a highly effective nitrogen fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 2007;173:168–180. doi: 10.1111/j.1469-8137.2006.01894.x. [DOI] [PubMed] [Google Scholar]
- 12.Flores M, Mavingui P, Girard L, Perret X, Broughton WJ, Martínez-Romero E, Dávila G, Palacios R. Three replicons of Rhizobium sp. strain NGR234 harbor symbiotic gene sequences. J Bacteriol. 1998;180:6052–6053. doi: 10.1128/jb.180.22.6052-6053.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galibert F, Finan TM, Long SR, Pühler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 14.Goormachtig S, Capoen W, Holsters M. Rhizobium infection: lessons from the versatile nodulation behaviour of water-tolerant legumes. Trends Plant Sci. 2004;9:518–522. doi: 10.1016/j.tplants.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Hotter GS, Scott DB. Exopolysaccharide mutants of Rhizobium loti are fully effective on a determinate nodulating host but are ineffective on an indeterminate nodulating host. J Bacteriol. 1991;173:851–859. doi: 10.1128/jb.173.2.851-859.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002;9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 18.Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology. 2001;147:981–993. doi: 10.1099/00221287-147-4-981. [DOI] [PubMed] [Google Scholar]
- 19.Lavin M, Pennington RT, Klitgaard BB, Sprent JI, de Lima HC, Gasson PE. The dalbergioid legumes (Fabaceae): delimitation of a pantropical monophyletic clade. Am J Bot. 2001;88:503–533. [PubMed] [Google Scholar]
- 20.Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- 21.MacLean AM, Finan TM, Sadowsky MJ. Genomes of the symbiotic nitrogen-fixing bacteria of legumes. Plant Physiol. 2007;144:615–622. doi: 10.1104/pp.107.101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun. 2010;1:10. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manassila M, Nuntagij A, Kotepong S, Boonkerd N, Teaumroong N. Characterization and monitoring of selected rhizobial strains isolated from tree legumes in Thailand. Afr J Biotechnol. 2007;6:1393–1402. [Google Scholar]
- 24.McRae D, Miller R, Berndt W, Joy K. Transport of C4-dicarboxylates and amino acids by Rhizobium meliloti bacteroids. Mol. Plant-Microbe Interact. 1989;2:273–278. [Google Scholar]
- 25.Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, Catrice O, Mausset A.-E, Barloy-Hubler F, Galibert F, Kondorosi A. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proc Natl Acad Sci USA. 2006;103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miché L, Balandreau J. Effects of rice seed surface sterilization with hypochlorite on inoculated Burkholderia vietnamiensis. Appl Environ Microbiol. 2001;67:3046–3052. doi: 10.1128/AEM.67.7.3046-3052.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miché L, Moulin L, Chaintreuil C, Contreras Jimenez JL, Munive Hernández JA, Del Carmen M, Hernandez Villegas, Crozier F, Béna G. Diversity analyses of Aeschynomene symbionts in Tropical Africa and Central America reveal that nod independent stem nodulation is not restricted to photosynthetic bradyrhizobia. Environ Microbiol. 2010;12:2152–2164. doi: 10.1111/j.1462-2920.2009.02090.x. [DOI] [PubMed] [Google Scholar]
- 28.Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B, Gillis M, de Lajudie P, Masson-Boivin C. Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulated species and form a separate 16S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol. 1999;65:3084–3094. doi: 10.1128/aem.65.7.3084-3094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noisangiam R, Teamtisong K, Tittabutr P, Boonkerd N, Toshiki U, Minamisawa K, Teaumroong N. Genetic diversity, symbiotic evolution, and proposed infection process of Bradyrhizobium strains isolated from root nodules of Aeschynomene americana L. in Thailand. Appl. Environ. Microbiol. 2012;78:6236–6250. doi: 10.1128/AEM.00897-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nzoué A, Miché L, Klonowska A, Laguerre G, de Lajudie P, Moulin L. Multilocus sequence analysis of bradyrhizobia isolated from Aeschynomene species in Senegal. Syst Appl Microbiol. 2009;32:400–412. doi: 10.1016/j.syapm.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Oke V, Long SR. Bacteroid formation in the Rhizobium legume symbiosis. Curr Opin Microbiol. 1999;2:641–646. doi: 10.1016/s1369-5274(99)00035-1. [DOI] [PubMed] [Google Scholar]
- 32.Oono R, Schmitt I, Sprent JI, Denison RF. Multiple evolutionary origins of legume traits leading to extreme rhizobial differentiation. New Phytol. 2010;187:508–520. doi: 10.1111/j.1469-8137.2010.03261.x. [DOI] [PubMed] [Google Scholar]
- 33.Plazinski J, Cen YH, Rolfe BG. General method for the identification of plasmid species in fast-growing soil microorganisms. Appl Environ Microbiol. 1985;49:1001–1003. doi: 10.1128/aem.49.4.1001-1003.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Vol. 2. Cold spring harbor laboratory press; New York: 1989. [Google Scholar]
- 35.Schmeisser C, Liesegang H, Krysciak D, Bakkou N, Le Quéré A, Wollherr A, Heinemeyer I, Morgenstern B, Pommerening-Röser A, Flores M. Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl Environ Microbiol. 2009;75:4035–4045. doi: 10.1128/AEM.00515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuldes J, Orbegoso MR, Schmeisser C, Krishnan HB, Daniel R, Streit WR. Complete genome sequence of the broad-host-range strain Sinorhizobium fredii USDA257. J Bacteriol. 2012;194:4483–4483. doi: 10.1128/JB.00966-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somasegaran P, Hoben HJ. Handbook for rhizobia: methods in legume-Rhizobium technology. Springer-Verlag New York Inc; New York: 1994. [Google Scholar]
- 38.Sprent JI. Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytol. 2007;174:11–25. doi: 10.1111/j.1469-8137.2007.02015.x. [DOI] [PubMed] [Google Scholar]
- 39.Stowers MD, Elkan G. Criteria for selecting ineffective and efficient strains of Rhizobium for use intropical agriculture. Technical Bulletin 1980 [Google Scholar]
- 40.Thomas P, Golly K, Zyskind J, Virginia R. Variation of clonal, mesquite-associated rhizobial and bradyrhizobial populations from surface and deep soils by symbiotic gene region restriction fragment length polymorphism and plasmid profile analysis. Appl Environ Microbiol. 1994;60:1146–1153. doi: 10.1128/aem.60.4.1146-1153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Berkum P, Tully RE, Keister DL. Nonpigmented and bacteriochlorophyll-containing bradyrhizobia isolated from Aeschynomene indica. Appl Environ Microbiol. 1995;61:623–629. doi: 10.1128/aem.61.2.623-629.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weidner S, Becker A, Bonilla I, Jaenicke S, Lloret J, Margaret I, Pühler A, Ruiz-Sainz JE, Schneiker-Bekel S, Szczepanowski R. Genome sequence of the soybean symbiont Sinorhizobium fredii HH103. J Bacteriol. 2012;194:1617–1618. doi: 10.1128/JB.06729-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuttavanichakul W, Lawongsa P, Wongkaew S, Teaumroong N, Boonkerd N, Nomura N, Tittabutr P. Improvement of rhizobail inoculants by incorporation biocontrol of seed borne pathogenic fungi of peanut. Biol Control. 2012;63:87–97. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.