Abstract
The domestic ferret (Mustela putorius furo) is an important animal model for multiple human respiratory diseases. It is considered the ‘gold standard’ for modeling human influenza virus infection and transmission1–4. Here we describe the 2.41 Gb draft genome assembly of the domestic ferret, constituting 2.28 Gb of sequence plus gaps. We annotate 19,910 protein-coding genes on this assembly using RNA-seq data from 21 ferret tissues. We characterize the ferret host response to two influenza virus infections by RNA-seq analysis of 42 ferret samples from influenza time courses, and show distinct signatures in ferret trachea and lung tissues specific to 1918 or 2009 human pandemic influenza virus infections. Using microarray data from 16 ferret samples reflecting cystic fibrosis (CF) disease progression, we show that transcriptional changes in the CFTR-knockout ferret lung reflect pathways of early disease that cannot be readily studied in human infants with CF disease.
We performed whole-genome sequencing with DNA from an individual female sable ferret (Mustula putorius furo) and created a genome assembly using ALLPATHS-LG5. The draft assembly is 2.41 Gb including gaps, has a contig N50 size of 44.8 kb, a scaffold N50 size of 9.3 Mb, and quality metrics comparable to other genomes sequenced using Illumina technology (Table 1, Supplementary Note). RNA-seq data for annotation were obtained from poly-adenylated transcripts using RNA from 24 samples of 21 tissues from male and female ferrets, including both developmental and adult tissues (Supplementary Table 1). The genome assembly was annotated using the Ensembl gene annotation system6 (Ensembl release 70). Protein-coding gene models were annotated by combining alignments of Uniprot7 mammal and other vertebrate protein sequences and the aforementioned RNA-seq data. The ferret genome can be viewed as unanchored scaffolds along with the Ensembl genome models in both the UCSC and Ensembl genome browser interfaces. We also used the tool Lift Over to map the coordinates from the ferret assembly onto the well-finished genome sequence of its phylogenetic neighbor, the domestic dog (Canis familiaris) (V3.1); this mapping is a useful resource for a genome based on short-read sequencing and facilitates browsing the ferret genome in the surrogate context of dog chromosomes (Supplementary Fig. 1).
Table 1.
Summary details of the ferret genome assembly and associated Ensembl annotation.
| Mustela putorius furo genome | |
|---|---|
| Assembly | MusPutFur1.0 |
| Date | June 2011 |
| Total Assembly Length | 2,41 Gb |
| Total Sequence | 2.28 Gb |
| Short Read Coverage | 162 X |
| Organization | 7,783 unplaced scaffolds |
| Scaffold N50 | 9.3 Mb |
| Ensembl annotation | |
|---|---|
| Database version | 70.1 |
| Date | Aug 2012 |
| Coding genes | 19,910 |
| Noncoding genes | 3,614 |
| Pseudogenes | 287 |
| Gene transcripts | 23,963 |
Supporting evidence: 1.1×109 mRNA-seq reads (Paired end 2 × 100; 220 GBases) from 21 individual tissues including developmental stages, and respiratory tissues from 2009 SOIV infected ferrets.
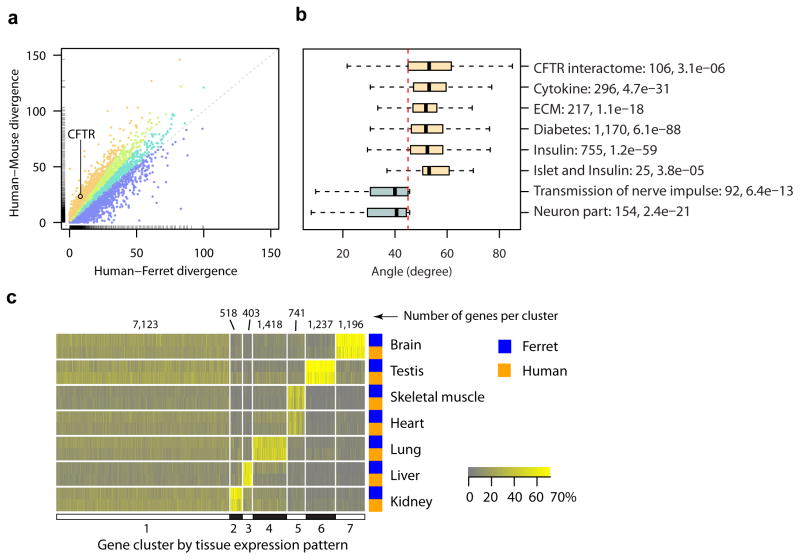
Using the annotated ferret protein sequences, we constructed a highly resolved phylogenetic tree (Supplementary Fig. 2). As expected, ferret falls within the Caniformia suborder of the Carnivores, as represented by the domestic dog, cat, giant panda and walrus, and the support values are high for most clades (Supplementary Tables 2 and 3, Methods). Although the clade containing the ferret diverged from a common ancestor before the divergence of the rodent and human/primate lineages, branch lengths in the tree indicate rapid evolution in the rodent clade, which has resulted in less genetic divergence between humans and ferrets than between humans and mice. Indeed, in comparing protein sequences between the species, we found that for 75% of all orthologous triplets, ferret proteins are closer than mouse proteins to human proteins (Figure 1a, Supplementary Table 4). For example, the ferretcystic fibrosis trans membrane conductance regulator (CFTR) protein is considerably closer to the human than is its mouse counterpart (%-identities [PAM distance] for ferret to human = 92% [8.1]; mouse to human = 79% [23.3]). Overall, basic cell physiology related Gene Ontology (GO) terms tend to be enriched among the genes residing in the angular sector representing the top 25% of genes where the ferret sequence is closer to human than the mouse ortholog. The enriched GO terms include nucleic acid metabolism, nuclear division, regulation of expression, and protein modification and localization (Supplementary Fig. 3, Supplementary Tables 5 and 6). Extending this comparison from CFTR to 106 CFTR-interacting proteins, we found that the ferret-to-human protein sequence similarity is significantly greater than the corresponding mouse ortholog (Wilcoxon test p-value = 3.1×10−6, Figure 1b). In additional comparisons, we examined gene sets pertinent to CF disease processes including inflammation, lung and pancreatic remodeling, and the regulation of insulin and diabetes, and in all cases found the encoded human proteins to be better conserved in ferret than in mouse (Figure 1b, Supplementary Fig. 4). In contrast, proteins encoded by some nervous system related genes appear to be more divergent from human in ferrets than mouse (Figure 1b). In summary, the overall high sequence similarity between ferret and human proteins shown by these genome-level analyses indicates many ferret proteins have likely evolved to conserve similar molecular functions as their human protein orthologs.
Figure 1.
Cross-species comparisons show that ferret protein sequence and tissue-specific expression are similar to that of human. a. Scatter plot of human vs. mouse protein divergence in Point Accepted Mutation (PAM) metric (y-axis) against the corresponding human vs. ferret protein divergence (x-axis). Proteins appear above the 45° diagonal (grey dashes) when the ferret sequence is closer to the human sequence than the corresponding mouse sequence. The angle of the line to each protein from the origin is directly related to the ratio of mouse divergence from human sequence and ferret divergence from the human sequence. A greater angle from the origin indicates greater divergence. The quartiles of the distribution of these ratios are displayed in different colors (blue being the least conserved in ferret relative to mouse, and orange-brown being the most conserved). Hatched lines on the axes show the metric distributions for the individual species (Supplementary Table 4). b. Box plots of the angles represented in panel a for proteins in eight selected biological functions. For gene sets related to CF (light yellow), human protein sequence is better conserved in ferret than in mouse. For two nervous system related gene sets (blue), human protein sequence tended to be more conserved in mouse. Next to each function are the number of proteins in the function and the p-value from one-sided Wilcoxon signed rank test comparing the human-ferret (x-axis in a) vs. human-mouse (y-axis in a) divergence in PAM metric. c. K-means clustering of ferret-human orthologous genes by their tissue expression patterns reveals similarities in tissue-specificity. The color scale represents relative abundance across all tissues within each species and is saturated at 70%. Vertical partitions correspond to the seven clusters of genes from the optimal clustering, numbers of genes per cluster appearing on the top. Horizontal groupings are organized by tissue with ferret and human pairings denoted by the color bar at the side, and highlight the tissue-specificity of clusters 2 through 7.
Next, we investigated whether ferret and human genes exhibit similar tissue expression. We compared the patterns of relative transcript abundance across seven tissues in common between our data set and previously reported human RNA-seq data8 (Figure 1c). First, we determined the genes with highest relative abundance across all tissues within each species and found that the intersection of these tissue-specific sets between human and ferret was highly significant (Chi-squared test p-values < 10−186, Supplementary Table 7). To refine the sets of tissue-specific genes, we clustered genes with similar expression patterns across the 14 tissue samples (7 from ferret and 7 from human) into 7 disjoint clusters (Figure 1c, Supplementary Table 8). This clustering analysis revealed that many ferret and human genes exhibited highly concordant, tissue-specific expression patterns. The assignment of a gene cluster to a specific tissue was evident by its significantly increased expression in that tissue relative to the rest of the tissues of the same species for all comparisons except between skeletal muscle and heart (Supplementary Table 9). The similarity between skeletal muscle and heart may be attributed to the presence of striated muscle cells in both tissues. The clusters include transcription factors (TFs) with known tissue specificities in human samples, such asOLIG2 and NEUROD2 (brain); OVOL1 (testis); MYF6 (heart, skeletal muscle), and lung-specific Iroquois-class homeodomain TFsIRX2, IRX3 and IRX59, and the same specificity was seen for the ferret tissues. Sequence comparisons showed that ferret TFs in brain, skeletal muscle/heart, lung and kidney gene clusters exhibited even greater similarity to human orthologs as compared to the rest of the ferret genome, suggesting strong conservation of functional regulation (Supplementary Tables 10 and 11). Some genes related to immune and inflammatory functions, including TFs associated with Th17 cells (BATF, IRF4, AHR)10, showed increased expression in ferret and human lungs, which is likely a consequence of the greater proportion of immune cells in this compartment and the possible presence of bronchus-associated lymphoid tissue. The broad similarity between ferret and human tissue-specific gene expression suggests the regulation of gene expression in tissue compartments is also highly conserved.
Ferrets are frequently used as a model for human influenza virus infection, in part due to the similar distribution of viral attachment receptors in the respiratory tract of humans and ferrets2, 11. We used our genome sequence to profile the transcriptional response of ferrets to pandemic influenza virus. To this end, we infected ferrets with either of two human pandemic influenza viruses—the H1N1 2009 pandemic virus A/CA/04/2009 (CA04) or the reconstructed H1N1 1918 pandemic virus (1918)—and collected samples from both the upper (trachea) and the lower (lung) respiratory tract at 1, 3 and 8 days post-infection (dpi) for transcriptome analysis (Supplementary Table 12, Supplementary Fig. 5 and 6). To increase the coverage of our transcriptome analysis beyond standard Ensembl annotated genes, including non-polyadenylated transcripts, we performed RNA-seq on total RNA after ribosomal RNA depletion (Methods). To augment standard Ensembl annotation, we predicted additional transcript models using the RNA-seq data collected from both the lung and trachea samples from these infected animals and the tissue samples described in the previous paragraph (Methods, Supplementary Table 13). Additional analyses indicate that the RNA-seq derived transcripts are enriched with novel protein-coding isoforms, and polyadenylated and non-polyadenylated intergenic non-coding RNAs (Supplementary Fig. 7 and 8). To make these genomic resources more accessible for gene expression profiling, we also designed and validated two versions of ferret-specific oligonucleotide microarrays: version 1 interrogates 23,582 Ensembl annotated transcripts plus13,368 intergenic transcripts derived from RNA-seq analysis of ferret mRNAs; version 2 provides broader coverage with probes for an additional 27,288 intergenic transcripts from RNA-seq analysis of ferret total RNAs (Supplementary Table 14, Supplementary Fig. 9 – 16, Supplementary Note).
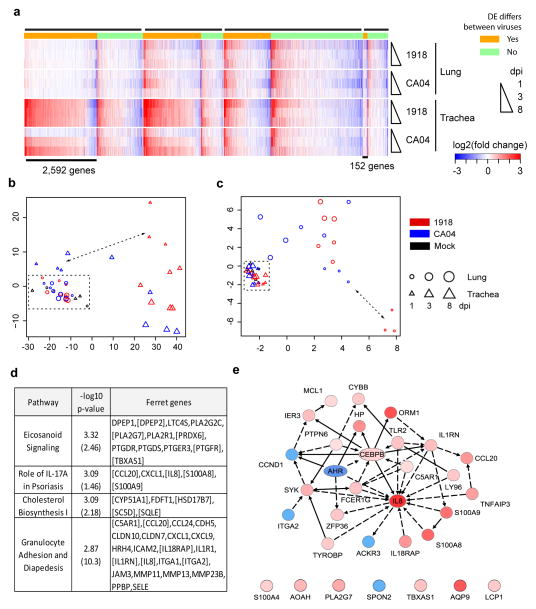
As quantified by RNA-seq, host transcriptional changes were much more extensive in infected trachea (9,869 differentially expressed (DE) genes, adjusted p-value < 0.01) than in infected lung (4,646 DE genes), and the kinetics of the response differed by virus and compartment (Supplementary Fig. 17). In the trachea, the 1918 virus induced a pronounced transcriptional response, both in the number of DE genes and in the magnitude of their changes, that commenced at 1 dpi and was largely sustained through day 8; in contrast, infection with the CA04 virus resulted in a gradual escalation of overall transcriptional changes in these same genes, resulting in peak expression by 8 dpi. Different kinetics occurred in the lung, where both viruses induced a similar number of DE genes at 1 dpi, followed by a decline to far fewer DE genes by day 8. A detailed tissue-by-virus comparison revealed distinct transcriptional signatures that differentiated the response to the 1918 and CA04 viruses in the two respiratory tissues (Figure 2a, Supplementary Fig. 18, Supplementary Table 15). Within the trachea-specific host response, a subset of 2,592 ferret genes distinguished the two viruses, with extensive perturbation at 1 dpi in response to the 1918 virus and minimal alteration in response to CA04 (Figure 2b). This gene set has an over-representation of diverse biological processes such as Apoptosis Signaling, NGF Signaling, and Ceramide Signaling (one-sided Fisher exact test p-values of 4.28×10−7, 1.61×10−6, 3.76×10−6, respectively, Supplementary Table 16). Related lipid-receptor signaling systems, such as sphingosine-1-phosphate (S1P) receptor signaling, can protect the host from influenza virus-induced “cytokine storm” by inhibiting pro-inflammatory responses12, 13. Similarly, some DE transcripts were exclusively observed within the lung compartment, with a subset of 152 ferret genes that differentiated the two virus infections (Figure 2c). Within this subset, we observed enrichment of Prothrombin Activation Pathway and differential expression of Il13 and Il20, associated with Role of Cytokines in Mediating Communication between Immune Cells (p-value of 1.53×10−2), that are produced by pulmonary innate lymphoid cells14 and maturing dendritic cells15, respectively. In summary, the ferret genomic resources described here enabled a side-by-side comparison of ferret transcriptional responses to two human pandemic influenza viruses. The results revealed that the host response to the two pandemic viruses differs in a tissue compartment–dependent manner.
Figure 2.
Transcriptomic analyses of the host response to influenza virus infection and CF disease progression in ferrets. a. Heat map visualization shows distinct gene expression changes in lung and trachea samples from ferrets infected with either the 2009 pandemic H1N1 influenza A/CA/04/2009 virus (CA04) or the 1918 pandemic H1N1 influenza A/Brevig Mission/1/1918 virus (1918). Each row shows the log2 (fold-change) for three infected animals relative to corresponding tissue from three mock-infected ferrets. The heat map is organized by the specificity of the changes with respect to tissue or virus. From left to right black bars at the top of the panel indicate four groups of genes: specific to trachea; distinct profiles in trachea and lung; similar profiles in trachea and lung; specific to lung (for additional details see Supplementary Fig. 18); within each group orange subsections differ between the virus strains, green subsections do not. b. Multidimensional scaling (MDS) representation of the distances among samples based on the indicated cluster of 2,592 genes from a that distinguish viruses in trachea but not in lung. Points show individual animals as indicated on the far right. The x- and y-axes represent a conceptual 2-dimensional space to which the MDS algorithm projected individual lung and trachea samples of high-dimensionality; i.e., the number of genes in the block associated with each sample, while preserving the distances/dissimilarities between samples as closely as possible. Double arrow illustrates that the gene signature distinguishes the two virus infections in trachea at 1 dpi, while lung samples show no separation (dotted rectangle). c. As in b, for the indicated cluster of 152 genes that is differentially regulated in lung but not trachea tissues and separates the two virus strains on 1 dpi. d and e. Differential transcriptional responses in an experiment comparing lung samples from 15-day-old CF ferrets (n=3) vs. non-CF ferrets (n=5). d. Similar pathways enriched in genes differentially expressed in 15-day-old CF ferret lung samples and CF human bronchial brushings, derived from Ingenuity Pathway Analysis. The values in parenthesis are the enrichment p-values for the corresponding pathways in the genes differentially expressed in CF human bronchial brushing19. In brackets are genes which were differentially expressed in both ferret and human CF/non-CF comparisons. e. Network illustration of 32 genes of the function ‘inflammatory response’, which were differentially expressed in the same direction in ferret and human CF datasets (for additional details see Supplementary Fig. 20). Red and blue shading reflects the extent of increased or decreased expression, respectively, in CF relative to non-CF individuals. A solid line between two genes indicates direct interaction(s) among them and a dotted line for indirect interaction(s), as documented in the literature.
Genetically engineered cystic fibrosis (CF) ferrets model two key components of disease not observed in CF mice, namely lung disease16, 17 and diabetes18. To investigate CF disease progression in the ferret model, we performed microarray expression analysis on lung specimens from newborn and 15-day-old CFTR knockout (CF) and normal (non-CF) ferrets. In newborn animals, genotypic differences in transcriptomes were limited; 472 DE protein-coding genes were identified using a relaxed threshold (absolute fold-change ≥ 1.4, p-value ≤0.1). Nonetheless, functional analyses of these DE genes showed disturbances in several canonical pathways, including Coagulation System, Primary Immunodeficiency Signaling, Serotonin Receptor Signaling, and Signaling in T Helper Cells (Supplementary Table 17). Genotype-dependent gene expression differences between 15-day-old animals were much more extensive (1,468 DE protein-coding genes, absolute fold-change ≥1.5, false discovery rate ≤0.05) and included expression changes in genes of pathways involved in Cholesterol Biosynthesis, Eicosanoid Signaling, Granulocyte Adhesion and Diapedesis, and IL8 regulation (Figure 2d and 2e, Supplementary Table 17). In a previous study, gene expression in these pathways was also significantly perturbed in human bronchial brushings from adult CF patients19 (Supplementary Table 17). Further, in CF ferrets, changes in the expression of most of these genes were highly positively correlated (ANOVA p-value 4.9e-5, Pearson correlation coefficient 0.63) with that in human CF samples, consistent with the overall positively correlated expression changes between day 15 ferret and human CF samples (Supplementary Fig. 19). The exception of some cholesterol biosynthesis pathway genes may be the result of variation in epithelia sampled in the ferret (intact lung) and human (conducting larger airways), or differences in CF disease status between infants and adults.
Similar expression changes in the CF ferret and human datasets were also evident at the level of broader biological functions such as Chronic Inflammatory Disorder, Cell Movement of Phagocytes, and Inflammatory Response (Supplementary Table 18). As anticipated, IL8 gene expression was one of the most significantly increased inflammatory genes in older CF ferret (14.6-fold up-regulated) and CF human (11.7-fold up-regulated) samples (Supplementary Table 17), consistent with a dominant role of IL8in CF lung disease20. Indeed, many genes associated with IL8 regulation, including CCL20, S100A8, S100A9, IL18RAP, IL1RN, and ITGA2, were differentially regulated concordantly in CF ferret and human lung samples (Figure 2e, Supplementary Fig. 20). Although these findings suggest commonalities in the pathways of CF inflammation between the two species, it is worth noting that the dominant bacterial pathogens of the lung are distinctly different between CF ferret and humans with this disease—Pseudomonas aeruginosa21 in humans and enteric pathogens in both young and old CF ferrets16, 17. Thus, the predominant gene pathways involved in CF inflammatory responses appear to be conserved across ferrets and humans and largely independent of the pathogen’s taxa. Of the DE genes that changed in opposite directions between ferret and human datasets, one of the most significant functional pathways included 19 genes associated with Cell Movement (Supplementary Fig. 20). This suggests that there are differences in the extent of injury, repair, and/or migratory inflammatory cell infiltrates between the ferret and human datasets. Such differences are not surprising given the larger number of DE genes associated with Granulocyte Adhesion and Diapedesis (Supplemental Table 17) and inflammation (Supplemental Table 18) in the older human CF samples. Despite these differences, the overall positively correlated expression changes, especially the high concordance in key CF-related pathways and functions between 15-day-old ferret and adult human CF samples, suggest that many disease changes associated with adult CF in humans may begin in infancy. Thus, the CF ferret represents a tractable model by which to systemically address disease progression–related changes in gene expression at anatomical sites not possible in humans.
Ferrets are extensively used to study human diseases such as influenza virus infection and cystic fibrosis, but the lack of genome sequence information has limited the ability to understand ferret transcriptional responses. Our transcriptomic analyses of the host response to human pandemic influenza virus infection and of CF disease progression in ferrets illustrate how the availability of the ferret genome sequence can enhance the sophistication of ferret respiratory-disease models. The analyses revealed high protein-sequence similarity and shared tissue-expression patterns between ferret and human, suggesting the potential utility of ferret models in a broader set of diseases. The ferret genome will also prove valuable to investigators exploring the conservation genomics of the highly imperiled North American black-footed ferret (M. nigripes), which is a congener to M. putorius furo22. The black-footed ferret underwent a population bottleneck in the 1980s leading to greatly diminished genetic diversity, and resulting congenital defects include reduced immune capacity and anomalies in male fertility23. The genomic resources presented here can aid genetic analysis of these defects and the ongoing captive breeding program necessary for the survival of the species24.
Online Methods
Animal usage was performed under protocols approved by the Institutional Animal Care and Use Committees (IACUCs) at the University of Wisconsin School of Veterinary Medicine or the University of Iowa. Appropriate biosafety containment was utilized in the course of infections with the indicated influenza strains.
Genome sequencing and assembly
Three adult sable female ferrets (Mustela putorius furo) (421 days old) obtained from Marshall Farms (via John Engelhardt, Iowa) were sacrificed and specimens sent to the Broad Institute for heterozygosity testing. The individual ID#1420 was selected for sequencing due to its low heterozygosity. The ferret DNA was sequenced to 90X total coverage by Illumina sequencing technology, and was comprised of 45X coverage using 180 bp fragment libraries, 42X coverage using 3kb sheared jumping libraries, 2X coverage using 6–14kb shread jumping libraries, and 1X coverage using ShARC jumping libraries. The reads were assembled into MusPutFur1.0 (Accession # AEYP00000000.1) using ALLPATHS-LG5. The M. putorius furo genome has previously been reported to have a karyotype of 40 chromosomes25. The draft assembly is 2.41 Gb in size and is composed of 2.28 Gb of sequence plus gaps between contigs. The ferret genome assembly has a contig N50 size of 48.8 kb, a scaffold N50 size of 9.3 Mb, and quality metrics comparable to other Illumina genomes.
RNA sequencing and assembly
A panel of 24 ferret samples from multiple tissues were RNA sequenced to aid with genome annotation. Developmental (3 staged embryos) and uninfected adult tissues (the individual used for genome sequencing) were obtained from sable ferrets (obtained by the Engelhardt laboratory, Iowa). Two pooled RNA samples were prepared from ferrets that had been infected with strains of 2009 pandemic H1N1; one pool was generated from lung and trachea specimens collected in the laboratory of T. Tumpey (CDC) from two ferrets sacrificed 3 days after infection with A/Mexico/4482/2009 (H1N1). The other RNA pool was generated from materials from laboratory of Y. Kawaoka (Wisconsin), using spleens harvested at days 3 and 6 following infection of two ferrets with A/Wisconsin/WSLH049/2009 (H1N1). All RNAs were extracted at the University of Washington and the RNA-seq libraries were then produced at the Broad Institute by the strand-specific dUTP method from Oligo dT polyA-isolated RNA26. The libraries were sequenced by Hi-Seq Illumina machines, producing 101 bp reads (3–6 Gb of sequence/tissue). All 24 RNA-seq datasets were assembled via the genome-independent RNA-seq assembler Trinity27.
Ensembl Gene Annotation
The genome assembly was annotated by the Ensembl gene annotation system6 (Ensembl release 70, August 2012). Protein-coding gene models were annotated by combining alignments of Uniprot7 mammal and other vertebrate protein sequences and RNA-seq data. RNA seq models were generated from a survey of adult ferret and embryonic ferret tissues, including tissues from an influenza-infected ferret. This pipeline produced 23,963 transcripts arising from 19,910 protein coding genes and 3,614 short non-coding genes. The ferret gene annotation is available on the Ensembl website (http://www.ensembl.org/Mustela_putorius_furo/), including orthologues, gene trees, and whole-genome alignments against human, mouse and other mammals. Also included are the tissue-specific mRNA-seq transcript models, indexed BAM files, and the complete set of splice junctions identified by our pipeline. Further information about the annotation process can be found at http://www.ensembl.org/Mustela_putorius_furo/Info/Annotation - assembly as well as PDF giving a detailed description of the ferret genebuild.
Comparative Genomics
Orthology inference: orthology among the ferret genome and 33 other genomes was inferred using the OMA pipeline28. This yielded pairwise orthologs between all species and orthologous groups. The latter were used for 3-way human/mouse/ferret comparisons and species tree inference. The number of genes conserved across mammals and carnivores was computed from hierarchical orthologous groups identified using GETHOGs29.
Species tree
The 789 orthologous groups covering at least 31 of the 34 species considered were aligned individually and then concatenated. Missing data was represented as “X” characters. Alignment was performed using MAFFT’s local-alignment based L-INS-i algorithm. Phylogenetic inference was performed using PhyML, under the JTT, WAG, and LG models, and also as a partitioned analysis in RAxML30, 31. Support values were calculated using bootstrapping (RAxML and PhyML), and approximate Bayesian support (PhyML aBayes).
Scatterplot analysis
To contrast the divergence between human-ferret genes and their human-mouse counterparts, we extracted triplets of orthologs between the three species from all OMA groups computed above. Divergence was computed using two measures: (i) point accepted mutation (PAM) unit estimated by pairwise Maximum Likelihood distance estimation using Gonnet matrices32; and (ii) nucleotide divergence calculated in PhyML from triplet alignments using the general time-reversible model.
Gene Ontology (GO) annotations and enrichment analyses
GO terms were assigned to OMA groups by propagating experimental GO annotations (GO evidence codes EXP, IDA, IPI, IMP, IGI, IEP) of any group member to the rest of the group. This procedure assigned 13,509 GO terms, most of them from the Biological Process ontology, to 9,117 OMA groups, resulting in 541,220 GO annotations.
To perform the GSEA analysis33, we created a list by ordering our data points in Figure 1B according to the angle to the x-axis and determined, for each GO term separately, whether data points are randomly distributed throughout the list or are primarily found at the top or at the bottom. To perform a two-tailed Fisher’s exact test, we partitioned the data points in the scatterplot (Figure 1B) into 4 quantiles according its angle to the x-axis, thereby accounting for the relative evolutionary distance between ferret and mouse. For each GO term, we contrasted each quartile with the other three fourths of the data. We used a two-tailed Fisher’s exact test as implemented in R. In both statistical analyses we adjusted the p-values for multiple testing using the Benjamini & Hochberg method as implemented in R. To organize the enriched GO terms, the terms that passed the enrichment criteria were processed with REVIGO to remove redundant GO terms and cluster semantically similar terms. Very generic GO terms (e.g. “macromolecular complex”, “organelle part”) were excluded, as were singleton terms that were not aggregated into clusters.
Comparison of gene sets specific to models of human health and disease
Increased conservation in gene subsets (Figure 1b) were determined by Wilcoxon Signed-Rank test in R (version 2.14.1) by the wilcox. test function. Interactions with CFTR (gene subset ‘CFTR interactome’ in Figure 1b) were obtained from a previously published CFTR IP - mass spec dataset34. Other five CF-related gene subsets in Figure 1b were obtained from www.GeneCards.org35 using relevancy score cutoffs at the point of greatest Euclidean distance. Two nervous system related GO terms in Figure 1b were from the enrichment analysis of proteins with mouse sequences closer to human when tested with Fisher’s exact test and Gene Set Enrichment Analysis (false-discovery rate < 0.05) (Supplementary Tables 5 and 6).
Expanded custom ferret genome annotation for differential expression analysis
We assembled ferret transcript contigs de novo using Trinity27 from ferret RNA-seq data with default parameters. The RNA-seq data used included mRNA-seq data of the panel of 24 tissue samples for 21 different tissues or tissue mixes (each sample was assembled separately), as well as both mRNA-seq and Total RNA-seq data from the influenza study (all 21 virus infected or control lung samples (each condition and protocol was assembled separately). All assembled transcript contigs were aligned to the ferret reference genome (MusPutFur1.0) using GMAP36 with default parameters. For those uniquely aligned ferret transcript contigs, their alignments across all mRNA-seq data (transcript contigs from lung Total RNA-seq data were not included here) were merged using cuffmerge (Cufflinks version 2.0.2)37 to remove redundant alignments and to predict novel genes and transcripts. Predicted transcripts were checked against Ensembl annotation (version 69) to identify: 1) putative intergenic transcripts - those did not overlap with any Ensembl annotated transcripts directly (with class code ‘u’) and indirectly through any predicted transcripts overlapping Ensembl annotated transcripts, and 2) putative novel isoforms of Ensembl annotated transcripts (with class code ‘j’, at least one splice junction is shared with a reference transcript). We removed all single exon transcripts or any transcript with the alignment to reference genome shorter than 200nt to minimize unspliced precursor fragments. For putative intergenic transcripts, we also removed a small number of predicted transcripts located within introns of other predicted transcripts. For putative novel isoforms, we removed those predicted transcripts that spanned two or more Ensembl annotated genes to minimize mis-assembled transcripts. Similarly, we predicted intergenic transcripts from lung Total RNA-seq data, which were then filtered against both Ensembl annotation and the newly predicted transcripts from mRNA-seq data. We did not require intergenic transcripts from Total RNA-seq have to be spliced, but the length of alignment to reference genome had to be at least 120nt which was intended to capture ncRNAs which would be longer than small RNAs like miRNAs. For Total RNA-seq data we did not predict novel isoforms to avoid unspliced transcripts. After filtering we combined all predicted transcripts with Ensembl annotation into one annotation, which was used for all downstream gene quantification and differential expression analysis. This expanded annotation was used for Agilent ferret microarray design.
To investigate if Total RNA-seq captured non-polyadenylated transcripts, we performed both Total RNA-seq and mRNA-seq analysis of 21 ferret lung samples. We reasoned that for the same gene in the same sample if Total RNA-seq analysis collected much more short reads than mRNA-seq analysis that gene likely transcribed non-polyadenylated transcripts, since by polyT priming mRNA-seq analysis selected against non-polyadenylated transcripts. To facilitate the comparison, the raw gene read counts were first preprocessed as follows: i) any gene with less than 50 raw read counts in all 42 RNA-seq measurements were removed to ensure genes to be compared were robustly detected at least once in the samples used here, and ii) all gene raw read counts were scaled by the total read counts of remaining genes in each RNA-seq analysis for each sample. Next, for each gene we counted the number of samples (out of 21 samples in total) in which the scaled read count from Total RNA-seq analysis was much larger (1.5 fold or more) than that from the corresponding mRNA-seq analysis.
Comparative analysis of tissue expression patterns
The panel of ferret tissue RNA-seq data generated for genome annotation was used to quantify ferret gene expressions in each tissue and was processed the same way as described in the influenza study section below; depending on the tissue type, the data was from a single individual or from 2 – 4 individuals. For human dataset, alignment files of RNA-Seq read alignment of 24 human tissues and cell types were downloaded from Human lincRNA Catalog website: http://www.broadinstitute.org/genome_bio/human_lincrnas/?q=home).
This data derived from specimens collected from 9 individuals, with 1 – 2 contributing individuals per tissue. Similarly we quantified the expression of human genes (Ensembl 69) in each tissue using HT-seq(http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html). We selected the set of 7 tissues (brain, testis, skeletal muscle, heart, lung, liver and kidney) that were common between two datasets for further comparative analysis. We limited the comparison to 15,597 ferret-human gene pairs which had 1:1 ortholog relationships as defined by Ensembl 69. The normalized read count in counts per million (cpm) for each tissue was obtained using edgeR38; in instances when there was more than one tissue donor, the resulting data is the average. To focus on genes that tended to be robustly detected in both ferret and human datasets, we applied two ad hoc filters on genes based on the observed abundances. First, before read count normalization we thresholded for genes with a raw read count of at least 100 in at least one of the ferret tissues and at least one of the human tissues (not necessarily the same tissue between ferret and human). Second, to account for the differences in sequencing depth, in the normalized read count we further required each gene to have at least 5 cpm in at least one of the ferret tissues and at least one of the human tissues (again, not necessarily the same tissue between ferret and human). The final working set was 12,636 genes. For each gene within each species, we calculated the relative abundance in each tissue as the ratio between its cpm in the tissue and the sum of cpms across all tissues. To evaluate if orthologous genes tend to exhibit concordant tissue-specific expression across ferret and human, we determined the number of genes with highest relative abundance across all tissues of the same species, as well as the intersection of these tissue-specific sets between human and ferret, and assessed the significance of the intersections using a chi-squared test.
To identify groups of genes with similar expression patterns across tissues and species we clustered genes using K-Means partitioning. We iteratively assessed the number of centers (k) to be used for clustering as following: for a given k, we calculated the difference between the converged, total within-cluster sum of squares vs. that from 500 random datasets generated by random permutation of the actual data matrix; for the series of k tested, k=7 had the maximum difference for the final gene clustering. For the results of the k=7 clustering, we used Mann-Whitney test to evaluate if the overall expression of a cluster of genes was significantly higher in one tissue relative to the rest tissues of the same species, based on the relative abundances in the normalized count matrix.
Cells and influenza viruses
The 2009 pandemic influenza A/California/04/2009 (H1N1) virus, referred to as ‘CA04’, and the 1918 pandemic influenza A/Brevig Mission/1/1918 (H1N1) virus, referred to as ‘1918’, were generated by reverse genetics, as previously described39–41. Madin-Darby canine kidney (MDCK) cells for virus titer measurements were from ATCC, and were grown in Eagle’s minimum essential medium (MEM) with 5% newborn bovine calf serum (HyClone, Thermo Fisher Scientific) and penicillin/streptomycin. Cell stocks are periodically restarted from early passage aliquots and routinely monitored for mycoplasma contamination.
Ferret infections
Twenty-one four- to eight-month-old female ferrets were obtained from Triple F Farms Inc. (Sayre, PA, USA), confirmed serologically negative by hemagglutination inhibition assay for currently circulating influenza viruses, were randomly assigned to experimental groups. Individual animals were intramuscularly anesthetized with ketamine and xylazine (5 mg and 0.5 mg per kg of body weight, respectively), followed by intranasal inoculation with 500 μl of phosphate-buffered saline (PBS; n = 3) alone, PBS containing 1 × 106 plaque forming units (PFU) of the CA04 virus (n = 9), or PBS containing 1 × 106 PFU of the 1918 virus (n = 9). On day 1 post-infection (p.i.), 3 animals from each infection group were euthanized, and tracheal and lung tissues were harvested for virological, immunohistochemical staining for influenza A virus antigen and RNA sequencing analysis. Tissues were similarly harvested from 3 additional CA04- or 1918-infected ferrets on days 3 and 8 p.i. Tracheal tissues harvested for each individual analysis were derived from the same general region in each ferret, and lung tissues for all analyses were derived from the same lung lobe. We previously examined pathologic lesions in 1918 virus-infected ferret lung, observing macroscopic pathologic changes by day 3 p.i. that included severe lesions and hemorrhage40. Since the primary purpose of the present study was to measure gene expression changes in regions of the lung where macroscopic lesions are known to develop, we carefully selected the lung lobe and lung region based on our previous study and then collected samples from the same region for all animals in the study to be consistent. Sample sizes of 3 animals per condition were in keeping with prior reports for exploratory animal models to characterize influenza infection when models require serial sacrifice. While the sample sizes were not the result of a power analysis for a pre-specified effect size, the evaluation of the gene expression differences between conditions is performed with statistical stringencies suitable for exploratory assessment, hypothesis generation, and reproducibility by alternate techniques such as qPCR. All procedures with ferrets were approved by the University of Wisconsin School of Veterinary Medicine Animal Care and Use Committee, and were performed in an enhanced biosafety level 3 Agriculture (BSL3-Ag) containment suite. All samples derived from influenza virus-infected ferret tissues and containing infectious virus were manipulated in BSL3-Ag containment. Ensuing analysis of samples from the influenza model was performed without blinding, with the exception of histopathology scoring.
Virus quantification and immunohistochemical staining
Ferret tracheal and lung tissues, frozen at −80°C at the time of excision, were thawed and homogenized in PBS containing penicillin/streptomycin. Cleared supernatants were titrated on MDCK cells using standard methods. For virus antigen immunohistochemical (IHC) analysis, tissues were preserved by immersion in 10% phosphate-buffered formalin (Sigma-Aldrich). Preserved tissues were paraffin embedded and several 5-μm-thick sections were cut for ferret tracheal and lung tissues. Sections were stained with standard hematoxylin and eosin and then processed for IHC staining with an in-house rabbit anti-influenza virus polyclonal antibody (R309) raised against influenza A/WSN/1933 (H1N1) virus40.
Quantitative reverse transcription (RT-PCR)
Quantitative RT-PCR was performed to assess viral mRNA transcripts from infected ferret tracheal and lung samples used for sequencing. Total RNAs were treated with DNase using DNA-free DNase Treatment and Removal Reagents (Ambion, Inc, Austin, TX). cDNAs from total RNAs were generated using the QuantiTect reverse transcription kit (Qiagen Inc.). A custom-designed TaqMan gene expression assay for influenza Matrix (M) sequence (MPCONS2010), with primers that have complete homology with both 1918 and CA04 M sequences, was ordered from Applied Biosystems, Inc. Taqman experiments were performed on the ABI 7500 Real-Time PCR System platform and each sample was run in quadruplicate. Ribosomal RNA (18S) was used as endogenous control to normalize quantification of each target within tissues using Applied Biosystems Sequence Detections Software version 1.3. The relative amount of viral mRNA (log 10) is presented in the final results.
RNA extraction and library preparation
Tissues used for RNA sequencing were excised and immediately immersed in RNA Later (Ambion) for 24 h at 4°C, subsequently frozen at −80°C, and later thawed and homogenized in TRIzol (Life Technologies); RNA was isolated using QIAGEN miRN easy protocols.
Total RNA from each sample was divided into two pools for whole transcriptome and mRNA library construction. RNA for whole transcriptome analysis was depleted of rRNA using the Epicentre RiboZero Gold protocol (Epicentre), designed for human, mouse and rat samples but was effective in reducing rRNA amounts for ferret total RNA samples. The presence of 18S and 28S rRNA peaks was checked using the Agilent 2100 Bioanalyzer instrument (Agilent). The rRNA depleted RNA was then used to make strand specific whole transcriptome libraries26. Strand-specific mRNA libraries were constructed using the Illumina TruSeq RNA Preparation Kit (Illumina) according to the manufacturer’s guide. Both libraries were quality controlled and quantitated using the Agilent 2100 Bioanalyzer instrument and qPCR (Kapa Biosystems).
Transcriptome sequencing, read mapping and differential expression analysis
Constructed libraries were sequenced using Illumina platform with stranded paired end reads, 2×100nt for all mRNA-seq data, 2×100nt for lung total RNA-seq data and 2×50nt for trachea total RNA-seq data. Lung datasets were assembled via the genome-independent RNA-seq assembler Trinity, with each set of three biological replicates assembled into a single transcriptome assembly. However the quantitative analysis used the ferret genome as a reference, mapping short reads to the ferret genome using RNA-seq aligner STAR42 with default parameters. The index used for STAR included splicing junctions from the expanded custom annotation constructed below, genome sequences of influenza viruses used in this study, and human and mouse ribosomal RNA sequences. Gene level quantification was based on the Ensembl gene annotations combined with the expanded list of transcribed genomic regions that were identified using the 63 RNA-seq data sets generated from the influenza model (cf. Supplemental Materials. Quantification was performed using HT-seq. The differential expression analysis was performed using edgeR38. Clustering and other statistical analyses were performed using R (http://www.r-project.org/).
Influenza model - transcriptomic analysis details
The expression data from both tissues were combined and processed together, using the generalized linear model approach provided by edgeR. Stages in the analysis are outlined in Supplementary Figure 18 in Supplementary Materials that genes were differentially regulated vs. mock in any of the infection conditions were partitioned into disjointed clusters reflecting their tissue or virus specificity. Both Ensembl annotated genes and the expanded list of transcribed genomic regions were quantified using mapped RNA-seq reads. Differential analysis using count data was called significant for adjusted p-values ≤ 0.01.
Functional analysis of differential gene expression data
Functional analysis was performed using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Inc). The software tool analyzes the experimental dataset in the context of known biological functions and pathways within the Ingenuity Pathways Knowledge Base, a curated repository of biological interactions and functional annotations. Analysis of the datasets used human annotations, based on the Ensembl listing of human-ferret orthologs. The p-values associated with functions or pathways were calculated using the right-tailed Fisher’s exact test.
Ferret microarray design and performance assessment
We designed two versions of oligonucleotide microarray using Agilent eArray Web portal (https://earray.chem.agilent.com/earray) to profile both Ensembl annotated transcripts and intergenic transcripts derived from ferret RNA-seq data as described above. In both cases, the longest isoform of each locus was selected for probe design with the ‘Design with 3’ Bias’ checked, and probe length was set to 60nt. For first version of microarray (design ID: 048471) 36,950 probes were selected to target Ensembl annotated genes and intergenic transcripts uncovered from mRNA-seq data, and is intended to work with conventional experimental protocols using polyA priming for cDNA synthesis. The second version (design ID: 048472) has 64,238 probes selected to target Ensembl annotated genes as well as intergenic transcripts uncovered from both mRNA-seq and Total RNA-seq data. It is intended to work with experimental protocols using random priming for cDNA synthesis to capture both polyA and non-polyA transcripts. The performance of designed microarray was evaluated by comparing the microarray measurements vs. RNA-seq measurements on the same influenza infected ferret samples.
Microarray measurements and data analysis
Cy3-labeled cRNA probes were prepared using standard approaches as provided by Agilent Technologies. For the ferret array design ID 048471, Agilent kit 5190–2305 yields probes derived from poly-adenylated RNAs in the starting sample. For design ID 048472, labeled probes were generated with the whole transcriptome labeling kit (part number 5190–2943). Hybridizations were performed as per manufacturer instructions and the slides read on an Agilent Technologies model G2565C high-resolution scanner with extended dynamic range. Image files were processed with Agilent Feature Extraction Software, yielding background-corrected fluorescence intensities with flags for those features deemed not significantly different from background. Statistical analyses to determine differentially regulated genes were performed with the Bioconductor package limma, as described above.
Transcriptional profiling of CF and non-CF ferrets
Homozygous CFTR knockout ferrets were generated and reared as described previously16. Non-CF ferrets were either homozygous or heterozygous for functional CFTR genes. Lung samples were collected after sacrifice at birth or at 15 days postpartum and flash frozen. For RNA isolation, lung specimens were ground in liquid nitrogen and then immediately suspended in TRIzol (Life Technologies) and RNA isolated with QIAGEN RN easy protocols. Animals were assigned to groups solely on the basis of age and genotype (i.e., CF or non-CF). At age 15 days, the comparisons used three CF ferrets (1 F, 2M) and 5 non-CF animals (3 F, 2 M); comparisons of newborn animals used four animals of each phenotype (sexes unknown) [NOTE: sexes are difficult to determine in newborn ferrets, and prior work has not shown any phenotypic differences between the sexes in CF ferrets]. Experiments were performed under protocols approved by the Institutional Animal Care and Use Committee at the University of Iowa; statistical considerations for sample sizes were described earlier in the context of the influenza infection model. Array measurements for poly-adenlyated transcripts (design 048471) were performed as described above and statistical comparisons of CF vs. non-CF animals were done as t-tests as implemented in limma; procedures were performed without blinding. Differences between CF vs. non-CF newborn animal were quite limited and were determined without the use of a multiple test correction. The threshold for differential expression was absolute fold-change ≥1.4 and un-adjusted p-value ≤0.1. Expression differences for CF vs. non-CF 15 day old animals did use a multiple test correction (Benjamini-Hochberg False Discovery Rate). The threshold for differential expression was absolute fold-change ≥1.5 and false discovery rate ≤0.05. These analyses were limited to those array probes that interrogated protein-coding genes within the Ensembl annotation for the ferret genome, and functional interpretation utilized the corresponding human gene symbols based on the Ensembl mapping of ferret-to-human orthologues. See each supplemental tables and figures for specifics of filtering criteria for the results of the statistical tests.
Comparative analysis of transcriptional changes in human cystic fibrosis bronchial epithelium
We downloaded the gene expression data on human cystic fibrosis (CF) CF and non-CF bronchial epithelium samples from Array Express (E-MTAB-360)19. The Illumina HumanRef-8 Expression Bead Chips summary expression data was statistically analyzed using limma with default settings. We filtered out two CF samples (“127 CF BBr”, “129 CF BBr”) and two non-CF samples (“112 control BBr”, “113 control BBr”) as potential outliers, due to their relative large deviations from other replicates upon inspecting Multidimensional scaling (MDS) plots and the overall expression changes. The final dataset included 17 non-CF samples and 10 CF samples. The statistical analysis of differential expression was done at the probe level, and we applied the same multiple test correction (Benjamini-Hochberg False Discovery Rate) as we did for day 15 ferret CF vs. non-CF comparison. The functional enrichment analysis of differentially expressed genes was performed using Ingenuity Pathway Analysis (IPA), similarly for differentially expressed ferret genes.
To highlight genes concordantly or discordantly differentially expressed in lung samples from day 15 CF ferrets and human CF bronchial epithelium samples, we first identified biological functions enriched in CF/non-CF differentially expressed genes, separately for day 15 ferret CF/non-CF comparison and human CF/non-CF comparison using IPA analysis. From one given function (or a subset of related functions) enriched in both comparisons, we gathered genes differentially expressed in either comparison to identify concordantly or discordantly differentially expressed genes between two comparisons. We applied the following steps to identify genes with concordant expression changes between two comparisons. First, the gene was significantly (fold change ≥1.5 and adjusted p-value ≤0.05) differentially expressed in both comparisons. Second, the gene had the same direction of expression changes in two comparisons. Third, we ranked these genes selected from steps 2 and 3 by their absolute log2 fold changes within each comparison, i.e. the gene with the largest fold change had a rank of 1 and the gene with the smallest fold change had a rank equal to the total number of these selected genes. This way each gene was assigned with two corresponding ranks, one from the day 15 ferret CF/non-CF comparison and one from the human CF/non-CF comparison. Fourth, we ranked these selected genes by the sum of 1) the difference between their two ranks and 2) the maximum of two ranks. The top genes from this process tended to have both large expression changes in both comparisons and their expression changes tended to be close in magnitude. Similarly we applied the following steps to identify genes with discordant expression changes between two comparisons. First, the gene was significantly (fold change ≥1.5 and adjusted p-value ≤0.05) differentially expressed in both comparisons. Second, the gene had the different direction of expression changes in two comparisons. Third, we ranked these genes selected from steps 2 and 3 by the absolute value of the difference between their two log2 fold changes, one from the day 15 ferret CF/non-CF comparison and one from the human CF/non-CF comparison. The top gene had the largest difference in fold changes between two comparisons, and with opposite expression changes.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Department of Health and Human Services, under Contract Nos. HHSN272200800060C and HHSN272201400005C and Public Health Service Grant P51OD010425 (M.G.K). For the Broad Institute of MIT and Harvard, this project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200900018C.D.J.G. also received training grants 9T32OD010423-06 and 5T32RR023916-05 from the NIH Office of the Director. J.F.E, S.R.T., X.S., Y.Y. (University of Iowa) were supported under the NIH National Institute of Diabetes and Digestive and Kidney Diseases grants R37 DK047967, R24 DK096518, P30 DK054759, and National Heart, Lung and Blood Institute grant R01 HL108902. C.D. acknowledges support by SNSF advanced researcher fellowship (#136461). A.S. receives support under NIH National Institute of General Medical Sciences R01 GM102192. For S.S., B.A. and T.H., the Wellcome Trust Sanger Institute is operated by Genome Research Limited, a charity registered in England with number 1021457 and a company registered in England with number 2742969. K.L-T. also receives support under EURYI ERC.
Footnotes
Author Contributions X.P., R.E.P., J.F.E., J.T.-G., A.J.E. and S.M.W. wrote the paper with input from other authors. F.D.P., J.A., B.W.B., K.L.-T. oversaw genome and transcriptome sequencing, and related computational efforts at the Broad Institute. D.B., P.R. and J.T-M. performed transcriptome analysis, initial genome annotation and initial expression analysis. J.J. assisted in coordinating samples and sequencing data at the Broad. M.L. and R.S. performed library construction for sequencing. A.M.B. and S.Y. generated the genome assembly. T.T. and Y.K. provided influenza infected ferret tissues listed in Table S1. T.H., B.A. and S.S. were responsible for Ensembl annotation pipeline. Y.K., A.J.E. oversaw the influenza model at the University of Wisconsin. M.H. implemented the ferret infection protocol and generated primary virological data. D.J.G and M.S. did immunohistochemical staining for influenza A virus antigen. Y.K., A.J.E and D.J.G interpreted overall biological outcomes of the influenza model. J.F.E., X.S. and Y.Y. provided normal ferret tissues for genome and transcriptome sequencing, and RNA samples from the CF ferret model. S.M.K. isolated and characterized RNA from normal ferret tissues. J.C. and M.T. isolated RNA from samples from the ferret influenza infection model and generated ribosomally depleted RNA. M.T. generated total RNA-seq libraries for RNA from ferret trachea and coordinated the generation of trachea total RNA-seq data. L.L. oversaw sample handling and coordinated data generation for the ferret influenza model. K.G. and C.D. performed the phylogenetic and ferret-to-human and mouse-to-human comparisons. N.S. generated the GO term enrichments for the differing angular quadrants of Fig. 1a. S.R.T. evaluated relative conservation of human genes between ferret or mouse for gene sets associated with biomedical models. A.S. advised on comparative genomics analyses. X.P. did the final analysis of all the RNA-seq data, including generating the expanded annotation, evaluating the tissue specificity, and performing the differential expression analysis for the ferret influenza model. J.T.-G. and R.E.P. provided the associated functional interpretation. X.P. also generated the designs for the ferret microarrays. L.L. coordinated sample handling and data generation with the ferret microarrays; J.C. generated the microarray data; X.P. performed the statistical comparisons; R.E.P., X.P., J.F.E., and S.R.T performed the functional interpretation of transcriptional changes between CF ferret and human samples. R.E.P. and M.G.K. coordinated contributions between the collaborating laboratories.
Competing Financial Interests The authors declare no competing financial interests.
Supplementary Information is available in the online version of the paper. Genome assemblies have been deposited in GenBank/EMBL (http://www.ncbi.nlm.nih.gov/genbank). The M. putorius furo genome has been deposited under the accession AEYP00000000, which provides access to assembled contigs and the derived unplaced genomic scaffolds. The assembly can also be found at http://uswest.ensembl.org/Mustela_putorius_furo/Info/Index. Genomic and RNA-seq datasets have been deposited in the NCBI Short Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra), connected to the following BioProjects: Genomic reads – PRJNA59869; mRNA-seq reads for tissue survey and lung samples from the influenza model – PRJNA78317. Trachea RNA-seq data is deposited under SRA accession SRX389385. Microarray datasets are deposited under Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) accession numbers GSE49060 (influenza arrays) and GSE49061 (CF arrays). Agilent array using the developed designs (IDs 048471 and 048472) can be ordered via the Agilent eArray utility (https://earray.chem.agilent.com). Additional community resources related to the paper, including the genomic coordinates for the intergenic and non-polyadenylated transcripts, and results for the ferret to dog Lift Over can be found at: http://ucsc.viromics.washington.edu/genomes/ferretGenome
References
- 1.Tripp RA, Tompkins SM. Animal models for evaluation of influenza vaccines. Curr Top Microbiol Immunol. 2009;333:397–412. doi: 10.1007/978-3-540-92165-3_19. [DOI] [PubMed] [Google Scholar]
- 2.van Riel D, et al. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belser JA, Katz JM, Tumpey TM. The ferret as a model organism to study influenza A virus infection. Dis Model Mech. 2011;4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrauwen EJ, et al. Possible increased pathogenicity of pandemic (H1N1) 2009 influenza virus upon reassortment. Emerg Infect Dis. 2011;17:200–208. doi: 10.3201/eid1702.101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curwen V, et al. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium U. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Riel D, et al. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006;312:399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 12.Walsh KB, et al. Suppression of cytokine storm with a sphingosine analog provides protection against pathogenic influenza virus. Proc Natl Acad Sci U S A. 2011;108:12018–12023. doi: 10.1073/pnas.1107024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsolais D, et al. A critical role for the sphingosine analog AAL-R in dampening the cytokine response during influenza virus infection. Proc Natl Acad Sci U S A. 2009;106:1560–1565. doi: 10.1073/pnas.0812689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neill DR, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolk K, et al. Maturing dendritic cells are an important source of IL-29 and IL-20 that may cooperatively increase the innate immunity of keratinocytes. J Leukoc Biol. 2008;83:1181–1193. doi: 10.1189/jlb.0807525. [DOI] [PubMed] [Google Scholar]
- 16.Sun X, et al. Disease phenotype of a ferret CFTR-knockout model of cystic fibrosis. J Clin Invest. 2010;120:3149–3160. doi: 10.1172/JCI43052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X, et al. Lung phenotype of juvenile and adult cystic fibrosis transmembrane conductance regulator-knockout ferrets. Am J Respir Cell Mol Biol. 2014;50:502–512. doi: 10.1165/rcmb.2013-0261OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olivier AK, et al. Abnormal endocrine pancreas function at birth in cystic fibrosis ferrets. J Clin Invest. 2012;122:3755–3768. doi: 10.1172/JCI60610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogilvie V, et al. Differential global gene expression in cystic fibrosis nasal and bronchial epithelium. Genomics. 2011;98:327–336. doi: 10.1016/j.ygeno.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Bonfield TL, et al. Inflammatory cytokines in cystic fibrosis lungs. Am J Respir Crit Care Med. 1995;152:2111–2118. doi: 10.1164/ajrccm.152.6.8520783. [DOI] [PubMed] [Google Scholar]
- 21.Ciofu O, Hansen CR, Hoiby N. Respiratory bacterial infections in cystic fibrosis. Curr Opin Pulm Med. 2013;19:251–258. doi: 10.1097/MCP.0b013e32835f1afc. [DOI] [PubMed] [Google Scholar]
- 22.Ouborg NJ, Pertoldi C, Loeschcke V, Bijlsma RK, Hedrick PW. Conservation genetics in transition to conservation genomics. Trends Genet. 2010;26:177–187. doi: 10.1016/j.tig.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Wisely SM, Buskirk SW, Fleming MA, McDonald DB, Ostrander EA. Genetic diversity and fitness in black-footed ferrets before and during a bottleneck. The Journal of heredity. 2002;93:231–237. doi: 10.1093/jhered/93.4.231. [DOI] [PubMed] [Google Scholar]
- 24.Wisely SM, McDonald DB, Buskirk SW. Evaluation of the genetic management of the endangered black-footed ferret (Mustela nigripes) Zoo Biol. 2003;22:287–298. [Google Scholar]
- 25.Cavagna P, Menotti A, Stanyon R. Genomic homology of the domestic ferret with cats and humans. Mamm Genome. 2000;11:866–870. doi: 10.1007/s003350010172. [DOI] [PubMed] [Google Scholar]
- 26.Levin JZ, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 2011;39:D289–294. doi: 10.1093/nar/gkq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altenhoff AM, Gil M, Gonnet GH, Dessimoz C. Inferring hierarchical orthologous groups from orthologous gene pairs. PLoS One. 2013;8:e53786. doi: 10.1371/journal.pone.0053786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 31.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 32.Gonnet GH, Cohen MA, Benner SA. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 35.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13:163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- 36.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 37.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozawa M, et al. Impact of amino acid mutations in PB2, PB1-F2, and NS1 on the replication and pathogenicity of pandemic (H1N1) 2009 influenza viruses. J Virol. 2011;85:4596–4601. doi: 10.1128/JVI.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe T, et al. Viral RNA polymerase complex promotes optimal growth of 1918 virus in the lower respiratory tract of ferrets. Proc Natl Acad Sci U S A. 2009;106:588–592. doi: 10.1073/pnas.0806959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann G, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.