Abstract
A pluripotent state of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) is maintained through the combinatorial activity of core transcriptional factors (TFs) such as Oct4, Sox2, and Nanog in conjunction with many other factors including epigenetic regulators. Proteins rarely act alone, and knowledge of protein-protein interaction network (interactome) provides an extraordinary resource about how pluripotency TFs collaborate and crosstalk with epigenetic regulators in ESCs. Recent advances in affinity purification coupled with mass spectrometry (AP-MS) allow for efficient, high-throughput identification of hundreds of interacting protein partners, which can be used to map the pluripotency landscape. Here we review recent publications employing AP-MS to investigate protein interaction networks in ESCs, discuss how protein-protein connections reveal novel pluripotency regulatory circuits and new factors for efficient reprogramming of somatic cells.
Introduction
Pluripotency, the capacity to differentiate into all cell types, is a defining property of embryonic stem cells (ESCs). The undifferentiated state of ESCs is maintained by a set of pluripotency factors [1]. Forced expression of these factors (i.e., the Yamanaka factors [2] Oct4, Sox2, Klf4, and c-Myc, OSKM) can reprogram lineage-committed cells back to an ESC-like state (called induced pluripotent stem cells, iPSCs), providing extraordinary potential for regenerative medicine [3,4]. Transcriptional cooperation and their regulatory networks among the pluripotency factors such as Oct4, Sox2, and Nanog have been extensively studied in ESCs [5,6]. Given that the proteins rarely act alone, the physical protein-protein interaction (PPI) networks of pluripotency factors should provide valuable information about how the pluripotent state is established and maintained. Here we review the recent advances in pluripotency interactome studies on understanding the intricate protein interaction networks and protein complexes surrounding several critical pluripotency factors. Three aspects are discussed in detail: the technology behind protein mass spectrometry to investigate PPIs, the emerging insights on the extended ESC protein interactome, and how the pluripotency interactome reveals novel factors for efficient somatic cell reprogramming.
Methods to study protein-protein interactions
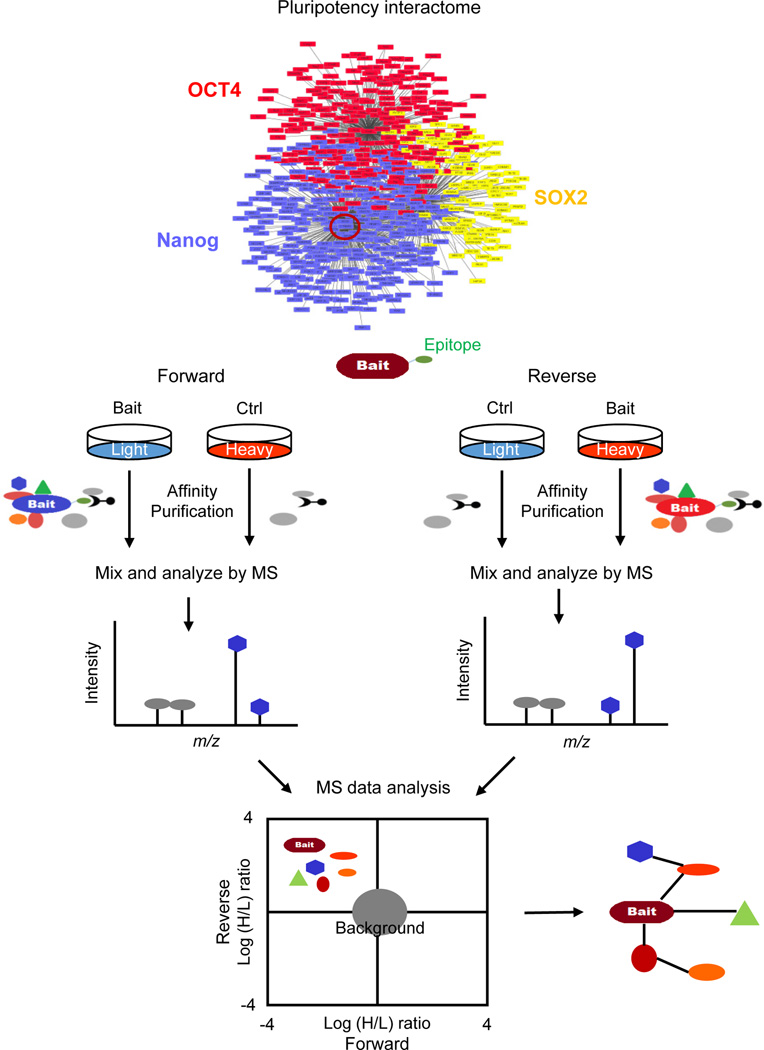
A number of methods have been developed to examine the binary PPIs in ESCs (reviewed in ref [7]). Among those approaches, affinity purification coupled with mass spectrometry (AP-MS) has become the method of choice [8]. The conceptual view of an interactome study using AP-MS is described in Figure 1. Five years ago, label-free approaches were used to compare the abundance of purified proteins by counting the number of detected peptides. Recently, due to the advent of high-accuracy MS, stable isotope labeling approaches (such as stable isotope labeling by amino acids in cell culture, SILAC) are being employed, yielding increasing robustness and information content of quantitative proteomics data [8]. Biological replicates with forward and reverse (swapped labeling) SILAC experiments are usually needed to further enhance confidence of protein interactions [9]. Conventional protein co-immunoprecipitation assays are also necessary to validate the interacting candidates.
Figure 1.
Strategy of interactome study using affinity purification coupled with mass spectrometry (AP-MS). A bait protein (indicated as the red circle) in pluripotency interactome with an epitope (either endogenous or a tag-conjugated protein) can be specifically recognized and purified in AP. Salt concentration during purification is important. High salt concentration will remove relatively weak protein-protein interactions or protein-mediated indirect interactions. Endonuclease such as benzonase is used to destroy the DNA/RNA-mediated protein-protein interactions. The abundance of purified protein is determined by MS in two strategies. In label-free method, protein abundance is quantified by counting the number of detected peptides. In stable isotope labeling method (shown in the figure), the lysates are combined together after AP in order to prevent potential heavy to light exchange of specific transiently interacting partners during the purification. Then protein abundance is quantified from the profiles of co-eluting light and heavy peptides, representing the relative amount of proteins from the bait- and control-AP, respectively. A biological replicate is recommended with label swapping (reverse experiment). Specific interactors by definition show a negative log heavy-versus-light (H/L) ratio in forward and a positive log (H/L) ratio in reverse experiments, whereas unspecific or background binders show a log (H/L) ratio close to 0 in both experiments. Finally, a protein-protein interaction network is constructed from the interactors surrounding the bait protein using a visualization program such as Cytoscape.
Pluripotency protein interactome in ESCs
The published pluripotency interactome centered on TFs and epigenetic regulators in ESCs is summarized in Table 1. In 2006, the first comprehensive interactome was conducted on a number of pluripotency proteins with a particular focus on Nanog in mouse ESCs [10]. Unlike the other pluripotency factors such as Oct4 and Sox2, which are uniform in all undifferentiated ESCs, Nanog expression is relatively heterogeneous [11]. Depletion of Nanog immediately decreases self-renewal efficiency of ESCs and leads to cellular differentiation [12]. With a high-affinity biotin/streptavidin (Bio/SA) purification combined with a high-salt eluting method, Wang et al identified 17 Nanog interactors with high confidence. The interaction network was expanded for the identified Nanog interactors such as Dax1 (Nr0b1), Nac1, Zfp281, Oct4, and Rex1 with the same AP-MS strategy [10]. The Nanog interactome was further extended in later studies by using biotin-, Flag tag-, and endogenous Nanog antibody-based affinity approaches [13–15]. With an improved tagging method and less stringent purification, Gagliardi et al [14] identified 130 Nanog interactors and dramatically expanded our knowledge of the Nanog interactome in ESCs. Many of the proteins identified are components of protein complexes involved in different machineries, especially in epigenetic regulation and chromatin remodeling. Interestingly, protein complexes with opposing functions are observed to interact with Nanog, such as histone acetyltransferase complexes (Tip60-p400, also called NuA4-HAT) and deacetylase complexes (NuRD, N-CoR, Sin3a); the histone 3 lysine 4 methyltransferase (MLL) complex and the LSD1 demethylase complex. All these data suggest a Nanog-dependent epigenetic regulation of distinct activated and repressed loci in pluripotent cells.
Table 1.
Summary of the published interactome studies in ESCs.
| Study | Bait proteins | Cell resource | Affinity Purification (AP) methods |
Important interactors and its function |
|---|---|---|---|---|
| Wang et al, 2006 [10] | Nanog, Dax1, Nac1, Zfp281, Oct4, Rex1 | Mouse ESCs | Biotin/streptavidin (Bio/SA) -AP | Survey study |
| van den Berg et al, 2008 [16] | Oct4 | Mouse ESCs | Flag-AP | Esrrb; positively regulate Nanog expression |
| Liang et al, 2008 [15] | Nanog | Mouse ESCs | Endogenous antibody-AP | NODE complex; represses ESC differentiation |
| Ho et al, 2009 [24] | Brg1 | Mouse ESCs | Endogenous antibody-AP | esBAF complex; a ES-sepcific BAF complex essential for self-renewal |
| Shen et al, 2009 [64] | Ezh1, Ezh2, Eed | Mouse ESCs | Double-step Flag-and Bio/SA-AP | Jmj (Jarid2); interacts with PRC2 complex and fine-tunes H3K27me3 |
| Pasini et al, 2010 [65] | Suz12, Jarid2 | AP-MS in Hela and 293T cells, validated in ESCs | Flag-AP (Suz12), double-step Flag- and HA-AP (Jarid2) | Jarid2; interacts with PRC2 complex and maintains H3K27me3 |
| Kim et al, 2010 [22] | Myc, Max, Dmap1, Tip60, Gcn4, E2F4 | Mouse ESCs | Bio/SA-AP | Survey study; defines three independent (core, PRC, and Myc) modules in ESCs |
| van den Berg et al, 2010 [18] | Oct4, Sall4, Dax1, Tcfcp2l1, Esrrb | Mouse ESCs | Flag-AP | Survey study |
| Pardo et al, 2010 [19] | Oct4 | Mouse ESCs | Flag-AP | Survey study |
| Mallanna et al, 2010 [66] | Sox2 | Early differentiating mouse ESCs | Flag-AP | Sox21; triggers differentiation |
| McDonel et al, 2012 [42] | Sin3a | Mouse ESCs | Flag-AP | Survey study |
| Ding et al, 2012 [20] | Oct4 | Mouse ESCs | Bio/SA-AP | Survey study |
| Fidalgo et al, 2012 [38] | Zfp281 | Mouse ESCs | Endogenous antibody-AP | NuRD complex; mediates Nanog autorepression |
| Gao et al, 2012 [67] | Sox2 | Mouse ESCs | Flag-AP | Survey study; Smarcd1, represses ESCs differentiation |
| Lai et al, 2012 [68] | Sox2 | Mouse ESCs | Flag-AP | Parp1; negatively regulates Sox2 in response to FGF signaling |
| Costa et al, 2013 [13] | Nanog | Mouse ESCs | Single-step Flag-AP, Bio/SA-AP, or Endogenous antibody-AP | Tet1; facilitates reprogramming |
| Gagliardi et al, 2013 [14] | Nanog | Mouse ESCs | Flag-AP | Survey study; domain-mapping of Nanog/Sox2 interaction |
| Chen et al, 2013 [41] | Tet2, Tet3 | AP-MS in 293T cells, validated in ESCs | SA-binding peptide/SA-AP | Ogt; mediates histone 2B Ser112 GlcNAcylation |
| Shi et al, 2013 [40] | Tet1 | Mouse ESCs | Endogenous antibody-AP | Ogt; modifies Tet1 by GlcNAC and positively regulates Tet1 expression |
| Vella et al, 2013 [43] | Ogt | Mouse ESCs | Double-step Flag-and Bio/SA-AP | Tet1; recruits Ogt to chromatin |
| Yakulov et al, 2013 [69] | β-Catenin | Mouse ESCs | Endogenous antibody-AP | Lsd1; is recruited by β-Catenin to repress Lefty1 |
Oct4 is another key pluripotency factor in the transcriptional regulatory network in ESCs. Orphan nuclear receptor estrogen-related beta (Esrrb or Err2) was initially identified as an Oct4 partner [16]. Esrrb can recruit the Oct4/Sox2 heterodimer to the Nanog proximal promoter, and positively regulates Nanog expression. Together with other reports demonstrating that Nanog interacts with Esrrb and regulates its transcription [10,17], a feedback regulatory loop has been suggested wherein Oct4/Esrrb modulates Nanog expression and ESC pluripotency. An interactome centered by Oct4 was recently reported in three independent studies (van der Berg et al [18], Pardo et al [19], and Ding et al [20]). Because the levels of Oct4 are critical in controlling the undifferentiated state, ectopic expression of tagged Oct4 may affect the self-renewal capacity of ESCs [21]. Both Ding’s and van den Berg’s studies used ZHBTc4 mESCs, in which both Oct4 alleles have been replaced and Oct4 expression is directed from a doxycycline-suppressible transgene [21], followed by re-introduction of either biotin- [20] or Flag-tagged [18] Oct4 in ZHBTc4 cells. There are 18 proteins identified consistently in three Oct4 interactomes, increasing the overall biological significance of those proteins. They are either involved in the NuRD (Chd4, Gatad2a, Gatad2b, Mta2, Mta3, Mbd3, Hdac1), SWI/SNF (also called BAF complex, Brg1, Baf155), or LSD1 (Lsd1, Rcor2) complexes, or they are individual TFs (Sall1, Sall4, Hcfc1, Hells, etc) playing important roles in ESCs. Interestingly, there is a high degree of consistency and numerous common transcriptional and epigenetic regulators revealed in both Oct4 and Nanog interactomes, which raises an important issue on how ubiquitous chromatin modifiers interact with pluripotency TFs to modulate an ESC-specific gene expression profile to maintain pluripotency.
In addition to the core pluripotency interactome pivoted by Oct4 and Nanog, Kim et al identified a distinct PPI network associated with c-Myc [22]. A notable finding from the Myc interactome is the interaction with the Tip60-p400 complex. An RNA interference (RNAi) screen revealed that Tip60-p400 is essential to maintain the pluripotent state, whereas inhibition of the components in the Tip60-p400 complex leads to differentiation [23]. Interestingly, the gene expression profiles upon Tip60 and p400 knockdown (KD) highly overlap with that of Nanog KD and are enriched for developmental regulators [23]. Together with the evidence that the Tip60-p400 complex is a Nanog [14] but probably not an Oct4 interactor (the Tip60-p400 complex was identified only in one of the three Oct4 interactome studies, and wasn’t validated by co-immunoprecipitation [18]), a unique Nanog function of epigenetic regulation that differs from Oct4 is suggested.
ESC-specific protein complexes identified by AP-MS
Several structurally specialized protein complexes in ESCs were explored because AP-MS is able to identify individual protein components. A striking finding is the identification of the ESC-specific BAF complex (esBAF) [24], which has been shown to be critical for self-renewal and maintenance of the ESC state. Using a stringent AP condition, it was demonstrated that esBAF contains Brg1, BAF155, BAF60A, but not other known BAF components such as Brm, BAF170, and BAF60C. The esBAF complex was shown to functionally interact with Oct4 and Sox2 and repress developmental genes by co-localizing genes with Oct4/Sox2 at the genomic loci, thus refining the core pluripotency circuitry [24,25]. BAF155 is also a component of the first Nanog interactome in mouse ESCs [10].
Another important ESC-specific protein complex is the Nanog/Oct4-associated deacetylase (NODE) complex identified by Nanog- and Oct4 AP-MS studies [15]. NODE lacks Chd4 and Mbd3, which are considered to be essential subunits of the canonical NuRD complex. However, both Chd4 and Mbd3 were later identified in the Oct4 interactome [18–20] as well as in the Nanog interactome ([14] and our unpublished data). Discrepancy of observations may be due to the fact that the NODE complex was purified by an endogenous antibody with low affinity and specificity, while the following studies used tagged Oct4 or Nanog for high affinity purification. Importantly, histone deacetylase activity is preserved in Mbd3−/− ESCs. KD of Mta1, a common subunit of NuRD and NODE complexes, resulted in upregulation of differentiation genes, which is distinct from that of Mbd3 KD [15]. Although it is suggested that both complexes are functionally important in ESCs, whether NODE is simply an experimental artifact, and if not, how the NODE and NuRD complexes assemble and distinctly interact with Nanog and Oct4, remains to be determined.
In addition, a nucleotide excision repair (NER) complex containing XPC, RAD23B, and CETN2 was uncovered as an Oct4/Sox2-dependent stem cell coactivator (SCC) complex, which is necessary for transcriptional activation of Nanog in vitro [26]. The SCC/XPC complex was isolated from a high-salt fraction of a multi-step chromatography purification procedure and identified by MS. It is worth noting that although interaction of Oct4 and SCC was validated in 293T cells by overexpression, a stable interaction in ESCs with endogenous levels of those proteins was unable to be reproduced [26]. Consistently, none of the subunits in the SCC complex was identified in the interactome studies of Oct4, suggesting that functional coactivator-activator interactions can often be weak and transient [27].
Pluripotency PPI network guides efficient somatic cell reprogramming (SCR)
Since the initial discovery of iPSCs induced by forced expression of "Yamanaka factors" Oct4, Sox2, Klf4, and c-Myc [2], the dynamics and molecular mechanisms of SCR have been extensively elucidated [28,29]. It is believed that forced expression of these genes perturbs various epigenetic processes leading to activation of the core pluripotency genes in reprogramming cells. Therefore, pluripotency interactome studies could strongly implicate how the ESC-like state is established and uncover many “necessities”, “drivers”, and “enhancers” of cellular reprogramming. For instance, the SCC/XPC complex is a requisite for reprogramming, as SCR efficiency is highly compromised in MEFs derived from XPC and RAD23B knockout mice [26]. Esrrb, an Oct4/Nanog interactor, was demonstrated as a driver of SCR capable of replacing Klf4 and c-Myc in the OSKM cocktail of reprogramming [30]. Moreover, combined overexpression of the Oct4/Nanog interactor esBAF components Brg1 and BAF155 has been reported to enhance OSK-induced reprogramming of fibroblasts [31]. Mechanistically, it was shown that Brg1 physically interacts with a conserved linker region between the POU-specific domain and the POU homeodomain of Oct4. A point mutation at this linker region abolishes Oct4 activity during reprogramming, suggesting an important role of Oct4 in recruiting key epigenetic regulators to the genomic sites occupied by Oct4 [32].
The Mbd3/NuRD complex also associates with Oct4 and Nanog, as well as other important pluripotency factors such as Nac1, Sall4, and Zfp281 [10]. NuRD is required to modulate ESC heterogeneity by repressing the pluripotency genes; therefore it is considered a barrier of SCR [33,34]. Depletion of Mbd3 significantly enhances the reprogramming efficiency to a deterministic extend [35]. In stark contrast, a recent study by dos Santos et al [36] revealed an opposite functional contribution of NuRD complex to the reprogramming process. Depletion of Mbd3 abrogates epiblast stem cell (EpiSC) and pre-iPSC reprogramming but has a minimal effect on MEF reprogramming. Ectopic Mbd3 together with Nanog expression synergistically promotes EpiSC/pre-iPSC reprogramming [36]. The positive function of NuRD in reprogramming also finds its support from another study showing that the NuRD complex is recruited by Sox2 to repress mTOR expression and induce autophagy in promoting reprogramming [37]. It is currently unclear what the underlying cause for such discrepancies is, and thus future studies are needed to clarify the function of Mbd3/NuRD complex in the context of reprogramming. Interestingly, the Nanog-associated protein Zfp281 was shown to recruit the NuRD complex to the Nanog locus to restrict its expression in maintaining optimal self-renewal of ESCs and during late stages of the pre-iPSC to iPSC transition. KD of Zfp281 enhances the efficiency in this pre-iPSC reprogramming model by upregulating Nanog expression [38].
In an extended Nanog interactome study, association of Nanog and the ten-eleven translocation (TET) family of dixoygenase proteins uncovered a critical role of Nanog in recruiting Tet1 to the core pluripotency network [13]. Tet1-mediated demethylation is linked to the activation of pluripotency loci in the late phase of reprogramming, in accordance with the reorganization of the DNA methylation landscape at this stage [29]. Recently, a set of interactome studies on TET family proteins (Tet1/2/3) revealed a number of important interactors related to TET functions, such as the Sin3a-Hdac complex and O-GlcNAc transferase (Ogt) [39–41]. These interactions were also confirmed in the Sin3a interactome [42] and the Ogt interactome [43], respectively. Although discrepancy of observations was reported, it is believed that all TET family members interact with Ogt [39,44]. Ogt was shown to positively regulate SCR, while KD of Ogt decreases efficiency of OSK-driven MEF reprogramming [45]. Furthermore, Ogt interacts with and O-GlcNAcylates Oct4 and Sox2 post-transcriptionally. Modification of O-GlcNAc at T228 of Oct4 is functionally critical, as a point mutation at this site abolishes the effects of Oct4 in MEF reprogramming [45,46].
Perspectives
PPI is context sensitive and detection of interacting partners heavily dependents on the experimental procedures (i.e., sample treatment, salt concentration in AP) and the intrinsic abundance of interactors. Proteins can directly contact, or indirectly associate with each other through DNA or RNA. For instance, the Sox2-Oct4 interaction is DNA-dependent and must be stabilized by UV cross-linking [47,48]. Furthermore, it was reported that the long intergenic noncoding RNAs (lincRNAs) play an important role in recruiting histone modifying complexes to genomic loci and mediating protein interaction [49,50]. Therefore, pretreating samples with benzonase is recommended only if the protein-protein interaction is investigated. Since an AP-MS experiment may not be able to discover all interactors of the bait protein, alternative methods can be used to complement the PPI network, such as high throughput yeast two-hybrid assays, domain-domain interactions, and protein microarray [51].
Given that most pluripotency factors are TFs, one urgent task for interactome studies is to ascertain transcriptional control in ESCs [52]. Together with the technique of chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq), interactomes interrogate how pluripotency TFs recruit co-activators/repressors and change the local epigenetic pattern and chromatin structure [53]. Genome-wide co-localizations of many interacting partners have been reported in ESCs, such as Nanog and its interactors Nac1, Dax1, and Zfp281 [53], Oct4 and Wdr5 [54], Tet1 and Sin3a [55], Tet1/2 and Ogt [39,41,43], and Hdac1/2 and NuRD [56]. Interestingly, ChIP-Seq also tells how a shared subunit functions in different protein complexes. For instance, Hdac1/2 are the subunits in both NuRD and Sin3a complexes. In mouse ESCs, Hdac1/2 showed a higher level of genomic co-localization with the NuRD than with the Sin3a complex (our unpublished data). NuRD represses the pluripotency TFs, while Sin3a was shown to positively regulate Sox2 and Nanog expression [57]. Therefore the Hdac activity mainly associated with the NuRD complex, a reprogramming barrier, is in line with the evidence that treatment of Hdac inhibitors accelerates SCR [58].
By now, most of the interactome studies on the pluripotency network are in mouse ESCs. Although the mouse is the most important organism for generating hypotheses in the field of stem cell biology, differences across species exist. Human ESCs are more similar to mouse EpiSCs in a primed state of pluripotency [59]. Obstacles in studying the human interactome include the high cost of maintaining human ESCs, lacking high affinity endogenous protein antibodies, and difficulty in genomic engineering of human cells. The advance of zinc-finger nuclease (ZFN) [60] and transcription activator-like effector nuclease (TALEN) [61] techniques comprise a powerful tool to manipulate human genes. Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system has shown great promise and flexibility for genetic engineering by multiplexed disruption and targeted integration of human genes [62,63]. High affinity pull-down is feasible by fusing epitope tags to the pluripotency factors at their endogenous loci.
As the functional players, protein expression and their interaction network delineate how they act individually and together in the processes of self-renewal and maintenance of pluripotency (Figure 2). Pluripotency protein interactome also provides a framework for exploring the new factors that may permit faithful reprogramming of somatic cells. Technically, further developments of interactome studies will continue to focus on increasing the sensitivity of AP-MS and reducing the required amount of proteins from a rare population of cells. A better understanding of the pluripotency machinery in ESCs awaits efforts on both systematic discovery of new interactors and sophisticated functional studies for those candidates.
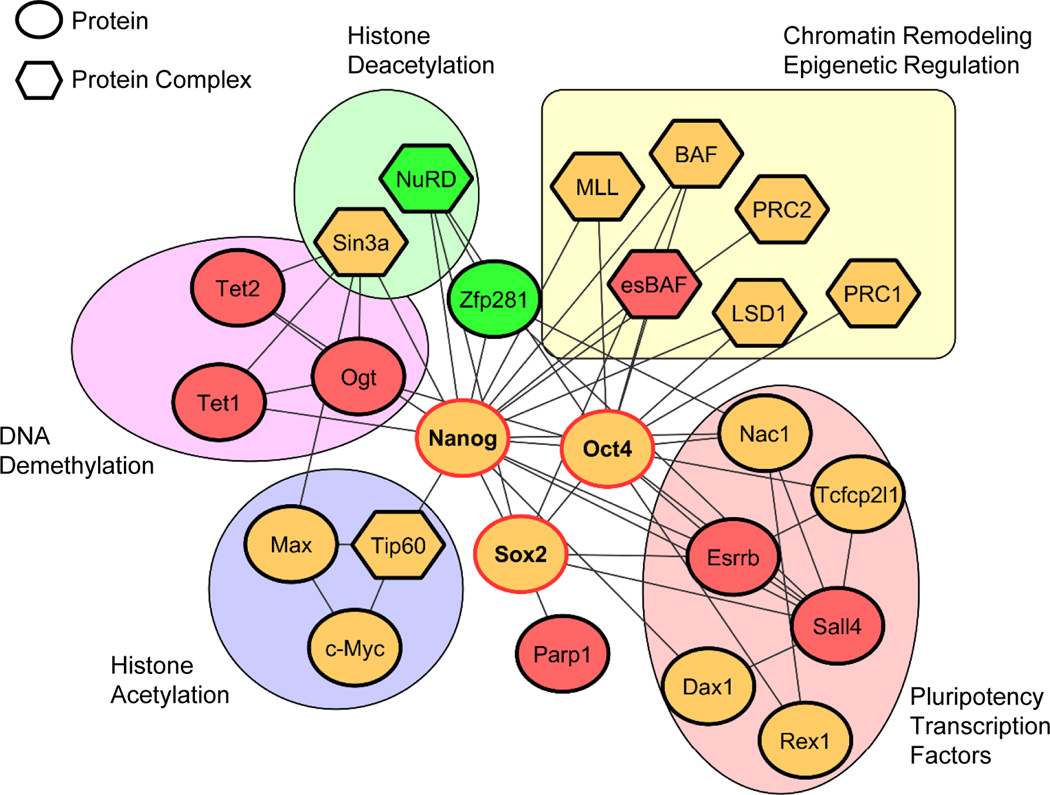
Figure 2.
An extended pluripotency interactome in ESCs. The network is an integrated view from multiple published interactome studies in mouse ESCs. Proteins (eclipses) and/or protein complexes (hexagons) are connected with solid lines, indicating physical associations with each other. The core pluripotency factors are indicated with bold text and red borders. Red-color filled shapes indicate the proteins that facilitate or replace one of the Yamanaka factors during somatic cell reprogramming. Green-color filled shapes indicate the barrier proteins of reprogramming. The proteins and protein complexes in the pluripotency interactome are classified into groups such as pluripotency transcription factors, histone acetylation, histone deacetylation, DNA demethylation, and other chromatin remodeling and epigenetic regulation complexes.
Acknowledgements
We are grateful to Arven Saunders and Francesco Faiola in our lab for critical reading and comments on the manuscript. The research in the Wang laboratory was funded by grants from the National Institutes of Health (NIH 1R01-GM095942) and the Empire State Stem Cell Fund through New York State Department of Health (NYSTEM C028103, C028121). J.W. is also a recipient of Irma T. Hirschl and Weill-Caulier Trusts Career Scientist Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132(4):532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2011;12(4):253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherry AB, Daley GQ. Reprogramming cellular identity for regenerative medicine. Cell. 2012;148(6):1110–1122. doi: 10.1016/j.cell.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Trowbridge JJ, Rao S, Orkin SH. Proteomic studies of stem cells. 2008 [PubMed] [Google Scholar]
- 8.Wang J. Deciphering Protein Complexes and Protein Interaction Networks for Stem Cell Pluripotency. In: Ma'ayan A, MacArthur BD, editors. New Frontiers of Network Analysis in Systems Biology. Netherlands: Springer; 2012. pp. 97–118. [Google Scholar]
- 9.Spruijt CG, Baymaz HI, Vermeulen M. Identifying specific protein-DNA interactions using SILAC-based quantitative proteomics. Methods Mol Biol. 2013;977:137–157. doi: 10.1007/978-1-62703-284-1_11. [DOI] [PubMed] [Google Scholar]
- 10. Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–368. doi: 10.1038/nature05284. •• This work is the first comprehensive interactome study in mouse ESCs with a particular focus on Nanog. Affinity purification coupled with mass spectrometry was performed for a number of biotinylated pluripotency factors Nanog, Dax1 (Nr0b1), Nac1, Oct4, Zfp281, and Rex1. A protein interaction network was constructed dedicating a cellular module of pluripotency.
- 11.Festuccia N, Osorno R, Wilson V, Chambers I. The role of pluripotency gene regulatory network components in mediating transitions between pluripotent cell states. Curr Opin Genet Dev. 2013;23(5):504–511. doi: 10.1016/j.gde.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma'ayan A, Boyer LA, Troyanskaya OG, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462(7271):358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, Das S, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495(7441):370–374. doi: 10.1038/nature11925. •• This work extends the Nanog interactome to include Tet1 and Tet2 as well as a number of novel partner proteins. Forced expression of Tet1 and Tet2, in synergy with Nanog, enhances the programming efficiency in a late-phase pre-iPSC reprogramming model.
- 14. Gagliardi A, Mullin NP, Ying Tan Z, Colby D, Kousa AI, Halbritter F, Weiss JT, Felker A, Bezstarosti K, Favaro R, Demmers J, et al. A direct physical interaction between Nanog and Sox2 regulates embryonic stem cell self-renewal. EMBO J. 2013;32(16):2231–2247. doi: 10.1038/emboj.2013.161. • This work further extends the known list of Nanog-interacting proteins to a total of 130 proteins. Sox2 was identified as a robust interacting partner of Nanog.
- 15.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 16.van den Berg DL, Zhang W, Yates A, Engelen E, Takacs K, Bezstarosti K, Demmers J, Chambers I, Poot RA. Estrogen-related receptor beta interacts with Oct4 to positively regulate Nanog gene expression. Mol Cell Biol. 2008;28(19):5986–5995. doi: 10.1128/MCB.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11(4):477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6(4):369–381. doi: 10.1016/j.stem.2010.02.014. •• This paper describes the first interactome surrounding Oct4 in mouse ESCs. Affinity purification was performed for the Oct4-interacting proteins and subsequent purifications of Oct4 partners Sall4, Tcfcp2l1, Dax1, and Esrrb were performed. A protein interaction network was constructed surrounding Oct4 and its associated proteins.
- 19.Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6(4):382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ding J, Xu H, Faiola F, Ma'ayan A, Wang J. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 2012;22(1):155–167. doi: 10.1038/cr.2011.179. •• This studiy employed an improved biotin/strapavidin purification method for AP-MS and identied additional Oct4 partners important for self-revewal and pluripotency in mouse ESCs. The novel Oct4 interactors are associated with multiple chromatin-modifying complexes. This study also provided a framework for exploring pluripotency factor-based reprogramming strategies.
- 21.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 22. Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. •• A Myc-centered regulatory network was investiaged in mouse ESCs by combining the protein-protein and protein-DNA interaction studies. Myc interacts with the Tip60-p400 acetylation complex. This work defines three separated functional modules (core, Polycomb, and Myc) in ESCs. The Myc module is active in various cancers and predicts cancer outcome.
- 23.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134(1):162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106(13):5181–5186. doi: 10.1073/pnas.0812889106. • This paper demonstrates the identification of ES-specific composition of the SWI/SNF chromatin remodeling complex esBAF. This complex is essential for ESC slef-renewal and pluripotency.
- 25.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106(13):5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell. 2011;147(1):120–131. doi: 10.1016/j.cell.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fong YW, Cattoglio C, Yamaguchi T, Tjian R. Transcriptional regulation by coactivators in embryonic stem cells. Trends Cell Biol. 2012;22(6):292–298. doi: 10.1016/j.tcb.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14(6):427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, Bar-Nur O, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151(7):1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, Vega VB, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11(2):197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 31.Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141(6):943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 32. Esch D, Vahokoski J, Groves MR, Pogenberg V, Cojocaru V, Vom Bruch H, Han D, Drexler HC, Arauzo-Bravo MJ, Ng CK, Jauch R, et al. A unique Oct4 interface is crucial for reprogramming to pluripotency. Nat Cell Biol. 2013;15(3):295–301. doi: 10.1038/ncb2680. • This work identifies a unique linker region in Oct4 as an interface for protein-protein interactions. This region was shown to interact with Smarca4 and Chd4, two important subunits of BAF and NuRD complexes, respectively. Point mutations of this region abolish the Oct4 activity in somatic cell reprogramming.
- 33.Luo M, Ling T, Xie W, Sun H, Zhou Y, Zhu Q, Shen M, Zong L, Lyu G, Zhao Y, Ye T, et al. NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem Cells. 2013;31(7):1278–1286. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O'Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T, Costello I, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10(5):583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, Maza I, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502(7469):65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 36.Dos Santos RL, Tosti L, Radzisheuskaya A, Caballero IM, Kaji K, Hendrich B, Silva JC. MBD3/NuRD Facilitates Induction of Pluripotency in a Context-Dependent Manner. Cell Stem Cell. 2014;15(1):102–110. doi: 10.1016/j.stem.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Xia P, Ye B, Huang G, Liu J, Fan Z. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell. 2013;13(5):617–625. doi: 10.1016/j.stem.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 38. Fidalgo M, Faiola F, Pereira CF, Ding J, Saunders A, Gingold J, Schaniel C, Lemischka IR, Silva JC, Wang J. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc Natl Acad Sci U S A. 2012;109(40):16202–16207. doi: 10.1073/pnas.1208533109. • This study identifies an important role of Zfp281 in fine-tuning the activity of Nanog. Zfp281 interacts with the NuRD deacetylation complex and recruits the NuRD complex onto the Nanog locus to mediate Nanog autorepression. Functionally, Zfp281 is a Nanog-dependent roadblock to efficient somatic cell reprogramming.
- 39.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32(5):645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi FT, Kim H, Lu W, He Q, Liu D, Goodell MA, Wan M, Songyang Z. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288(29):20776–20784. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493(7433):561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonel P, Demmers J, Tan DW, Watt F, Hendrich BD. Sin3a is essential for the genome integrity and viability of pluripotent cells. Dev Biol. 2012;363(1):62–73. doi: 10.1016/j.ydbio.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, Pasini D. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49(4):645–656. doi: 10.1016/j.molcel.2012.12.019. • This work establishes an Ogt interactome using Flag-Biotin-tagged Ogt. Tet1 and Tet2 were identified and validated as Ogt interactors. Genome-wide promoter loci with Ogt and Tet1 colocalization display enriched histone H3K4me3 and low methylated CpG in mouse ESCs.
- 44.Zhang Q, Liu X, Gao W, Li P, Hou J, Li J, Wong J. Differential Regulation of the Ten-Eleven Translocation (TET) Family of Dioxygenases by O-Linked beta-N-Acetylglucosamine Transferase (OGT) J Biol Chem. 2014;289(9):5986–5996. doi: 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jang H, Kim TW, Yoon S, Choi SY, Kang TW, Kim SY, Kwon YW, Cho EJ, Youn HD. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012;11(1):62–74. doi: 10.1016/j.stem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Myers SA, Panning B, Burlingame AL. Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2011;108(23):9490–9495. doi: 10.1073/pnas.1019289108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Rayner S, Katoku-Kikyo N, Romanova L, Kikyo N. Successful co-immunoprecipitation of Oct4 and Nanog using cross-linking. Biochem Biophys Res Commun. 2007;361(3):611–614. doi: 10.1016/j.bbrc.2007.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam CS, Mistri TK, Foo YH, Sudhaharan T, Gan HT, Rodda D, Lim LH, Chou C, Robson P, Wohland T, Ahmed S. DNA-dependent Oct4-Sox2 interaction and diffusion properties characteristic of the pluripotent cell state revealed by fluorescence spectroscopy. Biochem J. 2012;448(1):21–33. doi: 10.1042/BJ20120725. [DOI] [PubMed] [Google Scholar]
- 49.Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19(4):429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yousefi M, Hajihoseini V, Jung W, Hosseinpour B, Rassouli H, Lee B, Baharvand H, Lee K, Salekdeh GH. Embryonic stem cell interactomics: the beginning of a long road to biological function. Stem Cell Rev. 2012;8(4):1138–1154. doi: 10.1007/s12015-012-9400-9. [DOI] [PubMed] [Google Scholar]
- 52.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145(6):835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145(2):183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482(7384):221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baltus GA, Kowalski MP, Tutter AV, Kadam S. A positive regulatory role for the mSin3A-HDAC complex in pluripotency through Nanog and Sox2. J Biol Chem. 2009;284(11):6998–7006. doi: 10.1074/jbc.M807670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26(7):795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4(6):487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 60.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, Meng X, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27(9):851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi: 10.1016/j.cell.2009.12.003. • This study investigated the polycomb repressive complex 2 (PRC2) interactome in mouse ESCs. Jmj (Jarid2) was identifed as an important interactor of PRC2. It is revealed Jmj colocalizes with PRC2 on chromatin and fine-tunes histone H3K27me3 in cell-fate transition.
- 65.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 66.Mallanna SK, Ormsbee BD, Iacovino M, Gilmore JM, Cox JL, Kyba M, Washburn MP, Rizzino A. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells. 2010;28(10):1715–1727. doi: 10.1002/stem.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao Z, Cox JL, Gilmore JM, Ormsbee BD, Mallanna SK, Washburn MP, Rizzino A. Determination of protein interactome of transcription factor Sox2 in embryonic stem cells engineered for inducible expression of four reprogramming factors. J Biol Chem. 2012;287(14):11384–11397. doi: 10.1074/jbc.M111.320143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai YS, Chang CW, Pawlik KM, Zhou D, Renfrow MB, Townes TM. SRY (sex determining region Y)-box2 (Sox2)/poly ADP-ribose polymerase 1 (Parp1) complexes regulate pluripotency. Proc Natl Acad Sci U S A. 2012;109(10):3772–3777. doi: 10.1073/pnas.1108595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yakulov T, Raggioli A, Franz H, Kemler R. Wnt3a-dependent and -independent protein interaction networks of chromatin-bound beta-catenin in mouse embryonic stem cells. Mol Cell Proteomics. 2013;12(7):1980–1994. doi: 10.1074/mcp.M112.026914. [DOI] [PMC free article] [PubMed] [Google Scholar]