Abstract
Diffuse gliomas are the most common type of malignant primary brain tumor, and their initiation and/or progression are often associated with alternative splicing. They produce an enormous economic burden on society and greatly impair the quality of life of those affected. The aim of the current study was to explore the differentially expressed genes (DEGs) observed in glioblastoma (GBM) and oligodendroglioma (OD) at the splicing level, and to analyze their functions in order to identify the underlying molecular mechanisms of gliomas. The exon-level expression profile data GSE9385 was downloaded from the Gene Expression Omnibus database, and included 26 GBM samples, 22 OD samples and 6 control brain samples. The differentially expressed exon-level probes were analyzed using the microarray detection of alternative splicing algorithm combined with the splicing index method, and the corresponding DEGs were identified. Next, a Gene Ontology enrichment analysis of the DEGs was performed. Additionally, the protein-protein interaction (PPI) networks were constructed based on the depth-first search algorithm. A total of 300 DEGs were identified to be shared by GBM and OD, including 97 upregulated and 203 downregulated DEGs. Furthermore, screening with a defined threshold identified 6 genes that were highly expressed in GBM, including AFF2, CACNA2D3 and ARPP21, while the 6 highly expressed genes in OD notably included CNTN2. The TP53 and HIST1H3A genes were the hub nodes in the PPI network of DEGs from GBM, while CNTN2 was linked to the highest degree in the OD PPI network. The present study provides a comprehensive bioinformatics analysis of DEGs in GBM and OD, which may provide a basis for understanding the initiation and/or progression of glioma development.
Keywords: glioblastomas, oligodendrogliomas, alternative splicing, differentially expressed genes, protein-protein interaction network
Introduction
Diffuse gliomas are the most common type of intracranial malignant neoplasm, and account for >60% of all primary brain tumors (1). Based on the classification of nervous system tumors by the World Health Organization, diffuse gliomas are classified into seven principal categories: Diffuse astrocytoma (grade II), oligodendroglioma (OD; grade II), oligoastrocytoma (grade II), anaplastic astrocytoma (grade III), anaplastic oligodendroglioma (grade III), anaplastic oligoastrocytoma (grade III) and glioblastoma (GBM, grade IV) (1). Of these, GBM is the most common and aggressive type of primary brain tumor, accounting for 80% of malignant astrocytomas (2). GBM may develop rapidly without the diagnosis of a less malignant precursor lesion, and this is termed primary or de novo GBM. It may also develop slowly through progression from a pre-existing low-grade glioma, in which case it is termed a secondary GBM (3). Although GBMs are considered as primarily astrocytic gliomas (4), a subset of GBMs exhibit OD-like tumor cell differentiation (5). OD is a well-differentiated, slowly grown and diffusely infiltrated tumor observed in adults, and is typically located in the cerebral hemispheres (6). Despite advances in neurosurgery, chemotherapy and radiotherapy, glioma commonly has a poor prognosis (7). Therefore, it is critical that the genetic pathways underlying the development of this type of cancer are defined.
A previous study indicated that tumor-specific alternative splicing is important in the regulation of gene expression and corresponding protein functions during cancer development (8). Multiple alternative splicing transcripts have been identified as progression markers, including generalized splicing abnormalities and tumor- and stage-specific events (9–10). A number of studies have documented that the initiation and/or progression of glial brain tumors is influenced by aberrant splice isoforms, including epidermal growth factor receptor, phosphatase and tensin homolog, tumor protein p53 (TP53), proliferation-related Ki-67 antigen, murine double-minute 2, mutS homolog 2, platelet-derived growth factor α and Kruppel-like transcription factor (11–15). However, the molecular mechanisms associated with the alternative splicing that may lead to the development and progression of GBM and OD remain to be clearly demonstrated.
In the current study, the gene expression profiles of GBM, OD and patient-matched normal brain tissues were downloaded from the Gene Expression Omnibus (GEO) database. The significant differentially expressed exon-level probes and their corresponding genes were identified using a combination of the splicing index (SI) method and the microarray detection of alternative splicing (MIDAS) algorithm. In addition, the screened differentially expressed genes (DEGs) were further analyzed with bioinformatics methods. The current study aims to improve the understanding of molecular mechanisms of GBM and OD and may clarify the processes involved in the development of gliomas.
Materials and methods
Affymetrix microarray analysis
The gene expression profile data GSE9385 (7) was obtained from the National Center for Biotechnology Information and GEO database (http://www.ncbi.nlm.nih.gov/geo/), which is based on the GeneChip Human Exon 1.0 ST Array (GPL5188) platform (Affymetrix, Santa Clara, CA, USA). A total of 54 specimens were available, including 26 GBM, 22 OD and 6 control brain samples.
Data preprocessing
The background correction and data normalization were performed by the robust multiarray average (RMA) algorithm based on the Affymetrix Power Tools (http://www.affymetrix.com/) program (16). Additionally, the probe sets were filtered according to the methods described by Gardina et al (17). To reduce the false positive rate, only the genes with a PLIER signal of >200 and corresponding detection above background (DABG) with a P-value of ≤0.05 were accepted. Probe sets with cross-hybridization type were also removed (18).
DEG analysis
The differences in exon-level expression can result from one of two factors, namely, differential splicing or differential gene expression (18). To detect differential splicing, the gene expression level was normalized. Following normalization, the exon-level expression value (I) was calculated for each exon to reflect the actual exon-level expression. For exon i in gene j, Ii,j is denoted as follows:
G is the average gene expression value of all specimens and Gj is the gene expression value of specimen j. Ei,j represents the expression values of exon i in gene j. Analysis using this formula is known as the SI method (18,19). The exon- and gene-level expression values were computed using the RMA algorithm.
Based on the SI method, the differentially expressed exon-level probes between GBM/OD, GBM/normal and OD/normal samples were identified with Student’s t-test. Only exon-level probes with P-values <0.01 and differential regulation of RMA signals >2-fold were selected. Since the RMA-generated signals were reported as the value of log2 transformation, the geometric mean >1 represented an RMA signal that was upregulated >2-fold. Also, in order to improve the accuracy of the results, the MIDAS algorithm (http://www.affymetrix.com/) (17) was used to identify differentially expressed exon-level probes based on the analysis of variance. P<0.05 was selected as the cut-off criterion. Only the exons identified by the two methods were used. A hierarchical clustering of the screened DEGs was performed based on their expression values using the Hclust package of R software (20).
Gene ontology (GO) enrichment analysis
GO analysis is a commonly used approach for functional studies of large-scale genomics (21). The Database for Annotation, Visualization and Integrated Discovery (DAVID), a high-throughput and integrated data-mining environment, analyzes gene lists derived from high-throughput genomic experiments (22). The current study used the DAVID to identify which represented GO categories were significantly enriched in DEGs.
Protein-protein interaction (PPI) network construction
The Search Tool for the Retrieval of Interacting Genes (STRING) (23) database was used to annotate functional interactions between differentially expressed proteins that regulate alternative splicing and other proteins by calculating their confidence score. Interactions with a score >0.8 were selected. In order to remove the irrelevant nodes, reduce the network size and restrict the search space into a realistic and meaningful one, the depth first search algorithm was employed to construct the PPI network with the path length <3.
Results
Identification of DEGs
Exon-level expression data were compared between GBM/OD samples, GBM/normal samples and OD/normal samples. Using the SI method, 1,343 differentially expressed exon-level probes with P-values <0.01 and differential regulation of RMA signals >2-fold were identified. Based on the MIDAS method, 5,290 differentially expressed exon-level probes with the P-value <0.05 were selected. A total of 982 DEGs were identified by both methods simultaneously.
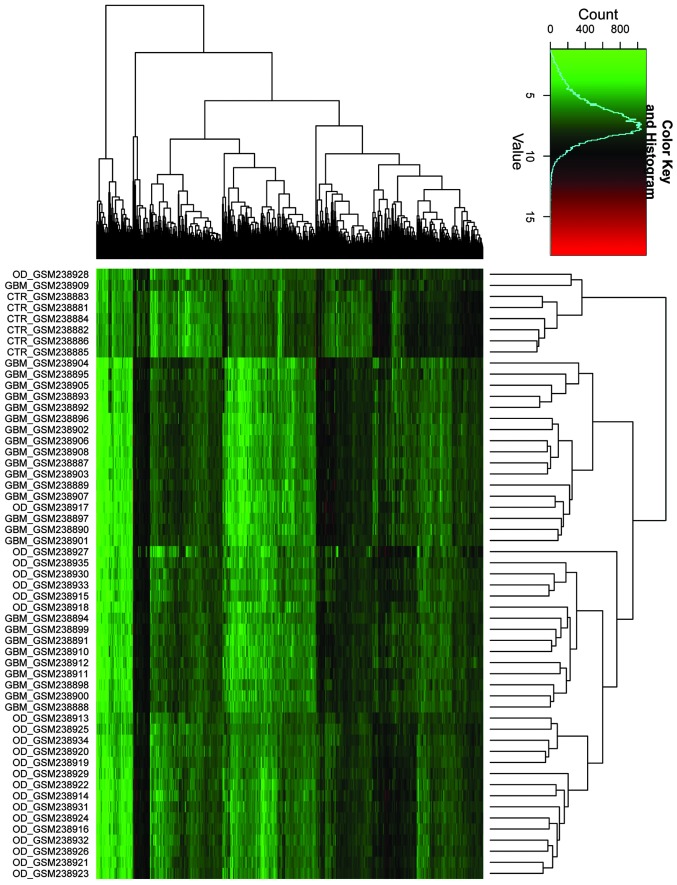
The expression values of these 982 DEGs were hierarchically clustered by the Hclust package of R software (Fig. 1). Compared with normal brain tissue, 617 and 498 DEGs were identified in GBM and OD, respectively (Fig. 2A). A total of 97 upregulated and 203 downregulated DEGs were identified to be present in both GBM and OD (Fig. 2B), and 236 DEGs were obtained between GBM and OD, 94 of which were upregulated and 142 downregulated in GBM (Fig. 2A).
Figure 1.
Hierarchical cluster analysis of 982 differentially expressed genes by the hclust package of R software.
Figure 2.
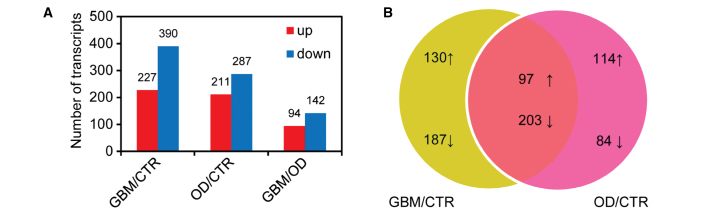
Identification of DEGs using the SI method and MIDAS algorithm. (A) DEGs between GBM/OD samples, GBM/normal samples and OD/normal samples. (B) A total of 617 and 498 differentially expressed exon-level genes were identified in GBM and OD compared with the control, respectively. A total of 97 upregulated and 203 downregulated genes were identified in both GBM and OD. DEG, differentially expressed gene; SI, splicing index; MIDAS, microarray detection of alternative splicing; GBM, glioblastoma; OD, oligodendroglioma; CTR, control.
With the strict threshold of RMA signals that were upregulated at least 4-fold and 2-fold compared with normal brain tissue and OD respectively, a total of 6 highly expressed genes were identified in GBM, including AFF2, GNAL, ARPP21, CACNA2D3, HIST1H3A~HIST1H3J, and RGS7 (Table I). Similarly, at the cut-off criteria of RMA signals upregulated at least 4-fold and 2-fold compared with normal brain tissue and GBM respectively, a total of 6 highly expressed genes were identified in OD, including CNTN2, ABCA6, MEGF11, DOCK5, MOXD1, and TRIM67. There were 7 genes downregulated at least 8-fold compared with controls in both GBM and OD. These included APBA2, MAP4, NUF2, INPP5F and TOP2A. These candidate genes were suggested as markers of alternative splicing in GBM and OD.
Table I.
Identification of differentially expressed genes between GBM/OD samples, GBM/normal samples and OD/normal samples.
| GBM/CTR | OD/CTR | GBM/OD | |||||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| Type | Gene name | log2 transformation | P-value | log2 transformation | P-value | log2 transformation | P-value |
| GBM | AFF2 | 2.18 | 2.86E-03 | −0.36 | 5.66E-01 | 2.55 | 2.48E-07 |
| GBM | GNAL | 2.46 | 2.08E-05 | 0.73 | 8.30E-02 | 1.74 | 3.74E-07 |
| GBM | ARPP21 | 2.75 | 3.26E-07 | 1.1 | 4.11E-02 | 1.65 | 2.85E-06 |
| GBM | CACNA2D3 | 3.37 | 1.10E-09 | 1.76 | 4.31E-05 | 1.61 | 1.98E-05 |
| GBM | HIST1H3A~HIST1H3J | 2.91 | 1.92E-08 | 1.48 | 8.97E-04 | 1.43 | 3.67E-07 |
| GBM | RGS7 | 3.36 | 3.08E-07 | 1.97 | 9.74E-05 | 1.39 | 1.17E-04 |
| OD | CNTN2 | 1.72 | 2.84E-03 | 2.84 | 1.40E-07 | −1.11 | 1.22E-03 |
| OD | ABCA6 | 1.42 | 6.40E-03 | 2.6 | 1.38E-04 | −1.18 | 1.00E-03 |
| OD | MEGF11 | 1.11 | 1.37E-02 | 2.26 | 1.17E-06 | −1.15 | 1.78E-05 |
| OD | DOCK5 | 0.91 | 2.19E-02 | 2.03 | 6.19E-05 | −1.12 | 1.59E-04 |
| OD | MOXD1 | −0.24 | 4.79E-01 | 2.45 | 2.01E-05 | −2.69 | 5.29E-10 |
| OD | TRIM67 | −0.37 | 6.15E-01 | 2.54 | 1.62E-04 | −2.91 | 9.58E-09 |
| GBM + OD | APBA2 | −4.49 | 1.89E-08 | −3.12 | 1.93E-05 | −1.36 | 1.86E-03 |
| GBM + OD | MAP4 | −4.1 | 1.69E-08 | −3.13 | 3.54E-06 | −0.97 | 1.17E-02 |
| GBM + OD | NUF2 | −3.98 | 1.86E-09 | −3.16 | 3.67E-05 | −0.82 | 3.31E-02 |
| GBM + OD | INPP5F | −3.17 | 2.41E-09 | −3.33 | 1.64E-09 | 0.16 | 5.27E-01 |
| GBM + OD | TOP2A | −8.68 | 4.11E-15 | −7.87 | 4.60E-08 | −0.81 | 1.85E-01 |
GBM, glioblastoma; OD, oligodendroglioma; CTR, control.
GO enrichment analysis
To investigate the functional changes in the initiation and/or progression of glial brain tumors, the 300 overlapping DEGs in GBM and OD were mapped to the GO database. A P-value ≤0.01 and fold change >2 were used for the threshold. The significant GO terms of the 97 upregulated DEGs shared by GBM and OD included various processes, including cell adhesion, extracellular structure organization, collagen biosynthetic process, neuron development and the regulation of small GTPase-mediated signal transduction (Table II). For the 203 downregulated DEGs shared by GBM and OD, the significant GO terms included processes such as the regulation of small GTPase-mediated signal transduction, transmission of nerve impulses and positive regulation of apoptosis (Table II).
Table II.
Significant GO terms of 97 upregulated and 203 downregulated DEGs shared by GBM and OD.
| Terms | Description | Count | P-value | FDR |
|---|---|---|---|---|
| Upregulated DEGs | ||||
| GO:0007155 | Cell adhesion | 15 | 2.02E-06 | 3.06E-03 |
| GO:0043062 | Extracellular structure organization | 7 | 8.96E-05 | 0.14 |
| GO:0032964 | Collagen biosynthetic process | 3 | 1.98E-04 | 0.30 |
| GO:0048666 | Neuron development | 8 | 7.96E-04 | 1.20 |
| GO:0051056 | Regulation of small GTPase-mediated signal transduction | 7 | 9.28E-04 | 1.40 |
| GO:0031175 | Neuron projection development | 7 | 1.01E-03 | 1.51 |
| GO:0030030 | Cell projection organization | 8 | 1.28E-03 | 1.92 |
| GO:0008624 | Induction of apoptosis by extracellular signals | 5 | 1.62E-03 | 2.42 |
| GO:0048667 | Cell morphogenesis involved in neuron differentiation | 6 | 2.47E-03 | 3.68 |
| GO:0048812 | Neuron projection morphogenesis | 6 | 2.69E-03 | 3.98 |
| GO:0008088 | Axon cargo transport | 3 | 2.92E-03 | 4.33 |
| GO:0043065 | Positive regulation of apoptosis | 8 | 3.10E-03 | 4.58 |
| GO:0030182 | Neuron differentiation | 8 | 3.43E-03 | 5.06 |
| GO:0000904 | Cell morphogenesis involved in differentiation | 6 | 4.79E-03 | 7.01 |
| GO:0048858 | Cell projection morphogenesis | 6 | 4.88E-03 | 7.12 |
| GO:0000902 | Cell morphogenesis | 7 | 5.23E-03 | 7.62 |
| GO:0032990 | Cell part morphogenesis | 6 | 5.86E-03 | 8.50 |
| Downregulated DEGs | ||||
| GO:0051056 | Regulation of small GTPase-mediated signal transduction | 11 | 2.57E-05 | 0.04 |
| GO:0019226 | Transmission of nerve impulses | 12 | 8.44E-05 | 0.13 |
| GO:0043065 | Positive regulation of apoptosis | 11 | 1.85E-03 | 2.92 |
| GO:0007268 | Synaptic transmission | 9 | 2.22E-03 | 3.49 |
| GO:0007267 | Cell-cell signaling | 13 | 2.34E-03 | 3.68 |
| GO:0001662 | Behavioral fear response | 3 | 5.87E-03 | 8.978 |
| GO:0046578 | Regulation of Ras protein signal transduction | 7 | 5.87E-03 | 8.98 |
GO, gene ontology; DEG, differentially expressed gene; GBM, glioblastoma; OD, oligodendroglioma; FDR, false discovery rate.
The DEGs that were upregulated ≥2-fold in one subgroup compared with the other were also mapped to the GO database. A total of 94 upregulated DEGs in GBM were significantly enriched into 10 GO terms, including neuron differentiation, exocytosis and regulation of neurotransmitter secretion (Table III). Additionally, 42 upregulated DEGs in OD were significantly enriched into 7 GO terms, which included the transmission of nerve impulses, cell-cell signaling and synaptic transmission (Table IV).
Table III.
Significant GO terms of DEGs in GBM that were upregulated at least 2-fold compared with OD.
| Term | Description | Count | P-value | FDR |
|---|---|---|---|---|
| GO:0030182 | Neuron differentiation | 9 | 2.44E-04 | 0.36 |
| GO:0006887 | Exocytosis | 5 | 9.80E-04 | 1.44 |
| GO:0046928 | Regulation of neurotransmitter secretion | 3 | 2.90E-03 | 4.21 |
| GO:0031175 | Neuron projection development | 6 | 2.94E-03 | 4.25 |
| GO:0006904 | Vesicle docking during exocytosis | 3 | 3.79E-03 | 5.45 |
| GO:0048278 | Vesicle docking | 3 | 4.44E-03 | 6.36 |
| GO:0051588 | Regulation of neurotransmitter transport | 3 | 4.44E-03 | 6.36 |
| GO:0060627 | Regulation of vesicle-mediated transport | 4 | 5.95E-03 | 8.44 |
| GO:0016192 | Vesicle-mediated transport | 8 | 6.20E-03 | 8.78 |
| GO:0022406 | Membrane docking | 3 | 6.28E-03 | 8.88 |
GO, gene ontology; DEG, differentially expressed gene; GBM, glioblastoma; OD, oligodendroglioma; FDR, false discovery rate.
Table IV.
Significant GO terms of DEGs in OD that were upregulated at least 2-fold compared with GBM.
| Term | Description | Count | P-value | FDR |
|---|---|---|---|---|
| GO:0019226 | Transmission of nerve impulses | 11 | 8.06E-05 | 0.12 |
| GO:0007267 | Cell-cell signaling | 14 | 1.13E-04 | 0.17 |
| GO:0007268 | Synaptic transmission | 10 | 1.23E-04 | 0.19 |
| GO:0006836 | Neurotransmitter transport | 6 | 1.92E-04 | 0.29 |
| GO:0007016 | Cytoskeletal anchoring at plasma membrane | 3 | 1.12E-03 | 1.69 |
| GO:0007269 | Neurotransmitter secretion | 4 | 1.33E-03 | 2.01 |
| GO:0003001 | Generation of a signal involved in cell-cell signaling | 5 | 2.17E-03 | 3.26 |
GO, gene ontology; DEG, differentially expressed gene; GBM, glioblastoma; OD, oligodendroglioma; FDR, false discovery rate.
PPI network construction
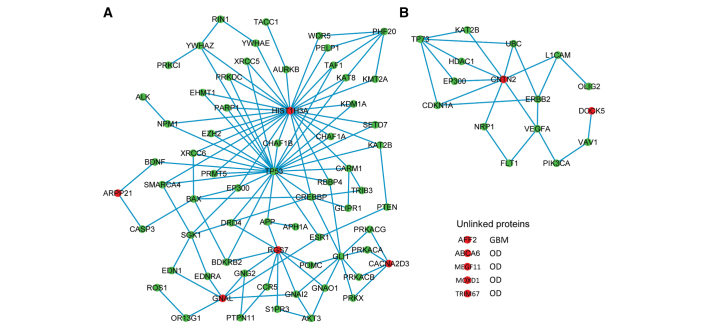
In order to construct the PPI network, the depth-first search algorithm was employed to obtain the PPI data from the STRING database. PPI networks of highly expressed genes in GBM (Fig. 3A) and OD (Fig. 3B) were constructed with the path length <3. In the PPI network of GBM, the genes HIST1H3A and TP53 contained the highest degrees. In addition, in the PPI network of OD, GNTN2 acted as hub nodes.
Figure 3.
Protein-protein interaction network of highly expressed genes in (A) glioblastoma and (B) oligodendroglioma.
Discussion
Formation and malignant progression of diffuse gliomas are associated with alterations in a variety of genes that regulate the normal homeostasis of cell proliferation, differentiation and apoptosis (24). The connection between abnormal regulation of alternative splicing and tumor development has emerged as a novel aspect of cancer biology. The identification of alternatively spliced genes in GBM and OD may provide novel molecular markers for the diagnosis and treatment of the two subtypes of glioma.
In the present study, 300 overlapping DEGs were identified in GBM and OD, compared with normal control tissue. These included 97 upregulated and 203 downregulated DEGs. Notably, 117 of these 300 DEGs were associated with alternative splicing. With the strict threshold, 6 highly expressed genes were screened in GBM, including AFF2, GNAL, ARPP21, CACNA2D3 and RGS7, in addition to 6 highly expressed genes in OD, including CNTN2, ABCA6, MEGF11, DOCK5, MOXD1 and TRIM67. Finally, by constructing a PPI network of DEGs, it was demonstrated that TP53 and HIST1H3A were the hub nodes in the PPI network of GBM and CNTN2 was the hub node in OD.
AFF2/FMR2 is extended over >600 kb in Xq27.3–q28, composed of 22 exons with a complex pattern of alternative splicing (25). It has been demonstrated that AFF2 mutations are associated with breast tumors (26). Recently, an excess of non-synonymous missense variants in FMR2 has been reported in males with autism spectrum disorders (27), indicating the role of FMR2 in normal brain function. The silencing of the AFF2 gene can lead to Fragile XE syndrome (28). In agreement with a previous study, the current study indicated that the AFF2 gene with differentially spliced exons was highly expressed in GBM. Bensaid et al (29) reported that as an RNA-binding protein, the AFF2 protein serves an essential role in alternative splicing regulation via the interaction with the G-quartet RNA-forming structure. CACNA2D3 protein is an auxiliary member of the α-2/δ subunit family of the voltage-dependent calcium channel complex (30). The CACNA2D3 gene has been suggested as a putative tumor suppressor gene in lung cancer, renal cell cancer neuroblastoma and squamous cell esophageal cancer (31), and has been identified as an indicator of prognosis in gastric cancer (32). CACNA2D3 is highly expressed in neuroblasts and neuroblastomas with a favorable prognosis, while its expression is downregulated in those with a poor prognosis (33). Additionally, ARPP21 encodes a 21-kDa cAMP-regulated phosphoprotein termed regulator of calmodulin signaling, which is enriched in the brain and may serve as a candidate tumor suppressor gene (34). ARPP21 is frequently downregulated, as is miR-128-2, in human breast cancer (35,36). Notably, miR-128 expression may significantly reduce glioma cell proliferation in vitro and glioma xenograft growth in vivo (37). In addition, Donzelli et al (36) reported that mutant TP53 is able to bind the putative promoter of the miR128-2 host gene (ARPP2), which determines the concomitant induction of ARPP21 mRNA expression in lung cancer cells, and thus inhibits apoptosis (36). In the present study, TP53 acted as the hub node in the GBM network, indicating its important role in the initiation and/or progression of GBM. A previous study suggested that TP53 regulates the proliferation, differentiation and survival of stem cells, which further highlights the importance of TP53 in GBM suppression (38). Additionally, TP53 mutations have been noted in 5–15% of cases of OD (39). TP53 mutations are the most frequent type of gene-specific alteration identified in human cancers (40). Shiraishi et al (41) studied the different locations of TP53 mutations between anaplastic astrocytoma and GBM, and suggested that the TP53 mutation may contribute to tumorigenesis and also to the progression of malignancy in gliomas.
It has been suggested that CNTN2 (also knows as contactin-2 or axonal glycoprotein TAG-1) is involved in the cell adhesion process and serves a critical function in the early stages of hepatocellular carcinoma (42). Adair et al (43) have also demonstrated that the CNTN2 gene is expressed in a variety of tumor cell lines, including those from the brain, breast and lung, and particularly in an unusually high percentage of melanoma cells. In the present study, CNTN2, with a high expression level, was the hub node in the PPI network of OD, suggesting that it may serve an essential function in the pathogenesis of OD.
GO enrichment analysis indicated a close relationship between the two subsets of gliomas and cell adhesion, which is in agreement with previous studies (44,45). An essential step in tumor progression is tumor invasion, which is dependent on the preservation of a delicate balance between cell adhesion and cell detachment. Chen et al (42) have suggested that the cell adhesion pathway is important for cancer cell invasion and metastasis. Abnormality of cell adhesion pathways is considered as a characteristic of advanced cancer. Another study demonstrated that alterations in several classes of adhesion molecules were implicated in the progression of various forms of cancer, including GBM (46).
In conclusion, the current data provide a comprehensive bioinformatics analysis of the alternatively spliced genes that may be involved in the pathogenesis of GBM and OD. A total of 300 overlapping DEGs were identified between GBM and OD. In addition, compared with normal brain tissue and OD, 6 highly expressed genes were identified in GBM, while 6 were identified in OD compared with normal brain tissue and GBM. The present analysis provides a basis for the understanding of the molecular mechanism of GBM and OD. However, further experimental studies with larger sample sizes are required to confirm these observations.
References
- 1.Wang H, Wang H, Zhang W, Huang HJ, Liao WS, Fuller GN. Analysis of the activation status of Akt, NFκB, and Stat3 in human diffuse gliomas. Lab Invest. 2004;84:941–951. doi: 10.1038/labinvest.3700123. [DOI] [PubMed] [Google Scholar]
- 2.Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Li S, Chen L, et al. Glioblastoma with an oligodendroglioma component: distinct clinical behavior, genetic alterations, and outcome. Neuro Oncol. 2012;14:518–525. doi: 10.1093/neuonc/nor232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraus JA, Lamszus K, Glesmann N, et al. Molecular genetic alterations in glioblastomas with oligodendroglial component. Acta Neuropathol. 2001;101:311–320. doi: 10.1007/s004010000258. [DOI] [PubMed] [Google Scholar]
- 6.Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci. 2009;100:2235–2241. doi: 10.1111/j.1349-7006.2009.01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.French PJ, Peeters J, Horsman S, et al. Identification of differentially regulated splice variants and novel exons in glial brain tumors using exon expression arrays. Cancer Res. 2007;67:5635–5642. doi: 10.1158/0008-5472.CAN-06-2869. [DOI] [PubMed] [Google Scholar]
- 8.Song SW, Fuller GN, Zheng H, Zhang W. Inactivation of the invasion inhibitory gene IIp45 by alternative splicing in gliomas. Cancer Res. 2005;65:3562–3567. doi: 10.1158/0008-5472.CAN-04-3392. [DOI] [PubMed] [Google Scholar]
- 9.LeFave CV, Squatrito M, Vorlova S, et al. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 2011;30:4084–4097. doi: 10.1038/emboj.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venables JP, Klinck R, Koh C, et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16:670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 11.Camacho-Vanegas O, Narla G, Teixeira MS, et al. Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int J Cancer. 2007;121:1390–1395. doi: 10.1002/ijc.22809. [DOI] [PubMed] [Google Scholar]
- 12.Ebert C, von Haken M, Meyer-Puttlitz B, et al. Molecular genetic analysis of ependymal tumors: NF2 mutations and chromosome 22q loss occur preferentially in intramedullary spinal ependymomas. Am J Pathol. 1999;155:627–632. doi: 10.1016/S0002-9440(10)65158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 14.Miller CR, Perry A. Glioblastoma. Arch Pathol Lab Med. 2007;131:397–406. doi: 10.5858/2007-131-397-G. [DOI] [PubMed] [Google Scholar]
- 15.Tortosa A, Viñolas N, Villà S, et al. Prognostic implication of clinical, radiologic, and pathologic features in patients with anaplastic gliomas. Cancer. 2003;97:1063–1071. doi: 10.1002/cncr.11120. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 17.Gardina PJ, Clark TA, Shimada B, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu Y, He F, Chen Y. Different effects of the probe summarization algorithms PLIER and RMA on high-level analysis of Affymetrix exon arrays. BMC Bioinformatics. 2010;11:211. doi: 10.1186/1471-2105-11-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laajala E, Aittokallio T, Lahesmaa R, Elo LL. Probe-level estimation improves the detection of differential splicing in Affymetrix exon array studies. Genome Biol. 2009;10:R77. doi: 10.1186/gb-2009-10-7-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. John Wiley & Sons, Ltd; New York: 2009. [Google Scholar]
- 21.Hulsegge I, Kommadath A, Smits MA. Globaltest and GOEAST: Two different approaches for Gene Ontology analysis. BMC Proc. 2009;3(Suppl 4):S10. doi: 10.1186/1753-6561-3-s4-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shostak K, Labunskyy V, Dmitrenko V, et al. HC gp-39 gene is upregulated in glioblastomas. Cancer Lett. 2003;198:203–210. doi: 10.1016/S0304-3835(03)00310-0. [DOI] [PubMed] [Google Scholar]
- 25.Gecz J. The FMR2 gene, FRAXE and non-specific X-linked mental retardation: clinical and molecular aspects. Ann Hum Genet. 2000;64:95–106. doi: 10.1046/j.1469-1809.2000.6420095.x. [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mondal K, Ramachandran D, Patel VC, et al. Excess variants in AFF2 detected by massively parallel sequencing of males with autism spectrum disorder. Hum Mol Genet. 2012;21:4356–4364. doi: 10.1093/hmg/dds267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stettner GM, Shoukier M, Höger C, Brockmann K, Auber B. Familial intellectual disability and autistic behavior caused by a small FMR2 gene deletion. Am J Med Genet A. 2011;155A:2003–2007. doi: 10.1002/ajmg.a.34122. [DOI] [PubMed] [Google Scholar]
- 29.Bensaid M, Melko M, Bechara EG, et al. FRAXE-associated mental retardation protein (FMR2) is an RNA-binding protein with high affinity for G-quartet RNA forming structure. Nucleic Acids Res. 2009;37:1269–1279. doi: 10.1093/nar/gkn1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Zhu CL, Nie CJ, et al. Investigation of tumor suppressing function of CACNA2D3 in esophageal squamous cell carcinoma. PloS One. 2013;8:e60027. doi: 10.1371/journal.pone.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmieri C, Rudraraju B, Monteverde M, et al. Methylation of the calcium channel regulatory subunit α2δ-3 (CACNA2D3) predicts site-specific relapse in oestrogen receptor-positive primary breast carcinomas. Br J Cancer. 2012;107:375–381. doi: 10.1038/bjc.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wanajo A, Sasaki A, Nagasaki H, et al. Methylation of the calcium channel-related gene, CACNA2D3, is frequent and a poor prognostic factor in gastric cancer. Gastroenterology. 2008;135:580–590. doi: 10.1053/j.gastro.2008.05.041. [DOI] [PubMed] [Google Scholar]
- 33.Thorell K, Bergman A, Carén H, et al. Verification of genes differentially expressed in neuroblastoma tumours: a study of potential tumour suppressor genes. BMC Med Genomics. 2009;2:53. doi: 10.1186/1755-8794-2-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha S, Singh RK, Alam N, Roy A, Roychoudhury S, Panda CK. Frequent alterations of hMLH1 and RBSP3/HYA22 at chromosomal 3p22.3 region in early and late-onset breast carcinoma: clinical and prognostic significance. Cancer Sci. 2008;99:1984–1991. doi: 10.1111/j.1349-7006.2008.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian P, Banerjee A, Wu ZS, et al. Loss of SNAIL regulated miR-128-2 on chromosome 3p22.3 targets multiple stem cell factors to promote transformation of mammary epithelial cells. Cancer Res. 2012;72:6036–6050. doi: 10.1158/0008-5472.CAN-12-1507. [DOI] [PubMed] [Google Scholar]
- 36.Donzelli S, Fontemaggi G, Fazi F, et al. MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ. 2012;19:1038–1048. doi: 10.1038/cdd.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouimet CC, Hemmings HC, Jr, Greengard P. ARPP-21, a cyclic AMP-regulated phosphoprotein enriched in dopamine-innervated brain regions. II. Immunocytochemical localization in rat brain. J Neurosci. 1989;9:865–875. doi: 10.1523/JNEUROSCI.09-03-00865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar HR, Zhong X, Sandoval JA, Hickey RJ, Malkas LH. Applications of emerging molecular technologies in glioblastoma multiforme. Expert Rev Neurother. 2008;8:1497–1506. doi: 10.1586/14737175.8.10.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe T, Nakamura M, Kros JM, et al. Phenotype versus genotype correlation in oligodendrogliomas and low-grade diffuse astrocytomas. Acta Neuropathol. 2002;103:267–275. doi: 10.1007/s004010100464. [DOI] [PubMed] [Google Scholar]
- 40.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 41.Shiraishi S, Tada K, Nakamura H, et al. Influence of p53 mutations on prognosis of patients with glioblastoma. Cancer. 2002;95:249–257. doi: 10.1002/cncr.10677. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Wang L, Xu H, Liu X, Zhao Y. Exome capture sequencing reveals new insights into hepatitis B virus-induced hepatocellular carcinoma at the early stage of tumorigenesis. Oncol Rep. 2013;30:1906–1912. doi: 10.3892/or.2013.2652. [DOI] [PubMed] [Google Scholar]
- 43.Adair SJ, Carr TM, Fink MJ, Slingluff CL, Jr, Hogan KT. The TAG family of cancer/testis antigens is widely expressed in a variety of malignancies and gives rise to HLA-A2-restricted epitopes. J Immunother. 2008;31:7–17. doi: 10.1097/CJI.0b013e318159f797. [DOI] [PubMed] [Google Scholar]
- 44.Giese A, Laube B, Zapf S, et al. Glioma cell adhesion and migration on human brain sections. Anticancer Res. 1998;18:2435–2447. [PubMed] [Google Scholar]
- 45.Cappellari AR, Vasques GJ, Bavaresco L, et al. Involvement of ecto-5′-nucleotidase/CD73 in U138MG glioma cell adhesion. Mol Cell Biochem. 2012;359:315–322. doi: 10.1007/s11010-011-1025-9. [DOI] [PubMed] [Google Scholar]
- 46.Shashidhar S, Lorente G, Nagavarapu U, et al. GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene. 2005;24:1673–1682. doi: 10.1038/sj.onc.1208395. [DOI] [PubMed] [Google Scholar]