Abstract
In this issue of Science Translational Medicine, the Butte Research group provides a concrete example of how reinterpreting and comparing genome-wide metrics may allow us to effectively hypothesize which drugs from one disease indication can be used for another. Here we discuss the basis of this shift toward genomic computational integrative approaches that has precedence in scalar theories of biological information and is aptly warranted for exploitation in drug repurposing.
“… it is undeniable that molecular biologists now have at their disposal the tools that are needed to unravel biological complexity and overcome the limitations of reductionism. Given our failures in developing drugs and vaccines against a wide range of debilitating diseases, this move away from the reductionist viewpoint and toolset is a high priority for both biological and biomedical research.”
– M.H.V. Van Regenmortel, 2004 (1)
In this issue of Science Translational Medicine, the Butte Research group provides a concrete and elegant example of how reinterpreting and comparing genome-wide metrics may enable effectively hypothesize which drugs from one disease indication can be used for another. Of particular significance, they demonstrate the their ability to derive novel therapeutic indications for approved drugs (2), i.e. drug “repurposing”, entirely from in-silico studies and to validate these in an animal model. It is even more remarkable that this straightforward model seamlessly incorporates genome-wide complexity and thus fundamentally differs from previous multiscale combinatorial approaches that reduce drug repositioning to prioritized molecular mechanisms. The basis for this shift toward computational integrative approaches has precedence in scalar theories of biological information and is aptly warranted for exploitation in drug repurposing.
Scalar Theories of Biological Information and Genomic Measures
In 1957, Francis Crick introduced a theory of information flow across biological scales from different molecular sequences: his “central dogma of molecular biology [that] deals with the detailed residue-by-residue transfer of sequential information.” (3). This theory of information transfer remains salient to genomic medicine and genome-based drug therapy where, in its most simplistic interpretation, DNA is transcribed into RNA and then translated into protein. While the critical concepts of information transfer and the emergence of new properties across biological scales are fundamental to this theory, they are often lost. In other words, just as DNA is transcribed and translated into protein, information irreversibly flows forward up a biological hierarchy, unveiling emergent functions as it progresses.
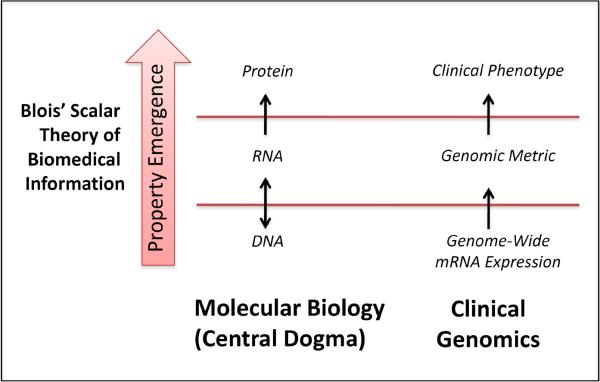
In 1984, these observations were further codified by Marsden Blois (4) in his scalar theory of biomedical information: “Emergent attributes come into existence with each increase of level, and contribute to the behavior of objects at the level of emergence and at all levels above, although they subsequently become embedded. A property emerging at a lower level…may participate in a process that itself only emerges at a still higher level…As we examine objects still higher in the hierarchy, new properties continue to emerge and still higher-level languages are required.” As shown in Figure 1, Blois's scalar theory is at once strikingly simple and profound. Indeed, Crick's “central dogma” clearly adheres to this paradigm. From strings of four nucleic acids emerge complex geometric protein structures that have properties and functions completely different than their progenitors.
Figure 1. Scalar information flow in molecular biology and present day genomics.
The central dogma of molecular biology was explicitly coined by Crick as an information model and is encompassed by a Blois' broader subsequent theory of biomedical information. These scalar theories remain salient to genome-based therapy as they appropriately frame the information intermediaries between molecular mechanisms and clinical-level phenotypes. Above, the flow of information from lower scales to higher ones is shown for the central dogma and for gene expression data. Conventional gene expression signature classifiers predictive of therapeutic utility correspond to an intermediate emergent property, measured by a genomic metric, consisting of a selected number of genes. In contrast, Butte proposes that a drug is a potential candidate for a disease treatment when their respective global genomic metrics are in opposition.
This scalar model also underlies the framework of Butte's group's methodology for drug repurposing. As shown in Figure 1, we can view mRNA expression as a lower-level function and its associated sample phenotypes (e.g. tumor vs. non-tumor, drug treated vs. non-drug treated) as significantly higher-level biological scales. The global genomic metric proposed by Butte represents an intermediate scale phenotype that has new intrinsic properties relevant to clinical practice that are distinct from its underlying microarray data. Genome-wide metrics differ from similarly scaled prioritized gene expression signature classifiers that have been increasingly used for molecular reclassification of diseases and targeting drugs.
Computational Biology and Bioinformatics for Predicting Response to Therapy
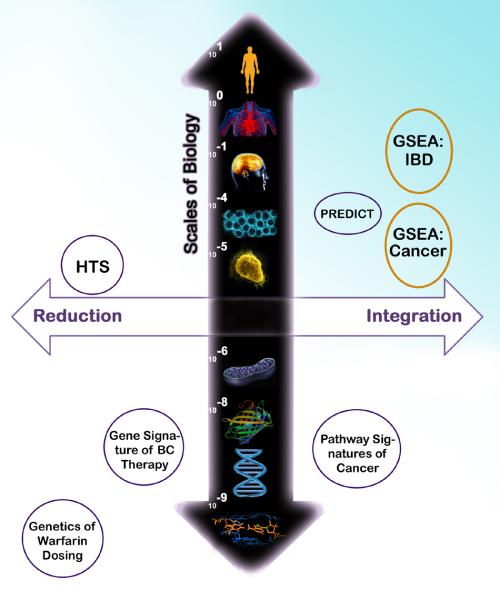
One of the great focuses in bioinformatics has been to develop multigene expression signature classifiers. Initially motivated by the success of single gene biomarkers in predicting response to therapy, considerable analytical development has been deployed to reduce genome-wide expression to classifiers containing a smaller number of genes (gene expression signatures). These classifiers function to provide sample classification based on expression levels of associated component genes and are trained to distinguish between different phenotypic states(5) and serve as archetype intermediate emergent properties (Figure 1, lower left quadrant of Figure 2). Indeed, gene expression signature classifiers have been described as “cancer phenotypes” in oncology(6,7) even though their constituent genes' mechanisms are not necessarily understood. To address this issue, pathway-specific signatures have also been explored. It thus follows that molecular pathways are emergent properties of gene expression signatures and provide deregulated pathways, such as expression of estrogen, progesterone and HER-2 receptors that may ultimately guide treatment for genetically selected patients (8) (lower right quadrant, Figure 2).
Figure 2. Biological Scales Contrasting Reductionist and Integrative Drug Repositioning Computations.
A few exemplar bioinformatics and computational biology studies of drug targeting or repositioning are used to illustrate the major differences in approaches: (i) reduction to the molecular function on the left quadrants, and (ii) integrated properties of multiple molecules to the right. The vertical axis discriminates among different scales of biological substrates for genetic or genomic assays. As shown at the DNA base pair scale, the genetics of warfarin dosage elucidated from a genome-wide association study is illustrated on the lower left quadrant (18). Gene expression signature classifiers follow at the mRNA scale (5). Integrated pathway-level scores of gene expression measures are shown in the lower right quadrant (19). High throughput screens (HTS) (11) of the upper left quadrant correspond to reductionism methods at the tissue level and contrast with increasingly integrative genome-wide tissular methods of the PREDICT algorithm (13) and that of Butte's research group conducted at the same biological scale (upper right quadrant, orange circles). Importantly, the genome-wide metric developed by Butte's group was applied to both a cellular-level disease (cancer) and a tissular one (IBD). As shown by the void in the uppermost part of the right quadrant, systemic diseases and multi-organ ones have not yet been explored by genome-wide metric and may require more comprehensive analyses than straightforward opposition of a genomic metric measured in surrogate tissues and drugs. Scales of biology (m): DNA base pairs, DNA, protein, organelle (mitochondria), cell, tissue, organ, system, organism.
An alternative view lies in the holistic approach of Gene Set Enrichment Analyses (GSEA)(9,10) which provides an unbiased genome-wide analysis of gene expression. The implied emergent property here is that of genome-wide similarity. In other words, if two genome metrics are established similar by GSEA, we may assume a degree of shared characteristics at higher biological scales, such as at the level of diseases. An advantage of GSEA is that evaluating genomic similarity is not subject to the same pre-existing knowledge constraint as that of pathways. This global integrative viewpoint allows GSEA-type approaches to take advantage of previously underexploited genome-wide patterns to reveal clinically relevant mechanisms and thus remain as a powerful tool for assessing similarity to archetypal expression profiles, such as motif gene sets and curated gene sets of online pathway annotations. Furthermore, the degree of genomic similarity can be quantified and is not merely a qualitative assessment that allows for far more exacting comparisons. As described below, Butte's group examines “genomic anti-similarity” between drug and disease's metrics.
Computations for Drug Repurposing
Traditionally, identifying drugs for repositioning has centered on biological methods that require a deep understanding of the deregulated biological mechanisms. These methods are implemented to identify diseases with appropriately related deregulations that could benefit from the same drug. For example, within the era of targeted therapy, imatinib, a tyrosine kinase inhibitor originally developed to target the BCR-ABL fusion protein constitutively expressed in chronic myelogenous leukemia (CML), was later repurposed to treat gastrointestinal stromal tumors (GIST) that harbored a mutated tyrosine kinase also targeted by imatinib. Alternatively, high-throughput drug screens have been utilized to identify novel targets in larger volumes (11)(upper left quadrant of Figure 2). Other methods use systematic clinical trial approaches that take advantage of post-marketing surveillance information designed to detect drug toxicities, yet may contain glimmers of unexpected potent positive effects (2).
Before the advent of the human genome, computational ligand- and structural-based approaches to in silico pharmacology offered opportunities for drug repurposing over specific targets. Subsequently, the maturation of computational biology and of compendium datasets from genome-wide measurements offered an automated, unbiased approach to drug repurposing. Indeed the assemblage of protein-drug, protein-protein interactions, and protein-disease networks has allowed statistical prioritization of new drug targets with retrospective validation using published cellular expression data and genome-wide association studies (8,12). For example, the PREDICT algorithm further extends this concept by incorporating chemical similarity of the drugs, disease-disease similarity metrics, along with protein-protein interaction distances in order to infer novel drug indications (13) (upper right quadrant of Figure 2); and was subsequently validated using data from tissue expression and ongoing clinical trials. Although these multiscale techniques are broad-ranging and provide some insight into the mechanism of imputed drug-disease associations, they all require a great amount of a priori knowledge of the diseases, drugs and deregulated pathway(s) of interest.
While the previous computational approaches arguably are multiplexed and reductionist, genome-level expression metrics between drugs have also been proposed to infer common mechanisms by Iorio et al (14). Likewise, the two related articles from the Butte Research Lab move us toward more encompassing and integrative approaches for drug repurposing. Rather than focusing on developing the best network and discriminatory algorithm, the Butte research group takes advantage of an emergent and comprehensive genome-wide metric that extends beyond carefully selected gene-expression signatures (15,16)(upper right quadrant, Figure 2). Interestingly, among all drug reposition or targeting studies illustrated in Figure 2, Butte's methods are the only ones exploiting measurements of a scale of biology (tissue) under environmental changes (i.e. drug exposure). Taking a progressive view of genome-wide expression data, these researchers posited whether or not global expression metrics have an emergent property of directionality. If so, it follows that drug and disease genome-wide expression metrics could be in opposite directions with one another. They then applied this emergent genome-scale property to a useful clinical scenario, that of drug repurposing.
Using a systematic computational approach, the researchers compared the patterns of differentially expressed genes from diseased versus non-diseased tissues to publically available reference drug expression signatures derived from the treatment of cancer cell lines. By quantifying the degree of anti-correlation, they were able to develop novel hypotheses in the utilization of existing drugs for other disease indications. In particular, Sirota et al. show that cimetidine, a histamine H2-receptor blocker and common over-the-counter anti-acid medication, slowed the rate of non-small cell lung adenocarcinoma (NSCLC) growth in mouse xenograft models. Additionally, they showed that topiramate, an anticonvulsant, might have a therapeutic role in alleviating inflammatory bowel disease flares in animal models. Excitingly, this emergent genome-wide property of directionality may ultimately translate into medical practice as a clinical finding analogous to an arrhythmia seen on EKG or that of a heart defect seen on a prenatal ultrasound. Arguably, physicians could incorporate these new and interpretable clinicogenomic findings in their practice to improve treatment decision-making process.
Equally striking is that these results were generated using surrogate tissue types. Drugs were tested from in vitro cancer cell lines whereas the disease profiling occurred in native diseased tissue. Although others have developed and clinically validated gene signatures from in vitro models (17), the ability of this anti-correlation algorithm to detect experimentally validated genome-wide differences highly suggests that such metrics are tissue origin-independent. An alternative hypothesis may be that any tissue specific noise may be drowned out by the powerful directionality property these genome-wide metrics appear to exhibit. Taken together, these two articles lend further credence to the notion that intermediary genomic states have emergent properties unseen at higher or lower hierarchical levels and have yet to be fully exploited.
The Future of Genome-Based Drug Repositioning
A decade ago as the human genome project unfolded, the limitations of oversimplified reductionism was contrasted by many with the opportunity for paradigm shifting integrative models of genomic complexity (1). By establishing the characteristics of emergent genome-wide metrics for drug repositioning, Butte's group establishes a milestone paving the way to fulfilling the promise of holism and integration in genome-based therapy modeling. Further, intermediate genomic phenotypes they describe can serve as a foundation for novel comprehensive models that incorporate more tissues for complex systemic clinical conditions such as diabetes mellitus type II. As an example, one could envision the search for approaches that include the strengths of both reductionism and integration, such as using a genome-wide metric conducted in multiple tissues and organs rather than in a single one for targeted drug development. These results rest on genome-wide metric measures at the mRNA scale, as these are perhaps the best-characterized genomic intermediary states. Alternatively, these genomic metrics could be combined with conventional clinical measurements for drug repurposing. Clearly, numerous other intermediary genome-wide metrics could be measured at other scales of biology, from microRNAs signatures to comparative genomic hybridization data. A conceptual framework for guiding the analysis of these scales' inputs and outputs is therefore of paramount importance for their accurate interpretation and application.
Acknowledgements
We thank Ms. Ellen Rebman and Ms. Kelly Regan for their contribution to the revisions and the illustrations.
References
- 1.Van Regenmortel MH. Reductionism and complexity in molecular biology. Scientists now have the tools to unravel biological and overcome the limitations of reductionism. EMBO Rep. 2004;5:1016–1020. doi: 10.1038/sj.embor.7400284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boguski MS, Mandl KD, Sukhatme VP. Drug discovery. Repurposing with a difference. Science. 2009;324:1394–1395. doi: 10.1126/science.1169920. [DOI] [PubMed] [Google Scholar]
- 3.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 4.Blois MS. Information and medicine : the nature of medical descriptions. University of California Press; Berkeley: 1984. [Google Scholar]
- 5.Simon R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J Clin Oncol. 2005;23:7332–7341. doi: 10.1200/JCO.2005.02.8712. [DOI] [PubMed] [Google Scholar]
- 6.Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M, D'Amico M, Pestell RG, West M, Nevins JR. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet. 2003;34:226–230. doi: 10.1038/ng1167. [DOI] [PubMed] [Google Scholar]
- 7.Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8:601–609. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins AL. Network pharmacology. Nat Biotechnol. 2007;25:1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 9.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 11.Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat Rev Drug Discov. 2009;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- 12.Hansen NT, Brunak S, Altman RB. Generating genome-scale candidate gene lists for pharmacogenomics. Clin Pharmacol Ther. 2009;86:183–189. doi: 10.1038/clpt.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlieb A, Stein GY, Ruppin E, Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol. 2011;7:496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, Murino L, Tagliaferri R, Brunetti-Pierri N, Isacchi A, di Bernardo D. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci U S A. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.S. M, Dudley JT, Shenoy M, Pai R, Roedder S, Chiang AP, Morgan AA, Sarwal M, Pasricha PJ, Butte AJ. Computational repositioning of the anticonvulstant topiramate for inflammatory bowel disease. Science Translational Medicine. 2011 doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D. J, Sirota M, Kim J, Morgan AA, Sweet-Cordero AS, Sage J, Butte AJ. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Science Translational Medicine. 2011 doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendiratta P, Mostaghel E, Guinney J, Tewari AK, Porrello A, Barry WT, Nelson PS, Febbo PG. Genomic strategy for targeting therapy in castration-resistant prostate cancer. J Clin Oncol. 2009;27:2022–2029. doi: 10.1200/JCO.2008.17.2882. [DOI] [PubMed] [Google Scholar]
- 18.Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, Limdi NA, Page D, Roden DM, Wagner MJ, Caldwell MD, Johnson JA. Estimation of the warfarin dose with clinical and pharmacogenetic data. The New England journal of medicine. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamoto T, Bianchini G, Booser D, Qi Y, Coutant C, Shiang CY, Santarpia L, Matsuoka J, Hortobagyi GN, Symmans WF, Holmes FA, O'Shaughnessy J, Hellerstedt B, Pippen J, Andre F, Simon R, Pusztai L. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. Journal of the National Cancer Institute. 2011;103:264–272. doi: 10.1093/jnci/djq524. [DOI] [PubMed] [Google Scholar]