Letter to the Editor
Epigenetic dysregulation represents an emerging paradigm in the pathogenesis of myeloid malignancies, and the pharmacologic targeting of pathways involved in regulating epigenetic modifications is a promising therapeutic strategy1. Potential targets include recurrently mutated genes encoding epigenetic modifiers (e.g. DNMT3A, IDH1/2, TET2, EZH2), and altered epigenetic modifiers (like MLL fusion genes) that are known to initiate acute myeloid leukemia (AML)2. Some leukemias with mixed lineage leukemia (MLL) translocations are dependent on the activity of DOT1L, a histone methyltransferase responsible for trimethylation of histone 3 on lysine 79 (H3K79); small-molecule inhibitors of DOT1L, like EPZ004777, can disrupt leukemic progression3–5. DOT1L inhibitors are in various stages of clinical development for MLL-rearranged leukemias, but it is currently unclear whether these inhibitors may also be active in AML cases that lack MLL rearrangements.
We recently developed an in vitro culture system for primary AML cells that captures the genetic complexity of primary AML samples, and which can be used to test the impact of novel pharmacologic agents on AML cells6. In vitro testing of primary leukemic samples has been very challenging for DOT1L inhibitors, which require exposure to cells for more than 1 week. However, with our system, primary human AML cells can be expanded for up to 2 weeks. We used this approach to test the effects of DOT1L inhibition against a set of cryopreserved, genomically characterized de novo AML patient samples, collected through a study approved by the Human Research Protection Office at Washington University School of Medicine after patients provided informed consent.
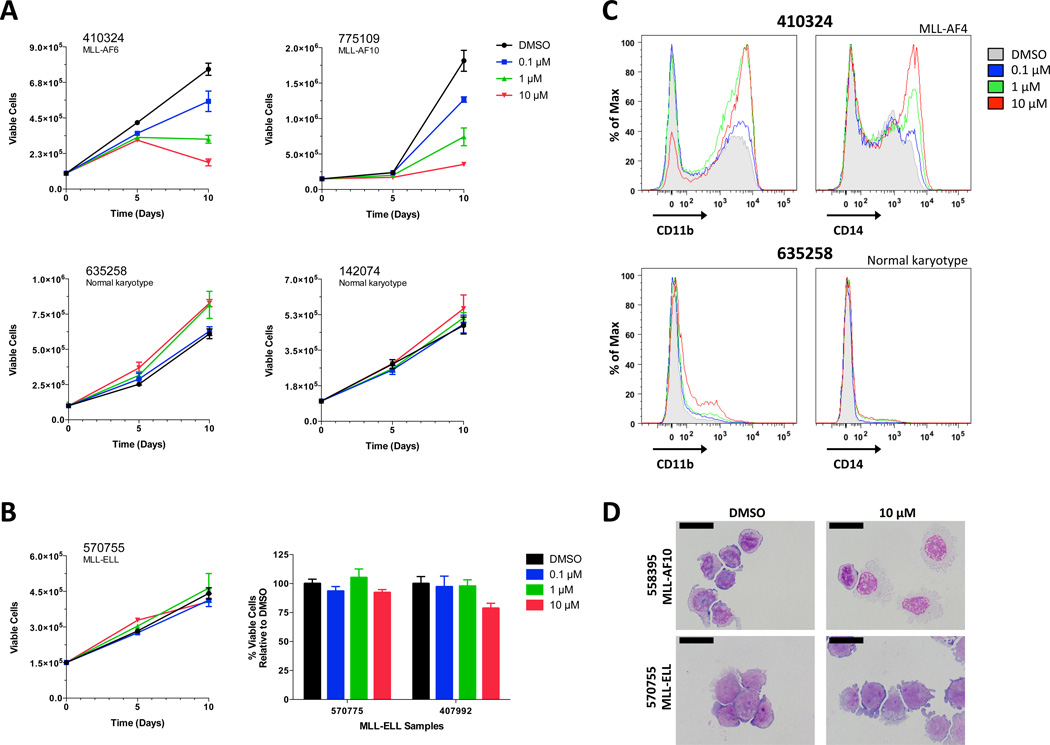
We first verified the sensitivity of MLL-rearranged primary AML cells to the DOT1L inhibitor EPZ004777. Cryopreserved peripheral blood or bone marrow specimens collected at diagnosis were thawed and expanded for 2–4 days in vitro to allow for cells to recover, and were then incubated with increasing concentrations (0.1 µM to 10 µM) of EPZ004777 (or DMSO vehicle control) for a period of 10 days. We initially selected 6 adult AML samples (including 4 with MLL translocations and 2 without) to monitor the influence of EPZ004777 on cell growth. Vehicle treated cells from each patient expanded at variable rates; however, all samples achieved a minimum of two-fold expansion by day 10. Dose-dependent growth inhibition was observed for 2 of the 4 MLL rearranged samples (one with MLL-AF6 and one with MLL-AF10) in the presence of EPZ004777. AML cell growth was minimally influenced at the maximum drug concentration in the non-MLL control samples (Figure 1a). Previous studies performed on cells expressing MLL fusions demonstrated that EPZ004777 reduced cell growth with delayed kinetics4 ; indeed, reduced growth was not evident in the responsive samples for at least 6–7 days. Global levels of H3K79 methylation were reduced following exposure to EPZ004777, and expression of HOXA cluster genes was also decreased, as expected (Figure S1). Each of the two MLL-rearranged samples that failed to exhibit a strong response to EPZ004777 had an MLL-ELL fusion (Figure 1b and Supplemental Table 1). The lack of response to EPZ004777 in these cases may relate to the reported absence of DOT1L in the MLL-ELL elongation complex7,8.
Figure 1. EPZ004777 Alters Growth and Differentiation of Primary AML cells with MLL rearrangements.
(A) Impact of EPZ004777 treatment on the growth of primary AML cells with (top panels) and without MLL rearrangements (bottom panels). Absolute cell numbers on y-axis; note different scales due to different rates of growth. Cells were expanded using a previously described stromal co-culture technique6 (also see methods in supplemental material) in the presence of DMSO or increasing concentrations of EPZ004777 (0.1 µM, 1 µM, 10 µM) over a 10-day period. Data represent mean values from two experiments assayed in duplicate ± SD (error bars). (B) Representative growth curve of MLL-ELL primary cells treated with increasing concentrations of EPZ004777 (left panel). Summary graph of cell growth results from two MLL-ELL samples treated with different doses of EPZ004777 (right panel); results are plotted as percent relative to DMSO control. Data represent mean values from two experiments assayed in duplicate ± SD (left) or SEM (right). (C) Histograms depicting the cell surface expression of myeloid markers CD11b (left panels) and CD14 (right panels) in two representative patient samples (one with a MLL translocation and one without). Cells were incubated in the presence of 10 µM EPZ004777 for 10 days and analyzed by flow cytometry. (D) Wright-Giemsa-stained cytospins of representative MLL and non-MLL rearranged patient samples treated for 10 days with DMSO or 10 µM EPZ004777. Scale bars represent 20 µm.
We also examined the effects of EPZ004777 on myeloid differentiation. Cell surface expression of CD11b and/or CD14 increases as myeloid cells differentiate toward neutrophils or mature monocytes, respectively. Expression of these markers increased in a dose-dependent manner after 10 days of drug treatment in the sensitive samples (Figure 1c, top). In contrast, the MLL-ELL and non-MLL control samples displayed only modest surface expression changes in the presence of drug (Figure 1c, bottom). Morphologic examination of the cells corroborated the flow cytometric findings (Figure 1d). Collectively, these data suggested that this in vitro culture system would be suitable for testing the sensitivity of an expanded panel of primary AML samples to EPZ004777.
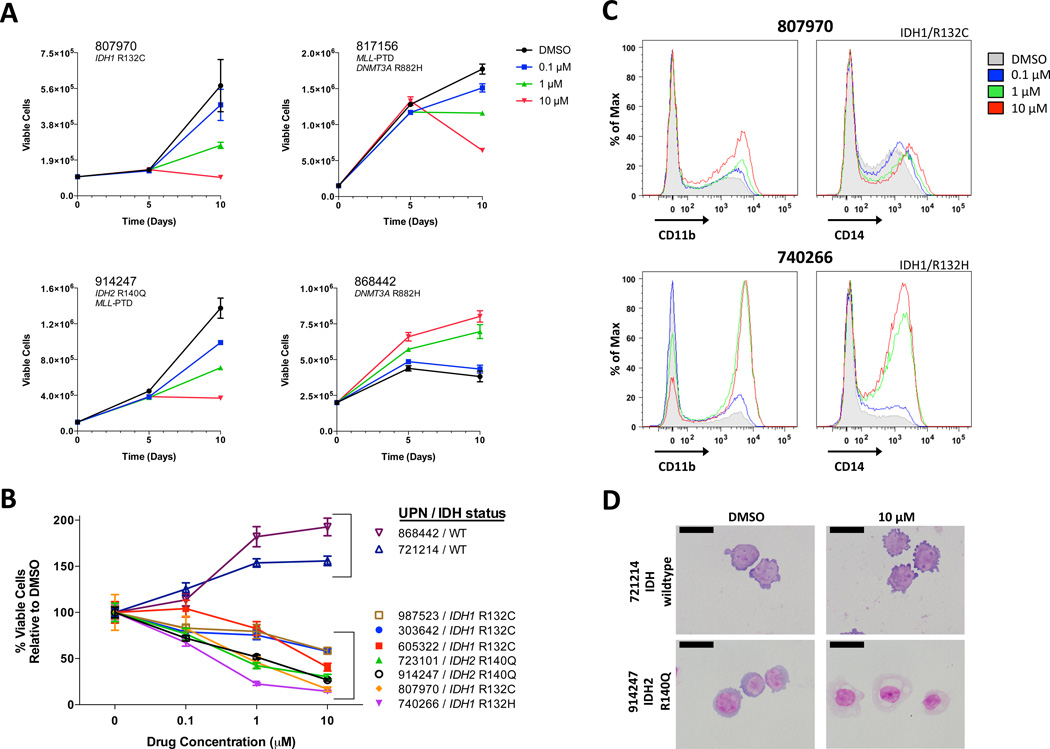
We therefore evaluated AML cells with mutations in other epigenetic modifiers (e.g. IDH1, IDH2, DNMT3A) or the MLL partial tandem duplication (MLL-PTD). All 4 cases with MLL-PTD alterations were sensitive to EPZ004777, consistent with a recent report9. Additionally, we also observed a potent response to EPZ004777 in an AML sample with a PICALM-MLLT10 fusion, which was predicted from a murine model3. Surprisingly, many AML samples without MLL fusions or MLL-PTDs were also highly sensitive to treatment with EPZ004777. While EPZ004777 did not slow the growth of AML samples with isolated DNMT3A mutations (e.g. UPN 721214 and 868442), 7/7 samples with canonical mutations in IDH1 (R132H/C) or IDH2 (R140Q) were sensitive. All cases demonstrated a dose-dependent decrease in viable cell numbers (compared to the vehicle control) after 10 days of drug treatment (Figures 2a and 2b). Of note, two of the seven samples also had MLL-PTD mutations (Supplemental Table 1). For the IDH-mutated samples, treatment with EPZ004777 for 10 days induced greater surface expression of CD11b and/or CD14 than vehicle alone (Figure 2c). Morphologic differentiation was also apparent (Figure 2d) in the sensitive samples. In total, 14/23 AML samples were sensitive to EPZ004777. All of the responders had an MLL rearrangement (that was not MLL-ELL), a PICALM-MLLT10 fusion, MLL-PTDs, and/or mutations in IDH1 or IDH2.
Figure 2. EPZ004777 Impairs the Growth and Differentiation of Genetically Defined, Non-MLL Rearranged Primary AML Cells.
(A) Growth curves of primary cells with an IDH1 mutation (UPN 807970), a MLL-PTD mutation (UPN 817156), both mutations (UPN 914247), or neither mutation (UPN 868442) incubated in the presence of DMSO or increasing concentrations of EPZ004777 (0.1 µM, 1 µM, 10 µM) over a 10-day period. Refer to Supplementary Table S1 for more detailed information on individual primary samples. Absolute cell numbers on y-axis; note different scales due to different rates of growth. Cells were expanded using the stromal co-culture technique (see methods). Data represent mean values from two experiments assayed in duplicate ± SD (error bars). (B) Impact of EPZ004777 on the proliferation of IDH wildtype (WT; UPN 868442 & 721214) and IDH mutant (all others) primary cells; refer to Supplementary Table S1 for more details on individual samples. Results represent number of viable cells counted after 10 days, and are expressed as a percentage relative to DMSO controls, to adjust for differences in growth rates between different primary samples. Data represent mean values from two experiments assayed in duplicate ± SEM (error bars). (C) Cell surface expression of CD11b (left panels) and CD14 (right panels) in two representative IDH mutant patient samples. Cells were incubated in the presence of 10 µM EPZ004777 for 10 days and analyzed by flow cytometry. (D) Wright-Giemsa-stained cytospins of representative IDH wildtype and mutant patient samples treated for 10 days with DMSO or 10 µM EPZ004777. Scale bars represent 20 µm.
In sum, our data suggest that a wider spectrum of AMLs may be responsive to DOT1L inhibitors than originally predicted. While the observed responses of AML samples with MLL-PTDs or the PICALM-MLLT10 fusion were anticipated, the responses of IDH-mutant samples were not. The neomorphic enzymatic activity of mutated IDH1/2 produces 2-hydroxyglutarate (2-HG), an oncometabolite capable of inhibiting enzymes that are involved in a variety of epigenetic processes, including histone and DNA methylation10. While IDH-mutated AMLs typically do not show dysregulation of HOXA cluster gene expression (which is common in MLL-rearrangements and MLL-PTDs), global levels of H3K79 dimethylation (H3K79me2) have been reported to be modestly increased when either IDH1 or IDH2 are mutated10–12; the precise locations of altered sites of H3K79 methylation are not yet known. Regardless, the observations presented here provide an impetus to expand studies of DOT1L inhibitors to AML samples with canonical IDH1 or IDH2 mutations, and to define the mechanisms by which it acts in these cases.
Supplementary Material
ACKNOWLEDGMENTS
Technical assistance was provided by the Alvin J. Siteman Cancer Center Tissue Procurement Core, the High Speed Cell Sorter Core, and the Molecular and Genomic Analysis Core at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO, which are all supported by the National Cancer Institute Cancer Center Support Grant P30CA91842. This work was supported by National Institutes of Health grant R01CA162086 (T.J.L.), PO1CA101937 (T.J.L.), K08HL116605 (J.M.K.), and the Barnes Jewish Hospital Foundation (T.J.L.) as well as the Doris Duke Charitable Foundation to Washington University (S.M.S., Clinical Research Fellow).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary information is available at Leukemia's website.
REFERENCES
- 1.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Deshpande AJ, Banka D, Bernt KM, Dias S, Buske C, et al. Abrogation of MLL-AF10 and CALM-AF10-mediated transformation through genetic inactivation or pharmacological inhibition of the H3K79 methyltransferase Dot1l. Leukemia. 2013;27:813–822. doi: 10.1038/leu.2012.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klco JM, Spencer DH, Lamprecht TL, Sarkaria SM, Wylie T, Magrini V, et al. Genomic impact of transient low-dose decitabine treatment on primary AML cells. Blood. 2013;121:1633–1643. doi: 10.1182/blood-2012-09-459313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KSJ, Allis CD, et al. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci. 2011;108:15751–15756. doi: 10.1073/pnas.1111498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn MWM, Hadler M, Daigle SR, Chen C, Sinha AU, Krivtsov AV, et al. Myeloid Leukemia Cells With MLL partial Tandem Duplication Are Sensitive To Pharmacological Inhibition Of The H3K79 Methyltransferase DOT1L. Blood. 2013;122:1256. [Google Scholar]
- 10.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P, Dong Q, Zhang C, Kuan P-F, Liu Y, Jeck WR, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.