Summary
Redox control of protein function involves oxidation and reduction of amino acid residues, but mechanisms and regulators involved are insufficiently understood. Here, we report that methionine-R-sulfoxide reductase B1 (MsrB1) regulates, in conjunction with Mical proteins, mammalian actin assembly via stereoselective methionine oxidation and reduction in a reversible, site-specific manner. Two methionine residues in actin are specifically converted to methionine-R-sulfoxide by Mical1 and Mical2 and reduced back to methionine by selenoprotein MsrB1, supporting actin disassembly and assembly, respectively. Macrophages utilize this redox control during cellular activation by stimulating MsrB1 expression and activity as a part of innate immunity. We identified the regulatory role of MsrB1 as a Mical antagonist in orchestrating actin dynamics and macrophage function. More generally, our study shows that proteins can be regulated by reversible site-specific methionine-R-sulfoxidation.
Introduction
Nearly every major cellular process is now known to be redox regulated, but the majority of associated mechanisms remain elusive. Much of the cellular redox control involves reactive oxygen species (ROS) causing the reversible oxidation of cysteine (Cys) residues (e.g., to disulfide, nitrosothiol, sulfenic and sulfinic acid forms), whereas redox modifications of other residues are largely thought to represent protein damage. Methionine (Met) is a sulfur-containing residue that, like Cys, is highly susceptible to oxidation. Oxidized Met residues are repaired by methionine sulfoxide reductases A (MsrA) and B (MsrB) (Lee et al., 2009; Weissbach et al., 2002), which are present in the majority of living organisms across the three domains of life. ROS oxidize Met to a mixture of two diastereomers, methionine-S-sulfoxide (Met-S-SO) and methionine-R-sulfoxide (Met-R-SO), that are stereoselectively reduced back to Met by MsrA and MsrB, respectively (Stadtman et al., 2002; Stadtman et al., 2003; Ugarte et al., 2010). Random Met oxidation typically inhibits protein function and Msr enzymes can restore it; they may also influence protein function and activity by restoring the reduced form of Met in proteins (Drazic et al., 2013; Lim et al., 2013; Erickson et al., 2008; Santarelli et al., 2006; Xiong et al., 2006).
Many reversible posttranslational modifications, such as phosphorylation, acetylation, and ubiquitination, control protein function and regulate diverse processes, such as signaling, central metabolism, cellular migration, transcription, and cell cycle control (Ahearn et al., 2011; Bhoj & Chen, 2009; Chiarugi & Buricchi, 2007; Xiong & Guan, 2012). Likewise, targeted reversible Met oxidation and reduction by Msr enzymes might regulate biological processes through reversible posttranslational modifications. For example, CaMK II is activated by oxidation of two consecutive Met residues in the absence of Ca2+/CaM, but further reduction by MsrA reverses this oxidation, protecting heart from oxidative damage (Erickson et al., 2008). MsrA can also serve as a stereoselective Met oxidase that regulates calmodulin (Lim et al., 2013). In addition, potassium channels were shown to be regulated by Met oxidation and subsequent Msr-based reduction (Ciorba et al., 1997; Santarelli et al., 2006). A recent study reported that transcription factor HypT is activated by Met oxidation and inactivated by a Msr (Drazic et al., 2013). Bacteria utilize such redox control to survive in a hostile environment.
In this regard, two Met residues in actin were recently found to be oxidized by a monooxygenase Mical under control of the semophorine-plexin complex in Drosophila (Hung et al., 2011; Hung et al., 2010), and this oxidation inhibited filamentous actin (F-actin) assembly. Actin is an essential protein and its regulated transition between the G-actin (soluble monomer) and F-actin (component of insoluble polymer microfilaments) states is involved in many biological processes, such as cell division, motility, and signaling. Various actin-binding proteins (ABPs) and small molecules control this transition and actin dynamics, highlighting the importance and complexity of actin organization (Hild et al., 2010). In support of the role of Mical in actin cytoskeleton organization, human Mical homologs were found to regulate actin stress fibers, although the molecular mechanisms remain to be elucidated (Giridharan et al., 2011). The finding of Mical-dependent oxidation provides a new paradigm for regulation of actin dynamics, but how this regulation occurs remains unclear. Here, we report that MsrB1 functions as an antagonist to Mical, that the pair acts in a stereospecific manner in oxidizing and reducing the target protein and that it regulates actin disassembly and reassembly by targeted Met oxidation and reduction. Thus, MsrB1 has a crucial role in regulating the innate immunity response through the redox control of actin.
Results
Mouse Mical1 and Mical2 support actin disassembly
Drosophila Mical is a monooxygenase that disassembles F-actin, but whether this function applies to other organisms, including mammals, is not known. In contrast to a single Mical in Drosophila, mammals have three Micals containing conserved flavin-containing monooxygenase (FMO), calponin homology actin-binding (CH) and LIM domains. These proteins include highly homologous Mical2 and Mical3, and a more distantly related Mical1 (Fig. 1A and Fig. S1A,B). We found that the recombinant mouse Mical1 (mMical1) and Mical2 (mMical2) forms containing FMO and CH domains depolymerized F-actin in a NADPH-dependent manner as revealed by changes in fluorescence intensity of the pre-assembled actin (Fig. 1B,C). The sedimentation/Coomassie staining assay, which enables the analysis of actin assembly and disassembly by taking advantage of differential solubility of G-actin and F-actin, further showed that most of the G-actin monomer treated with mMical1/NADPH or mMical2/NADPH did not assemble into the F-actin polymer (Fig. 1D). On the other hand, Micals did not affect microtubule polymerization or oxidation of dabsylated Met (Fig. S1C-E), suggesting specificity of Micals towards actin, in agreement with a previous report of Drosophila Mical specificity towards actin (Hung et al., 2011; Hung et al., 2010).
Figure 1.
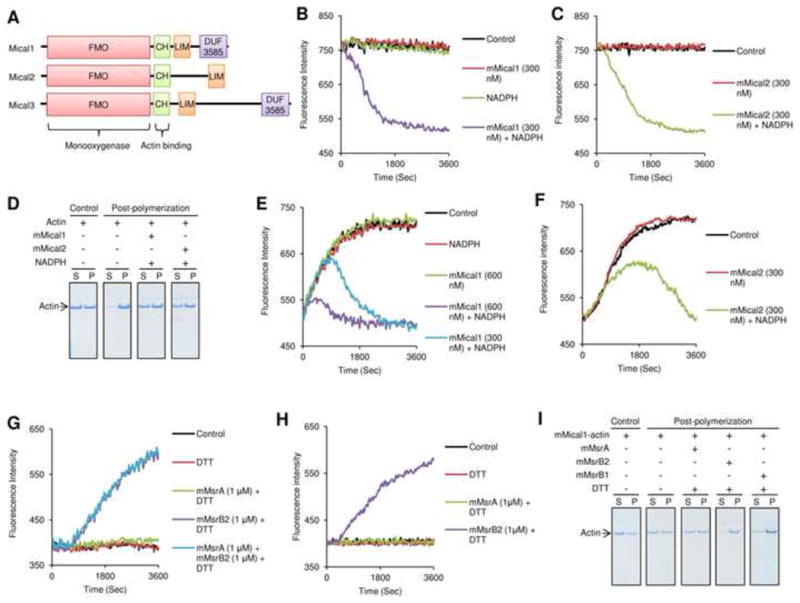
Mammalian Micals depolymerize F-actin, and MsrBs repolymerize it. (A) Mammals have three Mical genes that code for proteins composed of monooxygenase (FMO), actin-binding CH, LIM, and DUF3585 domains. (B, C) Pre-assembled pyrene-labeled actin (F-actin) was assayed for changes in fluorescence at 407 nm (excitation at 365 nm) in the presence of mMical1 (300 nM), mMical2 (300 nM) and/or NADPH (200 μM). As a control, fluorescence intensity of the pre-assembled pyrene-labeled actin was monitored in the absence of mMical1/NADPH and mMical2/NADPH. (D) Sedimentation/Coomassie staining assay was performed with G-actin before and after polymerization in the presence of mMical1 (1 μM)/NADPH (200 μM) or mMical2 (1 μM)/NADPH (200 μM). S refers to supernatant, and P represents both pellet and the remaining supernatant fraction. Fluorescence at 407 nm (excitation at 365 nm) was monitored for polymerization of pyrene-labeled G-actin incubated with or without (E) mMical1 (300 or 600 nM) and/or NADPH (200 μM) or (F) mMical2 (300 nM) and/or NADPH (200 μM). Actin alone was used for polymerization as a Control in (E) and (F). Then, (G) the mMical1/NADPH- or (H) mMical2/NADPH-treated pyrene-labeled actin monomer was monitored for repolymerization in the presence of mMsrA (1 μM), mMsrB2 (1 μM), and/or DTT (3 mM), or their absence as a control. (I) Sedimentation/Coomassie staining assay was performed with the mMical1 (1 μM)/NADPH (200 μM)-treated actin before and after repolymerization in the presence of mMsrA (1 μM)/DTT (3 mM), mMsrB2 (1 μM)/DTT (3 mM), or mMsrB1-Cys (10 μM)/DTT (3 mM). The mMical1-treated actin alone was used as a control and then this protein was used for further post-polymerization assay. All in vitro biochemical assays used 2.38 μM pyrene-labeled G-actin and all sedimentation/Coomassie staining assays used 4.76 μM G-actin. In the repolymerization assay, pyrene-labeled G-actin (2.38 μM) was first treated with mMicals/NADPH, and the actin monomer was repolymerized following buffer exchange. See also Figure S1.
MsrB reassemble the G-actin disassembled by Mical
We further tested if Micals disassemble F-actin by oxidation of its Met residues and if various Msrs could reverse this process. In the presence of NADPH, mMical1 and mMical2 blocked polymerization of G-actin (Fig. 1E,F), but mouse MsrB2 (mMsrB2) rescued G-actin assembly, whereas mouse MsrA (mMsrA) did not. MsrB1 (mMsrB1), used in this assay in the form of a low-activity Cys mutant of a natural selenoprotein (Kim & Gladyshev, 2005), also rescued G-actin assembly (Fig. 1G,H, Fig. S1F-J). The sedimentation/Coomassie staining assay confirmed that the mMicals/NADPH-treated G-actin monomer was reassembled when either mMsrB2 or mMsrB1 were added to the reaction mixture (Fig. 1I, Fig. S1K). We also found that jasplakinolide, a potent inducer of actin polymerization in vitro, was ineffective in stabilizing the mMical1/NADPH-treated actin in the polymer form. The addition of mMsrB2 to the mMical1/NADPH-treated actin enhanced the formation of the polymer by jasplakinolide, whereas the addition of mMsrA did not. The data suggested that the direct modification of actin by mMical1 blocked polymer formation and that mMsrB2 restored this process (Fig. S1L,M). We conclude that MsrB is the antagonist to Mical that acts on the modified form of actin and supports its reassembly.
Mical1 and Mical2 are stereospecific monooxygenases that convert Met to Met-R-SO in actin
If MsrBs could promote reassembly of the Micals-treated actin, why could not MsrA carry out this function? To address this question, we analyzed the redox modifications of Met residues in actin by Micals and Msrs. Mass spectrometry analyses showed that mMical1 and mMical2 oxidized Met46 and Met49 in actin (detected in the form of a tryptic peptide 42HQGVM*VGM*GQK52). Both mMsrB2 and mMsrB1 could reduce these two Met sulfoxide residues, while mMsrA could not (Fig. 2A,B and Fig. S2A-E). Moreover, mMsrB1 and mMsrB2 almost completely reduced the oxidized Met46 and Met49 in the Micals-treated actin, suggesting stereospecificity of the Mical monooxygenase activity. In support of the stereoselective reduction by MsrBs, we performed western blot analysis of the reduced and oxidized actin samples with the antibodies that we developed against the actin peptide containing Met in both Met46 and Met49 positions. The actin signal detected with these antibodies decreased upon incubation with either mMical1 or hydrogen peroxide, while the actin signal detected with common actin antibodies remained unchanged. In contrast, the signal of the mMical1-treated actin increased upon incubation with mMsrB2, whereas no change in the signal was observed upon treatment with mMsrA (Fig. 2C,D). Moreover, the signal of the hydrogen peroxide-treated actin increased upon incubation with either mMsrA or mMsrB2, indicating that mMsrA could reduce the Met-S-SO form of the two conserved Met residues in actin, but only when they were oxidized with hydrogen peroxide (Fig. S2F,G). Since MsrBs are stereoselective Met-R-SO reductases, these results demonstrate that mMicals stereoselectively oxidized Met46 and Met49 in mammalian actin to their Met-R-SO forms that disassembled actin, whereas MsrBs reduced the two Met-R-SO residues back to Met to promote actin assembly.
Figure 2.
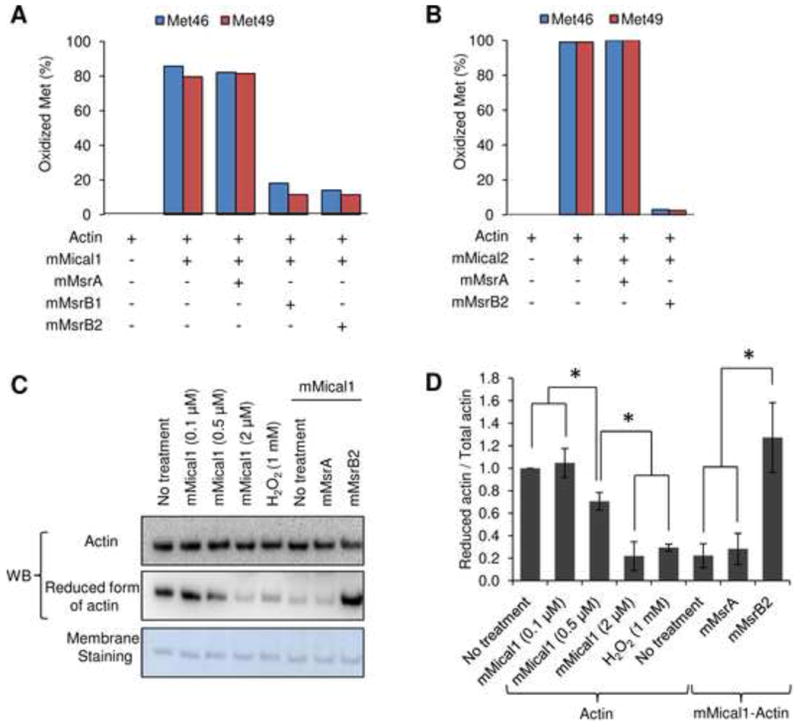
mMical is a stereospecific monooxygenase that converts two conserved Met to Met sulfoxide residues in actin, which can be further reduced by MsrB. (A, B) Oxidation status of two conserved Met residues (Met46 and Met49 in rabbit muscle actin) was analyzed by mass spectrometry. The fraction of oxidized Met was calculated based on the ratio of Met sulfoxide residues to total Met and Met sulfoxide residues in the detected peptides of actin. (A) Actin was incubated with mMical1 (1 μM)/NADPH (200 μM) for 1 h and further treated with mMsrA (1 μM)/DTT (3 mM), mMsrB1 (10 μM)/DTT (3 mM), or mMsrB2 (1 μM)/DTT (3 mM) for 1 h (following buffer change with G-actin buffer). (B) Actin was incubated with mMical2 (1 μM)/NADPH (200 μM) for 3 h and further treated with mMsrA (1 μM)/DTT (3 mM) or mMsrB2 (1 μM)/DTT (3 mM) for 3 h following buffer change with free G-actin buffer. Other Met residues were not oxidized by Micals (three Met residues were not detected). Actin incubated with mMical1 (0.1, 0.5, 2 μM)/NADPH (200 μM) or H2O2 (1 mM) and the Mical (2 μM)/NADPH (200 μM)-treated actin incubated with mMsrA (1 μM) or mMsrB2 (1 μM) with DTT (3 mM) were subjected to (C) western blotting with the antibodies specific for the reduced form of actin. Membrane was also stained with Amido Black. Then, (D) the ratio of reduced to total actin was calculated based on the quantification of band density using ImageJ. All data were normalized to No treatment (control actin), and this experiment was independently repeated three times and statistically analyzed by Student's t-test (*: p < 0.05). Error bars represent SD. All experiments used 9.5 μM or 4.76 μM G-actin for further mass spectrometry and western blot analyses. See also Figure S2.
Co-localization of mMsrB1 and actin
The biochemical assays showed that the Mical/MsrB pair stereoselectively regulates actin assembly via site-specific Met redox modification. We further found that mMsrB1 co-localized with actin in NIH 3T3 cells when mMical1 was overexpressed (Fig. 3A), but co-localization was not observed in the absence of mMical1 expression. We also examined endogenous MsrB1 localization in LPS-stimulated bone marrow derived macrophages (BMDMs) by using anti-MsrB1 antibodies and rhodamine-phalloidine actin staining. MsrB1 co-localized with actin in lamellapodia in BMDMs (Fig. 3B), suggesting that MsrB1 is needed for protrusion of actin in lamellapodia stimulated by LPS or serum. In addition, we analyzed relative fluorescence intensity of F-actin stained with rhodamine-phalloidine in HeLa cells transfected with mMical1 and/or mMsrB1 (Fig. 3C). Overexpression of mMsrB1 did not influence F-actin in HeLa cells, whereas overexpression of mMical1 reduced F-actin amounts up to 20%, suggesting that F-actin was depolymerized by mMical1 in vivo. Expression of mMsrB1 in mMical1-overexpressing cells rescued F-actin amounts. Overall, the data support importance of MsrB1 in F-actin formation, particularly with regard to the actin depolymerized by Mical.
Figure 3.
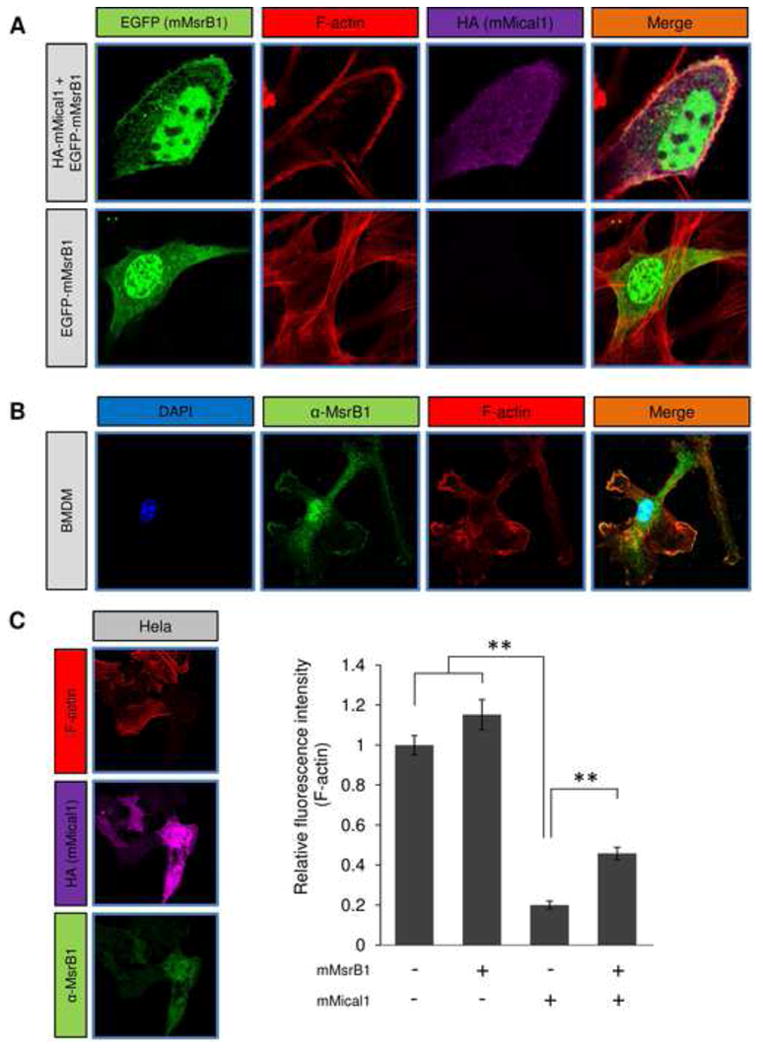
Localization of Mical1/MsrB1 and their effect on F-actin staining in mammalian cells. (A)Localization of mMsrB1 and actin was visualized in NIH 3T3 cells overexpressing EGFP-mMsrB1 and/or HA-tagged mMical1. Rhodamine-phalloidine was used to visualize F-actin. (B) Mouse bone marrow derived macrophages following 20 h LPS stimulation were stained for endogenous MsrB1 and F-actin. (C) HeLa cells were transfected with mMsrB1, mMical1 or both. Following standard immunocytochemistry analyses, we quantified F-actin staining intensity in individual cells. This experiment was independently repeated twenty times and then statistically analyzed by Student's t-test (**: p < 0.01). Error bars represent SD. See also Figure S3.
Regulation of actin polymerization-dependent processes by selenoprotein MsrB1 in macrophages
The response of macrophages to pathogens and pathogen-associated molecules like LPS depends on actin cytoskeleton reorganization (Aderem & Underhill, 1999; Buccione et al., 2004; Kleveta et al., 2012; Mooren et al., 2012). We found that BMDMs show a dramatic increase in MsrB1 expression (Fig. 4A and Fig. S3) and a two-fold increase in total MsrB activity (Fig. 4B). Like actin, MsrB1 is a cytosolic protein, whereas MsrB2 resides in mitochondria. Therefore, among MsrBs, changes in the MsrB1 expression are expected to regulate actin cytoskeleton organization in response to LPS stimulation. To test this possibility, we generated BMDMs from MsrA knockout (KO) and MsrB1 KO mice (Fomenko et al., 2009; Novoselov et al., 2010), as well as from MsrA/MsrB1 double KO mice that we developed by crossing the two mouse models. The LPS-stimulated BMDMs prepared from these animal models were analyzed for actin polymerization-dependent processes, including filopodia formation, macropinocytosis, and cytokine release. Upon LPS treatment, the number of filopodia per cell per circumference increased ∼1.5 fold in wild type (p<0.05) and MsrA KO BMDMs (p<0.05), but no effect was observed in MsrB1 KO and MsrA/MsrB1 KO BMDMs (Fig. 4C,D and Fig. S4). The macropinocytosis assays in which M-CSF was used to induce uptake of lucifer yellow fluorescent dye also revealed that MsrB1 KO and MsrA/MsrB1 KO BMDMs had reduced ability in internalizing the fluorescent dye, compared with wild type and MsrA KO BMDMs (p<0.05) (Fig. 4E,F). In addition, the release of three pro-inflammatory cytokines, MCP-1, IL-6, and TNF-α, was dramatically reduced in MsrB1 KO and MsrA/MsrB1 KO BMDM following LPS stimulation (p<0.05) (Fig. 4G-I). Thus, only MsrB1 deficiency led to a defect in actin polymerization-dependent processes in macrophages in response to LPS stimulation, suggesting that MsrB1 is required for the regulation of actin polymerization in macrophages in response to pathogen-associated molecules (Fig. 4J). These data provide another evidence for the regulatory function of Msrs. Met-R-SO can now be added to the list of reversible, site-specific posttranslational modifications that regulate protein function and biological processes.
Figure 4.
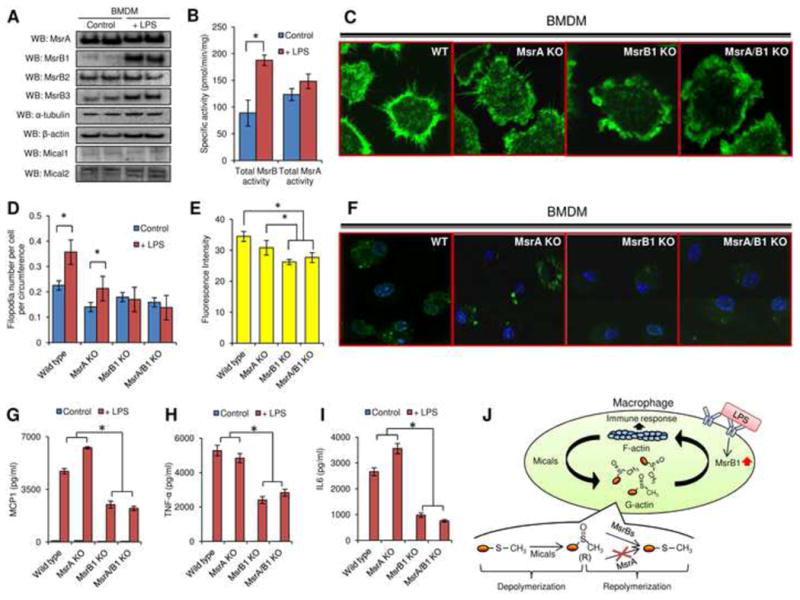
Regulation of actin polymerization-dependent processes by MsrB1 in bone marrow derived macrophages. (A) MsrA, MsrB1, MsrB2, MsrB3, α-tubulin, β-actin, Mical1, and Mical2 expression was analyzed by Western blots in control and LPS-stimulated cells. (B) Total MsrB and MsrA activities were measured by HPLC in bone marrow derived macrophages before and after LPS stimulation (n=3). (C) Filopodia formation and (F) Macropinocytosis were evaluated in LPS-primed macrophages generated from the bone marrow of wild type, MsrA KO, MsrB1 KO, and MsrA/MsrB1 KO mice. For macropinocytosis assays, M-CSF was used to stimulate the internalization of lucifer yellow fluorescence and only LPS-primed macrophages were included since unprimed macrophages did not exhibit detectable dye uptake. Quantitation of (D) filopodia number per cell per circumference (n=7) and (E) internalized lucifer yellow before in LPS primed macrophages is shown for wild type, MsrA KO, MsrB1 KO, MsrA/MsrB1 KO mice (n=4). (*: p < 0.05). Calculation of filopodia number is described in detail in Fig. S4C. (G) MCP1, (H) TNF-α, and (I) IL6 secretion before and after LPS stimulation from bone marrow derived macrophages generated from wild type, MsrA KO, MsrB1 KO, MsrA/MsrB1 KO mice was analyzed (n=4) (*: p < 0.05). (J) A model of redox regulation of actin function via reversible stereoselective site-specific Met oxidation and reduction and its importance for the macrophage immune response. All used error bars represent SD. See also Figures S3 and S4.
Discussion
Mical, one of recently characterized ABPs, supports a new regulatory mechanism of actin disassembly involving Met oxidation, implicating this oxidoreductase in diverse biological processes, such as neuronal growth, immune function, and wound healing. However, prior to our study the fate of the oxidized actin was unclear. In this regard, identifying the antagonist to Mical in reducing the oxidized Met in actin demonstrates the true potential of this redox regulation in controlling biological processes, i.e., this regulation acts in a reversible, site-specific manner like many other regulatory posttranslational modifications (e.g., phosphorylation). An additional feature of the regulation by Met oxidation is stereospecificity of this process, i.e., Met-R-sulfoxidation. Met oxidation in actin by ROS may also be random and harmful, as this process would interfere with the biological function of this protein. For example, it was reported that the oxidation of the two conserved Met residues (Met46 and Met49) was increased in aged muscle actin (Fedorova et al., 2010). Perhaps, increased oxidative stress leads to oxidative damage, thereby disrupting the F-actin filament. Thus, it appears that Msr proteins can contribute not only to the regulation of actin function, but also to the repair of damaged actin. Both overactivity of Mical and oxidative stress represent the risk factors, considering abundance and importance of actin in cells. As we show in the experiments with macrophages, mammalian MsrB1 is necessary for the regulation of macrophage function, but this regulatory mechanism is likely not limited to this function.
MsrB1 is a ubiquitous selenoenzyme and its expression is reduced by dietary selenium deficiency and as a function of age (Novoselov et al., 2010). With regard to actin regulation, suppressed MsrB1 expression may be a clue in explaining some open questions. For example, selenium deficiency and low selenoprotein status have long been associated with poor immune function, but the mechanisms are not known (Rayman et al., 2012). It appears that the decreased MsrB1 expression may contribute to the observed phenotypes via decreased efficiency of actin polymerization-dependent processes. There are also many reports of the age-related decline in actin polymerization in lymphocytes (Cheung et al., 1987; Rao et al., 1992), implicating MsrB1 in this process.
Altogether, we show for the first time that actin is redox regulated via stereoselective, reversible, site-specific oxidation of its two Met residues. Oxidation of these Met residues is carried out by Mical1 and Mical2 and their reduction by MsrB1. These findings establish the biological importance of MsrBs in regulating protein function, in addition to their role in protein damage repair. More generally, our study establishes a new mode of regulation of protein function, i.e., methionine-R-sulfoxidation.
Experimental Procedures
Constructs and Antibodies
We prepared recombinant mouse Mical1(NP_612188.1) and mouse Mical2 (NP_796256.1) constructs and also used previously developed mouse MsrB1 (Kim & Gladyshev, 2005), mouse MsrB2 (Kim & Gladyshev, 2005), mouse MsrA (Kim & Gladyshev, 2005) constructs. MsrA, MsrB1, MsrB2, MsrB3, Mical1 (S-14, Santa Cruz Biotechnology, Inc.), Mical2 (L-12, Santa Cruz Biotechnology, Inc.), α-tubulin (Sigma-Aldrich), β-actin (Sigma-Aldrich), and actin (Cytoskeleton, Inc.) antibodies were used for western blotting and/or immunohistochemistry analyses. In addition, actin antibodies were raised in rabbits against the peptide RHQGVMVGMGQKDS and used for western blotting. Detailed information on cloning, proteins, constructs, western blotting and immunohistochemistry can be found in Supplemental Information.
Mice
MsrA KO and MsrB1 KO mice have been previously described (Fomenko et al., 2009; Novoselov et al., 2010; Moskovitz et al., 2001). We backcrossed both strains onto a C57BL/6 (Jackson Lab) background for 7 generations, and then MsrA KO and MsrB1 KO mice were crossed to generate the MsrA/MsrB1 double KO mice.
In vitro actin polymerization, depolymerization, and repolymerization assays with mass spectrometry and western blot analyses
Actin polymerization, depolymerization, and repolymerization assays were carried out with modifications as described previously (Hung et al., 2011). Briefly, 1 mg of purified rabbit skeletal muscle actin labeled with or without pyrene was solubilized in a G-actin buffer and then used for polymerization and depolymerization assays in the presence of mMical1 or mMical2 and NADPH. Then, the actin depolymerized by mMicals was repolymerized in the presence of mMsrA, mMsrB1, and/or mMsrB2 (in the presence of DTT). Pyrene-labed actin was monitored for changes in fluorescence intensity, and the protein was subjected to the sedimentation/Coomassie staining assay for the analysis of F-actin assembly. Actin samples were used for further mass spectrometry analysis and western blotting. Detailed information can be found in Supplemental Information.
Macrophage cultures for macropinocytosis, filopodia formation, and cytokine release analyses
Bone marrow derived macrophages were cultured as previously described (Huang et al., 2012). Briefly, macrophage cells were prepared from bone marrow from 8-week-old male wild type, MsrA KO, MsrB1 KO, and MsrA/MsrB1 KO mice. On day 6, experiments were carried out on the BMDM, including studies in which cells were stimulated with LPS (0111:B4; 100 ng/ml; Sigma) for 18 h. These BMDMs were used for further macropinocytosis and filopodia studies, and the supernatants from stimulated BMDMs were used for the analysis of cytokine release. Detailed information can be found in Supplementary information.
Supplementary Material
Highlights.
Mical and MsrB regulate actin through stereospecific methionine oxidation/reduction
Met oxidation depolymerizes actin and reduction promotes actin repolymerization
Selenoprotein MsrB 1 regulates actin polymerization in response to LPS stimulation
Proteins can be regulated by reversible site-specific methionine-R-sulfoxidation
Acknowledgments
This research was supported by NIH grants AG021518, GM061603, RR017675, and AI089999.
Footnotes
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 2011;(1):39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;(7237):430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol. 2004;(8):647–657. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- Cheung HT, Rehwaldt CA, Twu JS, Liao NS, Richardson A. Aging and lymphocyte cytoskeleton: age-related decline in the state of actin polymerization in T lymphocytes from Fischer F344 rats. J Immunol. 1987;(1):32–36. [PubMed] [Google Scholar]
- Chiarugi P, Buricchi F. Protein tyrosine phosphorylation and reversible oxidation: two cross-talking posttranslation modifications. Antioxid Redox Signal. 2007;(1):1–24. doi: 10.1089/ars.2007.9.1. [DOI] [PubMed] [Google Scholar]
- Ciorba MA, Heinemann SH, Weissbach H, Brot N, Hoshi T. Modulation of potassium channel function by methionine oxidation and reduction. Proc Natl Acad Sci U S A. 1997;94:9932–9937. doi: 10.1073/pnas.94.18.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A, Miura H, Peschek J, Le Y, Bach NC, Kriehuber T, Winter J. Methionine oxidation activates a transcription factor in response to oxidative stress. Proc Natl Acad Sci U S A. 2013;110:9493–9498. doi: 10.1073/pnas.1300578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O'Donnell SE, Aykin-Burns N, et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;(3):462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova M, Kuleva N, Hoffmann R. Identification of cysteine, methionine and tryptophan residues of actin oxidized in vivo during oxidative stress. J Proteome Res. 2010;(3):1598–1609. doi: 10.1021/pr901099e. [DOI] [PubMed] [Google Scholar]
- Fomenko DE, Novoselov SV, Natarajan SK, Lee BC, Koc A, Carlson BA, Lee TH, Kim HY, Hatfield DL, Gladyshev VN. MsrB1 (methionine-R-sulfoxide reductase 1) knock-out mice: roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J Biol Chem. 2009;(9):5986–5993. doi: 10.1074/jbc.M805770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giridharan SS, Rohn JL, Naslavsky N, Caplan S. Differential regulation of actin microfilaments by human MICAL proteins. J Cell Sci. 2012;(Pt3):614–624. doi: 10.1242/jcs.089367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hild G, Bugyi B, Nyitrai M. Conformational dynamics of actin: effectors and implications for biological function. Cytoskeleton (Hoboken) 2010;67:609–629. doi: 10.1002/cm.20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Hoffmann FW, Fay JD, Hashimoto AC, Chapagain ML, Kaufusi PH, Hoffmann PR. Stimulation of unprimed macrophages with immune complexes triggers a low output of nitric oxide by calcium-dependent neuronal nitric-oxide synthase. J Biol Chem. 2012;(7):4492–4502. doi: 10.1074/jbc.M111.315598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, Yazdani U, Yoon J, Wu H, Yang T, Gupta N, Huang Z, van Berkel WJ, Terman JR. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823–827. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine-and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;(12):e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Role of structural and functional elements of mouse methionine-S-sulfoxide reductase in its subcellular distribution. Biochemistry. 2005;(22):8059–8067. doi: 10.1021/bi0501131. [DOI] [PubMed] [Google Scholar]
- Kleveta G, Borzecka K, Zdioruk M, Czerkies M, Kuberczyk H, Sybirna N, Sobota A, Kwiatkowska K. LPS induces phosphorylation of actin-regulatory proteins leading to actin reassembly and macrophage motility. J Cell Biochem. 2012;(1):80–92. doi: 10.1002/jcb.23330. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci U S A. 2002;(7):4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Lee BC, Marino SM, Zhang Y, Fomenko DE, Kaya A, Hacioglu E, Kwak GH, Koc A, Kim HY, Gladyshev VN. Functional analysis of free methionine-R-sulfoxide reductase from Saccharomyces cerevisiae. J Biol Chem. 2009;(7):4354–4364. doi: 10.1074/jbc.M805891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BC, Dikiy A, Kim HY, Gladyshev VN. Functions and evolution of selenoprotein methionine sulfoxide reductases. Biochim Biophys Acta. 2009;(11):1471–1477. doi: 10.1016/j.bbagen.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JC, Kim G, Levine RL. Stereospecific oxidation of calmodulin by methionine sulfoxide reductase A. Free Radic Biol Med. 2013;61C:257–264. doi: 10.1016/j.freeradbiomed.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci U S A. 2001;(23):12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselov SV, Kim HY, Hua D, Lee BC, Astle CM, Harrison DE, Friguet B, Moustafa ME, Carlson BA, Hatfield DL, Gladyshev VN. Regulation of selenoproteins and methionine sulfoxide reductases A and B1 by age, calorie restriction, and dietary selenium in mice. Antioxid Redox Signal. 2010;(7):829–838. doi: 10.1089/ars.2009.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao KM, Currie MS, Padmanabhan J, Cohen HJ. Age-related alterations in actin cytoskeleton and receptor expression in human leukocytes. J Gerontol. 1992;(2):B37–44. doi: 10.1093/geronj/47.2.b37. [DOI] [PubMed] [Google Scholar]
- Rayman MP. Selenium and human health. Lancet. 2012;379:1256–1268. doi: 10.1016/S0140-6736(11)61452-9. [DOI] [PubMed] [Google Scholar]
- Santarelli LC, Wassef R, Heinemann SH, Hoshi T. Three methionine residues located within the regulator of conductance for K+ (RCK) domains confer oxidative sensitivity to large-conductance Ca2+-activated K+ channels. J Physiol. 2006;571:329–348. doi: 10.1113/jphysiol.2005.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman ER, Moskovitz J, Berlett BS, Levine RL. Cyclic oxidation and reduction of protein methionine residues is an important antioxidant mechanism. Mol Cell Biochem. 2002;(1-2):3–9. [PubMed] [Google Scholar]
- Stadtman ER, Moskovitz J, Levine RL. Oxidation of methionine residues of proteins: biological consequences. Antioxid Redox Signal. 2003;5:577–582. doi: 10.1089/152308603770310239. [DOI] [PubMed] [Google Scholar]
- Ugarte N, Petropoulos I, Friguet B. Oxidized mitochondrial protein degradation and repair in aging and oxidative stress. Antioxid Redox Signal. 2010;(4):539–549. doi: 10.1089/ars.2009.2998. [DOI] [PubMed] [Google Scholar]
- Weissbach H, Etienne F, Hoshi T, Heinemann SH, Lowther WT, Matthews B, St John G, Nathan C, Brot N. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch Biochem Biophys. 2002;(2):172–178. doi: 10.1006/abbi.2001.2664. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Chen B, Smallwood HS, Urbauer RJ, Markille LM, Galeva N, Williams TD, Squier TC. High-affinity and cooperative binding of oxidized calmodulin by methionine sulfoxide reductase. Biochemistry. 2006;(49):14642–14654. doi: 10.1021/bi0612465. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Guan KL. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J Cell Biol. 2012;(2):155–164. doi: 10.1083/jcb.201202056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


