Abstract
Adverse environmental conditions faced by an individual early during its life, such as gestational hypoxia, can have a profound influence on the risk of diseases, such as neurological disorders, in later life. Clinical and preclinical studies suggest that epigenetic programming of gene expression patterns in response to maternal stress have a crucial role in the fetal origins of neurological diseases. Herein, we summarize recent studies regarding the role of epigenetic mechanisms in the developmental programming of neurological diseases in offspring, primarily focusing on DNA methylation/demethylation and miRNAs. Such information could increase our understanding of the fetal origins of adult diseases and help develop effective prevention and intervention against neurological diseases.
Keywords: brain, hypoxia, epigenetic programming, DNA methylation/demethylation, micro RNAs, neurological disease
Introduction
During the late 1980s, Barker and colleagues uncovered the correlation between the nutrition condition of early life (during the prenatal and postnatal periods) and the increased risk of adult ischemic heart disease [1], known as ‘Barker hypothesis’. Thereafter, numerous retrospective and prospective clinical and preclinical studies have supported this ‘fetal origins of adult disease’ theory [2–4], showing a substantial correlation between an adverse maternal environment and the development of various diseases in later life, including cardiovascular disease, diabetes and neurological disease [5–9]. One general mechanism by which maternal stress can be linked to phenotypic changes later in life is the epigenetic programming of genes, which has a central role in determining the functional output of the information that is stored in the genome.
Epigenetics is defined as heritable changes in gene expression that are not associated with concomitant alterations in the DNA sequence. The genetic information for the transcriptional program of gene expression is controlled by epigenetic mechanisms, including methylation/demethylation of DNA, post-translational modifications of histone, and noncoding RNAs (ncRNAs) such as miRNAs [10]. Epigenetic events are highly responding process[LM1] to alterations in endogenous and environmental signals; therefore, epigenetic instructions have crucial roles in the regulation of the expression of appropriate sets of genes, especially in particular tissues at specific time windows [10–12]. Accumulating evidence from both human and animal studies indicates that epigenetic modifications serve as a memory of early life events and can induce long-term changes in gene expression profiles, which potentially result in disease in later life [8,13,14] (Figure 1). For example, in a recent study, epigenetic programming of the glucocorticoid receptor (GR) gene after maternal hypoxia was found to be related to the enhanced susceptibility of neonate hypoxic-ischemic encephalopathy (HIE) brain injury [15].
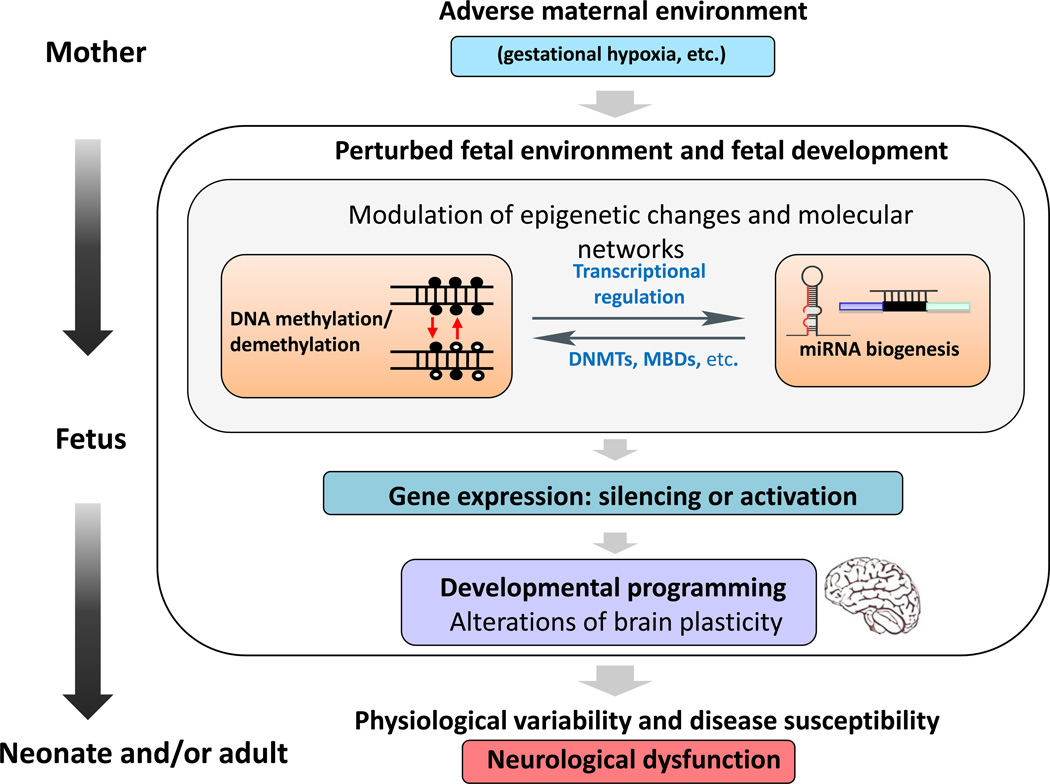
Figure 1.
Schematic diagram showing the correlation between adverse maternal environment (e.g., gestational hypoxia) and epigenetic mechanisms in developmental programming of neurological diseases in the offspring. A tightly controlled gene expression profile has a crucial role during brain development and plasticity. Exposure of the mother to adverse factors during pregnancy, such as maternal hypoxia, results in a perturbed maternal–fetal environment and can lead to modulated epigenetic changes that alter the gene expression profile as an adaption of the developing fetus to the adverse environment. Different epigenetic mechanisms corporately [LM6]orchestrate this process and also regulate each other. For example, DNA methylation/demethylation transcriptionally regulates miRNA biogenesis, and is conversely regulated by miRNAs modulating the expression of DNA methylation-related enzymes and proteins. Changes in physiology and neuroplasticity of the developing brain can permanently affect the structure and/or functionality, and result in increased disease susceptibility in the offspring. Abbreviations: DNMT, DNA methyltransferases; MBD, methyl-CpG binding protein.
Hypoxia is a common form of intrauterine stress, and the fetus might experience a period of hypoxic stress under a variety of conditions, including pregnancy at high altitude, pregnancy with anemia, placental insufficiency, cord compression, pre-eclampsia, heart, lung and kidney disease, or with hemoglobinopathy. Intrauterine hypoxia contributes significantly to developmental malformations in the fetal tissues and/or organs, particularly the brain. The brain represents only 2% of the body weight, but consumes 20% of the oxygen requirements of the body. The immature fetal brain is particularly sensitive to changes in oxygen level. Low levels of oxygen result in neurovascular development malformations, such as cerebral palsy and periventricular leukomalacia, in the developing brain and increases risk of brain injury in the newborn [16,17]. Maternal hypoxia also delays neuronal migration and alters neurotransmitters expression during embryogenesis [18], subsequently compromising neuronal circuits and affecting neural organization in the brain tissue [19]. As a result, maternal hypoxia leads to increased susceptibility to seizures, epilepsy [20], and cerebral insults in affected offspring [15,19]. However, the molecular mechanisms of gestational hypoxia in the programming of brain disorders in postnatal life remain largely elusive. Thus, here we summarize recent findings of the role of epigenetic mechanisms, specifically DNA methylation/demethylation and miRNAs, in the neural development and pathogenesis of neurological disorders, with a view to addressing whether fetal hypoxia might contribute to these neurological disorders in affected offspring.
Epigenetic mechanisms and the machineries
DNA methylation and the machineries
DNA methylation refers to the biological process whereby a methyl group is added to a DNA nucleotide, and occurs almost exclusively on cytosines occurring directly before guanine molecules (i.e., CpG dinucleotides). The major function of DNA methylation is to silence transcriptionally the expression of related genes, and the repression effect is generally correlated with the methylation level within the gene promoter and the extent of interference of transcription factor binding [21,22]. DNA methylation can inhibit gene expression by several mechanisms. For example, it can interfere directly with the binding of transcription factors to the gene-regulating sequences. In other cases, DNA methylation does not directly inhibit transcription factor binding to the gene promoter. Instead, a family of proteins called methyl-CpG binding proteins (MBDs) bind to methylated CpG residues independent of the DNA sequence and mediate other epigenetic events, such as histone modifications that lead to further alteration of gene expression. MBDs can intimately link DNA methylation to patterns of histone post-translational modifications. Initially, methylated DNA promotes the recruitment of MBDs at specific sites in the genome, which in turn recruit histone-modifying enzymes and chromatin-remodeling complexes to the methylated DNA sites [23,24]. This helps to orchestrate DNA replication and repair, thus reinforcing and stabilizing transcriptional suppression.
DNA methylation is catalyzed by DNA methyltransferases (DNMTs); there are five DNMTs in mammals although only three (DNMT1, DNMT3a and DNMT3b) have methyltransferase activity and are generally grouped into de novo DNMTs (DNMT3a and DNMT3b) and maintenance DNMTs (DNMT1) [25]. The de novo DNMTs are highly expressed in embryonic stem (ES) cells and are downregulated in differentiated cells. DNMT3a and DNMT3b are believed to be responsible for establishing the pattern of methylation during development [23]. DNMT1, the most abundant DNMT in the cell, is transcribed chiefly during the S phase of the cell cycle, and is often needed to methylate hemimethylated sites that are generated during semiconservative DNA replication [26]. CpG islands are defined as short interspersed DNA sequences that deviate significantly from the average genomic pattern by a G+C content of at least 50% [27]. CpG islands are highly enriched at or near the gene promoter region. In mammalian genomes, approximately 60% of gene promoters are associated with CpG islands. In normal cells, most CpG islands are unmethylated, although some (approximately 6%) become methylated in a tissue-specific manner during early development or in differentiated tissues [28]. By contrast, 80% of CpGs located outside of CpG islands are methylated [29].
Although usually associated with transcriptional suppression, recent studies suggest that DNA methylation might also be involved in the transcriptional activation of some genes. However, these mechanisms remain to be determined. In mouse hypothalamus tissue, methyl CpG binding protein 2 (MeCP2) dysfunction was shown to induce changes in the expression levels of thousands of genes, and most genes (approximately 85%) appeared to be activated by MeCP2 [30]. MeCP2 was associated with the transcriptional activator CAMP responsive element binding protein 1 (CREB1) of an activated target but not a repressed target at the promoter [31].
DNA demethylation and the machineries
DNA demethylation is the reverse process of DNA methylation, leading to the loss of the methyl group from methylated cytosine (5meC) nucleotides. DNA demethylation, occurring through both passive and active mechanisms (Figure 2), has been observed in specific contexts, including both early development and somatic cells. Passive DNA demethylation refers to the loss of methyl group when the maintenance DNMT1 is absent or inhibited during DNA replication. By contrast, active DNA demethylation is the enzymatic process that results in the removal of the methyl group from 5meC by breaking a carbon–carbon bond. Different tissues can use different demethylation pathways depending on whether demethylation is genome wide or local and on its targets in the genome. Current evidence indicates that genome-wide demethylation occurs at specific times during early development, whereas gene-specific demethylation serves as a response to specific signals in somatic cells. During development, previous paternal-specific methylation marks are removed in the zygote by extensive genome-wide DNA demethylation shortly after fertilization. Although passive demethylation might also be involved in this process, it is likely that active demethylation has a major role in mediating this process, because loss of methylation is detected before the completion of the first cell division [32–34]. Both methylation and demethylation are resisted by imprinted genes, which are epigenetically modified at a different crucial period of gametogenesis [35]. On specific genomic loci in somatic cells, active DNA demethylation also has been observed in response to certain signals. For example, in unstimulated neurons, the brain-derived neurotrophic factor (BDNF) is maintained in a repressed state through methylation of promoter and binding of MeCP2. When neuron was depolarized with KCl, demethylation of the promoter occurred, and MeCP2 was released, thereby leading to upregulation of BDNF [36]. These findings indicate that, in addition to the long-term regulation of gene expression, methylation and demethylation also function in the dynamic regulation of gene expression as a rapid response to specific stimuli.
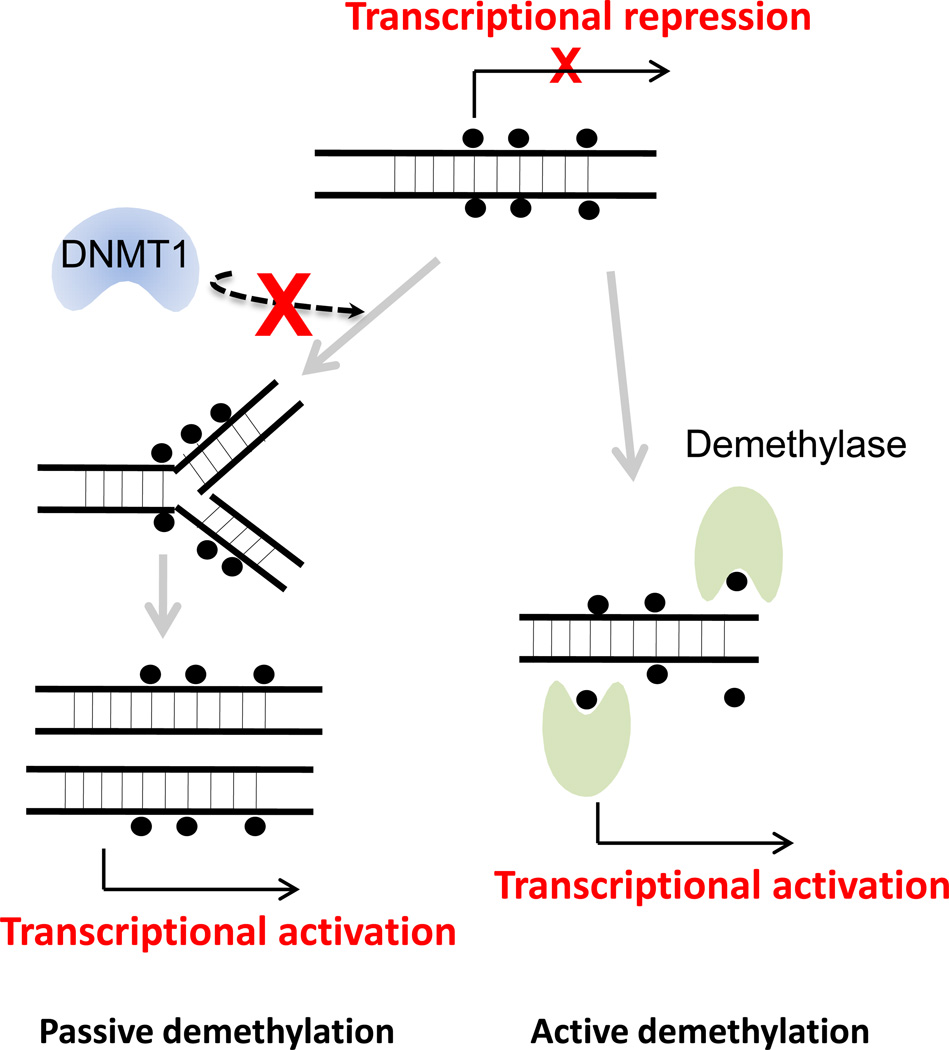
Figure 2.
Passive demethylation and active demethylation. Passive DNA demethylation occurs as a consequence of replication process in the absence of the maintenance DNA methyltransferase 1 (DNMT1). In contrast to passive demethylation, active demethylation is the enzymatic process by demethylases that results in the removal of the methyl group from 5-methylcytosine and subsequent transcriptional activation. Black filled circle: methylated CpG dinucleotides.
Based on various studies, several mechanisms have been proposed to address molecular mechanisms of active DNA demethylation, including enzymatic removal of the methyl group of 5meC, base excision repair (BER) through direct excise of 5meC, nucleotide excision repair (NER), deamination of 5meC to T followed by BER, and oxidative demethylation [37]. In plants, BER is used to achieve demethylation of DNA, although evidence supporting a similar mechanism in mammals is less compelling. Some proteins, including methyl-CpG binding domain protein 4 (MBD4) and a human T DNA glycosylase (TDG), have been shown to have glycosylase activity against 5meC in vivo in mammals. However, their excision activity against 5meC is comparatively low and their function in active demethylation remains to be elucidated. Decisive genetic and biochemical evidence supporting their roles in active DNA methylation in mammals remains to be determined. In vitro, activity-induced deaminase (AID) has been shown to be capable of delaminating 5meC to T and, thus, is implicated in the DNA demethylation process. In addition, a recent large-scale bisulfite sequencing study indicated a slightly increased level of methylation in primordial germ cells (PGCs) derived from AID-knockout mouse embryos. This suggests a contribution of AID in PGC demethylation. However, given the comparatively high levels of methylation in either ESCs or somatic cells (70–80%) compared with AID-null PGCs (approximately 20%), other mechanisms might be responsible for DNA demethylation.
Oxidative demethylation is another possible mechanism by which DNA demethylation can be achieved, and has recently received extensive attention. The discovery that members of the Ten-eleven translocation (Tet) family of proteins can convert 5-mC to 5-hydroxymethylcytosine (5hmC) provides further evidence to support this hypothesis. The Tet proteins family comprises three members, namely Tet1, Tet2 and Tet3, and shows broad expression patterns in different tissues [38]. In the mouse, all three Tets have the catalytic capacity to convert 5mC to 5hmC both in vitro and in vivo, and the presence of 5hmC depends on the presence of pre-existing of 5mC in vivo; this further suggests that 5mC is the only source of genomic 5hmC. Tet1 is expressed in ESCs and is required for the maintenance of embryonic status. The Tet1 mRNA level declines during ESC differentiation, accompanied by a decrease of the 5hmC levels [39]. Although Tet2 expression is observed in ESCs, the silencing of Tet2 fails to show obvious phenotypic changes in ESC biology. Tet3 expression is highest in oocytes and zygotes, where the 5hmC level increase dramatically in the male pronucleus. Moreover, zygotes with Tet3 depletion fail to demethylate the male pronucleus. Knockout of Tet1, but not Tet2 or Tet3, results in impaired ESC self-renewal and maintenance [40], and leads to embryonic lethality. The existence of three mammalian Tet enzymes raises the possibility that each has a distinct panel of genomic targets, such that their tissue specific expression might generate unique physiological effects.
DNA methylation and hypoxia
Under hypoxic conditions, hypoxia-induced transcription factors regulate a series of transcriptional responses, including cellular adaptation, angiogenesis, and cell survival. Accumulating evidence shows that these transcription factors, mainly hypoxia-inducible factor 1-alpha (HIF-1α), are controlled by, and cooperate with, epigenetic machineries, such as DNA methylation, and have an essential role in determining the cellular and molecular response to hypoxia [41]. Specifically, epigenetic changes at the DNA level influenced the HIF-1α expression level, its binding to target gene promoters and also HIF-1α protein stabilization [42].
The HIF-1α promoter contains CpG-rich regions, which might regulate HIF-1α expression by alteration of methylation status. In a hematopoietic cell line, Walczak-Drzewiecka and colleagues reported that the methylation of the HIF-1α promoter decreased its transcriptional activity, which was increased when cells were treated with a DNA demethylating agent, 5'-azacytidine [43]. Moreover, MeCP2 protein was also preferentially associated with the HIF-1α promoter in vivo [43]. Another study using tumor cells revealed a positive autoregulation mechanism of HIF-1α expression, in which demethylation of the HIF-1α promoter region enabled binding of HIF-1α to its own promoter, thereby increasing HIF-1α expression [44].
It is widely accepted that, in the presence of oxygen, HIF hydroxylases, prolyl hydroxylase domains (PHDs) destruct HIF-1α subunits by promoting von Hippel–Lindau tumor suppressor (VHL)-dependent ubiquitination and proteolysis. However, under hypoxic conditions, the HIF-1α degradation process is suppressed because of inactivation of the HIF hydroxylases complex [45]. Mounting evidence shows that an epigenetic mechanism is involved in controlling HIF-1α stability by affecting the activities of the degradation components. For example, in renal carcinomas, VHL gene expression was silenced when hypermethylation occurred in a normally unmethylated CpG island in the 5' region, resulting in the constitutive activation of HIF-1α [46]. Abnormal methylation in the CpG island of VHL has been observed in various disorders, including bone disease [47], retinoblastoma [48], and non-small cell lung cancer [49]. One member of the HIF hydroxylase group, prolyl hydroxylase 3 (PHD3), was also regulated by DNA methylation [50,51]. It has been reported that some cancer cell lines displayed a failure of PHD3 mRNA induction when placed in a hypoxic environment [51]. Further studies investigated the major mechanism in multiple tumors, showing that the suppression of PHD3 expression was the result of a change in methylation level of the CpG island [41,50]. Based on these findings, hypermethylation of PHD3 and VHL promoter could be an important epigenetic mechanism in HIF stabilization during hypoxia.
HIF-1α acts by binding to HIF-responsive elements (HREs) in the promoter of target genes; therefore, methylation of CpG islands in HREs might also interfere with the actions of HIF-1α. For example, the expression of erythropoietin [52] and the class III beta-tubulin [53] genes induced by hypoxia was dependent upon the tissue-specific methylation status of an HRE in the 3' flanking region. Indeed, chronic hypoxia can result in increases in global levels of DNA methylation, thus affecting the methylation level of HRE regions [54,55].
In addition to HIF-1α, transcriptional actions of other transcription factors under hypoxic conditions are also regulated by DNA methylation. For example, methylation of specific transcription factor binding sites (Egr-1 and Sp1[LM2]) at the PKCε promoter was increased in heart tissue of animals exposed to fetal hypoxia, leading to decreased expression of PKCε and loss of protection to the heart during ischemia-reperfusion injury [56–58]. In a cohort study, children exposed to maternal smoking had dramatically lower methylation of the DNA repetitive elements of the long interspersed element 1 (LINE-1) and AluYb8, which are normally hypermethylated under normal conditions, when compared with controls [59]. These studies provided evidence that prenatal exposure of the pregnant mother to tobacco smoke might be responsible for the increased risk of disease in their offspring later in life.
miRNAs
In humans, only 1–2% of the genome encodes protein, whereas ncRNAs that do not produce protein represent most human transcripts. Moreover, ncRNAs have important post-transcriptional regulatory roles in gene expression. miRNAs are one type of ncRNA, comprising 18–25 nucleotides and usually leading to suppression of their target genes. In mammals, approximately 50% of all genes are currently regulated by miRNAs. To date, mirBase, the primary miRNA information database, has over 2000 human, 1300 mouse, and 700 rat annotated mature miRNAs. A large number of mammalian-specific miRNAs have been described in the regulation of various biological processes, including development, differentiation, apoptosis, and cell proliferation [60–64].
miRNAs arise from either intergenic or intragenic regions of a host protein coding or noncoding gene [65]. In the nucleus, miRNAs are chiefly transcribed from those genomic regions by RNA polymerase II as primary miRNAs (pri-miRNAs). These are long segments and typically contain 1–6 precursors of mature miRNAs. The hairpin structure portions in the pri-miRNAs are trimmed by a microprocessor complex containing RNase type III, named Drosha, and the double-stranded (ds)RNA-binding protein DiGeorge syndrome critical region gene 8 (DGCR8) to pre-miRNAs. This latter contains a stem-loop structure and is approximately 70–100 nucleotides long [66]. With a nuclear transport receptor complex, exportin-5-RanGTP, pre-miRNAs are transported to the cytoplasm and further processed by a dsRNase type III, Dicer to an approximately 22-nt ds miRNA duplex, which is incorporated into a RNA-induced silencer complex (RISC)-loading complex (RLC) in an ATP-dependent manner [67]. The sense strand is then deleted from the RLC by a helicase, whereas the antisense strand remains in the complex to form a mature RNA-induced silencer complex (mature RISC) and serves as a template for capturing target mRNAs. Both the transcription of pri-miRNA and the post-transcriptional process leading to mature miRNA are finely regulated. Alterations of the processing molecules can result in a global change of mature miRNA levels.
The RISC is a complex containing multiple factors that work together to regulate target mRNA expression. Based on the extent of complementary between miRNAs and target mRNAs, RISC has different roles in regulating gene expression [68]. For highly complementary target mRNAs, the mature RISC complex cleaves and leads to degradation of the target mRNAs. For partially complementary targets, the RISC complex can decap or deadenylate target mRNAs, thereby decreasing the stability of target mRNAs. In fact, most animal miRNAs are only partially complementary to their targets, and several recent reports have indicated that miRNAs can also induce significant degradation of target mRNAs despite imperfect mRNA–miRNA base pairing [69,70]. Additionally, some components of the RISC complex can also inhibit translation of the target genes.
MiR-210 and hypoxia
HIFs are one of the most sensitive physiological sensors of hypoxia and control the cellular response to the low oxygen condition. A large number of miRNAs have been implicated in regulating HIF pathways. For instance, miR-199a, miR-17–92 clusters, and miR-20b regulate HIF-1α signals during hypoxia [71–73], whereas miR-23, miR-24, miR-26, miR-107, miR-210, and miR-373 have been shown to be downstream molecules of HIFs [74,75]. A recent study further confirmed the effect of hypoxia on miRNA expression, showing that the epidermal growth factor receptor (EGFR) is the upstream regulator of miRNA biogenesis through phosphorylation of argonaute 2 (AGO2) in response to hypoxia [76].
Although multiple miRNAs are regulated by hypoxia, miR-210 is regarded as the master of hypoxia-induced miRNA because it is progressively and consistently increased under hypoxia in various cell types [77]. The stemloop of miR-210 is located in an intron of a ncRNA, which is transcribed from AK123483 on chromosome 11p15.5. The predicted size of human pri-miR-210 is 2927 base pairs, which is almost the same as genomic AK123483 [78]. Additionally, expression of AK123483 is increased under hypoxia compared with normoxia conditions [79]. The highly preserved HRE is responsible for the expression of miR-210. The functional HRE element is located in the miR-210 promoter approximately 40 base pairs upstream of the predicted transcription starting site, and can directly bind HIF-1α under hypoxia [75].
In addition to HIFs, nuclear factor (NF)-κB can also induce miR-210 expression in response to hypoxia [80]. Genomic mapping of miR-210 showed a conserved κB binding site located on the 200-base pair core promoter region and NFκB p50 subunits can directly bind to and trans-activate the miR-210 promoter under hypoxia [80]. In hypoxic cardiomyocytes, Akt regulates miR-210 expression through an HIF-independent pathway [81]. A recent study also showed that DNA demethylation in neural progenitor cells (NPCs) regulates the expression of miR-210 in response to hypoxia through a HIF-1-independent pathway [82] (Figure 3).
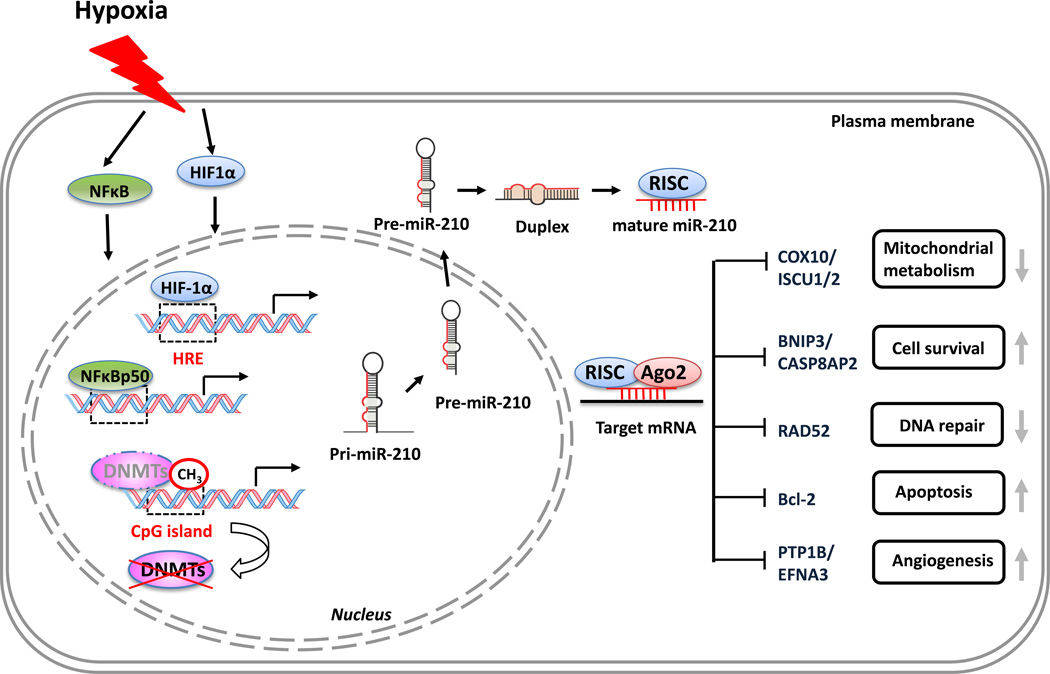
Figure 3.
Summary of miR-210 upregulation mechanisms under hypoxia and its target genes. Hypoxia induces miR-210 upregulation via hypoxia inducible factor (HIF)-1α-dependent and -independent pathways. HIF-1α binds to the hypoxia responsive element (HRE) region and initiates gene transcription: the so-called ‘HIF-1α- dependent pathway’. Hypoxia activates nuclear factor (NF)-κB, which regulates miR-210 transcription by binding to the promoter of miR-210. Hypoxia-induced demethylation by releasing DNA methyltransferases (DNMTs) from CpG islands or inactivating DNMTs also increases miR-210 expression. These represent the HIF- 1α-independent pathway. In the cytosol, pre-miRNA-210 is processed to mature miR-210, binds to 3’ untranslated region (UTR) and represses the expression of a subset of genes associated with a sequence of cellular functions, including mitochondrial metabolism (COX10/ISCU1/2), apoptosis (Bcl2/BNIP3/CASP8AP2), angiogenesis (PTP1B/EFNA3), and DNA repair (RAD52). Abbreviations: Ago2, Argonaute 2; RISC, RNA-induced silencer complex. For definitions of gene symbols, please see the main text.
One of the cellular responses to hypoxia is the upregulation of angiogenesis-related genes in endothelial cells to enhance new blood vessel formation and ensure survival. MiR-210 upregulation is a crucial element of the endothelial cell response to hypoxia, affecting cell survival, migration, and differentiation. Fasanaro and colleagues first found that miR-210 blockage inhibited the formation of capillary-like structures stimulated by hypoxia and decreased cell migration in response to vascular endothelial growth factor (VEGF). Further study showed that ephrin-A3 (EFNA3) was the direct target of miR-210 during hypoxia, and overexpression of EFNA3 significantly blocked the pro-angiogenic effect of miR-210 or hypoxia preconditioning [83]. Another study showed that miR-210 directly targeted protein-tyrosine PTP1B [84,85], which negatively regulated VEGF signaling in endothelial cells. Inconsistent with these results, bioinformatics analysis showed that miR-210 is correlated with hypoxia/VEGF and angiogenesis signaling in patients with breast cancer [86], indicating the involvement of miR-210 in angiogenesis.
Additionally, miR-210 has an important role in stem cell survival and differentiation under hypoxia conditions. It has been reported that ischemia preconditioning enhances bone marrow-derived mesenchymal stem cells (MSCs) survival via induced expression of miR-210, whereas miR-210 knockdown increases caspase-8-associated protein-2 (CASP8AP2) levels, a regulator in Fas-mediated apoptosis, resulting in enhanced cell apoptosis after ischemia [87]. Consistent with this finding, a recent study reported that upregulation of miR-210 during hypoxia decreased in NPC apoptosis as a result of inhibition of Bcl-2 adenovirus E1B 19kDa-interacting protein 3 (BNIP3) [88]. However, in contrast to the findings that miR-210 promotes cell survival, Chio and colleagues reported that miR- 210 directly targeted the 3' untranslated region (UTR) of the antiapoptotic gene Bcl-2, thereby enhancing the hypoxia-induced apoptosis of neuroblastoma cells treated with oxygen/glucose deprivation [89]. Thus, it is possible that miR-210 has different roles in different cell types, and further studies are needed to elucidate its role in cell survival.
MiR-210 is involved in DNA damage repair by repression of RAD52, a key protein for DNA ds break repair and homologous recombination cooperating with other DNA repair components [90]. Using tumor cells, Crosby and colleagues reported that RAD52 is downregulated under hypoxia and this hypoxic-induced downregulation can be partially reversed by antisense inhibition of miR-210 [74]. Further luciferase reporter gene assays showed that miR-210 was capable of binding to the 3′ UTR of RAD52 [74]. This study uncovers a new mechanism by which miR- 210 regulates the DNA damage repair and genetic instability that occurs under hypoxia.
MiR-210 represses mitochondrial respiration, which represents an evolutionarily ancient cellular adaptation to hypoxia and profoundly influences cell survival and function. Under hypoxia conditions, mitochondrial metabolism is suppressed and energy production is shifted from the tricarboxylic acid (TCA) cycle to glycolytic pathways. Recently, miR-210 has been suggested as a new player in regulating mitochondrial metabolism under hypoxia, through repressing the expression of iron-sulfur (FeS) cluster scaffold protein isoforms ISCU1 and ISCU2 [91] that facilitate the assembly of [4Fe-4S] and [2Fe-2S] iron-sulfur clusters. These are prosthetic groups that promote electron transport and oxidation–reduction reactions affecting various cellular processes, including ribosome biogenesis, DNA repair, and iron metabolism [92]. Moreover, miR-210 also disrupts the electron transport activity of mitochondria complex I under hypoxia by repressing ISCU1/2. Apart from ISCU1/2, miR-210 also targets cytochrome c oxidase assembly protein (COX10) [93] and SDHD, subunit D of succinate dehydrogenase complex (SDH) [94], which were key factors in electron transport chain.
Given the close correlation between miR-210 expression and hypoxia, circulating miR-210 could be a potential prognostic marker in patients. A recent study investigated miRNA levels derived from the placenta circulation in maternal blood in pregnancy complicated by fetal hypoxia, and found that several miRNAs, including miR-210, miR-424, miR-21, miR-373, miR-199a and miR-20b, were significantly increased throughout labor because of acute fetal hypoxia accompanied by each uterine contraction [95]. Levels of miR-210 increased 4.2-fold at the time of delivery compared with before the induction of labor in healthy term pregnancies. Moreover, a 3.6-fold upregulation of miR-210 was observed in pregnancy complicated by severe preterm fetal growth restriction, compared with gestation-matched controls. This study suggests that miR-210, with other miRNAs, such as miR-424, miR-199a, and miR-20b, could serve as non-invasive biomarkers for fetal hypoxia in utero [95]. Similarly, miR-210 can be detected in blood from patients with cancer and, thus, could act as a potential biomarker. For example, increased levels of miR-210 were detected in the blood of patients with pancreatic cancer [96] and diffuse large B-cell lymphoma [97].
Cross-talk between miRNA and DNA methylation/demethylation
Given that half of miRNA genes are associated with CpG islands, DNA methylation/demethylation is believed to have an important role in the regulation of miRNAs. Indeed, hypermethylation of CpG islands is a common feature of aberrant miRNA expression profiles. Administration of 5-aza-2′-deoxycytidine (5-Aza), a DNMT inhibitor, significantly induced the expression of miR-127, which is embedded in a CpG island promoter [98]. By contrast, miR-9-1, which is found to be hypermethylated and downregulated in breast cancer, can be induced by reduction of methylation and concomitant reactivation of expression by 5-Aza treatment [99]. The miR-34 family of miRNAs can be inactivated by the methylation of a CpG island at the transcription start site (TSS) of their host genes [100]. Similarly, methylation of a CpG island near the predicted TSS of the miR-200c/miR-141 gene is significantly correlated with its expression level [101]. Given that miRNA levels are tightly regulated by not only the transcription of pri-mRNA, but also the molecules involved in the post-transcriptional process, it is conceivable that epigenetic regulation of those processing proteins contributes to the alteration of miRNA profile.
miRNAs are involved in the establishment and/or maintenance of DNA methylation [102]. Intriguingly, a specific group of miRNAs, which is defined as epi-miRNAs, can regulate the expression of DNA methylation-related genes, such as those encoding DNMTs. miR-29 family members have been reported to downregulate directly DNMT3a and 3b by reactivating methylation-silenced tumor suppressor genes. This action can restore normal patterns of DNA methylation in non-small cell lung cancer cells [103]. Another study showed that short-hairpin RNA-mediated miR-148 repression led to an increase in DNMT3b1 expression, whereas overexpression of miR-148 resulted in decreased DNMT3b1 expression. Further experiments revealed that the miR-148 directly targeted DNMT3b1 coding sequences, because mutation of the putative miR-148 binding site in DNMT3b1 abolished the suppression by miR-148 [104].
Taken together, post-transcriptional regulation by miRNAs and transcriptional mechanisms by DNA methylation/demethylation cooperate with each other to organize the whole gene expression profile and to maintain cellular physiological functions. Once this epigenetic machinery is disrupted, normal physiological functions can be interfered with, thereby contributing to the development of various disease processes.
Environmental epigenetic programming and disease susceptibility
Traditionally, studies of disease susceptibility have often focused on the environmental influences on genetic factors, such as individual genotype variation, which is mostly a result of environmental exposure and germline mutations in the gene coding and promoter regions. However, environmental exposure encountered in early life also influences the health and risk of disease in adulthood. Thus, a general mechanism by which early environmental exposure could be linked to adult phenotypic changes is the alteration of epigenetic signatures, which are crucial in orchestrating the output of genomic information. Understanding of disease susceptibility will require consideration of both genetic and epigenetic information that indicates environmental interactions integrating with the genome. Indeed, it has been reported that prenatal and early postnatal environment factors, such as maternal hypoxia [15,105], maternal nutritional supplements [106], and xenobiotic chemicals [107], are responsible for altered reprogramming of epigenetic marks and subsequent changes of disease susceptibility in the developing fetus.
Some genomic targets, including gene promoter regions, transposable elements adjacent to genes with metastable epialleles, and regulatory elements of imprinted genes, are likely to be susceptible to gene-expression changes caused by environmental perturbations. These targets usually contain regions that are CpG-rich sequences with differentially methylation status, and some regions are also subject to histone modifications for the regulation of gene expression. Transposable elements are repetitive elements that are distributed throughout mammalian genomes, and comprise approximately 45% of the human genome. Although most of these are silenced by CpG methylation, the epigenetic state of a subset of transposable elements is metastable and could affect the expression of adjacent genes. Metastable epialleles are alleles that can be epigenetically modified in a variable and reversible manner and, therefore, are expressed variably in genetically identical individuals [108]. Importantly, epigenetic marks at metastable epialleles have been proved to be readily altered by gestational exposure to nutritional agents and other environmental factors [106,109]; thus, they could be associated with health and disease susceptibility in later life. Being more susceptible to environmental influence, epigenetic deregulation of genes with these marks tends to be involved in enhanced disease susceptibility. One good example of such epigenetically based phenotype viability is the Avy allele, the result of the insertion of a transposable element (transposon) upstream of the TSS of the mouse Agouti gene that encodes a paracrine signaling molecule involved in the production of a yellow pigment in mice melanocytes. Insertion of this transposon promotes constitutive expression of Agouti, leading to yellow fur, obesity, diabetes, and tumorigenesis. CpG methylation in the Avy correlates inversely with expression of Agouti. The degree of methylation varies dramatically among individual Avy/a mice, causing a wide distribution of coat color from yellow (unmethylated) to brown (methylated) [110]. Moreover, supplementation of pregnant mice with betaine, choline, folic acid, and vitamin B12 can lead to a shift in coat color from yellow to brown and provide protection against obesity in the offspring. This effect has been shown to result from an increase in DNA methylation of the CpG sites in the transposon [111,112].
Recent findings by Zucchi and colleagues have demonstrated that the gestational stress can induce miRNAome and transcriptome changes, which are related to the psychiatric and neurological diseases in the offspring. For example, miR-103, which is involved in brain pathologies, and miR-145, a marker of multiple sclerosis in human, are both upregulated in response to gestational stress [105]. Such findings not only improve the understanding of disease susceptibility formation by specifying the important role of epigenetic mechanisms in this process, but also indicate that environmental-induced epigenetic changes during early development are associated with diseases in adulthood.
Gestational hypoxia and vulnerability of neonate hypoxic-ischemic encephalopathy
Etiology of fetal hypoxia
Although low oxygen tension is important for normal fetal organogenesis and growth, additional hypoxia during gestation contributes to developmental malformations in the fetus. Intrauterine hypoxia can be related to various complications occurring in the mother, placenta, or fetus, which can manifest differently and lead to different consequences. There are three types of hypoxic condition during pregnancy: preplacental hypoxia, in which both the mother and her fetus are hypoxic; uteroplacental hypoxia, in which the maternal oxygenation is normal but the fetus is hypoxic because of impairment of uteroplacental circulation; and postplacental hypoxia, in which only the fetus is hypoxic [113]. Preplacental hypoxia represents the chronic hypoxia, mostly caused by high-altitude environments and pre-existing maternal cardiovascular disease, such as cyanotic heart disease, heart failure, or pulmonary hypertension [114]. Chronic hypoxia is a prominent risk factor of intrauterine growth restriction [115]. It limits the ability of the fetus to achieve its genetically determined growth potential and is responsible for multiple complications, such as cognitive dysfunction and cerebral palsy. It can also lead to brain morphological and functional alterations during development, or can potentially result in fetal brain injury via cell death or other cellular signals, owing to low oxygen. Uteroplacental hypoxia is associated with abnormal placental implantation and can be induced by gestational hypertension and preeclampsia. As a result, both mothers and fetus are at an increased risk for cardiovascular disease later in life [116,117]. Postplacental hypoxia is mainly related to fetal diseases, such as decreased uterine artery flow or genetic abnormalities, rather than to the direct effect of hypoxia on the fetus [114].
Hypoxia-induced brain injury occurs in both white matter and gray matter, which has been confirmed in animal models showing that severe acute hypoxia causes an equal injury in subcortical white matter and gray matter region in preterm fetal sheep at 0.6 and 0.7 of gestation [LM3][118–120]. Indeed, the cerebral cortex, hippocampus, and subventricular zone are the regions most vulnerable to hypoxic insults [121,122]. Brain injury caused by fetal hypoxia consequently leads to abnormal behavioral presentations. Using near-term intrauterine hypoxia in the spiny mouse, Ireland and colleagues reported that the acute hypoxic episode produced a mild neurological deficit during the early postnatal period [123]. A sequence of mechanisms is involved in fetal hypoxia-induced brain injury in the developing brain, including the loss of stem or progenitor cells, increased apoptotic cell death because of energy deprivation [124], accumulation of reactive oxygen species (ROS) and reactive nitrogen species (RNS), and immature oligodendrocytes in the periventricular leukomalacia region [125,126]. The inflammatory response is also associated with hypoxia-induced fetal brain injury by enhancing the expression of some pro-inflammatory cytokines and proapoptotic proteins [127].
Fetal hypoxia and neonate hypoxic-ischemic encephalopathy
Neonatal hypoxia-ischemia (HI) is a crucial perinatal event that is characterized by exposure of the individual to low oxygen (hypoxia) and decreased blood flow (ischemia) before, during, or after labor. HI is the leading cause of acute mortality and chronic disability in newborns with an incidence of 1–8 cases per 1000 births, ultimately affecting 60% of preterm infants [128,129]. The chronic complications of cerebral palsy, mental retardation, and epilepsy induced by HI affect a staggering 80% of the survivors [130]. If occurrence of HI is severe enough to damage the brain, it leads within 12–36 h to hypoxic-ischemic encephalopathy (HIE), which is also called perinatal asphyxia [131]. In addition to short-term medical complications, HIE is also associated with mental and neurological diseases with delayed clinical onset [132]. The underlying mechanisms remain elusive and the etiology of brain damage secondary to HIE is complicated and multifaceted. Compared with the adult brain, the neonatal brain shows some differences in physiological structure organization, ontogeny, function, cellular composition, and signaling pathways related to gene expression and, therefore, demonstrates more sensitive and plastic features to challenges [14]. The clinical signs of HIE manifest in the progression of various cellular and molecular responses, such as delayed cell death, suppression and/or overactivation of gene expression, triggered by initial insults that probably occurred during gestation [131,132]. Fetal hypoxia is one of these initial insults, and can lead to abnormal brain development and increased susceptibility of neonates to HIE at the perinatal stage. However, little is known about the role of epigenetic programming in this process. A recent study showed that epigenetic programming of the GR in response to fetal hypoxia resulted in enhanced brain vulnerability to hypoxic/ischemic brain injury in neonatal rats [133]. This finding demonstrated that fetal hypoxia resulted in repression of the GR via methylation of the GR promoter in the developing brain, which enhanced the vulnerability of neonates to HI brain injury. This suggests that fetal hypoxia-induced epigenetic programming of brain developing gene expression is, at least partly, responsible for neonate HIE. Using a gestational intermittent hypoxia rat model, Wang and colleagues observed a sex-dependent anxiety-like behavior in 90-day-old (P90) male offspring that were exposed to hypoxia during their prenatal stage. This behavior was linked to the activation of corticotropin-releasing hormone type-1 (CRHR1) transcription in P90 paraventricular nucleus and E19 embryo hypothalamus, which was controlled by the DNA methylation level at several specific CpG sites of the promoter [134]. These studies demonstrate that the epigenetic programming of gene expression induced by fetal hypoxia increases the risk of brain disorders in the offspring, further supporting the crucial role of epigenetic machinery in regulating brain development.
Epigenetic programming and neurological diseases
Epigenetic mechanisms and neural development
Epigenentic mechanisms together orchestrate CNS development and plasticity, as well as brain function throughout life, by contributing to promoting neural lineage commitment, and the determination or maintenance of neural cell identity. A range of evidence has shown that epigenetic factors, including DNMT [135], MBDs [136], histone- and chromatin-modifying enzymes [137], and miRNA biogenesis factors [138], are essential for the process of neurogenesis and gliogenesis. For exmple, epigenetic regulatory complexes, repressor for element-1 (RE1)- silencing transcription factor (REST) and co-RE1-silencing transcription factor (CoREST) complexes, are central players in regulating seminal neural cell fate decisions and neuronal or glial lineage specification, through targeting transcriptional and epigenetic regulation of developmental gene networks [139]. A large amount of miRNAs in the mammalian brain have specialized roles in neurodevelopment and adult homeostasis and/or plasticity. For instance, miR-184 promotes neural stem cell proliferation and inhibits neuronal differentiation. MiR-125b, miR-128, miR-132, miR134, and miR-138 are also associated with neuronal maturation and neural network integration [140].
Epigenetic mechanisms also regulate the synaptic plasticity, which is associated with long-term potentiation, depression, and learning and memory, via control of the expression of related genes. DNA methylation is responsible for silencing these genes, which are associated with pluripotency and alternative lineage programming, thereby regulating premature neural stem cell development and synaptic plasticity. DNMTs are expressed throughout brain development. Even in the adult brain, these enzymes also function in specific regions mediating ongoing neurogenesis. In animal models, genetic knockout of both the Dnmt1 and Dnmt3a genes reduced the DNA methylation levels, which led to plasticity abnormalities in the hippocampal CA1 region and consequently damage learning and memory in the affected animals. Moreover, hippocampal Dnmt3a2 levels determine cognitive abilities in both young adult and aged mice [141]. MiRNAs also target genes encoding synaptic proteins, and have important roles in activity-dependent synaptic plasticity and memory formation [142,143]. As is evident, the dysregulation of miRNAs lead to various neurological diseases.
Epigenetic programming of neurological disorders
Many epigenetic modifications have been associated with fetal brain development, and the abnormal expression of epigenetic factors results in neurological disease in adults. MeCP2 is a key transcriptional regulator of gene expression during brain development and is tightly regulated to ensure normal neurological function at different developing stages. For example, the expression level of MeCP2 is usually low at the fetal stage, but increases significantly during postnatal development [144]. X-linked Mecp2 mutations are the primary cause of an autism spectrum disorder, Rett syndrome (RTT). Mecp2 has multiple different types of mutation, including missense, nonsense, deletions, and insertions, all of which result in RTT-like symptoms [145]. In addition, Mecp2 mutations are also found in patients with other neurological conditions, such as learning disabilities, neonatal encephalopathy, autism, and X-linked mental retardation [146,147]. The important role of MeCP2 has been confirmed by genetic ablation experiments showing that the symptom in Mecp2-null mice is rescued by reintroduction of MeCP2 [147]. A recent study showed that miR-483-5p, an intragenic miRNA of the imprinted insulin-like growth factor 2 (IGF2) gene, has important roles in regulating MeCP2 expression in the fetal brain [148].
A growing body of evidence has demonstrated that alterations of DNA methylation are associated with the pathogenesis of Alzheimer’s disease (AD) [149,150]. Mastroeni and colleagues evaluated the expression of two DNA methylation markers and eight methylation maintenance factors in entorhinal cortex layer II, a region exhibiting substantial effects of AD. Their findings indicated that the neuronal epigenetic factors were significantly decreased in AD cases, particularly in neurofibrillary tangle-bearing neurons [150]. The extracellular amyloid β (Aβ) plaque is the main mark of both familial and sporadic forms of AD. It has been reported that the promoter region of the APP gene, which encodes the Aβ precursor protein, is regulated by at least two GC elements [151] and exhibits differential expression patterns in human brain [152]. These observations implicate a role of methylation patterns in AD pathogenesis via regulation of APP expression.
miRNAs are also associated with various neurological diseases. For example, miRNAs are regarded as key players in leucine-rich repeat kinase 2 (LRRK2)-mediated Parkinson disease (PD) pathogenesis. Through negative regulation of miRNA-mediated translational repression, LRRK2 mutations can cause familial and sporadic forms that are characteristic of PD. Blockage of let-7 and miR-184* regenerated the effects in wild type LRRK2 cells, whereas overexpression of let-7 and miR-184* alleviated LRRK2 mutant-induced pathology [153]. Compelling evidence now indicates that miRNAs are differentially expressed in the AD brain and might be associated with AD pathogenesis [154,155]. The relation between miRNAs and AD is initially recognized by postmortem analyses indicating that beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) protein level, but not mRNA level, was upregulated in brains from patients with AD [156]. Wang and colleagues also found that miR-107 was significantly decreased in patients with the earliest stages of AD pathology when BACE1 mRNA levels tended to be increased. Bioinformatics analysis predicted that the 3' UTR of BACE1 mRNA has multiple binding sites for miR- 107, and at least one of these sites was shown to be functionally regulated by miR-107 in reporter gene assays [155]. The miR-29a/b-1 cluster has also been reported to regulate BACE1 expression in brain development and in primary neuronal cells. Correspondingly, the miR-29a/b-1 cluster was significantly decreased in patients with AD and abnormally high levels of BACE1 protein [157]. APP was regulated by miR-101, which targeted the RNA responsive element (RE) at the 3' UTR of APP mRNA. Moreover, inhibition of endogenous miR-101 increased the level of APP, whereas overexpression of miR-101 significantly reduced APP and Aβ load in cultured hippocampal neurons [158].
Other neurological diseases, such as fragile X syndrome (FXS), an X chromosome-linked intellectual disability, resulted from the loss of fragile X mental retardation protein (FMRP), are also linked to miRNA machinery. FMRP interacts with many brain plasticity-associated miRNAs, including miR-132, miR-124, and miR-219, to control the spatial and temporal translation of mRNAs involved in synaptic plasticity [143]. Indeed, phosphorylation of FMRP is involved in the inhibition of postsynaptic density protein 95 (PSD-95), a key postsynaptic component of glutamatergic synapses via formation of a complex with miR-125a and Argonaute 2 (Ago2). FMRP dephosphorylation released PSD-95 mRNA from miR-125a-mediated repression and increased the PSD-95 protein level [159].
Environmental cues lead to activation of diverse epigenetic mechanisms, which mediate ESC maintenance and maturation, and neural circuit formation and synaptic plasticity. In addition to fetal hypoxia, as discussed above, maternal cigarette smoking is another well-known pathogen that contributes to the increased risk of neonatal morbidity. Although the underlying mechanisms remain unclear, the nicotine-induced downregulation of AT2R in the developing brain might be associated with increased brain vulnerability to HI injury of neonatal offspring. Moreover, AT2R downregulation is mediated by epigenetic mechanisms and methylation of a single CpG locus near the TATA-box at AT2R promoter resulted in decreased binding affinity of TATA-binding protein (TBP), leading to repression of the AT2R gene in the developing brain [160]. In addition, BDNF has a vital role in brain development and its expression is regulated by alternations in DNA methylation [161]. The BDNF-4 promoter, BDNF-6 promoter, and 5′UTR were modulated by stress hormones, antidepressant treatment, and DNA methyltransferase inhibitors [162,163]. Studies in humans showed that cigarette smoking is associated with a reduced blood level of BDNF protein [164]; however, the extent to which maternal smoking is related to BDNF methylation in adolescent offspring remains unknown. Toledo-Rodriguez and and colleagues conducted an epidemiologic study in adolescents whose mothers smoked during pregnancy and revealed that nicotine exposure during the prenatal period increased the methylation level of the BDNF-6 promoter, which could have led to changes in the plasticity and development of the brain [165].
In addition to environment stressors, a continuous supply of methyl group donors in food might also regulate nervous system development and functioning via epigenetic mechanisms. Maternal diets deficient of methyl donors (MDD) were shown to increase anxiety and learning ability in aged female offspring. However, the DNA methylation patterns of the genes encoding GR, 11β-HSD2, neuronatin, and reelin in hippocampus of rat offspring were not altered significantly when compared with offspring from mothers on a control diet [166]. Although a large number of candidate genes were related to the neurobehavioral function, only a small group of genes was investigated in this study; therefore, it is possible that other DNA regions or uninvestigated epigenetic mechanisms were involved. In rats, this notion was confirmed by a large-scale study in offspring exposed to maternal stress during gestational days 12–16. Over 700 genes in the frontal cortex and hippocampus were found to be differentially expressed following prenatal stress. The DNA methylation that is associated with behavioral changes in offspring rats was increased with both sex-dependent and region-specific profiles [167].
Potential therapeutic interventions for developmental programming disease
Understanding of epigenetic mechanisms in developmental programming of health and disease has a potential in revealing targets for therapeutic interventions and in for drug discovery. Recent laboratory studies have explored the reversibility of induced phenotypic effects by intervention on epigenetic factors. Several compounds have been shown to manipulate DNMT or histone deacetylase (HDAC) activity, and are capable of reversing programmed phenotypic effects. For example, infusion of the HDAC inhibitor trichostatin A (TSA) or the essential amino acid Lmethionine into the brain ventricles of adult animals reversed epigenetic modifications in the hippocampus, GR expression and HPA [LM4]response to stress in the female offspring of dams showing low levels of maternal care [168–170]. Another example is that of the plant-derived isoflavone genistein, which can reactivate methylationsilenced genes. Maternal dietary genistein supplementation during gestation increased methylation of six CpG sites in the transposon upstream of Agouti and led to a shift in the fur color of Avy/a mouse offspring to pseudoagouti [109]. Supplementation of the methyl donor folate to dams on a low-protein diet prevented the elevation of blood pressure, impairment of vasodilatation, and the decrease of nitric oxide synthase mRNA levels in the offspring. In addition, folate supplementation during pregnancy improved the cardiovascular function of offspring exposed to maternal protein restriction [171]. Exogenous leptin administration from day 3 to 13 in the neonatal offspring of undernourished rats rescued the phenotypic changes, gene expression, and associated methylation changes in the PPARα promoter, suggesting that developmental metabolic programming is potentially reversible by an intervention late during developmental plasticity [172]. In addition, Godfrey and colleagues revealed that the DNA methylation level of umbilical cord tissue at a specific site in the retinoid X receptor alpha (RXRA) promoter is associated with lower maternal carbohydrate intake in early pregnancy and higher childhood body fat [173]. RXRA is a transcription factor involved in fat metabolism and insulin sensitivity and, thus, the finding of epigenetic alteration to its promoter could provide utility in identifying individual vulnerability to later obesity and metabolic disease, and /or other conditions. The same principle has been extended to the developmental programming of brain disorders. Recent studies found that exposure to gestational hypoxia downregulated GR in the developing brain, which was associated with enhanced vulnerability of neonates to HI brain injury. Further experiments showed that gestational hypoxia-induced GR downregulation resulted from increased DNA methylation and decreased binding of transcription factors Egr-1 and Sp1 to GR gene exon 17 and 111 promoters [15]. These findings indicate that developmental epigenetics could provide new therapeutic targets against fetal programming of disease.
Recently, based on the extended studies of miRNAs, intervention on miRNAs has been developed step by step and has shown therapeutic potential [174–177]. Increasing amounts of modified oligonucleotide mimics or antisense oligonucleotide are being applied to diseases caused by epigenetic alterations [178]; moreover, newer biochemical modifications have been introduced to enhance the stability, specificity, and efficacy of these oligonucleotides. Locked nucleic acids (LNAs), a family of conformationally locked nucleotide analogs, have been designed and widely used to modulate miRNA function [179]. For instance, LNA-modified oligonucleotide (LNAantimiR) effectively antagonized the liver-expressed miR-122 in nonhuman primates, resulting in the depletion of mature miR-122 and dose-dependent lowering of plasma cholesterol [180]. In addition, other effects have been directed toward the development of small-molecule drugs that can intervene in the biogenesis process of miRNAs or directly influence their function [181]. However, there remains much uncertainty regarding the relative importance of developmental factors in the development of disease and, thus, more research needs to be done in contemporary cohorts to define a better early-life growth pattern.
Concluding remarks
Accumulating evidence from human epidemiological and laboratory animal studies suggests that one’s environment situation during early development sets the stage for one’s subsequent health. A growing body of evidence indicates that, at the molecular level, developmental programming is reflected by transcriptional and/or translational changes in neurodevelopment-related genes, and these changes are achieved, for the most part, through alteration of epigenetic modifications of the genes involved. Various complications during development, including maternal hypoxia and nutritional signals, are capable of altering the epigenetic marks surrounding the DNA and RNA. Although each epigenetic pattern has its own characteristics, they function as an orchestrated and complicated network to ensure integrated fine-tuning of gene regulation. Moreover, the detrimental effects, as well as altered epigenetic marks, resulting from prenatal insults can be inherited transgenerationally and affect more than one generation. Fortunately, the concept of developmental programming of health and disease has been demonstrated by numerous studies and has received extensive attention. In addition to avoiding possible prenatal insults, further understanding of the underlying molecular mechanisms of fetal programming is likely to promote timely intervention and valid therapy approaches to minimize the pathophysiological consequences induced by prenatal programming. Not only does such understanding have great implications for individual health, but the awareness of, and dedicated investigations for, preventative and therapeutic approaches also have enormous national and global relevance.
Highlights.
-
►
Epigenetic mechanisms are important in the brain development
-
►
Hypoxia is a common form of intrauterine stress
-
►
Hypoxic-ischemic brain injury is a leading cause of neonatal mortality and subsequent neurological disorders
-
►
Epigenetic mechanisms play a key role in programming of increased risk of ischemic brain injury
Acknowledgments
This work was supported in part by National Institutes of Health Grants HL082779 (L.Z.), HL083966 (L.Z.), HL089012 (L.Z.), HL110125 (L.Z.), HD031226 (L.Z.) and HL118861 (L.Z.). We apologize to those authors whose studies covered by the scope of this review we were unable to cite because of reference limits.
Biographies

Lubo Zhang
Lubo Zhang is professor of pharmacology and physiology and director of Center for Perinatal Biology at the Loma Linda University School of Medicine. He was president of the Western Pharmacology Society in 2008. He has been a member of various grant review study groups for the US National Institutes of Health and American Heart Association for more than 15 years. Lubo is also the (co)author of over 500 scientific articles, book chapters, and abstracts. His research interests focus on the molecular and epigenetic mechanisms in the regulation of uteroplacental circulation and developmental programming of health and disease.

Qingyi Ma
Qingyi Ma received his PhD in physiology from Loma Linda University in 2011. He did his postdoctoral training in Zilkha Neurogenetic Institute, Keck School of Medicine, of the University of Southern California. Currently, he is a postdoctoral fellow in the Center for Perinatal Biology at the Loma Linda University School of Medicine. His primary research interest is in the molecular mechanisms of brain injury and protection following stroke and neurodegeneration disorders.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Teaser: Epigenetic mechanisms are important in the brain development, and have a key role in the programming of increased risk of neurological diseases resulting from early-life stress.
References
- 1.Barker DJ, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986;1:1077–1081. doi: 10.1016/s0140-6736(86)91340-1. [DOI] [PubMed] [Google Scholar]
- 2.de Boo HA, Harding JE. The developmental origins of adult disease (Barker) hypothesis. Aust. N. Z. J. Obstet. Gynaecol. 2006;46:4–14. doi: 10.1111/j.1479-828X.2006.00506.x. [DOI] [PubMed] [Google Scholar]
- 3.Warner MJ, Ozanne SE. Mechanisms involved in the developmental programming of adulthood disease. Biochem. J. 2010;427:333–347. doi: 10.1042/BJ20091861. [DOI] [PubMed] [Google Scholar]
- 4.Langley-Evans SC, McMullen S. Developmental origins of adult disease. Med. Princ. Pract. 2010;19:87–98. doi: 10.1159/000273066. [DOI] [PubMed] [Google Scholar]
- 5.Barker DJ, et al. Fetal nutrition and cardiovascular disease in adult life. Lancet. 1993;341:938–941. doi: 10.1016/0140-6736(93)91224-a. [DOI] [PubMed] [Google Scholar]
- 6.Barker DJ, et al. Growth and chronic disease: findings in the Helsinki Birth Cohort. Ann. Hum. Biol. 2009;36:445–458. doi: 10.1080/03014460902980295. [DOI] [PubMed] [Google Scholar]
- 7.Harris A, Seckl J. Glucocorticoids, prenatal stress and the programming of disease. Horm. Behav. 2011;59:279–289. doi: 10.1016/j.yhbeh.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Gluckman PD, et al. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gluckman PD, Hanson MA. Living with the past: evolution, development, and patterns of disease. Science. 2004;305:1733–1736. doi: 10.1126/science.1095292. [DOI] [PubMed] [Google Scholar]
- 10.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 2013;20:282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- 11.Gicquel C, et al. Epigenetic regulation and fetal programming. Best Pract. Res. Clin. Endocrinol. Metab. 2008;22:1–16. doi: 10.1016/j.beem.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 12.John RM, Lefebvre L. Developmental regulation of somatic imprints. Differentiation. 2011;81:270–280. doi: 10.1016/j.diff.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Egger G, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 14.Chen M, Zhang L. Epigenetic mechanisms in developmental programming of adult disease. Drug Discov. Today. 2011;16:1007–1018. doi: 10.1016/j.drudis.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Rodriguez PJ, et al. Fetal hypoxia increases vulnerability of hypoxic-ischemic brain injury in neonatal rats: role of glucocorticoid receptors. Neurobiol. Dis. 2014;65:172–179. doi: 10.1016/j.nbd.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallak M, et al. Magnesium sulfate protection of fetal rat brain from severe maternal hypoxia. Obstet. Gynecol. 2000;96:124–128. doi: 10.1016/s0029-7844(00)00844-9. [DOI] [PubMed] [Google Scholar]
- 17.Volpe JJ. Neurobiology of periventricular leukomalacia in the premature infant. Pediatr. Res. 2001;50:553–562. doi: 10.1203/00006450-200111000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Golan MH, et al. Impaired migration signaling in the hippocampus following prenatal hypoxia. Neuropharmacology. 2009;57:511–522. doi: 10.1016/j.neuropharm.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 19.Herlenius E, Lagercrantz H. Neurotransmitters and neuromodulators during early human development. Early Hum. Dev. 2001;65:21–37. doi: 10.1016/s0378-3782(01)00189-x. [DOI] [PubMed] [Google Scholar]
- 20.Louzoun-Kaplan V, et al. Prenatal hypoxia down regulates the GABA pathway in newborn mice cerebral cortex; partial protection by MgSO4. Int. J. Dev. Neurosci. 2008;26:77–85. doi: 10.1016/j.ijdevneu.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Freitag M, Selker EU. Controlling DNA methylation: many roads to one modification. Curr. Opin. Genet. Dev. 2005;15:191–199. doi: 10.1016/j.gde.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum. Mol. Genet. 2007;16(Spec no. 1):R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. Br. J. Cancer. 2008;98:1881–1885. doi: 10.1038/sj.bjc.6604374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Portela A, Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 26.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Crews D, McLachlan JA. Epigenetics, evolution, endocrine disruption, health, and disease. Endocrinology. 2006;147(6 Suppl):S4–S10. doi: 10.1210/en.2005-1122. [DOI] [PubMed] [Google Scholar]
- 28.Straussman R, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 29.Trasler JM. Gamete imprinting: setting epigenetic patterns for the next generation. Reprod. Fertil. Dev. 2006;18:63–69. doi: 10.1071/rd05118. [DOI] [PubMed] [Google Scholar]
- 30.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen S, et al. Medicine. Activating a repressor. Science. 2008;320:1172–1173. doi: 10.1126/science.1159146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer W, et al. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 33.Kishigami S, et al. Epigenetic abnormalities of the mouse paternal zygotic genome associated with microinsemination of round spermatids. Dev. Biol. 2006;289:195–205. doi: 10.1016/j.ydbio.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 34.Wossidlo M, et al. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010;29:1877–1888. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reik W, et al. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 36.Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 37.Bhutani N, et al. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohr F, et al. TET genes: new players in DNA demethylation and important determinants for stemness. Exp. Hematol. 2011;39:272–281. doi: 10.1016/j.exphem.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen MP, et al. Epigenetic regulation of hypoxia inducible factor in diseases and therapeutics. Arch. Pharm. Res. 2013;36:252–263. doi: 10.1007/s12272-013-0058-x. [DOI] [PubMed] [Google Scholar]
- 42.Watson JA, et al. Epigenetics, the epicenter of the hypoxic response. Epigenetics. 2010;5:293–296. doi: 10.4161/epi.5.4.11684. [DOI] [PubMed] [Google Scholar]
- 43.Walczak-Drzewiecka A, et al. DNA methylation-dependent suppression of HIF1A in an immature hematopoietic cell line HMC-1. Biochem. Biophys. Res. Commun. 2010;391:1028–1032. doi: 10.1016/j.bbrc.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 44.Koslowski M, et al. Tumor-associated CpG demethylation augments hypoxia-induced effects by positive autoregulation of HIF-1alpha. Oncogene. 2011;30:876–882. doi: 10.1038/onc.2010.481. [DOI] [PubMed] [Google Scholar]
- 45.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell. Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 46.Herman JG, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatzimichael E, et al. Absence of methylation-dependent transcriptional silencing in TP73 irrespective of the methylation status of the CDKN2A CpG island in plasma cell neoplasia. Leuk. Res. 2009;33:1272–1275. doi: 10.1016/j.leukres.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 48.Livide G, et al. Epigenetic and copy number variation analysis in retinoblastoma by MS-MLPA. Pathol. Oncol. Res. 2012;18:703–712. doi: 10.1007/s12253-012-9498-8. [DOI] [PubMed] [Google Scholar]
- 49.Dmitriev AA, et al. Genetic and epigenetic analysis of non-small cell lung cancer with NotI-microarrays. Epigenetics. 2012;7:502–513. doi: 10.4161/epi.19801. [DOI] [PubMed] [Google Scholar]
- 50.Hatzimichael E, et al. The prolyl-hydroxylase EGLN3 and not EGLN1 is inactivated by methylation in plasma cell neoplasia. Eur. J. Haematol. 2010;84:47–51. doi: 10.1111/j.1600-0609.2009.01344.x. [DOI] [PubMed] [Google Scholar]
- 51.Place TL, et al. Aberrant promoter CpG methylation is a mechanism for impaired PHD3 expression in a diverse set of malignant cells. PLoS ONE. 2011;6:e14617. doi: 10.1371/journal.pone.0014617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rossler J, et al. Hypoxia-induced erythropoietin expression in human neuroblastoma requires a methylation free HIF-1 binding site. J. Cell. Biochem. 2004;93:153–161. doi: 10.1002/jcb.20133. [DOI] [PubMed] [Google Scholar]
- 53.Raspaglio G, et al. Hypoxia induces class III beta-tubulin gene expression by HIF-1alpha binding to its 3' flanking region. Gene. 2008;409:100–108. doi: 10.1016/j.gene.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 54.Watson JA, et al. Generation of an epigenetic signature by chronic hypoxia in prostate cells. Hum. Mol. Genet. 2009;18:3594–3604. doi: 10.1093/hmg/ddp307. [DOI] [PubMed] [Google Scholar]
- 55.Shahrzad S, et al. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics. 2007;2:119–125. doi: 10.4161/epi.2.2.4613. [DOI] [PubMed] [Google Scholar]
- 56.Lawrence J, et al. Foetal nicotine exposure causes PKCepsilon gene repression by promoter methylation in rat hearts. Cardiovasc. Res. 2011;89:89–97. doi: 10.1093/cvr/cvq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patterson AJ, et al. Chronic prenatal hypoxia induces epigenetic programming of PKC{epsilon} gene repression in rat hearts. Circ. Res. 2010;107:365–373. doi: 10.1161/CIRCRESAHA.110.221259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong F, et al. Norepinephrine causes epigenetic repression of PKCepsilon gene in rodent hearts by activating Nox1-dependent reactive oxygen species production. FASEB J. 2012;26:2753–2763. doi: 10.1096/fj.11-199422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Breton CV, et al. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am. J. Respir. Crit. Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 61.Pasquinelli AE, et al. MicroRNAs: a developing story. Curr. Opin. Genet. Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 63.Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol. Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- 64.Bartel F, et al. [HDMX amplification and high levels of HDMX-S splice variant are correlated with a poor prognosis in soft tissue sarcomas] Verh. Dtsch. Ges. Pathol. 2004;88:199–206. [PubMed] [Google Scholar]
- 65.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum. Mol. Genet. 2005;14(Spec no. 1):R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 66.Kim VN, et al. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 67.Yoda M, et al. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Behm-Ansmant I, et al. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb. Symp. Quant. Biol. 2006;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 69.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 70.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 71.Taguchi A, et al. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008;68:5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- 72.Rane S, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lei Z, et al. Regulation of HIF-1alpha and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS ONE. 2009;4:e7629. doi: 10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crosby ME, et al. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang X, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen J, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan YC, et al. miR-210: the master hypoxamir. Microcirculation. 2012;19:215–223. doi: 10.1111/j.1549-8719.2011.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang X, et al. MiR-210: micromanager of the hypoxia pathway. Trends Mol. Med. 2010;16:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Z, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y, et al. Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia: new insights into molecular mechanisms for the disease. J. Cell. Mol. Med. 2012;16:249–259. doi: 10.1111/j.1582-4934.2011.01291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mutharasan RK, et al. microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1519–H1530. doi: 10.1152/ajpheart.01080.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiong L, et al. DNA demethylation regulates the expression of miR-210 in neural progenitor cells subjected to hypoxia. FEBS J. 2012;279:4318–4326. doi: 10.1111/febs.12021. [DOI] [PubMed] [Google Scholar]
- 83.Fasanaro P, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fasanaro P, et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J. Biol. Chem. 2009;284:35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu S, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122(11 Suppl):S124–S131. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Foekens JA, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim HW, et al. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J. Biol. Chem. 2009;284:33161–33168. doi: 10.1074/jbc.M109.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang F, et al. miR-210 suppresses BNIP3 to protect against the apoptosis of neural progenitor cells. Stem Cell Res. 2013;11:657–667. doi: 10.1016/j.scr.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 89.Chio CC, et al. MicroRNA-210 targets antiapoptotic Bcl-2 expression and mediates hypoxia-induced apoptosis of neuroblastoma cells. Arch. Toxicol. 2013;87:459–468. doi: 10.1007/s00204-012-0965-5. [DOI] [PubMed] [Google Scholar]
- 90.Benson FE, et al. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 91.Chan SY, et al. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell. Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rouault TA, Tong WH. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407. doi: 10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen Z, et al. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 94.Puissegur MP, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell. Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Whitehead CL, et al. Circulating MicroRNAs in maternal blood as potential biomarkers for fetal hypoxia in-utero. PLoS ONE. 2013;8:e78487. doi: 10.1371/journal.pone.0078487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang J, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lawrie CH, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 98.Saito Y, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 99.Lehmann U, et al. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J. Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 100.Lodygin D, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 101.Neves R, et al. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res. Notes. 2010;3:219. doi: 10.1186/1756-0500-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 103.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Duursma AM, et al. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zucchi FC, et al. Maternal stress induces epigenetic signatures of psychiatric and neurological diseases in the offspring. PLoS ONE. 2013;8:e56967. doi: 10.1371/journal.pone.0056967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Waterland RA, et al. Maternal methyl supplements increase offspring DNA methylation at Axin Fused. Genesis. 2006;44:401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- 107.Ho SM, et al. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dolinoy DC, et al. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr. Res. 2007;61:30R–37R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- 109.Dolinoy DC, et al. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Duhl DM, et al. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 111.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wolff GL, et al. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 113.Kingdom JC, Kaufmann P. Oxygen and placental villous development: origins of fetal hypoxia. Placenta. 1997;18:613–621. doi: 10.1016/s0143-4004(97)90000-x. discussion 623-616. [DOI] [PubMed] [Google Scholar]
- 114.Hutter D, et al. Causes and mechanisms of intrauterine hypoxia and its impact on the fetal cardiovascular system: a review. Int. J. Pediatr. 2010;2010:401323. doi: 10.1155/2010/401323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Palmer SK, et al. Altered blood pressure course during normal pregnancy and increased preeclampsia at high altitude (3100 meters) in Colorado. Am. J. Obstet. Gynecol. 1999;180:1161–1168. doi: 10.1016/s0002-9378(99)70611-3. [DOI] [PubMed] [Google Scholar]
- 116.Ramsay JE, et al. Microvascular dysfunction: a link between pre-eclampsia and maternal coronary heart disease. Br. J. Obstet. Gynecol. 2003;110:1029–1031. [PubMed] [Google Scholar]
- 117.Haukkamaa L, et al. Risk for subsequent coronary artery disease after preeclampsia. Am. J. Cardiol. 2004;93:805–808. doi: 10.1016/j.amjcard.2003.11.065. [DOI] [PubMed] [Google Scholar]
- 118.George S, et al. Induced cerebral hypothermia reduces post-hypoxic loss of phenotypic striatal neurons in preterm fetal sheep. Exp. Neurol. 2007;203:137–147. doi: 10.1016/j.expneurol.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 119.Bennet L, et al. The effect of cerebral hypothermia on white and grey matter injury induced by severe hypoxia in preterm fetal sheep. J. Physiol. 2007;578:491–506. doi: 10.1113/jphysiol.2006.119602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dean JM, et al. Endogenous alpha2-adrenergic receptor-mediated neuroprotection after severe hypoxia in preterm fetal sheep. Neuroscience. 2006;142:615–628. doi: 10.1016/j.neuroscience.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 121.Northington FJ, et al. Early neurodegeneration after hypoxia-ischemia in neonatal rat is necrosis while delayed neuronal death is apoptosis. Neurobiol. Dis. 2001;8:207–219. doi: 10.1006/nbdi.2000.0371. [DOI] [PubMed] [Google Scholar]
- 122.Li Y, et al. Fetal stress and programming of hypoxic/ischemic-sensitive phenotype in the neonatal brain: mechanisms and possible interventions. Prog. Neurobiol. 2012;98:145–165. doi: 10.1016/j.pneurobio.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ireland Z, et al. Behavioural effects of near-term acute fetal hypoxia in a small precocial animal, the spiny mouse (Acomys cahirinus) Neonatology. 2010;97:45–51. doi: 10.1159/000227293. [DOI] [PubMed] [Google Scholar]
- 124.Rees S, et al. The biological basis of injury and neuroprotection in the fetal and neonatal brain. Int. J. Dev. Neurosci. 2011;29:551–563. doi: 10.1016/j.ijdevneu.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Riddle A, et al. Spatial heterogeneity in oligodendrocyte lineage maturation and not cerebral blood flow predicts fetal ovine periventricular white matter injury. J. Neurosci. 2006;26:3045–3055. doi: 10.1523/JNEUROSCI.5200-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Back SA, et al. Selective vulnerability of late oligodendrocyte progenitors to hypoxia-ischemia. J. Neurosci. 2002;22:455–463. doi: 10.1523/JNEUROSCI.22-02-00455.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guo R, et al. Brain injury caused by chronic fetal hypoxemia is mediated by inflammatory cascade activation. Reprod. Sci. 2010;17:540–548. doi: 10.1177/1933719110364061. [DOI] [PubMed] [Google Scholar]
- 128.Vannucci RC. Hypoxic-ischemic encephalopathy. Am. J. Perinatol. 2000;17:113–120. doi: 10.1055/s-2000-9293. [DOI] [PubMed] [Google Scholar]
- 129.Vannucci RC, et al. Rat model of perinatal hypoxic-ischemic brain damage. J. Neurosci. Res. 1999;55:158–163. doi: 10.1002/(SICI)1097-4547(19990115)55:2<158::AID-JNR3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 130.Derrick M, et al. A model of cerebral palsy from fetal hypoxia-ischemia. Stroke. 2007;38(2 Suppl):731–735. doi: 10.1161/01.STR.0000251445.94697.64. [DOI] [PubMed] [Google Scholar]
- 131.Fatemi A, et al. Hypoxic-ischemic encephalopathy in the term infant. Clin. Perinatol. 2009;36:835–858. vii. doi: 10.1016/j.clp.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Herrera-Marschitz M, et al. Perinatal asphyxia: CNS development and deficits with delayed onset. Front. Neurosci. 2014;8:47. doi: 10.3389/fnins.2014.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gonzalez-Rodriguez PJ, et al. Fetal hypoxia increases vulnerability of hypoxic-ischemic brain injury in neonatal rats: role of glucocorticoid receptors. Neurobiol. Dis. 2014 doi: 10.1016/j.nbd.2014.01.020. XX, YYY-ZZZ[LM5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang X, et al. Gestational hypoxia induces sex-differential methylation of Crhr1 linked to anxiety-like behavior. Mol. Neurobiol. 2013;48:544–555. doi: 10.1007/s12035-013-8444-4. [DOI] [PubMed] [Google Scholar]
- 135.Fan G, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- 136.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 137.Lee S, Lee SK. Crucial roles of histone-modifying enzymes in mediating neural cell-type specification. Curr. Opin. Neurobiol. 2010;20:29–36. doi: 10.1016/j.conb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jimenez-Mateos E, et al. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat. Med. 2012;18:1087–1094. doi: 10.1038/nm.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Qureshi IA, et al. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle. 2010;9:4477–4486. doi: 10.4161/cc.9.22.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Qureshi IA, Mehler MF. Understanding neurological disease mechanisms in the era of epigenetics. JAMA Neurol. 2013;70:703–710. doi: 10.1001/jamaneurol.2013.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Oliveira AM, et al. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 2012;15:1111–1113. doi: 10.1038/nn.3151. [DOI] [PubMed] [Google Scholar]
- 142.Lambert TJ, et al. MicroRNA132 modulates short-term synaptic plasticity but not basal release probability in hippocampal neurons. PLoS ONE. 2010;5:e15182. doi: 10.1371/journal.pone.0015182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Balmer D, et al. Elevated methyl-CpG-binding protein 2 expression is acquired during postnatal human brain development and is correlated with alternative polyadenylation. J. Mol. Med. 2003;81:61–68. doi: 10.1007/s00109-002-0396-5. [DOI] [PubMed] [Google Scholar]
- 145.Goffin D, et al. Rett syndrome mutation MeCP2 T158A disrupts DNA binding, protein stability and ERP responses. Nat. Neurosci. 2012;15:274–283. doi: 10.1038/nn.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Neul JL, Zoghbi HY. Rett syndrome: a prototypical neurodevelopmental disorder. Neuroscientist. 2004;10:118–128. doi: 10.1177/1073858403260995. [DOI] [PubMed] [Google Scholar]
- 147.Guy J, et al. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Han K, et al. Human-specific regulation of MeCP2 levels in fetal brains by microRNA miR-483-5p. Genes Dev. 2013;27:485–490. doi: 10.1101/gad.207456.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Adwan L, Zawia NH. Epigenetics: a novel therapeutic approach for the treatment of Alzheimer's disease. Pharmacol. Ther. 2013;139:41–50. doi: 10.1016/j.pharmthera.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mastroeni D, et al. Epigenetic changes in Alzheimer's disease: decrements in DNA methylation. Neurobiol. Aging. 2010;31:2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pollwein P, et al. The expression of the amyloid precursor protein (APP) is regulated by two GC-elements in the promoter. Nucleic Acids Res. 1992;20:63–68. doi: 10.1093/nar/20.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rogaev EI, et al. The upstream promoter of the beta-amyloid precursor protein gene (APP) shows differential patterns of methylation in human brain. Genomics. 1994;22:340–347. doi: 10.1006/geno.1994.1393. [DOI] [PubMed] [Google Scholar]
- 153.Gehrke S, et al. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Provost P. MicroRNAs as a molecular basis for mental retardation, Alzheimer's and prion diseases. Brain Res. 2010;1338:58–66. doi: 10.1016/j.brainres.2010.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang WX, et al. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Holsinger RM, et al. Increased expression of the amyloid precursor beta-secretase in Alzheimer's disease. Ann. Neurol. 2002;51:783–786. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- 157.Hebert SS, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/betasecretase expression. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vilardo E, et al. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J. Biol. Chem. 2010;285:18344–18351. doi: 10.1074/jbc.M110.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Muddashetty RS, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li Y, et al. Promoter methylation represses AT2R gene and increases brain hypoxic-ischemic injury in neonatal rats. Neurobiol. Dis. 2013;60:32–38. doi: 10.1016/j.nbd.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dennis KE, Levitt P. Regional expression of brain derived neurotrophic factor (BDNF) is correlated with dynamic patterns of promoter methylation in the developing mouse forebrain. Brain Res. Mol. Brain Res. 2005;140:1–9. doi: 10.1016/j.molbrainres.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 162.Dwivedi Y, et al. Antidepressants reverse corticosterone-mediated decrease in brain-derived neurotrophic factor expression: differential regulation of specific exons by antidepressants and corticosterone. Neuroscience. 2006;139:1017–1029. doi: 10.1016/j.neuroscience.2005.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Aid T, et al. Mouse and rat BDNF gene structure and expression revisited. J. Neurosci. Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim TS, et al. Increased plasma brain-derived neurotrophic factor levels in chronic smokers following unaided smoking cessation. Neurosci. Lett. 2007;423:53–57. doi: 10.1016/j.neulet.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 165.Toledo-Rodriguez M, et al. Maternal smoking during pregnancy is associated with epigenetic modifications of the brain-derived neurotrophic factor-6 exon in adolescent offspring. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B:1350–1354. doi: 10.1002/ajmg.b.31109. [DOI] [PubMed] [Google Scholar]
- 166.Konycheva G, et al. Dietary methyl donor deficiency during pregnancy in rats shapes learning and anxiety in offspring. Nutr. Res. 2011;31:790–804. doi: 10.1016/j.nutres.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 167.Mychasiuk R, et al. Prenatal stress produces sexually dimorphic and regionally specific changes in gene expression in hippocampus and frontal cortex of developing rat offspring. Dev. Neurosci. 2011;33:531–538. doi: 10.1159/000335524. [DOI] [PubMed] [Google Scholar]
- 168.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 169.Weaver IC, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J. Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Weaver IC, et al. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Torrens C, et al. Folate supplementation during pregnancy improves offspring cardiovascular dysfunction induced by protein restriction. Hypertension. 2006;47:982–987. doi: 10.1161/01.HYP.0000215580.43711.d1. [DOI] [PubMed] [Google Scholar]
- 172.Vickers MH, et al. Neonatal leptin treatment reverses developmental programming. Endocrinology. 2005;146:4211–4216. doi: 10.1210/en.2005-0581. [DOI] [PubMed] [Google Scholar]
- 173.Godfrey KM, et al. Epigenetic gene promoter methylation at birth is associated with child's later adiposity. Diabetes. 2011;60:1528–1534. doi: 10.2337/db10-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu XQ, et al. Targeted delivery of antisense inhibitor of miRNA for antiangiogenesis therapy using cRGD-functionalized nanoparticles. Mol. Pharm. 2011;8:250–259. doi: 10.1021/mp100315q. [DOI] [PubMed] [Google Scholar]
- 175.Wiggins JF, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Takeshita F, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol. Ther. 2010;18:181–187. doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mehler MF. Epigenetics and the nervous system. Ann. Neurol. 2008;64:602–617. doi: 10.1002/ana.21595. [DOI] [PubMed] [Google Scholar]
- 179.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 181.Li Y, et al. Emergence of chemical biology approaches to the RNAi/miRNA pathway. Chem. Biol. 2010;17:584–589. doi: 10.1016/j.chembiol.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]