Abstract
Hippocampus-dependent learning is known to induce changes in gene expression, but information on gene expression differences between different learning paradigms that require the hippocampus is limited. The bulk of studies investigating RNA expression after learning use the contextual fear conditioning task, which couples a novel environment with a footshock. Although contextual fear conditioning has been useful in discovering gene targets, gene expression after spatial memory tasks has received less attention. In this study, we used the object-location memory task and studied gene expression at two time points after learning in a high-throughput manner using a microfluidic qPCR approach. We found that expression of the classic immediate-early genes changes after object-location training in a fashion similar to that observed after contextual fear conditioning. However, the temporal dynamics of gene expression are different between the two tasks, with object-location memory producing gene expression changes that last at least 2 hours. Our findings indicate that different training paradigms may give rise to distinct temporal dynamics of gene expression after learning.
Keywords: transcription, spatial memory, fear conditioning, object-location memory, hippocampus
1. Introduction
Long-term memory is critical to our lives, yet the molecular mechanisms that create and stabilize memories are still poorly understood. The hippocampus, which encodes contextual information, has been heavily studied in an effort to better understand these mechanisms. Transcription is required to convert labile short-term memories into stable long-term memories during the period of memory consolidation (Agranoff et al, 1967; Igaz et al, 2002). The expression of many genes is regulated within the first hour after learning in the hippocampus (Hawk et al, 2012; Keeley et al, 2006; Lemberger et al, 2008; Levenson et al, 2004a; Lonergan et al, 2010; Ramamoorthi et al, 2011). Epigenetic mechanisms, such as histone acetylation, can modulate this transcription to enhance or dampen long-term memory formation (Alarcon et al, 2004; Guan et al, 2009; Korzus et al, 2004; Levenson et al, 2004b; McQuown et al, 2011; Vecsey et al, 2007; Wood et al, 2006; Wood et al, 2005).
Most research into transcriptional regulation in the hippocampus has used contextual fear conditioning as the paradigm to test learning and memory (Barnes et al, 2012; Keeley et al, 2006; Levenson et al, 2004a; Mei et al, 2005). This is primarily because contextual fear conditioning produces a robust memory that has a well-defined time of acquisition due to the requirement of only a single training session (Abel & Lattal, 2001). Although this task has proven useful for dissecting the phases of memory and mapping the transcriptional landscape after learning, it also introduces a footshock that can be stressful to the animal. It is therefore important to study gene expression in other memory tasks that are more similar to the learning events that occur in daily life.
Spatial learning requires the hippocampus and can be measured using the Morris water maze, Barnes maze, or object-location memory (OLM) tasks that do not require a footshock. These spatial tasks are also known to regulate transcription in the hippocampus, including many of the same genes and processes required for contextual fear memory (Bousiges et al, 2010; Cavallaro et al, 2002; Florian et al, 2006; Fordyce et al, 1994; Haettig et al, 2011; Hawk et al, 2011; Klur et al, 2009; McNulty et al, 2012; Pittenger et al, 2002; Vogel-Ciernia et al, 2013). There is evidence that contextual and spatial learning in the hippocampus can utilize different molecular pathways (Mizuno & Giese, 2005), so gene expression may also differ after these two tasks. Like contextual fear memory, OLM is a hippocampus-dependent task (Oliveira et al, 2010). However, the targets and temporal resolution of the gene expression changes after OLM have not been thoroughly studied. The goal of this study was to investigate the transcriptional profile that occurs within the first transcriptional wave after OLM learning (Bourtchouladze et al, 1998; Igaz et al, 2002) and compare this transcriptional profile to that of contextual fear conditioning. Gene expression changes within this window after fear conditioning are typically highest 30 minutes after training and return to baseline by 2 hours (Hawk et al, 2012; Keeley et al, 2006; Peixoto et al, 2013). Using a Fluidigm HD microfluidic high-throughput qPCR system, we examined expression of 96 different candidate genes at both 30 minutes and 2 hours after OLM training in a single experiment. We found that the most commonly studied genes after fear conditioning show a similar profile after OLM. However, OLM produces long-lasting expression changes in a number of genes that are not observed after fear conditioning.
2. Methods
2.1. Subjects
Forty-two C57BL/6J mice were maintained under standard conditions with food and water available ad libitum. Adult male mice 3 months of age were kept on a 12-hr light/12-hr dark cycle with lights on at 7AM. All behavioral and biochemical experiments were performed during the light cycle with training starting at approximately 7AM (ZT0). All procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
2.2. Behavior
Object-location memory (OLM) was carried out as previously described (Hawk et al, 2011; Oliveira et al, 2010). Briefly, naïve three month old male C57Bl/6J mice were singly housed for a week and handled for 2 min/day for five consecutive days prior to tissue collection. One animal per behavioral group was trained and dissected each day for 10 total days to allow all animals to be dissected at the same circadian time. Exploration was normal in all mice used in this experiment (data not shown). One animal per training session was tested in a 24hr retrieval test the following day to ensure the training proceeded correctly. Half of the handled animals received OLM training, and half of the animals were left undisturbed on training day and were sacrificed at the same circadian time points as trained animals. On the day of training, OLM mice were given a single block of four 6 min trials with an inter-trial interval of 3 min. The animals were habituated to an empty arena with a black and white striped spatial cue on one wall in the first trial, followed by three trials of object exposure. Each mouse was exposed to three distinct objects: a rectangular metal tower, a glass bottle, and a white plastic cylinder that were arranged in a V-shaped spatial pattern in the arena. Objects were positioned in the arena with at least two inches of spacing around each object to allow free exploration of all objects. During the ITI, animals were gently removed from the arenas, and the arenas and objects were cleaned with 70% ethanol. Objects were not moved during the ITI. Immediately following the final trial, animals were gently placed in their home cage, and returned to the colony room until tissue collection.
Fear conditioning was performed as previously described (Hawk et al, 2012; Vecsey et al, 2007) with handling for 3 days prior to conditioning. Briefly, the conditioning protocol entailed a single 2-sec, 1.5mA footshock terminating at 2.5 minutes after placement of the mouse in the novel chamber. Mice were left in the chamber for an additional 30 seconds and then returned to their homecage.
2.3 RNA isolation
Hippocampi were dissected 30 minutes and 2 hours after the last training session into RNAlater (Qiagen, Valencia, CA) and frozen on dry ice. Tissue was homogenized using a TissueLyser system and RNA was extracted using the miRNeasy kit (Qiagen) according to the manufacturer's instructions.
2.4 cDNA synthesis and high-throughput qPCR
RNA concentration was determined using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA) and 1μg RNA was used in each RETROscript (Ambion, Austin, TX) cDNA synthesis reaction with random decamers, 10x RT Buffer and no heat denaturation according to the manufacturer's protocol. Concentrated cDNA was used in a specific target reaction following the manufacturer's recommendations (Fluidigm Corp. South San Francisco, CA). Briefly, Taqman assays for all 96 probes were pooled to a concentration of 0.2X (1:100) and 1.25ul of the pooled assay mix was combined with 2.5ul 2X Taqman Preamp Master Mix (Life technologies, Carlsbad, CA) and 1.25μl cDNA. Taqman probe IDs can be found in Table S1. The preamplification reaction was cycled using the following protocol in a 7500 Fast Real-Time PCR system: 10 min at 95C, then 14 cycles of 95C for 15s followed by 60C for 4min. Preamplified samples were diluted 1:5 using 1X TE. Samples were then delivered to the Molecular Profiling Core at the University of Pennsylvania, where they were run on a 96.96 Dynamic Array IFC on the Biomark HD machine (Fluidigm Corp).
For validation and the 120 minute fear conditioning experiment, cDNA reactions were diluted to 2 ng/ul in water, and real-time RT-PCR reactions were prepared in 384-well optical reaction plates with optical adhesive covers (Life technologies). Each reaction was composed of 2.25μl cDNA (2 ng/ul), 2.5μl 2x Taqman Fast Universal Master Mix (Life Technologies), and 0.25μl of Taqman probe. Reactions were performed in triplicate on the Viia7 Real-Time PCR system (Life Technologies, Carlsbad, CA).
2.5 Data analysis
High-throughput qPCR was analyzed using the Fluidigm Real Time PCR Analysis program and Microsoft Excel. Genes with at least one sample having an average Ct ≥20 were discarded as being non-expressed or failed reactions. This included Dnmt3b, Erbb2, Esrrg, Fosb, Hdac1, Hdac4, Jun, Nr6a1, Pparg, and Trdmt1, which brought the total number of genes tested to 86. Relative quantification of gene expression between groups was performed using the ΔΔCt method as described previously (Vecsey et al, 2007). The difference between each Ct and the average Ct for that gene was subtracted from the average of three housekeeper genes treated in the same manner. A p-value of 0.01 was used for significance to control for the number of t-tests performed. This p-value cutoff was chosen because we selected genes for analysis that we expected to change, and thus Bonferroni correction is too strict. This 1% chance of a type I error corresponds to one false positive per 100 t-tests. Because 86 t-tests were performed, this p-value would suggest less than 1 false positive in the data, limiting the amount of type I errors introduced by multiple testing.
3. Results
3.1 Immediate Early Genes Are Regulated 30 minutes after OLM training
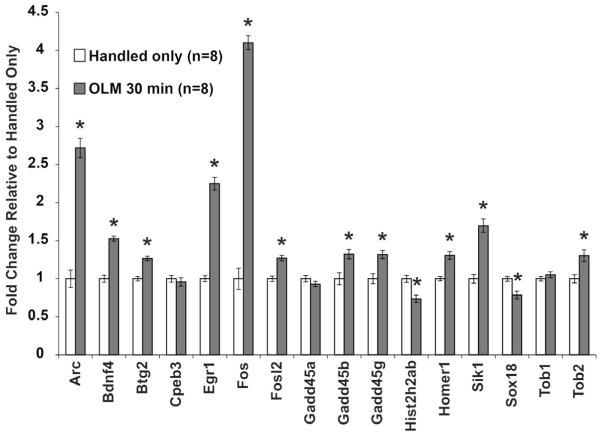
We chose sixteen representative genes that have been studied 30 minutes after fear conditioning to examine expression profiles 30 minutes after OLM training. The genes were chosen for well-studied expression changes (Arc, Bdnf4, Egr1, Fos, Homer1), genes our lab has previously studied [Fosl2 (Hawk et al, 2012), Gadd45 family (Leach et al, 2012)], or from microarray data (Btg2, Cpeb3, Histh2hab, Sik1, Sox18, Tob1, Tob2 (Peixoto et al, 2013)). cDNA samples underwent specific target amplification and were run on a 96.96 Fluidigm Biomark HD plate in triplicate (96 genes, 32 samples). The full list of Taqman assays is available in Table S1. Ten genes were excluded due to too low expression or a failed reaction, bringing the total number of genes tested to 86 (See Methods for genes). In all cases, immediate early gene (IEG) expression after OLM mirrored expression after fear conditioning (Figure 1). Previously studied genes including Arc, Bdnf4, Egr1, Fos and Homer1 were upregulated as anticipated (Arc 272% p=4.7×10−8; Bdnf4 53% p=3.3×10−7; Egr1 225% p=4.4×10−8; Fos 410% p=1.2×10−10; Homer1 31% p=1.4×10−4) (Keeley et al, 2006; Lonergan et al, 2010; Mahan et al, 2012; Mizuno et al, 2012). The probe against Homer1 recognizes both Homer1a and Homer1c, but research from our lab and others suggests that this effect is primarily due to Homer1a (Mahan et al, 2012). Further investigation is required to investigate specific Homer1 isoforms regulated by OLM. Genes that our lab discovered to be regulated after contextual fear conditioning using microarrays (Peixoto et al, 2013), including Btg2 (27% p=8.9×10−6), Hist2h2ab (−26% p=8.8×10−4), Sik1 (70% p=1.3×10−5), Sox18 (−21% p=0.002), and Tob2 (30% p=0.004) showed similar changes after OLM. The genes Gadd45b and Gadd45g showed increased expression (32% p=0.004, 32% p=0.001) while Gadd45a did not (p=0.20), as has been reported previously by our lab and others (Leach et al, 2012; Sultan et al, 2012). This observation suggests that the most commonly studied genes after contextual fear conditioning are similarly regulated after spatial behavioral tasks such as object-location memory.
Figure 1. Classic IEGs Show Expected Expression Changes after OLM Training.
Sixteen genes that are known to be induced 30 minutes after contextual fear conditioning were studied 30 minutes after OLM training. Each gene tested displayed the expression change that would be expected after contextual fear conditioning indicating these genes may represent a common transcriptional response to learning. All error bars denote s.e.m. and * indicates p<0.01.
3.2 Nuclear Hormone Receptors Display a Limited Response to OLM
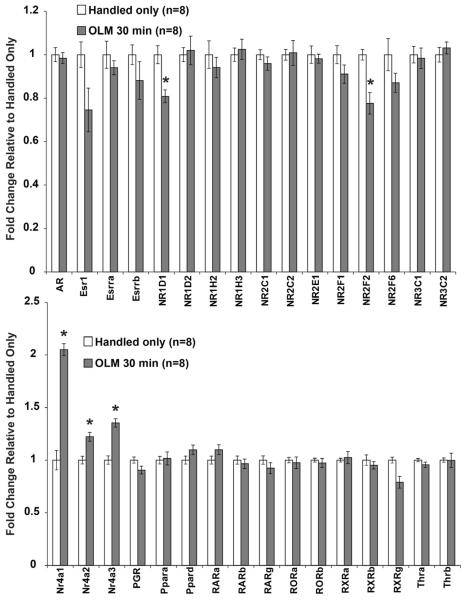
A subset of nuclear hormone receptors are known to be regulated 30 minutes after fear conditioning, including the Nr4a family of orphan nuclear receptors (Hawk et al, 2012). We tested all 37 nuclear hormone receptors that are expressed in the hippocampus for changes after OLM training (Figure 2). The Nr4a family of nuclear receptors (Nr4a1, Nr4a2, Nr4a3), which are known to be necessary for long-term fear memory (Hawk et al, 2012; McNulty et al, 2012; McQuown et al, 2011), all displayed increased expression at 30 minutes after OLM. Rev-ErbA (NR1D1), COUP-TFII (NR2F2), and retinoid X receptor gamma (Rxrg) all showed decreased expression at 30 minutes. No other nuclear receptors were observed to respond to spatial learning at this time point or at 120 minutes after OLM (data not shown). This contrasts with the large number of nuclear receptors that our lab observed to change after fear conditioning training in our previous study (Hawk et al, 2012), which included increased expression of 13 nuclear receptor genes between 30 and 120 minutes after training. These results may indicate transcriptional regulation of this class of genes depends on the training paradigm.
Figure 2. Limited Expression Changes of Nuclear Receptors after OLM training.
Because of the known involvement of the Nr4a nuclear receptor family in memory, we tested expression of all nuclear receptors expressed in the hippocampus 30 minutes after OLM training. The Nr4a family displayed increased expression after OLM, while NR1D1, NR2F2, and RXRg had reduced expression. All error bars denote s.e.m. and * indicates p<0.01.
3.3 Regulators of Transcription Show Limited Changes in Response to OLM
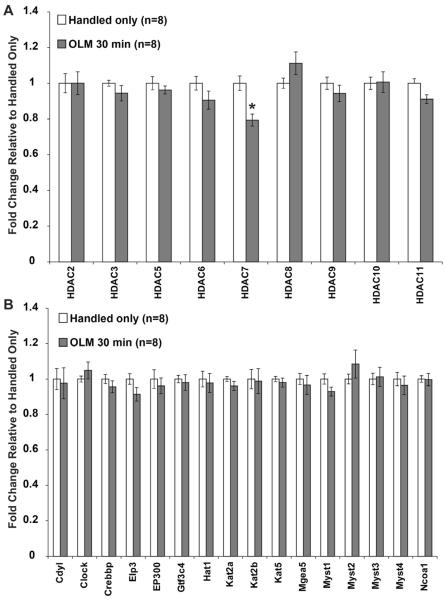
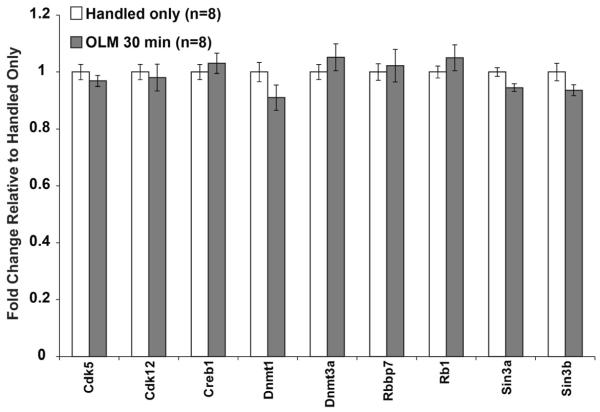
Histone acetylation is known to be a crucial regulator of transcription during memory consolidation (Alarcon et al, 2004; Barrett et al, 2011; Haettig et al, 2011; Korzus et al, 2004; Levenson et al, 2004b; McQuown et al, 2011; Vecsey et al, 2007; Wood et al, 2006; Wood et al, 2005). To test whether expression levels of histone acetylation modifying enzymes are regulated by OLM, we tested all histone deacetylases (HDACs, Figure 3A) and 16 histone acetyltransferases (HATs, Figure 3B) representing each class of enzyme, including the HATs CBP and p300 that have been shown to be essential for memory formation. The probes against Hdac1 and Hdac4 did not amplify and were discarded. None of the HATs tested showed a gene expression change, in contrast to previous reports showing changes in expression of CBP, p300 and PCAF after the Morris Water Maze (Bousiges et al, 2010). However, Hdac7 displayed reduced expression after OLM. HDAC7 is a class IIa HDAC that has not been previously linked to memory formation. This may suggest a novel role for HDAC7 in hippocampus-dependent memory formation. In addition to the regulators of histone acetylation, we chose ten genes that are known to regulate transcription in other ways. None of these genes showed any changes in transcription at 30 minutes after OLM training (Figure 4).
Figure 3. Modifiers of Histone Acetylation Display Limited Regulation after OLM Training.
Histone modifying enzymes were tested for expression changes 30 minutes after OLM training. (A) Hdac7, a class IIa HDAC, was the only family member found to change expression after OLM. (B) No HATs were observed to change expression after OLM. All error bars denote s.e.m. and * indicates p<0.01.
Figure 4. No Changes in Other Transcriptional Regulators after OLM Training.
Other genes that can regulate gene expression, including DNMTs, were tested 30 minutes after OLM training. No differences in any gene were observed. All error bars denote s.e.m. and * indicates p<0.01.
3.4 OLM Induces Longer Lasting Gene Expression Changes than Fear Conditioning
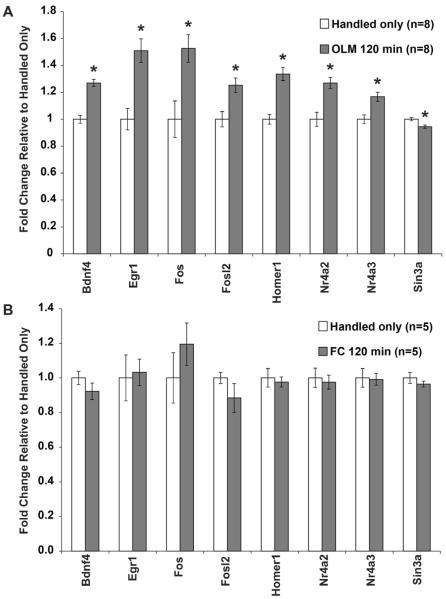
In addition to the 30 minute timepoint that has shown such robust changes after fear conditioning, we also tested hippocampal samples taken 2 hours after OLM training to investigate the persistence of these transcriptional changes. Interestingly, a number of genes that are upregulated at 30 minutes remain elevated 2 hours after OLM training. This includes highly induced genes that appear to be slowly returning to baseline, such as Egr1 and Fos, but also genes that maintain a similar level of induction as observed at 30 minutes such as Bdnf4, Fosl2, Homer1, Nr4a2 and Nr4a3 (Figure 5A). Sin3a was not changed at 30 minutes, but shows a selective change at 2 hours. The gene expression profiles at 30 minutes and 2 hours for Arc, Egr1, Fos, Nr4a1, and Nr4a2 were confirmed by standard 384-well qPCR (data not shown). To test whether these same genes show transcriptional changes after fear conditioning, we prepared cDNA from samples that were collected 2 hours after fear conditioning. None of the genes determined to change 2 hours after OLM showed a significant change 2 hours after fear conditioning (Figure 5B), indicating a long-lasting gene expression response specific to OLM.
Figure 5. OLM Training Induces Long-Lasting Changes in Gene Expression Not Seen after Fear Conditioning.
(A) Every gene was also tested 2 hours after OLM training to observe the maintenance of transcription. Genes shown in this figure are those that were changed at 2 hours after OLM, all other genes were unchanged. Sin3a was the only gene uniquely regulated at the 2 hour time point. (B) These same genes do not show gene expression changes 2 hours after contextual fear conditioning. All error bars denote s.e.m. and * indicates p<0.01.
4. Discussion
In this study, we investigated the transcriptional changes that occur in response to OLM training using powerful high-throughput qPCR technology and compared these changes to fear conditioning. In a single run, we were able to study 96 different genes in 2 different time points after OLM training with n=8 mice per group using microfluidic high-throughput qPCR. This type of throughput, flexibility, and consistency is not possible with any other qPCR technology. In addition to requiring more pipetting steps, standard qPCR would have required the same housekeepers to be run on each individual plate and limited the number of targets that could be tested. Using a high-throughput approach allowed us to reliably determine that gene expression changes after OLM last longer than similar expression changes after contextual fear.
Our study discovered that commonly studied IEGs, such as Fos and Arc, show similar expression differences after fear conditioning and after OLM, indicating overlap between contextual and spatial learning. In a previous study from our lab (Hawk et al, 2012), we found that a number of nuclear receptors exhibit increased expression after contextual fear conditioning. Our current findings suggest a more limited regulation of this class of genes after OLM. It is unclear whether the wider regulation after fear conditioning is in response to the footshock or whether the timecourse of expression after OLM is different. As seen after fear conditioning, all 3 members of the Nr4a family of orphan nuclear receptors were upregulated after OLM. However, while Nr4a1 returned to baseline by 2 hours, Nr4a2 and Nr4a3 did not, suggesting that different processes may regulate Nr4a1 than the other two family members. Future studies will aim to determine how expression increases of Nr4a2 and Nr4a3 are maintained after OLM training.
It is interesting to note that Hdac7 and Sin3a are regulated by OLM while HATs are not. This may suggest that relieving the negative repression of histone acetylation is a crucial step for long-term memory formation. Although class I HDACs have been heavily implicated in learning and memory (Bahari-Javan et al, 2012; Guan et al, 2009; Hawk et al, 2011; McQuown et al, 2011), class IIa HDACs have received less attention. A study by Agis-Balboa et al. demonstrated that loss of the class IIa member HDAC5 impairs spatial memory (Agis-Balboa et al, 2013), but those experiments used a complete knockout mouse line that has the potential for developmental or extrahippocampal effects. Our study is the first to observe changes in Hdac7 in response to learning in the hippocampus.
The most intriguing finding of this study was the long-lasting regulation of gene expression 2 hours after OLM, something that is not seen after fear conditioning. It might be expected that the fear of a footshock would produce a stronger transcriptional response in the hippocampus than would the spatial rearrangement of objects. There are a number of potential causes for this disparity, although the most likely explanation is that the multiple training sessions required for OLM induce a stronger response than the single shock training used by our lab for fear conditioning. It would be inter esting to test whether a multiple shock fear conditioning protocol induces longer lasting gene expression changes. Also, there could be an association between the novel context and the novel objects formed during OLM training that is not present in fear conditioning. Testing mice in the context only, introducing novel objects, or altering the number of training trials could determine whether these changes are sufficient to elicit gene expression changes. Further, different molecular mechanisms may regulate contextual and spatial learning (Mizuno & Giese, 2005). Future studies can test for changes at the protein level, although mRNA and protein levels generally agree after learning (Stanciu et al, 2001; Steward et al, 1998). Additional investigation into later time points after OLM training will be required to see if gene expression changes that occur well after fear conditioning (Mizuno et al, 2012) also exist after OLM. It is interesting that not all genes with increased expression at the 30 minute timepoint remain elevated for 2 hours after OLM. Future studies will determine whether specific epigenetic modifications regulate this longer term maintenance of gene expression at particular genes.
Supplementary Material
Highlights.
Microfluidic technology allows for large-scale qPCR experiments
Object-location memory increases IEG expression similarly to fear conditioning
Object-location memory induces gene expression changes that last at least 2 hours
Acknowledgments
We thank the Molecular Profiling Core at UPENN, particularly Kathakali Addya, for loading and running the Fluidigm plate. We thank Morgan Bridi and Robbert Havekes for constructive discussions and critical reading of the manuscript. This research was supported by NIH R01MH087463 and S10RR027374-01A1 to T.A. The Fluidigm loading and running was supported by the Penn Center for Musculoskeletal Disorders, Award Number P30AR050950 from the National Institute of Arthritis, Musculoskeletal and Skin Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Arthritis, Musculoskeletal and Skin Diseases or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abel T, Lattal KM. Molecular mechanisms of memory acquisition, consolidation and retrieval. Curr Opin Neurobiol. 2001;11:180–187. doi: 10.1016/s0959-4388(00)00194-x. [DOI] [PubMed] [Google Scholar]
- Agis-Balboa RC, Pavelka Z, Kerimoglu C, Fischer A. Loss of HDAC5 impairs memory function: implications for Alzheimer's disease. Journal of Alzheimer's disease : JAD. 2013;33:35–44. doi: 10.3233/JAD-2012-121009. [DOI] [PubMed] [Google Scholar]
- Agranoff BW, Davis RE, Casola L, Lim R. Actinomycin D blocks formation of memory of shock-avoidance in goldfish. Science. 1967;158:1600–1601. doi: 10.1126/science.158.3808.1600. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Bahari-Javan S, Maddalena A, Kerimoglu C, Wittnam J, Held T, Bahr M, Burkhardt S, Delalle I, Kugler S, Fischer A, Sananbenesi F. HDAC1 regulates fear extinction in mice. J Neurosci. 2012;32:5062–5073. doi: 10.1523/JNEUROSCI.0079-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P, Kirtley A, Thomas KL. Quantitatively and qualitatively different cellular processes are engaged in CA1 during the consolidation and reconsolidation of contextual fear memory. Hippocampus. 2012;22:149–171. doi: 10.1002/hipo.20879. [DOI] [PubMed] [Google Scholar]
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA. Hippocampal Focal Knockout of CBP Affects Specific Histone Modifications, Long-Term Potentiation, and Long-Term Memory. Neuropsychopharmacology. 2011 doi: 10.1038/npp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchouladze R, Abel T, Berman N, Gordon R, Lapidus K, Kandel ER. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn Mem. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- Bousiges O, Vasconcelos AP, Neidl R, Cosquer B, Herbeaux K, Panteleeva I, Loeffler JP, Cassel JC, Boutillier AL. Spatial memory consolidation is associated with induction of several lysine-acetyltransferase (histone acetyltransferase) expression levels and H2B/H4 acetylation-dependent transcriptional events in the rat hippocampus. Neuropsychopharmacology. 2010;35:2521–2537. doi: 10.1038/npp.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro S, D'Agata V, Manickam P, Dufour F, Alkon DL. Memory-specific temporal profiles of gene expression in the hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16279–16284. doi: 10.1073/pnas.242597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian C, Mons N, Roullet P. CREB antisense oligodeoxynucleotide administration into the dorsal hippocampal CA3 region impairs long- but not short-term spatial memory in mice. Learn Mem. 2006;13:465–472. doi: 10.1101/lm.249306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordyce DE, Bhat RV, Baraban JM, Wehner JM. Genetic and activity-dependent regulation of zif268 expression: association with spatial learning. Hippocampus. 1994;4:559–568. doi: 10.1002/hipo.450040505. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haettig J, Stefanko DP, Multani ML, Figueroa DX, McQuown SC, Wood MA. HDAC inhibition modulates hippocampus-dependent long-term memory for object location in a CBP-dependent manner. Learn Mem. 2011;18:71–79. doi: 10.1101/lm.1986911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk JD, Bookout AL, Poplawski SG, Bridi M, Rao AJ, Sulewski ME, Kroener BT, Manglesdorf DJ, Abel T. NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. The Journal of clinical investigation. 2012;122:3593–3602. doi: 10.1172/JCI64145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk JD, Florian C, Abel T. Post-training intrahippocampal inhibition of class I histone deacetylases enhances long-term object-location memory. Learn Mem. 2011;18:367–370. doi: 10.1101/lm.2097411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Vianna MR, Medina JH, Izquierdo I. Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. J Neurosci. 2002;22:6781–6789. doi: 10.1523/JNEUROSCI.22-15-06781.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley MB, Wood MA, Isiegas C, Stein J, Hellman K, Hannenhalli S, Abel T. Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus. Learn Mem. 2006;13:135–142. doi: 10.1101/lm.86906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klur S, Muller C, Pereira de Vasconcelos A, Ballard T, Lopez J, Galani R, Certa U, Cassel JC. Hippocampal-dependent spatial memory functions might be lateralized in rats: An approach combining gene expression profiling and reversible inactivation. Hippocampus. 2009;19:800–816. doi: 10.1002/hipo.20562. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach PT, Poplawski SG, Kenney JW, Hoffman B, Liebermann DA, Abel T, Gould TJ. Gadd45b knockout mice exhibit selective deficits in hippocampus-dependent long-term memory. Learn Mem. 2012;19:319–324. doi: 10.1101/lm.024984.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberger T, Parkitna JR, Chai M, Schutz G, Engblom D. CREB has a context-dependent role in activity-regulated transcription and maintains neuronal cholesterol homeostasis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22:2872–2879. doi: 10.1096/fj.08-107888. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Choi S, Lee SY, Cao YA, Ahn HJ, Worley KC, Pizzi M, Liou HC, Sweatt JD. A bioinformatics analysis of memory consolidation reveals involvement of the transcription factor c-rel. J Neurosci. 2004a;24:3933–3943. doi: 10.1523/JNEUROSCI.5646-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. The Journal of biological chemistry. 2004b;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Lonergan ME, Gafford GM, Jarome TJ, Helmstetter FJ. Time-dependent expression of Arc and zif268 after acquisition of fear conditioning. Neural plasticity. 2010;2010:139891. doi: 10.1155/2010/139891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan AL, Mou L, Shah N, Hu JH, Worley PF, Ressler KJ. Epigenetic modulation of Homer1a transcription regulation in amygdala and hippocampus with pavlovian fear conditioning. J Neurosci. 2012;32:4651–4659. doi: 10.1523/JNEUROSCI.3308-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty SE, Barrett RM, Vogel-Ciernia A, Malvaez M, Hernandez N, Davatolhagh MF, Matheos DP, Schiffman A, Wood MA. Differential roles for Nr4a1 and Nr4a2 in object location vs. object recognition long-term memory. Learn Mem. 2012;19:588–592. doi: 10.1101/lm.026385.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, Wood MA. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei B, Li C, Dong S, Jiang CH, Wang H, Hu Y. Distinct gene expression profiles in hippocampus and amygdala after fear conditioning. Brain Res Bull. 2005;67:1–12. doi: 10.1016/j.brainresbull.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Dempster E, Mill J, Giese KP. Long-lasting regulation of hippocampal Bdnf gene transcription after contextual fear conditioning. Genes, brain, and behavior. 2012;11:651–659. doi: 10.1111/j.1601-183X.2012.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Giese KP. Hippocampus-dependent memory formation: do memory type- specific mechanisms exist? Journal of pharmacological sciences. 2005;98:191–197. doi: 10.1254/jphs.crj05005x. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Hawk JD, Abel T, Havekes R. Post-training reversible inactivation of the hippocampus enhances novel object recognition memory. Learn Mem. 2010;17:155–160. doi: 10.1101/lm.1625310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto L, Wimmer M, Poplawski SG, N.R. Z, Abel T. Neuroscience 2013 Abstracts. San Diego, CA: 2013. Transcriptome analysis reveals differences in processes that regulate gene expression during memory consolidation and retrieval. [Google Scholar]
- Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S, Kandel ER. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron. 2002;34:447–462. doi: 10.1016/s0896-6273(02)00684-0. [DOI] [PubMed] [Google Scholar]
- Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, Otto T, Lin Y. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334:1669–1675. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanciu M, Radulovic J, Spiess J. Phosphorylated cAMP response element binding protein in the mouse brain after fear conditioning: relationship to Fos production. Brain Res Mol Brain Res. 2001;94:15–24. doi: 10.1016/s0169-328x(01)00174-7. [DOI] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic deletion of Gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J Neurosci. 2012;32:17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel-Ciernia A, Matheos DP, Barrett RM, Kramar EA, Azzawi S, Chen Y, Magnan CN, Zeller M, Sylvain A, Haettig J, Jia Y, Tran A, Dang R, Post RJ, Chabrier M, Babayan AH, Wu JI, Crabtree GR, Baldi P, Baram TZ, Lynch G, Wood MA. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat Neurosci. 2013;16:552–561. doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Mem. 2006;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.