Abstract
Pseudomonas fluorescens Pf0-1 exhibited chemotactic responses to l-malate, succinate, and fumarate. We constructed a plasmid library of 37 methyl-accepting chemotaxis protein (MCP) genes of P. fluorescens Pf0-1. To identify a MCP for l-malate, the plasmid library was screened using the PA2652 mutant of Pseudomonas aeruginosa PAO1, a mutant defective in chemotaxis to l-malate. The introduction of Pfl01_0728 and Pfl01_3768 genes restored the ability of the PA2652 mutant to respond to l-malate. The Pfl01_0728 and Pfl01_3768 double mutant of P. fluorescens Pf0-1 showed no response to l-malate or succinate, while the Pfl01_0728 single mutant did not respond to fumarate. These results indicated that Pfl01_0728 and Pfl01_3768 were the major MCPs for l-malate and succinate, and Pfl01_0728 was also a major MCP for fumarate. The Pfl01_0728 and Pfl01_3768 double mutant unexpectedly exhibited stronger responses toward the tomato root exudate and amino acids such as proline, asparagine, methionine, and phenylalanine than those of the wild-type strain. The ctaA, ctaB, ctaC (genes of the major MCPs for amino acids), Pfl01_0728, and Pfl01_3768 quintuple mutant of P. fluorescens Pf0-1 was less competitive than the ctaA ctaB ctaC triple mutant in competitive root colonization, suggesting that chemotaxis to l-malate, succinate, and/or fumarate was involved in tomato root colonization by P. fluorescens Pf0-1.
Keywords: chemotaxis, Pseudomonas fluorescens, root colonization, methyl-accepting chemotaxis protein, plant-microbe interaction
Chemotaxis involves the movement of an organism toward chemical attractants and away from chemical repellents (2). Since many chemical attractants are growth substrates (15, 21, 22), chemotaxis is believed to assist bacterial cells in moving to areas suitable for growth. Bacterial chemotaxis can be also viewed as an important prelude to ecological interactions such as symbiosis, infection, and root colonization (7). Previous studies demonstrated that chemotaxis was involved in nodulation by Rhizobium leguminosarum (20), plant infection by Ralstonia solanacearum (38), and root colonization by Pseudomonas fluorescens (9, 25).
Certain strains of P. fluorescens belong to plant growth-promoting rhizobacteria (PGPR) (17, 24). They exert beneficial effects on plants by preventing the growth or actions of plant-pathogenic microorganisms such as Pythium ultimum (1, 30), Gaeumannomyces graminis (36, 37), and Fusarium oxysporum (6). Efficient root colonization by PGPR strains is assumed to be essential for the biocontrol of these root pathogens (35). The roles of motility and chemotaxis in root colonization by P. fluorescens have been reported previously. Barahona et al. demonstrated that a hyper motile mutant of P. fluorescens F113 was more competitive for rhizosphere colonization than the wild-type strain and exhibited improved biocontrol activity against F. oxysporum (5). Conversely, de Weert et al. reported that a general chemotaxis mutant (the cheA mutant) of P. fluorescens WCS365 colonized the tomato root tip less efficiently than the wild-type strain (9). Plant root exudates contain various organic compounds and the major components of the tomato root exudate are amino acids (glutamic acid, aspartic acid, leucine, isoleucine, and lysine as the major components [32]), organic acids (especially citric acid, malic acid, and succinic acid [13]), and sugars (glucose and xylose as the major components [18]). P. fluorescens strains have been shown to exhibit chemotactic responses toward plant seeds, root exudates, and their components (9, 25, 33, 35). Therefore, chemotaxis to these components may play a role in effective root colonization.
Methyl-accepting chemotaxis proteins (MCPs) are chemotaxis sensory proteins that are responsible for the detection of chemotactic ligands (10). Chemotactic ligands bind to the periplasmic domains of MCPs and their binding initiates chemotactic signaling. A genome sequence analysis of P. fluorescens Pf0-1 (accession number: CP000094) suggested the presence of 37 MCPs. We previously identified CtaA (Pfl01_4431), CtaB (Pfl01_0124), and CtaC (Pfl01_0354) as MCPs for amino acids in P. fluorescens Pf0-1 (25). Chemotaxis toward 18 naturally-occurring amino acids was found to be defective in the ctaA ctaB ctaC triple mutant of P. fluorescens Pf0-1 (designated FLD3), but it still showed decreased, but significant responses to proline and cysteine. In competitive tomato root colonization assays, FLD3 was less competitive than the wild-type strain, whereas this strain was more competitive than the cheA mutant of P. fluorescens Pf0-1, which is non-chemotactic, but motile. These findings suggested that chemotaxis to amino acids was involved in root colonization and there were still chemoattractants other than the 18 amino acids involved in root colonization by P. fluorescens Pf0-1.
We assumed that chemotaxis to root exudate components other than amino acids would also be involved in effective root colonization in P. fluorescens; however, no studies have investigated the relationship between root colonization and chemotaxis to sugars and organic acids in soil bacteria including P. fluorescens. Thus, we first measured chemotactic responses to various organic acids and sugars and found that P. fluorescens Pf0-1 showed strong responses to l-malate, succinate, and fumarate. We then identified MCPs for these dicarboxylic acids in P. fluorescens Pf0-1. We also assessed the involvement of chemotaxis to organic acids in tomato root colonization by competitive root colonization assays using P. fluorescens Pf0-1 mutant strains defective in chemotaxis to l-malate, succinate, and fumarate.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli JM109 (27) and S17-1 (31) were used for plasmid construction and transconjugation, respectively. P. fluorescens, Pseudomonas aeruginosa, and E. coli strains were grown with shaking in 2×YT medium (27) supplemented with appropriate antibiotics. P. aeruginosa and E. coli strains were cultivated at 37°C, while P. fluorescens strains were grown at 28°C.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference (s) |
|---|---|---|
| Pseudomonas fluorescens | ||
| Pf0-1 | wild-type strain | (8) |
| Pf01Rif | Pf0-1 derivative, spontaneous rifampicin-resistant mutant | (25) |
| FLD3 | Pf0-1 derivative, ΔctaA (Pfl01_4431) ΔctaB (Pfl01_0124) ΔctaC (Pfl01_0354) | (25) |
| FLD3Rif | FLD3 derivative, spontaneous rifampicin-resistant mutant, ΔctaA ΔctaB ΔctaC | (25) |
| KPF09 | Pf0-1 derivative, ΔmcpS (Pfl01_0728) | This study |
| KPF21 | Pf0-1 derivative, ΔmcpT (Pfl01_3768) | This study |
| OX1 | Pf0-1 derivative, ΔmcpS ΔmcpT | This study |
| FLD5 | Pfl0-1 derivative, ΔctaA ΔctaB ΔctaC ΔmcpS ΔmcpT | This study |
| FLD5Rif | FLD5 derivative, spontaneous rifampicin-resistant mutant, ΔctaA ΔctaB ΔctaC ΔmcpS ΔmcpT | This study |
| Pf0-1 ΔcheA | Pf0-1 derivative, ΔcheA (Pfl01_1566) | (25) |
| Pseudomonas aeruginosa | ||
| PAO1 | wild-type strain | (12) |
| RPc | PAO1 derivative, PA2652 gene knockout mutant | This study |
| Escherichia coli | ||
| JM109 | recA1, endA1, gyrA96, thi-1, hsdR17 (rk− mk+), e14− (mcrA−), supE44, relA1, Δ (lac-proAB)/F′[traD36, proAB+, lacIq, lacZ ΔM15] | (27) |
| S17-1 | MM294, RP4-2 Tc::Mu-Km::Tn7 chromosomally integrated | (31) |
| Plasmids | ||
| pUCP18 | Escherichia-Pseudomonas shuttle vector; Cbr | (29) |
| pFLCP09 | pUCP18 with a 2.5 kb PCR fragment containing mcpS (Pfl01_0728); Cbr | This study |
| pFLCP21 | pUCP18 with a 2.1 kb PCR fragment containing mcpT (Pfl01_3768); Cbr | This study |
| pK18mobsacB | Kmr pUCP18 derivative, lacZα, mob site, sacB | (28) |
| pNMFL09 | pK18mobsacB with a 1.1-kb PCR fragment upstream of mcpS and 0.9-kb PCR fragment downstream of mcpS; Kmr | This study |
| pNMFL21 | pK18mobsacB with a 1.5-kb PCR fragment upstream of mcpT and 1.2-kb PCR fragment downstream of mcpT; Kmr | This study |
| pNMPAR | pK18mobsacB with a 1.6-kb PCR fragment upstream of the PA2652 gene and 1.2-kb PCR fragment downstream of the PA2652 gene; Kmr | This study |
Cbr, carbenicillin resistance; Kmr, kanamycin resistance.
Chemotaxis assay
The computer-assisted capillary assay method was performed as described previously (23). Cells in a 10-μL suspension were placed on a coverslip, and the assay was started by placing the coverslip upside down on the U-shaped spacer to fill the chemotaxis chamber with the cell suspension. Cells were videotaped over 1.5 min. Digital image processing was used to count the number of bacteria accumulating toward the mouth of a capillary containing a known concentration of an attractant plus 1% (w/v) agarose. The strength of the chemotactic response was determined by the number of bacterial cells per frame. The chemotaxis buffer was 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-ethanesulfonic acid) buffer (pH 7.0). We selected test compounds for chemotactic responses by P. fluorescens Pf0-1 based on previous findings by Kamilova et al. (13), which included 21 organic acids and 5 sugars (Table 2).
Table 2.
Chemotactic responses of P. fluorescens Pf0-1 to plant-associated compounds
| Compoundsa | Chemotactic responsesb |
|---|---|
| l-(−)-malic acid | + + + |
| d-(+)-malic acid | − |
| fumaric acid | + + |
| maleic acid | − |
| succinic acid | + + |
| oxaloacetic acid | + |
| trans-aconitic acid | − |
| citric acid | + |
| acetic acid | − |
| benzoic acid | − |
| o-hydroxy benzoic acid | − |
| m-hydroxy benzoic acid | − |
| p-hydroxy benzoic acid | − |
| l-pyroglutamic acid | − |
| phenoxyacetic acid | − |
| syringic acid | − |
| protocatechuic acid | − |
| d-(−)-quinic acid | − |
| trans-ferulic acid | − |
| shikimic acid | − |
| γ-aminobutyric acid | − |
| glucose | − |
| fructose | − |
| maltose | − |
| ribose | − |
| xylose | − |
| Serine | +++ |
| Cysteine | +++ |
Compounds other than trans-ferulic acid were used at a concentration of 5 mM. Trans-ferulic acid was provided as a saturated solution in 10 mM HEPES buffer (pH 7.0).
Videotape frames were analyzed at the initiation of observations and 1 min later. Normalized cell numbers were calculated by dividing the number of bacteria at 1 min by that at the initiation of observations. The value of the normalized cell number was represented by the symbols, as follows: + + + > 4; 4 ≥ + + > 2; 2 ≥ + > 1.5; 1.5 ≥ − > 0. Serine and cysteine were positive controls.
DNA manipulation
Standard procedures were used for plasmid DNA preparations, restriction enzyme digestions, ligations, transformations, and agarose gel electrophoresis (27). PCR was conducted using KOD Plus DNA polymerase (Toyobo, Tokyo, Japan) according to the manufacturer’s instructions. The oligonucleotides used for PCR are listed in Table S1. P. aeruginosa was transformed by electroporation as described previously (19). Plasmids were introduced to P. fluorescens strains by transconjugation using E. coli S17-1 (31).
Plasmid construction and construction of deletion mutants of P. fluorescens Pf0-1
The Pfl01_0728 and Pfl01_3768 genes were amplified from the P. fluorescens Pf0-1 genome by PCR using the FLCP09f/FLCP09r and FLCP21f/FLCP21r primer sets, and then cloned into broad-host-range plasmid pUCP18 (29) to construct pFLCP09 and pFLCP21, respectively. Suicide plasmids pNMFL09 and pNMFL21 were constructed for unmarked gene deletion in P. fluorescens Pf0-1. PCR using the primer sets NM09Uf/NM09Ur and NM09Df/NM09Dr was conducted to amplify a 1.1-kb upstream region and 0.9-kb downstream region of Pfl01_0728 from the P. fluorescens Pf0-1 genome, respectively. The amplified upstream and downstream regions were digested with SalI-BamHI and XhoI-EcoRI, respectively, and ligated with the backbone of BamHI-EcoRI-digested pK18mobsacB (28) to obtain pNMFL09. PCR using the primer sets NM21Uf/NM21Ur and NM21Df/NM21Dr was conducted to amplify a 1.5-kb upstream region and 1.2-kb downstream region of Pfl01_3768 from the P. fluorescens Pf0-1 genome, respectively. The amplified upstream and downstream regions were digested with XbaI-EcoRI and HindIII-XbaI, respectively, and ligated with the backbone of EcoRI-HindIII-digested pK18mobsacB to obtain pNMFL21. The chromosomal Pfl01_0728 and Pfl01_3768 genes were deleted by an unmarked gene deletion technique (28) using suicide plasmids pNMFL09 and pNMFL21, respectively.
Preparation of the tomato root exudate
An exudate was prepared from the plant species tomato (Solanum lycopersicum cv. Oogatafukuju). Tomato seeds were sterilized by gentle shaking for 10 min in a solution of 8.75% (v/v) sodium hypochlorite supplemented with 0.1% (v/v) Tween 20. The sterilized seeds were soaked six times for 15 min in sterile deionized water. Nine sterile seeds were placed in 3 mL of SSE medium (4) in glass tubes (22 mm inner diameter, 25 mm outer diameter, 12 cm length), consisting of 5 mM KH2PO4, 4 mM CaSO4, 2 mM MgCl2, 2.5 mM NH4NO3, 0.5 mM KOH, 2.5 mM NaOH, and 0.02 mM Fe (as FeEDTA), and were allowed to grow in a climate-controlled growth chamber (NK system, Osaka, Japan) at 28°C with 16 h of daylight. Root exudates were collected after 18 d, evaporated to dryness at 45°C under a vacuum, dissolved in 1 mL of water, and sterilized by membrane filtration (0.45-μm pore size).
Gnotobiotic root colonization assays
Twenty grams of quartz sand (0.1 to 0.3 mm) was placed into glass tubes (22 mm inner diameter, 25 mm outer diameter, 12 cm length) and compacted by gentle shaking. The open end of the tube was plugged with a silicone resin stopper. The tube was then autoclaved for 15 min at 121°C. Five mL PNS (plant nutrient solution) (11), consisting of 1.25 mM Ca(NO3)2, 1.25 mM KNO3, 0.5 mM MgSO4, 0.25 mM KH2PO4, and trace elements (in mg L−1): Fe (as FeEDTA), 4.6; B, 0.5; Zn, 0.05; Cu, 0.02; Mo, 0.01, was added to an autoclaved sand column. Tomato seeds (S. lycopersicum cv. Oogatafukuju) were sterilized as described in the “Preparation of tomato root exudate” section. To synchronize germination, seeds were placed on Petri dishes containing PNS solidified with 1.5% (w/v) BactoTM Agar (Becton, Dickinson and Company, New Jersey, USA) and incubated overnight in the dark at 4°C, followed by incubation at 28°C for 2 d. A germinated seed was aseptically placed at the center of a growth tube and 5 mm below the surface of quartz sand. Bacterial cells were grown for 14 h in 2x YT medium, centrifuged (3,300 × g, 2 min), washed three times with sterile deionized water, and adjusted to 107 CFU mL−1 in PNS. In the competitive colonization assay, 50 μL of a 1:1 (v/v) mixture of the tested strain and the competitor was mixed and inoculated to the edge of a plant growth tube. The plant growth tubes were incubated in a climate-controlled growth chamber (28°C, 16 h daylight) to allow the plantlets to grow. After 3 d of growth in the plant growth tubes, the root systems of the tomato were mostly unbranched. The root tip (1 to 2 cm in length) was removed and shaken vigorously in the presence of adhering sand particles in 0.5 mL of sterile deionized water to remove bacteria. The bacterial suspension was diluted and 100 μL was plated on 2x YT agar plates. In the competitive colonization assay, the bacterial suspension was spread on 2x YT agar plates with and without rifampicin. The nonparametric Wilcoxon-Mann-Whitney test was used for statistical analyses (34).
Results and Discussion
Chemotactic responses of P. fluorescens Pf0-1 to organic acids and sugars
To identify chemoattractants other than the 18 amino acids involved in root colonization, we first measured the chemotactic responses of P. fluorescens Pf0-1 to components in tomato root exudates. P. fluorescens Pf0-1 showed the strongest response to l-malate among the compounds tested. Responses to l-malate were similar to those to serine and cysteine, the strongest attractants among amino acids (25). It also exhibited strong responses to succinate and fumarate, while citrate was a weak attractant. We did not detect any chemotactic responses of P. fluorescens Pf0-1 to the sugars examined.
Identification of MCPs for l-malate
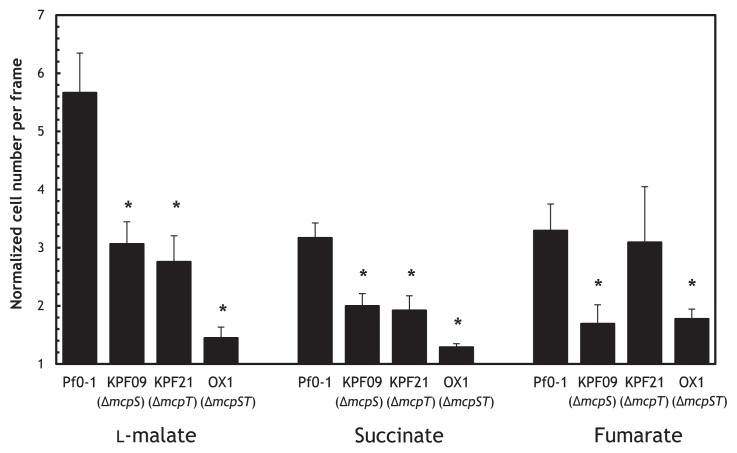
To identify a MCP(s) for organic acids, we constructed a plasmid library of thirty-seven P. fluorescens Pf0-1 putative mcp genes by cloning their PCR products into pUCP18. P. aeruginosa PAO1 PA2652 has been identified as a MCP for malate (3). Therefore, we introduced the plasmid library to the PA2652 deletion mutant of P. aeruginosa PAO1 and examined the resulting recombinant strains for their chemotactic responses to l-malate. Only the introduction of the P. fluorescens Pf0-1 Pfl01_0728 and Pfl01_3768 genes restored the ability of the P. aeruginosa PAO1 PA2652 mutant to respond to l-malate (Fig. 1). To confirm the functions of Pfl01_0728 and Pfl01_3768 as MCPs for l-malate, we disrupted the chromosomal Pfl01_0728 and Pfl01_3768 genes in P. fluorescens Pf0-1 to construct Pfl01_0728 and Pfl01_3768 single mutants (designated KPF09 and KPF21, respectively) and tested them for chemotactic responses to l-malate. The responses of both mutants to l-malate were weaker than that of the P. fluorescens Pf0-1 wild-type strain (Fig. 2). Moreover, the Pfl01_0728 and Pfl01_3768 double mutant (designated OX1) did not show any responses to l-malate, confirming that Pfl01_0728 and Pfl01_3768 were the major MCPs for l-malate in P. fluorescens Pf0-1. Both of the single mutants showed significantly weaker responses to succinate than the wild-type strain while the double mutant exhibited no response to succinate (Fig. 2). The Pfl01_0728 single mutant (KPF09) showed markedly decreased responses to fumarate, while the deletion mutation of Pfl01_3768 did not significantly affect chemotactic responses to fumarate (Fig. 2). We demonstrated that Pfl01_0728 acted as the major MCP for succinate and fumarate as well as l-malate while Pfl01_3768 was the MCP for l-malate and succinate in P. fluorescens Pf0-1.
Fig. 1.
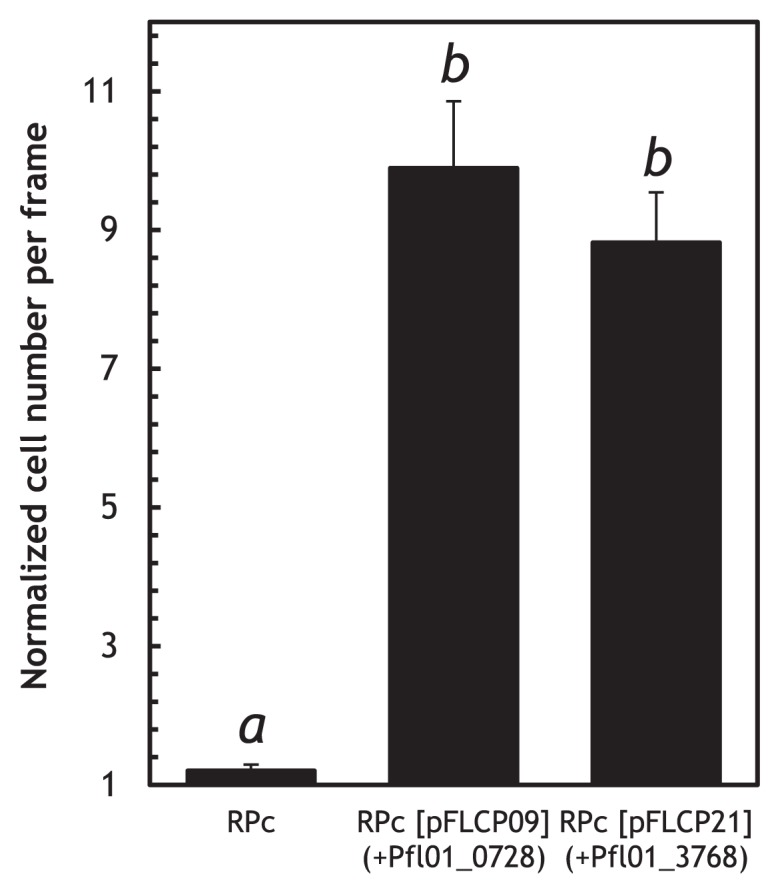
Chemotactic responses to 5 mM l-malate by P. aeruginosa RPc (PA2652 mutant), RPc [pFLCP09], and RPc [pFLCP21]. Digital image processing was used to count the number of bacteria around the mouth of a capillary containing 5 mM l-malate and 1% (w/v) agarose. Videotape frames were analyzed at the initiation of observations and 1 min later. Normalized cell numbers were calculated by dividing the number of bacterial cells at 1 min by that at the initiation of observations. Vertical bars represent the standard errors of measurements from two independent experiments conducted in triplicate. Different letters indicate significant differences as calculated by the Student’s t-test (P < 0.01).
Fig. 2.
Chemotactic responses to 5 mM l-malate, 5 mM succinate, and 5 mM fumarate by P. fluorescens Pf0-1 wild-type, KPF09 (the mcpS single mutant), KPF21 (the mcpT single mutant), and OX1 (the mcpS mcpT double mutant).
Videotape frames were analyzed at the initiation of observations and 1 min later. Vertical bars represent the standard errors of measurements from at least two independent experiments conducted in triplicate. Asterisks indicate the chemotactic responses in the wild-type and mutants were significantly different (Student’s t-test, P < 0.05).
MCPs are membrane-spanning homodimers and the typical features of MCPs are as follows: a positively charged N terminus followed by a hydrophobic membrane-spanning region, a hydrophilic periplasmic domain, a second hydrophobic membrane-spanning region, and a hydrophilic cytoplasmic domain (10). Chemotactic ligands bind to the periplasmic domains of MCPs and their binding initiates chemotactic signaling. The diverse ligand specificities among MCPs reflect amino acid sequence diversities in the periplasmic domains of MCPs. Both Pfl01_0728 and Pfl01_3768 are typical MCPs and showed the features described above. We performed a BLASTP analysis on the protein database in the National Center for Biotechnology Information using the putative periplasmic domains of Pfl01_0728 (249 amino acids, residues 42 to 290 of Pfl01_0728) and Pfl01_3768 (171 amino acids, residues 33 to 203) as query sequences. The BLASTP analysis revealed that there was no significant similarity between the periplasmic domains of Pfl01_0728 and Pfl01_3768 and other P. fluorescens strains including F113 (accession number of the genomic sequence: CP003150), SBW25 (AM181176), WH66 (AEAZ00000000), and A506 (CP003041) possess orthologs of both Pfl01_0728 and Pfl01_3768 with up to 86% identity. The periplasmic domain of Pfl01_0728 showed 49 and 48% identities to Pseudomonas putida KT2440 McpS (PP_4658) and P. putida F1 McfS (Pput_4520), respectively, both of which were previously reported to be MCPs for malate, succinate, and fumarate (16, 26). The Pfl01_3768 periplasmic domain shares 57% identity with that of P. aeruginosa PAO1 PA2652, which has already been identified as a MCP for malate (3). Based on these findings, the Pfl01_0728 and Pfl01_3768 genes were designated mcpS and mcpT, respectively. All these MCPs sensed malate. P. putida KT2440 McpS sensed succinate and acetate, while P. putida F1 McfS also sensed succinate, citrate, and fumarate as well as malate. P. aeruginosa PAO1 did not possess an ortholog of McpS/McfS while P. putida KT2440 and P. putida F1 did not possess an ortholog of PA2562. P. fluorescens Pf0-1 had orthologs of both McpS/McfS and PA2562. Genomic data indicated that other P. fluorescens strains also had both McpS/McfS and PA2562 orthologs. These results imply that chemotaxis to organic acids plays an important role in P. fluorescens strains.
The mcpS mcpT double mutant exhibited enhanced chemotactic responses to the tomato root exudate and specific amino acids
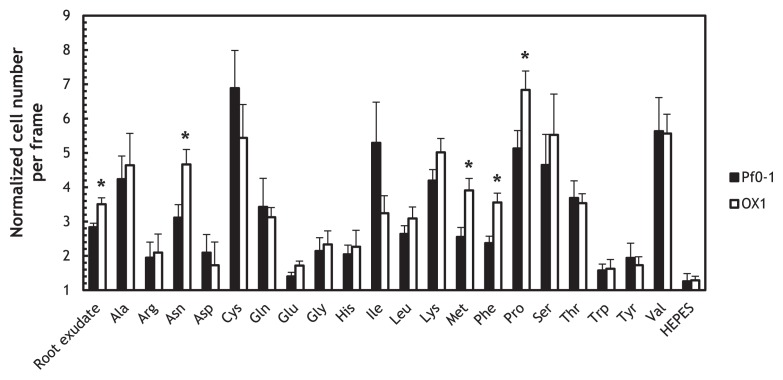
We previously showed that the ctaA ctaB ctaC triple mutant exhibited decreased chemotactic responses to the tomato root exudate (25). A previous study reported that organic acids including l-malate and succinate were components in the root exudate (13); therefore, we predicted that the mcpS mcpT mutant (OX1) would also show decreased responses to the tomato root exudate. However, the responses of OX1 to the tomato root exudate were significantly stronger than those of wild-type Pf0-1 (P < 0.05) (Fig. 3). To determine whether chemotactic responses were generally enhanced in the mcpS mcpT mutant, we measured the chemotactic responses of OX1 to 20 naturally-occurring amino acids (Fig. 3). The results obtained indicated that OX1 only showed significantly stronger responses to specific amino acids, including proline, phenylalanine, methionine, and asparagine, that those of wild-type Pf0-1 (P < 0.05). We previously demonstrated that CtaA, CtaB, and CtaC sensed 16 amino acids, 16 amino acids, and 5 amino acids, respectively (25). Only CtaA sensed proline, phenylalanine, methionine, and asparagine. However, since the mcpS mcpT double mutation did not affect chemotactic responses to 16 amino acids, it is currently difficult to explain the enhanced responses to four amino acids in OX1 only by the upregulation of ctaA. We speculate that the mcpS mcpT mutation may have enhanced the expression of MCP(s) for amino acids other than CtaA, CtaB, and CtaC, and that it may be responsible for the increased responses observed to these four amino acids in OX1.
Fig. 3.
Chemotactic responses of P. fluorescens Pf0-1 wild-type (solid bars) and OX1 (open bars) to the root exudate, 0.5 mM naturally-occurring amino acids, and 10 mM HEPES (control).
Videotape frames were analyzed at the initiation of observations and 1 min later. Vertical bars represent the standard errors of measurements from at least two independent experiments conducted in triplicate. Asterisks indicate the chemotactic responses in the wild-type and OX1 were significantly different (Student’s t-test, P < 0.05).
Involvement of chemotaxis to malate, succinate, and/or fumarate in tomato root colonization by P. fluorescens Pf0-1
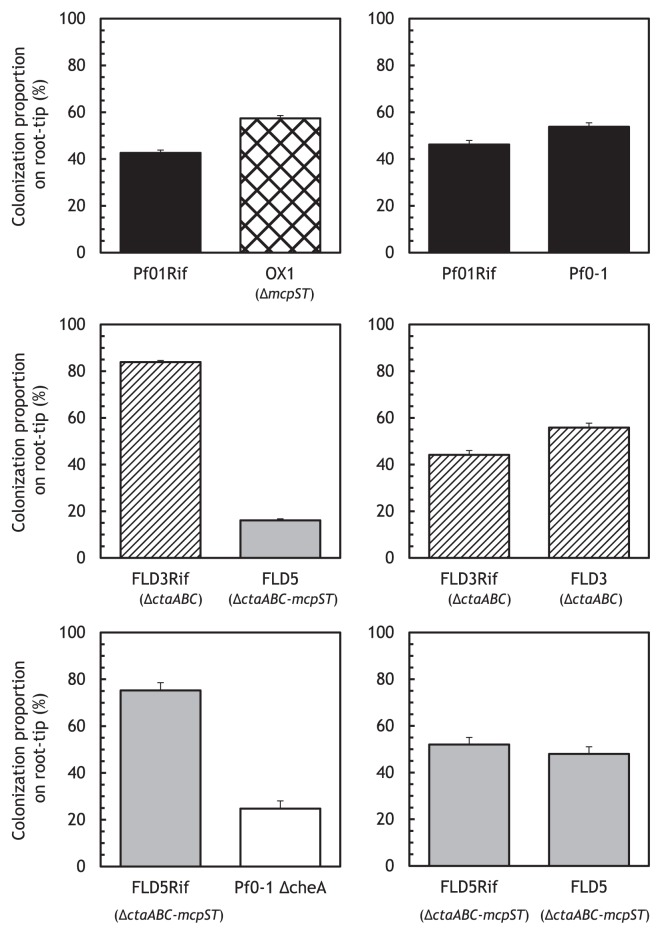
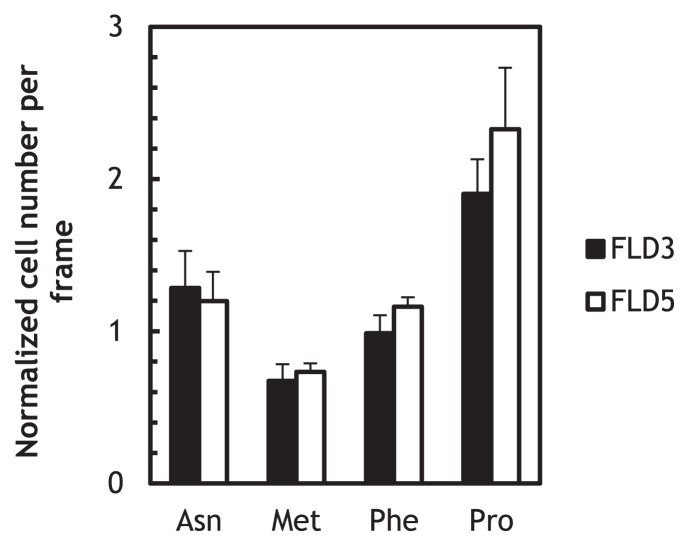
To investigate whether chemotaxis to l-malate, succinate, and/or fumarate were involved in tomato root colonization, we performed competitive root colonization assays by inoculating tomato seedlings with a 1:1 mixture of a test strain and competitor strain in a gnotobiotic root colonization system. We confirmed that there were no significant differences in growth in PNS medium supplemented with glucose between mutants and the wild-type Pf0-1 (Fig. S1). Since Pf01Rif and FLD3Rif (spontaneous rifampicin-resistant mutants of Pf0-1 and FLD3 [the ctaA ctaB ctaC mutant of Pf0-1]) competed fully with Pf0-1 and FLD3, respectively (Fig. 4), we used Pf01Rif and FLD3Rif as competitor strains in competitive colonization assays to distinguish competitor strains from test strains. OX1 (the mcpS mcpT mutant) was significantly more competitive than Pf01Rif (Fig. 4). Since OX1 exhibited enhanced chemotactic responses to four amino acids, it was not possible to assess whether chemotaxis to organic acids was involved in root colonization based on this result. We found that the mcpS mcpT double mutation did not affect chemotactic responses to the four amino acids in the ctaA ctaB ctaC mutant background (i.e. the ctaABC-mcpS mcpT quintuple mutant, designated FLD5) (Fig. 5). Therefore, we evaluated the contribution of chemotaxis toward dicarboxylic acids to root colonization in competitive root colonization assays between the ctaABC triple mutant and ctaABC-mcpS mcpT quintuple mutant (FLD5) (Fig. 4). The quintuple mutant FLD5 showed a significantly reduced ability to colonize tomato root in competitive root colonization assays using the ctaABC triple mutant (FLD3) as the competitor strain (P < 0.01) (Fig. 4). This result indicated that chemotaxis to l-malate, succinate, and/or fumarate was involved in effective root colonization by P. fluorescens Pf0-1. However, the quintuple mutant was more competitive than the cheA mutant in the competitive root colonization assay, suggesting that there were chemotactic compounds other than the 18 amino acids and dicarboxylic acids (l-malate, succinate, and fumarate) that were involved in root colonization by P. fluorescens Pf0-1. Kamilova et al. reported that F. oxysporum altered the metabolism of organic acids in tomato plants and increased the amount of succinate (14). In such a case, chemotaxis to succinate may contribute more to the migration of P. fluoresces to plant roots in soil.
Fig. 4.
Tomato root tip colonization by P. fluorescens strains in competition with Rifr spontaneous mutants. Root systems were sampled in at least three independent experiments conducted in triplicate. Vertical bars represent the standard errors of measurements. Significant differences were observed in colonization between Pf01Rif and OX1 (P < 0.05), FLD3Rif and FLD5 (P < 0.01), and FLD5Rif and Pf0-1 ΔcheA (P < 0.01). The nonparametric Wilcoxon-Mann-Whitney test was used for statistical analyses.
Fig. 5.
Chemotactic responses of P. fluorescens FLD3 (solid bars) and FLD5 (open bars) to 0.5 mM l-asparagine, 0.5 mM l-methionine, 0.5 mM l-phenylalanine, and 0.5 mM l-proline.
Videotape frames were analyzed at the initiation of observations and 1 min later. Vertical bars represent the standard errors of measurements from at least two independent experiments conducted in triplicate.
To identify the compounds involved in root colonization by P. fluorescens Pf0-1, we mainly focused on the strong chemoattractants of this bacterium in our present and previous studies (25). Strong chemoattractants were selected based on chemotaxis assay data using P. fluorescens Pf0-1 cells grown in basal salt medium (T0 medium). However, the chemotactic response profile by cells grown in T0 medium may not necessarily be identical to that by cells existing in the gnotobiotic root colonization assay system due to a difference in the mcp gene expression profile. mcp gene expression profiles in soil environments should be taken into consideration when identifying additional chemotactic compounds involved in root colonization by P. fluorescens.
Supplementary Information
Acknowledgements
This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry in Bio-oriented Technology Research Advancement Institution (BRAIN), Japan, and Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry in Ministry of Agriculture, Forestry and Fisheries of Japan.
References
- 1.Aarons S, Abbas A, Adams C, Fenton A, O’Gara F. A regulatory RNA (Prrb RNA) modulates expression of secondary metabolite genes in Pseudomonas fluorescens F113. J Bacteriol. 2000;182:3913–3919. doi: 10.1128/jb.182.14.3913-3919.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler J. Chemotaxis in bacteria. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Ortega C, Harwood CS. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl Environ Microbiol. 2007;73:7793–7795. doi: 10.1128/AEM.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angle JS, McGrath SP, Chaney RL. New culture medium containing ionic concentrations of nutrients similar to concentrations found in the soil solution. Appl Environ Microbiol. 1991;57:3674–3676. doi: 10.1128/aem.57.12.3674-3676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barahona E, Navazo A, Martínez-Granero F, Zea-Bonilla T, Pérez-Jiménez RM, Martín M, Rivilla R. Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl Environ Microbiol. 2011;77:5412–5419. doi: 10.1128/AEM.00320-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolwerk A, Lagopodi AL, Wijfjes AH, Lamers G, Chin-A-Woeng TF, Lugtenberg BJ, Bloemberg GV. Interactions in the tomato rhizosphere of two Pseudomonas biocontrol strains with the phytopathogenic fungus Fusarium oxysporum f. sp. radicislycopersici. Mol Plant Microbe Interact. 2003;16:983–993. doi: 10.1094/MPMI.2003.16.11.983. [DOI] [PubMed] [Google Scholar]
- 7.Chet I, Mitchell R. Ecological aspects of microbial chemotactic behavior. Annu Rev Microbiol. 1976;30:221–239. doi: 10.1146/annurev.mi.30.100176.001253. [DOI] [PubMed] [Google Scholar]
- 8.Compeau G, Al-Achi BJ, Platsouka E, Levy SB. Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl Environ Microbiol. 1988;54:2432–2438. doi: 10.1128/aem.54.10.2432-2438.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Weert S, Vermeiren H, Mulders IH, Kuiper I, Hendrickx N, Bloemberg GV, Vanderleyden J, De Mot R, Lugtenberg BJ. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant Microbe Interact. 2002;15:1173–1180. doi: 10.1094/MPMI.2002.15.11.1173. [DOI] [PubMed] [Google Scholar]
- 10.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffland E, Findenegg GR, Nelemans JA. Solubilization of rock phosphate by rape. Plant Soil. 1989;113:161–165. [Google Scholar]
- 12.Holloway BW, Krishnapillai V, Morgan AF. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamilova F, Kravchenko LV, Shaposhnikov AI, Azarova T, Makarova N, Lugtenberg B. Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol Plant Microbe Interact. 2006;19:250–256. doi: 10.1094/MPMI-19-0250. [DOI] [PubMed] [Google Scholar]
- 14.Kamilova F, Kravchenko LV, Shaposhnikov AI, Makarova N, Lugtenberg B. Effects of the tomato pathogen Fusarium oxysporum f. sp. radicis-lycopersici and of the biocontrol bacterium Pseudomonas fluorescens WCS365 on the composition of organic acids and sugars in tomato root exudate. Mol Plant Microbe Interact. 2006;19:1121–1126. doi: 10.1094/MPMI-19-1121. [DOI] [PubMed] [Google Scholar]
- 15.Kato J, Ito A, Nikata T, Ohtake H. Phosphate taxis in Pseudomonas aeruginosa. J Bacteriol. 1992;174:5149–5151. doi: 10.1128/jb.174.15.5149-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacal J, Alfonso C, Liu X, Parales RE, Morel B, Conejero-Lara F, Rivas G, Duque E, Ramos J, Krell T. Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands. J Biol Chem. 2010;285:23126–23136. doi: 10.1074/jbc.M110.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lugtenberg BJJ, de Weger LA, Bennett JW. Microbial stimulation of plant growth and protection from disease. Curr Opin Biotechnol. 1991;2:457–464. [Google Scholar]
- 18.Lugtenberg BJJ, Kravchenko LV, Simons M. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ Microbiol. 1999;1:439–446. doi: 10.1046/j.1462-2920.1999.00054.x. [DOI] [PubMed] [Google Scholar]
- 19.Masduki A, Nakamura J, Ohga T, Umezaki R, Kato J, Ohtake H. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J Bacteriol. 1995;177:948–952. doi: 10.1128/jb.177.4.948-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller LD, Yost CK, Hynes MF, Alexandre G. The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol Microbiol. 2007;63:348–362. doi: 10.1111/j.1365-2958.2006.05515.x. [DOI] [PubMed] [Google Scholar]
- 21.Moench TT, Konetzka WA. Chemotaxis in Pseudomonas aeruginosa. J Bacteriol. 1979;133:427–429. doi: 10.1128/jb.133.1.427-429.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moulton RC, Montie TC. Chemotaxis by Pseudomonas aeruginosa. J Bacteriol. 1978;137:274–280. doi: 10.1128/jb.137.1.274-280.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikata T, Sumida K, Kato J, Ohtake H. Rapid method for analyzing bacterial behavioral responses to chemical stimuli. Appl Environ Microbiol. 1992;58:2250–2254. doi: 10.1128/aem.58.7.2250-2254.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Sullivan DJ, O’Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oku S, Komatsu A, Tajima T, Nakashimada Y, Kato J. Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ. 2012;27:462–469. doi: 10.1264/jsme2.ME12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parales RE, Luu RA, Chen GY, Liu X, Wu V, Lin P, Hughes JG, Nesteryuk V, Parales JV, Ditty JL. Pseudomonas putida F1 has multiple chemoreceptors with overlapping specificity for organic acids. Microbiology. 2013;159:1086–1096. doi: 10.1099/mic.0.065698-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; New York, NY: 1989. [Google Scholar]
- 28.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 29.Schweizer HP. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 30.Shanahan P, Borro A, O’Gara F, Glennon JD. Isolation, trace enrichment and liquid chromatographic analysis of diacetylphloroglucinol in culture and soil samples using UV and amperometric detection. J Chromatogr. 1992;606:171–177. [Google Scholar]
- 31.Simon R, Prifer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]
- 32.Simons M, Permentier HP, de Weger LA, Wijffelman CA, Lugtenberg BJJ. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol Plant Microbe Interact. 1997;10:102–106. [Google Scholar]
- 33.Singh T, Arora DK. Motility and chemotactic response of Pseudomonas fluorescens toward chemoattractants present in the exudate of Macrophomina phaseolina. Microbiol Res. 2001;156:343–351. doi: 10.1078/0944-5013-00119. [DOI] [PubMed] [Google Scholar]
- 34.Sokal RR, Rohlf FJ. Biometry. Freeman and Co; San Francisco: 1981. [Google Scholar]
- 35.Suslow TV. Role of root-colonizing bacteria in plant growth. In: Mount MS, Lacy GH, editors. Phytopathogenic Prokaryotes. Vol. 1. Academic Press; London: 1982. pp. 187–223. [Google Scholar]
- 36.Thomashow LS, Weller DM. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomashow LS, Weller DM, Bonsall RF, Pierson LS. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol. 1990;56:908–912. doi: 10.1128/aem.56.4.908-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao J, Allen C. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J Bacteriol. 2006;188:3697–3708. doi: 10.1128/JB.188.10.3697-3708.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.