Abstract
Promoter CpG island hypermethylation is an important mechanism for inactivating key cellular enzymes that mediate epigenetic processes in hepatitis-related hepatocellular carcinoma (HCC). The ubiquitin-fold modifiers 1 (Ufm1) conjugation pathway (Ufmylation) plays an essential role in protein degradation, protein quality control and signal transduction. Previous studies showed that the Ufmylation pathway was down regulated in alcoholic hepatitis (AH), non alcoholic steatohepatitis (NASH) and in mice fed DDC, resulting in the formation of Mallory-Denk bodies (MDBs). In this study, we further discovered that betaine, a methyl donor, fed together with DDC significantly prevents the increased expression of Ufmylation in drug-primed mice fed DDC. Betaine significantly prevented transcript silencing of Ufm1, Uba5 and UfSP1 where MDBs developed and also prevented the increased expression of FAT10 and LMP7 caused by DDC re-fed mice. Similar down regulation of Ufmylation was observed in multiple AH and NASH biopsies which had formed MDBs. The DNA methylation levels of Ufm1, Ufc1 and UfSP1 in the promoter CpG region were significantly increased both in AH and NASH patients compared to normal subjects. DNA (cytosine-5-)-methyltransferase 1 (DNMT1) and DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B) mRNA levels were markedly up regulated in AH and NASH patients, implying that the maintenance of Ufmylation methylation might be mediated by DNMT1 and DNMT3B together. These data show that MDB formation results from Ufmylation expression epigenetically in AH and NASH patients. Promoter CpG methylation may be a major mechanism silencing Ufmylation expression.
Keywords: Promoter CpG methylation, Ufmylation, Mallory-Denk bodies (MDBs), DNMT1
Introduction
The ubiquitin-fold modifier 1 (Ufm1) is a crucial post-translational modifier that belongs to the ubiquitin-like protein (UBL) family. The Ufm1 cascade (Ufmylation) has been implicated in endoplasmic reticulum (ER) homeostasis (Lemaire et al., 2011; Zhang et al., 2012) and cell cycle control (Kwon et al., 2010; Shiwaku et al., 2010). The Ufm1 cascade is also involved in various diseases including cancer (Kwon et al., 2010; Lemaire et al., 2011; Kim et al., 2013). Recently, we reported that the Ufmylation pathway is down regulated both in the livers of DDC re-fed mice and in the livers of human alcoholic hepatitis (AH), non alcoholic steatohepatitis (NASH) with Mallory-Denk Bodies (MDBs) present, and cirrhosis, where protein quality control was down regulated (Liu et al., 2014). MDBs are found in various hepatic diseases such as hepatitis B and C, chronic cholestasis, nonalcoholic fatty liver disease (NAFLD) and hepatocellular carcinoma (HCC) (Basaranoglu et al., 2011). This pathological phenomenon, the Mallory body consists of abnormally ubiquitylated, misfolded proteins, and cross-linked keratins or non-keratin components in ballooned hepatocytes (Caldwell et al., 2010; Basaranoglu et al., 2011). In the DDC fed mouse model where liver cells proliferate, MDBs form and later, after DDC withdrawal (DDC primed hepatocytes), HCC develops (Oliva et al., 2008). The MDBs mostly disappear after 1 month of DDC withdrawal (Li et al., 2008). SAMe, a methyl donor, prevents MDB formation by preventing the switch of SAMe metabolism from the methylation pathway to the decarboxylating methylthioadenosine pathway, thus preventing the demethylation of histones (Bardag-Gorce et al., 2008). SAMe prevents the induction of the immunoproteasome and preserves the 26S proteasome in the DDC induced MDB mouse model, leading to the accumulation of undigested proteins and MDB formation (Bardag-Gorce et al., 2010). Betaine, another methyl donor, also prevents MDB formation and FAT10 positive hepatocyte proliferation in drug-primed mice by epigenetic mechanism (Oliva et al., 2009).
Epigenetic mechanisms are important for human carcinogenesis. Epigenetic abnormalities are involved in the early stages of tumorigenesis and may trigger genetic events leading to tumor development (Dumitrescu, 2009). DNA methylation is the most commonly studied epigenetic mechanism and is crucial in the development of nearly all types of cancer (Jaenisch & Bird, 2003). CpG island hypermethylation, histone modification, and transmitted chromatin structure are the underlying mechanisms for epigenetic transmission. CpG island hypermethylation is a key component for altered gene expression associated with human cancers (Kang, 2012). Promoter CpG island hypermethylation is found in virtually all human cancer tissue types and acts as an important mechanism for the inactivation of tumor suppressor genes and tumor-related genes (Esteller et al., 2001). DNA methylation does not change genetic information, but alters the readability of the DNA and results in the inactivation of genes by subsequent repression of transcription (Dong & Wang, 2014). Tumors often possess decreased genomic DNA methylation levels and hypermethylated CpG islands (Liu et al., 2012).
Methylation of DNA at the 5-position of cytosine, catalyzed by DNA methyltransferases (DNMTs), is the predominant epigenetic modification in mammals. In mammals, five members of the DNMT family have been reported, DNMT1, DNMT2, DNMT3A, DNMT3B and DNMT3L (Ren et al., 2011). Among these proteins, only DNMT1, DNMT3A and DNMT3B exhibit methyltransferase activity (Kim et al., 2009). DNMT1 is the most abundant DNMT involved in the maintenance of methylation (Miremadi et al., 2007). DNMT3 functions as a de novo methyltransferase and consists of two related proteins encoded by distinct genes, DNMT3A and DNMT3B (Ferguson-Smith & Greally, 2007). In human hepatocarcinogenesis, DNMT1, DNMT3A and DNMT3B show a progressively increasing expression from normal liver, to chronic hepatitis/cirrhosis, to HCC (Oh et al., 2007). In the early and late stages of HCC development, global DNA hypomethylation and aberrant expression of DNMT1 and DNMT3B were identified in a glycine N-methyltransferase gene knockout mouse model for HCC (Liao et al., 2009). Recently, a novel target of DNMT3B, metastasis suppressor 1 (MTSS1), was found to act as a tumor suppressor in HCC (Fan et al., 2012).
In this study, we found that betaine, a methyl donor, can significantly prevent the down regulation of Ufmylation in the livers of DDC re-fed mice. We further showed a good correlation between promoter methylation with transcriptional silencing of Ufmylation in multiple AH and NASH biopsies. Importantly, DNMT1 and DNMT3B mRNA levels were significantly up regulated in these biopsies.
Materials and Methods
Biopsies
Human archived formalin-fixed paraffin-embedded (FFPE) liver biopsies from patients who had alcoholic hepatitis (AH; n=3–5) and non-alcoholic steatohepatitis (NASH; n=3–5) were obtained from Harbor UCLA hospital archives. In all the cases liver forming MDBs were present except in the normal control livers (Control; n=3). The biopsy sections were cut 4 µm thick.
Mouse livers
Diethyl 1, 4-dihydro-2, 4, 6-trimethyl-3, 5-pyridine-dicarboxylate (DDC) was used as a model to induce the formation of Mallory-Denk bodies (MDBs) in mice. One-month-old C3H male mice were fed 0.1% DDC added to the control diet and a second group (n=4) were fed control diet for 10 weeks (Li et al., 2008). The mice were then withdrawn from the drug for 1 month, as withdrawn controls and then re-fed DDC with or without betaine (2% in the drinking water) for 7 days as was previously done (Oliva et al., 2009). Three mice were used in each of three groups as follows: 1) control, 2) DDC+2% Betaine. 3) DDC withdrawal. Mice livers were placed in isopentane and were then fast frozen with liquid nitrogen and stored at −80°C. The livers used had been used in a prior study (Oliva et al., 2009). All mice were treated in a humane manner as approved by the Animal Care Committee at Harbor UCLA Laboratory Bio Medical Research Institute according to the Guidelines of the National Academy of Science.
Tissue sectioning
Archived mice liver frozen sections (control, DDC+2% Betaine, DDC withdrawn) were performed as standard protocol. Mice liver frozen sections were cut (4 µm thickness) at −20°C and immediately transferred to a micro slide box kept on dry ice and stored at −80°C. These slides were placed under the hood to dry for 1–2 hour and subsequently stored in a micro slide box at −80°C.
DNA isolation
For DNA isolation of FFPE tissue sections, paraffin was first removed from tissue samples when human liver biopsies were assayed. The paraffin-embedded tissues sections were mounted on a glass slide and dried at 60°C for 30 minutes. The slides were then submerged in xylene at room temperature for 30 minutes changing the xylene once after 30 minutes. The samples were hydrated by washing progressively for 2 minutes in 100%, 70%, 50% ethanol, and then pure DNase-free water before air-drying the samples on the slides for approximately 10 minutes. DNA isolation was processed using the Pinpoint™ Slide DNA isolation System (ZYMO, Irvine, CA) by adding Pinpoint Solution directly to the region of the tissue section to allow the solution to dry completely at the room temperature. The embedded tissue was then removed from the slide using a sterile blade or scalpel to scrape tissues from the slides followed by transferring the tissues to a micro-centrifuge tube for subsequent proteinase K digestion. The DNA was then extracted and purified according to the manufacturer’s protocol (ZYMO, Irvine, CA).
Bisulfite Conversion (C to T conversion) and Methylation-Specific PCR (MSP)
Bisulfite conversion was performed with the above mentioned DNA (600ng) for C to T conversion using EZ DNA Methylation™ Kit following the instruction (ZYMO, Irvine, CA). Then the bisulfite treated DNA was used as a template for methylation-specific PCR (MSP). Several pairs of MSP primers targeting the methylated or unmethylated alleles of the promoter region were designed to amplify promoter CpG sites containing possible methylation sites as previously described (Li & Dahiya, 2002). MSP was performed, and the reaction condition consisted of 95°C for 5 min, followed by 40 cycles of 95°C for 30 sec, 60 °C for 30 sec, 72 °C for 1 min, and extended at 72 °C 10 min. MSP primer sequences and related gene Accession Number are listed in Table I Methylation ratio of biopsies in each MSP was calculated with Methylated intensity/ (Methylated intensity+Unmethylated intensity) %.
Table I.
Sequences of the left and right methylation specific PCR (MSP) primers. M, methylated; U, unmethylated
| Gene name (Species) |
Accession Number |
Sequences of MSP primers |
|---|---|---|
| Ufm1 (Human) |
NM_001256799 | Left M Primer:: 5’- TAG GGT TAA TTT TTA GGT AGT GCG T -3’ Right M Primer: 5’- CGA CTT CCT CTA ATA TAC TAA AAA CCG -3’ |
| Ufm1 (Human) |
NM_001256799 | Left U Primer: 5’- TAG GGT TAA TTT TTA GGT AGT GTG T -3’ Right U Primer: 5’- CAC AAC TTC CTC TAA TAT ACT AAA AAC CAC -3’ |
| Ufc1 (Human) |
NM_016406 | Left M Primer: 5’- GGT TTA TTG TAA TTT TCG TTT TTC G -3’ Right M Primer: 5’- ACC AAC CTA ACT AAT AAA ACC CGT C -3’ |
| Ufc1 (Human) |
NM_016406 | Left U Primer: 5’- GTT TAT TGT AAT TTT TGT TTT TTG G -3’ Right U Primer: 5’- ACC AAC CTA ACT AAT AAA ACC CAT C -3’ |
| UfSP1 (Human) |
NM_001015072 | Left M Primer: 5’- GAG TTA GGG GTA GAG GTA GAA CGT C -3’ Right M Primer: 5’- TAA CCT ACC TAC GAA CCT CAT TAC G -3’ |
| UfSP1 (Human) |
NM_001015072 | Left U Primer: 5’- AGT TAG GGG TAG AGG TAG AAT GTT G -3’ Right U Primer: 5’- ACC TAC CTA CAA ACC TCA TTA CAC A -3’ |
RNA isolation and cDNA synthesis
RNA isolation of FFPE sections of human liver biopsies were performed as we previously described (Liu et al., 2014). The process described above was also followed for mice frozen liver tissues, except for the deparaffinization step. The quality and yield of the resulting total RNAs were assessed with an absorbance reading at 260 nm (A260) using a spectrophotometer (Thermo, Waltham, MA). Synthesis of first-strand cDNAs was performed with the above mentioned total RNA (250ng), and random hexamer primers using SuperSript III First-Strand Synthesis SuperMix following the instruction (Invitrogen, San Diego, CA).
Quantitative Real-time PCR analysis
Real-time PCR was performed using the Fast SYBR Green Master Mix on a StepOnePlus™ Real-time PCR System (Applied Biosystems) with a primer concentration of 200 nM. Primer sequences of Ufmylation components including Ufm1, Uba5, Ufc1, Ufl1, UfSP1 and UfSP2 in mice used had been used in a prior study (Liu et al., 2014). Primer sequences of human DNMT1, DNMT3A and DNMT3B are as followed: DNMT1 (NM_001379): Forward Primer 5’-CGA GTT GGT GAT GGT GTG T-3’ and Reverse Primer: 5’-GTG GAC TTG TGG GTG TTC TC-3’. DNMT3A (NM_175629): Forward Primer 5’-CTG TGA TGA TTG ATG CCA AA-3’ and Reverse Primer: 5’-GAC TGG GAA ACC AAA TAC CC-3’. DNMT3B (NM_006892): Forward Primer 5’-ATA AGA CAC CCC CTC AAA CC-3’ and Reverse Primer 5’-TTC CCG TTC TCC CTA AAA AC-3’. Reaction conditions consisted of 95°C for 20 sec, followed by 40 cycles of 95°C for 3 sec, 60 °C for 30 sec. Single PCR product was confirmed with the heat dissociation protocol at the end of the PCR cycles. Human α-tubulin and mice β-actin was used as control to normalize the starting quantity of RNA. The target mRNA abundance in each sample was normalized to its endogenous control level and the relative mRNA expression levels were analyzed using the ΔΔCT method. Reaction of each sample was performed in triplicate.
Western blot
For protein isolation of FFPE tissue sections, paraffin was first removed from tissue samples as we previously described (Liu et al., 2014). Tissues samples were further pelleted and washed five times with RIPA buffer for 10 minutes at room temperature. The washed pellets were then suspended with reducing sample buffer quickly and denatured by heating at 100°C for 10 min. Then 50 µg of each cell lysate was used for PAGE. Proteins were fractionated by electrophoresis through 10% SDS-PAGE and electrophoretically transferred onto a PVDF blot membrane (Bio-Rad, Hercules, CA). Membranes were blocked with 5% BSA and TBST at room temperature for 2 h and washed three times with TBST. Anti-Ufm1 polyclonal antibodies were diluted according to the company’s recommendation in TBST and incubated with the membranes overnight at 4°C. After washes three times with TBST, the membranes were incubated with the corresponding secondary antibody. Proteins were visualized using the enhanced chemilumines-cence (ECL) detection system.
Statistical analysis
All data were presented as the mean ±S.E.M and were representative of at least two-independent experiments done in triplicate. Statistical differences were carried out using t-test with SigmaStat software. P < 0.05 indicated a statistically significant difference.
Results
Betaine restores the Ufmylation expression in the livers of drug-primed mice
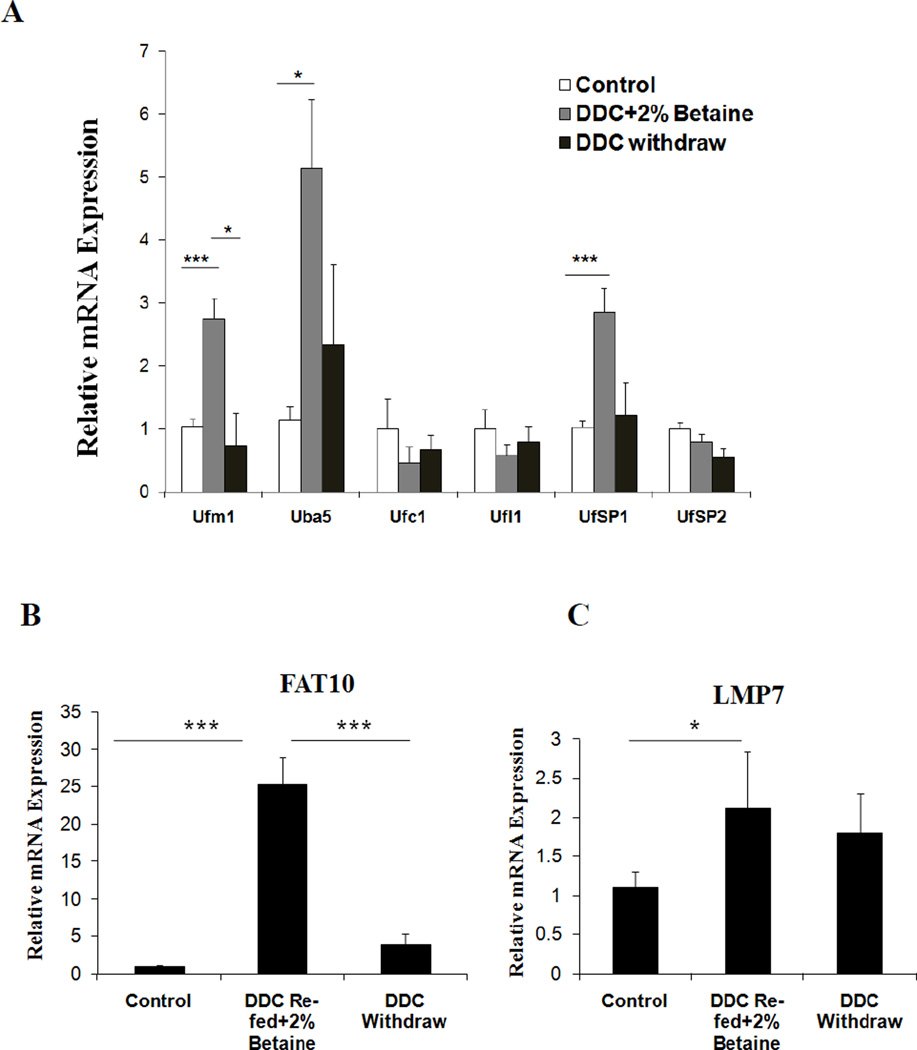
Epigenetic changes can be reversed by pharmacological intervention (Subramaniam et al., 2014). Betaine, a methyl donor, can prevent MDB formation and FAT10 positive hepatocyte proliferation in drug-primed mice by epigenetic mechanisms (Oliva et al., 2009). Mallory body forming cells express the pre-neoplastic hepatocyte phenotype in the DDC fed model (Nan et al., 2006). Our recent study showed that the Ufmylation pathway was down regulated in alcoholic hepatitis (AH) and non alcoholic steatohepatitis (NASH) patients’ liver biopsies in which MDBs (Liu et al., 2014). The down regulation of Ufmylation results in MDB formation by causing accumulation of undigested proteins in the cell. The mRNA expression of Ufmylation components were further investigate in the livers of DDC re-fed mice plus betaine compared with the controls and DDC withdrawal groups. Several mRNAs of Ufmylation components were restored in DDC re-fed plus betaine group (Fig. 1A) when compared to the prominent down regulation in the DDC refed group as was previously reported (Liu et al., 2014). This prevented the DDC refed group from forming MDBs (due to the reduced protein quality control mechanism). mRNA expression of Ufm1 and Uba5 were induced to approximately 3- and 5-fold levels respectively (p<0.05) in this test when compare to other mRNAs. UfSP1 showed a 3-fold increased expression (Fig. 1A). Other components of the Ufm1 cascade including Ufc1, Ufl1 and UfSP2 were also slightly up regulated in this test (Fig. 1A) compared to previous observations. The expression of other mRNAs were down regulated in the livers of DDC re-fed mice (Liu et al., 2014). In parallel, the mRNA expression of Ufmylation was not different from the control levels and after 1 month withdrawal (Fig.1A).
Figure 1.
Betaine prevents the down regulation of Ufmylation in the livers of DDC re-fed mice. (A) Gene expression levels of different Ufmylation components (B) FAT10 and (C) LMP7 in DDC refed plus betaine group and DDC withdrawal group. Data represent mean values ±S.E.M. Statistical significance was determined using the SigmaStat software. * p<0.05 and ***p<0.001 by One Way ANOVA test.
FAT10 mRNA was increased in the DDC re-fed plus betaine group compared to the control group and the DDC withdrawn group (Fig. 1B). The catalytic subunit LMP7 was increased compared to control group (Fig.1C). This data clearly shows that betaine prevents the down regulation of Ufmylation expression in the livers of drug-primed mice.
Promoter CpG methylation of Ufmylation was significantly increased in AH and NASH
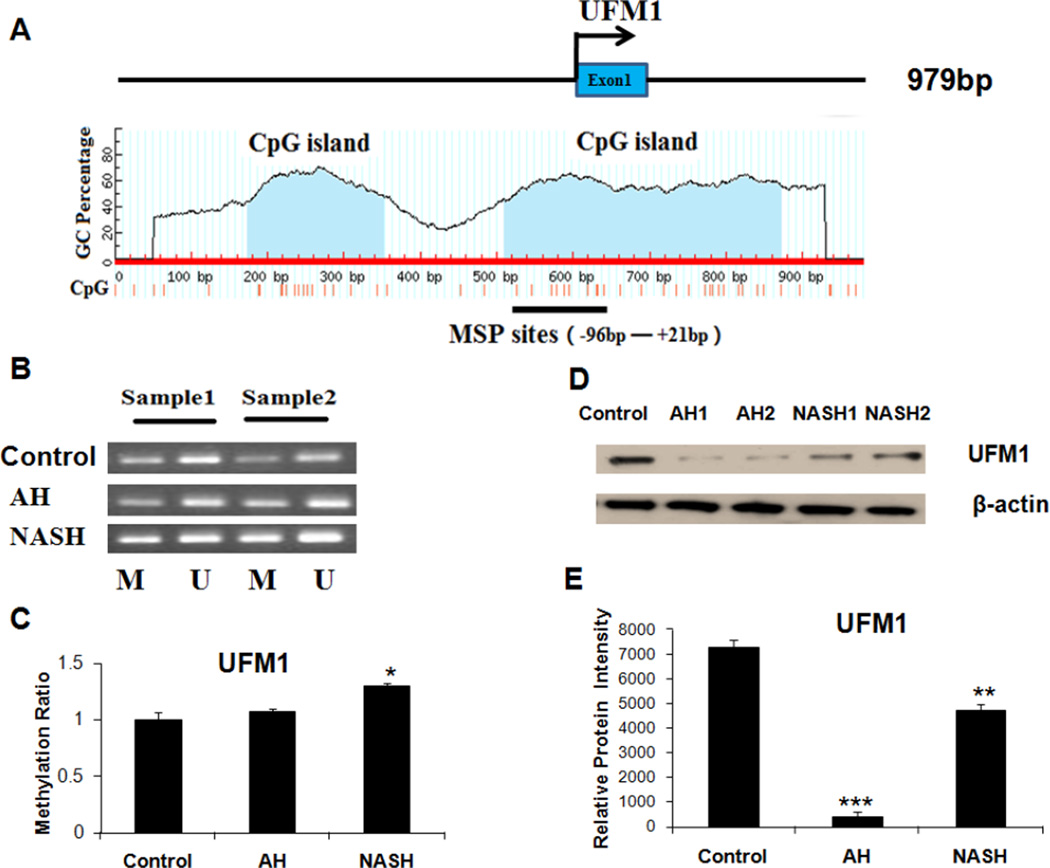
Sequence analysis revealed that almost all members of the Ufmylation pathway contained CpG islands (CGI) in the promoter region except Uba5, indicating CpG methylation may be a major mechanism in silencing its expression in MDB-forming liver tissue. Ufm1 has a small CGI and a bigger CGI in the promoter region, which is located in −426bp to −248bp and −90bp to +272bp sites respectively (Fig. 2A). MSP analysis showed that the Ufm1 promoter had increased methylated levels in the AH and NASH biopsies examined (Fig. 2B), with a 7% and 30% (respectively) increase of the methylation ratio in AH and NASH patients compared to normal liver tissues (Fig. 2C). The protein level of Ufm1 was frequently silenced or reduced in multiple AH and NASH biopsies by western blot analysis (Fig. 2D and Fig. 2E).
Figure 2.
A, The Ufm1 CGI. Transcription start site is indicated by an arrow. The CGI and MSP regions analyzed are indicated. B, Representative analyses of Ufm1 promoter methylation in two AH and NASH patients. M, methylated; U, unmethylated. C, The methylation ratio of Ufm1 in AH and NASH was normalized to that in normal liver tissues. D, Western blot analysis of Ufm1 protein in AH and NASH biopsies with MDBs present. E, Protein expression intensity of Ufm1 in AH and NASH. * p<0.05, * * p<0.01 and ***p<0.001 by t-test with SigmaStat software.
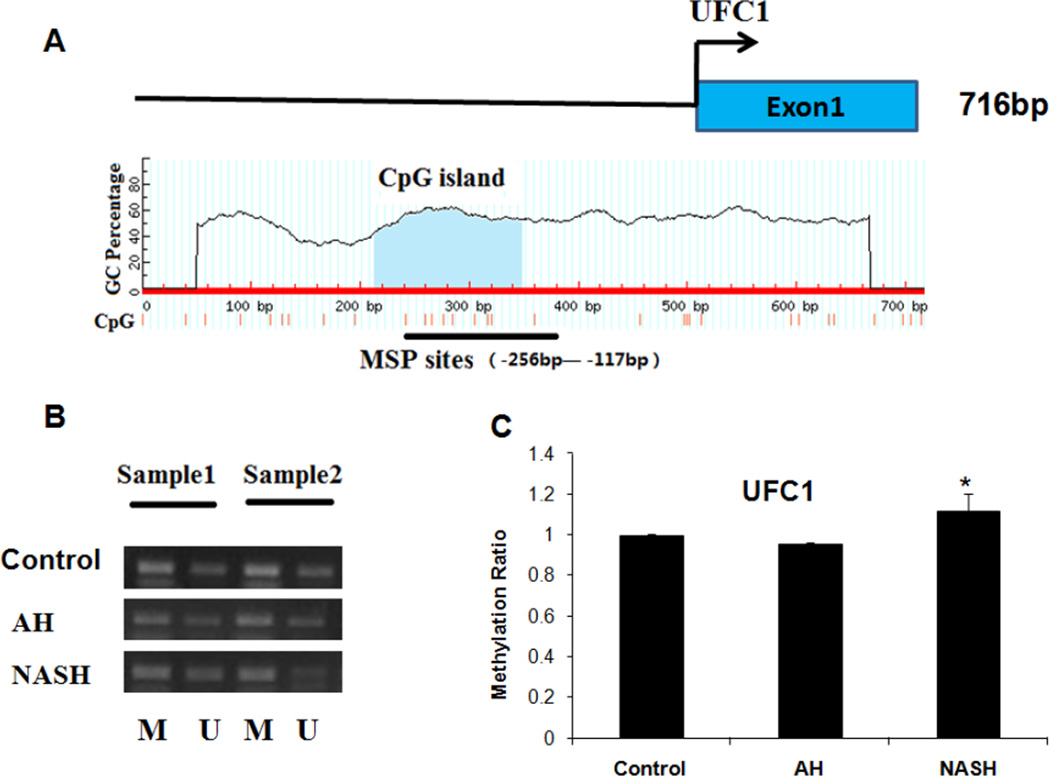
Ufc1, an E2 enzyme of Ufmylation, with a CpG island (−287bp to −117bp) in the promoter region (Fig. 3A) also was detected (an 8% increased methylation ratio) in NASH patients by MSP analysis (Fig. 3B), while no obvious methylation was detected in AH biopsies (Fig. 3C) compared to normal subjects.
Figure 3.
Analyses of Ufc1 methylation in normal liver tissues, AH and NASH by MSP. A, the Ufc1 CGI. Transcription start site is indicated by an arrow. The CGI and MSP regions analyzed are indicated. B, Representative analyses of Ufc1 promoter CpG methylation in AH and NASH patients. M, methylated; U, unmethylated. C, The methylation ratio of Ufc1 in AH and NASH was normalized to that in normal liver tissues. * p<0.05 by t-test with SigmaStat software.
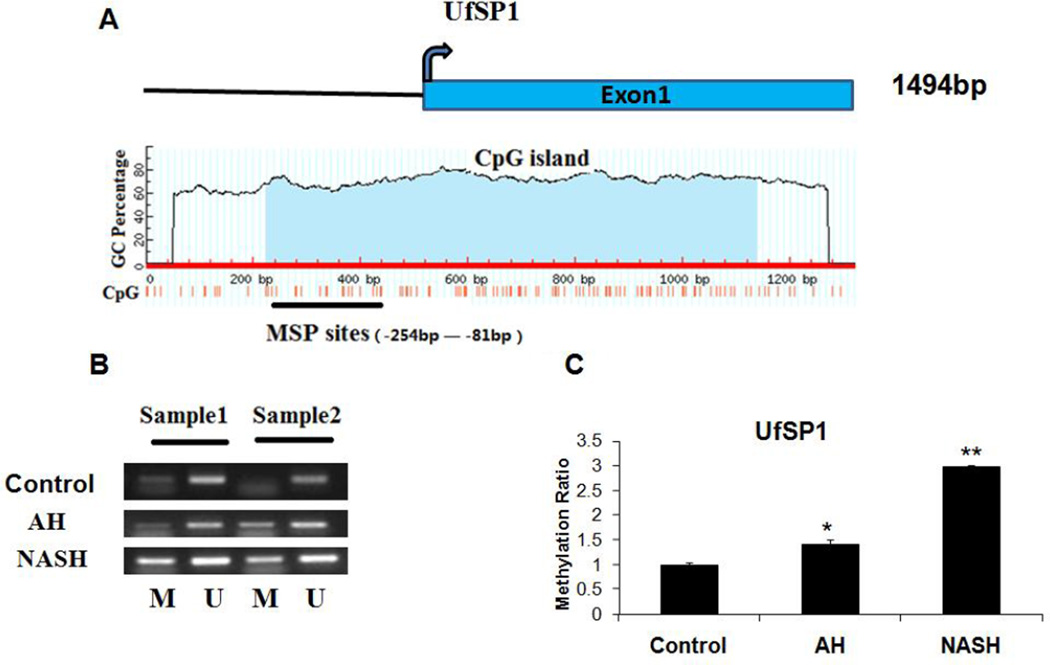
UfSP1, another member of Ufm1-specific proteases, can cleave both the C terminus of pro-Ufm1 and several Ufm1 conjugates. It had a large CpG island (−376bp to +539bp) in its promoter site (Fig. 4A). MSP showed that there was an increase in promoter methylation both in AH and NASH biopsies (Fig. 4B). Among these, the methylation ratio of UfSP1 was increased by 40% in AH biopsies, and a nearly 3-fold increase in NASH biopsies (Fig. 4C). These results clearly indicated that promoter methylation of Ufmylation was increased, and it was a common event in multiple AH and NASH biopsies.
Figure 4.
Analyses of UfSP1 methylation in normal tissues, AH and NASH by MSP. A, the UfSP1 CGI. Transcription start site is indicated by a curved arrow. The CGI and MSP regions analyzed are indicated. B, Representative analyses of UfSP1 promoter methylation in AH and NASH patients. M, methylated; U, unmethylated. C, The methylation ratio of UfSP1 in AH and NASH was normalized to that in normal liver tissues. * p<0.05 and **p<0.01 by t-test with SigmaStat software.
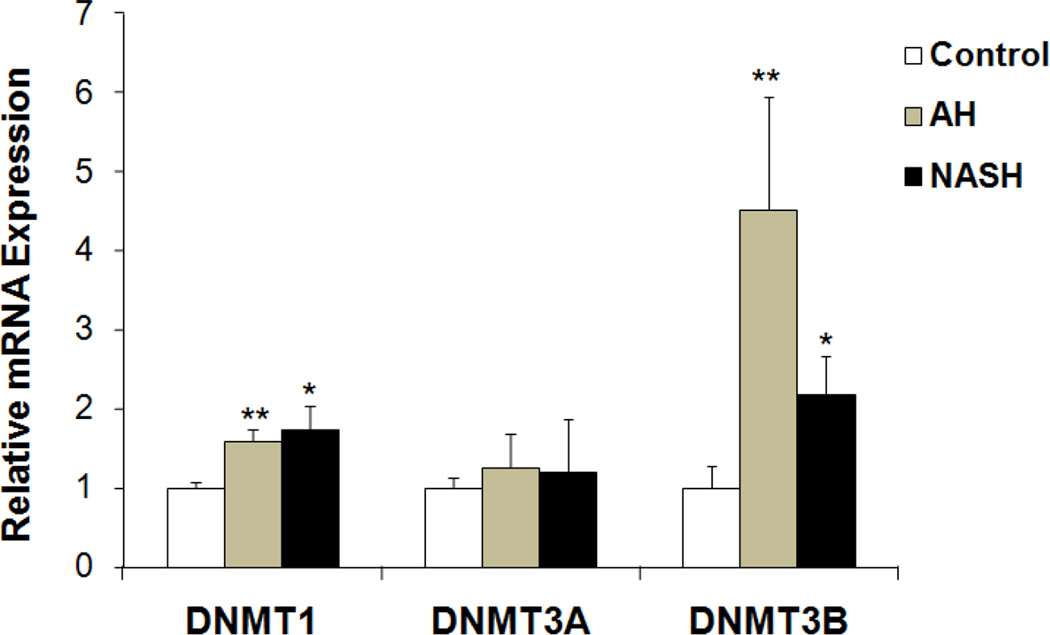
DNMT1 and DNMT3B are significantly up regulated in the livers of AH and NASH
Methylation of mammalian genomic DNA is catalyzed by DNMTs. Among these proteins, only DNMT1, DNMT3A and DNMT3B exhibit methyltransferase activity (Dong & Wang, 2014). The mRNA expression of DNMT1, DNMT3A and DNMT3B were analyzed by real-time PCR. The mRNA levels of DNMT1, DNMT3A and DNMT3B were all up regulated to various degrees (Fig. 5). DNMT1 mRNA in AH and NASH liver biopsies was induced to 60% and 80% levels (respectively), while DNMT3B mRNA was significantly up regulated in AH and NASH biopsies with a 4.5- and 2.2 fold induction respectively (Fig. 5). The DNMT3A mRNA, however, was not increased (Fig. 5). These results suggested that the maintenance of methylation in AH and NASH was mainly mediated by both DNMT1 and DNMT3B.
Figure 5.
Transcriptional expression of human DNMT1, DNMT3A and DNMT3B in patient livers from archived biopsies including AH, NASH and normal liver tissue. Data represent mean values ±S.E.M. * p<0.05 and **p<0.01 by One Way ANOVA with SigmaStat software.
Discussion
Aberrant epigenetic modification of tumor cells is important in initiating carcinogenesis (Herman, 1999). Promoter CpG methylation causes the loss of key cellular enzyme functions, which occur frequently during tumor development and progression (Jones & Baylin, 2002). Currently, it is accepted that epigenetic alterations can precede genetic changes during tumorigenesis (Jones & Baylin, 2002). In this report, it has been shown that betaine, a methyl donor, can restore the down regulation of the Ufm1 conjugation pathway in the livers of DDC re-fed mice where betaine had also prevented MDB formation. This supported the concept that MDB formation is an epigenetic phenomenon. It was also found that there was a good correlation between promoter methylation and transcriptional silencing of Ufmylation in multiple AH and NASH biopsies where MDBs had formed. This is the first clinical evidence of direct involvement of the Ufm1 conjugation system with promoter CpG methylation in liver MDB formation.
Post-translational modifications are cellular mechanisms that enable a rapid response to internal and external changes. The Ufm1 conjugation system was recently identified as an Ubl system which seems to be involved in various human diseases development including cancer (Daniel & Liebau, 2014). It was reported by our group that the Ufmylation pathway is down regulated both in the livers of DDC re-fed mice and in the livers of AH and NASH patients with MDBs present (Liu et al., 2014). However, the molecular mechanism resulting in transcript down regulation of the Ufmylation pathway is still unknown. By bioinformatics we discovered that almost all members of Ufmylation have CpG islands in their promoter region, indicating that CpG methylation might be a major mechanism of silencing Ufmylation expression in the liver of AH and NASH patients. Promoter CpG island hypermethylation is an important mechanism for inactivation of tumor suppressor genes or tumor-related genes in human cancers and occurs in virtually all human cancer types (Baylin & Chen, 2005). Our MSP analysis further revealed that the promoter methylation levels of Ufm1, Ufc1 and UfSP1 involved in Ufmylation were increased in AH and NASH patients when compared to normal subjects. It has been observed that the methylation ratio of UfSP1 in its promoter region was significantly increased both in AH and NASH patients. UfSP1 plays an important role in the reversal of protein modification by Ufm1 as well as in the processing of the Ufm1 precursor (Kang et al., 2007). A series of CpG island methylation alterations have been observed in the HCC cell lines such as Hep3B, HepG2, PLC/RPF/5/RPF/5, SMMC-7721, BEL-7402, MHCC97-H, MHCC97-L, HCCLM3 and HCCLM6. CpG island hypermethylation of tumor suppressor genes leads to a decrease in their expression (Liu et al., 2010). Interestingly, we found that the methylation levels of Ufm1, Ufc1 and UfSP1 in NASH patients are all higher than that in AH patients, suggesting a progressive increase of promoter methylation compared to normal liver.
Aberrant DNA methylation of CpG islands is catalyzed by DNA methyltransferases (DNMTs). Thus, abnormal variations of DNMTs can contribute to hepatocarcinogenesis. The human DNMT1, DNMT3A and DNMT3B coordinate mRNA expression in normal tissues and over expression in tumors (Robertson et al., 1999). The expression levels of these DNMTs are reportedly elevated in HCC (Oh et al., 2007). The role of altered expression of DNMTs in DNA hypomethylation and hypermethylation in cancer is uncertain and may involve changes in mRNA or protein expression. In human fibroblasts, sustained over expression of DNMT1 leads to the processive time- dependent hypermethylation of a number of CpG islands (Vertino et al., 1996). Conversely, reduction of DNMT1 levels appears to have protective effects. Reduction of DNMT1 through an antisense approach also blocks tumorigenesis (Ramchandani et al., 1997; Rountree et al., 2001). Like DNMT1, the aforementioned DNMT3A and DNMT3B enzymes also appear to be modestly over expressed in cancer (Li et al., 2006).
In the present study, the mRNA levels of DNMT1 and DNMT3B were all up regulated to various degrees. DNMT1 mRNA was induced by up to a 2- and 4-fold (respectively) in the liver of AH and NASH biopsies, and DNMT3B mRNA was induced by up to 4.5- and 2.3 fold in AH and NASH respectively. This result suggested that the maintenance of methylation in AH and NASH with MDBs present was mediated by DNMT1 and DNMT3B. These observations point to a complicated network of connections between DNMTs and Ufmylation involved in gene regulation and epigenetic signaling during liver MDB formation. However, we did not observe an increase in DNMT3A mRNA. It is possible that this enzyme appears to function as a de novo methyltransferase with a dispersed distribution throughout the nucleus (Xu et al., 1999).
Currently, epigenetic alterations are increasingly recognized as valuable targets for the development of cancer therapies. In the DDC re-fed mouse model, betaine, a methyl donor, can prevent MDB formation and FAT10 positive hepatocyte proliferation in drug-primed mice by an epigenetic mechanism (Oliva et al., 2009). FAT10 over expression is induced in DDC fed mice by interferon gamma binding to the interferon stimulating response element on the FAT10 promoter (Oliva et al., 2010). FAT10 is a marker for liver preneoplasia change in the DDC mouse model (Oliva et al., 2008). Betaine may function through removing homocysteine and S-adenosylhomocysteine (SAH) in the liver (Purohit et al., 2007). Betaine feeding can also prevent the blood alcoholic cycle seen in rats fed ethanol intragastrically (Li et al., 2011). The transcription expression of Ufmylation (Ufm1, Uba5, Ufc1, Ufl1, UfSP1 and UfSP2) was further investigated by real-time PCR analysis in the livers of DDC re-fed mice, plus betaine fed and DDC withdrawal groups. As expected, almost all mRNAs expression of Ufmylation components were normalized in the DDC re-fed plus betaine group compared to the prominent down regulation seen in the DDC re-fed group (Liu et al., 2014). The mRNA expression of Uba5 was induced by approximately 5-fold (p<0.05). However, we did not detect a CpG island in the promoter region of Uba5 by sequence analysis.
The mRNA expression of Ufmylation was similar to the control levels in the group of 1 month DDC withdrawal. At the same time, we found that mRNA levels of LMP7 and FAT10 tended to decrease when betaine was added to the DDC re-fed group compared to the DDC re-fed group, indicating that betaine prevented the formation of MDBs epigenetically as previously reported (Oliva et al., 2009; French et al., 2010).
In summary, provided here for the first time is evidence of transcription down regulation of the Ufm1 conjugation due to hypermethylation of the promoter CpG in AH and NASH patients when MDBs form. The maintenance of Ufmylation methylation may be mediated by DNMT1 and DNMT3B together, which would explain the down regulation of Ufmylation observed in Nash and Ash liver biopsies.
Acknowledgments
This work was supported by grants from NIH (AAU01021898-02). We would like to thank Adriana Flores for typing this manuscript.
Abbreviations
- AH
alcoholic hepatitis
- CGI
CpG island
- DDC
diethyl 1, 4-dehydro-2, 4, 6-trimethyl-3, 5-pyridine-dicarboxylate
- DNMT1
DNA (cytosine-5-)-methyltransferase 1
- ER
Endoplasmic reticulum
- HCC
hepatocellular carcinoma
- MDB
Mallory-Denk body
- NASH
non alcoholic steatohepatitis
- Ubl
Ubiquitin-like proteins
- Ufm1
ubiquitin fold modifier 1
- Ufmylation
Ufm1-conjugating system.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Bardag-Gorce F, Oliva J, Li J, French BA, French SW. SAMe prevents the induction of the immunoproteasome and preserves the 26S proteasome in the DDC-induced MDB mouse model. Exp Mol Pathol. 2010;88:353–362. doi: 10.1016/j.yexmp.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardag-Gorce F, Oliva J, Villegas J, Fraley S, Amidi F, Li J, Dedes J, French B, French SW. Epigenetic mechanisms regulate Mallory Denk body formation in the livers of drug-primed mice. Exp Mol Pathol. 2008;84:113–121. doi: 10.1016/j.yexmp.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basaranoglu M, Turhan N, Sonsuz A, Basaranoglu G. Mallory-Denk Bodies in chronic hepatitis. World J Gastroenterol. 2011;17:2172–2177. doi: 10.3748/wjg.v17.i17.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Chen WY. Aberrant gene silencing in tumor progression: implications for control of cancer. Cold Spring Harb Symp Quant Biol. 2005;70:427–433. doi: 10.1101/sqb.2005.70.010. [DOI] [PubMed] [Google Scholar]
- Caldwell S, Ikura Y, Dias D, Isomoto K, Yabu A, Moskaluk C, Pramoonjago P, Simmons W, Scruggs H, Rosenbaum N, Wilkinson T, Toms P, Argo CK, Al-Osaimi AM, Redick JA. Hepatocellular ballooning in NASH. J Hepatol. 2010;53:719–723. doi: 10.1016/j.jhep.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J, Liebau E. The ufm1 cascade. Cells. 2014;3:627–638. doi: 10.3390/cells3020627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Wang A. Aberrant DNA methylation in hepatocellular carcinoma tumor suppression (Review) Oncol Lett. 2014;8:963–968. doi: 10.3892/ol.2014.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu RG. Epigenetic targets in cancer epidemiology. Methods Mol Biol. 2009;471:457–467. doi: 10.1007/978-1-59745-416-2_23. [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- Fan H, Chen L, Zhang F, Quan Y, Su X, Qiu X, Zhao Z, Kong KL, Dong S, Song Y, Chan TH, Guan XY. MTSS1, a novel target of DNA methyltransferase 3B, functions as a tumor suppressor in hepatocellular carcinoma. Oncogene. 2012;31:2298–2308. doi: 10.1038/onc.2011.411. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Greally JM. Epigenetics: perceptive enzymes. Nature. 2007;449:148–149. doi: 10.1038/449148a. [DOI] [PubMed] [Google Scholar]
- French SW, Bardag-Gorce F, Li J, French BA, Oliva J. Mallory-Denk body pathogenesis revisited. World J Hepatol. 2010;2:295–301. doi: 10.4254/wjh.v2.i8.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG. Hypermethylation of tumor suppressor genes in cancer. Semin Cancer Biol. 1999;9:359–367. doi: 10.1006/scbi.1999.0138. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Kang GH. CpG island hypermethylation in gastric carcinoma and its premalignant lesions. Korean J Pathol. 2012;46:1–9. doi: 10.4132/KoreanJPathol.2012.46.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Kim GR, Seong M, Baek SH, Seol JH, Bang OS, Ovaa H, Tatsumi K, Komatsu M, Tanaka K, Chung CH. Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J Biol Chem. 2007;282:5256–5262. doi: 10.1074/jbc.M610590200. [DOI] [PubMed] [Google Scholar]
- Kim CH, Nam HS, Lee EH, Han SH, Cho HJ, Chung HJ, Lee NS, Choi SJ, Kim H, Ryu JS, Kwon J. Overexpression of a novel regulator of p120 catenin, NLBP, promotes lung adenocarcinoma proliferation. Cell Cycle. 2013;12:2443–2453. doi: 10.4161/cc.25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell Mol Life Sci. 2009;66:596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J, Cho HJ, Han SH, No JG, Kwon JY, Kim H. A novel LZAP-binding protein, NLBP, inhibits cell invasion. J Biol Chem. 2010;285:12232–12240. doi: 10.1074/jbc.M109.065920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire K, Moura RF, Granvik M, Igoillo-Esteve M, Hohmeier HE, Hendrickx N, Newgard CB, Waelkens E, Cnop M, Schuit F. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS One. 2011;6:e18517. doi: 10.1371/journal.pone.0018517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Rauch T, Chen ZX, Szabo PE, Riggs AD, Pfeifer GP. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem. 2006;281:19489–19500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- Li J, Bardag-Gorce F, Dedes J, French BA, Amidi F, Oliva J, French SW. S-adenosylmethionine prevents Mallory Denk body formation in drug-primed mice by inhibiting the epigenetic memory. Hepatology. 2008;47:613–624. doi: 10.1002/hep.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li XM, Caudill M, Malysheva O, Bardag-Gorce F, Oliva J, French BA, Gorce E, Morgan K, Kathirvel E, Morgan T, French SW. Betaine feeding prevents the blood alcohol cycle in rats fed alcohol continuously for 1 month using the rat intragastric tube feeding model. Exp Mol Pathol. 2011;91:540–547. doi: 10.1016/j.yexmp.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Liao YJ, Liu SP, Lee CM, Yen CH, Chuang PC, Chen CY, Tsai TF, Huang SF, Lee YH, Chen YM. Characterization of a glycine N-methyltransferase gene knockout mouse model for hepatocellular carcinoma: Implications of the gender disparity in liver cancer susceptibility. Int J Cancer. 2009;124:816–826. doi: 10.1002/ijc.23979. [DOI] [PubMed] [Google Scholar]
- Liu BB, Zheng D, Liu YK, Kang XN, Sun L, Guo K, Sun RX, Chen J, Zhao Y. Array-based profiling of the differential methylation status of CpG islands in hepatocellular carcinoma cell lines. Oncol Lett. 2010;1:815–820. doi: 10.3892/ol_00000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li J, Tillman B, French BA, French SW. Ufmylation and FATylation pathways are downregulated in human alcoholic and nonalcoholic steatohepatitis, and mice fed DDC, where Mallory-Denk bodies (MDBs) form. Exp Mol Pathol. 2014;97:81–88. doi: 10.1016/j.yexmp.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WR, Shi YH, Peng YF, Fan J. Epigenetics of hepatocellular carcinoma: a new horizon. Chin Med J (Engl) 2012;125:2349–2360. [PubMed] [Google Scholar]
- Miremadi A, Oestergaard MZ, Pharoah PD, Caldas C. Cancer genetics of epigenetic genes. Hum Mol Genet. 2007;16(Spec No 1):R28–R49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]
- Nan L, Bardag-Gorce F, Wu Y, Li J, French BA, French SW. Mallory body forming cells express the preneoplastic hepatocyte phenotype. Exp Mol Pathol. 2006;80:109–118. doi: 10.1016/j.yexmp.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Oh BK, Kim H, Park HJ, Shim YH, Choi J, Park C, Park YN. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int J Mol Med. 2007;20:65–73. [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, French BA, Li J, McPhaul L, Amidi F, Dedes J, Habibi A, Nguyen S, French SW. Fat10 is an epigenetic marker for liver preneoplasia in a drug-primed mouse model of tumorigenesis. Exp Mol Pathol. 2008;84:102–112. doi: 10.1016/j.yexmp.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, Li J, French BA, Nguyen SK, Lu SC, French SW. Betaine prevents Mallory- Denk body formation in drug-primed mice by epigenetic mechanisms. Exp Mol Pathol. 2009;86:77–86. doi: 10.1016/j.yexmp.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, Lin A, French BA, French SW. The role of cytokines in UbD promoter regulation and Mallory-Denk body-like aggresomes. Exp Mol Pathol. 2010;89:1–8. doi: 10.1016/j.yexmp.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit V, Abdelmalek MF, Barve S, Benevenga NJ, Halsted CH, Kaplowitz N, Kharbanda KK, Liu QY, Lu SC, McClain CJ, Swanson C, Zakhari S. Role of S-adenosylmethionine, folate, and betaine in the treatment of alcoholic liver disease: summary of a symposium. Am J Clin Nutr. 2007;86:14–24. doi: 10.1093/ajcn/86.1.14. [DOI] [PubMed] [Google Scholar]
- Ramchandani S, MacLeod AR, Pinard M, von Hofe E, Szyf M. Inhibition of tumorigenesis by a cytosine-DNA, methyltransferase, antisense oligodeoxynucleotide. Proc Natl Acad Sci U S A. 1997;94:684–689. doi: 10.1073/pnas.94.2.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Singh BN, Huang Q, Li Z, Gao Y, Mishra P, Hwa YL, Li J, Dowdy SC, Jiang SW. DNA hypermethylation as a chemotherapy target. Cell Signal. 2011;23:1082–1093. doi: 10.1016/j.cellsig.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;20:3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- Shiwaku H, Yoshimura N, Tamura T, Sone M, Ogishima S, Watase K, Tagawa K, Okazawa H. Suppression of the novel ER protein Maxer by mutant ataxin-1 in Bergman glia contributes to non-cell-autonomous toxicity. EMBO J. 2010;29:2446–2460. doi: 10.1038/emboj.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertino PM, Yen RW, Gao J, Baylin SB. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc'his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang M, Wu J, Lei G, Li H. Transcriptional regulation of the Ufm1 conjugation system in response to disturbance of the endoplasmic reticulum homeostasis and inhibition of vesicle trafficking. PLoS One. 2012;7:e48587. doi: 10.1371/journal.pone.0048587. [DOI] [PMC free article] [PubMed] [Google Scholar]