Abstract
The physiological function of Ataxin-3 (ATXN3), a deubiquitylase (DUB) involved in Machado–Joseph Disease (MJD), remains elusive. In this study, we demonstrate that ATXN3 is required for neuronal differentiation and for normal cell morphology, cytoskeletal organization, proliferation and survival of SH-SY5Y and PC12 cells. This cellular phenotype is associated with increased proteasomal degradation of α5 integrin subunit (ITGA5) and reduced activation of integrin signalling and is rescued by ITGA5 overexpression. Interestingly, silencing of ATXN3, overexpression of mutant versions of ATXN3 lacking catalytic activity or bearing an expanded polyglutamine (polyQ) tract led to partially overlapping phenotypes. In vivo analysis showed that both Atxn3 knockout and MJD transgenic mice had decreased levels of ITGA5 in the brain. Furthermore, abnormal morphology and reduced branching were observed both in cultured neurons expressing shRNA for ATXN3 and in those obtained from MJD mice. Our results show that ATXN3 rescues ITGA5 from proteasomal degradation in neurons and that polyQ expansion causes a partial loss of this cellular function, resulting in reduced integrin signalling and neuronal cytoskeleton modifications, which may be contributing to neurodegeneration.
INTRODUCTION
The importance of ubiquitin signalling in the nervous system is becoming increasingly recognized (1–3). Impairment of the ubiquitin-proteasome pathway (UPP) and mutations in some of its components have been linked to both neurodevelopmental and neurodegenerative disorders, the later including Alzheimer's, Parkinson's and Huntington's diseases (4–6). In the context of the nervous system, deubiquitylases (DUBs) are central players in the regulation of protein ubiquitylation in processes, such as (i) axon guidance and establishment of neuronal connectivity (7), (ii) dendritic and axon pruning (8,9), (iii) regulation of synaptic number and size (10,11), (iv) regulation of synaptic plasticity (11) and (v) modulation of the postsynaptic structure (7,12).
Ataxin-3 (ATXN3) is a protein with DUB activity known to be mutated in Machado–Joseph Disease (MJD), an autosomal dominant neurodegenerative disorder caused by a polyglutamine (polyQ) tract expansion within the C-terminus of this protein (13). PolyQ expansions are thought to cause deleterious effects in neurons by conferring toxic properties to the proteins into which they are inserted (gain of function model) and by perturbing some of the biological activities of these proteins (partial loss of function model) (14–16).
Although the physiological role and substrates of ATXN3 are mostly unknown, functional analyses in different cell and animal models have shed some light on its biological functions. Evidence supports ATXN3 involvement in protein quality control pathways: (i) DUB activity conferred by cysteine 14 (C14) within the N-terminal Josephin-domain, which is essential for its protease activity (17–19); (ii) interaction with ubiquitin, polyubiquitin chains, ubiquitylated proteins (20–22) and proteasome subunits (21,23); (iii) interaction with the ubiquitin-like protein NEDD8 and deneddylase activity (24) and (iv) binding to and regulating the activity of VCP/p97, which is involved in shuttling substrates for proteasomal degradation (25,26) and binding to UBXN-5, an adaptor of substrate binding to VCP (27). In addition to its involvement in the regulation of protein degradation, the numerous molecular partners of ATXN3 known to date suggest that it is involved in other cellular processes (28–31). Although mouse and nematode knockouts (KO) for this gene are viable and show no gross phenotype, our previous results showed that the absence of ataxin-3 in Caenorhabditis elegans affects the expression of several transcripts related to cell structure/motility (32) and that ataxin-3 regulates the degradation of integrin subunits such as α5 integrin subunit (ITGA5), a molecular partner of ATXN3 (33). These regulatory functions were shown to be important for the cytoskeleton organization of different cell types (31,33).
Integrins are the major family of transmembrane cell surface receptors that mediate cell-to-cell and cell-to-extracellular matrix (ECM) interactions, regulating many cellular functions (34,35). Integrins are implicated in many aspects of neuronal development and function, such as proliferation, survival, adhesion, cytoskeletal organization, process outgrowth and synaptic function (36–40). Furthermore, cumulative evidence suggests that a disruption of the neuronal cytoskeleton network may be a common feature contributing to several neurodegenerative diseases (41,42). Data suggest that cytoskeletal deregulation initiates a cascade of intracellular events that may underlie the loss of synaptic connectivity, the decreased ability to transmit incoming axonal information and the cell death that is observed in these disorders (43–47).
In this work, we demonstrate that ATXN3 depletion deregulates ITGA5 levels through increased proteasomal degradation, which leads to decreased cell adhesion and disorganization of the neuronal cytoskeleton. Loss of function of ATXN3 also has a negative impact on neuronal differentiation and on the associated exit from the cell cycle, promoting continued proliferation. Our data also indicate that an expanded polyQ tract leads to a partial loss of the cellular function of ATXN3 that may be relevant to neurodegeneration.
RESULTS
ATXN3 knock down results in altered morphology, proliferation, migration and cell death in differentiating SH-SY5Y cells
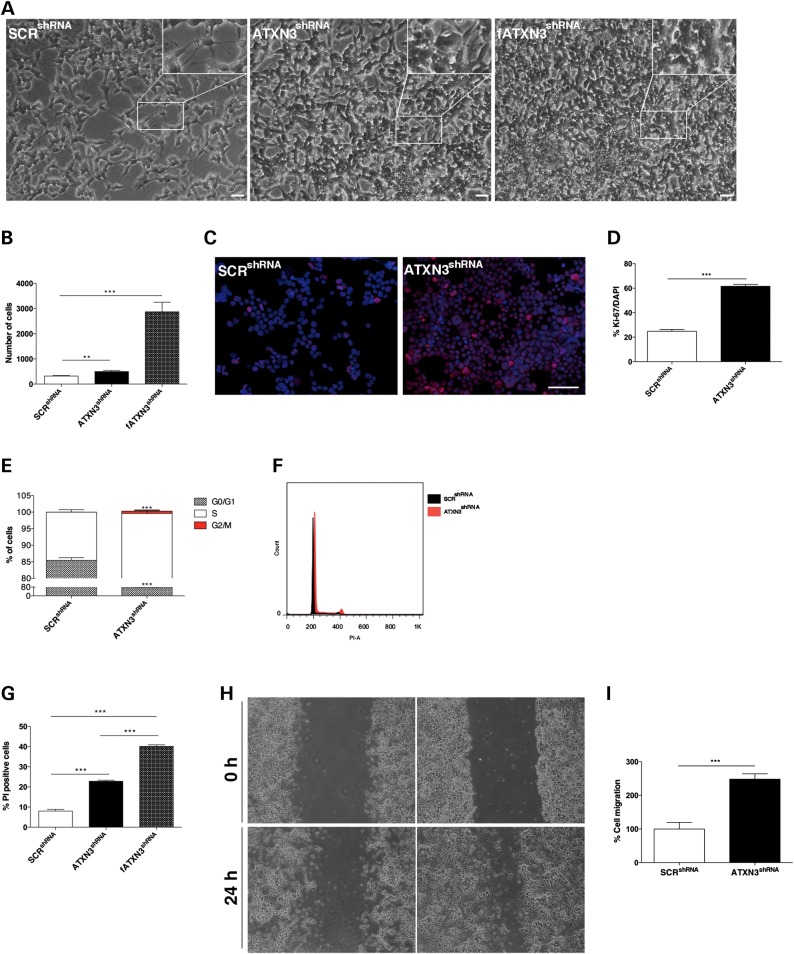
We evaluated the effects of ataxin-3 silencing in SH-SY5Y human neuroblastoma cells, widely used as a model for neuronal function and differentiation (1). After lentiviral infection and puromycin selection, we obtained monoclonal stable SH-SY5Y cell lines containing a shRNA sequence targeting ATXN3 (ATXN3shRNA), the empty vector pLKO.1 (SH-SY5Y-pLKO.1) or a scrambled shRNA (SCRshRNA) sequence. A western blot analysis of protein extracts confirmed that ATXN3 expression was markedly suppressed and almost undetectable in ATXN3shRNA clones when compared with SH-SY5Y-pLKO.1 and SCRshRNA controls (Supplementary Material, Fig. S1A and B). We confirmed by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) that the shRNA sequence used for silencing ATXN3 did not interfere with similar mRNAs, as the expression levels of the transcripts encoding the ATXN3-like (ATXN3L) and Josephin-domain proteins (JOSD1 and JOSD2) were not significantly different between ATXN3shRNA and SCRshRNA cells (Supplementary Material, Fig. S1C). ATXN3 depletion in non-differentiated SH-SY5Y cells had a mild impact on cell morphology (Supplementary Material, Fig. S2A) and statistically significant effect in migration (Supplementary Material, Fig. S2B); however, it did not affect cell cycle progression (Supplementary Material, Fig. S2C) or survival (Supplementary Material, Fig. S2E). Subsequently, we sought to analyze the effect of ATXN3 depletion on retinoic acid (RA)-differentiated SH-SY5Y cells. RA has previously been shown to induce differentiation and inhibit the cellular growth of cultured human SH-SY5Y cells (48). As shown in Figure 1A, after 7 days of RA-induced differentiation, SCRshRNA control cells exhibited branching neuritic networks with small rounded bodies and an inhibition of cell proliferation, whereas cells in which ATXN3 had been silenced displayed large, flat cell bodies with very few extensions. These striking morphological changes were observed in the different clonal cell lines and were evident in cells with only a few passages after silencing (fATXN3shRNA) as well as cells with long-term silencing of ATXN3, albeit in a more attenuated way in the latter. Such partial recovery in cell morphology might be explained by compensatory mechanisms, as has been observed in different ATXN3 KO animal models (32,49).
Figure 1.
Characterization of a SH-SY5Y neuronal cell line stably silenced for ATXN3. (A) ATXN3shRNA cells were less elongated and showed thin cell extensions as compared with the SCRshRNA controls after RA treatment. This phenotype was more pronounced in fATXN3shRNA cells. Scale bar: 100 μm. (B) RA treatment did not inhibit proliferation of ATXN3shRNA cells. (C and D) Ki-67 staining (red) showed that RA did not inhibit proliferation of ATXN3shRNA cells. Nuclei were counterstained with DAPI (blue). Scale bar: 200 μm. (E and F) Flow cytometry analysis of the cell cycle showed an increase in the percentage of cells in S phase in ATXN3shRNA cultures. (G) Flow cytometry analysis using PI showed an increased cell death in ATXN3shRNA cultures, which was even higher in fATXN3shRNA cultures. (H and I) ATXN3 knockdown increased SH-SY5Y cell migration in a wound scratch assay. Bars represent migration rate expressed as a percentage of control and calculated as the proportion of the distance between the borderlines caused by scratching, to the distance that remained cell-free after 24 h. n ≥ 3 independent biological replicates in all experiments. **P < 0.01; ***P < 0.001.
After RA treatment, there were significantly more ATXN3shRNA cells when compared with SCRshRNA control cultures (P = 0.0019) (Fig. 1B). Consistent with a higher proliferation rate, ATXN3shRNA cultures showed an increased number of Ki-67-positive cells (61.57% versus 24.24% in control cells; P = 9.46 × 10−6) (Fig. 1C and D). Accordingly, in comparison to the control SCRshRNA cells, the ATXN3shRNA cell population had a greater number of cells in S phase (24.4% compared with 14.5%; P = 0.0018) (Fig. 1E and F). This higher cell proliferation was accompanied by an increased cell death in RA-treated ATXN3shRNA cultures as determined by flow cytometry analysis (22.8% versus 8.02% of PI-positive cells; P = 0.0001) that was even higher in fATXN3shRNA cultures (40.1%; P = 5.60 × 10−5) (Fig. 1G). Because it has previously been demonstrated that ataxin-3 interacts with and regulates the levels of integrins (33) and because adhesion to the substrate is key for cell migration, we analyzed collective cell movement using a wound healing assay (50). Cells were monitored for 24 h starting immediately after the initial wound. As shown in Figure 1H and I, ATXN3shRNA cells had partially recolonized the scratched area and exhibited a 50% increase in the cell migration rate when compared with SCRshRNA cells (P = 4.42 × 10−7), which suggests that ATXN3shRNA cells did not adhere as well to the substrate. The same was observed in two other clonal cell lines with similar degrees of ATXN3 silencing. Interestingly, these effects of ATXN3 absence seem to be generalized to different neuronal cell types, as similar phenotypes were observed in PC12 cells upon ATXN3 silencing (PC12_ATXN3shRNA), in the context of nerve growth factor (NGF)-induced differentiation (Supplementary Material, Figs S1D–F and S6).
ATXN3shRNA cells fail to progress towards a mature neuronal phenotype
RA treatment increases the synthesis of neuron-specific enzymes, neurotransmitters, neuropeptide hormones, growth factors and cell surface receptors and induces changes in cytoskeleton markers in SH-SY5Y cells (51). Because we observed that neuronal differentiation was impaired in ATXN3shRNA cells, we wanted to further characterize the differentiation status of these cells. For this, we evaluated several neuronal markers associated with RA-induced differentiation (52).
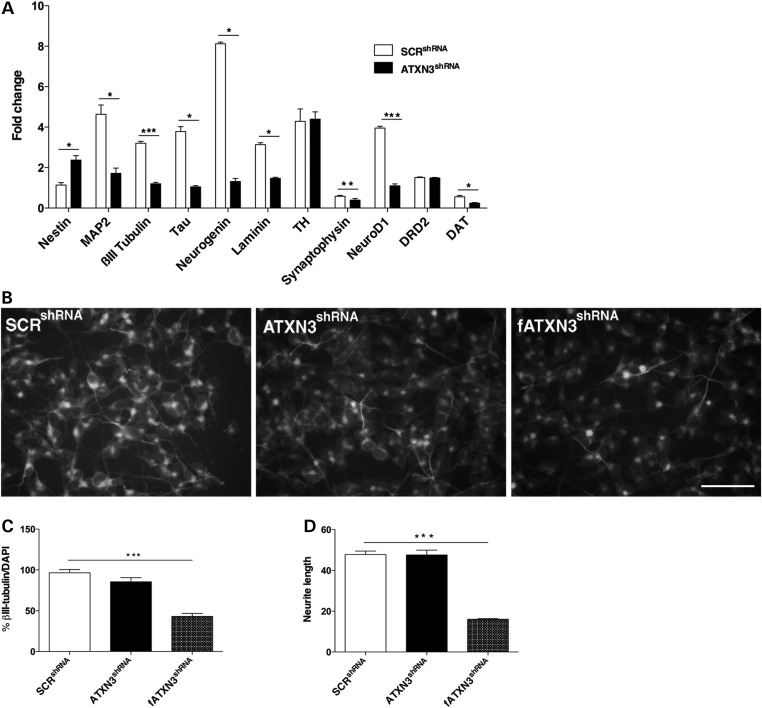
As expected, we observed increased mRNA levels for most of the neuronal markers in the SCRshRNA cells after RA treatment and decreased expression of nestin, a marker of undifferentiated cells (Fig. 2A). In contrast, the mRNA expression pattern in RA-treated ATXN3shRNA cells was consistent with immature stages of neuronal differentiation, with increased expression of nestin (P = 0.02) and decreased expression of βIII-tubulin (P = 0.0002), laminin (P = 0.02), synaptophysin (P = 0.01), neurogenin (P = 0.03), neuroD1 (P = 7.63 × 10−5) and dopamine transporter (DAT) (P = 0.04) (Fig. 2A). Consistently, decreased expression of some neuronal differentiation markers, including βIII-tubulin (P = 0.01), was also observed in NGF-treated PC12_ATXN3shRNA cells (Supplementary Material, Fig. S6F). Immunocytochemistry confirmed that the percentage of βIII-tubulin positive cells, a marker for mature neurons, was lower in ATXN3shRNA cultures (86.26% versus 97.26%) upon RA treatment and more significantly so in fATXN3shRNA cells (41.96%; P = 4.12 × 10−8) (Fig. 2B and C). Additionally, the average neurite length was significantly reduced in RA-treated fATXN3shRNA cells (P = 1.18 × 10−15) when compared with control cells (Fig. 2D), which is compatible with the immature filopodia phenotype observed in the absence of ATXN3.
Figure 2.
Impairment of the differentiation process in ATXN3shRNA cultures. (A) qRT-PCR analysis of the variation of neuronal markers after RA treatment showed that the mRNA level was not altered for TH and DRD2, was increased for nestin and decreased for the other markers analyzed in ATXN3shRNA cultures. Transcript levels were normalized to levels in undifferentiated cells and to the HBMS gene. (B and C) ATXN3shRNA cultures presented a decreased number of βIII-tubulin positive cells. (D) The average length of the neurites was reduced in fATXN3shRNA cells. This morphological alteration was attenuated after long-term silencing of ATXN3. *P < 0.05; n ≥ 3 independent biological replicates in all experiments. **P < 0.01; ***P < 0.001.
These results indicate that ATXN3-deficient cells fail to progress towards a mature neural phenotype upon RA treatment. Nevertheless, we verified that these cells are able to respond to RA, as demonstrated by the induction of the RA-responsive tissue transglutaminase (tTG) gene (53) (Supplementary Material, Fig. S3A).
ATXN3 depletion disrupts the actin cytoskeleton network in SH-SY5Y cells
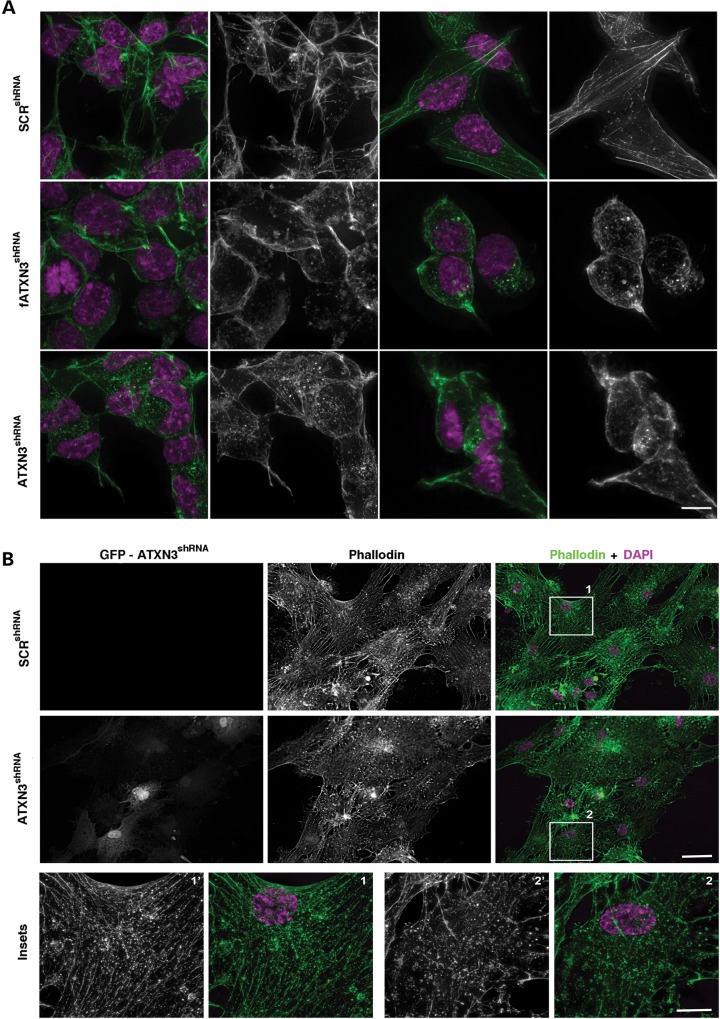
Neuronal development is highly dependent on controlled cytoskeleton dynamics and reorganization, and it has been shown that in several neurodegenerative diseases, this reorganization is defective (41,42). Phalloidin staining of actin polymers revealed the presence of disarrayed actin filaments and aggregates in ATXN3shRNA and fATXN3shRNA cells (Fig. 3A) as well as in PC12_ATXN3shRNA cells (Supplementary Material, Fig. S6G). Additionally, the ATXN3shRNA cells exhibited a higher number of small filopodia (microspikes) and the PC12_ATXN3shRNA cells grew mostly in multilayers, which were not observed in the control cultures. These cytoplasmic projections are normally present in migrating cells and at initial stages of neurogenesis (54). This finding further supports the hypothesis that in the absence of ATXN3, cells fail to properly complete differentiation and to establish mature neuronal processes. Very importantly, primary neurons with silenced ATXN3 expression also show a highly disrupted cytoskeleton network as compared with the scrambled controls, providing further evidence for the role of ataxin-3 in the regulation of the cytoskeleton (Fig. 3B).
Figure 3.
ATXN3 depletion affects the actin cytoskeleton network in neuronal cells. (A) Phalloidin staining (green) showed unparallel actin filaments and abnormal filopodia in ATXN3shRNA cells. The right panel shows a detailed view of isolated cells. (B) Primary neurons silenced for ATXN3 (GFP positive cells) showed a disrupted network of actin filaments as assessed by phalloidin staining (green). The lower panel shows a detailed view of isolated cells. Nuclei were counterstained with DAPI (purple). n ≥ 3 independent biological replicates in all experiments. Scale bar is 5 μm.
Absence of ATXN3 leads to downregulation of α5 integrin signalling
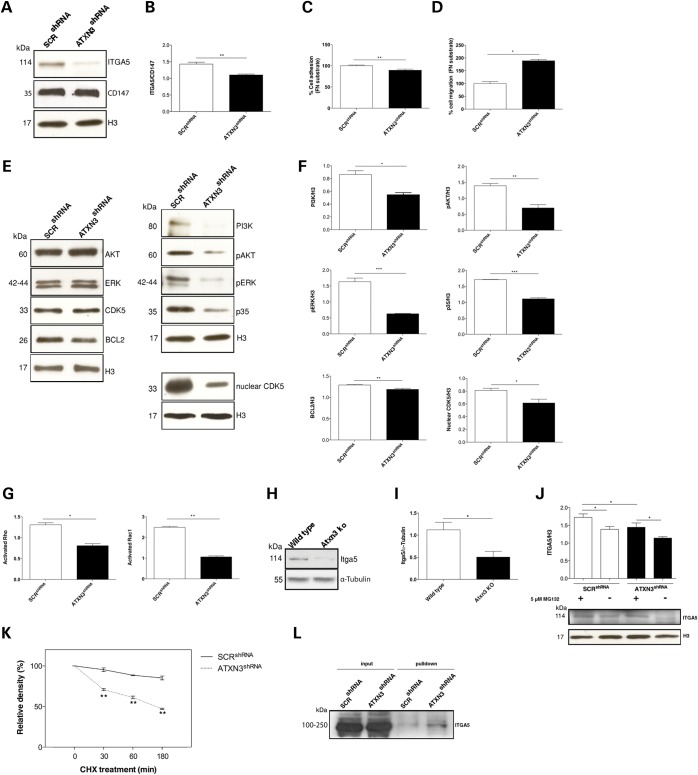
Integrins play an important role in mediating cell adhesion to the ECM proteins and activating signalling cascades that control cytoskeletal organization and cell motility (55,56). We have previously reported that ataxin-3 regulates the degradation of integrin subunits such as the ITGA5 (33). Accordingly, we observed that depletion of ATXN3 downregulates ITGA5 protein levels in SH-SY5Y (P = 0.001) and PC12 (P = 0.0047) cells induced to differentiate (with RA and NGF treatment, respectively) (Fig. 4A and B, Supplementary Material, Fig. S6H–J, respectively), but does not affect those of other integrin subunits, such as the α1 integrin subunit (Supplementary Material, Fig. S4A). Because α5β1 is the receptor for fibronectin (FN), which has been implicated in neuronal cell migration, adhesion, proliferation and differentiation both in vitro and in vivo (36), we next assessed the FN-binding capacity of ATXN3shRNA cells using a CultreCoat®Fibronectin 96 well adhesion assay (Ambsio). As expected, the adhesion of ATXN3shRNA cells to FN was significantly reduced (89.5%; P = 0.004) in relation to SCRshRNA controls (Fig. 4C). We subsequently assessed cell migration on an FN substrate. For this, we coated the plates with FN and performed the wound healing assay as described above. As shown in Figure 4D, RA-treated ATXN3shRNA cells had almost fully recolonized the scratched area with an 89% increase in the wound-healing rate when compared with the RA-treated SCRshRNA control cells (P = 0.0009). Interestingly, this phenotype was specific for FN, because on laminin or poly-d-lysine substrates, no significant differences between cells with or without ATXN3 were observed (Supplementary Material, Fig. S4B). Additionally, it has been suggested that integrins modulate CDK5 activity, which in turn modulates the activity of the ERK and PI3K/AKT pathways (57) affecting neuronal development and survival (58). Immunoblotting analysis of RA-treated ATXN3shRNA cells showed a decreased expression of p35 (P = 5.94 × 10−5), which is known to be associated with a decrease in CDK5 activity (Fig. 4E and F). The decrease in CDK5/p35 complex activity is associated with reduced phosphorylation-mediated activation of ERK1/2 (P = 0.001), AKT (P = 0.005) and PI3K (P = 0.012) in ATXN3shRNA cells, with no alteration in the total levels of these proteins, and, with a downregulation of BCL2 (P = 0.007) (Fig. 4E and F). BCL2 inhibits BAX and BAK apoptotic functions (59); therefore, decreased levels of BCL2 may contribute to the increased neuronal death observed in ATXN3shRNA cultures (Fig. 1G). Interestingly, the levels of nuclear CDK5 were also significantly reduced in ATXN3shRNA cells (Fig. 4E and F), which correlates well with the increased proliferation (Fig. 1C and D) observed upon RA treatment (60). Knowing that integrin cell signalling can modulate the activity of Rho and Rac1 GTPases (61), we next investigated if the decreased levels of ITGA5 affected the activity of these regulatory proteins in neuronal cells lacking ATXN3. We found that the activities of both Rho and Rac1 proteins were significantly reduced in RA-treated ATXN3shRNA cells (Fig. 4G). To verify the relevance of our findings in vivo, we assessed the levels of Itga5 in the cerebellum of Atxn3 KO mice (62), confirming that they were reduced as observed in the ATXN3 knockdown neuronal cell cultures (Fig. 4H and I).
Figure 4.
Decrease in adhesion proteins and decreased activation of regulatory molecules in ATXN3shRNA cells. (A and B) Western blot analysis showed a decrease of ITGA5 levels in ATXN3shRNA cells. (C) Depletion of ATXN3 leads to decreased FN responsiveness in SH-SY5Y cells. (D) ATXN3shRNA cells showed increased cell migration on a FN substrate. Bars represent migration rate relative to control cells and expressed as the proportion of the mean distance that remained cell free after 24 h. (E and F) Deregulation of the ERK and PI3K/AKT pathways, with decreased levels of p35, pERK1/2, pAKT and PI3K, and decreased levels of nuclear CDK5 in ATXN3shRNA cells. The results were normalized for H3 levels. (G) ATXN3shRNA cells showed significantly decreased levels of active Rac1 and Rho proteins. (H and I) Western blot analysis showed decreased levels of Itga5 in the cerebellum of Atxn3 KO mice. The results were normalized for α-tubulin levels. (J) Levels of ITGA5 were increased upon treatment with 5 μM MG132 for 24 h after induction of differentiation with RA for 7 days in ATXN3shRNA cells. (K) Relative amounts of ITGA5 in SCRshRNA and ATXN3shRNA cells at various cycloheximide treatment times. (L) Pulldown of polyubiquitylated proteins showed an increase in ubiquitylated forms of ITGA5 in ATXN3shRNA cells. The results were normalized for H3 or CD147 levels. n ≥ 3 independent biological replicates in all experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Based on previous observations in non-neuronal cells (33) and considering that ATXN3 is a DUB enzyme, we hypothesized that ATXN3 could modify ubiquitylation and regulate the degradation of ITGA5 through the UPP. To test this hypothesis, we assessed the levels of ITGA5 upon proteasome inhibition with 5 μM MG132 for 24 h in cultures treated with RA and found them to be significantly increased in ATXN3shRNA cells (Fig. 4J). Additionally, inhibition of protein synthesis by cycloheximide (CHX) treatment showed a decrease in ITGA5 half-life in these cells (Fig. 4K); this suggests that ATXN3 normally acts to inhibit ITGA5 degradation also in neurons. We found that inhibition of the proteasome with MG132 increased the levels of ubiquitylated ITGA5 in ATXN3shRNA cells (P = 0.02) (Fig. 4L). We confirmed that ATXN3 affects the steady-state levels of ITGA5 by acting at the protein level, as qRT-PCR analysis revealed no significant differences in ITGA5 transcript levels between ATXN3shRNA and SCRshRNA cells (Supplementary Material, Fig. S3B). Together, these results suggest that excessive degradation of ITGA5 in ATXN3shRNA cells induced to differentiate caused defects in adhesion to FN substrates, deregulation of the ERK and PI3K/AKT pathways and a decrease in the activity of small GTPases Rho and Rac1, leading to abnormal cell morphology, motility, proliferation and differentiation.
Restoring the levels of ITGA5 ameliorates the phenotype of ATXN3shRNA cells
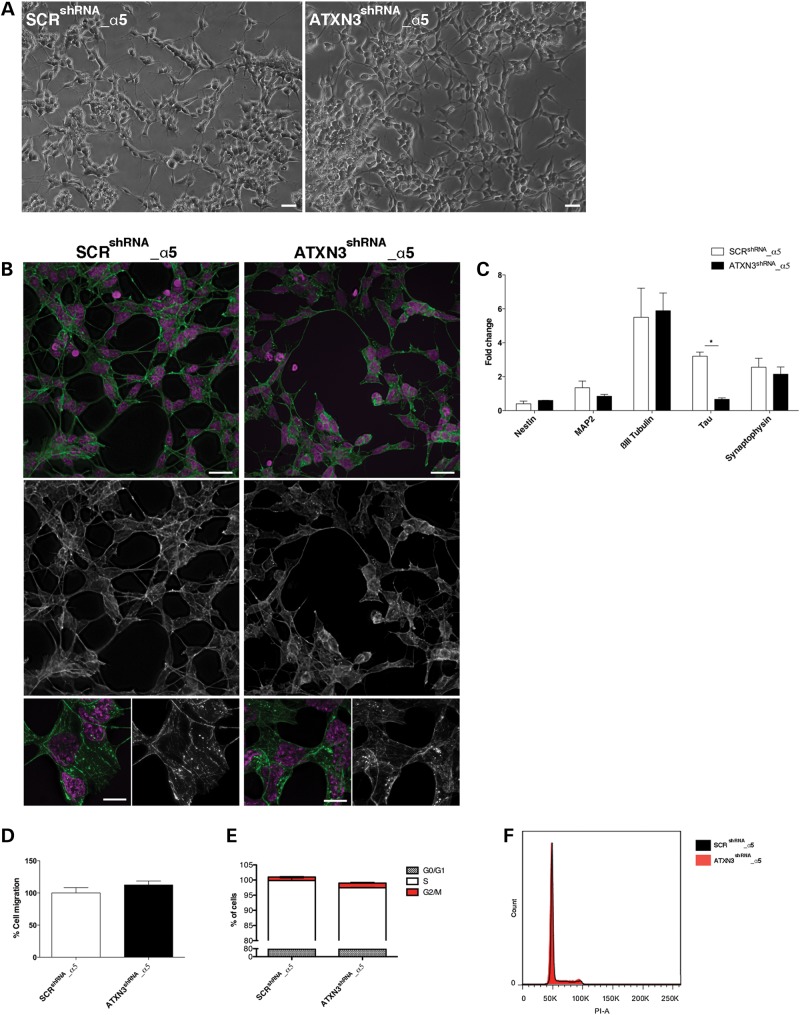
To test if the ATXN3 depletion phenotype could be reverted by the normalization of ITGA5 levels, we overexpressed ITGA5 in ATXN3shRNA cells (ATXN3shRNA_α5) and evaluated their phenotype upon RA treatment. We found that not only were the cellular morphology (Fig. 5A) and cytoskeletal organization (Fig. 5B) significantly restored but also the ATXN3shRNA_α5 cells expressed normal levels of the majority of the neuronal markers tested (Fig. 5C). In addition, the migration rate (Fig. 5D) and cell cycle progression (Fig. 5E and F) were comparable to the SCRshRNA_α5 control cells. Therefore, we conclude that most aspects of the phenotype observed in ATXN3-deficient cells were caused by the reduction in the ITGA5 levels.
Figure 5.
The phenotype observed in ATXN3shRNA is dependent on α5 integrin subunit levels. (A) Overexpression of ITGA5 in ATXN3shRNA cells (ATXN3shRNA_α5) restored their cellular morphology upon RA treatment. Scale bar: 100 μm. (B) ATXN3shRNA_α5 cells no longer presented detectable actin filament disorganization, as shown by phalloidin staining (green). Nuclei were counterstained with DAPI (purple). Upper panel scale bar: 20 μm. Lower panel scale bar: 5 μm. (C) mRNA levels of the neuronal markers analyzed by qRT-PCR in ATXN3shRNA_α5 cells were normal and comparable to the SCRshRNA_α5 control cells. mRNA levels were normalized to undifferentiated cells and HMBS gene. (D) Normalized ITGA5 levels abolished their increased migration of ATXN3shRNA cells in a wound scratch assay, 24 h after scratching. Bars represent migration rate expressed as a percentage of control and calculated calculated as the proportion of the distance between the borderlines caused by scratching, to the distance that remained cell-free after 24 h. (E and F) Normal cell cycle in ATXN3shRNA_α5 cells. n ≥ 3 independent biological replicates in all experiments. *P < 0.05.
Interference with the DUB activity of ATXN3 leads to an abnormal neuronal phenotype that parallels the loss of expression of this protein
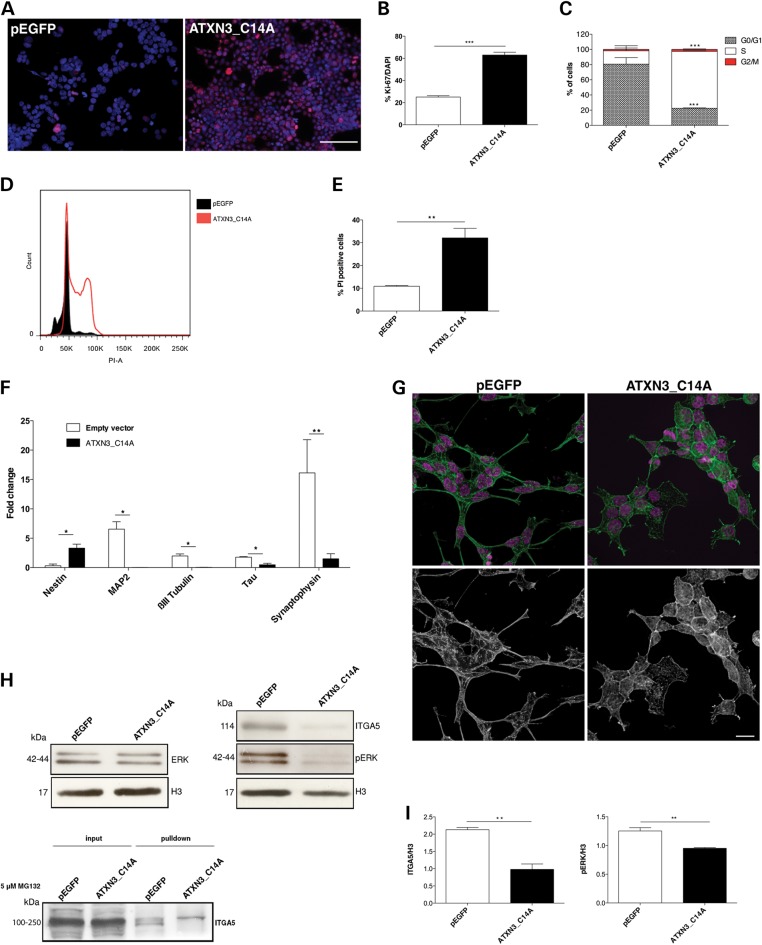
To clarify whether the regulation of ITGA5 levels by ATXN3 was dependent on its DUB activity, we generated a SH-SY5Y cell line that stably overexpressed a catalytically inert version of ATXN3 (Supplementary Material, Fig. S5A), obtained by mutating the catalytic cysteine residue (C14) (ATXN3_C14A). Interestingly, cells expressing ATXN3_C14A recapitulated many characteristics of ATXN3-deficient neurons: (i) a round, flat shape with very few and short extensions in comparison to the controls (Supplementary Material, Fig. S5B); (ii) a lack of proliferative inhibition after RA treatment as assessed by Ki-67 staining (62.97% Ki-67-positive cells in ATXN3_C14A cultures versus 25.03% for the pEGFP controls; P = 1.10 × 10−14) (Fig. 6A and B) and cell cycle analysis (74.9% in S phase versus 17.5%; P = 0.0018) (Fig. 6C and D); (iii) an increased cell death (32.1% versus 10.9%; P = 0.002) (Fig. 6E); (iv) a significant decrease in mRNA levels for several neuronal markers, indicating an immature differentiation status (Fig. 6F) and (v) misaligned actin with randomly distributed filaments (Fig. 6G). ATXN3_C14A cells also showed decreased levels of ITGA5 (P = 0.0017) and downstream targets of CDK5 (the expression of p35 protein was not detectable) (Fig. 6H and I). These results suggest that ATXN3 with the C14A mutation interferes with the function of normal (endogenous) ATXN3 through a dominant negative effect, provoking a more severe phenotype as compared with the silencing, probably due to the lack of the compensatory mechanisms. Importantly, as found in ATXN3shRNA cells (Fig. 4J), 5 μM MG132 treatment for 24 h increased the levels of polyubiquitylated ITGA5 in ATXN3_C14A cultures treated with RA (Fig. 6H and I), supporting the hypothesis that ITGA5 is a substrate of the DUB activity of ATXN3 in neurons.
Figure 6.
Effects of ATXN3 silencing is likely related to its DUB activity. (A and B) RA treatment did not inhibit proliferation of ATXN3_C14A cells, as assessed by Ki-67 labelling (red). Nuclei were counterstained with DAPI (blue). Scale bar: 200 μm. (C and D) Flow cytometry analysis of the cell cycle showed an increase in S phase in ATXN3_C14A cultures. (E) ATXN3_C14A cultures presented high percentage of cell death as compared with the pEGFP controls, as assessed by PI staining followed by flow cytometry analysis. (F) ATXN3_C14A cells presented increased expression of nestin and decreased mRNA levels of neuronal markers. mRNA levels were normalized to undifferentiated cells and HMBS gene expression. (G) Phalloidin staining (green) showed that actin filaments were disorganized and not parallel in ATXN3_C14A cells. Nuclei were counterstained with DAPI (purple). Scale bar: 20 μm. (H and I) Western blot analysis showed a decrease of ITGA5 levels and downstream targets of CDK5 in ATXN3_C14A cells. The results were normalized for H3 levels. n ≥ 3 independent biological replicates in all experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
PolyQ expansion in ATXN3 causes a similar but milder neuronal phenotype than the absence of this protein
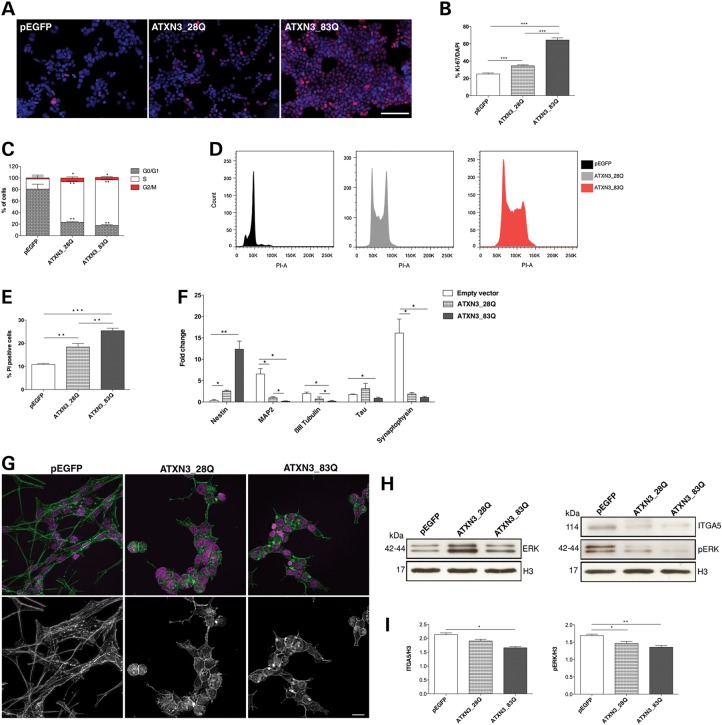
To determine whether the presence of an expanded polyQ tract within ATXN3 would lead to a gain or loss of function, we generated a SH-SY5Y cell line expressing an ATXN3 protein bearing 83 glutamines (ATXN3_83Q) (Supplementary Material, Fig. S5A). We found that, although expression of the polyQ did not cause visible aggregation of the expanded ATXN3, it led to a similar but slightly milder neuronal phenotype than the one observed in cells lacking this protein. Upon RA treatment, the cells displayed: (i) an abnormal morphology without extensions (Supplementary Material, Fig. S5B); (ii) maintenance of their proliferative activity (64.39% Ki-67-positive cells; P = 1.03 × 10−13) (Fig. 7A and B); an increased proportion of cells in the S phase (80.5% versus 17.5% for the pEGFP controls; P = 0.0008) (Fig. 7C and D); (iii) increased cell death (25.4% versus 18.4% for ATXN3_28Q; P = 0.009 and 10.9% for the pEGFP controls; P = 1.43 × 10−5) (Fig. 7E); (iv) reduced expression of several neuronal differentiation markers (Fig. 7F) and (v) disorganization of the actin cytoskeleton (Fig. 7G). ATXN3_83Q cells also had reduced levels of ITGA5 (P = 0.01) and decreased activation of its downstream targets (Fig. 7H and I). While overexpression to similar levels, i.e. 2.5-fold, of wild-type (WT) ATXN3 (with 28 glutamines) also caused some degree of perturbation in differentiation and cytoskeleton organization, the effects were much milder than those of ATXN3 loss of function or polyQ expansion (Fig. 7). Together, these results suggest that the dose of ATXN3 needs to be strictly balanced and that polyQ expansion perturbs the normal function of ATXN3 in neuronal cells.
Figure 7.
Partial loss of function of expanded ATXN3. (A and B) Ki-67 staining (red) showed that ATXN3_83Q cells kept their proliferative activity upon RA treatment. Nuclei were counterstained with DAPI (blue). Scale bar: 200 μm. (C and D) ATXN3_83Q cultures showed an increase in S phase of the cell cycle as assessed by flow cytometry analysis. (E) Increased cell death in ATXN3_83Q cultures as assessed by flow cytometry analysis. (F) mRNA level of nestin was increased and expression of neuronal differentiation markers was significantly decreased in ATXN3_83Q cells. mRNA levels were normalized to undifferentiated cells and HMBS gene expression. (G) Actin filaments network was severely affected in ATXN3_83Q cells with unparalleled actin polymers, as shown by phalloidin staining. Nuclei were counterstained with DAPI (purple). Scale bar: 20 μm. (H and I) Western blot analysis showed a decrease of ITGA5 levels and its downstream targets in ATXN3_83Q cells. The results were normalized for H3 levels. n ≥ 3 independent biological replicates in all experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
In vivo evidence for downregulation of α5 integrin signalling in the context of Machado–Joseph disease
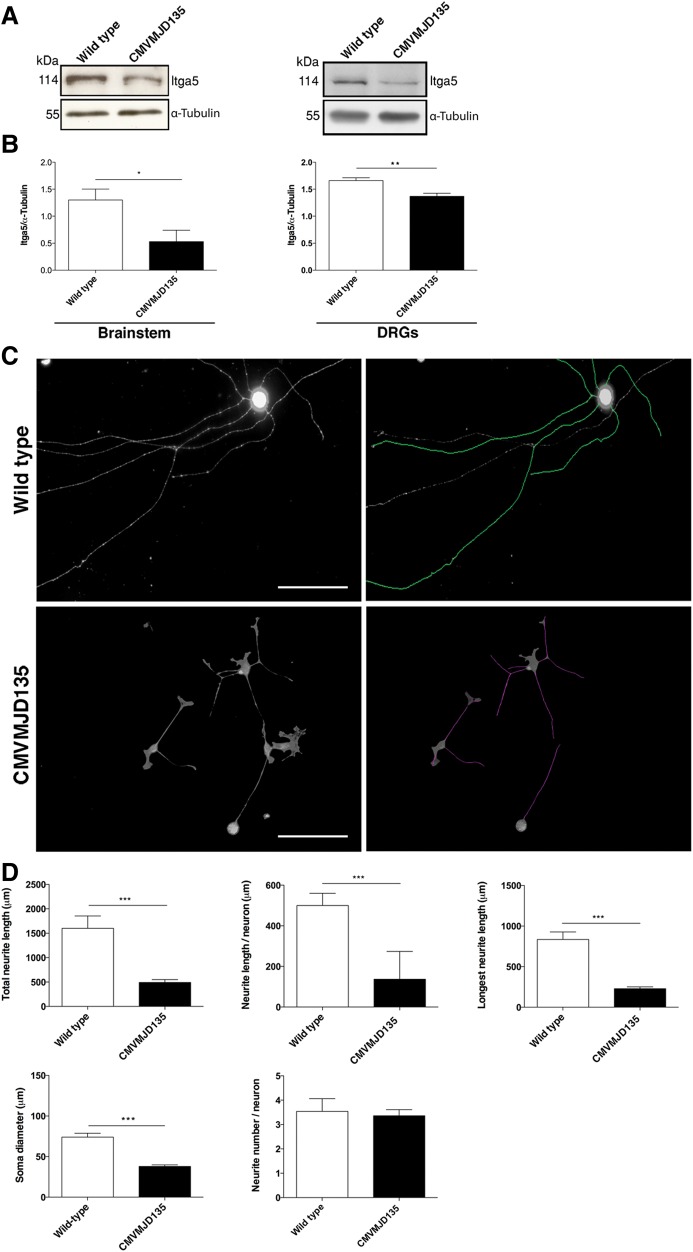
To investigate whether the partial loss of function of expanded ATXN3 could contribute to the pathogenesis of MJD, we analyzed the levels of Itga5 in the nervous system of an MJD mouse model (CMVMJD135) generated in our lab, expressing an expanded human ATXN3 with 135 glutamines and mimicking the neurological and neuropathological phenotype of the human disease, as described elsewhere (63). As shown in Figure 8A and B, CMVMJD135 mice had significantly reduced levels of Itga5 in the brainstem (an affected brain region) (P = 0.04) and dorsal root ganglia (DRG) (P = 0.0019). Considering the cytoskeletal disorganization and reduced branching phenotype observed in the absence of ATXN3, we analyzed neurons isolated from the DRGs of CMVMJD135 mice. We found a marked difference in the morphology of these neurons (Fig. 8C and D), with a drastic reduction in both the total (P = 1.4 × 10−7) and mean (P = 1.6 × 10−12) neurite length when compared with WT littermate controls (Fig. 8C and D). Additionally, the cell bodies of the DRGs from transgenic animals had a reduced diameter in comparison to those from WT controls (P = 3.63 × 10−10) (Fig. 8C and D). This supports the hypothesis that a partial loss of function of ATXN3 due to the expansion of the polyQ tract may contribute to neuronal dysfunction in MJD.
Figure 8.
The polyQ expansion in ATXN3 affects the integrin signalling pathway. (A and B) Western blot analysis showed decreased levels of Itga5 in the nervous system of CMVMJD135 mice (n = 5 animals/genotype). The results were normalized for α-tubulin levels. (C and D) DRG neurons from CMVMJD135 mouse (two pools of three animals for each genotype) showed no differences in the number of neurites and decreased neurite length and soma diameter as compared with the WT controls. Neurite length was measured using NeuronJ software and Feret's diameter was used to measure the soma (n = 50 neurons/genotype). Scale bar: 200 μm. n ≥ 3 independent biological replicates in all experiments. *P < 0.05; ***P < 0.001.
DISCUSSION
Given the significance of DUBs to nervous system function and homeostasis, we focussed here on characterization of the normal function of ATXN3 in neuronal cells. Stable depletion of ATXN3 in SH-SY5Y cells by lentiviral transduction of shRNA inhibited neuronal differentiation following exposure to RA. ATXN3-depleted cells displayed a markedly abnormal morphology, becoming flat and with very few extensions, and showing an altered structure of the actin cytoskeleton. Furthermore, the proliferative activity of these cells was not inhibited by RA treatment, their motility was increased and cell survival was significantly reduced.
Although ATXN3 KO animals do not show an overt phenotype, including any neuronal differentiation abnormalities, which is likely due to compensatory mechanisms (32,49,62), the current findings concur with previous studies showing abnormal differentiation, adhesion and morphology in other, non-neuronal, cellular models of ATXN3 loss of function (31,33). This is interesting, as cumulative evidence suggests that disruption of the neuronal cytoskeleton network may be a common feature contributing to neurodegeneration in several diseases, including polyQ disorders (41,64–66). In addition, ATXN3shRNA cultures showed reduced levels of many neuronal markers while maintaining expression of immature cell markers and a significantly decreased neurite length, which is compatible with an immature filopodia phenotype. Filopodia play an important role in cell migration, neurite outgrowth and wound healing, and their elongation depends on tightly regulated actin polymerization (54). Thus, disruption of the actin filament network impairs neurite elongation and ultimately cell differentiation, as we observed in ATXN3-deficient neuronal cells. Interestingly, when ATXN3shRNA cells were kept in culture for long, we observed a partial recovery of several effects seen with silencing of ATXN3, which might be explained by compensatory mechanisms similar to those occurring in ATXN3 KO animal models. This recovery was less evident in cells expressing the catalytically silent version of the protein. In the disease model (ATXN3_83Q), the compensatory mechanisms also seem to be prevented by the presence of the expanded protein, which has a dominant negative effect.
Neuronal process outgrowth and maturation, as well as neuronal survival, are at least partially regulated by the integrin signalling pathway. We found that the phenotype observed in ATXN3shRNA cells was due to a decrease in the expression of ITGA5, associated with a decrease in the CDK5/p35 complex activity and, consequently, to a deregulation of the PI3K/AKT and ERK pathways. This had a negative impact on neurite outgrowth, cytoskeleton regulation, cell adhesion and motility and survival. CDK5 plays an important role in neuronal differentiation (60), morphogenesis (67), proliferation (68,69), survival, axon guidance, synaptic transmission, neurocytoskeletal dynamics and neuronal degeneration (70–75). It has been shown that inhibition of CDK5 either pharmacologically (with roscovitine) or by expression of a dominant negative form, causes a dramatic decrease in RA-induced cell differentiation (73). Moreover, Cdk5-deficient neurons are significantly arrested or delayed in their developmental program, both in vitro and in vivo (60), and homozygous Cdk5 KO mice mutants die in utero with neuronal migration deficits throughout the brain (70). Additionally, we observed decreased activation of small GTPases, both Rac1 and the Rho family, which are key actin cytoskeleton regulators, further compromising actin dynamics. Interestingly, a recent large-scale huntingtin–protein interaction study revealed that Rho family GTPases and actin remodelling play an important role in huntingtin function and Huntington's disease pathogenesis (76).
The fact that cells expressing the catalytically inactive ATXN3 also show a reduction of ITGA5 protein levels and that inhibition of the proteasome leads to an accumulation of polyubiquitylated ITGA5 species in both ATXN3shRNA and ATXN3_C14A cells suggests that ITGA5 protein levels are regulated through the DUB activity of ATXN3 that defines the extent of ITGA5 degradation by the proteasome. This proposed mechanism is compatible with the previous finding that the ITGA5 is a molecular partner of ataxin-3 (33). Confirming the relevance of ATXN3 for regulation of ITGA5 in vivo, the levels of this protein were significantly reduced in the nervous system of Atxn3 KO mice.
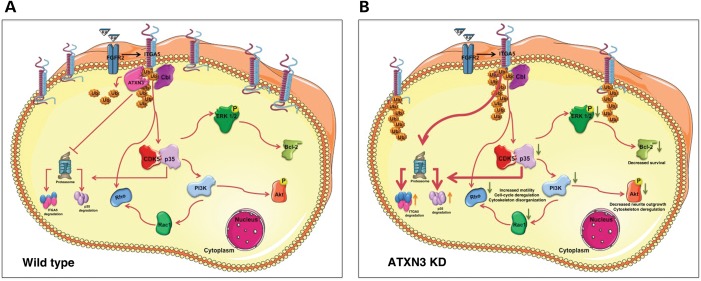
To date, the type(s) of ubiquitylation of ITGA5 and the amino acid position of this modification are not known. However, comparing the cytoplasmic tails of all human α integrins, Lobert and Stenmark observed that the first lysine residue was strictly conserved among human integrins and suggested that this might be the site at which ubiquitylation occurs (77). Although it has been described that ITGA5 may be degraded in lysosomes (78), there are also reports demonstrating that it is degraded by the proteasome through the action of the E3 ligase Cbl (33,79). Indeed, Kaabeche and colleagues showed that Cbl recruitment induced by FGFR2 activation triggers ITGA5 proteasomal degradation (79). These observations, together with the fact that FGFR2 expression is known to be upregulated by RA (80), lead us to propose a mechanism that may explain the phenotype of ATXN3shRNA cells upon RA treatment (Fig. 9). In this model, ATXN3 prevents the degradation of ITGA5 triggered by the RA-induced activation of FGFR2 (Fig. 9A). When ATXN3 is silenced, RA treatment causes ITGA5 degradation to an extent that leads to a deregulation of the CDK5, PI3K/AKt and ERK pathways and affects neuronal morphology and cell adhesion, proliferation, differentiation and survival (Fig. 9B). In agreement with this model, overexpression of a catalytically inactive form of ATXN3 led to similar morphological and biochemical changes and the same differentiation impairment as the silencing of ATXN3, suggesting that the regulation of ITGA5 levels in neurons is dependent on the DUB activity of ATXN3. Furthermore, the fact that proteasomal inhibition prevents the decrease of ITGA5 in ATXN3shRNA cells indicates that ATXN3 rescues ITGA5 from degradation by the proteasome. Importantly, restoring the levels of ITGA5 rescued most of the abnormal features of ATXN3shRNA cells.
Figure 9.
Model of α5 integrin subunit modulation of the PI3K/AKT and ERK cascades to insure neuronal differentiation and survival. (A) In normal functioning neurons, ATXN3 is preventing degradation of ITGA5 triggered by the RA induced activation of FGFR2. (B) When ATXN3 is silenced, RA treatment causes ITGA5 degradation to an extent that leads to a deregulation of the PI3K/AKt and ERK pathways affecting neuronal morphology and cell adhesion and survival.
Given that this DUB is involved in the human neurodegenerative disease MJD, we conducted experiments to determine the relevance of this abnormal phenotype and impairment in neuronal differentiation to MJD pathogenesis. Interestingly, similar but milder alterations were found in cultures expressing expanded ATXN3, which suggests a partial loss of normal function of the protein in the presence of this type of mutation. Overexpression of the WT ATXN3 also caused some degree of toxicity, suggesting the importance of a tight regulation of ATXN3 expression levels, as has been described elsewhere (81–85).
Confirming the perturbation of ATXN3 normal function by polyQ expansion, we observed a downregulation of Itga5 in the brainstem and DRGs of CMVMJD135 transgenic mice. Cultures of DRG neurons from these animals showed that transgenic neurons were smaller and displayed a drastic decrease in the neurite length, as observed in neuronal cultures depleted for ATXN3. This finding suggests that the loss of this normal cellular function of ATXN3 might be relevant for the neurodegeneration caused by the polyQ tract. Although the in vitro assays performed so far suggest that the polyQ expansion does not significantly affect ATXN3 DUB activity (86), these assays were performed using artificial substrates that may not mirror the biological substrates of ATXN3, or the post-translational modifications, co-factors and key partners that may be lacking. Improved assays addressing DUB activity within a cellular environment based on specific substrates and/or addressing ATXN3 as part of multi-protein complexes would be important to adequately assess WT and mutant ATXN3 activity.
Overall, our data support the importance of ATXN3 in neuronal cells and the link between its biological function and MJD. Loss of function of ATXN3 in neuronal cells affects degradation of ITGA5 through its DUB activity, which affects many processes that are important for neuronal development and function, such as cell adhesion, differentiation, cytoskeleton organization and neurite length. PolyQ expansion imparts loss of this cellular function of ATXN3 in neurons, with effects on neuronal morphology and process extension, which chronically may contribute to neurodegeneration.
MATERIALS AND METHODS
Cell culture
SH-SY5Y cell cultures:human neuroblastoma SH-SY5Y cell line (ATCC, CRL-2266) was cultured in Dulbecco's modified eagle medium: nutrient mixture (DMEM)/F-12 (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (FBS) (Biochrom), 2 mm glutaMAX (Invitrogen), 100 U/ml penicillin, 100 µg/ml streptomycin and 25 ng/ml puromycin (Sigma Aldrich). Medium was changed every 2 days. Differentiation was induced by 0.1 µm all-trans-retinoic acid (Sigma Aldrich) in opti-MEM (Invitrogen) supplemented with 0.5% FBS. Medium was replaced every 2 days. PC12 cell cultures: rat PC12 cells were cultured in DMEM (Invitrogen) supplemented with 10% (v/v) FBS (Biochrom), 5% (v/v) horse serum (HS) (Biochrom), 2 mm GlutaMAX (Invitrogen), 100 U/ml penicillin, 100 µg/ml streptomycin and 25 ng/ml puromycin (Sigma Aldrich). Medium was changed every 2 days. Differentiation was induced by 1 µg/ml NGF (Invitrogen) in DMEM supplemented with 0.75% FBS and 0.75% HS. Medium was replaced every 2 days. Primary cultures of dorsal root ganglion neurons: for dorsal root ganglion (DRG) neuron culture, cells isolated from 8 weeks-old mice were plated in poly-d-lysine (Sigma) and laminin (Sigma) pre-coated coverslips at a density of 5 × 103 cells/ml in DMEM/F12 supplemented with 10% (v/v) FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 50 ng/ml NGF (Millipore), 1 × B27 (Invitrogen) and 1.176 g/L l-glutamine (Invitrogen) for 72 h. Primary cultures of hipoccampal neurons: hippocampal neuron cultures were prepared from P4 Wister rats. Briefly, upon dissection, hippocampi were submitted to a trypsin-based enzymatic digestion followed by mechanical dissociation. Isolated cells were then plated on coverslips previously coated with poly-d-lysine (Sigma) at a density of 40 000 cells/cm2 using Neurobasal A medium (Gibco) supplemented with 1 mm GlutaMAX (Gibco), 10 ng/ml bFGF (Gibco), 0.1 mg/ml kanamycin (Gibco) and 1 × B27 (Gibco) for 7 days. Cells were incubated in a humidified 37°C/95% air/5% CO2 incubator.
Vectors and lentivirus packaging
HEK293T packaging cells were plated at a density of 3 × 105 cells/well in 6-well plates, cultured in opti-MEM supplemented with 10% FBS and transfected on the following day with the scrambled sequence vector (CAACAAGATGAAGAGCACCAA), the pLKO.1/shRNA-ATXN3 vector (CCGGGCAGGGCTATTCAGCTAAGTACTCGAGTACTTAGCTGAATAGCCCTGCTTTTT) or the empty vector pLKO-1 (TRC1; Open Biosystems) for virus production, following the RNAi Consortium High-Throughput Lentiviral production protocol (87). For overexpression the following plasmids were used: PPCB7/ITGA5 (Addgene plasmid 16 041), pEGFP/ATXN3_28Q, pEGFP/ATXN3_83Q and pEGFP/ATNX3_C14A. Medium was changed in the next day and cells were cultured for 48 h. Conditioned medium was then collected and stored at −80°C.
Transduction of target cells
A total of 2.5 × 105 SH-SY5Y or PC12 cells were seeded on a 6-well plate in complete DMEM/F-12 medium and transduced by the lentiviral vectors. Medium was changed 24 h after, and cells were incubated for 72 h. For selection of clones with stable shRNA expression, fresh complete DMEM/F-12 or DMEM medium containing 25 µg/ml puromycin was added to the cells. During the selection period, medium was replaced by fresh medium containing puromycin every 2 days. After 5 days, cells that formed colonies were selected and sub-cultured in 96-well plates under puromycin selective pressure (2.5 µg/ml puromycin) for subsequent expansion, with passaging every 3–5 days. The percentage of silencing of each clone was monitored by immunoblotting.
High-throughput high-content functional imaging
SH-SY5Y cells were seeded at a density of 4 × 103 cells/well in flat bottom 96-well plates previously coated with Matrigel (BD, Biosciences), and 10 μm all-trans-retinoic acid (Sigma Aldrich) was added the day after plating in DMEM/F-12 with 1% FBS. After 5 days, cells were washed with DMEM/F-12 and incubated with 50 ng/ml BDNF (Peprotech) in DMEM/F-12 without serum for 3 days. Cells were then labelled for βIII-tubulin (1:1000, R&D Systems), scanned at different locations of each well and the quantitative analysis of total number of cells, number of βIII-tubulin positive cells and neurite length were automatically done using the automatic imaging system Thermo Scientific Cellomics® ArrayScan® VTI.
Pulldown
RA-treated SH-SY5Y cells were washed in ice-cold PBS, incubated 5 min on ice in FISH buffer (50 mm Tris–HCl, pH 7.4, 2 mm MgCl2, 10% glycerol, 1% NP-40, 100 mm NaCl and protease inhibitors cocktail) and centrifuged for 5 min, 21 000 g, at 4°C. Aliquots were taken from the supernatant to compare protein amounts. The supernatant was incubated with bacterially produced GST-PAK-CD or GST-RHOTEKIN-RBD fusion proteins bound to glutathione-coupled sepharose beads (GE Healthcare) at 4°C for 30 min. The beads and proteins bound to the fusion protein were washed three times in an excess of FISH buffer, eluted in Laemmli buffer and analyzed for bound Rho and Rac1 by immunoblotting. For the pulldown of polyubiquitylated proteins, RA-treated cells were lysed in lysis buffer (50 mm HEPES, pH 7.5, 0.15 m NaCl, 1 mm EDTA, 1% NP-40, 10% glycerol, 50 μM PR-619 and protease inhibitors cocktail) and centrifuged for 10 min, 14 000 g, at 4°C, or treated with 5 μM MG132 (Calbiochem) for 24 h prior to lysis. One milligram of the supernatant were incubated with 100 μl of Agarose-TUBEs (Lifesensors) and incubated 1 h at 4°C. Beads were washed three times with TBS-T and bound proteins were eluted in Laemmli buffer.
Protein synthesis inhibition and proteasome inhibition
RA-treated SH-SY5Y cells were incubated with 5 μm CHX (Merck) during 30, 60 or 180 min. For proteasome inhibition, RA-treated cells were incubated with 5 μM MG132 (Calbiochem) for 24 h prior to lysis.
Immunoblotting
RA-treated SH-SY5Y cells or NGF-treated PC12 cells were pelleted and frozen in liquid nitrogen. For cellular and brain tissue extracts, 50 µg of total protein isolated in RIPA buffer [150 mm NaCl, 50 mm Tris–HCl, pH 7.6, 0.5% NP-40, 1 mm Phenylmethylsulfonyl fluoride, protease inhibitors (Roche)] were resolved in 10% SDS-PAGE gels and transferred to a nitrocellulose membrane (Bio-Rad). After incubation with the primary antibodies against ATXN3 (1H9, 1:2000, Millipore), α5 integrin (1:5000, Millipore), CDK5 (1:1000, Millipore), p35 (1:1000, cell signalling), pERK1/2 (1:2000, cell signalling), ERK1/2 (1:1000, cell signalling), PI3K (1:1000, cell signalling), pAKT (1:1000, cell signalling), AKT (1:3000, cell signalling), BCL2 (1:100, Abcam), pan-Rho (1:1500, Millipore) and Rac1 (1:2000, Millipore), overnight at 4°C, membranes were incubated with secondary antibodies for 1 h at room temperature (anti-rabbit or anti-mouse, 1:10 000, Bio-Rad). Antibody affinity was detected by chemiluminescence (Clarity kit, Bio-Rad). Histone H3 (1:7500, Millipore), MCT4 (1:500, Santa Cruz), CD147 (1:500, Santa Cruz) and α-tubulin (1:500, DSHB) were assessed as loading controls.
Flow cytometry
For propidium iodide (PI) staining, RA-treated SH-SY5Y cells or NGF-treated PC12 cells were collected and fixed using ice-cold 70% ethanol for 1 h on ice. Pelleted cells were washed with PBS and incubated with staining solution (0.1% Triton-X100, 20 μg/ml PI solution, 250 µg/ml RNase in PBS) for 1 h at 50°C. For cell cycle analysis, cells were starved for 6 h before RA treatment. Samples were washed with PBS and analyzed using a FACSCaliber2 flow cytometer (BD-Biosciences) with a 568-nm excitation laser. Signals from 30 000 cells/sample were captured in FL3 (>670 nm) at a flow rate of 1000 cells/s. Offline data were processed with the FlowJo (Tree Star) software and quantified with WinList software.
Wound healing assay
SH-SY5Y cells were grown to confluent monolayers on 6-well plates. After RA treatment, monolayers were wounded by a pipette tip. This initial wounding and the movement of the cells in the scratch were photographically monitored using the Olympus IX-51 inverted microscope equipped with a TH4-200 camera for 24 h. This time window has been chosen because it is shorter than the doubling time reported for SH-SY5Y cell line (88). Eight different fields were considered for quantitative estimation of the distance between the borderlines and in each image five different equidistant points were measured. The migration rate was calculated as the distance between the borderlines caused by scratching (defined at 0 h) minus the width that remained cell-free after 24 h and expressed as a percentage of the control.
Real-time quantitative reverse transcription polymerase chain reaction
One microgram of total RNA purified from RA-treated SH-SY5Y cells or NGF-treated PC12 cells was reverse transcribed using the One-step SuperScript kit (Bio-Rad). qRT-PCR reaction was performed using the Quantitec SYBR Green kit (Qiagen) and the primers previously described (52), in a CFX96 real-time PCR detection system (Bio-Rad). Gene expression was normalized to HMBS levels. The results are presented as fold change.
Immunocytochemistry
Cells cultured on poly-d-lysine (Sigma Aldrich) and gelatin, Collagen (BD Biosciences) or Ply-d-lysine pre-coated glass coverslips (SH-SY5Y, PC12 or primary neurons, respectively) were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. Cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min. Next, cells were incubated with 10% fetal calf serum blocking buffer for 1 h, followed by overnight incubation with primary antibody against Ki-67 (1:300, Millipore) or βIII-tubulin (1:1000, R&D systems) at 4°C, or by incubation with phalloidin (1:500, Sigma Aldrich) for 45 min and counterstained with DAPI (1:2000, Sigma Aldrich) for 10 min at room temperature. Alexa Fluor 568 (A11004) conjugated antibody was used at 1:2000 (Molecular probes).
Animals
The MJD mouse model (CMVMJD135) was generated as described (63). Wister Rats used for the primary cultures were purchased from Charles River, Spain. For the mouse primary cultures, two pools of three male animals for each genotype at approximately 2 months of age were used. Animals were sacrificed by decapitation and the DRGs were removed along the spinal cord. For the rat primary cultures, six P4 rats per experiment were used. Animals were sacrificed by decapitation and brains were dissected. All animal procedures were conducted in accordance with European regulations (European Union Directive 86/609/EEC) and approved by the joint Animal Ethics Committee of the Life and Health Sciences Research Institute, University of Minho. Health monitoring was performed according to FELASA guidelines (89). All animals were housed and maintained in a controlled environment at 22–24°C and 55% humidity, on 12 h light/dark cycles and fed with regular rodent's chow and tap water ad libitum. Animal facilities and the people directly involved in animal experiments (A.N.-C., S.D.-S. and A.S.-F.) were certified by the Portuguese regulatory entity—Direcção Geral de Alimentação e Veterinária. Animals were sacrificed by decapitation at 8 weeks of age. Their brains were immediately dissected and stored at −80°C.
Microscopy
Image acquisition of fixed cells was carried out in a Zeiss AxioImager Z1 equipped with a Axiocam MR. Images represent maximum-intensity projections of all Z planes acquired with PLANAPO ×40 or ×63 objectives and following blind deconvolution with a AutoquantX (Media Cybernetics). Adobe photoshop CS5 (adobe Systems) was used for image processing.
Statistical analysis
Comparison between the different cell lines was performed using the t-test in the GraphPad prism version 5.0 software, assuming the homogeneity of the variances. For immunoblottings, the mean density and area of each band were measured using at least three experiments in TINA 2.0 software according to manufacturer's instructions. For qRT-PCR data, results were presented using the ΔΔCt method, as described before (90). A critical value for significance of two-tailed P < 0.05 was used throughout the study.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by Fundação para a Ciência e Tecnologia and COMPETE through the project ‘(PTDC/SAU-GMG/101572/2008)’ and by National Institutes of Health (NIH) ‘(R01NS038712)’. A.N.-C., S.D.-S. and A.S.-F. were supported by the FCT fellowships SFRH/BD/51059/2010, SFRH/BD/78388/2011 and SFRH/BPD/91562/2012, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Shushant Jain for the help with generation of cell lines, Dr António Salgado for the help with the primary cultures and Dr Belém Marques for the support with the flow cytometry analysis.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Chen P.C., Bhattacharyya B.J., Hanna J., Minkel H., Wilson J.A., Finley D., Miller R.J., Wilson S.M. Ubiquitin homeostasis is critical for synaptic development and function. J. Neurosci. 2011;31:17505–17513. doi: 10.1523/JNEUROSCI.2922-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baptista M.S., Duarte C.B., Maciel P. Role of the ubiquitin-proteasome system in nervous system function and disease: using C. elegans as a dissecting tool. Cell. Mol. Life Sci. 2012;69:2691–2715. doi: 10.1007/s00018-012-0946-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kowalski J.R., Juo P. The role of deubiquitinating enzymes in synaptic function and nervous system diseases. Neural Plast. 2012;2012:892749. doi: 10.1155/2012/892749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Vrij F.M., Fischer D.F., van Leeuwen F.W., Hol E.M. Protein quality control in Alzheimer's disease by the ubiquitin proteasome system. Progr. Neurobiol. 2004;74:249–270. doi: 10.1016/j.pneurobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Upadhya S.C., Hegde A.N. Ubiquitin-proteasome pathway components as therapeutic targets for CNS maladies. Curr. Pharm. Des. 2005;11:3807–3828. doi: 10.2174/138161205774580651. [DOI] [PubMed] [Google Scholar]
- 6.Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 7.Bingol B., Schuman E.M. Synaptic protein degradation by the ubiquitin proteasome system. Curr. Opin. Neurobiol. 2005;15:536–541. doi: 10.1016/j.conb.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 8.Watts R.J., Hoopfer E.D., Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/s0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- 9.Kuo C.T., Zhu S., Younger S., Jan L.Y., Jan Y.N. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Hegde A.N., DiAntonio A. Ubiquitin and the synapse. Nat. Rev. Neurosci. 2002;3:854–861. doi: 10.1038/nrn961. [DOI] [PubMed] [Google Scholar]
- 11.Hegde A.N. Ubiquitin-proteasome-mediated local protein degradation and synaptic plasticity. Progr. Neurobiol. 2004;73:311–357. doi: 10.1016/j.pneurobio.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Colledge M., Snyder E.M., Crozier R.A., Soderling J.A., Jin Y., Langeberg L.K., Lu H., Bear M.F., Scott J.D. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I., et al. CAG expansions in a novel gene for Machado–Joseph disease at chromosome 14q32.1. Nat. Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 14.Cattaneo E., Zuccato C., Tartari M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat. Rev. Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 15.Lim J., Crespo-Barreto J., Jafar-Nejad P., Bowman A.B., Richman R., Hill D.E., Orr H.T., Zoghbi H.Y. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orr H.T. Cell biology of spinocerebellar ataxia. J. Cell Biol. 2012;197:167–177. doi: 10.1083/jcb.201105092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chow M.K., Mackay J.P., Whisstock J.C., Scanlon M.J., Bottomley S.P. Structural and functional analysis of the Josephin domain of the polyglutamine protein ataxin-3. Biochem. Biophys. Res. Commun. 2004;322:387–394. doi: 10.1016/j.bbrc.2004.07.131. [DOI] [PubMed] [Google Scholar]
- 18.Mao Y., Senic-Matuglia F., Di Fiore P.P., Polo S., Hodsdon M.E., De Camilli P. Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc. Natl. Acad. Sci. USA. 2005;102:12700–12705. doi: 10.1073/pnas.0506344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicastro G., Menon R.P., Masino L., Knowles P.P., McDonald N.Q., Pastore A. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc. Natl. Acad. Sci. USA. 2005;102:10493–10498. doi: 10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burnett B., Li F., Pittman R.N. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- 21.Doss-Pepe E.W., Stenroos E.S., Johnson W.G., Madura K. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol. Cell. Biol. 2003;23:6469–6483. doi: 10.1128/MCB.23.18.6469-6483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chai Y., Berke S.S., Cohen R.E., Paulson H.L. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J. Biol. Chem. 2004;279:3605–3611. doi: 10.1074/jbc.M310939200. [DOI] [PubMed] [Google Scholar]
- 23.Tsai Y.C., Fishman P.S., Thakor N.V., Oyler G.A. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J. Biol. Chem. 2003;278:22044–22055. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- 24.Ferro A., Carvalho A.L., Teixeira-Castro A., Almeida C., Tome R.J., Cortes L., Rodrigues A.J., Logarinho E., Sequeiros J., Macedo-Ribeiro S., et al. NEDD8: a new ataxin-3 interactor. Biochim. Biophys. Acta. 2007;1773:1619–1627. doi: 10.1016/j.bbamcr.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q., Li L., Ye Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J. Cell Biol. 2006;174:963–971. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong X., Pittman R.N. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum. Mol. Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues A.J., Neves-Carvalho A., Ferro A., Rokka A., Corthals G., Logarinho E., Maciel P. ATX-3, CDC-48 and UBXN-5: a new trimolecular complex in Caenorhabditis elegans. Biochem. Biophys. Res. Comm. 2009;386:575–581. doi: 10.1016/j.bbrc.2009.06.092. [DOI] [PubMed] [Google Scholar]
- 28.Li F., Macfarlan T., Pittman R.N., Chakravarti D. Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. J. Biol. Chem. 2002;277:45004–45012. doi: 10.1074/jbc.M205259200. [DOI] [PubMed] [Google Scholar]
- 29.Evert B.O., Araujo J., Vieira-Saecker A.M., de Vos R.A., Harendza S., Klockgether T., Wullner U. Ataxin-3 represses transcription via chromatin binding, interaction with histone deacetylase 3, and histone deacetylation. J. Neurosci. 2006;26:11474–11486. doi: 10.1523/JNEUROSCI.2053-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woelk T., Sigismund S., Penengo L., Polo S. The ubiquitination code: a signalling problem. Cell Div. 2007;2:11. doi: 10.1186/1747-1028-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues A.J., do Carmo Costa M., Silva T.L., Ferreira D., Bajanca F., Logarinho E., Maciel P. Absence of ataxin-3 leads to cytoskeletal disorganization and increased cell death. Biochim. Biophys. Acta. 2010;1803:1154–1163. doi: 10.1016/j.bbamcr.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Rodrigues A.J., Coppola G., Santos C., Costa Mdo C., Ailion M., Sequeiros J., Geschwind D.H., Maciel P. Functional genomics and biochemical characterization of the C. elegans orthologue of the Machado–Joseph disease protein ataxin-3. FASEB J. 2007;21:1126–1136. doi: 10.1096/fj.06-7002com. [DOI] [PubMed] [Google Scholar]
- 33.do Carmo Costa M., Bajanca F., Rodrigues A.J., Tome R.J., Corthals G., Macedo-Ribeiro S., Paulson H.L., Logarinho E., Maciel P. Ataxin-3 plays a role in mouse myogenic differentiation through regulation of integrin subunit levels. PLoS One. 2010;5:e11728. doi: 10.1371/journal.pone.0011728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hynes R.O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 35.Berrier A.L., Yamada K.M. Cell-matrix adhesion. J. Cell. Physiol. 2007;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 36.George E.L., Georges-Labouesse E.N., Patel-King R.S., Rayburn H., Hynes R.O. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 37.Chun D., Gall C.M., Bi X., Lynch G. Evidence that integrins contribute to multiple stages in the consolidation of long term potentiation in rat hippocampus. Neuroscience. 2001;105:815–829. doi: 10.1016/s0306-4522(01)00173-7. [DOI] [PubMed] [Google Scholar]
- 38.Chan C.S., Weeber E.J., Kurup S., Sweatt J.D., Davis R.L. Integrin requirement for hippocampal synaptic plasticity and spatial memory. J. Neurosci. 2003;23:7107–7116. doi: 10.1523/JNEUROSCI.23-18-07107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheng L., Leshchyns'ka I., Sytnyk V. Cell adhesion and intracellular calcium signaling in neurons. Cell Commun. Signal. 2013;11:94. doi: 10.1186/1478-811X-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denda S., Reichardt L.F. Studies on integrins in the nervous system. Methods Enzymol. 2007;426:203–221. doi: 10.1016/S0076-6879(07)26010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffner G., Kahlem P., Djian P. Perinuclear localization of huntingtin as a consequence of its binding to microtubules through an interaction with beta-tubulin: relevance to Huntington's disease. J. Cell Sci. 2002;115:941–948. doi: 10.1242/jcs.115.5.941. [DOI] [PubMed] [Google Scholar]
- 42.Gunawardena S., Goldstein L.S. Polyglutamine diseases and transport problems: deadly traffic jams on neuronal highways. Arch. Neurol. 2005;62:46–51. doi: 10.1001/archneur.62.1.46. [DOI] [PubMed] [Google Scholar]
- 43.McMurray C.T. Neurodegeneration: diseases of the cytoskeleton? Cell Death Differ. 2000;7:861–865. doi: 10.1038/sj.cdd.4400764. [DOI] [PubMed] [Google Scholar]
- 44.Schafer D.P., Jha S., Liu F., Akella T., McCullough L.D., Rasband M.N. Disruption of the axon initial segment cytoskeleton is a new mechanism for neuronal injury. J. Neurosci. 2009;29:13242–13254. doi: 10.1523/JNEUROSCI.3376-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balaratnasingam C., Morgan W.H., Bass L., Kang M., Cringle S.J., Yu D.Y. Axotomy-induced cytoskeleton changes in unmyelinated mammalian central nervous system axons. Neuroscience. 2011;177:269–282. doi: 10.1016/j.neuroscience.2010.12.053. [DOI] [PubMed] [Google Scholar]
- 46.Duan Y., Dong S., Gu F., Hu Y., Zhao Z. Advances in the pathogenesis of Alzheimer's disease: focusing on tau-mediated neurodegeneration. Transl. Neurodegener. 2012;1:24. doi: 10.1186/2047-9158-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Florenzano F. Localization of axonal motor molecules machinery in neurodegenerative disorders. Int. J. Mol. Sci. 2012;13:5195–5206. doi: 10.3390/ijms13045195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pahlman S., Ruusala A.I., Abrahamsson L., Mattsson M.E., Esscher T. Retinoic acid-induced differentiation of cultured human neuroblastoma cells: a comparison with phorbolester-induced differentiation. Cell Differ. 1984;14:135–144. doi: 10.1016/0045-6039(84)90038-1. [DOI] [PubMed] [Google Scholar]
- 49.Schmitt I., Linden M., Khazneh H., Evert B.O., Breuer P., Klockgether T., Wuellner U. Inactivation of the mouse Atxn3 (ataxin-3) gene increases protein ubiquitination. Biochem. Biophys. Res. Commun. 2007;362:734–739. doi: 10.1016/j.bbrc.2007.08.062. [DOI] [PubMed] [Google Scholar]
- 50.Liang C.C., Park A.Y., Guan J.L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 51.Melino G., Thiele C.J., Knight R.A., Piacentini M. Retinoids and the control of growth/death decisions in human neuroblastoma cell lines. J. Neuro-Oncol. 1997;31:65–83. doi: 10.1023/a:1005733430435. [DOI] [PubMed] [Google Scholar]
- 52.Constantinescu R., Constantinescu A.T., Reichmann H., Janetzky B. Neuronal differentiation and long-term culture of the human neuroblastoma line SH-SY5Y. J. Neural Transm. Suppl. 2007;72:17–28. doi: 10.1007/978-3-211-73574-9_3. [DOI] [PubMed] [Google Scholar]
- 53.Tucholski J., Lesort M., Johnson G.V. Tissue transglutaminase is essential for neurite outgrowth in human neuroblastoma SH-SY5Y cells. Neuroscience. 2001;102:481–491. doi: 10.1016/s0306-4522(00)00482-6. [DOI] [PubMed] [Google Scholar]
- 54.Mattila P.K., Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 55.Legate K.R., Wickstrom S.A., Fassler R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 2009;23:397–418. doi: 10.1101/gad.1758709. [DOI] [PubMed] [Google Scholar]
- 56.Huttenlocher A., Horwitz A.R. Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 2011;3:a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B.S., Ma W., Jaffe H., Zheng Y., Takahashi S., Zhang L., Kulkarni A.B., Pant H.C. Cyclin-dependent kinase-5 is involved in neuregulin-dependent activation of phosphatidylinositol 3-kinase and Akt activity mediating neuronal survival. J. Biol. Chem. 2003;278:35702–35709. doi: 10.1074/jbc.M302004200. [DOI] [PubMed] [Google Scholar]
- 58.Harada T., Morooka T., Ogawa S., Nishida E. ERK induces p35, a neuron-specific activator of Cdk5, through induction of Egr1. Nat. Cell Biol. 2001;3:453–459. doi: 10.1038/35074516. [DOI] [PubMed] [Google Scholar]
- 59.Dlugosz P.J., Billen L.P., Annis M.G., Zhu W., Zhang Z., Lin J., Leber B., Andrews D.W. Bcl-2 changes conformation to inhibit Bax oligomerization. EMBO J. 2006;25:2287–2296. doi: 10.1038/sj.emboj.7601126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cicero S., Herrup K. Cyclin-dependent kinase 5 is essential for neuronal cell cycle arrest and differentiation. J. Neurosci. 2005;25:9658–9668. doi: 10.1523/JNEUROSCI.1773-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwartz M.A., Shattil S.J. Signaling networks linking integrins and rho family GTPases. Trend. Biochem. Sci. 2000;25:388–391. doi: 10.1016/s0968-0004(00)01605-4. [DOI] [PubMed] [Google Scholar]
- 62.Reina C.P., Zhong X., Pittman R.N. Proteotoxic stress increases nuclear localization of ataxin-3. Hum. Mol. Genet. 2010;19:235–249. doi: 10.1093/hmg/ddp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Silva-Fernandes A., Duarte-Silva S., Neves-Carvalho A., Amorim M., Soares-Cunha C., Oliveira P., Thirstrup K., Teixeira-Castro A., Maciel P. Chronic treatment with 17-DMAG improves balance and coordination in a new mouse model of Machado–Joseph disease. Neurotherapeutics. 2014;19:235–249. doi: 10.1007/s13311-013-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berke S.J., Chai Y., Marrs G.L., Wen H., Paulson H.L. Defining the role of ubiquitin-interacting motifs in the polyglutamine disease protein, ataxin-3. J. Biol. Chem. 2005;280:32026–32034. doi: 10.1074/jbc.M506084200. [DOI] [PubMed] [Google Scholar]
- 65.Vickers J.C., King A.E., Woodhouse A., Kirkcaldie M.T., Staal J.A., McCormack G.H., Blizzard C.A., Musgrove R.E., Mitew S., Liu Y., et al. Axonopathy and cytoskeletal disruption in degenerative diseases of the central nervous system. Brain Res. Bull. 2009;80:217–223. doi: 10.1016/j.brainresbull.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 66.Wu C.H., Fallini C., Ticozzi N., Keagle P.J., Sapp P.C., Piotrowska K., Lowe P., Koppers M., McKenna-Yasek D., Baron D.M., et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nikolic M., Dudek H., Kwon Y.T., Ramos Y.F., Tsai L.H. The cdk5/p35 kinase is essential for neurite outgrowth during neuronal differentiation. Genes Dev. 1996;10:816–825. doi: 10.1101/gad.10.7.816. [DOI] [PubMed] [Google Scholar]
- 68.Kimura T., Ishiguro K., Hisanaga S. Physiological and pathological phosphorylation of tau by Cdk5. Front. Mol. Neurosci. 2014;7:65. doi: 10.3389/fnmol.2014.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niu Y., Li H., Herrup K., Zhang J. Neuronal cell cycle regulation of Cdk5 in Alzheimer's disease. Brain Disord. Ther. 2012;S1:004. [Google Scholar]
- 70.Gilmore E.C., Ohshima T., Goffinet A.M., Kulkarni A.B., Herrup K. Cyclin-dependent kinase 5-deficient mice demonstrate novel developmental arrest in cerebral cortex. J. Neurosci. 1998;18:6370–6377. doi: 10.1523/JNEUROSCI.18-16-06370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka T., Veeranna, Ohshima T., Rajan P., Amin N.D., Cho A., Sreenath T., Pant H.C., Brady R.O., Kulkarni A.B. Neuronal cyclin-dependent kinase 5 activity is critical for survival. J. Neurosci. 2001;21:550–558. doi: 10.1523/JNEUROSCI.21-02-00550.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li B.S., Zhang L., Takahashi S., Ma W., Jaffe H., Kulkarni A.B., Pant H.C. Cyclin-dependent kinase 5 prevents neuronal apoptosis by negative regulation of c-Jun N-terminal kinase 3. EMBO J. 2002;21:324–333. doi: 10.1093/emboj/21.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith D.S., Tsai L.H. Cdk5 behind the wheel: a role in trafficking and transport? Trend. Cell Biol. 2002;12:28–36. doi: 10.1016/s0962-8924(01)02181-x. [DOI] [PubMed] [Google Scholar]
- 74.Gupta A., Tsai L.H. Cyclin-dependent kinase 5 and neuronal migration in the neocortex. Neuro-Signals. 2003;12:173–179. doi: 10.1159/000074618. [DOI] [PubMed] [Google Scholar]
- 75.Kesavapany S., Li B.S., Pant H.C. Cyclin-dependent kinase 5 in neurofilament function and regulation. Neuro-Signals. 2003;12:252–264. doi: 10.1159/000074627. [DOI] [PubMed] [Google Scholar]
- 76.Annweiler C., Beauchet O. Vitamin D-mentia: randomized clinical trials should be the next step. Neuroepidemiology. 2011;37:249–258. doi: 10.1159/000334177. [DOI] [PubMed] [Google Scholar]
- 77.Lobert V.H., Stenmark H. Ubiquitination of alpha-integrin cytoplasmic tails. Commun. Integr. Biol. 2010;3:583–585. doi: 10.4161/cib.3.6.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lobert V.H., Brech A., Pedersen N.M., Wesche J., Oppelt A., Malerod L., Stenmark H. Ubiquitination of alpha 5 beta 1 integrin controls fibroblast migration through lysosomal degradation of fibronectin–integrin complexes. Dev. Cell. 2010;19:148–159. doi: 10.1016/j.devcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 79.Kaabeche K., Guenou H., Bouvard D., Didelot N., Listrat A., Marie P.J. Cbl-mediated ubiquitination of alpha5 integrin subunit mediates fibronectin-dependent osteoblast detachment and apoptosis induced by FGFR2 activation. J. Cell Sci. 2005;118:1223–1232. doi: 10.1242/jcs.01679. [DOI] [PubMed] [Google Scholar]
- 80.Johannesson M., Stahlberg A., Ameri J., Sand F.W., Norrman K., Semb H. FGF4 and retinoic acid direct differentiation of hESCs into PDX1-expressing foregut endoderm in a time- and concentration-dependent manner. PLoS One. 2009;4:e4794. doi: 10.1371/journal.pone.0004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fujigasaki H., Uchihara T., Koyano S., Iwabuchi K., Yagishita S., Makifuchi T., Nakamura A., Ishida K., Toru S., Hirai S., et al. Ataxin-3 is translocated into the nucleus for the formation of intranuclear inclusions in normal and Machado–Joseph disease brains. Exp. Neurol. 2000;165:248–256. doi: 10.1006/exnr.2000.7479. [DOI] [PubMed] [Google Scholar]
- 82.Takahashi J., Tanaka J., Arai K., Funata N., Hattori T., Fukuda T., Fujigasaki H., Uchihara T. Recruitment of nonexpanded polyglutamine proteins to intranuclear aggregates in neuronal intranuclear hyaline inclusion disease. J. Neuropathol. Exp. Neurol. 2001;60:369–376. doi: 10.1093/jnen/60.4.369. [DOI] [PubMed] [Google Scholar]
- 83.Uchihara T., Fujigasaki H., Koyano S., Nakamura A., Yagishita S., Iwabuchi K. Non-expanded polyglutamine proteins in intranuclear inclusions of hereditary ataxias—triple-labeling immunofluorescence study. Acta Neuropathol. 2001;102:149–152. doi: 10.1007/s004010100364. [DOI] [PubMed] [Google Scholar]
- 84.Broderick J., Wang J., Andreadis A. Heterogeneous nuclear ribonucleoprotein E2 binds to tau exon 10 and moderately activates its splicing. Gene. 2004;331:107–114. doi: 10.1016/j.gene.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 85.Seilhean D., Takahashi J., El Hachimi K.H., Fujigasaki H., Lebre A.S., Biancalana V., Durr A., Salachas F., Hogenhuis J., de The H., et al. Amyotrophic lateral sclerosis with neuronal intranuclear protein inclusions. Acta Neuropathol. 2004;108:81–87. doi: 10.1007/s00401-004-0855-x. [DOI] [PubMed] [Google Scholar]
- 86.Winborn B.J., Travis S.M., Todi S.V., Scaglione K.M., Xu P., Williams A.J., Cohen R.E., Peng J., Paulson H.L. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moffat J., Grueneberg D.A., Yang X., Kim S.Y., Kloepfer A.M., Hinkle G., Piqani B., Eisenhaure T.M., Luo B., Grenier J.K., et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 88.Aguirre P., Valdes P., Aracena-Parks P., Tapia V., Nunez M.T. Upregulation of gamma-glutamate-cysteine ligase as part of the long-term adaptation process to iron accumulation in neuronal SH-SY5Y cells. Am. J. Phys. Cell Physiol. 2007;292:C2197–C2203. doi: 10.1152/ajpcell.00620.2006. [DOI] [PubMed] [Google Scholar]
- 89.Nicklas W., Baneux P., Boot R., Decelle T., Deeny A.A., Fumanelli M., Illgen-Wilcke B. Recommendations for the health monitoring of rodent and rabbit colonies in breeding and experimental units. Lab. Anim. 2002;36:20–42. doi: 10.1258/0023677021911740. [DOI] [PubMed] [Google Scholar]
- 90.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.