Abstract
A growing body of work has raised concern that many human pluripotent stem cell (hPSC) lines possess tumorigenic potential following differentiation to clinically relevant lineages. In this review, we highlight recent work characterizing the spectrum of cancer-like epigenetic derangements in human embryonic stem cells (hESC) and human induced pluripotent stem cells (hiPSC) that are associated with reprogramming errors or prolonged culture that may contribute to such tumorigenicity. These aberrations include cancer-like promoter DNA hypermethylation and histone marks associated with pluripotency, as well as aberrant X-chromosome regulation. We also feature recent work that suggests optimized high-fidelity reprogramming derivation methods can minimize cancer-associated epigenetic aberrations in hPSC, and thus ultimately improve the ultimate clinical utility of hiPSC in regenerative medicine.
Introduction
Human pluripotent stem cells (hPSC) are stable cell lines that can be indefinitely propagated in culture and have enormous potential for use in regeneration and repair of human disease and injury. The discovery of methods to isolate human embryonic stem cells (hESC) from pre-implantation embryos [1], and the derivation of human induced pluripotent stem cell (hiPSC) lines from human differentiated cells with defined factors [2,3] inaugurated the practical development of that potential. However, from the beginning, concern existed regarding the degree to which these artificially-derived hPSC lines truly recapitulated the normally-regulated embryonic pluripotent state.
Most hPSC lines share remarkably similar superficial measures of pluripotency (such as cell surface markers and teratoma formation in immunocompromised mice), but possess distinct cell line-dependent variations and lineage skewing in their potency of differentiation. This has been observed among both hESC [4–6] and hiPSC lines [7–12]. In efforts to understand the mechanisms underlying this skewing in differentiation potency, hPSC were found to have significant variation in transcriptomes and epigenomes [13–15]. In particular, the reactivation of self-renewal and de-differentiation inherent in the reprogramming process of hiPSC induces aberrations in patterns of transcription, methylation [16–19] and hydroxymethylation [20,21] that are not observed in hESC derived directly from pre-implantation human embryos.
This review synthesizes research suggesting that the aberrant epigenetic regulation observed in many hPSC lines may potentially confer increased tumorigenic potential in their use for regeneration and repair of diseased tissues. We detail the growing evidence of parallels between aberrant epigenetic regulation in cancer, and epigenetic aberrations which arise during establishment and subsequent propagation of hPSC cell lines that are generated with methods involving ectopic expression of defined pluripotency factors which are also oncogenes. We also highlight emerging evidence of aberrant X-chromosome regulation in many hPSC lines that may further have cancer-related implication. Finally, we feature recent research suggesting the potential of optimizing derivation conditions to minimize or avoid these cancer-associated epigenetic aberrations. Together, these emerging findings strongly indicate the need for further research to more completely understand the mechanisms underlying the development (or avoidance) of hPSC-associated epigenetic aberrations. The development of derivation methods that produce hPSC lines that more faithfully recapitulate the normal, non-cancerous pluripotent state is needed.
Cancer-associated promoter hypermethylation, histone modification, and hPSC tumorigenic safety
Concern regarding reprogramming-associated epigenetic aberrations in hPSC initially focused on risks introduced by hiPSC derivation with viral constructs. The most commonly employed methods of hiPSC derivation utilized overexpression of reprogramming transcription factors (e.g., MYC, KLF4, OCT4) that also have established roles in oncogenesis [22]. Methods of hiPSC generation employing viral vectors to express these defined factors, posed oncogenic risks associated with the viral integration and constitutive expression of proto-oncogenes [23]. Thus, hiPSC generated by such standard methodology might theoretically trigger a latent oncogenic potential, which could manifest as formation of tumors upon differentiation and implantation in clinical contexts.
Recent reports have provided validation of these theoretical concerns. For example, when 21 hiPSC lines derived from five different hiPSC induction methods were differentiated in parallel to cartilage, four of the 21 human iPSC lines generated cartilage in vitro that contained abnormal tumor-like glandular histology with expression of CEA and CA19-9 tumor markers, as well as forming glandular epithelial cells following transplantation into SCID mice [24]. Similarly, foci of malignant-like characteristics are more consistently found in teratomas generated by incompletely-reprogrammed and partially-reprogrammed hiPSC, as assessed by blinded histologic comparisons [25]. These data correlated with previous findings of overexpression of cancer-associated genes in hPSC-derived hepatocytes, endothelial cells, and neural crest cells, vs corresponding primary tissues [26]. Finally, parallel differentiation of 40 hiPSC lines into dopaminergic neurons revealed seven “differentiation-defective” clones that formed teratomas after transplantation into NOD/SCID mouse brains [27]. Together, these studies demonstrate that among hiPSC sharing similar superficial measures of pluripotency, at least some hiPSC harbor the potential to form tumors upon differentiation and transplantation in vivo. Understanding the molecular basis for this inherent oncogenic potential harbored by hiPSC is critical both to enable screening of iPSC for safety, and for refinement of derivation methods to reduce this oncogenic potential.
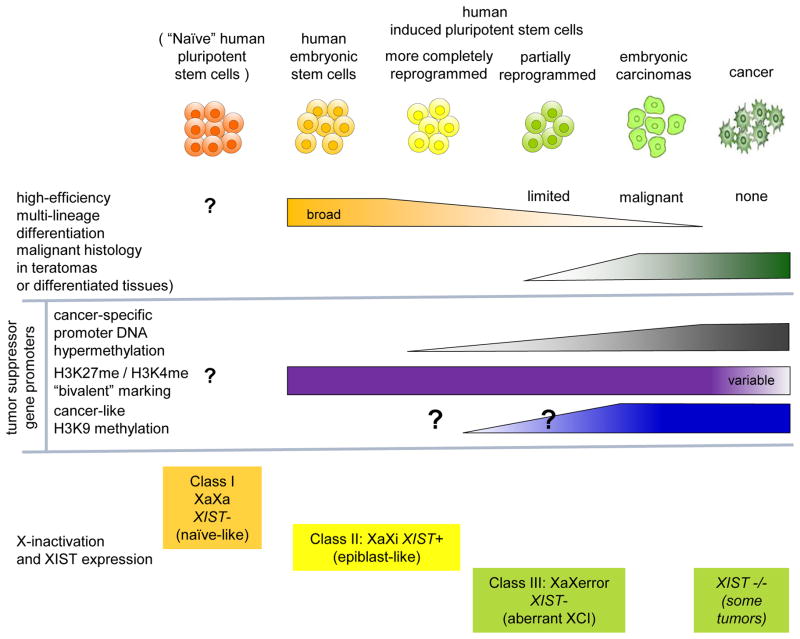
Human iPSC derivation methods that achieved only partial reprogramming to a bona fide pluripotent state resulted in hiPSC lines that exhibit malignant histology [25][28]. Likewise, in mouse models, premature termination of reprogramming leads to malignant tumor formation [29]. Dissection of the underlying epigenetics reveal an emerging model that suggests individual hiPSC lines may exist epigenetically on a continuum between normal embryonic cells (and hESC) on one end, and frankly aberrant embryonic carcinoma or cancer lines on the other (summarized in Fig 1). This model synthesizes together mechanistically related studies of epigenetic changes in cancer and associated reprogramming.
Figure 1. Schematic model for the constellation of epigenetic derangements that stratify human pluripotent stem cells (hPSC) on a continuum of tumorigenic potential.
Among the best characterized of the hPSC derangements to date include variant degrees of cancer-specific CpG DNA hypermethylation and H3K9 histone methylation (especially on stem cell-like “bivalent” marked gene promoters), as well as aberrant regulation of X chromosome inactivation. Despite similarities in superficial measures of molecular pluripotency, hPSC exhibit a spectrum of variation in differentiation potency or “functional pluripotency” (orange bar), varying from a high-quality naïve ground state or primed epiblast stem cell (EpiSC)-like hPSC with broad differentiation potential, to hPSC with more limited differentiation potential and formation of more cancer-like histology upon differentiation. Growing evidence suggests that this spectrum of differentiation potency inversely correlates with degree of cancer-like epigenetic derangements. Associated cancer-like epigenetic derangements include cancer-specific promoter hypermethylation (grey), especially in association with bivalent marking (purple); degree of cancer-like H3K9 methylation (blue), and aberrant X-chromosome regulation (XIST- XaXerror, see bottom).
Central to this model has been the discovery that reprogramming can establish epigenetic states resembling those seen in cancer. For example, abnormal DNA hypermethylation at gene promoters established persistent silencing of tumor-suppressor genes and other key regulators leading to tumorigenesis [30]. Systematic comparison of normal stem cells, hiPSC lines, and cancer lines revealed a spectrum of aberrant, cancer-like promoter DNA hypermethylation and gene silencing, from normal hESC lacking cancer-like aberrations, to hiPSC that displayed progressively greater degrees of cancer-like promoter hypermethylation and gene silencing abnormalities [25,31]. This pattern of promoter hypermethylation in cancer cell lines and some hiPSC, but reduced in other hiPSC and hESC, has been recapitulated in subsequent independent comparisons [27]. These findings clearly suggest that the process of inducing self-renewal and de-differentiation to convert differentiated cells to hiPSC, may also steer those same cells towards an aberrant cancer-like epigenetic state not seen in hESC.
Further studies have directly linked aberrant, cancer-like DNA promoter hypermethylation with epigenetic states central to pluripotency. Pluripotent stem cells are characterized by a set of key developmental genes bearing “bivalent” chromatin marking with repressive H3K27me3 and activating H3K4me3 histone marks. This balance of repressive and activating “bivalent” chromatin marks upon these key promoters, and its associated occupancy by Polycomb-repressive complexes, leaves stem cells “poised” for rapid reconfiguration upon appropriate developmental signals to establishment of a terminal differentiated state [32]. A succession of papers demonstrated links between cancer-associated promoter hypermethylation and pluripotent stem cell epigenetic marks. For example, Polycomb-mediated H3K27me3 was found to pre-mark genes for de-novo DNA methylation in a colon cancer cell line [33], and Polycomb group targets were found to be 12 times more likely to have cancer-specific promoter DNA hypermethylation [34]. Building upon these earlier studies, a more direct, explicit connection between these two fundamental epigenetic mechanisms was recently established through integration of genome-wide epigenetic data between stem cells and cancer cells. Over 75% of genes bearing CpG-island DNA hypermethylation specifically in cancers, were found to be “bivalently” marked in stem cells. This aberrant hypermethylation among “bivalent” marked genes was enriched among developmental regulators. The methylation status of the genes within this intersecting cancer/stem cell hypermethylation module, when correlated with whole-genome methylation data from hundreds of patient samples, crisply segregated colon and breast cancer patients with previously established subtypes with better and worse prognoses [35]. Together, these findings explicitly correlate cancer as an epigenetically-deranged state that intersects with an underlying stem cell self-renewal program.
Further information was derived from comparisons between hESC, cancer, and human embryonic carcinoma cells. Such analyses revealed that partially-reprogrammed hiPSC represent malignant counterparts of normal hESC that although retaining minor degrees of spontaneous differentiation and multi-lineage commitment, primarily exist in an intermediate state between hESC and adult cancer cells. Those comparisons again demonstrated the association between a stem cell H3K27/H3K4 methylated “bivalent” chromatin state, and subsequent promoter DNA hypermethylation in cancer. Furthermore, embryonic carcinoma cells which exist in an undifferentiated but malignant developmental state were also found to possess such intermediate epigenetic states. Promoters for key tumor suppressor genes progressed from an umethylated, “bivalent” marked state in hESC, to an intermediate state in embryonic carcinomas characterized by the addition of cancer-like patterns of H3K9 di and tri methylation, before finally additionally acquiring cancer-like DNA promoter hypermethylation in adult cancer cell lines [36]. This is particularly relevant because enrichment of H3K9me3 was likewise enriched in human iPSC in regions of DNA with aberrent patterns of methylation vs. hESC [16], thus lending additional support to the notion that many iPSC may exist in an intermediate epigenetic state between hESC, and embryonic carcinomas and cancer. Proper regulation of H3K9 methylation have been shown to play a central role in reprogramming to pluripotency in mouse iPSC systems [37,38], and derangement of H3K9 methylation has been observed in a wide variety of cancers [39]. Thus, it is reasonable to hypothesize that different derivation methods of iPSC generation may vary the effectiveness of proper H3K9 reprogramming, leading to iPSC lines that exist along a continuum of epigenetic derangement between hESC and malignant embryonic carcinomas.
These works collectively highlight the importance of future dissection of mechanisms leading to establishment of cancer-like epigenetic signatures of aberrant promoter DNA methylation (especially in association with H3K27/H3K4 “bivalent” marks), and aberrant H3K9 methylation. Given the constellation of other aberrant histone and other epigenetic modifications involved in cancer [39], it is certain additional histone and other epigenetic modifications will also be found that stratify hiPSC (and even hESC) on a continuum from normal pluripotency to tumorigenic potential. These will be revealed as more comprehensive characterization of histone modifications in human iPSC lines emerge to parallel recent such studies among mouse iPSC [38]. Understanding the epigenetic mechanisms underlying these cancer-like epigenetic signatures that arise during reprogramming will be critical to the identification and development of hiPSC that are as free as possible from tumorigenic potential.
X-chromosome regulation and PSC tumorigenic safety
In addition to connecting aberrant promoter hypermethylation and chromatin modifications in hPSC to tumorigenic potential, a growing body of work has implicated aberrant regulation of X-chromosome methylation, specifically X-chromosome inactivation (XCI), as an important contributor to hPSC tumorigenicity. In differentiated somatic female cells there is normally inactivation of one of the two X chomosomes (XaXi). This process is mediated by a complex of regulators including the non-coding RNA XIST [40].
Initially, many female hESC and hiPSC lines were also found to exist in a XaXi state. (This is in contrast to the initially isolated mouse ESC and iPSC, the implications of which to be discussed further below) [41]. However, systematic analysis of degrees of XCI and associated X-chromosome hypermethylation and XIST expression, revealed a substantial range in variation among hESC and hiPSC lines in these measures of normal X-chromosome regulation [17,42,43]. Importantly, many female hPSC lines were found (especially with increasing passage) to gradually lose XIST expression, and/or aberrantly reactivate the previously silenced X-chromosome (extensively reviewed in [44]). Human iPSC with this XaXerror XCI status had distinct corresponding transcriptional and phenotypic abnormalities, including upregulation of oncogenes, accelerated growth, and poorer quality differentiation [43,45]. Furthermore, at least some male hiPSCs were found to have patterns of aberrant differential gene expression that more closely resembled female hiPSC with deranged XCI [43]. This implies that the (yet unknown) mechanisms underlying aberrant loss of normal XCI and XIST expression may not be avoidable simply by using male hPSC, but may instead be symptomatic of a more global underlying epigenetic derangement.
This aberrancy persisted in differentiated derivatives, since hPSC which lost XIST expression resulted in differentiated progeny that likewise lacked normal, expected XIST expression [43]. This is of particular concern given recent demonstration that hematopoetic stem cell targeted deletion of Xist in female mice led to rapid development of a highly aggressive myeloproliferative neoplasm and myelodysplastic syndrome. Transplanting Xist −/− HSC into irradiated wild-type mice recapitulated the hematologic neoplasm seen in HSC Xist −/− mice, while transplanting wild-type HSC into HSC Xist −/− mice cured them of malignancy, clearly demonstrating the neoplasm was HSC autonomous [46]. This raises obvious concerns regarding the therapeutic safety of clinical therapies based on differentiated derivatives of hPSC with unstable XCI and XIST expression.
Efficient derivation methods can diminish the degree of acquisition of cancer-associated epigenetic derangements
Recent research has shown that highly optimized derivation methods can generate next-generation hiPSC lines that lessen or avoid acquisition of cancer-like epigenetic derangements seen in standard reprogramming methodologies. For example, a refined non-viral, non-integrating method of generating hiPSC from myeloid precursors with unprecedented (~ 50%) efficiency [47] resulted in hiPSC that largely lack the cancer-like aberrant hypermethylation and gene silencing (Zambidis, Baylin et al, manuscript in preparation) seen in previous generation hiPSC [25]. These same myeloid-derived hiPSC were also capable of differentiating into lineages with characteristics not previously reported, including vascular progenitors capable of long-term engraftment in vivo [48] and three-dimensional retinal cups with light-sensistive photoreceptors [49]. These data imply a connection between higher efficiency of derivation, lack of cancer-like epigenetic derangements, and higher differentiation potency, as also suggested by others [18]. Subsequent work by others has confirmed that myeloid progenitors represent a “privileged” somatic donor type that can be more efficiently reprogrammed [50], suggesting that myeloid precursors may possesses an epigenetically plastic state more amenable to pluripotency induction.
Micro-environmental modification of the quality and hierarchy of the pluripotent state may also minimize aberrant regulation of X-chromosome function and development of an XaXerror state seen in some hPSC, and thus diminish associated potential tumorigenic risks. As noted previously, the first isolated mouse ESC and iPSC showed full X reactivation (XaXa), in contrast to the XaXi status or the aberrant partially reactivated XaXerror state seen to date in most hESC and hiPSC. Subsequent work has revealed this discrepancy to be in part symptomatic of a larger divide in classes of pluripotent stem cells, between developmentally earlier “naïve” ground state PSC (as represented by the XaXa mouse naïve PSCs) and developmentally more mature “epiblast” PSC (as represented by XaXi mouse epiblast PSC and most previous human PSC) (extensively reviewed in [51]). Modifications of epiblast hPSC derivation can begin to shift the balance of X-inactivation status towards a more naïve-like ground state of pluripotency [52]. More recently, methods of either fully converting existing “epiblast-like” hPSC to the “naïve” state, or deriving “naïve” hPSC de novo have been reported [53–55]. This might enable avoidance of all of the XCI-associated epigenetic regulation concerns noted above.
Taken together, these recent works demonstrate that acquisition of cancer-like aberrant epigenetic signatures is not an absolutely inevitable consequence of generating artificial hPSC lines. Rather, these findings demonstrate that hPSC to be used for regeneration and repair must be screened for tumorigenic potential, that epigenetic marks can provide important tools for this screening, and that derivation methods engineered to produce hPSC free of either autosomal or X-linked cancer-like epigenetic derangements is both a possible and a necessary objective to enable hPSC to be confidently used in regeneration and repair.
Conclusions
Despite sharing largely indistinguishable performance in superficial measures of pluripotency (such as expression of cell surface markers, expression of pluripotency genes, and formation of teratomas), it has become increasingly obvious that different hPSC lines, derived or isolated under differing circumstances, have more subtle epigenetic variation that has substantial implications for their tumorigenic potential (especially after differentiation). The degree of aberrant stem cell-gene associated DNA hypermethylation, associated cancer-like signatures of histone modifications, and quality of X-chromosome regulation are symptomatic of underlying differences in the quality of hPSC that are the product of different derivation methods, and potentially confer greater vulnerability to tumorigenic conversion. Future efforts to more completely understand these mechanisms, and develop optimized derivation methods will be critical to producing hPSC whose safety for use in regeneration and repair can be confidently asserted.
Acknowledgments
This work was supported by grants from the NIH/NHLBI (U01HL099775 and U01HL100397 (ETZ, SBB); PCBC2012Pilot_01 (ETZ), the NIH/NCI (CA60441 (JSH)), and the Maryland Stem Cell Research Fund (2011-MSCRF-II-0008-00 (ETZ); 2013-MSCRFII-0032 (ETZ); 2013-MSCRFF-033 (JSH)). We thank Dr. Alan Friedman for careful reading of the manuscript. We apologize to our colleagues whose work we may have inadvertently omitted due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 5.Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Chang KH, Nelson AM, Fields PA, Hesson JL, Ulyanova T, Cao H, Nakamoto B, Ware CB, Papayannopoulou T. Diverse hematopoietic potentials of five human embryonic stem cell lines. Exp Cell Res. 2008;314:2930–2940. doi: 10.1016/j.yexcr.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, Zhang SC. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajiwara M, Aoi T, Okita K, Takahashi R, Inoue H, Takayama N, Endo H, Eto K, Toguchida J, Uemoto S, et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mills JA, Wang K, Paluru P, Ying L, Lu L, Galvao AM, Xu D, Yao Y, Sullivan SK, Sullivan LM, et al. Clonal genetic and hematopoietic heterogeneity among human-induced pluripotent stem cell lines. Blood. 2013;122:2047–2051. doi: 10.1182/blood-2013-02-484444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasu A, Ikeya M, Yamamoto T, Watanabe A, Jin Y, Matsumoto Y, Hayakawa K, Amano N, Sato S, Osafune K, et al. Genetically matched human iPS cells reveal that propensity for cartilage and bone differentiation differs with clones, not cell type of origin. PLoS One. 2013;8:e53771. doi: 10.1371/journal.pone.0053771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hynes K, Menicanin D, Mrozik K, Gronthos S, Bartold PM. Generation of Functional Mesenchymal Stem Cells from Different Induced Pluripotent Stem Cell Lines. Stem Cells Dev. 2014 doi: 10.1089/scd.2013.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman AM, Cooper JB. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell. 2010;7:258–262. doi: 10.1016/j.stem.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 16*.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. Whole-genome profiles of DNA methylation and histone modification reveal systematic differences unique to iPSC vs hESC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC, Boscolo FS, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. Human pluripotent stem cells possess a range of aberrations in genomic imprinting, X-inactivation and XIST expression, which persist on directed differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz S, Diep D, Gore A, Panopoulos AD, Montserrat N, Plongthongkum N, Kumar S, Fung HL, Giorgetti A, Bilic J, et al. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:16196–16201. doi: 10.1073/pnas.1202352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang K, Shen Y, Xue Z, Bibikova M, April C, Liu Z, Cheng L, Nagy A, Pellegrini M, Fan J-B, et al. A Panel of CpG Methylation Sites Distinguishes Human Embryonic Stem Cells and Induced Pluripotent Stem Cells. Stem Cell Reports. 2013;2:36–43. doi: 10.1016/j.stemcr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Wu H, Li Y, Szulwach KE, Lin L, Li X, Chen IP, Goldlust IS, Chamberlain SJ, Dodd A, et al. Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency. Nat Cell Biol. 2013;15:700–711. doi: 10.1038/ncb2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar R, DiMenna L, Schrode N, Liu TC, Franck P, Munoz-Descalzo S, Hadjantonakis AK, Zarrin AA, Chaudhuri J, Elemento O, et al. AID stabilizes stem-cell phenotype by removing epigenetic memory of pluripotency genes. Nature. 2013;500:89–92. doi: 10.1038/nature12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suvà ML, Riggi N, Bernstein BE. Epigenetic Reprogramming in Cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee AS, Tang C, Rao MS, Weissman IL, Wu JC. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19:998–1004. doi: 10.1038/nm.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Yamashita A, Liu S, Woltjen K, Thomas B, Meng G, Hotta A, Takahashi K, Ellis J, Yamanaka S, Rancourt DE. Cartilage tissue engineering identifies abnormal human induced pluripotent stem cells. Sci Rep. 2013;3:1978. doi: 10.1038/srep01978. Demonstration of a spectrum of oncogenic potential among iPSC through in vivo development of tumor-like histology and secretion of tumor markers by some human iPSC subjected to terminal differentiation to cartiliage and subsequent in vivo implantation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Ohm JE, Mali P, Van Neste L, Berman DM, Liang L, Pandiyan K, Briggs KJ, Zhang W, Argani P, Simons B, et al. Cancer-related epigenome changes associated with reprogramming to induced pluripotent stem cells. Cancer Res. 2010;70:7662–7673. doi: 10.1158/0008-5472.CAN-10-1361. The authors reveal a spectrum of cancer-like derangements in iPSC absent in hESC, including malignant-like foci in teratomas, aberrent gene silencing events, and cancer-like aberrant responses to epigenetic-modifying drugs and gene promoter DNA methylation alterations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh Z, Huang M, Hu S, Wilson KD, Dey D, Wu JC. Dissecting the Oncogenic and Tumorigenic Potential of Differentiated Human Induced Pluripotent Stem Cells and Human Embryonic Stem Cells. Cancer Research. 2011;71:5030–5039. doi: 10.1158/0008-5472.CAN-10-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Koyanagi-Aoi M, Ohnuki M, Takahashi K, Okita K, Noma H, Sawamura Y, Teramoto I, Narita M, Sato Y, Ichisaka T, et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci U S A. 2013;110:20569–20574. doi: 10.1073/pnas.1319061110. Aberrant, cancer-like transcriptional and epigenetic signatures are associated with decreased potency of iPSC differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griscelli F, Feraud O, Oudrhiri N, Gobbo E, Casal I, Chomel JC, Bieche I, Duvillard P, Opolon P, Turhan AG, et al. Malignant germ cell-like tumors, expressing Ki-1 antigen (CD30), are revealed during in vivo differentiation of partially reprogrammed human-induced pluripotent stem cells. Am J Pathol. 2012;180:2084–2096. doi: 10.1016/j.ajpath.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, et al. Premature Termination of Reprogramming In Vivo Leads to Cancer Development through Altered Epigenetic Regulation. Cell. 2014;156:663–677. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 34.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 35**.Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, Aryee MJ, Joyce P, Ahuja N, Weisenberger D, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Research. 2012;22:837–849. doi: 10.1101/gr.131169.111. The authors use an integrated genomics approach to characterize in depth the functional association between stem-cell “bivalent” chromatin state and abnormal cancer-like gene promoter DNA hypermethylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. The authors identify histone H3K9 methylation as an epigenetic mark intermediate in the progression from normal pluripotency to cancer-like aberrant chromatin states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Wu Y, Guo L, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 38.Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geutjes EJ, Bajpe PK, Bernards R. Targeting the epigenome for treatment of cancer. Oncogene. 2012;31 :3827–3844. doi: 10.1038/onc.2011.552. [DOI] [PubMed] [Google Scholar]
- 40.Barakat TS, Gribnau J. X chromosome inactivation in the cycle of life. Development. 2012;139:2085–2089. doi: 10.1242/dev.069328. [DOI] [PubMed] [Google Scholar]
- 41.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 42.Bruck T, Benvenisty N. Meta-analysis of the heterogeneity of X chromosome inactivation in human pluripotent stem cells. Stem Cell Res. 2011;6:187–193. doi: 10.1016/j.scr.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 43*.Anguera MC, Sadreyev R, Zhang Z, Szanto A, Payer B, Sheridan SD, Kwok S, Haggarty SJ, Sur M, Alvarez J, et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. The spectrum of derangement in X-inactivation among hiPSC is correlated with poorer differentiation, upregulation of X-linked oncogenes, and accelerated growth rate in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lessing D, Anguera MC, Lee JT. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet. 2013;14:85–110. doi: 10.1146/annurev-genom-091212-153530. [DOI] [PubMed] [Google Scholar]
- 45.Bruck T, Yanuka O, Benvenisty N. Human pluripotent stem cells with distinct X inactivation status show molecular and cellular differences controlled by the X-Linked ELK-1 gene. Cell Rep. 2013;4 :262–270. doi: 10.1016/j.celrep.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 46.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park TS, Huo JS, Peters A, Talbot CC, Jr, Verma K, Zimmerlin L, Kaplan IM, Zambidis ET. Growth Factor-Activated Stem Cell Circuits and Stromal Signals Cooperatively Accelerate Non-Integrated iPSC Reprogramming of Human Myeloid Progenitors. PLoS One. 2012;7:e42838. doi: 10.1371/journal.pone.0042838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park TS, Bhutto I, Zimmerlin L, Huo JS, Nagaria P, Miller D, Rufaihah AJ, Talbot C, Aguilar J, Grebe R, et al. Vascular progenitors from cord blood-derived induced pluripotent stem cells possess augmented capacity for regenerating ischemic retinal vasculature. Circulation. 2014;129:359–372. doi: 10.1161/CIRCULATIONAHA.113.003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Peters A, Zambidis ET, Meyer J, Gamm DM, Gamm K-W, et al. Human-iPSC-Derived, 3-Dimensional Retinal Cups with Functional Photoreceptors. Submitted for review. 2013 [Google Scholar]
- 50.Guo S, Zi X, Schulz VP, Cheng J, Zhong M, Koochaki SH, Megyola CM, Pan X, Heydari K, Weissman SM, et al. Nonstochastic reprogramming from a privileged somatic cell state. Cell. 2014;156:649–662. doi: 10.1016/j.cell.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Los Angeles A, Loh Y-H, Tesar PJ, Daley GQ. Accessing naïve human pluripotency. Current Opinion in Genetics & Development. 2012;22:272–282. doi: 10.1016/j.gde.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomoda K, Takahashi K, Leung K, Okada A, Narita M, Yamada NA, Eilertson KE, Tsang P, Baba S, White MP, et al. Derivation conditions impact x-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell. 2012;11:91–99. doi: 10.1016/j.stem.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 54.Honda A, Hatori M, Hirose M, Honda C, Izu H, Inoue K, Hirasawa R, Matoba S, Togayachi S, Miyoshi H, et al. Naive-like conversion overcomes the limited differentiation capacity of induced pluripotent stem cells. J Biol Chem. 2013;288:26157–26166. doi: 10.1074/jbc.M113.502492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez-Caliani AJ, Deng X, Cavanaugh C, Cook S, et al. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]