Abstract
STUDY QUESTION
Are single-nucleotide polymorphisms (SNPs) at the interleukin 1A (IL1A) gene locus associated with endometriosis risk?
SUMMARY ANSWER
We found evidence for strong association between IL1A SNPs and endometriosis risk.
WHAT IS KNOWN ALREADY
Genetic factors contribute substantially to the complex aetiology of endometriosis and the disease has an estimated heritability of ∼51%. We, and others, have conducted genome-wide association (GWA) studies for endometriosis, which identified a total of nine independent risk loci. Recently, two small Japanese studies reported eight SNPs (rs6542095, rs11677416, rs3783550, rs3783525, rs3783553, rs2856836, rs1304037 and rs17561) at the IL1A gene locus as suggestively associated with endometriosis risk. There is also evidence of a link between inflammation and endometriosis.
STUDY DESIGN, SIZE, DURATION
We sought to further investigate the eight IL1A SNPs for association with endometriosis using an independent sample of 3908 endometriosis cases and 8568 controls of European and Japanese ancestry. The study was conducted between October 2013 and July 2014.
PARTICIPANTS/MATERIALS, SETTING, METHODS
By leveraging GWA data from our previous multi-ethnic GWA meta-analysis for endometriosis, we imputed variants in the IL1A region, using a recent 1000 Genomes reference panel. After combining summary statistics for the eight SNPs from our European and Japanese imputed data with the published results, a fixed-effect meta-analysis was performed. An additional meta-analysis restricted to endometriosis cases with moderate-to-severe (revised American Fertility Society stage 3 or 4) disease versus controls was also performed.
MAIN RESULTS AND THE ROLE OF CHANCE
All eight IL1A SNPs successfully replicated at P < 0.014 in the European imputed data with concordant direction and similar size to the effects reported in the original Japanese studies. Of these, three SNPs (rs6542095, rs3783550 and rs3783525) also showed association with endometriosis at a nominal P < 0.05 in our independent Japanese sample. Fixed-effect meta-analysis of the eight SNPs for moderate-to-severe endometriosis produced a genome-wide significant association for rs6542095 (odds ratio = 1.21; 95% confidence interval = 1.13–1.29; P = 3.43 × 10−8).
LIMITATIONS, REASONS FOR CAUTION
The meta-analysis for moderate-to-severe endometriosis included results of moderate-to-severe endometriosis cases from our European data sets and all endometriosis cases from the Japanese data sets, as disease stage information was not available for endometriosis cases in the Japanese data sets.
WIDER IMPLICATIONS OF THE FINDINGS
SNP rs6542095 is located ∼2.3 kb downstream of the IL1A gene and ∼6.9 kb upstream of cytoskeleton-associated protein 2-like (CKAP2L) gene. The IL1A gene encodes the IL1a protein, a member of the interleukin 1 cytokine family which is involved in various immune responses and inflammatory processes. These results provide important replication in an independent Japanese sample and, for the first time, association of the IL1A locus in endometriosis patients of European ancestry. SNPs within the IL1A locus may regulate other genes, but if IL1A is the target, our results provide supporting evidence for a link between inflammatory responses and the pathogenesis of endometriosis.
STUDY FUNDING/COMPETING INTEREST(S)
The research was funded by grants from the Australian National Health and Medical Research Council and Wellcome Trust. None of the authors has competing interests for the study.
Keywords: endometriosis, genome-wide association studies, interleukin 1A, inflammation, immune responses
Introduction
Endometriosis (MIM 131200) is a common gynaecological disorder that affects women in their reproductive years and is primarily characterized by the presence of normal endometrial tissue in the peritoneal cavity. The most common symptoms include severe pelvic pain, heavy or irregular menstrual bleeding and pain during intercourse and exercise, however some women remain asymptomatic. Endometriosis is also associated with infertility, lower abdominal and back pain, diarrhoea and/or constipation and chronic fatigue. Current epidemiological estimates indicate that endometriosis occurs in 6–10% of women of reproductive age (Treloar et al., 1999) and in 20–50% of women with infertility (Gao et al., 2006). Since it affects younger working age women, there are severe economic consequences arising from loss of income, treatment costs, depression, anxiety and other disease-related disorders, posing a significant burden on patients and the society. The aetiology of the disease is complex, and is likely to result from a combined interplay of genetic, health, lifestyle and reproductive risk factors. Endometriosis has an estimated heritability of ∼51% (Treloar et al., 1999).
Genome-wide association (GWA) studies for endometriosis to date have implicated nine single-nucleotide polymorphism (SNP) loci at a genome-wide significant level (P < 5 × 10−8) (Uno et al., 2010; Painter et al., 2011; Nyholt et al., 2012; Albertsen et al., 2013). The first GWA study was conducted in a Japanese population comprising 1907 cases and 5292 controls, and this identified an SNP (rs1096523) in CDKN2BAS at 9p21.3 (Uno et al., 2010). Subsequently, a European GWA study conducted by the International Endogene Consortium (IEC), including 3194 endometriosis cases and 7060 controls from Australian and UK case–control data, identified an intergenic SNP (rs12700667) at 7p15.2 (Painter et al., 2011). The study also provided genome-wide significant evidence for another SNP (rs7521902) near WNT4 at 1p36.12, after combining published results from Uno et al. (2010). In 2012, the IEC carried out a formal multi-ethnic GWA meta-analysis (Nyholt et al., 2012) of summary results from the Japanese (Uno et al., 2010) and European (Painter et al., 2011) GWA studies, providing strong evidence for association of seven SNP loci with endometriosis. The results further established association for three previously reported loci (1p36.12, 7p15.2 and GREB1 association signal at 2p25.1) and identified four novel SNP loci (rs10859871 near VEZT at 12q22, rs4141819 at 2p14, rs7739264 near ID4 at 6p22.3 and rs1537377 near CDKN2B-AS1 at 9p21.3). Of the four novel loci, three SNP loci (at 2p14, 6p22.3 and 9p21.3) were uncovered when analysis was performed after excluding endometriosis cases with minimal or mild [revised American Fertility Society (rAFS) (American Society for Reproductive Medicine, 1997) stage 1 or 2 disease] endometriosis. Furthermore, the GREB1 signal at 2p25.1 was implicated in an additional meta-analysis of European and Japanese GWA data after combining published results for rs1339416 from Adachi et al. (2010) which was a small GWA study comprising 696 endometriosis cases and 825 controls of Japanese descent.
Even though Adachi et al. (2010) did not find any SNP loci with genome-wide significant evidence, the authors reported four SNPs (rs6542095, rs11677416, rs3783550 and rs3783525; hereafter referred to as ‘common tagSNPs’) located within or near the interleukin 1A gene (IL1A) as suggestively associated with endometriosis (P < 10−5). Since these SNPs were not genotyped in the European and Japanese GWA data, they were not previously analysed by the IEC in the multi-ethnic GWA meta-analysis for endometriosis (Nyholt et al., 2012). More recently, the same (Adachi et al., 2010) group sequenced all the exons of IL1A in 377 endometriosis cases and 457 healthy controls and provided strong evidence for association of four additional SNPs [rs17561, rs1304037, rs2856836 and rs3783553 (indel variant); hereafter referred to as ‘coding SNPs’] in IL1A with endometriosis (Hata et al., 2013). The strongest association signal was reported for a non-synonymous variant (rs17561) in IL1A (P = 2.5 × 10−7), after meta-analysing with an independent replication set of 524 endometriosis cases and 533 controls. The protein (IL1a) encoded by IL1A is a member of the interleukin 1 cytokine family and is involved in primarily pro-inflammatory immune processes and haematopoiesis. Studies have reported associations of endometriosis with increased serum and peritoneal fluid inflammatory markers (Barbieri et al., 1986; Abrao et al., 1997; Vercellini et al., 2011) and some researchers have reported co-occurrences of autoimmune diseases and endometriosis (Mathur, 2000; Barrier, 2010). The primary symptom of endometriosis, pelvic pain, is also found to be relieved by the use of anti-inflammatory drugs (Vercellini et al., 2011; Vincent, 2011).
Encouraged by several lines of evidence supporting a potential link between inflammation and endometriosis, as well as two genetic case–control studies reporting association of IL1A variants with endometriosis in a Japanese population, we sought to investigate variants at the IL1A locus for involvement with endometriosis susceptibility by analysing the independent European and Japanese GWA data utilized in our previous multi-ethnic GWA meta-analysis for endometriosis, after imputation using a recent panel of 1000 Genomes Project (March 2012 release). Due to a denser coverage of markers from imputation, such an analysis would allow us to investigate the previously reported eight IL1A variants by the Japanese group while interrogating additional variants in the region for their possible associations with susceptibility to endometriosis.
Materials and Methods
Data sets
In this study, we utilized data from three individual GWA data sets of endometriosis, which were included in our recent multi-ethnic meta-analysis (Nyholt et al., 2012); a brief summary of these has been provided in Table I. Of these, two GWA samples (QIMR and OX) (Painter et al., 2011; Nyholt et al., 2012) are of European origin and one (BioBank of Japan, BBJ) (Nyholt et al., 2012) is of Japanese descent. Endometriosis cases (n = 3181) in the European GWA sample were recruited by the IEC [Australia (QIMR) = 2262; UK (OX) = 919] and a detailed description is provided elsewhere (Painter et al., 2011; Nyholt et al., 2012). Briefly, all cases in the QIMRHCS and OX GWA data had a surgically confirmed diagnosis of endometriosis and their disease stage was assessed from surgical records using the rAFS classification system (American Society for Reproductive Medicine, 1997). Cases were grouped into minimal or mild (rAFS stage 1 or 2 or some ovarian disease with a few adhesions; n = 1680) endometriosis (‘Grade_A’), moderate-to-severe (rAFS stage 3 or 4; n = 1357) endometriosis (‘Grade_B’) or unknown stage (n = 144). Ancestry-matched population controls (n = 8075) in the European GWA data were from an Australian adolescent twin study, the Hunter Community Study (HCS) (McEvoy et al., 2010) and the Wellcome Trust Case–Control Consortium 2 (WTCCC2) [combined Australia-HCS (QIMRHCS) = 2924; UK (OX) = 5151]. The QIMRHCS and OX samples were genotyped on Illumina 670Quad (cases) and 610Quad (controls) using the services from deCODE genetics. HCS controls were genotyped at the University of Newcastle using Illumina 610Quad Beadchips. The WTCCC2 controls were genotyped at the Welcome Trust Sanger Institute using Illumina HumanHap1M Beadchips. Cases (n = 1423) in the Japanese GWA data were obtained from the BBJ, Institute of Medical Science at the University of Tokyo and were diagnosed with endometriosis by the presence of multiple clinical symptoms, physical examinations and/or laparascopy or imaging tests. Female controls (n = 1318) were healthy volunteers from the Osaka-Midosuji Rotary Club, Osaka, Japan, and women from BBJ with no prior history of endometriosis. All cases and controls in the BBJ GWA data were genotyped on Illumina 550K Beadchips.
Table I.
Summary of case–control data used in this study.
| Data sets | Endometriosis cases (‘Grade_B’) | Controls | Ethnicity |
|---|---|---|---|
| QIMRHCS | 2262 (905) | 2924 | European |
| OX | 919 (452) | 5151 | European |
| BBJ | 1423 | 1318 | Japanese |
| Adachi et al. (2010) | 696 | 825 | Japanese |
| Hata et al. (2013) | 377 | 457 | Japanese |
QIMRHCS, Queensland Institute of Medical Research and Hunter Community Study; OX, Oxford (UK); BBJ, BioBank of Japan.
Standard quality control procedures were applied in individual QIMRHCS, OX and BBJ genotyped GWA data, as described earlier (Nyholt et al., 2012).
Genotype imputation
Using a similar approach adopted in our previous multi-ethnic GWA meta-analysis of endometriosis (Nyholt et al., 2012), we defined the IL1A region by extending 2500 kb 5′ and 3′ of the best SNP (rs3783525; P = 1.4 × 10−6) reported by Adachi et al. (2010). We then imputed genotypes within the defined IL1A region using a reference panel of 1000 Genomes project (March 2012 release). Imputation was carried out separately in the combined QIMRHCS-OX and BBJ genotyped data with only the overlapping genotyped SNPs, using the MaCH and minimac programs (Li et al., 2009, 2010) and following the two-step approach outlined in the online Minimac: 1000 Genomes Imputation Cookbook (http://genome.sph.umich.edu/wiki/Minimac:_1000_Genomes_Imputation_Cookbook). The quality of the imputed genotypes was assessed by R2 metric, which estimates the squared correlation between true and imputed genotypes. Poorly imputed SNPs indicated by R2 < 0.3 were excluded from the downstream analyses.
Association meta-analysis
For SNPs passing quality control, analysis of imputed genotype dosage scores was performed using mach2dat and PLINK programs (Purcell et al., 2007) in individual QIMRHCS, OX and BBJ imputed data. After combining summary results from individual QIMRHCS, OX and BBJ imputed data comprising all endometriosis cases (‘All’) and controls with published results from Adachi et al. (2010) and Hata et al. (2013), we performed a meta-analysis for the eight IL1A SNPs, using a fixed-effect (inverse variance-weighted) model implemented in the GWAMA program (Magi and Morris, 2010). Heterogeneity of allelic associations was examined using the Cochran's Q statistic P < 0.1 (Cochran, 1954), as well as the I2 index (Ioannidis et al., 2007), which indicates the proportion of variance attributable to between-study heterogeneity. A meta-analysis of SNPs associated in the fixed-effect model showing evidence of heterogeneity (P < 0.1) was also carried out using the Han–Eskin random-effects model (RE2) (Han and Eskin, 2011) implemented in METASOFT program. Compared with the conventional random-effects model, the RE2 model offers greater power under heterogeneity (Han and Eskin, 2011).
Considering the relatively greater genetic loading (or liability) of ‘Grade_B’ endometriosis compared with ‘Grade_A’ endometriosis (Painter et al., 2011; Nyholt et al., 2012), a meta-analysis of moderate-to-severe endometriosis cases in the QIMRHCS and OX imputed data, and all endometriosis cases in the BBJ imputed data, Adachi et al. (2010) and Hata et al. (2013) versus controls, was also performed. As shown previously, such an analysis has the potential to enrich true genetic effects, despite the reduced sample size (Painter et al., 2011; Nyholt et al., 2012).
Conditional analysis
Given that three coding SNPs (rs17561, rs1304037 and rs2856836) are in a weak LD [r2∼0.42 in Japanese and ∼0.11 in Caucasians, as calculated from 1000G data using --r2 option in PLINK (Purcell et al., 2007)] with the other three common tagSNPs (rs6542095, rs3783550 and rs3783525), we first explored whether these two sets of IL1A SNPs represent two independent association signals, by performing an additional analysis conditioned on rs6542095 using the --condition option in PLINK (Purcell et al., 2007). SNPs rs3783550 and rs3783525 are in a perfect LD (r2 = 1) with rs6542095 in the Japanese and Caucasians 1000G populations. Due to availability, conditional analysis was performed using the combined QIMRHCS + OX European imputed data only.
Results
All eight SNPs were accurately imputed with R2 > 0.98 in the individual QIMRHCS, OX and BBJ data. Furthermore, we also compared imputed genotypes (dosage scores) with true genotypes available for two IL1A SNPs (rs6542095 and rs1761) in a subset of QIMRHCS samples (2213 endometriosis cases and 1375 controls). Genotype concordances (as measured by Pearson's correlation coefficient) between the two sets of genotypes for rs6542095 and rs1761 were 0.99 (P < 2.2 × 10−16) and 1 (P < 2.2 × 10−16), respectively.
Our primary meta-analysis of the eight IL1A SNPs including individual QIMRHCS, OX, BBJ imputed data and published results of coding SNPs and the common tagSNPs provided further evidence for implication of the IL1A locus in endometriosis (Table II). Overall, effect direction of the eight SNPs in both ‘All’ and ‘Grade_B’ endometriosis and in all three individual imputed data sets were in line with the published results of the coding SNPs and common tagSNPs. One common tagSNP (rs11677416) and three coding SNPs (rs2856836, rs1304037 and rs17561) showed nominally significant association (P < 0.05) with ‘All’ and ‘Grade_B’ endometriosis in QIMRHCS imputed data. However, these SNPs were not nominally significant in OX imputed data. The other three common tagSNPs (rs6542095, rs3783550 and rs3783525) and a coding SNP (rs3783553) were associated with both ‘All’ and ‘Grade_B’ endometriosis at a nominal P < 0.05 in OX imputed data but not in QIMRHCS imputed data. Importantly, all eight SNPs were associated (P < 0.014) in the combined European imputed data (QIMRHCS + OX) for both ‘All’ and ‘Grade_B’ endometriosis. Of these, three common tagSNPs (rs6542095, rs3783550 and rs3783525) also showed nominally significant (P < 0.05) association with endometriosis in BBJ imputed data. Data for the indel variant (rs3783553) were not available in the BBJ imputed data.
Table II.
Summary of association results for the IL1A variants in individual case–control data sets.
| Chr | SNP | Position (bp) | RA | OA | QIMRHCS (‘All’) |
QIMRHCS (‘Grade_B’) |
OX (‘All’) |
OX (‘Grade_B’) |
QIMRHCS+OX (‘All’) |
QIMRHCS + OX (‘Grade_B’) |
BBJ |
Adachi et al. (2010) |
Hata et al. (2013) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | |||||
| 2 | rs6542095 | 113 529 183 | C | T | 1.06 (0.98–1.15) | 1.70E−01 | 1.07 (0.95–1.19) | 2.70E−01 | 1.18 (1.06–1.31) | 1.91E−03 | 1.32 (1.14–1.52) | 1.58E−04 | 1.11 (1.04–1.18) | 2.77E−03 | 1.15 (1.06–1.26) | 1.48E−03 | 1.16 (1.02–1.31) | 2.14E−02 | 1.50 (1.27–1.78) | 2.90E−06 | — | — |
| 2 | rs11677416 | 113 529 240 | T | C | 1.13 (1.04–1.23) | 5.17E−03 | 1.14 (1.01–1.28) | 3.34E−02 | 1.02 (0.92–1.14) | 6.84E−01 | 1.13 (0.97–1.32) | 1.13E−01 | 1.09 (1.02–1.16) | 1.43E−02 | 1.13 (1.03–1.24) | 7.96E−03 | 0.98 (0.82–1.17) | 8.13E−01 | 2.00 (1.50–2.67) | 2.30E−06 | — | — |
| 2 | rs3783550 | 113 532 885 | G | T | 1.06 (0.98–1.15) | 1.66E−01 | 1.07 (0.95–1.19) | 2.69E−01 | 1.18 (1.06–1.31) | 1.83E−03 | 1.32 (1.14–1.52) | 1.53E−04 | 1.11 (1.04–1.18) | 2.61E−03 | 1.15 (1.06–1.26) | 1.45E−03 | 1.16 (1.02–1.31) | 2.30E−02 | 1.51 (1.27–1.79) | 2.70E−06 | — | — |
| 2 | rs3783525 | 113 541 819 | T | A | 1.06 (0.98–1.15) | 1.62E−01 | 1.07 (0.95–1.19) | 2.70E−01 | 1.18 (1.06–1.31) | 1.86E−03 | 1.32 (1.14–1.52) | 1.52E−04 | 1.11 (1.04–1.18) | 2.56E−03 | 1.15 (1.06–1.26) | 1.44E−03 | 1.15 (1.02–1.31) | 2.70E−02 | 1.52 (1.28–1.81) | 1.40E−06 | — | — |
| 2 | rs3783553 | 113 531 715 | — | TGAA | 1.07 (0.98–1.16) | 1.24E−01 | 1.07 (0.96–1.20) | 2.26E−01 | 1.19 (1.07–1.32) | 1.57E−03 | 1.32 (1.14–1.52) | 1.54E−04 | 1.11 (1.04–1.19) | 1.58E−03 | 1.16 (1.06–1.27) | 1.08E−03 | — | — | — | — | 1.40 (1.10–1.76) | 6.10E−03 |
| 2 | rs2856836 | 113 532 083 | A | G | 1.13 (1.04–1.23) | 5.10E−03 | 1.14 (1.01–1.28) | 3.34E−02 | 1.02 (0.92–1.14) | 6.85E−01 | 1.13 (0.97–1.32) | 1.13E−01 | 1.09 (1.02–1.16) | 1.42E−02 | 1.13 (1.03–1.24) | 7.98E−03 | 0.98 (0.82–1.17) | 8.13E−01 | — | — | 1.94 (1.30–2.89) | 1.40E−03 |
| 2 | rs1304037 | 113 532 236 | T | C | 1.13 (1.04–1.23) | 5.39E−03 | 1.14 (1.01–1.28) | 3.23E−02 | 1.02 (0.92–1.14) | 6.67E−01 | 1.13 (0.97–1.32) | 1.09E−01 | 1.09 (1.02–1.16) | 1.42E−02 | 1.13 (1.03–1.24) | 7.96E−03 | 0.98 (0.82–1.17) | 8.13E−01 | — | — | 1.88 (1.26–2.82) | 2.40E−03 |
| 2 | rs17561 | 113 537 223 | C | A | 1.13 (1.04–1.23) | 5.00E−03 | 1.14 (1.01–1.28) | 3.34E−02 | 1.02 (0.92–1.14) | 6.85E−01 | 1.13 (0.97–1.32) | 1.13E−01 | 1.09 (1.02–1.16) | 1.40E−02 | 1.13 (1.03–1.25) | 7.48E−03 | 0.98 (0.82–1.17) | 8.13E−01 | — | — | 1.88 (1.26–2.82) | 2.40E−03 |
Chr, chromosome; Position, chromosomal position (bp) based on Human Build 37 (GRCh37/hg19); RA, risk allele; OA, other allele; OR, odds ratio; CI, confidence interval.
After including results from Adachi et al. (2010) and Hata et al. (2013) with QIMRHCS, OX and BBJ imputed data using fixed-effect meta-analysis, we observed a near genome-wide significant association signal of rs6542095 with ‘All’ endometriosis [odds ratio (OR) = 1.15, 95% confidence interval (CI) = 1.09–1.22, P = 3.45 × 10−7] (Table III and Fig. 1). Two other proxy SNPs (rs3783525 and rs3783550) of rs6542095 also showed similar strengths of association [rs3783525: OR = 1.15, 95% CI = 1.09–1.22, P = 4.78 × 10−7 and rs3783550: OR = 1.15, 95% CI = 1.09–1.22, P = 5.21 × 10−7]. The remaining five IL1A SNPs showed reduced association (P < 7.65 × 10−3). Between-study heterogeneity of allelic effects were significant for all the eight SNPs (Cochran's Q statistic P < 0.06, 0.64 ≥ I2≤ 0.85) and hence additional meta-analysis using the RE2 model was performed. The results from the RE2 model were comparable with the ones from fixed-effect meta-analysis (Table III).
Table III.
Summary of meta-analysis results for the IL1A variants.
| Chr | SNP | Position (bp) | RA | OA | Meta-analysis (fixed-effect) |
Meta-analysis (RE2) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ‘All’ |
‘Grade_B’ |
‘All’ | ‘Grade_B’ | |||||||||||||
| OR (95% CI) | P-value | Q statistic P | I2 | Direction | OR (95% CI) | P-value | Q statistic P | I2 | Direction | P-value | P-value | |||||
| 2 | rs6542095 | 113 529 183 | C | T | 1.15 (1.09–1.22) | 3.60E−07 | 3.45E−03 | 0.78 | ++++ | 1.21 (1.13–1.29) | 3.43E−08 | 4.47E−03 | 0.77 | ++++ | 1.03E−07 | 8.87E−09 |
| 2 | rs11677416 | 113 529 240 | T | C | 1.10 (1.04–1.17) | 1.54E−03 | 1.46E−04 | 0.85 | ++−+ | 1.15 (1.06–1.25) | 5.38E−04 | 5.80E−04 | 0.83 | ++−+ | 8.25E−05 | 5.66E−05 |
| 2 | rs3783550 | 113 532 885 | G | T | 1.15 (1.09–1.22) | 5.21E−07 | 4.00E−03 | 0.77 | ++++ | 1.20 (1.13–1.29) | 5.89E−08 | 4.57E−03 | 0.77 | ++++ | 1.06E−07 | 9.43E−09 |
| 2 | rs3783525 | 113 541 819 | T | A | 1.15 (1.09–1.22) | 4.78E−07 | 3.04E−03 | 0.78 | ++++ | 1.20 (1.13–1.29) | 5.47E−08 | 3.46E−03 | 0.78 | ++++ | 7.44E−08 | 6.54E−09 |
| 2 | rs3783553 | 113 531 715 | - | TGAA | 1.13 (1.06–1.20) | 1.59E−04 | 6.23E−02 | 0.64 | ++?+ | 1.19 (1.09–1.29) | 5.49E−05 | 3.06E−02 | 0.71 | ++?+ | 1.84E−04 | 2.99E−05 |
| 2 | rs2856836 | 113 532 083 | A | G | 1.09 (1.02–1.16) | 7.15E−03 | 9.88E−03 | 0.74 | ++−+ | 1.12 (1.04–1.22) | 4.29E−03 | 2.37E−02 | 0.68 | ++−+ | 1.01E−02 | 6.18E−03 |
| 2 | rs1304037 | 113 532 236 | T | C | 1.09 (1.02–1.16) | 7.65E−03 | 1.51E−02 | 0.71 | ++−+ | 1.12 (1.04–1.22) | 4.44E−03 | 3.38E−02 | 0.65 | ++−+ | 1.09E−02 | 6.44E−03 |
| 2 | rs17561 | 113 537 223 | C | A | 1.09 (1.02–1.16) | 7.58E−03 | 1.45E−02 | 0.72 | ++−+ | 1.12 (1.04–1.22) | 4.71E−03 | 3.39E−02 | 0.65 | ++−+ | 1.07E−02 | 6.82E−03 |
Chr, chromosome; Position, chromosomal position (bp) based on Human Build 37 (GRCh37/hg19); RA, risk allele; OA, other allele; OR, odds ratio; CI, confidence interval; RE2, Han–Eskin's random effects model meta-analysis; Q statistic P, Cochran's Q between-study heterogeneity test P-value; I2, percentage of variance attributable to between-study heterogeneity.
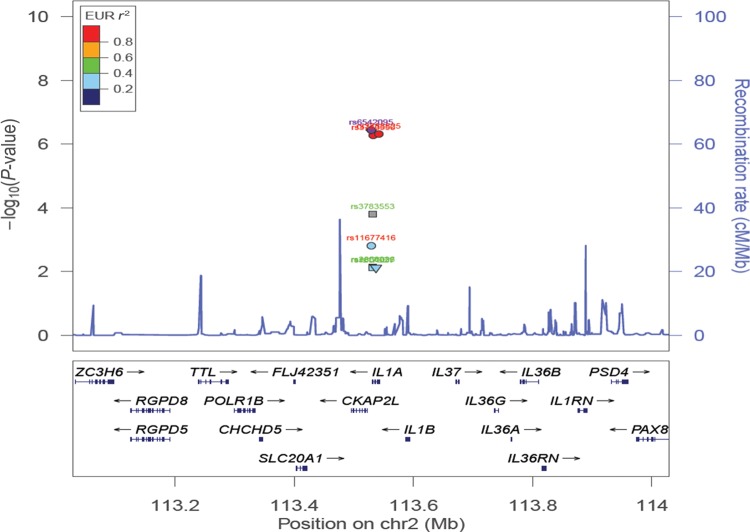
Figure 1.
Evidence of association in ‘All’ endometriosis from the fixed-effect meta-analysis of the eight IL1A variants, using imputed data from QIMRHCS, OX and BBJ and published results from Adachi et al. (2010) and Hata et al. (2013). Circles in the plot represent SNPs with no functional annotations. Squares represent SNPs located in either coding or untranslated regions. Inverted triangles denote non-synonymous SNPs. SNP names in red font are reported by Adachi et al. (2010) and SNP names in green font are reported by Hata et al. (2013). The most significant SNP (rs6542095) is represented by a purple circle. All other SNPs are colour coded according to the strength of LD with the best SNP (as measured by r2 in the European 1000 Genomes data).
In a meta-analysis of ‘Grade_B’ endometriosis cases versus controls, we observed genome-wide significant association of rs6542095 (OR = 1.21, 95% CI = 1.13–1.29, P = 3.43 × 10−8) with endometriosis (Table III and Fig. 2). The associations of rs3783525 (OR = 1.20, 95% CI = 1.13–1.29, P = 5.47 × 10−8) and rs3783550 (OR = 1.20, 95% CI = 1.13–1.29, P = 5.89 × 10−8) with endometriosis also persisted with stronger evidence than in ‘All’ endometriosis. The other five IL1A SNPs also remained associated with endometriosis (P < 6.82 × 10−3). All eight SNPs produced evidence for between-study effect heterogeneity (Cochran's Q statistic P < 0.03, 0.65 ≥ I2≤ 0.83); however, after appropriate modelling of this heterogeneity, association signals of the top three common tagSNPs became slightly stronger (rs6542095: P = 8.87 × 10−9, rs3783525: P = 6.54 × 10−9 and rs3783550: P = 9.43 × 10−9) in the RE2 model.
Figure 2.
Evidence of association in ‘Grade_B’ endometriosis from the fixed-effect meta-analysis of the eight IL1A variants, using imputed data from QIMRHCS, OX and BBJ and published results from Adachi et al. (2010) and Hata et al. (2013). Circles in the plot represent SNPs with no functional annotations. Squares represent SNPs located in either coding or untranslated regions. Inverted triangles denote non-synonymous SNPs. SNP names in red font are reported by Adachi et al. (2010) and SNP names in green font are reported by Hata et al. (2013). The most significant SNP (rs6542095) is represented by a purple circle. All other SNPs are colour coded according to the strength of LD with the best SNP (as measured by r2 in the European 1000 Genomes data).
In an additional meta-analysis for all other remaining variants in the IL1A region using individual QIMRHCS, OX and BBJ imputed data, a total of four SNPs (rs1969294, rs13000462, rs3783543 and rs2071376 that were present in all three data sets) showed marginally stronger association P-values than at least one of the top three common tagSNPs in either ‘All’ or ‘Grade_B’ endometriosis (Supplementary data, Table SI, Supplementary data, Figs 1 and 2). However, these SNPs are in a strong LD (r2 > 0.87) with rs6542095 in all the Japanese and Caucasians 1000G data and hence are likely to represent the same association signal of rs6542095. Furthermore, when we conditioned on rs6542095, the association signals, for three of the IL1A coding SNPs reported by Hata et al. (2013) and one common tagSNP reported by Adachi et al. (2010), vanished (P > 0.14) (Supplementary data, Table SII) in both ‘All’ and ‘Grade_B’ endometriosis.
Discussion
Endometriosis is considered to be associated with inflammatory responses, which are in part initiated by a network of pro-inflammatory proteins of the IL1 cytokine family. IL1A encodes IL1a, a member of the IL1 cytokine family that possesses a strong pro-inflammatory effect. A small Japanese GWA study reported four common tagSNPs in and around IL1A that showed suggestive associations with endometriosis (Adachi et al., 2010). The same group, subsequently, sequenced all the exons of IL1A in 377 endometriosis cases and 457 healthy controls of Japanese ancestry (Hata et al., 2013). The authors reported a total of 11 SNPs, of which four coding SNPs were suggestively associated with the risk of endometriosis. Herein, we carefully examined those eight IL1A SNPs in the context of endometriosis, using imputed data from an independent sample of 3908 cases and 8568 controls of European and Japanese ancestry. The imputed data were accurate and robust with imputation R2 > 0.98 for the eight IL1A SNPs and genotype concordance >0.99 between imputed and true genotypes of two SNPs in a subset of samples. Our results provide important replication in an independent Japanese sample and association of the IL1A locus in endometriosis patients of European Ancestry.
All eight IL1A SNPs were associated with endometriosis in at least one European data set at a nominal P < 0.05 (Table II). After combining the two European imputed data sets (QIMRHCS and OX), these SNPs remained significant at P < 0.014. Effect sizes for all eight IL1A SNPs were similar and in the same direction as the published results. Furthermore, the associations were stronger in moderate-to-severe (‘Grade_B’) endometriosis than in all endometriosis (‘All’), in line with our previous reports supporting greater genetic loading for moderate-to-severe endometriosis (Painter et al., 2011; Nyholt et al., 2012). Three common tagSNPs (rs6542095, rs3783525, rs3783550) and the indel variant (rs3783553) that are in a strong LD with each other (r2 > 0.94 in Japanese and Caucasian 1000G data) showed slightly stronger associations than the remaining SNPs. The indel variant was not present in the BBJ imputed data, but the other results were consistent (association P < 0.027) in the BBJ results. The association evidence for the three common tagSNPs (rs6542095, rs3783525 and rs3783550) became stronger in the fixed-effect meta-analysis, with a genome-wide evidence (P = 3.4 × 10−8) for rs6542095 in ‘Grade_B’ endometriosis (Table III and Fig. 2). These SNPs were genome-wide significant in the meta-analysis using the RE2 model, taking into account between-study heterogeneity observed in the fixed-effect meta-analysis.
Even though Hata et al. (2013) reported rs17561, a non-synonymous coding variant, as the strongest IL1A SNP for endometriosis in a Japanese population, our results do not support their findings. Instead, our meta-analysis results more strongly implicate another SNP in the IL1A locus, rs6542095, which is in weak LD with rs17561. Notably, after conditioning on rs6542095, no residual association signal (P > 0.14) was observed for rs17561 or for two other coding SNPs (rs2856836 and rs1304037) and a common tagSNP (rs11677416) that are in perfect LD (r2 = 1 in Japanese and Caucasian 1000G data) with rs17561 (Supplementary data, Table SII). Further meta-analysis of other IL1A variants in the European and BBJ imputed data also did not implicate additional association signals for endometriosis, independent of rs6542095 (Supplementary data, Table SI). Consequently, our data do not suggest the IL1A locus harbours multiple independent association signals for endometriosis (in addition to rs6542095), at least in the European populations.
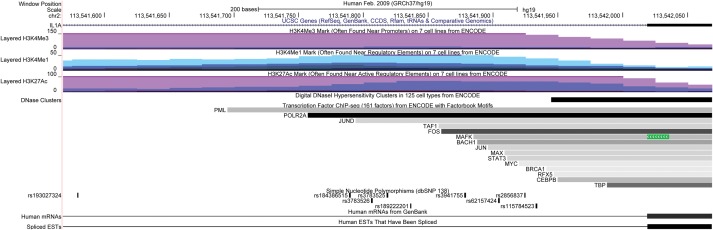
SNP rs6542095 is intergenic, located ∼2.3 kb downstream of IL1A gene and ∼6.9 kb upstream of the cytoskeleton associated protein 2-like (CKAP2L) gene. The two other common tagSNPs, rs3783525 and rs3783550, which are in perfect LD with rs6542095 (r2 = 1 in Japanese and Caucasian 1000G data), are intronic SNPs located at intron 1 and intron 6 of the IL1A gene, respectively. IL1A is a well-characterized gene that encodes the IL1a protein of the interleukin 1 cytokine family and is involved in a wide range of inflammatory activities and immune responses. A small GWA study (McClay et al., 2011) of treatment outcomes of antipsychotic drugs in schizophrenia reported two IL1A SNPs, rs11677416 and rs17561, as significantly associated with olanzapine and working memory (P = 6.67 × 10−7 and 1.05 × 10−6, respectively). The study utilized GWA data generated from 738 schizophrenia patients of European American and African American ancestries. Furthermore, based on the ENCODE data queried through the UCSC genome browser, rs3783525 directly overlaps with several regulatory regions, such as the H3K4Me1 mark (often found near regulatory elements), the H3K4Me3 mark (often found near promoters) and the H3K27Ac mark (often found near enhancers) (Fig. 3). The data indicated that rs3783525 also overlaps with binding sites of three transcription factors, including RNA Polymerase II (POLR2A), suggesting a potential regulatory or functional role of rs3783525.
Figure 3.
UCSC Genome Browser snapshot of rs3783525, with 250 bp flanking regions on its either side. Tracks displayed from top to bottom include genome base position (hg19), UCSC Genes, Layered H3K4Me3 mark on seven cell lines from ENCODE, Layered H3K4Me1 mark on seven cell lines from ENCODE, Layered H3K27Ac mark on seven cell lines from ENCODE, DNaseI Hypersensitivity Clusters in 125 cell types from ENCODE, Transcription factor CHIP-seq (161 factors) from ENCODE, All SNPs (138), Human mRNAs and Spliced ESTs, respectively.
IL1A encodes one of two related pro-inflammatory cytokines (IL-1a and IL-1b) that signal through the same receptors (Gabay et al., 2010). The two genes IL1A and IL1B are located next to each other on chromosome 2 and adjacent to interleukin 37 (IL37). While our results implicate the IL1A locus, variants associated with endometriosis risk could affect one or more these genes. Tissue-specific studies measuring RNA transcripts and proteins are needed to identify the target gene(s). In addition, fine mapping with better coverage of genomic variation at this locus and functional studies are required to identify the causal variant(s). While results from these studies may not be translated immediately into the clinic, they serve as a starting point to understand biological mechanisms underlying the role of causal variants at the IL1A locus increasing the risk of endometriosis.
In conclusion, these study results provide genome-wide significant evidence implicating the IL1A locus in endometriosis susceptibility. The original IL1A signals reported in a Japanese population were robustly replicated in European population. Consistent with our previous reports, the observed associations at the IL1A locus were stronger in moderate-to-severe endometriosis than in all endometriosis cases. SNPs within the IL1A locus may regulate other genes in the region, but if IL1A is the target, our results provide supporting evidence for a link between inflammatory responses and the pathogenesis of endometriosis.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors' roles
The authors were responsible for the following aspects of the study: Y.S.: design (QIMR part), analysis and interpretation, drafting manuscript and final approval; S.-K.L.: design and analysis (BBJ part), revision for critical content and final approval. J.A.: design (HCS part), revision for critical content and final approval; S.D.G.: design and analysis (QIMR part), revision for critical content and final approval; A.K.H.: design (QIMR part), revision for critical content and final approval; E.G.H.: design and analysis (HCS part), revision for critical content and final approval; S.M.: design (QIMR part), revision for critical content and final approval; N.G.M.: design and sample collection (QIMR part), revision for critical content and final approval; M.M.: design (HCS part), revision for critical content and final approval; A.P.M.: design and analysis (OX part), revision for critical content and final approval; A.T.: design (BBJ part), revision for critical content and final approval; R.J.S.: design (HCS part), revision for critical content and final approval; M.K.: conception and design (BBJ part), revision for critical content and final approval; K.T.Z.: conception and design (OX part), revision for critical content and final approval; G.W.M.: conception and design (QIMR part), revision for critical content and final approval; D.R.N.: conception and design (QIMR part), analysis and interpretation, revision for critical content and final approval.
Funding
The QIMR study was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (241944, 339462, 389927, 389875, 389891, 389892, 389938, 443036, 442915, 442981, 496610, 496739, 552485 and 552498), the Cooperative Research Centre for Discovery of Genes for Common Human Diseases (CRC), Cerylid Biosciences (Melbourne) and donations from N. Hawkins and S. Hawkins. The work presented here was supported by a grant from the Wellcome Trust (WT084766/Z/08/Z) and makes use of WTCCC2 control data generated by the Wellcome Trust Case–Control Consortium. A full list of the investigators who contributed to the generation of these data is available from http://www.wtccc.org.uk. Funding for the WTCCC project was provided by the Wellcome Trust under awards 076113 and 085475. D.R.N. was supported by an NHMRC Fellowship (613674) and ARC Future Fellowship (FT0991022) schemes. E.G.H. (631096) and G.W.M. (339446, 619667) were supported by the NHMRC Fellowships Scheme. The HCS was funded by the University of Newcastle, the Gladys M Brawn Fellowship scheme and the Vincent Fairfax Family Foundation in Australia. A.P.M. was supported by a Wellcome Trust Senior Research Fellowship. K.T.Z. is supported by a Wellcome Trust Research Career Development Fellowship (WT085235/Z/08/Z).
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We would like to acknowledge all of the study participants in the QIMR, OX, HCS, WTCCC2 and BBJ endometriosis studies that provided an opportunity for current study. We also thank many hospital directors and staff, gynaecologists, general practitioners and pathology services in Australia, UK and Japan who provided assistance with confirmation of diagnoses.
References
- Abrao MS, Podgaec S, Filho BM, Ramos LO, Pinotti JA, de Oliveira RM. The use of biochemical markers in the diagnosis of pelvic endometriosis. Hum Reprod. 1997;12:2523–2527. doi: 10.1093/humrep/12.11.2523. [DOI] [PubMed] [Google Scholar]
- Adachi S, Tajima A, Quan J, Haino K, Yoshihara K, Masuzaki H, Katabuchi H, Ikuma K, Suginami H, Nishida N, et al. Meta-analysis of genome-wide association scans for genetic susceptibility to endometriosis in Japanese population. J Hum Genet. 2010;55:816–821. doi: 10.1038/jhg.2010.118. [DOI] [PubMed] [Google Scholar]
- Albertsen HM, Chettier R, Farrington P, Ward K. Genome-wide association study link novel loci to endometriosis. PLoS One. 2013;8:e58257. doi: 10.1371/journal.pone.0058257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Society for Reproductive Medicine. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril. 1997;67:817–821. doi: 10.1016/s0015-0282(97)81391-x. [DOI] [PubMed] [Google Scholar]
- Barbieri RL, Niloff JM, Bast RC, Jr., Scaetzl E, Kistner RW, Knapp RC. Elevated serum concentrations of CA-125 in patients with advanced endometriosis. Fertil Steril. 1986;45:630–634. doi: 10.1016/s0015-0282(16)49333-7. [DOI] [PubMed] [Google Scholar]
- Barrier BF. Immunology of endometriosis. Clin Obstet Gynecol. 2010;53:397–402. doi: 10.1097/GRF.0b013e3181db7c33. [DOI] [PubMed] [Google Scholar]
- Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- Gabay C, Lamacchia C, Palmer G. IL-1 pathways in inflammation and human diseases. Nat Rev Rheumatol. 2010;6:232–241. doi: 10.1038/nrrheum.2010.4. [DOI] [PubMed] [Google Scholar]
- Gao X, Outley J, Botteman M, Spalding J, Simon JA, Pashos CL. Economic burden of endometriosis. Fertil Steril. 2006;86:1561–1572. doi: 10.1016/j.fertnstert.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Nakaoka H, Yoshihara K, Adachi S, Haino K, Yamaguchi M, Nishikawa N, Kashima K, Yahata T, Tajima A, et al. A nonsynonymous variant of IL1A is associated with endometriosis in Japanese population. J Hum Genet. 2013;58:517–520. doi: 10.1038/jhg.2013.32. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One. 2007;2:e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur SP. Autoimmunity in endometriosis: relevance to infertility. Am J Reprod Immunol. 2000;44:89–95. doi: 10.1111/j.8755-8920.2000.440204.x. [DOI] [PubMed] [Google Scholar]
- McClay JL, Adkins DE, Aberg K, Bukszar J, Khachane AN, Keefe RS, Perkins DO, McEvoy JP, Stroup TS, Vann RE, et al. Genome-wide pharmacogenomic study of neurocognition as an indicator of antipsychotic treatment response in schizophrenia. Neuropsychopharmacology. 2011;36:616–626. doi: 10.1038/npp.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy M, Smith W, D'Este C, Duke J, Peel R, Schofield P, Scott R, Byles J, Henry D, Ewald B, et al. Cohort profile: the Hunter Community Study. Int J Epidemiol. 2010;39:1452–1463. doi: 10.1093/ije/dyp343. [DOI] [PubMed] [Google Scholar]
- Nyholt DR, Low SK, Anderson CA, Painter JN, Uno S, Morris AP, MacGregor S, Gordon SD, Henders AK, Martin NG, et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat Genet. 2012;44:1355–1359. doi: 10.1038/ng.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter JN, Anderson CA, Nyholt DR, Macgregor S, Lin J, Lee SH, Lambert A, Zhao ZZ, Roseman F, Guo Q, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Genet. 2011;43:51–54. doi: 10.1038/ng.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treloar SA, O'Connor DT, O'Connor VM, Martin NG. Genetic influences on endometriosis in an Australian twin sample. sueT@qimr.edu.au. Fertil Steril. 1999;71:701–710. doi: 10.1016/s0015-0282(98)00540-8. [DOI] [PubMed] [Google Scholar]
- Uno S, Zembutsu H, Hirasawa A, Takahashi A, Kubo M, Akahane T, Aoki D, Kamatani N, Hirata K, Nakamura Y. A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Genet. 2010;42:707–710. doi: 10.1038/ng.612. [DOI] [PubMed] [Google Scholar]
- Vercellini P, Crosignani P, Somigliana E, Vigano P, Frattaruolo MP, Fedele L. ‘Waiting for Godot’: a commonsense approach to the medical treatment of endometriosis. Hum Reprod. 2011;26:3–13. doi: 10.1093/humrep/deq302. [DOI] [PubMed] [Google Scholar]
- Vincent K. Pelvic pain in women: clinical and scientific aspects. Curr Opin Support Palliat Care. 2011;5:143–149. doi: 10.1097/SPC.0b013e3283460b05. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.