Abstract
Non-typeable Haemophilus influenzae (NTHi), a human respiratory tract pathogen can form colony biofilms in vitro. Bacterial cells and the amorphous extracellular matrix (ECM) constituting the biofilm can be separated using sonication. The ECM from 24 hr and 96 hr NTHi biofilms contained polysaccharides and proteinaceous components as detected by NMR and FTIR spectroscopy. More conventional chemical assays on the biofilm ECM confirmed the presence of these components and also DNA. Proteomics revealed eighteen proteins present in biofilm ECM that were not detected in planktonic bacteria. One ECM protein was unique to 24 hr biofilms, two were found only in 96 hr biofilms, and fifteen were present in the ECM of both 24 hr and 96 hr NTHi biofilms. All proteins identified were either associated with bacterial membranes or were cytoplasmic proteins. Immunocytochemistry showed two of the identified proteins, a DNA-directed RNA polymerase and the outer membrane protein OMP P2, associated with bacteria and biofilm ECM. Identification of biofilm-specific proteins present in immature biofilms is an important step in understanding the in vitro process of NTHi biofilm formation. The presence of a cytoplasmic protein and a membrane protein in the biofilm ECM of immature NTHi biofilms suggests that bacterial cell lysis may be a feature of early biofilm formation.
Keywords: nuclear magnetic resonance, electron microscopy, proteomics, immunocytochemistry, diagnostics
INTRODUCTION
Biofilms are aggregations of sessile bacteria cells embedded in an extracellular matrix, one function of which is to attach bacteria to one another and to biotic and abiotic substrates (Stoodley et al., 2002). Depending on the species of bacteria and growth conditions, this matrix is composed of varying amounts of exopolysaccharides, nucleic acids, lipids and proteins (Flemming & Wingender, 2001). We have previously examined biofilms formed by non-typeable Haemophilus influenzae (NTHi) and shown the ECM to contain a large number of bacterial proteins (Gallaher et al., 2006), including a high molecular weight protein (HMW1), a known adhesin with a role in bacterial adhesion, in an extracellular location (Webster et al., 2006).
Biofilm-specific proteins have been demonstrated to play roles in adhesion and attachment (Kawakami et al., 1998, Swords et al., 2000, Ahren et al., 2001, Ahren et al., 2001, Fink et al., 2002, Berenson et al., 2005, Bookwalter et al., 2008, Ronander et al., 2008), biofilm-specific protein regulation, and control of transformation into the biofilm phenotype (Sauer & Camper, 2001, Oosthuizen et al., 2002, Sauer et al., 2002, Vilain et al., 2004, Southey-Pillig et al., 2005, Allegrucci et al., 2006). Proteins within the ECM may also assist in forming the complex structures reported previously within biofilms (Schaudinn et al., 2006, Baum et al., 2009, Pelzer et al., 2012, Schaudinn et al., 2014).
Extracellular proteins can be firmly bound to the cell surface, associated with the ECM, or be freely diffusing throughout the biofilm matrix (Hoffman & Decho, 1999). The presence and function of extracellular proteins has been reported for a wide range of Gram-positive and Gram-negative bacteria, both in cultures of free-swimming (planktonic) cells and in biofilms (Wingender et al., 1999). Extracellular proteins from planktonic cells are present in low concentration and are a minor component of a complex in vivo milieu. The confined microenvironment of the biofilm matrix accumulates extracellular molecules and prolongs the stability and activity of extracellular enzymes (Hoffman & Decho, 1999). The variety, abundance, and distribution of extracellular proteins within the ECM of biofilms formed by pathogenic bacteria have only recently attracted interest within the scientific community (Southey-Pillig et al., 2005, Allegrucci et al., 2006, Webster et al., 2006, Das et al., 2014).
Non-typeable Haemophilus influenzae (NTHi) bacteria are Gram-negative commensal organisms of humans that colonize the respiratory tract and can act as opportunistic pathogens, with roles in sinus infection and otitis media (Hall-Stoodley et al., 2006, Barkai et al., 2009). NTHi bacteria are also linked to infections of the lungs of cystic fibrosis patients (Starner et al., 2006), and morbidity in patients with chronic obstructive pulmonary disease (COPD) (Murphy et al., 2004, Moghaddam et al., 2011, Thanavala & Lugade, 2011). NTHi is a demonstrated biofilm-forming organism in humans (Murphy et al., 2005, Hall-Stoodley et al., 2006, Murphy et al., 2009), in animal model systems (Miyamoto & Bakaletz, 1996, Jurcisek et al., 2003, Jurcisek et al., 2005), and in vitro systems (Murphy & Kirkham, 2002, Swords et al., 2004, Webster et al., 2004, Webster et al., 2006, Moriyama et al., 2009).
In this study we used NTHi colony biofilms formed within the first 96 hr of incubation (Webster et al., 2004, Webster et al., 2006) to examine proteins present in the ECM. These biofilms are considered to be early, or immature, biofilms (Jurcisek & Bakaletz, 2007) yet they express extracellular proteins in the biofilm ECM (Gallaher et al., 2006, Webster et al., 2006). We have used NMR and FTIR spectroscopy to detect ECM proteins, proteomics to identify biofilm-specific proteins present in the biofilm ECM, and immunocytochemical methods to confirm an extracellular location of biofilm-specific proteins. One goal of this study was to determine if it was possible to identify biofilm-specific proteins that could be used as the basis for diagnostic tests to identify biofilm-related infections.
MATERIALS AND METHODS
Preparation of non-typeable Haemophilus influenzae (NTHi) planktonic culture
50 µl of NTHi frozen stock was inoculated onto chocolate agar plates and incubated at 37°C, 5% CO2, and 95% relative humidity. After 24hrs, bacterial colonies on the plate were used to inoculate sBHI (brain-heart-infusion broth supplemented with 20 ng/mL NAD and 100 ng/mL hemin); OD600 was adjusted to 0.09 before incubating at 37°C, 5% CO2, and 95% relative humidity for 24 hours. This 24-hr (overnight) culture was then used to inoculate fresh sBHI, with OD600 adjusted to 0.1 before placing in a shaker-incubator and grown at 200 rpm and 37°C until mid exponential growth phase was reached.
NTHi colony biofilms
Each NTHi colony biofilm was prepared on Millipore filters as previously described (Webster et al., 2006). Frozen aliquots of NTHi clone 9274 were thawed, plated onto chocolate agar plates (Hardy Diagnostics, Santa Maria, CA) and grown for 20 hr at 37°C in 5% CO2 and 95% relative humidity (culture conditions applied to all bacterial and biofilm incubations). BHI broth supplemented with hemin and NAD (Poje & Redford, 2003) was inoculated with colonies formed on the agar, and the suspension incubated for 24 hr (stationary phase or overnight culture). Relative numbers of bacteria in sBHI suspension were estimated by reading the optical density at 600 nm (OD600) (Poje & Redford, 2003).
Overnight culture was diluted 1:200 (1 × 107 per mL) in fresh sBHI broth and 170 µL inoculated onto 25 mm diameter filters (Millipore Corp., Billerica, MA, Catalog # GSWP 025 00) on chocolate agar plates (Webster et al., 2004, Webster et al., 2006). The filters with bacteria were incubated for increasing times from 1 hr to 96 hr. The NTHi colony biofilms on filters (Zahller & Stewart, 2002, Webster et al., 2004) were either prepared for examination by electron electron microscopy, proteomic analysis, biochemical analysis, or were used to estimate total numbers of culturable bacteria (CFU’s).
Estimation of colony-forming units (CFU’s) and dry weights
Culturable bacteria estimates (CFU’s) present in the colony biofilms were obtained using established counting methods (Herigstad et al., 2001). 25 colony biofilms for each time point, formed with identical amounts of overnight culture, were placed in sterile phosphate buffered saline (PBS) and sonicated in a sonicator bath for 6 min. The optical density of the resulting total biofilm suspension was read at 600 nm (OD600) to obtain an estimate of total biomass, and the suspension then serially diluted in sterile BHI broth, plated onto chocolate agar plates and incubated overnight. Estimates of total viable bacteria were calculated at increasing time-points for up to 96 hr and the results tabulated as numbers of viable bacteria per colony biofilm. CFU counts were replicated 3 times on 5 different occasions.
Preparation of biofilms for chemical assay and proteomic analysis
Planktonic bacteria and colony biofilms of NTHi strain 9274 biofilms were prepared as described above. Colony biofilms were formed on mixed cellulose filters, and dry weighs determined on triplicate batches dried under vacuum. Other NTHi colony biofilms were dislodged from their filter substrates by 3 cycles of washing in PBS using an ultrasonic bath at 35 kHz (Bransonic 5510R-DTH, 40kHz; Branson Ultrasonics Corp., Danbury, CT) and (4°C for 3 min followed by incubation on ice for another 3 minutes. The resulting slurry of bacterial biofilm material in PBS was transferred to a fresh tube. Biofilm components soluble in PBS (biofilm ECM) were separated from bacteria and associated insoluble components (non-ECM biofilm fraction) by centrifugation at 8,600 rcf at 4 °C for 5 minutes. For proteomic analysis NTHi colony biofilms were dislodged from growth substrate and biofilm ECM and non-ECM biofilm fractions were collected as decribed, but with the addition of a protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany, Catalog # 11836145001). The cocktail was used following manufacturers instructions where 1 tablet was added to 7 mL PBS. The non-ECM fraction was resuspended in protein extraction buffer (50mM Tris, 150mM NaCl, 0.1% Triton X-100, 0.1% SDS, pH 7.5) containing protease inhibitor cocktail (1 tablet for each 7 mL extraction buffer), processed in the same way as for planktonic bacteria described above, and the supernatant was collected as homogenised, non-ECM biofilm fraction.
Chemical Assays
NMR and FTIR analysis of biofilm ECM from 6.5 hr, 12 hr, 24 hr and 96 hr biofilms was carried out using previously described methods (Baum et al., 2009), total protein concentration was determined using the Pierce BCA protein assay (Thermo Scientific), and DNA concentration by DAPI conjugation and fluorometry. The DNA content of SDS-buffered samples was estimated according to the method described by Brunk et al. (Brunk et al., 1979) using a fluorescence spectrophotometer (F-4500, Hitachi, Schaumburg, IL) and a calibration curve made with deoxyribonucleic acid sodium salt from salmon testes (D1626, Sigma, Milwaukee, WI, 2.4 mg in 100 mL SDS-buffer). For measurements, 15 uL aliquots (4-6) of biofilm solution were incubated for 2 min in DAPI (100 ng/mL in buffer composed of 100 mM NaCl, 10 mM EDTA, and 10 mM Tris, pH 7.0). Ultraviolet-visible spectra of biofilm ECM were recorded on a Model 8452A diode array spectrophotometer (Hewlett-Packard/Agilent Technologies, Santa Clara, CA). Fluorescence spectra were acquired on a Model F-4500 fluorescence spectrometer (Hitachi High-Technologies, Pleasanton, CA). Infrared spectra of biofilm ECM were acquired on a Magna-IR 560 FTIR spectrometer as potassium bromide discs (Nicolet/Thermo Scientific, Madison, WI).
Nuclear Magnetic Resonance Spectroscopy (NMR)
NMR Spectra were acquired on the water-soluble and DMSO-d6-soluble fractions of the freeze-dried ECM from 96 hr biofilms using an Inova 600 NMR spectrometer with an inverse triple resonance probe (Varian/ Agilent Technologies, Santa Clara, CA). All experiments were done with standard pulse programs and parameter sets supplied with the Chempack extension of the VnmrJ applications software (Agilent Technologies, Santa Clara, CA).
Proteins from Planktonic Bacteria
Culture medium was removed from planktonic bacteria culture at exponential growth phase by centrifugation at 8,600 rcf for 3 min. The bacteria pellet was resuspened in 1XPBS containing protease inhibitors cocktail (Roche Applied Science, Mannheim, Germany, Catalog # 11836145001), repelleted at 8,600 rcf for 3 min., and the supernatant discarded. The pellet was then resuspended in extraction buffer (50mM Tris, 150mM NaCl, 0.1% Triton X-100, 0.1%SDS, pH 7.5) containing protease inhibitors cocktail, incubated on ice for 30 min., and subjected to 10 sonication cycles each consisting of 30 seconds sonication with a probe micro-ultrasonic cell-disruptor at maximum power (1,200 W per sq. in. of ultrasonic energy) (Kontes, Vineland, NJ) and 3 min. incubation on ice. Extracted proteins were prepared by centrifuging this solution at 16,800 rcf for 5 min. and collecting the supernatant.
Proteomic analysis
Biofilm ECM components were separated by 1-D SDS-PAGE (Webster et al., 2006) with 14 µg of protein suspension added to each lane. Two lanes from the gel of each developmental stage were divided into 10 separate bands and digested with trypsin using established methods (Gallaher et al., 2006). Protein digests of each band were analyzed using a nanoLC-2D system (Eksigent , Dublin, CA) interfaced with an autosampler and an ABI/MDS SCIEX 4000 QTRAP (ABI, Ontario, Canada). Nanoflow LC was performed using C-18 reverse phase chromatography (ZORBAX 300SB-C18) with an acetonitrile/water (containing 0.1% formate) gradient (5-85% over 75 min) at 300 nL min−1. Data analysis was first performed using MASCOT software (Matrix Science, London, England), and subsequently refined using Scaffold v. 2.0 or v. 3.0 (Proteome Software, Portland, OR). Final results met the major criteria outlined by Carr et al. (Carr et al., 2004). Proteins from the 24 hr and 96 hr biofilm ECM were compared with proteins identified from planktonic forms of NTHi bacteria.
Filters were subjected to proteomic analysis after 6.5 hr, 12 hr, 24 hr and 96 hr biofilms had been removed by sonication. Protein residues on washed filters were solubilized in lysis buffer (50 mM Tris-HCL, 150 mM NaCl, 1% Triton X-100, pH 7.4) and digested with trypsin as before (Gallaher et al., 2006). The protein digests were analyzed using an Agilent 1200 nanoLC system interfaced with an Agilent 6520 Q-TOF equipped with a Chip-cube source. A volume of 10 uL sample was loaded at 5uL/min onto a high capacity Agilent micro-fluidic device (chip) consisting of a 160 nL trapping column and a 150mm×75um analytical column, both packed with Zorbax 300SB-C18. Peptides were separated using an Agilent 1200 nanoLC system with an acetonitrile/water (containing 0.1% formic acid) gradient (3-85% over 46 min) at 300 nL/min. The mass spectrometer was set to scan at 4 scans per second with a m/z range of 300-2500.
After each scan, the instrument chose up to 6 ions exceeding a threshold of 1000 counts of signal intensity in the full scan and subjected them to MS/MS. Ions selected for MS/MS were required to have a measurable charge of at least 2 and were subject to a screening test to ensure they consisted primarily of a single component rather than multiple co-eluting components of the same approximate m/z. Ions selected for MS/MS were fragmented with a variable collision cell potential set to 3.6 V * precursor m/z/100 – 4.8 V. MS/MS spectra were collected from 50 to 3000 m/z at 2 scans per second, with the scan speed increased to avoid spectra with more than a calculated 50,000 ions per spectrum. After being selected for fragmentation, ions were placed on a temporary exclusion list for 12 seconds so they would not be re-analyzed immediately.
Tandem mass spectra were extracted by Masshunter (v 3.5). Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using X! Tandem (The GPM, thegpm.org, v. Tornado (2009.04.01.1)). X! Tandem was set up to search the Haemophilus influenzae NCBI and Uniprot databases with a fragment ion mass tolerance of 50 PPM and a parent ion tolerance of 25 PPM. Iodoacetamide derivative of cysteine was specified in X! Tandem as a fixed modification. Deamination of asparagine and glutamine, oxidation of methionine and tryptophan, sulphone of methionine and acetylation of the n-terminus were specified in X! Tandem as variable modifications. Results were imported and visualized into Scaffold (v.3, Proteome Software, Portland, OR) .
Scanning Electron Microscopy
Colony biofilms on filters were rapidly frozen by immersion in liquid propane and subsequently stored in liquid nitrogen (Webster et al., 2004). Frozen specimens were freeze-substituted in dry ethanol containing 1% glutaraldehyde at −80 °C, gradually warmed to 4 °C in 100% ethanol, and critical point dried. The dried biofilms were mounted onto specimen stubs, sputter-coated with 9 nm thick films of platinum, and examined in a scanning electron microscope (SEM, XL30 SFEG; FEI, Hillsboro, OR).
Filters where the biofilm had previously been removed for chemical analysis were also prepared for SEM examination using rapid freezing and freeze substitution as described above.
Transmission Electron Microscopy
Colony biofilms were rapidly frozen by high-pressure freezing (EMPACT2 HPF; Leica Microsystems Inc. Deerfield, IL) (McDonald et al., 2007) and freeze substituted in ethanol containing 1% osmium tetroxide (Webster et al., 2006). The freeze-substituted biofilms were embedded in Lowicryl HM20 (EMS, Hatfield, PA) at −50°C under UV light (AFS2, Leica Microsystems Inc.,) (Webster et al., 2004, Webster et al., 2006). Thin sections (60–80 nm) were labeled with specific mouse monoclonal [8RB13] to RNA polymerase beta (Abcam, ab81865; Cambridge, MA), which was used to detect the DNA-directed RNA polymerase subunit beta, rabbit anti-mouse antibodies and 10 nm protein A-gold (PAG; University of Utrecht, The Netherlands), or rabbit anti-OMP P2 (YKA) (Neary & Murphy, 2006) and 10 mn PAG using sequential labeling protocols (Webster et al., 2006). The rabbit anti-mouse antibodies were diluted in blocking solution containing 10% fish skin gelatin and 10 µg/mL bacterial protein suspension (prepared by heat denaturation, lysis and solubilization of NTHi and Listeria monocytogenes cells). Immunolabeled sections were imaged using a Tecnai G2 20 TEM (FEI Inc, Hillsboro, OR) and images were digitally recorded (XR41; AMT, MA). When required, brightness and contrast adjustments were applied to the whole image (Adobe®Photoshop, Adobe, San Jose, CA).
Western blotting
SDS-PAGE protein separation and western blot analyses were performed as described previously (Webster et al., 2006).
Quantification
All experiments were performed at least in triplicate. Experiments estimating protein and DNA content were performed in quadruplet. Estimates of viable bacterial and measurement of soluble biomass were performed 6 times each using 8 well rows on 96 well plates (48 replicas).
Quantification of gold particle label was performed on three different planktonic populations of cells, each sectioned in 4 regions chosen using systematic random sampling, and each section selected in 5 different regions at low magnification where gold particles could not be detected. The number of labeled cells was estimated by counting directly from images taken at high magnification and expressed as a percentage of the total population. Labeling densities of gold particles on cells (gold/µm2) and cell membranes (gold/µm) were estimated using the STEPanizer software package (Tschanz et al., 2011) installed on an iMac. Statistical significance was determined by comparing two groups using an unpaired t-test with Welch’s correction. Calculations for t-values were performed in MS Excel, p-values determined from a t-test table, and statistical significance defined as p= <0.05.
Proteomic analysis was performed on three different biofilm preparations and four different planktonic preparations. Planktonic proteins were identified using peptide thresholds set at 80% minimum and protein thresholds set at 80% minimum with a 1-peptide minimum (Scaffold software). Proteins from the biofilm ECM of 24 hr and 96 hr colony biofilms and from washed filters were identified using peptide thresholds set at 99.9% minimum and protein thresholds set at 95% minimum with a 3-peptide minimum. Proteins present in any one of three replicate samples were tabulated.
RESULTS
ECM was rapidly produced by early stage NTHi biofilms
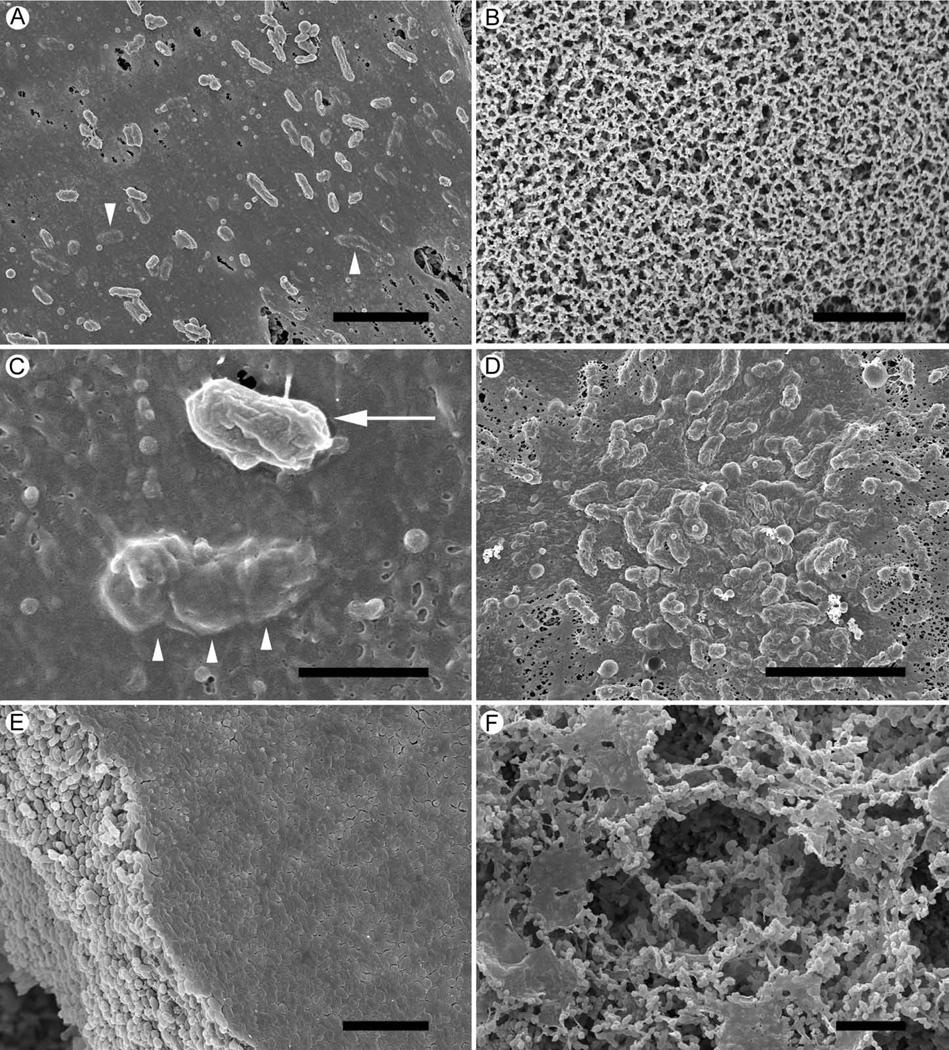
Cryo-preparation methods used to prepare NTHi colony biofilms for scanning electron microscopy (SEM) examination preserved extracellular material and resulted in the preservation of intact biofilms. Such preservation methods allowed for examination of early biofilms developing on mixed cellulose filters. After 1 hr incubation the growth surface was covered with extensive amounts of amorphous material (Fig 1A). No bacterial cells or amorphous material was detected on growth surfaces inoculated with NTHi-free suspensions (Fig 1B). The amorphous material produced by the bacteria wrapped the cells, covered them, and seemed to attach them to the growth surface (Fig 1C). By 2 hr incubation, bacteria were completely covered with amorphous material and had formed aggregates of increased numbers of cells (Fig 1D). At 6 hr, 12 hr and 24 hr, dense biofilms consisting mainly of bacterial cells were produced, all of which were similar in appearance and were represented by a 12 hr biofilm (Fig 1E). When left undisturbed and unfed for 96 hr, the bacteria arranged themselves into structures consisting of thin sheets and channels running through the biofilm (Fig 1F).
Figure 1. Scanning EM of NTHi 9274 colony biofilms.
A. A 1 hr incubation of NTHi bacteria on the growth surface resulted in the inoculated area being covered in a layer of amorphous material, which also covered the bacteria present (arrowheads)
B. A similar, sterile growth surface incubated for 1 hr after inoculation with BHI only, showed no evidence of material.
C. A close view of the 1 hr biofilm bacteria on the growth surface showed a bacterium attached to the amorphous material (arrow). In the same image is a bacterium covered with, or embedded in, the layer of amorphous material (arrowheads).
D. After 2 hr incubation the bacteria appeared to be forming a biofilm with aggregates of bacteria embedded under a layer of amorphous material.
E. By 12hr, the biofilm had developed into a mat of tightly packed bacteria with a layer of amorphous material covering the top surface of the mat. The growth surface was completely covered by this biofilm.
F. A 96 hr biofilm showing how the bacterial cells had arranged themselves into a structure consisting of sheets and spaces within the biofilm.
Scale bars = 5 µm.
Residual biomass remains attached to filters after biofilm removal
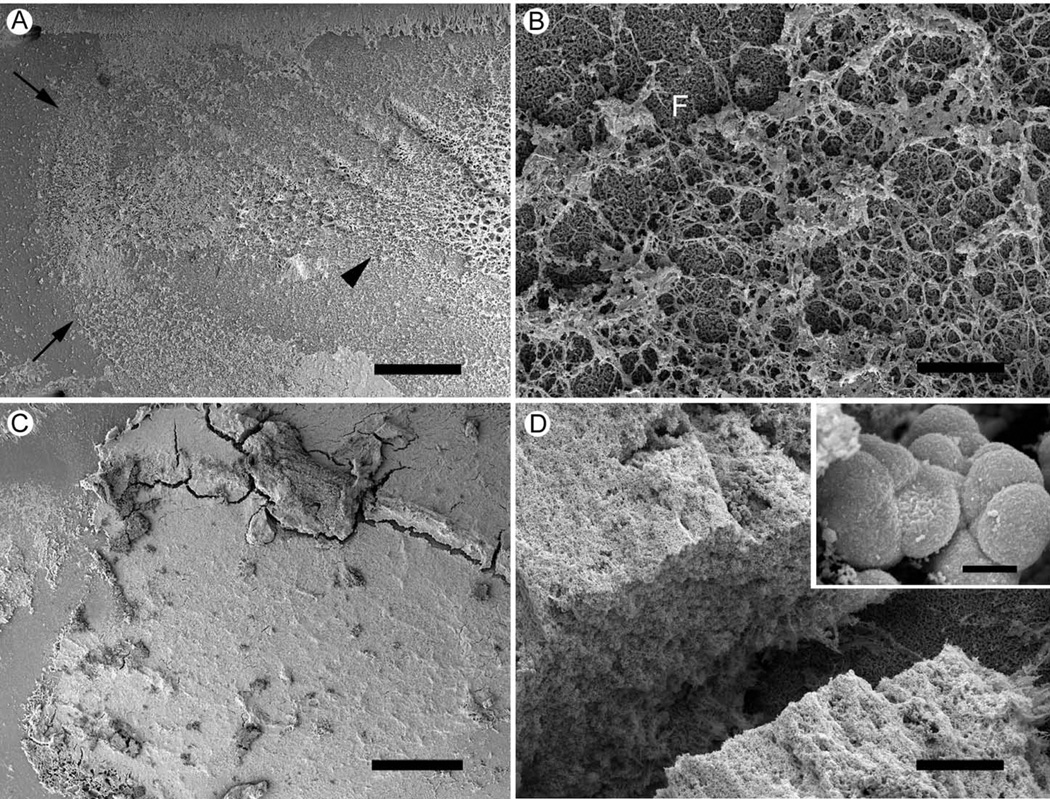
SEM examination performed on filters washed to remove NTHi biofilms for chemical analysis revealed traces of biofilm biomass remaining attached to the filter surface after extensive washing and sonication (Fig 2). Low magnification images revealed the biofilm biomass delineated the boundaries of where the 24 hr biofilm formed (Fig 2A). More central regions of where the biofilm was removed (Fig 2A) consisted of a fibrous mat of material attached to the filter (Fig 2B). Large amounts of biofilm biomass were present on filters where 96 hr biofilms had been removed (Figs 2C & 2D). The material appeared as a thick layer over the filter, and was only present where the biofilm had formed (Fig 2C). Higher magnification of the amorphous layer of material revealed the presence of embedded, misshaped bacterial cells (Fig 2D).
Figure 2. Scanning EM of filter surfaces after removal of 24 hr and 96 hr NTHi biofilms.
24 hr and 96 hr biofilms were removed from the growth surface by sonication in PBS and the washed filter surfaces were examined by SEM.
A. 24 hr , washed filter. The washing protocol almost completely removes 24 hr biofilms. At low magnification enough of the biofilm material remains on the filter to identify the outer border (arrows) and remnants of the biofilm (arrowhead) within the zone of biofilm formation. Scale bar = 500µm.
B. 24 hr washed filter. Within the zone of biofilm formation remains a thin network of material attached to the filter surface (F). Scale bar = 20µm.
C. 96 hr washed filter. Biofilm material remains attached to the surface of filters where 96 hr biofilms have been removed. Scale bar = 500µm.
D. 96 hr washed filter. The material left behind by the 96 hr biofilm resembles a network of fibrous material. Scale bar = 50µm.
Insert. Bacterial cells embedded in the 96 hr biofilm residue were more spherical than planktonic NTHi. Scale bar = 500nm.
Chemical analysis, FTIR and NMR
General chemical profiles of biofilm ECM from 24 hr and 96 hr biofilms were obtained by chemical and direct analyses. IR spectra of 24 hr and 96 hr non-ECM biofilm fractions were indistinguishable, but significantly different from the biofilm ECM at the corresponding time points. The main spectral bands for these samples are shown and assigned in Tables 1 and 2. The biofilm ECM from each time point seemed to contain a mixture of polysaccharides and proteinaceous material, while the non-ECM biofilm fractions consisted largely of proteinaceous material, with the 1160 and 1084 cm−1 bands characteristic of polysaccharides (Serra et al., 2007) being absent.
Table 1.
Principal IR-active bands in water-soluble biofilm samples.
| Band location (cm−1) | Band intensity | Assignment |
|---|---|---|
| 3433 | vs, br | νN-H (1) |
| 3320 | sh | νO-H (2); νN-H (1) |
| 3096 | w | aromatic νC-H (1); CH2 νC-H (1) |
| 2970 | m | N-CH3 νC-H (1); N-CH2 νC-H (1); CO2- νC=O (1); νC-N (1) |
| 2953 | m | νC-H (2-3) |
| 2900 | sh | νC-H (2-3) |
| 2648 | sh, br | νPO-H (1) |
| 1655 | vs | amide I νC=O (3-5) |
| 1590 | vs | amide II νC=O (3-5) |
| 1410 | vs | C-H bending in CH2 (6) |
| 1252 | m | O-acetyl ester νC=O (2-3, 6); nucleic acid νP=O (7) |
| 1160 | vs | polysaccharide νC-C and νC-O (2-4, 7) |
| 1084 | vs | polysaccharide νC-OH (2, 7) |
| 860 | s | polysaccharide νC-C, C-C-O & C-C-H bending (2); aromatic νC-H (1) |
| 544 | s | N/A |
vs, very strong; s, strong, m. medium; w, weak; sh, shoulder; br, broad
Table 2.
Principal IR-active bands in water-insoluble biofilm samples.
| Band location (cm−1) | Band intensity | Assignment |
|---|---|---|
| 3330 | vs | νN-H (1) |
| 3100 | sh | νO-H (2); νN-H (1) |
| 2968 | m | aromatic νC-H (1); CH2 νC-H (1) |
| 2933 | m | νC-H (2-3) |
| 2875 | sh | νC-H (2-3) |
| 1675 | vs | amide I νC=O (3-5) |
| 1552 | vs | amide II νC=O (3-5) |
| 1493 | m | CO2- νC=O (1); aromatic C-C (1) |
| 1404 | m | C-H bending in CH2 (6) |
| 1247 | vs | O-acetyl ester νC=O (2-3, 6); nucleic acid νP=O (7) |
| 1088 | vs | polysaccharide νC-OH (2, 7) |
| 995 | w | νC-O-H (1); νP-N (1) |
| 957 | w | νC=C (1); νP-N (1) |
| 661 | sh | N/A |
| 554 | m | N/A |
vs, very strong; s, strong, m. medium; w, weak; sh, shoulder; br, broad
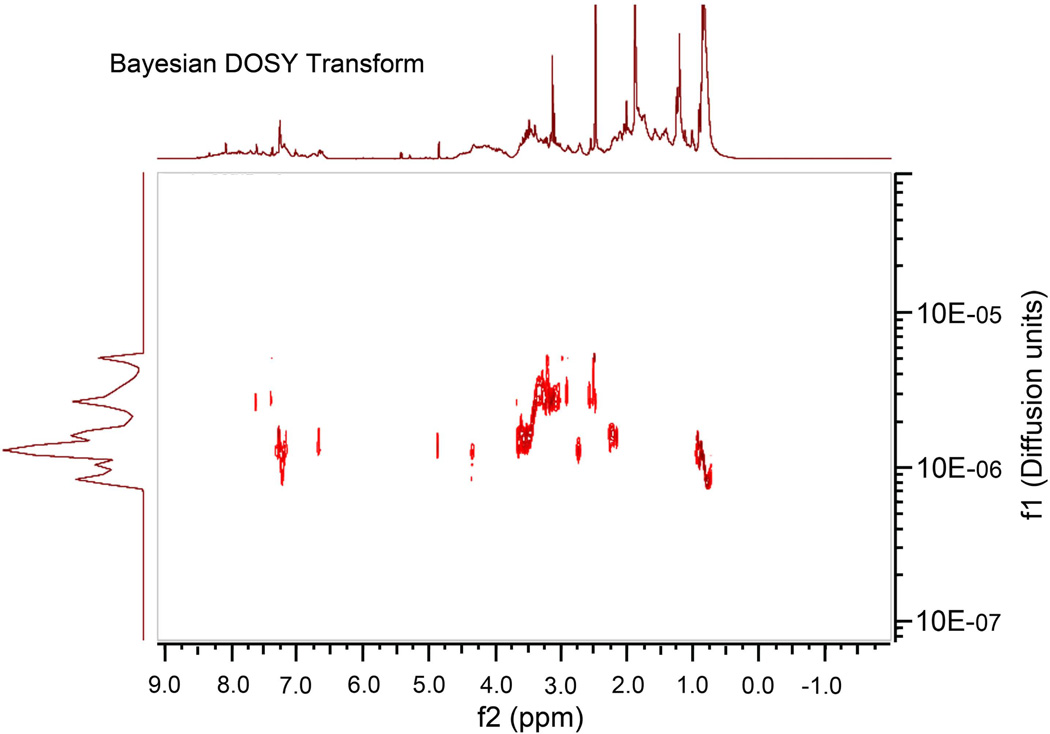
NMR spectra were obtained using DMSO-d6-soluble fractions of the freeze-dried ECM from 96 hr biofilms, representing the major portion of the material (>75%). The fractions were poorly water- and trifluoroacetic acid-soluble. Aqueous acids were not used to solubilize the ECM material as this could depolymerize a range of macromolecular biomolecules. The biofilm pellets and supernatants obtained at 24 hr and 96 hr produced similar 1H NMR spectra. The unchanged chemical composition over the four days of incubation supports observations using FTIR spectroscopy. Representative 1D 1H spectra are presented under Supporting Information (Supplemental Fig. 1). 1H spectra of the water-soluble fraction (Supplemental Fig. 1A) contained numerous peaks with narrow line-widths, suggesting the presence of small molecules (i.e., not biopolymers), in addition to numerous peaks in the 3.0-4.5 ppm range. These features were less abundant in 1H spectra of the water-insoluble material (Supplemental Fig. 1B). The above observations are supported by DOSY spectra, which show a significant distribution of molecular masses in the supernatant samples (Fig 3), while the pellet fractions did not (data not shown). The DOSY spectrum (Fig 3) appears to show the presence of lower molecular weight saccharides (δH 2.5 and 2.75-3.5 ppm), as well as proteins.
Figure 3. Baysian DOSY Transform Spectra.
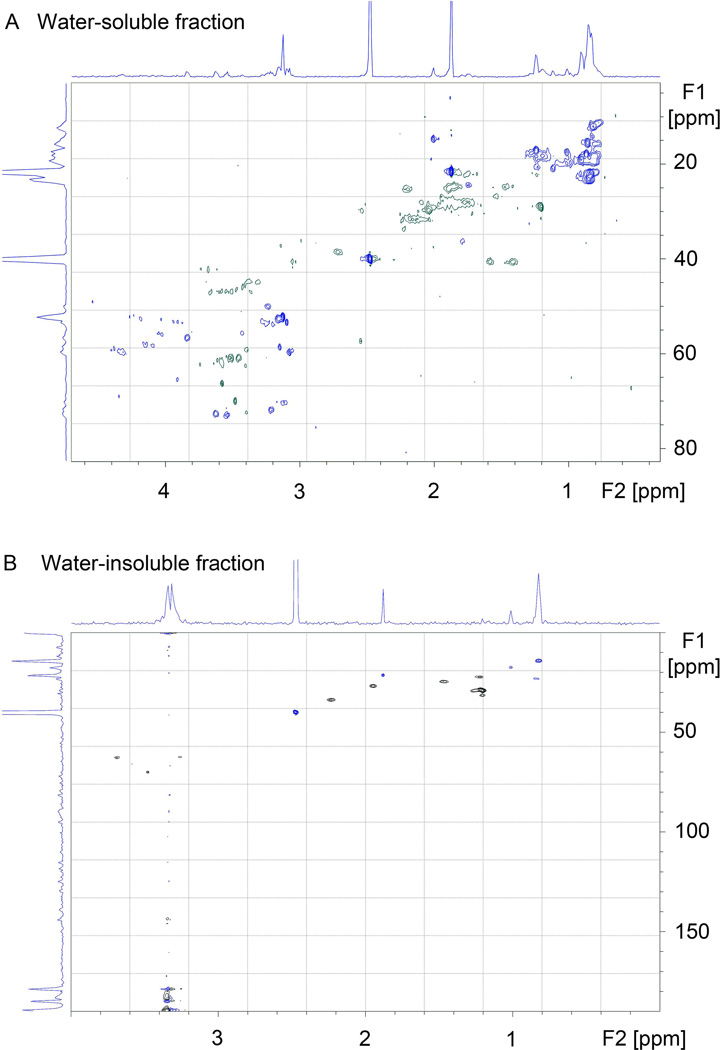
HSQC NMR spectra of the DMSO-soluble and -insoluble biofilm fractions from 24 hr biofilms (Fig 4) are typical for random coil polypeptides with the expected distribution of methyne, methylene, and methyl proton/carbon pairs along with characteristic δ-CH2 serine peaks (δH 3.6-3.9 ppm, δC 62-65 ppm). The spectrum for the water-soluble fraction (Fig 4A) is more complex and appears to contain both polypeptide and saccharide peaks, consistent with the IR results. The spectrum from the water-insoluble material (Fig 4B) is noticeably different from typical polysaccharide or nucleic acid HSQC spectra, which would be dominated by hydroxylated carbons mostly downfield of 70 ppm, a few downfield methyl groups, and in the latter case, sp2 hybridized carbons.
Figure 4. HSQC NMR spectra of DMSO-soluble and –insoluble ECM from NTHi 9274 biofilms.
ECM contains protein and DNA
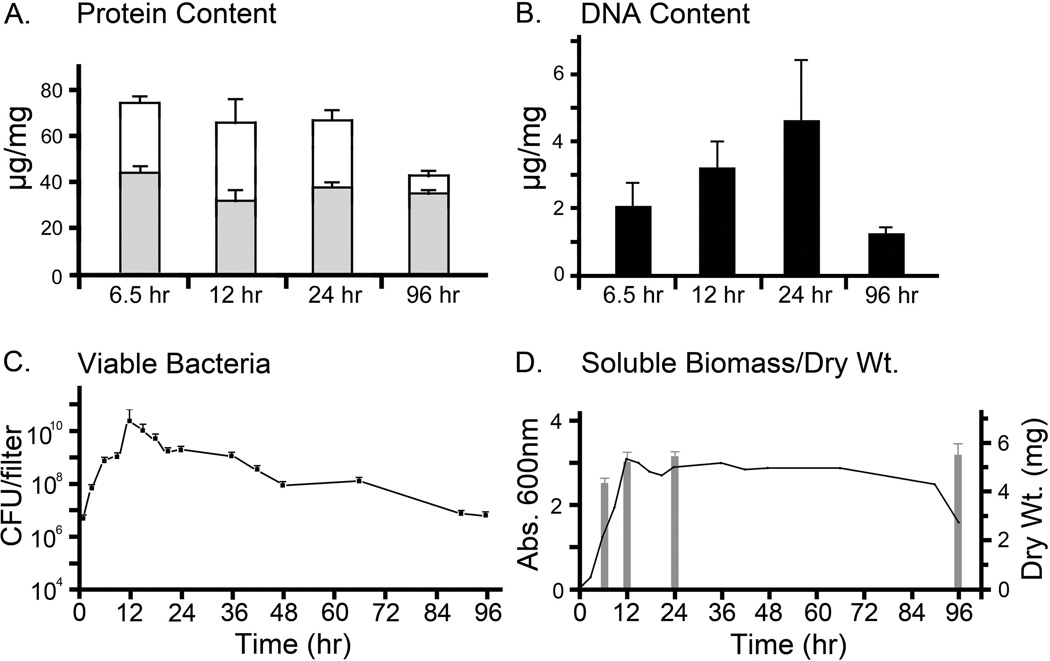
Protein assays on the biofilm revealed the presence of increased amounts of protein in the biofilm ECM than were extractable from the non-ECM biofilm fractions (Fig 5A). The protein content of biofilm ECM from the 24 hr and 96 hr biofilms was 66 µg protein/mg biofilm dry weight and 41 µg protein/mg biofilm dry weight, respectively (Fig 5A). We also estimated biofilm ECM protein content from 6.5 hr and 12 hr biofilms, which were 73 and 65 µg protein/mg biofilm dry weight, respectively (Fig 5A). Total extractable protein levels present in the non-ECM biofilm fractions, in µg protein/mg biofilm dry weight, were 44 (6.5 hr), 31 (12 hr), 37 (24 hr) and 34 (96 hr). DNA content, in µg DNA/mg biofilm dry weight, of the biofilm ECM increased from 2 (6.5 hr), through 3 (12 hr) to a max of 5 at 24 hr (Fig 5B). The DNA content of the biofilm ECM from the 96 hr biofilm dropped to 1 µg/mg biofilm dry weight (Fig 5B).
Figure 5. Protein, DNA and viable bacteria in the forming biofilms.
A. Protein content of non-ECM biofilm fraction (shaded bars) and of biofilm ECM (open bars) from NTHi biofilms. The biofilm ECM contained more protein than the non-ECM biofilm fraction, which contained mostly bacteria. The protein content of the non-ECM biofilm fraction remained mostly constant, while the protein content of the biofilm ECM dropped over time.
B. DNA content of ECM from NTHi biofilms. The amount of DNA in the ECM fraction of the biofilms increased until it peaked at 24 hr. DNA in the ECM from the 96 hr biofilm was much lower than DNA present in the 12 hr and 24 hr biofilms.
C. Colony biofilms were collected in PBS at increasing time intervals over a 96 hr period. The numbers of viable bacteria in these suspensions were estimated. Over the 96 hr period, the numbers of viable bacteria increased 12-fold by 12 hr but returned to initial inoculum levels by 96 hr. CFUs were 1 hr - 6.1x106; 9 hr - 1.3x109; 12 hr - 3.0x1010; 15 hr - 1.3x1010; 24 hr - 2.3x109; 96 hr - 8x106.
B. The OD600 of total biofilm material suspended in PBS (left scale) was used to estimate total biomass in the biofilms. The biofilm biomass peaked at 12 hr and remained at high levels but dropped at the last time point. Dry weights of forming biofilms, indicated with a bar histogram (and right scale), showed a rapid increase in biofilm biomass by 6.5 hr, increasing slowly to a maximum at 24 hr. The dry weight of the 96 hr biofilm was similar to that observed for the 12 hr and 96 hr biofilms.
Culturable bacteria were present in NTHi biofilms
Culturable NTHi in forming biofilms (as CFUs per biofilm before removal of ECM) over a 96 hr period were estimated (Fig 5C). The numbers of viable bacteria slowly increased from approx. 6.1 × 106 CFUs per biofilm at 1 hr after incubation, to 1.3 × 109 CFUs per biofilm after 9 hrs (Fig 5C). Between 9 hr and 12 hr, the numbers of viable bacteria increased, reaching a peak by 12 hr (approx. 3.0 × 1010 CFUs per biofilm). The numbers of viable bacteria per biofilm dropped to approx 1.3 × 1010 at 15 hr, continued to fall until at 24 hr, they had dropped to 2.3 × 109 CFUs per biofilm, a level similar to that observed at 9 hr (Fig 5C). Viable bacteria per biofilm continued to fall until at 96 hr (8.0 × 106) the number of viable bacteria was greater than were present in 1 hr biofilms (6.1 × 106). The 96 hr biofilms contained fewer culturable bacteria (8.0 × 106) than were present in 24 hr biofilms (2.3 × 109).
ECM contained biofilm-specific proteins
A proteomic analysis of the biofilm ECM of 24 hr and 96 hr biofilms was compared with a similar proteomic analysis of total proteins taken from planktonic bacteria to produce a list of 153 identified proteins (supplemental Table 1). All proteins common to both the planktonic and biofilm lists were disregarded and the remaining 18 proteins were designated as being biofilm- specific proteins (Table 3). Transketolase 2 and chaperone protein dnaK 2 were associated only with the 96 hr biofilms, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was associated only with the 24 hr biofilm. The remaining 15 identified proteins were present in extracellular matrix from 24 hr and 96 hr biofilms. Seven proteins were associated with the bacterial cell membranes (including OMP P1, OMP P2 and OMP P5), while the others were either enzymes involved with metabolism (e.g. transketolase 2, ornithine carbamoyltransferase), glycolysis (e.g GAPDH), or transcription (e.g. DNA-directed RNA polymerase), or were chaperones (Table 3).
Table 3.
Biofilm specific proteins identified by LC/MS/MS of soluble ECM fraction
| Identified Proteins | UniProt | mol wt (kDa) |
Gene | Taxonomy | Location | Peptide matches |
Number of matches |
Secondary Function |
|
|---|---|---|---|---|---|---|---|---|---|
| Biosynthesis/Metabolism/Catabolism | 24 hr | 96 hr | |||||||
| Oligopeptidase A | P44573 | 78 | prlC | Haemophilus influenzae | (Cytoplasm, presumed) | 3 | 2 | 2 | |
| Ornithine carbamoyltransferase, catabolic | P44770 | 38 | arcB | Haemophilus influenzae | Cytoplasm | 4 | 2 | 1 | adhesion |
| Probable 5'-nucleotidase | P44569 | 66 | NA | Haemophilus influenzae | (Cytoplasm, presumed) | 4 | 1 | 2 | adhesion |
| Purine nucleoside phosphorylase deoD-type | Q4QN30 | 26 | deoD | Haemophilus influenzae | (Cytoplasm, presumed) | 3 | 2 | 2 | temperature shock adaptation |
| Transketolase 2 | P57958 | 73 | tktB | Pasteurella multocida | (Cytoplasm, presumed) | 4 | 0 | 1 | |
| Cell membrane/transport/cell surface | |||||||||
| High-affinity zinc uptake system protein znuA | P44526 | 38 | znuA | Haemophilus influenzae | Periplasm | 4 | 3 | 3 | Intracellular survival |
| Iron-utilization periplasmic protein | P35755 | 36 | fbpA | Haemophilus influenzae | Periplasm | 3 | 4 | 5 | |
| Outer membrane protein P1 | P43838; Q9KHG0 | 49 | ompP1 | Haemophilus influenzae | Cell membrane | 4 | 3 | 2 | |
| Outer membrane protein P2 | Q48219; Q48024; P46027 | 40 | ompP2 | Haemophilus influenzae | Cell membrane | 4 | 5 | 4 | |
| Outer membrane protein P5 | P43840 | 38 | ompA | Haemophilus influenzae | Cell membrane | 4 | 3 | 4 | |
| Sialic acid-binding periplasmic protein siaP | P44542 | 37 | siaP | Haemophilus influenzae | Periplasm | 4 | 5 | 4 | |
| Uncharacterized periplasmic iron-binding protein HI0362 | Q57449 | 32 | NA | Haemophilus influenzae | Periplasm | 4 | 2 | 2 | |
| Chaparone/Protein folding/stress response | |||||||||
| Chaperone protein dnaK 2 | Q5N1J4 | 68 | dnaK2 | Synechococcus elongatus | (Cytoplasm, presumed) | 4 | 0 | 1 | adhesion |
| Probable FKBP-type peptidyl-prolyl cis-trans isomerase | P44760 | 26 | NA | Haemophilus influenzae | (Cytoplasm, presumed) | 4 | 1 | 1 | |
| Glycolysis/energy metabolism | |||||||||
| Dihydrolipoyl dehydrogenase | P43784 | 51 | pdA | Haemophilus influenzae | Cytoplasm | 4 | 1 | 1 | adhesion |
| Glyceraldehyde-3-phosphate dehydrogenase |
Q59906; P0C0G7 |
36 | gap | Streptococcus dysgalactiae; Streptococcus pyogenes | Cytoplasm | 4 | 2 | 0 | adhesion |
| Pyruvate kinase (Fragment) | P19680 | 22 | pyk | Spiroplasma citri | (Cytoplasm, presumed) | 4 | 1 | 1 | |
| Translation/Transcription | |||||||||
| DNA-directed RNA polymerase subunit beta' | Q4QN33 | 150 | rpoB | Haemophilus influenzae | (Cytoplasm, presumed) | 4 | 2 | 1 | adhesion |
A similar proteomic analysis was performed on proteins extracted from the filters from which the NTHi biofilms had been removed for other analysis. As before, the proteins obtained was compared with the proteins identified from planktonic bacteria and all common proteins excluded. A total of 60 proteins were identified on these washed filters. All 60 of them were on the filters from the 96hr biofilm, 46 were on filters from all the sampled time points, 11 on all filters except the 6.5 hr, 2 on 24 hr and 96 hr biofilm filters and one, a hypothetical protein (CGSHiEE_00395), identified only on the 96 hr biofilm filter (Table). The presence of bacterial cells in the residual material attached to the filters after biofilm removal made it impossible for us to determine if the proteins we identified were ECM-specific, or if they were also associated with biofilm bacterial cells. However, traces of 5 proteins identified in the more soluble ECM fraction were also found in the proteins remaining attached to the filter after washing. Two of the proteins were membrane-associated, OMP-P2 and OMP-P5. The other three proteins were cytoplasmic and involved with transport (periplasmic iron binding protein) or transcription (molecular chaperone DnaK and DNA-directed RNA polymerase). The DNA-directed RNA polymerase and OMP P2 proteins were chosen for further immunocytochemical analysis using criteria based on their location within the biofilm fractions as well as antibody availability and specificity.
DNA-directed RNA polymerase and OMP P2 were components of the NTHi biofilm ECM
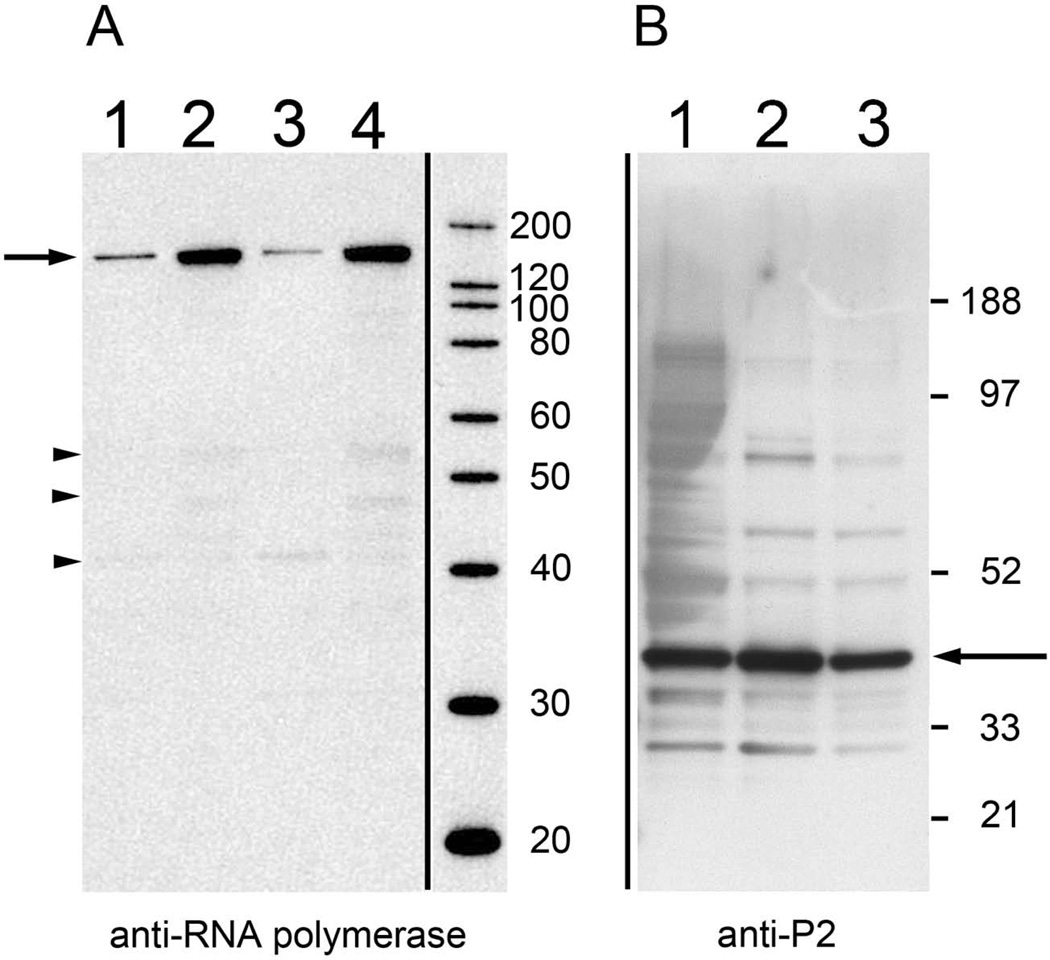
On western blots the anti-RNA polymerase antibodies bound to a protein band in the 150 kDa range (where the DNA-directed RNA polymerase would be expected to appear) and showed only minimal fragmentation of the protein (Fig 6A). The anti- RNA polymerase antibodies bound to 24 hr and 96 hr biofilm ECM, and 24 hr and 96 hr non-ECM biofilm fractions (Fig 6A). Anti-OMP P2 YKA antibodies bound to a protein band in the 42 kDa range (Fig 6) and recognized proteins in the biofilm ECM from 24 hr and 96 hr biofilms. The western blot analysis confirmed proteomic results that both proteins were present in biofilm ECM.
Figure 6. Anti-DNA-directed RNA polymerase and OMP P2 antibodies label NTHi biofilm ECM proteins.
A. Western blot using anti-DNA-directed RNA polymerase. A protein band in the ECM and in non-ECM biofilm fractions of 24 hr and 96 hr biofilms was detected in the 150kDa range (arrow). Weak binding bands in the range of 45 – 55 kDa (arrowheads) were also present.
Western blot showing label on:
Lane 1: ECM, 24 hr biofilm
Lane 2: Insoluble fraction, 24 hr biofilm
Lane 3: ECM, 96 hr biofilm
Lane 4: Insoluble fraction, 96 hr biofilm
B. Western blot using anti-OMP P2 antibodies
A protein band in the 42kDa range (arrow), corresponding to the OMP P2 protein, in the 24 hr and 96 hr biofilms was detected.
Lane 1: ECM, 24 hr biofilm
Lane 2: ECM, 96 hr biofilm
Lane 3: Total Extract, 96 hr biofilm
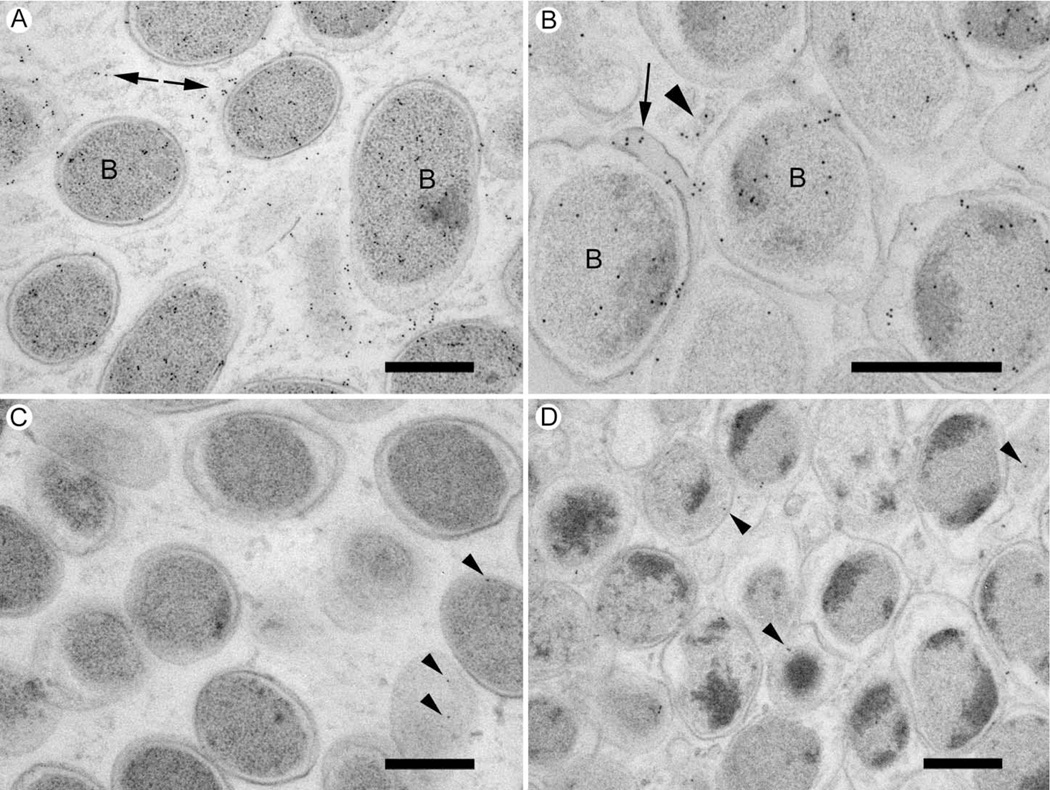
ImmunoEM was then used to screen for anti-RNA polymerase and anti-OMP P2 labeling on the ECM of in vivo formed biofilms. Anti-RNA polymerase label was observed over bacterial cells in the biofilms (Fig 7) and associated with the bacterial ECM (Fig 7). Extracellular label was present in 24 hr biofilms (Fig 7A), but there appeared to be more labeling on sections through the 96 hr biofilms (Fig 7B). In the 96 hr biofilms, the label was associated with the cell cytoplasm, extracellular regions of the biofilm, small extracellular vesicles, and with ECM that was in close association with the bacterial cells (Fig 7B). Negative controls, where specific anti-RNA polymerase antibodies were omitted, showed negligible amounts of gold labeling over the biofilm sections (Fig 7C & D).
Figure 7. DNA-directed RNA polymerase is located in the NTHi biofilm ECM.
Sections through cryo-preserved NTHi biofilms were labeled with specific monoclonal antibodies, rabbit anti-mouse bridging antibody, and 10nm protein A-gold.
A. 24 hr biofilm. Specific label is associated with the bacterial cells (some of which are labeled “B”) and with ECM (arrows).
Scale bar = 500nm.
B. 96 hr biofilm. Specific label is found over the bacterial profiles (B) as well as ECM around the bacteria (Arrow), and with extracellular vesicles (arrowhead).
Scale bar = 500nm.
C. 24 hr biofilm control. Specific primary antibody was omitted and labeled only with secondary antibody and protein A-gold. Some gold particles associate with bacterial profiles (arrowheads) but the labeling density is lower than when primary antibody is included.
Scale bar = 500nm.
D. 96 hr biofilm control labeled only with secondary antibody and protein A gold. Non-specific labeling is detectable over bacterial cells and ECM (arrowheads). The labeling density is lower than when primary antibody is included.
Scale bar = 500nm.
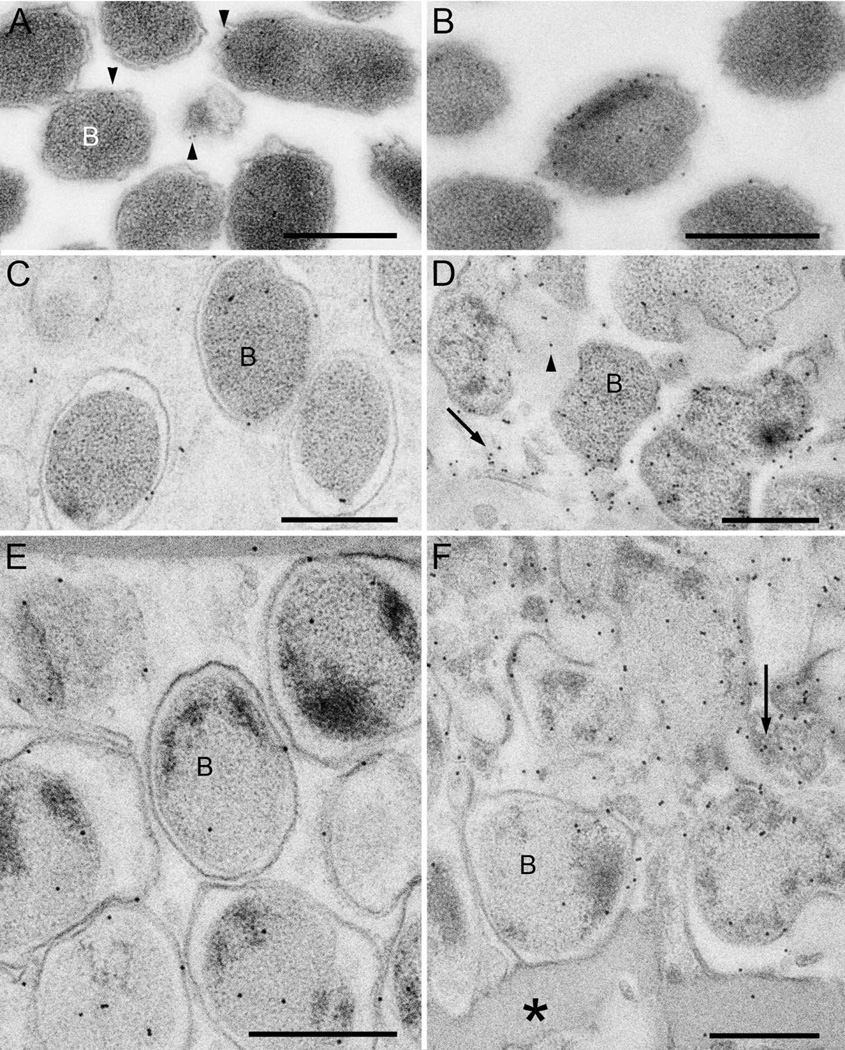
The anti-OMP P2 YKA antibodies labeled planktonic forms of bacteria but showed a low level of label over most cells (Fig 8A). Occasional cells were more densely labeled (more than 5 gold particles) with the antibodies (Fig 8B). When quantified, it was revealed that only 2% of the planktonic cells were heavily labeled with the anti-OMP P2 YKA antibodies (Table 4). Estimates of the mean labeling density of labeling over the whole cell population revealed a distribution of 2.8 gold/µm2 on the bacterial cell area and 0.3 gold/µm on membrane length (Table 4). On 24 hr NTHi biofilms anti-OMP P2 YKA labeling at the top of the biofilm was mostly associated with the cell membrane (Fig 8C). However, at the bottom of the biofilm there was more detectable label over the cell cytoplasm and the ECM (Fig 8D). The anti-OMP P2 YKA labeled cell membranes at the top of 96 hr NTHi biofilms (Fig 8E). At the bottom of the biofilm, the label was associated with bacteria and extracellular material consisting of what appeared to be cell fragments (Fig 8F). Some of the amorphous material at the bottom of the biofilm did not label with the anti-OMP P2 YKA antibodies (Fig 8F).
Figure 8. Outer membrane protein OMP P2 is located in the NTHi biofilm ECM.
Sections through cryo-preserved NTHi biofilms were labeled with specific antibodies and 10nm protein A-gold.
A. The planktonic bacteria (B) labeled with the anti-OMP P2 YKA antibody with a low level of label over most cells (arrowheads).
B. A higher labeling density with the anti-OMP P2 YKA antibody was observed over some bacteria.
C. Bacteria (B) at the top of the 24 hr biofilm were labeled mostly over the cell membrane.
D. Bacteria (B) at the bottom of the 24 hr biofilm label with the antibody. Labeling was also observed associated with the ECM (arrowhead), and with membrane structures in the ECM.
E. Labeling at the top of the 96 hr biofilm is associated with the cell membranes of the bacteria (B).
F. At the bottom of the biofilm the label was associated with bacteria (B), with ECM, and with membranes in the ECM (arrow). Label was not detected over amorphous material at the bottom of the biofilm (asterisk).
Table 4.
Quantification of anti-OMP P2 YKA labeling on sections of planktonic NTHi bacteria
| A) Percentage of heavily labeled cells | |
| OMP P2 positive | 2% |
| OMP P2 negative | 98% |
| p < 0.0001 | |
| B) Mean labeling densities of anti-OMP P2 YKA label on planktonic cell population | ||
| Protein A gold (PAG) labeling only | 0.03 ± 0.05 /µm | |
| Gold particles associated with bacteria cell | 2.8 ± 1.2 /µm2 | |
| Gold particles associated with cell wall | 0.3 ± 0.1 /µm | |
| p < 0.001 (compared with PAG only) | ||
DISCUSSION
The extracellular part of the biofilm is one of the most important components, contributing to bacterial attachment and antimicrobial tolerance (Costerton, 1999). The extracellular component of bacterial biofilms is often called the EPS, an abbreviation for either exopolysaccharides or extracellular polymeric matrix. The matrix of newly forming and immature biofilms contain more than polysaccharide, and the components are not fully polymerized so the term EPS does not adequately describe the biofilm component we are studying. A more generic term for the non-cellular components of a forming biofilm is the extracellular matrix (ECM) a term used previously to describe extracellular biofilm components (Hall-Stoodley & Lappin-Scott, 1998, Dongari-Bagtzoglou, 2008). Other studies of biofilm ECM has revealed it to be composed of varying amounts of polysaccharide, protein, nucleic acid and lipid, the actual proportions dependent on many different factors, two of which being the species of biofilm-forming bacteria and the developmental stage of the biofilm (Oosthuizen et al., 2002, Sauer et al., 2002, Sauer, 2003, Fux et al., 2005, Cao et al., 2011, Mueller et al., 2011).
In this work we extend our studies of ECM proteins of immature NTHi biofilms (Webster et al., 2004, Gallaher et al., 2006, Webster et al., 2006) where we documented proteins in the biofilm ECM with diverse metabolic and structural functions. Using proteomics, FTIR, NMR, microscopy, and a variety of biochemical approaches we have now clearly identified biofilm-specific proteins present in the ECM of NTHi biofilms formed after 24 hr and 96 hr. Most importantly, we have confirmed an extracellular location for two of the biofilm-specific ECM proteins using an immunocytochemical approach.
A list of proteins identified in the biofilm ECM (identified using a high stringency 99.9% probability) was compared with proteins from planktonic bacteria identified at lower stringency (80% probability). Of the18 biofilm-specific proteins we have identified, twelve had been detected previously in NTHi biofilm ECM (Gallaher et al., 2006), thus further strengthening their identification as biofilm-specific proteins.
The biofilm-specific proteins we identified have been linked to biofilms formed by NTHi and other bacteria. Some have roles in biofilm formation, such as the sialic acid-binding periplasmic protein (siaP) in NTHi biofilms (Swords et al., 2004), and RNA polymerase subunits in Streptococcus mutans and Mycobacterium smegmatis ((Mathew et al., 2006, Xue et al., 2010).
A number of the biofilm-specific proteins have previously reported to be expressed on bacterial cell surfaces and implicated in a secondary role of adhesion or attachment. Surface-associated proteins that have been implicated in biofilm formation include glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Pancholi & Fischetti, 1992, Bergmann et al., 2004), ornithine carbamoyltransferase (OCT) (Hussain et al., 1999, Hughes et al., 2002, Oosthuizen et al., 2002, Winterhoff et al., 2002, Alam et al., 2009), the chaperone protein DnaK 2 (Mangalappalli-Illathu et al., 2009, Singh et al., 2012), and 5'-nucleotidase (Davies et al., 2009, Fan et al., 2012). The presence of these proteins in the early, or immature biofilms, where initial attachment is important might be expected if they have a secondary role in adhesion. It is not surprising that proteins with multifunctional roles are present in bacteria given their preponderance in eukaryotic cells (Cassiday & Maher, 2002, Scott et al., 2011, Muresan & Muresan, 2012).
The biofilm ECM fraction was removed from colony biofilms using mild sonication, however, a thin fibrous net of biomass on filters where 24 hr biofilms had formed, and an amorphous layer of biomass on the 96 hr filters remained on the filters. A proteomic analysis of the washed filters revealed the attached biomass to contain proteins with a variety of cellular functions. The biofilm biomass remaining attached to the filter after extensive washing of the 96 hr biofilm might explain the drop in biofilm biomass observed in our chemical analysis. The presence of bacterial cells embedded in the attached biomass of the 96 hr biofilm filters suggests both a protective and adhesive role for this material.
The presence of many proteins in the biomass remaining attached to the filter after washing suggests that some may have adhesion properties. Five of the 60 proteins identified as being biofilm-specific were also identified in the biofilm ECM fraction, thus suggesting that the adhesion properties of these proteins was not strong. However, from the images of the washed biofilms and the results of the proteomic analysis it seems that adhesion to substrates is a process that develops over time. More material was found attached to the filters from biofilms formed after 96 hr than on filters from 24 hr biofilms. Similarly, more biofilm-specific proteins were identified on filters from 96 hr biofilms, an accumulation that increased over time.
We were concerned that the sonication protocol might potentially disrupt viable bacteria and the contents from disrupted cells could therefore contribute to the ECM in our analysis. However, biomass estimates and CFU data from the forming biofilm indicated a gradual increase in culturable bacteria during the first 12 hr of the experiment providing evidence that cell division was occurring. The drop in the numbers of culturable bacteria between 12 hr and 24 hr might suggest that significant bacterial lysis was occurring. However, the loss of culturable bacteria could also result from bacteria switching to an unculturable biofilm phenotype (Costerton et al., 2003). Our ultrastructural analysis of the NTHi biofilms showing the presence of cell fragments at the biofilm base in the 96 hr biofilms would support a role for cell death in biofilm development (Fagerlind et al., 2012). Whether lysed bacteria were the source of proteins identified in the ECM could not be definitively answered, indicating a need for further studies on this subject.
In order to confirm an extracellular location for the proteins identified in the EMC fraction, we performed an immunocytochemical analysis using specific antibodies applied to thin sections. We were limited in which proteins we could detect immunocytochemically because few commercial antibodies to the biofilm-specific proteins were available that targeted bacterial proteins. Most of the available antibodies were directed to mammalian homologues of our identified proteins. Moreover, many of the commercial primary antibodies detected multiple bands on western blots, suggesting multiple bacterial proteins were being recognized in addition to our target proteins.
Bacteria-specific activity was also detected by western blotting in polyclonal secondary, or bridging antibodies, directed to affinity purified mammalian immunoglobulins, a problem we had previously encountered when using polyclonal antibodies (Webster et al., 2006). We concluded that the bacterial activity was a possible consequence of the use of Freund’s adjuvant, an emulsion of mineral oil and mycobacteria commonly used to stimulate immunoglobulin production. The bacteria-specific activity present in secondary, or bridging antibodies could thus be removed by including bacterial protein suspension in blocking solution (Webster et al., 2006). However, this simple yet effective affinity purification approach could not be applied to the primary antibodies because of the possibility of it removing specific activity to the target proteins. Therefore we excluded commercial polyclonal antibodies raised to mammalian proteins from this study. The two antibodies chosen were a commercial monoclonal antibody to RNA polymerase, and a polyclonal antibody (anti-OMP P2 YKA) directed against the NTHi outer membrane protein P2 (Neary & Murphy, 2006).
The anti- RNA polymerase antibodies labeled cells, cell fragments and the biofilm ECM, thus confirming the proteomic result that the protein was present in an extracellular site. It is clear that immunolabeling was the most definitive way of showing that the DNA directed RNA-polymerase protein, a cytoplasmic protein, was free within the ECM and was not exclusively localized within bacterial cells. We strongly suggest that immunolabeling experiments are the best approach for confirming proteomic results, and unambiguously demonstrating the location of proteins in the ECM, or on structures associated with the ECM.
The presence of proteins associated with the cell membrane or periplasmic space, suggest that bacterial lysis was occurring in the biofilm. This speculation is supported by the presence of cell fragments at the base of both 24 hr and 96 hr biofilms. The anti-OMP P2 YKA antibody we used was a polyclonal rabbit antiserum to a peptide corresponding to the loop 6 region of the P2 porin protein (AKTKNYKAKHEKS), a part of the molecule with an intra-membrane location (Neary & Murphy, 2006). All current data on the OMP P2 protein indicate that it is not regulated by the cells, being constitutively expressed and is present in abundant amounts in the outer membrane. However, immunolabeling revealed a lower labeling density over planktonic cells when compared with bacterial cells in the biofilm. In addition, only about 1% of planktonic cells expressed large amounts of the protein. The increased labeling over biofilm bacteria could indicate the occurrence of a structural alteration to the NTHi bacteria as they take on a biofilm phenotype. Structural modifications to the bacterial membrane could result in higher labeling efficiency because of increased antibody accessibility to the antigen. The anti-OMP P2 YKA antibodies labeled intact cells at the top of the 24 hr and 96 hr biofilms with more label being present than that associated with planktonic bacteria. At the base of the biofilms the anti-OMP P2 YKA antibodies also labeled intact cells as well as membrane fragments. The absence of OMP P6 (Webster et al., 2006) in the proteomic analysis of the NHTi biofilm ECM suggest that cells or intact membranes, which contained the OMP P6 protein, were being removed during centrifugation. However, small OMP P2-positive membrane vesicles could remain in the biofilm ECM fraction, if they were low-density vesicles, and could not be removed by centrifugation.
Outer membrane vesicles (OMVs) formed by NTHi in planktonic culture have been recently purified and subjected to a proteomic analysis to reveal the presence of the OMPs P1 (OMP-P1), P2 (OMP-P2) and P5 (OMP-P5) (Sharpe et al., 2011). Similar vesicles in the ECM of 24 hr and 96 hr NTHi biofilms labeled with the anti-OMP-P2 YKA and anti-RNA polymerase in our studies. The iron utilization periplasmic protein we identified in the ECM of the NTHi biofilms was also present in the NTHi OMVs as was HMW1 (Sharpe et al., 2011), an adhesin we have previously immunolocalized in NTHi biofilms (Webster et al., 2006). Outer membrane proteins P2 (OMP-P2) and P5 (OMP-P5) have recently been identified as promising candidates for the detection of NTHI biofilms (Das et al., 2014). Positive detection in nasopharyngeal secretions of chinchillas experimentally infected with NTHi (Das et al., 2014) suggests such a diagnostic test might be feasible.
A preliminary analysis of the contents of the biofilm ECM from 24 hr (early stage) biofilm was performed using nondestructive spectroscopic methods. For this, we applied NMR and FTIR spectroscopy as these techniques provided a rapid and detailed means of chemically characterizing the biofilm suspensions. Analysis of biofilms formed by other bacteria using FTIR spectroscopy is a successful approach for identifying ECM components (Eboigbodin & Biggs, 2008, Baum et al., 2009, Cao et al., 2011); however, NMR analysis has only recently been applied to the study of components of bacterial and fungal biofilms (Jakobsen et al., 2011, Seo et al., 2012).
Infrared spectroscopy is a powerful tool for nondestructive analysis of the chemical composition of bacterial biofilms. Briefly, this technique makes use of fact that vibrational and rotational-vibrational changes within a molecule are associated with specific energies, in an analogous fashion to UV-visible spectroscopy, which probes electronic transitions within molecules. Excitation of the energy levels occurs in well-defined regions of the electromagnetic spectrum, usually in the range of 4000 to 600 cm−1. The energy of the band provides key structural information and can help identify the associated functional group, while the band intensity can be correlated quantitatively to their abundance in the sample.
Using FTIR spectroscopy we were able to identify differences between the biofilm ECM and the non-ECM biofilm fractions, which consisted mostly of intact bacteria (Gallaher et al., 2006). The non-ECM biofilm fractions were mostly proteinaceous with no evidence of polysaccharide, as might be expected from concentrated bacterial cells. The biofilm ECM from 24 hr and 96 hr biofilms showed evidence of both protein and polysaccharide. Although there were differences between the water-soluble and water-insoluble samples of the lyophilized biofilm ECM, the FTIR analysis did not detect differences in protein and polysaccharide levels between samples taken from the 24 hr and 96 hr biofilms.
The NMR spectra supported the FTIR findings in detecting differences between the biofilm ECM and non-ECM biofilm fractions, but no differences in samples taken from 24 hr or 96 hr biofilms. In addition to detecting the presence of polysaccharides and proteins in the water-soluble fractions of the biofilm ECM, the NMR spectra also revealed the presence of small molecules and molecules with a wide size distribution, which might suggest that proteolysis is occurring in the ECM. Together, the FTIR and NMR data provided compelling evidence to indicate that ECM is composed mostly of proteins. Determining the nature of these proteins required a different analytical approach.
While polypeptide and polysaccharide profiles were repeatedly detected by NMR and FTIR analysis, the data did not reveal nucleic acids, a possible limitation of this analytical approach. DNA in the biofilm ECM was detected using a specific DNA assay that we previously used to detect DNA in Pseudomonas fluorescens biofilms (Baum et al., 2009). Release of DNA into the ECM is a common occurrence in NTHI biofilms (Goodman et al., 2011) as well as in biofilms formed by other bacteria (Petersen et al., 2005, Steinberger & Holden, 2005, Allesen-Holm et al., 2006, Bockelmann et al., 2006, Spoering & Gilmore, 2006, Izano et al., 2008, Mann et al., 2009, Flemming & Wingender, 2010). Released DNA appears to play a structural role in biofilms formed by Burkholderia cenocepacia, a pathogen that colonizes the lungs of cystic fibrosis patients (Novotny et al., 2013). The mechanism for DNA release into the ECM is not well understood, but our use of static culture conditions, where bacterial cells were not put under mechanical stress, in contrast to previous studies where a sheer stress was imposed on bacterial cells (Jurcisek & Bakaletz, 2007, Izano et al., 2009, Goodman et al., 2011), suggests that DNA release is an active process, not a result of cell damage. This speculation is supported by observations in other bacterial species where DNA release appears to be an event controlled by biofilm-forming bacteria (Spoering & Gilmore, 2006).
In conclusion, we have observed the early events of NTHi biofilm formation using an in vitro system where bacteria rapidly attached to an abiotic substrate and produced extracellular material to cover attaching bacteria. FTIR and NMR detected protein and polysaccharide in the NTHi biofilm ECM. These analyses were able to detect differences between protein and polysaccharide levels in biofilm ECM and non-ECM fractions. Chemical analysis of the biofilm ECM showed a gradual decrease in protein content over time and an increase in DNA content up to 24hr. Viable bacteria in the early biofilms increased to a peak at 12 hr, and although the numbers fell over time, the 96 hr biofilms contained amounts of bacteria higher than in the initial inoculum.
Proteomics identified 18 proteins easily isolated proteins that were unique to the NTHi biofilm ECM, suggesting that, in vivo, the proteins could be present in fluids taken from infected regions. We have also identified 60 proteins present in the biomass remaining attached to the filter substrate after the biofilm had been removed by washing. We have demonstrated by immuno-electron microscopy that two biofilm proteins (DNA-directed RNA polymerase and OMP P2 YKA) were present in the ECM of the immature NTHi biofilms, confirming the ECM location obtained by proteomic analysis. Many of the biofilm-specific ECM proteins had secondary functions in cellular adhesion, indicating an important role in bacterial cell attachment, especially during the early stages of biofilm formation. The ECM proteins may also be important components of the complex biofilm structures constructed by NTHi and other species of bacteria. It is clear that no matter what role these extracellular proteins play in biofilm formation or maintenance, it will be important to determine how and why such large amounts of cytoplasmic and membrane proteins are released into the biofilm ECM.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are especially grateful to financial support provided by their respective organizations, and the generosity of the Ahmanson Foundation for their support in equipping the Imaging Center at the House Research Institute. Rapid freezing and freeze substitution was carried out using equipment purchased with funds from the National Science Foundation (NSF #0722354). The study was also supported in part by The Fritz Burns Foundation, The Hope for Hearing Foundation, the Deafness Research Foundation, the Hearst Foundation, and the Capita Foundation. A grant from the NIDCD (5 P-30 DC006276-03) supported the Ahmanson Imaging Core where a substantial part of this work was performed. Prof. T. F. Murphy, University at Buffalo, State University of New York provided polyclonal antibodies to the OMP P2 protein. Dr. Gabriel B. Gugiu of the Mass Spectrometry and Proteomics Core Facility at the Beckman Research Institute of the City of Hope performed the proteomic analysis of protein preparations from the 6.5 hr, 12 hr, 24 hr and 96 hr filters after biofilm removal. Tim Gallaher and Richard Johnson provided expert bioinformatics assistance and instruction. Tyler Woolsey and Nandini Girish assisted with early microscopy and bacterial culturing (NG: immunolabeling, TW viable bacteria and biomass estimates).
List of abbreviations used
- BHI
Brain Heart Infusion
- CFU
Colony Forming Units
- COPD
Chronic Obstructive Pulmonary Disease
- DAPI
4',6-diamidino-2-phenylindole
- DMSO
Dimethyl sulfoxide
- DNA
Deoxyribonucleic acid
- DOSY
Diffusion Ordered Spectroscopy
- ECM
Extracellular Matrix
- EPS
Exopolysaccharide
- FTIR
Fourier Transform Infrared Spectroscopy
- HSQC
Heteronuclear Single Quantum Coherence
- NAD
Nicotinamide adenine dinucleotide
- NMR
Nuclear magnetic resonance
- NTHi
non-typeable Haemophilus influenzae
- OD600
Optical Density read at 600 nm wavelength
- OMP
Outer Membrane Protein
- OMV
Outer Membrane Vesicle
- PBS
Phosphate Buffered Saline
- RNA
Ribonucleic acid
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEM
Scanning Electron Microscope/Microscopy
- TEM
Transmission Electron Microscope/Microscopy
Footnotes
The authors have stated no conflict of interest.
Contributor Information
Siva Wu, Email: sivawu@lbl.gov, Bioenergy/GTL & Structural Biology Department, Life Sciences Division, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA, Phone: (626) 318 6436.
Marc M. Baum, Email: m.baum@oak-crest.org, Department of Chemistry, Oak Crest Institute of Science, 2275 E. Foothill Blvd, Pasadena, CA 91107, USA, Phone: (626) 817 0883.
James Kerwin, Email: jlkerwin@hotmail.com, UCLA Radiology Department, 10833 LeConte Avenue, Los Angeles, CA 90095, USA, Phone: (310) 562 9016.
Debbie Guerrero-Given, Email: Debbie.guerrero-given@mpfi.org, Max Planck Florida Institute, 1 Max Planck Way, Jupiter, FL 33458, USA, Phone: (561) 972 9000.
Simon Webster, Email: swebs001@gmail.com, One Lamda (Thermo Fisher), 21001 Kittridge Street, Canoga Park, CA 91303, USA, Phone: (626) 643 2875.
Christoph Schaudinn, Email: schaudinnc@rki.de, Robert Koch Institute, Nordufer 20, 13353 Berlin, Germany.
David VanderVelde, Email: davidv@caltech.edu, Division of Chemistry and Chemical Engineering, California Institute of Technology, 1200 E. California Blvd, Pasadena, CA 91125, USA, Phone: (626) 395 3004.
Paul Webster, Center for Electron Microscopy and Microanalysis (CEMMA), University of Southern California, 814 Bloom Walk, Los Angeles, CA 90089, USA.
REFERENCES
- Ahren IL, Janson H, Forsgren A, Riesbeck K. Protein D expression promotes the adherence and internalization of non-typeable Haemophilus influenzae into human monocytic cells. Microb Pathog. 2001;31:151–158. doi: 10.1006/mpat.2001.0456. [DOI] [PubMed] [Google Scholar]
- Ahren IL, Williams DL, Rice PJ, Forsgren A, Riesbeck K. The importance of a beta-glucan receptor in the nonopsonic entry of nontypeable Haemophilus influenzae into human monocytic and epithelial cells. J Infect Dis. 2001;184:150–158. doi: 10.1086/322016. [DOI] [PubMed] [Google Scholar]
- Alam SI, Bansod S, Kumar RB, Sengupta N, Singh L. Differential proteomic analysis of Clostridium perfringens ATCC13124; identification of dominant, surface and structure associated proteins. BMC Microbiol. 2009;9:162. doi: 10.1186/1471-2180-9-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegrucci M, Hu FZ, Shen K, Hayes J, Ehrlich GD, Post JC, Sauer K. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J Bacteriol. 2006;188:2325–2335. doi: 10.1128/JB.188.7.2325-2335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- Barkai G, Leibovitz E, Givon-Lavi N, Dagan R. Potential contribution by nontypable Haemophilus influenzae in protracted and recurrent acute otitis media. Pediatr Infect Dis J. 2009;28:466–471. doi: 10.1097/inf.0b013e3181950c74. [DOI] [PubMed] [Google Scholar]
- Baum MM, Kainovic A, O'Keeffe T, Pandita R, McDonald K, Wu S, Webster P. Characterization of structures in biofilms formed by a Pseudomonas fluorescens isolated from soil. BMC Microbiol. 2009;9:103. doi: 10.1186/1471-2180-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenson CS, Sayles KB, Huang J, Reinhold VN, Garlipp MA, Yohe HC. Nontypeable Haemophilus influenzae-binding gangliosides of human respiratory (HEp-2) cells have a requisite lacto/neolacto core structure. FEMS Immunol Med Microbiol. 2005;45:171–182. doi: 10.1016/j.femsim.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Rohde M, Hammerschmidt S. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect Immun. 2004;72:2416–2419. doi: 10.1128/IAI.72.4.2416-2419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockelmann U, Janke A, Kuhn R, Neu TR, Wecke J, Lawrence JR, Szewzyk U. Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiol Lett. 2006;262:31–38. doi: 10.1111/j.1574-6968.2006.00361.x. [DOI] [PubMed] [Google Scholar]
- Bookwalter JE, Jurcisek JA, Gray-Owen SD, Fernandez S, McGillivary G, Bakaletz LO. A carcinoembryonic antigen-related cell adhesion molecule 1 homologue plays a pivotal role in nontypeable Haemophilus influenzae colonization of the chinchilla nasopharynx via the outer membrane protein P5-homologous adhesin. Infect Immun. 2008;76:48–55. doi: 10.1128/IAI.00980-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Shi L, Brown RN, Xiong Y, Fredrickson JK, Romine MF, Marshall MJ, Lipton MS, Beyenal H. Extracellular polymeric substances from Shewanella sp. HRCR-1 biofilms: characterization by infrared spectroscopy and proteomics. Environ Microbiol. 2011;13:1018–1031. doi: 10.1111/j.1462-2920.2010.02407.x. [DOI] [PubMed] [Google Scholar]
- Carr S, Aebersold R, Baldwin M, Burlingame A, Clauser K, Nesvizhskii A. The need for guidelines in publication of peptide and protein identification data: Working group on publication guidelines for peptide and protein identification data. Mol Cell Proteomics. 2004;3:531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- Cassiday LA, Maher LJ., 3rd Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW. Introduction to biofilm. Int J Antimicrob Agents. 1999;11:217–221. doi: 10.1016/s0924-8579(99)00018-7. discussion 237–219. [DOI] [PubMed] [Google Scholar]
- Das S, Rosas LE, Jurcisek JA, Novotny LA, Green KB, Bakaletz LO. Improving patient care via development of a protein-based diagnostic test for microbe-specific detection of chronic rhinosinusitis. Laryngoscope. 2014;124:608–615. doi: 10.1002/lary.24333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JR, Svensater G, Herzberg MC. Identification of novel LPXTG-linked surface proteins from Streptococcus gordonii. Microbiology. 2009;155:1977–1988. doi: 10.1099/mic.0.027854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A. Pathogenesis of mucosal biofilm infections: challenges and progress. Expert Rev Anti Infect Ther. 2008;6:201–208. doi: 10.1586/14787210.6.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eboigbodin KE, Biggs CA. Characterization of the extracellular polymeric substances produced by Escherichia coli using infrared spectroscopic, proteomic, and aggregation studies. Biomacromolecules. 2008;9:686–695. doi: 10.1021/bm701043c. [DOI] [PubMed] [Google Scholar]
- Fan J, Zhang Y, Chuang-Smith ON, Frank KL, Guenther BD, Kern M, Schlievert PM, Herzberg MC. Ecto-5'-nucleotidase: a candidate virulence factor in Streptococcus sanguinis experimental endocarditis. PLoS One. 2012;7:e38059. doi: 10.1371/journal.pone.0038059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink DL, Green BA, St Geme JW., 3rd The Haemophilus influenzae Hap autotransporter binds to fibronectin, laminin, and collagen IV. Infect Immun. 2002;70:4902–4907. doi: 10.1128/IAI.70.9.4902-4907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. Relevance of microbial extracellular polymeric substances (EPSs)--Part I: Structural and ecological aspects. Water Sci Technol. 2001;43:1–8. [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Gallaher TK, Wu S, Webster P, Aguilera R. Identification of biofilm proteins in non-typeable Haemophilus influenzae. BMC Microbiol. 2006;6:65. doi: 10.1186/1471-2180-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, Bakaletz LO. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011;4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Lappin-Scott H. Biofilm formation by the rapidly growing mycobacterial species Mycobacterium fortuitum. FEMS Microbiol Lett. 1998;168:77–84. doi: 10.1111/j.1574-6968.1998.tb13258.x. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Hu FZ, Gieseke A, et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herigstad B, Hamilton M, Heersink J. How to optimize the drop plate method for enumerating bacteria. J Microbiol Methods. 2001;44:121–129. doi: 10.1016/s0167-7012(00)00241-4. [DOI] [PubMed] [Google Scholar]
- Hoffman M, Decho AW. Extracellular enzymes within microbial biofilms and the role of the extracellular polymer matrix. In: Wingender J, Neu J, Flemming H-C, editors. Microbial Extracellular Polymeric Substances. Berlin, Germany: Springer-Verlag; 1999. pp. 217–230. [Google Scholar]
- Hughes MJ, Moore JC, Lane JD, et al. Identification of major outer surface proteins of Streptococcus agalactiae. Infect Immun. 2002;70:1254–1259. doi: 10.1128/IAI.70.3.1254-1259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Peters G, Chhatwal GS, Herrmann M. A lithium chloride-extracted, broad-spectrum-adhesive 42-kilodalton protein of Staphylococcus epidermidis is ornithine carbamoyltransferase. Infect Immun. 1999;67:6688–6690. doi: 10.1128/iai.67.12.6688-6690.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Shah SM, Kaplan JB. Intercellular adhesion and biocide resistance in nontypeable Haemophilus influenzae biofilms. Microb Pathog. 2009;46:207–213. doi: 10.1016/j.micpath.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen TH, Bragason SK, Phipps RK, et al. Food as a source for QS inhibitors: Iberin from horseradish revealed as a quorum sensing inhibitor of Pseudomonas aeruginosa. Appl Environ Microbiol. 2012;78:2410–2421. doi: 10.1128/AEM.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek J, Greiner L, Watanabe H, Zaleski A, Apicella MA, Bakaletz LO. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun. 2005;73:3210–3218. doi: 10.1128/IAI.73.6.3210-3218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–3875. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Durbin JE, Kusewitt DF, Bakaletz LO. Anatomy of the nasal cavity in the chinchilla. Cells Tissues Organs. 2003;174:136–152. doi: 10.1159/000071154. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Ahmed K, Utsunomiya Y, Rikitomi N, Hori A, Oishi K, Nagatake T. Attachment of nontypable Haemophilus influenzae to human pharyngeal epithelial cells mediated by a ganglioside receptor. Microbiol Immunol. 1998;42:697–702. doi: 10.1111/j.1348-0421.1998.tb02342.x. [DOI] [PubMed] [Google Scholar]
- Mangalappalli-Illathu AK, Lawrence JR, Korber DR. Cells in shearable and nonshearable regions of Salmonella enterica serovar Enteritidis biofilms are morphologically and physiologically distinct. Can J Microbiol. 2009;55:955–966. doi: 10.1139/w09-048. [DOI] [PubMed] [Google Scholar]
- Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Mukherjee R, Balachandar R, Chatterji D. Deletion of the rpoZ gene, encoding the omega subunit of RNA polymerase, results in pleiotropic surface-related phenotypes in Mycobacterium smegmatis. Microbiology. 2006;152:1741–1750. doi: 10.1099/mic.0.28879-0. [DOI] [PubMed] [Google Scholar]
- McDonald KL, Morphew M, Verkade P, Muller-Reichert T. Recent advances in high-pressure freezing: equipment- and specimen-loading methods. Methods Mol Biol. 2007;369:143–173. doi: 10.1007/978-1-59745-294-6_8. [DOI] [PubMed] [Google Scholar]
- Miyamoto N, Bakaletz LO. Selective adherence of non-typeable Haemophilus influenzae (NTHi) to mucus or epithelial cells in the chinchilla eustachian tube and middle ear. Microb Pathog. 1996;21:343–356. doi: 10.1006/mpat.1996.0067. [DOI] [PubMed] [Google Scholar]
- Moghaddam SJ, Ochoa CE, Sethi S, Dickey BF. Nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease and lung cancer. Int J Chron Obstruct Pulmon Dis. 2011;6:113–123. doi: 10.2147/COPD.S15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama S, Hotomi M, Shimada J, Billal DS, Fujihara K, Yamanaka N. Formation of biofilm by Haemophilus influenzae isolated from pediatric intractable otitis media. Auris Nasus Larynx. 2009;36:525–531. doi: 10.1016/j.anl.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Mueller RS, Dill BD, Pan C, Belnap CP, Thomas BC, Verberkmoes NC, Hettich RL, Banfield JF. Proteome changes in the initial bacterial colonist during ecological succession in an acid mine drainage biofilm community. Environ Microbiol. 2011;3:2279–2292. doi: 10.1111/j.1462-2920.2011.02486.x. [DOI] [PubMed] [Google Scholar]
- Muresan V, Muresan Z. Unconventional functions of microtubule motors. Arch Biochem Biophys. 2012;52:17–29. doi: 10.1016/j.abb.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TF, Kirkham C. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol. 2002;2:7. doi: 10.1186/1471-2180-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TF, Brauer AL, Schiffmacher AT, Sethi S. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;170:266–272. doi: 10.1164/rccm.200403-354OC. [DOI] [PubMed] [Google Scholar]
- Murphy TF, Kirkham C, Sethi S, Lesse AJ. Expression of a peroxiredoxin-glutaredoxin by Haemophilus influenzae in biofilms and during human respiratory tract infection. FEMS Immunol Med Microbiol. 2005;44:81–89. doi: 10.1016/j.femsim.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Murphy TF, Faden H, Bakaletz LO, Kyd JM, Forsgren A, Campos J, Virji M, Pelton SI. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr Infect Dis J. 2009;28:43–48. doi: 10.1097/INF.0b013e318184dba2. [DOI] [PubMed] [Google Scholar]
- Neary JM, Murphy TF. Antibodies directed at a conserved motif in loop 6 of outer membrane protein P2 of nontypeable Haemophilus influenzae recognize multiple strains in immunoassays. FEMS Immunol Med Microbiol. 2006;46:251–261. doi: 10.1111/j.1574-695X.2005.00033.x. [DOI] [PubMed] [Google Scholar]
- Novotny LA, Amer AO, Brockson ME, Goodman SD, Bakaletz LO. Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS One. 2013;8:e67629. doi: 10.1371/journal.pone.0067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosthuizen MC, Steyn B, Theron J, Cosette P, Lindsay D, Von Holy A, Brozel VS. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl Environ Microbiol. 2002;68:2770–2780. doi: 10.1128/AEM.68.6.2770-2780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V, Fischetti VA. A major surface protein on group A streptococci is a glyceraldehydes-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer ES, Allan JA, Theodoropoulos C, Ross T, Beagley KW, Knox CL. Hormone-dependent bacterial growth, persistence and biofilm formation--a pilot study investigating human follicular fluid collected during IVF cycles. PLoS One. 2012;7:e49965. doi: 10.1371/journal.pone.0049965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen FC, Tao L, Scheie AA. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol. 2005;187:4392–4400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poje G, Redford RJ. General methods for culturing Haemophilus influenzae. In: Herbert MA, Hood DW, Moxon ER, editors. Haemophilus influenzae protocols. Vol. 71. Totowa, New Jersey: Humana Press; 2003. pp. 51–56. [Google Scholar]
- Ronander E, Brant M, Janson H, Sheldon J, Forsgren A, Riesbeck K. Identification of a novel Haemophilus influenzae protein important for adhesion to epithelial cells. Microbes Infect. 2008;10:87–96. doi: 10.1016/j.micinf.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Sauer K. The genomics and proteomics of biofilm formation. Genome Biol. 2003;4:219. doi: 10.1186/gb-2003-4-6-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol. 2001;183:6579–6589. doi: 10.1128/JB.183.22.6579-6589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaudinn C, Stoodley P, Kainovic A, O’Keefe T, Costerton JW, Robinson D, Baum MM, Ehrlich G, Webster P. Bacterial biofilms, other structures seen as mainstream concepts. Microbe. 2006;2:231–237. [Google Scholar]
- Schaudinn C, Stoodley P, Hall-Stoodley L, et al. Death and transfiguration in static Staphylococcus epidermidis culture. PLoS One. 2014 doi: 10.1371/journal.pone.0100002. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Weldon S, Taggart CC. SLPI and elafin: multifunctional antiproteases of the WFDC family. Biochem Soc Trans. 2011;39:1437–1440. doi: 10.1042/BST0391437. [DOI] [PubMed] [Google Scholar]
- Seo H, Kim J, Jung J, Jin HM, Jeon CO, Park W. Complexity of cell-cell interactions between Pseudomonas sp. AS1 and Acinetobacter oleivorans DR1: metabolic commensalism, biofilm formation and quorum quenching. Res Microbiol. 2012;163:173–181. doi: 10.1016/j.resmic.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Serra D, Bosch A, Russo DM, Rodriguez ME, Zorreguieta A, Schmitt J, Naumann D, Yantorno O. Continuous nondestructive monitoring of Bordetella pertussis biofilms by Fourier transform infrared spectroscopy and other corroborative techniques. Anal Bioanal Chem. 2007;387:1759–1767. doi: 10.1007/s00216-006-1079-9. [DOI] [PubMed] [Google Scholar]
- Sharpe SW, Kuehn MJ, Mason KM. Elicitation of epithelial cell-derived immune effectors by outer membrane vesicles of nontypeable Haemophilus influenzae. Infect Immun. 2011;79:4361–4369. doi: 10.1128/IAI.05332-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Syring M, Singh A, Singhal K, Dalecki A, Johansson T. An insight into the significance of the DnaK heat shock system in Staphylococcus aureus. Int J Med Microbiol. 2012;302:242–252. doi: 10.1016/j.ijmm.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Southey-Pillig CJ, Davies DG, Sauer K. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187:8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering AL, Gilmore MS. Quorum sensing and DNA release in bacterial biofilms. Curr Opin Microbiol. 2006;9:133–137. doi: 10.1016/j.mib.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Starner TD, Zhang N, Kim G, Apicella MA, McCray PB., Jr Haemophilus influenzae forms biofilms on airway epithelia: implications in cystic fibrosis. Am J Respir Crit Care Med. 2006;174:213–220. doi: 10.1164/rccm.200509-1459OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger RE, Holden PA. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl Environ Microbiol. 2005;71:5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Swords WE, Moore ML, Godzicki L, Bukofzer G, Mitten MJ, VonCannon J. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect Immun. 2004;72:106–113. doi: 10.1128/IAI.72.1.106-113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords WE, Buscher BA, Ver Steeg Ii K, Preston A, Nichols WA, Weiser JN, Gibson BW, Apicella MA. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol Microbiol. 2000;37:13–27. doi: 10.1046/j.1365-2958.2000.01952.x. [DOI] [PubMed] [Google Scholar]
- Thanavala Y, Lugade AA. Role of nontypeable Haemophilus influenzae in otitis media and chronic obstructive pulmonary disease. Adv Otorhinolaryngol. 2011;72:170–175. doi: 10.1159/000324785. [DOI] [PubMed] [Google Scholar]
- Tschanz SA, Burri PH, Weibel ER. A simple tool for stereological assessment of digital images: the STEPanizer. J Microsc. 2011;243:47–59. doi: 10.1111/j.1365-2818.2010.03481.x. [DOI] [PubMed] [Google Scholar]
- Vilain S, Cosette P, Hubert M, Lange C, Junter GA, Jouenne T. Comparative proteomic analysis of planktonic and immobilized Pseudomonas aeruginosa cells: a multivariate statistical approach. Anal Biochem. 2004;329:120–130. doi: 10.1016/j.ab.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Webster P, Wu S, Webster S, Rich KA, McDonald K. Ultrastructural preservation of biofilms formed by non-typeable Hemophilus influenzae. Biofilms. 2004;1:165–182. [Google Scholar]
- Webster P, Wu S, Gomez G, Apicella M, Plaut AG, St Geme JW., 3rd Distribution of bacterial proteins in biofilms formed by non-typeable Haemophilus influenzae. J Histochem Cytochem. 2006 doi: 10.1369/jhc.6A6922.2006. [DOI] [PubMed] [Google Scholar]
- Wingender J, Jeaeger K-E, Flemming H-C. Interaction between extracellular polysaccharides and enyzmes. In: Wingender J, Neu J, Flemming H-C, editors. Microbial Extracellular Polymeric Substances. Berlin, Germany: Springer-Verlag; 1999. [Google Scholar]
- Winterhoff N, Goethe R, Gruening P, Rohde M, Kalisz H, Smith HE, Valentin-Weigand P. Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the Arginine Deiminase system of Streptococcus pyogenes. J Bacteriol. 2002;184:6768–6776. doi: 10.1128/JB.184.24.6768-6776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Tomasch J, Sztajer H, Wagner-Dobler I. The delta subunit of RNA polymerase, RpoE, is a global modulator of Streptococcus mutans environmental adaptation. J Bacteriol. 2010;192:5081–5092. doi: 10.1128/JB.00653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahller J, Stewart PS. Transmission electron microscopic study of antibiotic action on Klebsiella pneumoniae biofilm. Antimicrob Agents Chemother. 2002;46:2679–2683. doi: 10.1128/AAC.46.8.2679-2683.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.