Abstract
Background
GB virus C (GBV-C) may have a beneficial impact on HIV disease progression; however, the epidemiologic characteristics of this virus are not well characterized. Behavioral factors and gender may lead to differential rates of GBV-C infection; yet, studies have rarely addressed GBV-C infections in women or racial/ethnic minorities. Therefore, we evaluated GBV-C RNA prevalence and genotype distribution in a large prospective study of high-risk women in the US.
Results
438 hepatitis C virus (HCV) seropositive women, including 306 HIV-infected and 132 HIV-uninfected women, from the HIV Epidemiologic Research Study were evaluated for GBV-C RNA. 347 (79.2%) women were GBV-C RNA negative, while 91 (20.8%) were GBV-C RNA positive. GBV-C positive women were younger than GBV-C negative women. Among 306 HIV-infected women, 70 (22.9%) women were HIV/GBV-C co-infected. Among HIV-infected women, the only significant difference between GBV-negative and GBV-positive women was age (mean 38.4 vs. 35.1 years; p<0.001). Median baseline CD4 cell counts and plasma HIV RNA levels were similar. The GBV-C genotypes were 1 (n = 31; 44.3%), 2 (n = 36; 51.4%), and 3 (n = 3; 4.3%). The distribution of GBV-C genotypes in co-infected women differed significantly by race/ethnicity. However, median CD4 cell counts and log10 HIV RNA levels did not differ by GBV-C genotype. GBV-C incidence was 2.7% over a median follow-up of 2.9 (IQR: 1.5, 4.9) years, while GBV-C clearance was 35.7% over a median follow-up of 2.44 (1.4, 3.5) years. 4 women switched genotypes.
Conclusions
Age, injection drug use, a history of sex for money or drugs, and number of recent male sex partners were associated with GBV-C infection among all women in this analysis. However, CD4 cell count and HIV viral load of HIV/HCV/GBV-C co-infected women were not different although race was associated with GBV-C genotype.
Introduction
GB virus type C (GBV-C), first isolated in 1995, is a single-stranded RNA virus that belongs to the Flaviviridae family and is the closest known relative of the hepatitis C virus (HCV) [1]. GBV-C is transmitted efficiently via percutaneous and sexual routes. While male-to-male sex is a highly effective mode of GBV-C transmission [2], there is ample evidence that GBV-C can be transmitted heterosexually and perinatally as well [3]. The reported prevalence of GBV-C RNA ranges from 14 to 45% in HIV-infected persons [4]. However, the prevalence of GBV-C RNA is highly variable across distinct at-risk groups [4]. In a cross-sectional analysis of HIV-infected patients attending an urban HIV clinic, GBV-C RNA was most common among men who have sex with men (MSM) (35%) but less common among injection drug users (IDUs) (22%) or heterosexual men/women (22%) [5]. Furthermore, persistent GBV-C viremia is highest in MSM and is associated with an increased number of partners [2]. However, no trend between GBV-C persistence and number of partners was found among female sex workers. These data imply that gender and/or behavioral differences may lead to different rates of GBV-C infection and/or viral persistence. Several international studies have examined perinatal transmission of GBV-C or the impact of GBV-C infection on HIV transmission or disease progression in pregnant women [6]–[13]. For instance, in a large study of pregnant women, GBV-C infection was associated with increasing number of lifetime sexual partners, IDU, and HIV infection, while GBV-C clearance was associated with increasing age, more than 10 lifetime sexual partners, and no HIV infection [14]. Sexual transmission of GBV-C has been examined as well [2], [15], [16]. We reported previously that active GBV-C infection was more common in men than women and that more women than men had no evidence of GBV-C infection [17]. In health clinic attendees in the US, a higher prevalence of GBV-C occurred in subjects currently seeking STD treatment, while non-white race was associated with GBV RNA positivity [18]. GBV-C infection has been investigated in HIV-positive pregnant women in the US but did not include evaluation of factors associated with GBV-C infection [19].
Several groups have reported beneficial effects of GBV-C viremia on HIV disease [20]–[27]. At the population level, at least 7 GBV-C genotypes exist based upon phylogenetic analysis of the 5′ untranslated region (UTR) [28], [29]. The existence of multiple GBV-C genotypes has led several authors to suggest that differences in GBV-C strains circulating within populations might affect their impact on HIV disease [4], [30], [31]. For example, Muerhoff et al. reported that CD4 cell counts were lower in HIV co-infected patients infected with GBV-C genotype 2a than patients with genotype 2b [31]. We found that among a cohort with HIV/HCV/GBV-C triple infection, GBV-C genotype 2 was associated with higher CD4 cell counts compared to GBV-C genotype 1 [17]. While similar findings have also been reported in a Brazilian cohort [32], a comparison of genotype 2 versus non-2 infections in an Australian cohort observed no such difference in CD4 cell counts [33]. GBV-C viral load may also differ by genotype [34]. Thus, it is possible that GBV-C genotype could at least partially account for the beneficial association between GBV-C replication and HIV disease progression but requires further investigation in larger cohorts with multiple circulating GBV-C genotypes.
Investigations of GBV-C in women are generally limited to cross-sectional prevalence studies and perinatal transmission studies. Large longitudinal studies specifically addressing the prevalence and possible effects of GBV-C co-infection and GBV-C genotypes on the natural history of HIV infection in women are rare. Thus, there is limited information regarding the sociodemographic factors associated with GBV-C infection in women or racial minorities, particularly in the US. Therefore, the current study evaluated the rate of GBV-C RNA detection, GBV-C genotype distribution, and the GBV-C incidence and clearance rates in a large cohort study of HIV-infected and high-risk, HIV-uninfected women in the US with a substantial proportion of racial minorities. In understudied and/or disadvantaged populations such as women, ethnic minorities, and IDUs in which HIV-infected individuals may not have access to antiretroviral therapy, a better understanding of the epidemiology of GBV-C infection, as well as the anti-HIV effects of GBV-C infection, may ultimately result in novel therapeutic strategies.
Materials and Methods
Study population and sample selection
From 1993 until 2000, a prospective natural history study of HIV infection - the HIV Epidemiologic Research (HER) Study - was conducted in US women [35]. 871 HIV-infected women and 438 demographically similar, uninfected women were recruited from 4 sites: Baltimore, Maryland; Bronx, New York; Providence, Rhode Island; and Detroit, Michigan. By study design, one-half reported injection drug use (IDU), while the other half reported only sexual risk behavior for HIV. Women were assessed at 6-month intervals for up to 14 study visits. Women with a clinical AIDS diagnosis or any AIDS-defining opportunistic infections were ineligible for enrollment. Among the 871 HIV-infected women enrolled in the HERS cohort, 233 had a CD4 cell count of ≤200 cells/mm3 at baseline. At study entry, ∼30% of HIV-infected women received monotherapy or dual therapy, and none received HAART [36]. As described elsewhere, hepatitis C virus (HCV) serostatus was determined using the Abbott HCV EIA 2.0 or Ortho HCV version 3.0 ELISA [37]. In the original cohort, the seroprevalence of HCV was 56.5%, with rates of 48.0% and 60.8% among HIV-uninfected and HIV-infected women, respectively. Less than 1% received any HCV therapy; therefore, HCV treatment is not likely to confound analyses. Institutional review boards at each participating medical center and at the Centers for Disease Control and Prevention approved the HER study protocol. All participants provided voluntary informed consent prior to sample and clinical data collection.
To facilitate future analyses of the effects of GBV-C infection on liver disease during HIV/HCV co-infection, this analysis was restricted to 438 randomly selected women who were HCV seropositive and had available serum for analysis. The current study population did not differ at baseline from the remaining women in the HERS cohort with respect to race, HIV serostatus, or mean HIV RNA levels. However, women in the current analysis were older than the remaining women (37.0 years at baseline versus 34.0 years [p<0.001] for HIV-positive women and 36.8 years versus 33.1 years [p<0.001] for HIV-negative women). CD4 cell counts at baseline were also significantly different (411.8 cells/mm3 for HIV-positive women in the current study versus 365.4 cells/mm3 for the remaining HIV-positive HERS women, p = 0.021).
Detection of GBV-C RNA
Viral RNA was extracted from 140 µl of serum with the QIAmp Viral RNA Mini Kit (QIAGEN, Valencia, CA) following the manufacturer’s instructions. GBV-C RNA was detected by nested RT-PCR using primers corresponding to the 5′ untranslated region (UTR) as described previously [17]. PCR products were analyzed by agarose gel electrophoresis for the presence of a 256-nucleotide band corresponding to nucleotides 107–362 of GenBank accession number AY196904.
Phylogenetic analysis
GBV-C genotype was determined by population-based amplification of the 5′UTR region as described previously [17]. Sequences were aligned with a database reference using Clustal X 2.1 [38]. The reference sequences used to confirm GBV-C genotype included the following GenBank accession numbers: 1A: U59540 and U59543; 1B: U59549 and U59555; 2A: U59520 and U59521; 2B: U59529 and U59533; 3: U59538 and U59539; 4: AB018667 and AB021287; 5: AY949771 and AF092894; 6: AB003292 and AF177619. The statistical robustness and reliability of the branching order within the phylogenetic tree was confirmed by bootstrap analysis using 1,000 replicates. Additional phylogenetic inference was performed using a Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in the Bayesian Evolutionary Analysis by Sampling Trees (BEAST) v1.7.5 program [39] under an uncorrelated log-normal relaxed molecular clock and the generalized time reversible (GTR) model with nucleotide site heterogeneity estimated using a gamma distribution. The BEAST MCMC analysis was run for a chain length of 200,000,000. All effective sample size values were >100 indicating sufficient sampling. The maximum clade credibility tree was selected from the posterior tree distribution after a 10% burn-in using TreeAnnotator v1.7.5.
Statistical analysis
Categorical variables were evaluated for an association with GBV-C infection using the chi-squared test or Fisher’s exact test if any expected cell count was less than 5. The means of normally-distributed continuous variables were compared across GBV-C infection status using a t-test and across the three GBV-C genotypes using a one-way analysis of variance. The medians of non-normally distributed continuous variables were compared across GBV-C infection status using the Wilcoxon rank sum test and across genotypes using the Kruskal-Wallis test. All p-values are two-sided. P-values <0.10 are provided, although only p-values <0.05 are considered statistically significant.
Results
438 women, including 306 HIV-infected and 132 HIV-uninfected women, from the HERS cohort were evaluated for GBV-C RNA. 347 women were GBV-C RNA negative (79.2%), while 91 were GBV-C RNA positive (20.8%) as shown in Table 1. In univariate analysis, GBV-C positive women were younger (p<0.001), less likely to report a history of IDU (p = 0.036), and more likely to have sex partners in the past 6 months (p = 0.037) than GBV-C negative women. GBV-C positive women were also more likely to be HIV-positive than GBV-C negative women, although this difference did not reach statistical significance (p = 0.099).
Table 1. Demographic and clinical data for 438 women from the HERS cohort included in the current study.
| Characteristic | GBV-C RNAnegative(N = 347) | GBV-C RNApositive(N = 91) | P value |
| Mean age in years (SD) | 38.3 (6.0) | 34.7 (6.3) | <0.001 |
| Race (%) | NS | ||
| Black | 220 (63.4%) | 52 (57.1%) | |
| White | 75 (21.6%) | 22 (24.2%) | |
| Hispanic | 48 (13.8%) | 16 (17.6%) | |
| Native American/Asian/Other | 4 (1.2%) | 1 (1.1%) | |
| Non-white | 272 (78.4%) | 69 (75.8%) | |
| White | 75 (21.6%) | 22 (24.2%) | |
| Sexual and drug use behaviors | |||
| IDU since 1985 (%) | 327 (94.2%) | 80 (87.9%) | 0.036 |
| IDU in previous 6 months (%) | 128 (36.9%) | 35 (38.5%) | NS |
| ≥5 sex partners in past 6 months (%) | 109 (31.4%) | 23 (25.3%) | NS |
| Ever had sex with male IDU (%) [435] | 301/344 (87.5%) | 80/91 (87.9%) | NS |
| Ever had sex with partnerknown/suspected HIV+ (%) [435] | 179/344 (52.0%) | 48/91 (52.8%) | NS |
| Ever had sex for money or drugs (%) | 200 (57.6%) | 42(46.2%) | 0.050 |
| Male sex partners in previous 6 months (%)[436] | |||
| 0 | 107/345 (31.0%) | 19 (20.9%) | |
| 1–10 | 229/345 (66.4%) | 72 (79.1%) | |
| >10 | 9/345 (2.6%) | 0 | 0.037 |
| Currently using hormonal contraceptives (%) | 11 (3.2%) | 5 (5.5%) | NS |
| Currently using condoms (%)[309 sexually active] | 170/238 (71.4%) | 50/71 (70.4%) | NS |
| Ever been pregnant (%) | 326 (94.0%) | 87 (95.6%) | NS |
| Currently pregnant (%) [435] | 3/344 (0.9%) | 0 | NS |
| Currently using alcohol (%) | 200 (57.6%) | 52 (57.1%) | NS |
| All-cause mortality (%) | NS | ||
| 1993–1996 | 24 (6.9%) | 8 (8.8%) | |
| 1997–2000 | 39 (11.2%) | 11 (12.1%) | |
| HIV positive (%) | 236 (68.0%) | 70 (76.9%) | 0.099 |
| HBsAg positive (%) [345] | 13/278 (4.7%) | 3/67 (4.5%) | NS |
In parentheses are percentages unless otherwise noted. Numbers in brackets denote women with available data. P-values <0.10 are provided, although only p-values <0.05 are considered statistically significant. NS = not significant (p≥0.10); SD = standard deviation.
306 women with HIV were evaluated for GBV-C RNA, and 70 (22.9%) were HIV/GBV-C co-infected. As shown in Table 2, the only significant difference between HIV-infected/GBV-negative and HIV-infected/GBV-positive women was age (p<0.001) with dually infected women being younger. Median baseline CD4 cell count and plasma HIV RNA, as well as the proportion of women on antiretroviral therapy, were similar between HIV-positive women with and without GBV-C RNA.
Table 2. Demographic and clinical data for 306 HIV-positive women from the HERS cohort included in the current study.
| Characteristic | GBV-C RNAnegative(N = 236) | GBV-C RNApositive(N = 70) | P value |
| Mean age in years (SD) | 38.4 (5.8) | 35.1 (6.2) | <0.001 |
| Race (%) | NS | ||
| Black | 156 (66.4%) | 43 (61.4%) | |
| White | 43 (18.3%) | 14 (20.0%) | |
| Hispanic | 34 (14.5%) | 12 (17.1%) | |
| Native American/Asian/Other | 2 (0.8%) | 1 (1.4%) | |
| Non-white | 192 (81.7%) | 56 (80.0%) | |
| White | 43 (18.3%) | 14 (20.0%) | |
| Sexual and drug use behaviors | NS | ||
| IDU since 1985 (%) | 219 (93.2%) | 61 (87.1%) | |
| IDU in previous 6 months (%) | 77 (32.6%) | 24 (34.3%) | |
| ≥5 sex partners in past 6 months (%) | 64 (27.2%) | 15 (21.4%) | |
| Ever had sex with male IDU (%) [303] | 208/233 (89.3%) | 63/70 (90.0%) | |
| Ever had sex with partner known/suspected HIV+ (%) [304] | 134/234 (57.3%) | 42/70 (60.0%) | |
| Ever had sex for money or drugs (%) | 132 (56.2%) | 32 (45.7%) | |
| Male sex partners in previous 6 months (%) [304] | NS | ||
| 0 | 83 (35.2%) | 17 (24.3)% | |
| 1–10 | 149 (63.1%) | 53 (75.7)% | |
| >10 | 4 (1.7%) | 0 | |
| Currently using hormonal contraceptives (%) | 7 (3.0%) | 4 (5.7%) | NS |
| Currently using condoms (%) [206] | 119/153 (77.8%) | 39/53 (73.6%) | NS |
| Ever been pregnant (%) | 221 (94.0%) | 67 (95.7%) | NS |
| Currently pregnant (%) [304] | 2/234 (0.9%) | 0/70 (0%) | NS |
| Currently using alcohol (%) | 129 (54.7%) | 38 (54.3%) | NS |
| All-cause mortality (%) | NS | ||
| 1993–1996 | 23 (9.8%) | 8 (11.4%) | |
| 1997–2000 | 37 (15.7%) | 11 (15.7%) | |
| CD4 cell count (%) [296] | NS | ||
| <200 | 39/229 (17.0%) | 9/67 (13.4%) | |
| 200–499 | 117/229 (51.1%) | 40/67 (59.7%) | |
| ≥500 | 73/229 (31.9%) | 18/67 (26.9%) | |
| Median CD4 cell count in cells/uL (IQR) [296] | 382 (251, 571) | 413 (267, 536) | NS |
| Median log10 HIV RNA in copies/ml(IQR) - all women with data [301] | 3.24 (2.65, 3.90) | 3.46 (2.81, 3.85) | NS |
| Median log10 HIV RNA in copies/ml(IQR) - only women not on ART [202] | 3.24 (2.65, 4.06) | 3.47 (2.81, 3.83) | NS |
| Currently using ART (%) | 74 (31.4%) | 27 (38.6%) | NS |
| HBsAg positive (%) [246] | 7/192 (3.7%) | 2/54 (3.7%) | NS |
In parentheses are percentages unless otherwise noted. Numbers in brackets denote women with available data. P-values <0.10 are provided, although only p-values <0.05 are considered statistically significant. NS = not significant (p≥0.10); SD = standard deviation; IQR = interquartile range.
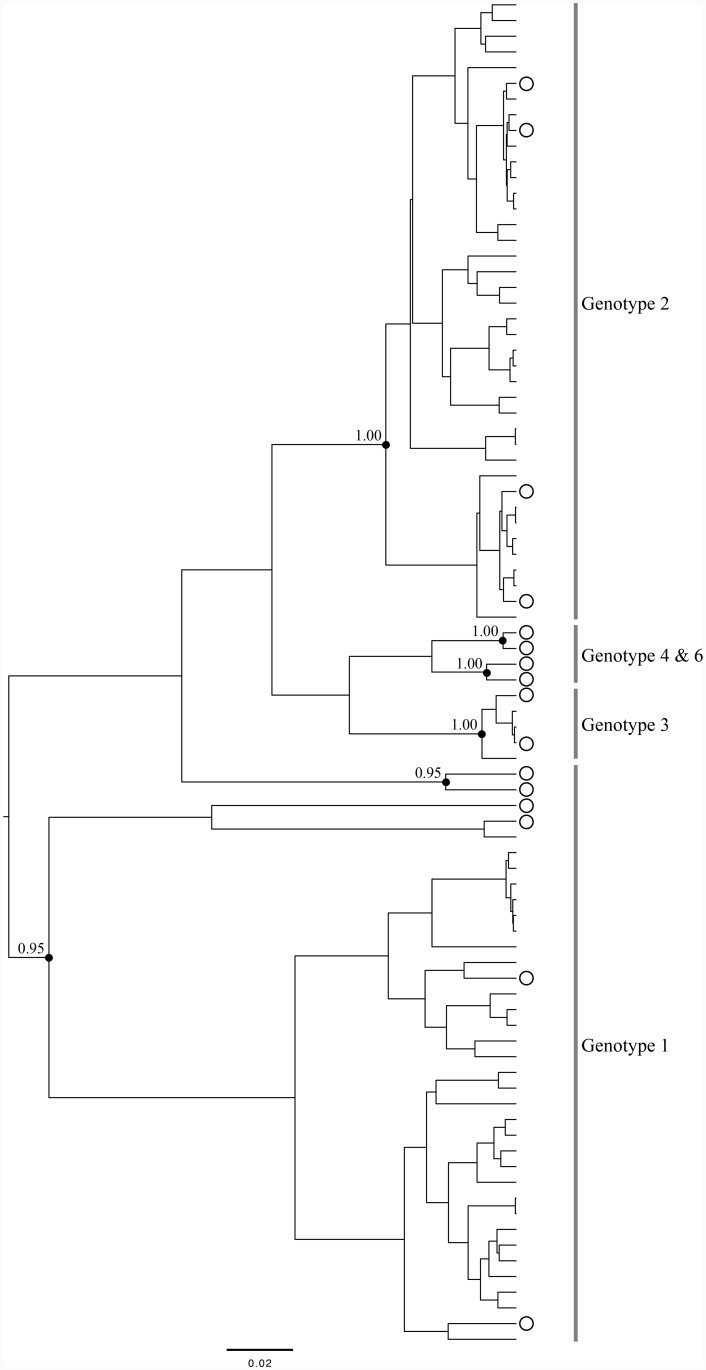
Previous studies suggest a potential role for GBV-C genotype in modulating HIV disease; therefore, we determined GBV-C genotype among HIV/GBV-C co-infected women (Fig. 1). Based on analysis of the 5′UTR, the GBV-C genotypes were 1 (n = 31; 44.3%), 2 (n = 36; 51.4%), and 3 (n = 3; 4.3%). The distribution of GBV-C genotypes in co-infected women was significantly different among racial groups (p = 0.003) with Hispanics being more likely to be infected with genotype 1 and whites being less likely to be infected with this genotype (Table 3). There was also a suggestion of distinct GBV-C genotype distributions in previous IDUs versus women that were never IDUs, with the latter being predominantly infected with genotype 2 (p = 0.080). However, median baseline CD4 cell counts (interquartile range [IQR]) were not markedly different across the three GBV-C genotypes: 400 (IQR: 277, 468), 408 (IQR: 267, 577), and 490 (IQR: 222, 490) cells/mm3 for women infected with genotypes 1, 2, and 3, respectively. Moreover, median baseline log10 HIV RNA levels were similar across genotypes: 3.32 (IQR: 2.79, 4.14), 3.48 (IQR: 2.78, 3.83), and 3.46 (IQR: 2.86, 4.56) copies/mL for genotypes 1, 2, and 3, respectively.
Figure 1. Phylogenetic tree based on consensus 5′UTR sequences for 70 HIV/GBV-C co-infected women.
GenBank reference sequences are indicated by open circles. Relevant posterior probabilities (out of 1.00) are shown. The scale bar indicates 0.02 nucleotide substitutions per site.
Table 3. Demographic and clinical data 70 HIV/GBV-C co-infected women from the HERS cohort included in the current study.
| Characteristic | GBV-C genotypes | |||
| Type 1 | Type 2 | Type 3 | P-value | |
| (n = 31) | (n = 36) | (n = 3) | ||
| Median age in years (SD) | 34.8 (5.1) | 35.4 (6.5) | 35.9 (13.8) | NS |
| Race (%) | ||||
| Black | 20 (64.5%) | 22 (61.1%) | 1 (33.3%) | 0.003 |
| White | 1 (3.2%) | 11 (30.6%) | 2 (66.7%) | |
| Hispanic | 9 (29.0%) | 3 (8.3%) | 0 | |
| Native American/Asian/Other | 1 (3.2%) | 0 | 0 | |
| IV drug use | 0.080 | |||
| Never | 1 (3.2%) | 8 (22.2%) | 0 | |
| Previously | 30 (96.8%) | 28 (77.8%) | 3 (100%) | |
| All-cause mortality | NS | |||
| 1993–1996 | 5 (16.1%) | 2 (5.6%) | 1 (33.3%) | |
| 1997–2000 | 4 (12.9%) | 7 (19.4%) | 0 | |
| CD4 cell count [69] | NS | |||
| <200 | 4 (12.9%) | 5/35 (14.3%) | 0 | |
| 200–499 | 19 (61.3%) | 18/35 (51.4%) | 3 (100.0%) | |
| ≥500 | 8 (25.8%) | 12/35 (34.3%) | 0 | |
| Median CD4 cell count in cells/uL(IQR) [67] | 400 (277, 468) | 408 (267, 577) | 490 (222, 490) | NS |
| Median log10 HIV RNA in copies/mL(IQR) [69] | 3.32 (2.79, 4.14) | 3.48 (2.78, 3.83) | 3.46 (2.86, 4.56) | NS |
| Median log10 HIV RNA in copies/mL(IQR) - only women not on ART [42] | 3.34 (2.81, 4.17) | 3.48 (2.78, 3.73) | 4.01 (3.46, 4.56) | NS |
| Currently using ART (%) | 14 (45.2%) | 12 (33.3%) | 1 (33.3%) | NS |
| HBsAg positive (%) [54] | 1 (4.2%) | 1 (3.7%) | 0 | NS |
In parentheses are percentages unless otherwise noted. Numbers in brackets denote women with available data. P-values <0.10 are provided, although only p-values <0.05 are considered statistically significant. NS = not significant (p≥0.10); SD = standard deviation; IQR = interquartile range.
193 women were tested for GBV-C RNA at 2 time points. Of the 151 women who were GBV-C negative at their first visit and had a subsequent GBV-C result, 4 (2.7%) became GBV-C RNA positive over a median follow-up of 2.9 (IQR: 1.5, 4.9) years. Of the 42 women who were initially GBV-C RNA positive, 15 (35.7%) became GBV-C RNA negative at the second test over a median follow-up of 2.44 (1.4, 3.5) years. Of the 27 women who were positive at both the first and second test points, 4 (14.8%) switched genotypes: 2 from genotype 1 to genotype 2b, 1 from genotype 2b to genotype 1, and 1 from genotype 2a to genotype 1.
Discussion
Multiple studies have demonstrated a beneficial effect of GBV-C viremia on HIV disease [20], [22]–[25]. For instance, Tillman et al. reported that longer AIDS-free survival, higher CD4 cell counts, and lower plasma HIV viral loads were associated with the presence of GBV-C RNA. Moreover, an inverse correlation between the GBV-C viral load and the HIV viral load – but not CD4 cell count – was observed [22]. Xiang et al. further demonstrated prolonged survival among HIV-positive persons co-infected with GBV-C [20]. HIV replication was also inhibited by GBV-C co-infection in peripheral blood mononuclear cell cultures. Using data from the Multicenter AIDS Cohort Study, Williams et al. examined the effect of GBV-C infection measured 12–18 months after HIV seroconversion in men but did not find a statistically significant protective effect [25]. However, when a similar analysis was performed using data on GBV-C infection measured 5–6 years after HIV seroconversion, a statistically significant protective effect of GBV-C co-infection was observed. Bjorkman et al. reported that loss of GBV-C RNA during follow-up was associated with the poorest survival [21], a finding also confirmed in other studies [25], [26]. Nevertheless, prolonged survival in GBV-C/HIV co-infected persons has not been observed in all studies [21], [26], [40]–[42]. Due to the potential importance of persistence of GBV-C viremia, a meta-analysis [43] was conducted to synthesize data from eight prospective studies of HIV-positive persons in which GBV-C RNA was determined and all-cause death was determined. There was no conclusive evidence for an association between survival and GBV-C infection early in HIV disease, but when GBV-C infection was present later in HIV disease, a significant reduction in mortality was observed. Studies in the highly active antiretroviral therapy (HAART) era have found that a complete virologic response to HAART was more frequent in patients co-infected with GBV-C, independent of baseline CD4 cell count and plasma HIV RNA level [27]. However, 87% of participants were male; therefore, whether GBV-C RNA is an independent predictor of the virologic response to HAART in women remains to be determined. Collectively, these studies suggest a beneficial effect of GBV-C co-infection even among HAART-treated individuals, although the beneficial effects of GBV-C have not been confirmed in all studies [44]. Although the mechanism remain unclear, a number of biological mechanisms could explain the impact of GBV-C on HIV disease, including downregulation of HIV entry receptors, increased chemokine expression, interference with HIV binding and/or fusion, activation of interferon-stimulated genes, cytokine polarization, altered expression of T lymphocyte activation markers, protection from Fas-mediated apoptosis, and (reviewed in [45]). Further is study is required to elucidate the mechanisms of interaction between HIV and GBV-C.
Male-to-male (MSM) sex is an effective mode of GBV-C transmission, although GBV-C is transmitted heterosexually and perinatally as well [2], [3]. Importantly, persistent GBV-C infection is highest in MSM, and increased partner number is associated with increased GBV-C infection [2]; however, the same association is not observed among female sex workers. In a cross-sectional analysis of HIV-infected patients, GBV-C RNA was more common among MSM than heterosexual men/women [5]. To date, investigations of GBV-C in women are generally limited to cross-sectional prevalence studies and/or perinatal transmission studies with the GBV-C prevalence ranging from 11% to 37% [2], [5]–[7], [9], [15], [16], [19].
In the current study, we examined the associations between GBV-C infection and HIV infection in HCV seropositive women with a significant proportion of racial/ethnic minorities. Our findings suggest that GBV-C infection is common among women with or at risk for HIV infection. Age, injection drug use, a history of sex for money or drugs, and number of recent male sex partners were associated with GBV-C infection among all women in this analysis. HIV positivity was associated with increased likelihood of GBV-C infection, although this did not reach statistical significance. These findings agree with that of Bhanich Supapol et al. who reported similar risk factors for GBV-C infection in a cohort of Thai women [14]. We also found that GBV-C genotype distribution differed by racial group and possibly by IDU status among HIV/GBV-C co-infected women. Surprisingly, we did not observe a significant association of GBV-C RNA or GBV-C genotype with HIV disease severity as measured by CD4 cell count or HIV viral load. These disparate results may reflect demographic differences between populations or insufficient statistical power to observe minor differences between GBV-C infected versus uninfected individuals. Moreover, while the majority of GBV-C genotyping studies have utilized population-based amplification of the 5′UTR [46], sequencing the 5′UTR cannot efficiently discriminate GBV-C subtypes (i.e., 1a versus 1b) in every instance.
This study has several limitations. First, the sample size may not provide sufficient power to detect small differences between GBV-C RNA positive and negative individuals. Moreover, given the genotype distribution within this population, we were only able to meaningfully compare GBV-C genotype 1 to genotype 2. Second, this analysis included GBV-C RNA testing at only two time points per woman and did not evaluate GBV-C at all time points available per woman. Thus, the number of GBV-C incident and/or cleared infections may be underestimated, and GBV-C testing of additional time points is warranted. Third, testing for anti-E2 antibodies – indicative of past infection – was not performed, although several studies suggest that E2 antibodies are not a reliable marker of previous GBV-C infection in immunocompromised individuals [17], [47]. Fourth, given the initial inclusion/exclusion criteria of the HER Study, it is unclear if our findings are generalizable to all women or to populations outside of the US. Fifth, GBV-C infection was not associated with decreased immunodeficiency in this analysis; however, this may reflect the low frequency of severe immunodeficiency in these women. As noted above, a meta-analysis concluded that there was limited evidence for an association between early GBV-C infection and HIV-related survival. In contrast, a significant reduction in mortality was observed when GBV-C infection was present later in HIV disease [43]. Additionally, loss of GBV-C RNA has also been associated with accelerated HIV disease [21], [25], [42]. Thus, longitudinal analyses in women with more advanced immunodeficiency are necessary to evaluate the effects of persistent GBV-C co-infection on HIV disease progression in women. Furthermore, epidemiologic characterization of GBV-C infection and its anti-HIV effects may ultimately result in novel therapeutic strategies in understudied and/or disadvantaged populations such as women, ethnic minorities, and IDUs.
Acknowledgments
The authors would like to thank the HER Study staff and participants. The HER Study group consists of Robert S. Klein, MD, Ellie Schoenbaum, MD, Julia Arnsten, MD, MPH, Robert D. Burk, MD, Penelope Demas, PhD, and Andrea Howard, MD, MSc, from Montefiore Medical Center and the Albert Einstein College of Medicine; Paula Schuman, MD, Jack Sobel, MD, Suzanne Ohmit, DrPH, William Brown PhD, Michael Long PhD, Wayne Lancaster PhD, and Jose Vazquez, MD, from the Wayne State University School of Medicine; Anne Rompalo, MD, David Vlahov, PhD and David Celentano, PhD, from the Johns Hopkins University School of Medicine; Charles Carpenter, MD, Kenneth Mayer, MD, Susan Cu-Uvin, MD, Timothy Flanigan, MD, Joseph Hogan, ScD, Valerie Stone, MD, Karen Tashima, MD, and Josiah Rich, MD from the Brown University School of Medicine; Ann Duerr, MD, PhD, Lytt I. Gardner, PhD, Chad Heilig, PhD, Scott D. Holmberg, MD, Denise J. Jamieson, MD, MPH, Janet S. Moore, PhD, Ruby M. Phelps, BS, Dawn K. Smith, MD, MPH and Dora Warren, PhD from the Centers for Disease Control and Prevention; and Katherine Davenny, MPH from the National Institute of Drug Abuse. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. These data were presented at 6th International HIV and Hepatitis Co-Infection Workshop held in 2010 in Tel Aviv, Israel.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work was supported by the National Institute on Allergy and Infectious Diseases (award AI081564 to JTB). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Simons JN, Leary TP, Dawson GJ, Pilot-Matias TJ, Muerhoff AS, et al. (1995) Isolation of novel virus-like sequences associated with human hepatitis. Nature Medicine 1:564–569. [DOI] [PubMed] [Google Scholar]
- 2. Berzsenyi MD, Bowden DS, Bailey MJ, White C, Coghlan P, et al. (2005) Male to male sex is associated with a high prevalence of exposure to GB virus C. Journal of Clinical Virology. 33:243–246. [DOI] [PubMed] [Google Scholar]
- 3. Stapleton J (2003) GB virus type C/Hepatitis G virus. Seminars in Liver Disease 23:137–148. [DOI] [PubMed] [Google Scholar]
- 4. Berzsenyi MD, Bowden DS, Roberts S (2005) GB virus C: insights into co-infection. Journal of Clinical Virology 33:257–266. [DOI] [PubMed] [Google Scholar]
- 5. Smith SM, Donio MJ, Singh M, Fallon JP, Jitendranath L, et al. (2005) Prevalence of GB virus type C in urban Americans infected with human immunodeficiency virus type 1. Retrovirology 2:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weintrob AC, Hamilton JD, Hahn C, Klinzman D, Moyo G, et al. (2004) Active or prior GB virus C infection does not protect against vertical transmission of HIV in coinfected women from Tanzania. Clinical Infectious Diseases 38:e46–48. [DOI] [PubMed] [Google Scholar]
- 7. Zanetti AR, Tanzi E, Romano L, Principi N, Zuin G, et al. (1998) Multicenter trial on mother-to-infant transmission of GBV-C virus. The Lombardy Study Group on Vertical/Perinatal Hepatitis Viruses Transmission. Journal of Medical Virology 54:107–112. [DOI] [PubMed] [Google Scholar]
- 8. Supapol WB, Remis RS, Raboud J, Millson M, Tappero J, et al. (2008) Reduced mother-to-child transmission of HIV associated with infant but not maternal GB virus C infection. Journal of Infectious Diseases 197:1369–1377. [DOI] [PubMed] [Google Scholar]
- 9. Sathar MA, York DF, Gouws E, Coutsoudis A, Coovadia H (2004) GB virus type C coinfection in HIV-infected African mothers and their infants, KwaZulu Natal, South Africa. Clinical Infectious Diseases 38:405–409. [DOI] [PubMed] [Google Scholar]
- 10. Hino K, Moriya T, Ohno N, Takahashi K, Hoshino H, et al. (1998) Mother-to-infant transmission occurs more frequently with GB virus C than hepatitis C virus. Archives of Virology 143:65–72. [DOI] [PubMed] [Google Scholar]
- 11. Lefrère JJ, Sender A, Mercier B, Mariotti M, Pernot F, et al. (2000) High rate of GB virus type C/HGV transmission from mother to infant: possible implications for the prevalence of infection in blood donors. Transfusion 40:602–607. [DOI] [PubMed] [Google Scholar]
- 12. Viazov S, Riffelmann M, Sarr S, Ballauff A, Meisel H, et al. (1997) Transmission of GBV-C/HGV from drug-addicted mothers to their babies. Journal of Hepatology 27:85–90. [DOI] [PubMed] [Google Scholar]
- 13. Ohto H, Ujiie N, Sato A, Okamoto H, Mayumi M (2000) Mother-to-infant transmission of GB virus type C/HGV. Transfusion 40:725–730. [DOI] [PubMed] [Google Scholar]
- 14. Bhanich Supapol W, Remis RS, Raboud J, Millson M, Tappero J, et al. (2011) Prevalence and correlates of GB virus C infection in HIV-infected and HIV-uninfected pregnant women in Bangkok, Thailand. Journal of Medical Virology 83:33–44. [DOI] [PubMed] [Google Scholar]
- 15. Sawayama Y, Hayashi J, Etoh Y, Urabe H, Minami K, et al. (1999) Heterosexual transmission of GB virus C/hepatitis G virus infection to non-intravenous drug-using female prostitutes in Fukuoka, Japan. Digestive Diseases and Sciences 44:1937–1943. [DOI] [PubMed] [Google Scholar]
- 16. Kao JH, Chen W, Chen PJ, Lai MY, Lin RY, et al. (1997) GB virus-C/hepatitis G virus infection in prostitutes: possible role of sexual transmission. Journal of Medical Virology 52:381–384. [PubMed] [Google Scholar]
- 17. Schwarze-Zander C, Blackard JT, Zheng H, Addo MM, Lin W, et al. (2006) GB virus-C (GBV-C) infection in hepatitis C virus (HCV)/HIV co-infected patients receiving HCV treatment: importance of the GBV-C genotype. Journal of Infectious Diseases 194:410–419. [DOI] [PubMed] [Google Scholar]
- 18. Frey SE, Homan SM, Sokol-Anderson M, Cayco MT, Cortorreal P, et al. (2002) Evidence for probable sexual transmission of the hepatitis g virus. Clinical Infectious Diseases 34:1033–1038. [DOI] [PubMed] [Google Scholar]
- 19. Handelsman E, Cheng I, Thompson B, Hershow R, Mofenson LM, et al. (2007) Impact of GB virus type C infection on mother-to-child HIV transmission in the Women and Infants Transmission Study Cohort. HIV Medicine 8:561–567. [DOI] [PubMed] [Google Scholar]
- 20. Xiang J, Wunschmann S, Diekema DJ, Klinzman D, Patrick KD, et al. (2001) Effect of coinfection with GB virus C on survival among patients with HIV infection. New England Journal of Medicine 345:707–714. [DOI] [PubMed] [Google Scholar]
- 21. Bjorkman P, Flamholc L, Naucler A, Molnegren V, Wallmark E, et al. (2004) GB virus C during the natural course of HIV-1 infection: viremia at diagnosis does not predict mortality. AIDS 18:877–886. [DOI] [PubMed] [Google Scholar]
- 22. Tillmann H, Heiken H, Knapik-Botor A, Heringlake S, Ockenga J, et al. (2001) Infection with GB virus C and reduced mortality among HIV-infected patients. New England Journal of Medicine 345:715–724. [DOI] [PubMed] [Google Scholar]
- 23. Lefrere JJ, Roudot-Thoraval F, Morand-Joubert L, Petit JC, Lerable J, et al. (1999) Carriage of GB virus C/hepatitis G virus RNA is associated with a slower immunologic, virologic, and clinical progression of human immunodeficiency virus disease in coinfected persons. Journal of infectious Diseases 179:783–789. [DOI] [PubMed] [Google Scholar]
- 24. Heringlake S, Ockenga J, Tillmann HL, Trautwein C, Meissner D, et al. (1998) GB virus C/hepatitis G virus infection: a favorable prognostic factor in human immunodeficiency virus-infected patients? Journal of Infectious Diseases 177:1723–1726. [DOI] [PubMed] [Google Scholar]
- 25. Williams CF, Klinzman D, Yamashita TE, Xiang J, Polgreen PM, et al. (2004) Persistent GB virus C infection and survival in HIV-infected men. New England Journal of Medicine 350:981–990. [DOI] [PubMed] [Google Scholar]
- 26. Van der Bij AK, Kloosterboer N, Prins M, Boeser-Nunnink B, Geskus RB, et al. (2005) GB virus C coinfection and HIV-1 disease progression: the Amsterdam cohort study. Journal of Infectious Diseases 191:678–685. [DOI] [PubMed] [Google Scholar]
- 27. Rodriguez B, Woolley I, Lederman MM, Zdunek D, Hess G, et al. (2003) Effect of GB virus C coinfection on response to antiretroviral treatment in human immunodeficiency virus-infected patients. Journal of Infectious Diseases 187:504–507. [DOI] [PubMed] [Google Scholar]
- 28. Muerhoff AS, Dawson GJ, Desai S (2006) A previously unrecognized sixth genotype of GB virus C revealed by analysis of 5′-untranslated region sequences. Journal of Medical Virology 78:105–111. [DOI] [PubMed] [Google Scholar]
- 29. Feng Y, Zhao W, Feng Y, Dai J, Li Z, et al. (2011) A novel genotype of GB virus C: its identification and predominance among injecting drug users in Yunnan, China. PLoS One 6:e21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaye S, Howard M, Alabi A, Hansmann A, Whittle H, et al. (2005) No observed effect of GB virus C coinfection on disease progression in a cohort of African woman infected with HIV-1 or HIV-2. Clinical Infectious Diseases 40:876–878. [DOI] [PubMed] [Google Scholar]
- 31. Muerhoff AS, Tillmann HL, Manns MP, Dawson GJ, Desai S (2003) GB virus C genotype determination in GB virus-C/HIV co-infected individuals. Journal of Medical Virology 70:141–149. [DOI] [PubMed] [Google Scholar]
- 32. Alcalde R, Nishiya A, Casseb J, Inocêncio L, Fonseca LA, et al. (2010) Prevalence and distribution of the GBV-C/HGV among HIV-1-infected patients under anti-retroviral therapy. Virus Research 151:148–152. [DOI] [PubMed] [Google Scholar]
- 33. Berzsenyi MD, Bowden DS, Roberts SK, Revill P (2009) GB virus C genotype 2 predominance in a hepatitis C virus/HIV infected population associated with reduced liver disease. Journal of Gastroenterology and Hepatology 24:1407–1410. [DOI] [PubMed] [Google Scholar]
- 34. Giret MT, Miraglia JL, Sucupira MC, Nishiya A, Levi JE, et al. (2011) Prevalence, incidence density, and genotype distribution of GB virus C infection in a cohort of recently HIV-1-infected subjects in Sao Paulo, Brazil. PLoS One 6:e18407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith D, Warrne D, Vlahov D, Schuman P, Stein M, et al. (1997) Design and baseline participant characteristics of the Human Immunodeficiency Virus Epidemiology Research (HER) Study: a prospective cohort of human immunodeficiency virus infection in US women. American Journal of Epidemiology 146:459–469. [DOI] [PubMed] [Google Scholar]
- 36. Mayer K, Hogan J, Smith D, Klein R, Schuman P, et al. (2003) Clinical and immunologic progression in HIV-infected US women before and after the introduction of highly active antiretroviral therapy. Journal of Acquired Immune Deficiency Syndromes 33:614–624. [DOI] [PubMed] [Google Scholar]
- 37. Stover C, Smith D, Schmid D, Pellett P, Stewart J, et al. (2003) Prevalence of and risk factors for viral infections among human immunodeficiency virus (HIV)-infected and high-risk HIV-uninfected women. Journal of Infectious Diseases 187:1388–1396. [DOI] [PubMed] [Google Scholar]
- 38. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 2007:21. [DOI] [PubMed] [Google Scholar]
- 39. Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quiros-Roldan E, Maroto MC, Torti C, Moretti F, Casari S, et al. (2002) No evidence of beneficial effect of GB virus type C infection on the course of HIV infection. AIDS 16:1430–1431. [DOI] [PubMed] [Google Scholar]
- 41. Toyoda H, Fukuda Y, Hayakawa T, Takamatsu J, Saito H (1998) Effect of GB virus C/hepatitis G virus coinfection on the course of HIV infection in hemophilia patients in Japan. Journal of Acquired Immune Deficiency Syndromes 17:209–213. [DOI] [PubMed] [Google Scholar]
- 42. Birk M, Lindback S, Lidman C (2002) No influence of GB virus C replication on the prognosis in a cohort of HIV-1-infected patients. AIDS 16:2482–2485. [DOI] [PubMed] [Google Scholar]
- 43. Zhang W, Chaloner K, Tillmann HL, Williams CF, Stapleton J (2006) Effect of early and late GB virus C viraemia on survival of HIV-infected individuals: a meta-analysis. HIV Medicine 7:173–180. [DOI] [PubMed] [Google Scholar]
- 44. Sheng WH, Hung CC, Wu RJ, Wang JT, Chen PJ, et al. (2007) Clinical impact of GB virus C viremia on patients with HIV type 1 infection in the era of highly active antiretroviral therapy. Clinical Infectious Diseases 44:584–590. [DOI] [PubMed] [Google Scholar]
- 45. Bhattarai N, Stapleton J (2012) GB virus C: the good boy virus? Trends in Microbiology 20:124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schlueter V, Schmolke S, Stark K, Hess G, Ofenloch-Haehnle B, et al. (1996) Reverse transcription-PCR detection of hepatitis G virus. Journal of Clinical Microbiology 34:2660–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berzsenyi MD, Bowden DS, Kelly HA, Watson KM, Mijch AM, et al. (2007) Reduction in hepatitis C-related liver disease associated with GB virus C in human immunodeficiency virus coinfection. Gastroenterology 133:1821–1830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.